Significance
Consumers often regard heirloom fruit varieties grown in the garden as more flavorful than commercial varieties purchased at the grocery store. While plant breeders have historically focused on improving producer-orientated traits such as yield, consumer-oriented traits such as flavor have regularly been neglected. This is, in part, due to the difficulty associated with measuring the sensory perceptions of flavor. Here, we combine fruit chemical and consumer sensory panel information to train machine learning models that can predict how flavorful a fruit will be from its chemistry. By increasing the throughput of flavor evaluations, these models will help plant breeders to integrate flavor earlier in the breeding pipeline and aid in the design of varieties with exceptional flavor profiles.
Keywords: flavor, fruit quality, artificial intelligence
Abstract
Although they are staple foods in cuisines globally, many commercial fruit varieties have become progressively less flavorful over time. Due to the cost and difficulty associated with flavor phenotyping, breeding programs have long been challenged in selecting for this complex trait. To address this issue, we leveraged targeted metabolomics of diverse tomato and blueberry accessions and their corresponding consumer panel ratings to create statistical and machine learning models that can predict sensory perceptions of fruit flavor. Using these models, a breeding program can assess flavor ratings for a large number of genotypes, previously limited by the low throughput of consumer sensory panels. The ability to predict consumer ratings of liking, sweet, sour, umami, and flavor intensity was evaluated by a 10-fold cross-validation, and the accuracies of 18 different models were assessed. The prediction accuracies were high for most attributes and ranged from 0.87 for sourness intensity in blueberry using XGBoost to 0.46 for overall liking in tomato using linear regression. Further, the best-performing models were used to infer the flavor compounds (sugars, acids, and volatiles) that contribute most to each flavor attribute. We found that the variance decomposition of overall liking score estimates that 42% and 56% of the variance was explained by volatile organic compounds in tomato and blueberry, respectively. We expect that these models will enable an earlier incorporation of flavor as breeding targets and encourage selection and release of more flavorful fruit varieties.
Plant breeders and geneticists have made continuous and substantial progress in the development of varieties that are more resilient and higher-yielding—much to the benefit of producers worldwide. Yet, during this extended period of progress, consumer-oriented quality traits such as flavor have been often neglected or treated as low-priority breeding targets, contributing to widespread consumer dissatisfaction with modern varieties of fruits and vegetables (1). An important reason for this low priority is a reward system that pays growers based on crop yield, leading to prioritization of breeding targets that are mainly producer-oriented. However, as consumer willingness to pay premiums for higher-quality products rises, demand for consumer-oriented traits in food production systems is increasing (2). Accordingly, a reemerging interest in fruit and vegetable flavor quality creates the need for high-yielding varieties with exceptional flavor profiles.
Fruit flavor is the product of the complex interactions between the chemical composition of a fruit and the taste, olfaction, and psychology of the consumer (3–5). To breed and develop varieties with improved flavor properties, the genetic complexities of fruit flavor must be captured and assessed. Flavor is currently evaluated by consumer sensory panels or individually by breeders. Field evaluations are generally subjective and error-prone as they typically consist of the sensory preferences of one or few individuals. However, field evaluation has an advantage in that many varieties can be evaluated in a given day. In contrast, population-based sensory panels are more objective, accurate, and well-established, but they can be costly, time-consuming, and difficult to scale to a large breeding program. The difficulties associated with accurate flavor phenotyping have contributed to the lack of selection for fruit flavor and thereby contributed to the widespread consumer belief that commercial fruit flavor has declined (6, 7). Cheap and scalable flavor selection methods would greatly benefit the breeding process.
The main driver of fruit flavor perception is its chemical composition. Fruits contain a diverse array of sugars, acids, and volatiles whose concentrations are driven by genetic and environmental effects. Sugars and acids are largely perceived by taste receptors on the tongue and the volatiles by receptors located in the olfactory epithelium (8). We hypothesized that by quantifying the chemical profile of a fruit and its corresponding consumer perception, models predicting consumer flavor preferences can be created. These prediction models can increase throughput of flavor phenotyping, allowing a breeder to make selections for improved flavor on hundreds of genotypes per season. This approach is analogous to the concept of genomic selection (9), where DNA markers are used in plant breeding programs to predict the genetic merit of individuals for highly complex traits. Here, we propose the use of statistical methods to model the metabolomic profile in a breeding population and predict flavor perception. Additionally, by leveraging the trained models for inference, specific metabolites that underlie consumer flavor preferences can be elucidated, identifying targets for marker-assisted selection (MAS) and for the food industry to enhance flavor in its products.
In flavor studies, the most widespread statistical modeling approaches to date include multiple linear regression and partial least squares (PLS) regression (10, 11). However, the process of developing metabolomic-based prediction models can be challenging due to the large number of chemical compounds present in a fruit and the fact that the concentrations of many of the flavor-associated chemicals are correlated with one another due to shared biosynthetic pathways. Fortunately, breeders and quantitative geneticists are already dealing with similar types of data in the area of genomic selection to make selection of complex traits using genomic information. With the advent of genomic selection, for example, a variety of Bayesian linear regression models with different priors were proposed to predict complex phenotypes using DNA markers. These models included Bayes A and Bayes B (9), Bayes Cπ (12), Bayesian LASSO (13), and Bayesian ridge regression (14, 15), among others. Recently, there has been increasing interest in machine learning models applied to genomic (16) and metabolomic data (11) as well as metabolomic data applied to trait biomarker research (17–21). However, few empirical studies have applied machine learning models at the metabolome level, or specifically for the enhancement of fruit flavor.
Here we address the limitations in flavor phenotyping and propose an indirect phenotyping approach that has significantly higher throughput compared to current standards. We assessed a range of statistical and machine learning models that take the chemical profile of a fruit and make predictions of its consumer flavor perception. To this end, we combined information at the metabolome and sensory panel level for two important horticultural crops, tomato and blueberry, and demonstrate that metabolomic prediction models can be employed in a breeding program to make simpler and more accurate selections for flavorful varieties. Additionally, we leverage the trained models to infer the contributions of volatiles, sugars, and acids to sensory perceptions and consumer likeability. Our results suggest that up to 56% of the variance associated with overall consumer liking can be attributed to volatile compounds. Furthermore, we demonstrate that machine learning approaches are generally the best predictors of consumer flavor preferences and metabolomic selection accuracies are superior to genomic selection models, highlighting the potential in breeding applications.
Results
Data.
In order to study the capacity of different prediction models and the importance of different metabolites in flavor perception, we performed an analysis of previously published data (4, 5, 10, 22) combined with new data added in this study for two fruit species: tomato (Solanum lycopersicum) and blueberry (Vaccinium spp.). For each fruit, targeted sets of sugars, acids, and volatiles were quantified in diverse accessions including commercial cultivars, heirloom varieties, and germplasm selections from the University of Florida tomato and blueberry breeding programs. The tomato population includes a greater range of genetically diverse materials than previously analyzed, while the blueberry population is more representative of an elite breeding population. Consumer sensory panels rated each accession for flavor attributes including sweetness, sourness, flavor intensity, and overall liking. Additionally, sensory perceptions of umami were quantified solely for tomato.
Network Analysis Recapitulates Metabolic Pathways.
A weighted correlation network analysis was performed on the metabolite concentrations across all fruit accessions for tomato and blueberry (Figs. 1A and 2A). The results are largely consistent with knowledge of the individual biosynthetic pathways and provide insights into the relationships between pathways. For example, there are strong associations between the apocarotenoid volatiles (e.g., geranial and β-cyclocitral) and the fatty acid-derived volatiles (e.g., 1-pentanol and E-2-heptenal). Apocarotenoid volatiles are derived from precursors localized in plastids and their contents substantially increase during the conversion of chloroplasts to chromoplasts (23), while the precursors of fatty-acid-derived volatiles are membrane lipids. TomloxC is an essential enzyme for the synthesis of five- and six-carbon fatty acid-derived volatiles (24, 25). Recently, a potential link between these pathways was proposed with a quantitative trait locus (QTL) analysis that implicated a role for TomloxC in apocarotenoid synthesis, possibly by a cooxidation mechanism (26).
Fig. 1.
(A) Weighted correlation network analysis of tomato metabolites and their assigned clusters based on their known biochemical classification. The size of each metabolite node indicates betweenness centrality, a measure of how often a node exists on the shortest path between other nodes. The thickness of the lines connecting metabolites is scaled relative to the correlation between the metabolites. The identity of each metabolite is denoted by number in the legend. (B) Distribution of metabolite concentrations for each volatile group across the tomato population. Volatile concentrations are reported in nanograms per gram fresh weight per hour (ng/gfw/h) on a log 10 scale.
Fig. 2.
(A) Weighted correlation network analysis of blueberry metabolites and their assigned clusters based on their known biochemical classification. The size of each metabolite node indicates betweenness centrality. The thickness of the lines connecting metabolites is scaled relative to the correlation between the metabolites. The identity of each metabolite is denoted by number in the legend. (B) Distribution of metabolite concentrations for each volatile group across the blueberry population. Volatile concentrations are reported in nanograms per gram fresh weight per hour (ng/gfw/h) on a log 10 scale.
Contributions of Sugars, Acids, and Volatiles to Flavor Perceptions.
In order to determine if the fruit metabolome could explain variation in consumer sensory panel ratings, we partitioned the metabolites into modules according to their biochemical classifications (Figs. 1 and 2 and Datasets S1 and S2). We then separated the consumer sensory variance into aggregated components explained by each module (Fig. 3, SI Appendix, Fig. S1, and Dataset S3). We further combined the individual variance components into two main groups for analysis: sugars/acids and volatiles (Fig. 3). In both tomato and blueberry, a large proportion of the variance was explained by sugars/acids and volatiles, while little variance was attributed to the residuals (Fig. 3). Furthermore, the proportion of variance explained by sugars/acids varied across the flavor attributes and contrasted between the two species. For instance, 77% of the tomato sourness variance was explained by the content of sugars/acids, while these compounds only explained 43% of blueberry sourness. Similarly, while sugars/acids predominantly (60%) explained blueberry sweetness, a larger portion of the tomato phenotypic variance (62%) could be explained by the volatiles. As previously described (3), the results indicate the large influence that volatile compounds can have on sensory attributes in both species, which in turn highlights how important these compounds are to breeding programs for improvement of fruit flavor. For example, the variance decomposition of overall liking score estimates that 42% and 56% of the variance was explained by volatile organic compounds in tomato and blueberry, respectively.
Fig. 3.
Variation in sensory panel ratings explained by sugars/acids and volatiles overall, and by groups of metabolites of known biochemical classification in tomato and blueberry.
To further understand how the fruit chemical profile affected consumer flavor, we analyzed the variance explained by each metabolite module. The sugar module was a strong driver of liking (43% in tomato and 18% in blueberry) and sweetness (29% in tomato and 27% in blueberry), while the module representing acids drove sourness (54% in tomato and 38% in blueberry) in both fruits. Some volatile modules were found to make large contributions to flavor ratings. For instance, phenylalanine-derived and lipid-derived compounds contributed to sweetness perception (34 and 16%, respectively) and overall liking score (16 and 13%, respectively) in tomato. Lipid-derived volatiles and compounds grouped as carotenoid/terpenes explained 15 and 21% of blueberry overall liking score, respectively. These results are consistent with previous results that showed strong positive correlations of specific volatiles with fruit sweetness (4, 10).
Predicting Consumer Preferences.
Eighteen statistical and machine-learning methods were employed to predict sensory traits from sugar, acid, and volatile concentrations. Each model was evaluated in a 10-fold cross-validation and each fold was assessed by the correlation between predicted and observed consumer taste panel ratings (Fig. 4A, SI Appendix, Fig. S1, and Datasets S4 and S5). The cross-validation was repeated 10 times and results were averaged for the final prediction accuracies. We observed the highest prediction accuracies from the XGBoost, gradient-boosting machines, and random-forest models. The XGBoost model showed an average improvement of 20% over the linear regression and 11% over PLS, models traditionally used in food science applications. The accuracy for the model that, on average, performed the best (XGBoost) ranged from 0.62 to 0.87 across all traits and in both species. We found the most predictable traits in tomato to be sweetness (0.8), flavor intensity (0.77), and sourness (0.69) and the most predictable traits in blueberry to be sourness (0.87) and sweetness (0.75). The improvement of the full model that accounted for all the compounds over the model that included only sugars and acids ranged from 3.2 to 36.7% (SI Appendix, Table S1).
Fig. 4.
(A) Accuracy of predicting flavor ratings from metabolome data across a range of statistical and machine learning models for tomato and blueberry. Averages and model rankings are inclusive of umami accuracies depicted in SI Appendix, Fig. S1 and Datasets S4 and S5. (B) Accuracy of predicting perception traits for tomato using 70 individuals with genomic and metabolomic data. (C) Accuracies for tomato flavor prediction with a consistent test set of 39 samples and increasing training set sizes ranging from 50 to 170 samples.
To further evaluate the opportunity to use metabolomic selection in breeding and to understand its prediction potential compared to genomic selection, we compiled information from 70 varieties of tomato for which we had whole-genome sequence, chemical profile, and sensory panel data (5). Using a 10-fold cross-validation, we applied the genomic selection gBLUP (genomic best linear unbiased prediction) method (27) to predict the consumer sensory ratings from a subset of 79,821 single-nucleotide polymorphisms (SNPs) (Fig. 4B and Dataset S6). We then used the metabolomic information for the same 70 varieties and the same cross-validation partitioning to predict the panel ratings. These 70 genotypes represent a subset of the total 147 accessions. We found that metabolomic selection outperformed genomic selection in the prediction of all these complex traits, especially for sweetness and overall flavor liking. For these traits, the accuracies of metabolomic selection using 70 genotypes were 0.68 and 0.45 for sweetness and overall flavor liking, respectively. These traits were poorly predicted by gBLUP with accuracies of 0.16 and −0.11 for sweetness and overall flavor liking, respectively. While these results are not surprising, given the small population size and the fact that the metabolite data are capturing both genetic and environmental components of fruit flavor, they highlight the complexity of flavor perception as a breeding target and the potential of metabolomic selection as a phenotyping tool to support breeding programs compared with other available methods such as genomic selection, for example.
Next, in order to test how many samples are needed to train metabolomic selection models, we performed a subsampling analysis in tomato. For this analysis we randomly selected 39 samples as the test set and trained the model with increasing training set sample sizes from 50 to 170 in steps of 10. We repeated this process 10 times and averaged the accuracies at each sample size (Fig. 4C and Dataset S7). We found that the accuracies predominantly increased with increasing sample sizes but note that the accuracies can be relatively high for certain traits using as few as 50 samples. Sourness was more accurately predicted with the gBLUP and Bayes A models, while gradient-boosting machines achieved higher accuracies when predicting the more complex traits like overall liking.
Metabolites Associated with Desirable Flavor.
In order to find sugars, acids, and volatiles that enhance or suppress consumer sensory perceptions of flavor, models for each fruit were trained using all samples for which we had metabolome and sensory panel data (209 for tomato and 244 for blueberry). Two contrasting modeling approaches, Bayes A and gradient-boosting machines, were chosen for further inference analysis. In Bayes A, the beta coefficients indicate the individual additive effect of that chemical free of interactions. This coefficient predicts if a chemical is important for enhancing the flavor attribute (positive value) or decreasing the flavor attribute (negative value). For gradient-boosting machines the variable importance represents the marginal effect of that chemical including the interaction effects with other chemicals. This value is scaled between 0 and 100 where 0 is a not an important predictor and 100 is an important predictor (Fig. 5).
Fig. 5.
Relative importance estimated with gradient-boost machine (x axis) and β coefficients estimated with Bayes A (y axis) of each metabolite in predicting ratings of flavor attributes in blueberry and tomato. Colors indicate the type of metabolite. TA, titratable acidity.
For sweetness in tomato, we found glucose and fructose to be the most important sensory perception enhancers. The gradient boosting machines also estimated 1-penten-3-one and 2-phenylethanol to be important for perceived sweetness, while the Bayes A model highly ranked two volatiles (E-2-pentenal and 4-carene) to be important for sweetness enhancement. E-2-pentenal was also found to be an important contributor to overall flavor intensity and umami (SI Appendix, Fig. S1). In blueberry, components important for liking included soluble solids, fructose, and glucose. Additionally, volatiles found to be important for enhancing liking included 2-undecanone, 2-hexenyl-butyrate, and ethyl propionate, while volatiles that were negative to liking included eucalyptol and phenylacetaldehyde. Interestingly, two lipid-derived volatiles (2-hexen-1-al and 2-pentenal) had a high positive contribution and the highest negative contributions to sourness in blueberry, respectively. Glutamic acid was highly ranked by both methods as influencing umami perception for tomato, which by definition represents the taste of the amino acid l-glutamate. Three phenylalanine-derived compounds (benzyl cyanide, 2-phenyl ethanol, and 1-nitro-2-phenylethane) were also highly ranked by gradient-boosting machines as umami influencers (SI Appendix, Fig. S1). It is important to note, however, that this targeted metabolomic panel is enriched for putative sugar-enhancing compounds and may be limited in the characterization of compounds affecting umami.
Discussion
Fruit flavor is a complex trait at the intersection between the fruit biochemistry and the consumer sensory perception. Quantification of sensory perception using consumer flavor panels is time- and resource-consuming and not readily amenable to a high-throughput assay, which has hindered plant breeders from selecting for fruit flavor for many years. This has contributed to the widespread decline of consumer satisfaction of many commercial fruit varieties (2, 7). Recently, different high-throughput phenotyping applications were proposed to use two-dimensional visible light imaging as proxies for plant biomass (28), reflectance ratios as proxies for yield (29), hyperspectral reflectance as proxies for leaf chlorophyll and nitrogen content (30), and canopy temperature as proxies for drought response (31). Here, in order to create higher-throughput flavor phenotyping methods, we applied statistical and machine learning models that can predict consumer sensory panel ratings from the chemistry of a fruit.
Chemical Profiling of a Fruit.
Although flavor is a complex trait, relatively simple metrics have historically been used to quantify flavor preferences in most breeding programs, including titratable acidity, soluble solids, firmness, and the breeder “bite tests” (32, 33). Using two independent fruit species we showed that volatiles play an important role in consumer flavor perception and should therefore be broadly assayed when selecting for enhanced flavor profiles. In this case, a metabolomic approach will achieve a higher selection accuracy by identifying metabolites with small but nonnegligible effects.
In recent years, the cost of targeted and untargeted metabolomics has decreased (34) and the throughput for metabolite profiling has increased. The largest cost in this system is labor to process the fruit and to analyze the data. For the profiling described here, we estimate an in-house cost per sample similar to the per-sample genotyping costs used for genomic selection in many species, which in turn can be an order of magnitude cheaper than sensory evaluations. While these estimates assume an in-house analysis and do not consider the capital expenditure to acquire the instrumentation, it highlights the per-sample cost reduction over the years and the feasibility of its high-throughput application in plant breeding programs.
Network analysis of tomato flavor compounds demonstrated correlations among biochemicals in the same biosynthetic pathways (Fig. 1). The associations are consistent with the postulated biochemical groupings identified by Buttery and Ling (35), Baldwin et al. (36), and Mathieu et al. (37). The chemistry of blueberry flavor has not been as extensively studied as that of tomato. Our results offer insights into the biochemical pathways for blueberry volatile synthesis. For instance, the long chain lipid–derived volatiles (denoted in Fig. 2 as numbers 34, 35, 36, 37, and 38) are found linked together within the lipid-derived volatile cluster. Also, linalool levels are not correlated with levels of other terpenes, suggesting that linalool biosynthesis may not be regulated in the same manner as other terpenes.
Applications of Metabolomic Selection to Breeding of Fruits and Vegetables.
One alternative to phenotype fruits and vegetables for flavor quality is the establishment of consumer sensory panels. This approach has low throughput, from a breeding standpoint, because a sensory panel can usually only taste a limited number of samples (4–6) per day. The tomato and blueberry breeding programs at the University of Florida have been using consumer sensory panels to guide breeding decisions for several years (5, 10). To do this, selections currently in development by the breeding programs are subjected to biochemical analysis and simultaneous consumer evaluations each year. However, due to the low throughput of the assay, sensory characterization is typically performed in the final stages of selection prior to release, when favorable alleles may no longer be segregating in the population.
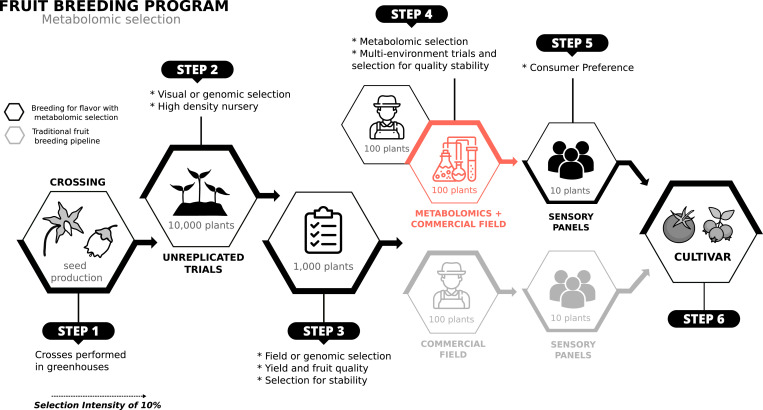
The use of metabolomic profiling as a phenotyping assay can enable accurate characterization of flavor profiles in earlier stages of a breeding program, when more genetic variability is available for selection (Fig. 6). Metabolic profiling at earlier stages of the breeding program opens up the possibility of identification of superior flavor genotypes that may otherwise have been discarded. Chemical profiling of fruits can capture the genetic potential of the variety as well as environmental variability. To reduce this variability and generate a phenotype that better represents the genetic potential of the individual, the breeder can characterize replicates from different environments and/or harvests. Replicates within plots in a single experiment, within harvest dates within a season, or even within environments can be pooled prior to running the instrument, maintaining the per-sample cost and resulting in an average prediction of the genotypic effect. Furthermore, in situations where fruit quality is under the influence of genotype-by-environment interactions, the breeder may choose to estimate fruit quality stability by profiling the chemical composition in each environment. While this additional analysis would increase the per-genotype cost of the analysis, the information would facilitate selection of stable genotypes that perform well across multiple environments.
Fig. 6.
Schematic representation of how the use of metabolomic selection could be applied in earlier stages of a breeding program, when compared to sensory panels.
Moreover, the use of metabolomic selection to estimate flavor perception complements a molecular breeding program. A by-product of applying metabolomic selection is the metabolomic profiling of many breeding lines, which in turn enables QTL mapping or genome-wide association study (GWAS) against metabolomic datasets. Thus, flavor-related metabolites identified by metabolomic selection could then be further used in GWAS analysis to identify the genes/loci contributing and create markers for molecular breeding (5, 22). This two-step approach can enable the use of MAS at the earliest stages of a breeding program and thereby speed up the genetic enrichment for flavor associated traits (Fig. 6, step 2). This approach is especially useful for fruit crops where there is much less available information on markers affecting flavor chemical composition. It is important to note that the chemical composition of a fruit can be highly affected by weather and agronomic practices. Like other quantitative phenotypes currently evaluated in breeding programs, flavor-related traits have large variability and low heritability and are subjected to complex interaction effects (38–40). With the availability of data from multiple environments, the MAS or genomic selection application can also be tailored to selecting early for stability of important metabolic classes. However, MAS alone cannot address the complexity of flavor perception. Hence, the value of metabolomic selection is derived from including all metabolites in the prediction models, even those with small effects, leading to better overall estimates of flavor perception. Thus, the most practical application of metabolomic selection is in the middle stages of a breeding program where genes involved in the biosynthesis of inferred volatiles still retain enough genetic variability to select flavorful cultivars (Fig. 6, steps 3 and 4). Finally, consumer sensory analyses can be restricted to the final stages, in which a few target genotypes will be subject to consumer evaluations prior to release (Fig. 6, step 5).
Machine Learning Models Can Accurately Predict Flavor Attributes.
The use of metabolomics to predict flavor attributes has important implications not only in plant breeding but also in food science and genetics research. Prediction using metabolomics is challenging due to the correlated nature of the metabolomic predictors since it requires a large number of sensory panels for model calibration. Flavor prediction has been attempted before using linear regression models (41, 42), random-forest models (39), and PLS regression (10), achieving variable levels of prediction accuracies. One of the objectives of our work was to evaluate a range of statistical and machine learning models to determine the best performers for metabolite-based phenotype prediction of flavor quality traits. Importantly, we wanted to access the predictive power of methods known to handle correlated features well and thus simultaneously predict the effect of all metabolites. Identification of the most accurate predictive models provides a simple way to improve phenotyping accuracy with the same available dataset. Here, machine learning models such as gradient-boosting machines and XGBoost were the most predictive across all the traits and in both species, whereas multiple linear regression and PLS methods were found to be the least predictive. Considering that PLS is still the standard in food science applications (43), these proposed predictive models show marked improvements with increases relative to PLS ranging from 3.3% for sweetness in blueberry to 44.6% for umami in tomato. Furthermore, the fact that the models worked well in two entirely different systems (blueberry and tomato; breeding population and diversity panel) supports the effectiveness of the models proposed.
To better understand the factors affecting flavor perception in each fruit species, we grouped compounds based on their biochemical classification and estimated a proportion of the phenotypic variance associated with each group. Biochemical pathways that were represented by a small number of chemicals were also grouped to minimize the effect of sampling variance in the creation of distance (variance/covariance) matrices (44). As would be expected, sugars (glucose, fructose, and soluble solids) were important predictors of sweetness as well as overall liking in both crops, while acids explained a large portion of the sourness variance. By grouping volatiles by their biochemical pathway, we were able to estimate a proportion of the total variance jointly explained by the chemicals within each group. Phenylalanine- and lipid-derived volatiles explained a large fraction of flavor variance in tomato, while lipid-derived, esters, and carotenoid/terpenoid volatiles explained most of the blueberry variance for liking score. Interestingly, ester compounds were shown to be negatively selected in red-fruited tomato as compared to related green-fruited species (45, 46), which potentially explains the lack of contribution to liking score in tomato contrasting to blueberry. Although tomato is botanically a fruit, it is not used as such in most cuisines. Thus, the fruity esters that are so important for flavor in most fruits do not serve the same function in a tomato.
The statistical models were used to infer which volatiles contribute to each flavor attribute (Fig. 5). Although many of these compounds have been shown to contribute to tomato liking and flavor intensity, our results show that several compounds including E-3-hexen-1-ol; (E,E)-2-,4-decadienal; and benzyl alcohol are important flavor components. Although benzyl alcohol and (E,E)-2-,4-decadienal were shown to contribute to flavor intensity, they did not contribute to overall liking when a simple regression or multivariate analysis was used (4, 5). Also, the contributions of methional and benzothiazole to sourness intensity are interesting, as these compounds have not typically been associated with sour flavor. Methional odor is described as malty or cooked potato-like, while benzothiazole odor is described as sulfurous or meaty. Multiple linear regression analysis of volatiles associated with sweetness identified three that contributed to sweetness independently of sugars, but the relative contribution of these volatiles was not determined (4). By grouping volatiles by biochemical pathway and using a linear mixed model, the important role of volatiles in sweetness perception was highlighted. These results also emphasize the need to include volatile detection in breeding programs because by focusing only on sugars and acids during breeding, part of the flavor profile may be lost (Fig. 3). On the other hand, the results suggest that the magnitude of the effect of each individual volatile is much smaller than the individual effect of sugar compounds, highlighting the complexity of breeding for fruit flavor and the challenges to improve flavor using MAS. The important contribution of volatiles to overall liking of tomatoes is illustrated by the negative effects of extended refrigeration on volatile contents and consumer preferences (47). Refrigeration substantially reduces volatile contents but not sugars or acids (48).
Conclusions and Future Directions.
In this work we demonstrate the comparison of different algorithms to predict consumer preferences. This information can benefit plant breeding programs to improve flavor perception of new varieties. It is important to note that while we believe that the approach outlined here is generally useful, the specific chemical contributions to overall liking will likely vary with the ethnic and geographic makeup of the consumer panel. Future extensions of this approach could include the modeling of information and parameters for each individual in the panel, such as the inclusion of demographic parameters to predict more nuanced variations in taste preferences. In summary, by creating predictive models for consumer perceptions of flavor we are able to increase the throughput of flavor phenotyping and provide new tools to make more informed, flavorful selections in breeding programs. Through inference, candidate flavor enhancers and suppressors were identified, indicating the possibility of their use as natural food additives in the food industry. Furthermore, genes involved in biosynthesis of these flavor enhancing/suppressing metabolites can now be targets for marker assisted selection or direct engineering of more flavorful fruit varieties.
Materials and Methods
Data.
Prediction analysis was carried in two fruit species: tomato (S. lycopersicum) and blueberry (Vaccinium spp.). For tomato, 68 sugars, acids, and volatiles were analyzed in 147 genotypes grown and evaluated in multiple seasons. A total of 209 samples were used, with 160 samples having been previously evaluated (4, 5). For blueberry, firmness and 55 sugars, acids, and volatiles were analyzed. Firmness was only available for a small number of genotypes in blueberry, but it was kept in the model since it is an important component of blueberry quality (49). Sixty-three genotypes were grown and evaluated in multiple seasons for a total of 244 samples, of which 164 were evaluated previously (10). Fruit flavor of tomato and blueberry accessions was assessed by consumer sensory panels. Our sensory panels averaged ∼80 participants sampled from a diverse university population (Datasets S8 and S9) with the intention to represent for potential person-to-person variation in flavor preferences. This study was approved by the University of Florida Institutional Review Board 2 (case #2003-U-0491). All participants provided informed consent. Panels were conducted in the Food Science and Human Nutrition Department at the University of Florida in Gainesville, FL. Flavor attributes including sweetness, sourness, flavor intensity, and overall liking were rated. Additionally, sensory perceptions of umami were quantified solely for tomato. Overall liking was rated on a scale from −100 to 100, while the remaining attributes were rated from 0 to 100 (3). All data were normalized to a mean of 0 and a variance of 1 for further analyses. Missing data were imputed by the mean value per metabolite. Volatile concentrations were quantified by gas chromatography as described in ref. 23, while sugars, soluble solids, and acids were quantified as described in ref. 50. Sensory analysis was conducted as described in Tieman et al. (4), and scaled data can be found in Datasets S1 and S2. All blueberry data collection was described in Gilbert et al. (10). Network analysis was performed using the R package WGCNA (51). Briefly, the pairwise Pearson correlation coefficient between each pair of metabolites was used to construct a weighted metabolite coexpression network. The process assumed an unsigned network and the network was visualized and represented using Cytoscape 3.7.1 (52). The network for each species is provided as a Cytoscape file in the GitHub repository.
Calculating Contributions of Metabolites to Flavor Ratings.
To estimate the proportion of variance in flavor ratings that each metabolite group explains, we divided the metabolites identified in tomato and blueberry in six (Nonaromatic Amino Acid-derived, Carotenoid/Terpenes, Lipid derived, Phe-derived, Sugars, and Acids) and seven groups (Nonaromatic Amino Acid-derived, Carotenoid/Terpenes, Lipid-derived, Ester, Phe-derived, Sugars, and Acids), respectively. In tomato, for example, we fit a linear model in which
where is the averaged consumer rating for cultivar , is the fixed model intercept, is a normally distributed and independent random residual effect; are design matrices for random effects associated to each biochemical group, and are the random terms associated to the chemical groups. For each random term , we assumed , where represents the Gaussian kernel matrices built as the pairwise Euclidean distance between each chemical in a given group (MVN; multivariate normal distribution). The proportion of the variance explained by a given metabolomic group was determined by , where is the variance component estimated for a given metabolomic group and the denominator is the variance represented by the sum of the variance explained by all other chemical groups () and the residual term () . To further represent the contribution of sugar/acids versus volatiles, we also presented it separately by summing the variance components estimated within each group. All analyses were carried out using the ASReml-R package (53).
Comparing Genomic and Metabolomic Selection.
In order to compare prediction performance between genomic and metabolomic selection models, we organized a group of 70 tomato accessions that had whole genome sequencing, metabolomic evaluation, and sensory panel information. The genomic data comprised 26,262,280 SNPs, which were mapped to the S. lycopersicum reference genome SL3.0 as described in ref. 5. We applied additional quality filters and retained only biallelic SNPs with minor alleles frequencies ≥0.1, excluded markers mapped on the chromosome 0 (unassigned scaffolds), and considered no more than 30% missing data and 20% heterozygosity rate. Using the SNPRelate R package (54). we removed redundant SNPs by pruning markers defined as r2 ≥ 0.9 in a 100-kilobase genome window. After this step, we retained 79,821 SNPs used in the genomic prediction steps.
Sensory traits were predicted using gBLUP models and compared to metabolomic predictions. The general model for genetic values is , where is the vector of observed values, is the fixed model intercept, and is a design matrix that relates the vector of random genetic effects. For genomic prediction, the random effect has null mean and a kernel covariance matrix () that represents the realized relationship among individuals computed as described by ref. 27. For metabolomic prediction, the kernel was defined in the Euclidean space as described by ref. 55. Residuals were defined as normally distributed and independent. As the number of accessions with metabolomic data are larger than the number of genotyped individuals, we considered the same number of individuals for the genomic and metabolomic prediction (70 individuals). To evaluate the prediction performance of each model, a 10-fold cross-validation was employed, described in the following section.
Cross-Validation.
To evaluate the predictive performance of each model, a 10-fold cross-validation was employed. In this way, the dataset was randomly split into 10 equal groups of varieties. For each of the ten iterations, nine groups of varieties were used to train the model (training set), and one unseen group of varieties was used as a “holdout” group to test the model (test set). During training, a secondary, nested 10-fold cross-validation was used to calibrate the tuning parameters of the machine learning models. The root-mean-squared error between predicted and observed flavor ratings in the secondary test set was minimized to obtain the optimal parameter values for the primary model. The trained models were then applied to the metabolite concentrations of the varieties within the primary test set and predicted flavor ratings were obtained. The correlation between predicted and observed flavor ratings is recorded. The average correlation of predicted and observed flavor ratings in the test set is referred to here as the accuracy of the model.
Statistical Models.
A diverse sample of 18 statistical and machine learning models representing a range of regression, regularization, genomic selection, decision tree, and neural network models were chosen for assessment. These include a linear model and PLS as our baseline models; regularization methods such as ridge regression, elastic net, and LASSO; kernel methods such as support vector machines, relevant vector machines, and reproducing kernel Hilbert space; neural network models such as a multilayer perceptron neural network and a Bayesian neural network; decision tree-based models such as random forest, gradient boosting machines, and XGBoost; and models frequently used in genomic selection such as Bayes A, Bayes B, and Bayes Cπ. Each model has its individual strengths, weaknesses, and assumptions. Here we assess which models are most useful for the application of flavor phenotyping by metabolomic selection. All models were implemented in R (56). Each model is described in more detail in SI Appendix. The Bayesian models were implemented in BGLR (57) and the machine learning models were implemented with caret (58) and a package specific to each model (Dataset S10).
Supplementary Material
Acknowledgments
We thank previous students who, as part of the University of Florida tomato and blueberry breeding program, supported the panel data collection and volatile extraction. This work was supported by the University of Florida royalty fund generated by the licensing of blueberry cultivars, by the NSF (award numbers IOS 1855585 to D.T. and H.K. and IOS 1564366 to D.T.), and by the National Institute of Food and Agriculture (award number SCRI 2018-51181-28419 to M.F.R.R.). We thank HiPerGator and University of Florida Research Computing for providing computational resources and support and Rebecca Key for drawing the tomato and blueberry artwork used in the figures.
Footnotes
Reviewers: E.B., Agricultural Research Service, US Department of Agriculture; and M.W., University of Arkansas, Fayetteville.
The authors declare no competing interest.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2115865119/-/DCSupplemental.
Data Availability
Sensory panel ratings and metabolite concentrations are provided in Datasets S1 and S2. Underlying data for Fig. 3 are provided in Dataset S3. Model accuracies in Fig. 4 are provided in Datasets S4–S7. Relevant scripts are provided in the GitHub repository at https://github.com/Resende-Lab/metabolomic_selection_for_enhanced_fruit_flavor. All other study data are included in the article and/or supporting information.
References
- 1.Fernqvist F., Hunter E., Who’s to blame for tasteless tomatoes? The effect of tomato chilling on consumers’ taste perceptions. Eur. J. Hortic. Sci. 77, 193–198 (2012). [Google Scholar]
- 2.Klee H. J., Tieman D. M., The genetics of fruit flavour preferences. Nat. Rev. Genet. 19, 347–356 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Bartoshuk L. M., Klee H. J., Better fruits and vegetables through sensory analysis. Curr. Biol. 23, R374–R378 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Tieman D., et al. , The chemical interactions underlying tomato flavor preferences. Curr. Biol. 22, 1035–1039 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Tieman D., et al. , A chemical genetic roadmap to improved tomato flavor. Science 355, 391–394 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Bruhn C. M., et al. , Consumer perceptions of quality: Apricots, cantaloupes, peaches, pears, strawberries, and tomatoes. J. Food Qual. 14, 187–195 (1991). [Google Scholar]
- 7.Klee H. J., Improving the flavor of fresh fruits: Genomics, biochemistry, and biotechnology. New Phytol. 187, 44–56 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Shepherd G. M., Smell images and the flavour system in the human brain. Nature 444, 316–321 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Meuwissen T. H., Hayes B. J., Goddard M. E., Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert J. L., et al. , Identifying breeding priorities for blueberry flavor using biochemical, sensory, and genotype by environment analyses. PLoS One 10, e0138494 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liebal U. W., Phan A. N. T., Sudhakar M., Raman K., Blank L. M., Machine learning applications for mass spectrometry-based metabolomics. Metabolites 10, 1–23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Habier D., Fernando R. L., Kizilkaya K., Garrick D. J., Extension of the Bayesian alphabet for genomic selection. BMC Bioinformatics 12, 186 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de los Campos G., et al. , Predicting quantitative traits with regression models for dense molecular markers and pedigree. Genetics 182, 375–385 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoerl A. E., Kennard R. W., Ridge regression: Biased estimation for nonorthogonal problems. Technometrics 42, 80–86 (2000). [Google Scholar]
- 15.Pérez P., de Los Campos G., Crossa J., Gianola D., Genomic-enabled prediction based on molecular markers and pedigree using the Bayesian linear regression package in R. Plant Genome 3, 106–116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zingaretti L. M., et al. , Exploring deep learning for complex trait genomic prediction in polyploid outcrossing species. Front. Plant Sci. 11, 25 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrmann A., Schauer N., “Metabolomics-assisted plant breeding” in The Handbook of Plant Metabolomics, Weckwerth W., Kahl G., Eds. (Wiley-VCH Verlag GmbH & Co. KGaA, 2013), pp. 245–254. [Google Scholar]
- 18.Fernie A. R., Schauer N., Metabolomics-assisted breeding: A viable option for crop improvement? Trends Genet. 25, 39–48 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Gemmer M. R., et al. , Can metabolic prediction be an alternative to genomic prediction in barley? PLoS One 15, e0234052 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toubiana D., et al. , Metabolic profiling of a mapping population exposes new insights in the regulation of seed metabolism and seed, fruit, and plant relations. PLoS Genet. 8, e1002612 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z., et al. , Non-invasive plant disease diagnostics enabled by smartphone-based fingerprinting of leaf volatiles. Nat. Plants 5, 856–866 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Ferrão L. F. V., et al. , Genome-wide association of volatiles reveals candidate loci for blueberry flavor. New Phytol. 226, 1725–1737 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Tieman D. M., et al. , Identification of loci affecting flavour volatile emissions in tomato fruits. J. Exp. Bot. 57, 887–896 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Chen G., et al. , Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. Plant Physiol. 136, 2641–2651 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen J., et al. , A 13-lipoxygenase, TomloxC, is essential for synthesis of C5 flavour volatiles in tomato. J. Exp. Bot. 65, 419–428 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao L., et al. , The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat. Genet. 51, 1044–1051 (2019). [DOI] [PubMed] [Google Scholar]
- 27.VanRaden P. M., Efficient methods to compute genomic predictions. J. Dairy Sci. 91, 4414–4423 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Golzarian M. R., et al. , Accurate inference of shoot biomass from high-throughput images of cereal plants. Plant Methods 7, 2 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Babar M. A., Van Ginkel M., Klatt A. R., Prasad B., Reynolds M. P., The potential of using spectral reflectance indices to estimate yield in wheat grown under reduced irrigation. Euphytica 150, 155–172 (2006). [Google Scholar]
- 30.Ge Y., et al. , High-throughput analysis of leaf physiological and chemical traits with VIS-NIR-SWIR spectroscopy: A case study with a maize diversity panel. Plant Methods 15, 66 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henry A., Gowda V. R. P., Torres R. O., McNally K. L., Serraj R., Variation in root system architecture and drought response in rice (Oryza sativa): Phenotyping of the OryzaSNP panel in rainfed lowland fields. Field Crop. Res. 120, 205–214 (2011). [Google Scholar]
- 32.Farneti B., et al. , Exploring blueberry aroma complexity by chromatographic and direct-injection spectrometric techniques. Front. Plant Sci. 8, 617 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kerr E. A., Note on selecting sweet corn for eating quality in early generations of inbreeding. Can. J. Plant Sci. 41, 438–439 (1961). [Google Scholar]
- 34.Goldansaz S. A., et al. , Livestock metabolomics and the livestock metabolome: A systematic review. PLoS One 12, e0177675 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buttery R. G., Ling L. C., Volatile Components of Tomato Fruit and Plant Parts (American Chemical Society, 1993), pp. 23–34. [Google Scholar]
- 36.Baldwin E. A., Scott J. W., Shewmaker C. K., Schuch W., Flavor trivia and tomato aroma: Biochemistry and possible mechanisms for control of important aroma components. HortScience 35, 1013–1022 (2000). [Google Scholar]
- 37.Mathieu S., et al. , Flavour compounds in tomato fruits: Identification of loci and potential pathways affecting volatile composition. J. Exp. Bot. 60, 325–337 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kouassi A. B., et al. , Estimation of genetic parameters and prediction of breeding values for apple fruit-quality traits using pedigreed plant material in Europe. Tree Genet. Genomes 5, 659–672 (2009). [Google Scholar]
- 39.Eggink P. M., et al. , Prediction of sweet pepper (Capsicum annuum) flavor over different harvests. Euphytica 187, 117–131 (2012). [Google Scholar]
- 40.Longin F., et al. , Aroma and quality of breads baked from old and modern wheat varieties and their prediction from genomic and flour-based metabolite profiles. Food Res. Int. 129, 108748 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Abegaz E. G., Tandon K. S., Scott J. W., Baldwin E. A., Shewfelt R. L., Partitioning taste from aromatic flavor notes of fresh tomato (Lycopersicon esculentum, Mill) to develop predictive models as a function of volatile and nonvolatile components. Postharvest Biol. Technol. 34, 227–235 (2004). [Google Scholar]
- 42.Schwieterman M. L., et al. , Strawberry flavor: Diverse chemical compositions, a seasonal influence, and effects on sensory perception. PLoS One 9, e88446 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wold S., Sjöström M., Eriksson L., PLS-Regression: A Basic Tool of Chemometrics in Chemometrics and Intelligent Laboratory Systems (Elsevier, 2001), pp. 109–130. [Google Scholar]
- 44.Yang J., et al. , Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goulet C., et al. , Role of an esterase in flavor volatile variation within the tomato clade. Proc. Natl. Acad. Sci. U.S.A. 109, 19009–19014 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goulet C., et al. , Divergence in the enzymatic activities of a tomato and Solanum pennellii alcohol acyltransferase impacts fruit volatile ester composition. Mol. Plant 8, 153–162 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Raffo A., et al. , Impact of different distribution scenarios and recommended storage conditions on flavor related quality attributes in ripening fresh tomatoes. J. Agric. Food Chem. 60, 10445–10455 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Zhang B., et al. , Chilling-induced tomato flavor loss is associated with altered volatile synthesis and transient changes in DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 113, 12580–12585 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gallardo R. K., et al. , Breeding trait priorities of the blueberry industry in the United States and Canada. HortScience 53, 1021–1028 (2018). [Google Scholar]
- 50.Vogel J. T., et al. , Carotenoid content impacts flavor acceptability in tomato (Solanum lycopersicum). J. Sci. Food Agric. 90, 2233–2240 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shannon P., et al. , Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Butler D. G., Cullis B. R., Gilmour A. R., Gogel B. J., ASReml-R Reference Manual (Queensland Department of Primary Industries, Fisheries and Forestry, Brisbane, Australia, 2009). [Google Scholar]
- 54.Zheng X., et al. , A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morota G., Gianola D., Kernel-based whole-genome prediction of complex traits: A review. Front. Genet. 5, 363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.R Core Team, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, 2020). [Google Scholar]
- 57.Pérez P., de los Campos G., Genome-wide regression and prediction with the BGLR statistical package. Genetics 198, 483–495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuhn M., caret: Classification and Regression Training. https://cran.r-project.org/package=caret (2020). Accessed 25 January 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sensory panel ratings and metabolite concentrations are provided in Datasets S1 and S2. Underlying data for Fig. 3 are provided in Dataset S3. Model accuracies in Fig. 4 are provided in Datasets S4–S7. Relevant scripts are provided in the GitHub repository at https://github.com/Resende-Lab/metabolomic_selection_for_enhanced_fruit_flavor. All other study data are included in the article and/or supporting information.