Evolutionary origin of pathogenic variants in human BRCA1 and BRCA2.
Abstract
BRCA1 and BRCA2 (BRCA) play essential roles in maintaining genome stability. BRCA germline pathogenic variants increase cancer risk. However, the evolutionary origin of human BRCA pathogenic variants remains largely elusive. We tested the 2,972 human BRCA1 and 3,652 human BRCA2 pathogenic variants from ClinVar database in 100 vertebrates across eight clades, but failed to find evidence to show cross-species evolution conservation as the origin; we searched the variants in 2,792 ancient human genome data, and identified 28 BRCA1 and 22 BRCA2 pathogenic variants in 44 cases dated from 45,000 to 300 yr ago; we analyzed the haplotype-dated human BRCA pathogenic founder variants, and observed that they were mostly arisen within the past 3,000 yr; we traced ethnic distribution of human BRCA pathogenic variants, and found that the majority were present in single or a few ethnic populations. Based on the data, we propose that human BRCA pathogenic variants were highly likely arisen in recent human history after the latest out-of-Africa migration, and the expansion of modern human population could largely increase the variation spectrum.
Introduction
Repairing the damaged DNA by environmental and metabolic factors is vital for all lives on earth. In eukaryotes, this is achieved by the DNA damage repair system composed of multiple pathways to repair different types of DNA damage (Jeggo et al, 2016). The homologous recombination pathway repairs double-strand DNA damage by joint activities of a group of genes to reach error-free repair of DNA double-strand break (Jasin & Rothstein, 2013). BRCA1 and BRCA2 (BRCA) are two of the essential genes in the homologous recombination pathway. BRCA1 was arisen in animal and plant kingdoms 1.2 billion yr ago, and BRCA2 was arisen in fungus, plant, and animal kingdoms 1.6 billion yr ago (Kojic et al, 2002; Pfeffer et al, 2017). Studies revealed that BRCA across wide-range species including most of the primates is evolutionarily highly conserved by negative selection in reflecting its essential roles in maintaining genome stability. In contrast, however, BRCA in humans and its closest living relatives of chimps and bonobos is rapidly evolving by positive selection (Huttley et al, 2000; Fleming et al, 2003; Abkevich et al, 2004; Pavlicek et al, 2004; Burk-Herrick et al, 2006; O’Connell, 2010; Lou et al, 2014). The positive selection likely resulted in the new function of BRCA gained in these three species, such as promoting immunity to counter viral infection (Lou et al, 2014), regulating gene expression and metabolism (Rosen et al, 2006; Chen et al, 2020), enhancing neural development (Pao et al, 2014), and increasing reproduction (Smith et al, 2013).
BRCA is one of the best-known genetic predisposition genes for cancer (Narod & Foulkes, 2004). Efforts made in the past decades have identified nearly 70,000 human BRCA variants (Cline et al, 2018) (https://brcaexchange.org/factsheet). A part of human BRCA variants is determined as “pathogenic” or “likely pathogenic” (BRCA PLP) in causing high cancer risk affecting mostly breast and ovary (Welcsh & King, 2001). BRCA PLP is widely used in clinical practice as the marker for cancer diagnosis, prevention, prognosis, and treatment through synthetic lethal mechanism (Anderson et al, 2008; Plon et al, 2008; Huen et al, 2010; George et al, 2017; Bhaskaran et al, 2019).
Human BRCA PLP provides an ideal system to study evolution origin of human disease susceptibility. While high evolutionary conservation across species suggests the possibility that human BRCA PLP could be originated from evolution conservation, the positive selection imposed in the humans, chimps, and bonobos but not in other species highlights another possibility that human BRCA PLP could be originated from human itself rather than from evolution conservation. However, there is no consent so far in determining which of the two possibilities could be the right origin for human BRCA PLP. We analyzed human BRCA variation data reported by previous evolutionary studies in BRCA (Huttley et al, 2000; Fleming et al, 2003; Burk-Herrick et al, 2006; Lou et al, 2014). The results showed that of the 111 human BRCA variants analyzed in these studies, 108 (94.7%) were non-pathogenic including 42 (37.8%) benign, 47 (42.3%) variants of uncertain significance (VUS), but only 6 (5.4%) were Pathogenic (Table S1). Therefore, the information from the previous studies reflects mainly evolutionary conservation of BRCA variation but not the origin of human BRCA PLP.
Table S1 Human BRCA variants used in previous BRCA evolution studies. (27.8KB, xlsx)
In the current study, we addressed the evolution origin of human BRCA PLP. The rapid progress of genomics studies provides rich DNA sequence data across a wide range of species for phylogenic study, and the recent anthropological studies have also generated abundant DNA sequence data from ancient humans. These rich resources provide unique opportunities to study the evolutionary origin of the human BRCA PLP on the scale and accuracy unimaginable before. Taking the advantages, we performed a comprehensive phylogenic and archeological study to investigate the evolution origin of human BRCA PLP. Data from our study provide evidence to show that human BRCA PLP was highly unlikely originated from cross-species evolutionary conservation, but most likely arisen during recent human history after the latest out-of-Africa migration and the great expansion of modern human population.
Results
Phylogenetic analysis of human BRCA PLPs in non-human vertebrates
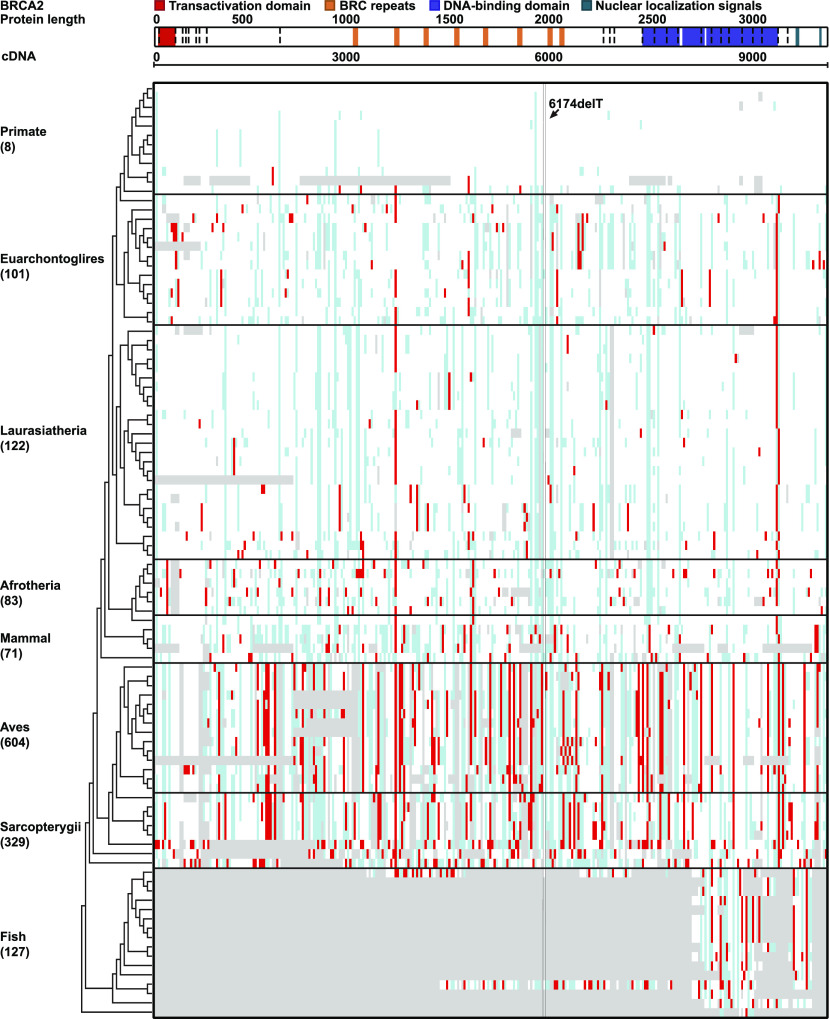
We identified a total of 6,624 BRCA PLP variants (2,972 in BRCA1 and 3,652 in BRCA2) from the ClinVar database for the study (Tables 1 and S2). We searched evidence for potential conservation of BRCA PLP variants between the humans and the 100 vertebrates distributed in eight clades of Primate, Euarchontoglires, Laurasiatheria, Afrotheria, Mammal, Aves, Sarcopterygii, and Fish. We identified 172 (5.8%) human BRCA1 PLP variants shared with 69 species, and 312 (8.6%) human BRCA2 PLP variants shared with 90 species (Figs 1, 2, and S1A and Tables S3 and S4).
Table 1.
Human BRCA PLP variants in 100 vertebrates.
| BRCA1 | BRCA2 | Total (%) | |
|---|---|---|---|
| A. Human BRCA PLP used in the study | |||
| Total | 2,972 | 3,652 | 6,624 |
| Pathogenic | 2,767 | 3,396 | 6,163 |
| Likely pathogenic | 110 | 155 | 265 |
| Pathogenic/likely pathogenic | 95 | 101 | 196 |
| Types of mutation | |||
| Frameshift deletion | 1,250 | 1,676 | 2,926 (44.2) |
| Stopgain/nonsense | 655 | 851 | 1,506 (22.7) |
| Frameshift insertion | 622 | 738 | 1,360 (20.5) |
| Splice site | 176 | 190 | 366 (5.5) |
| Nonsynonymous SNV | 116 | 47 | 163 (2.5) |
| Frameshift substitution | 88 | 114 | 202 (3.1) |
| Intron variant | 48 | 18 | 66 (1.0) |
| Nonframeshift deletion | 10 | 12 | 22 (0.3) |
| Nonframeshift substitution | 6 | 5 | 11 (0.2) |
| 5′/3′ UTR | 1 | 1 | 2 (0.03) |
| B. Human BRCA PLP shared with other species | |||
| Shared mutation | 172 (5.8) | 312 (8.6) | 484 (7.3) |
| Types of shared mutation | |||
| Stopgain/nonsense | 156 | 280 | 436 (6.6) |
| Splice site | 6 | 17 | 23 (0.3) |
| Nonsynonymous SNV | 5 | 8 | 13 (0.2) |
| Intron variant | 5 | 7 | 12 (0.2) |
| Frameshift deletion | — | — | — |
| Frameshift insertion | — | — | — |
| Nonframeshift deletion | — | — | — |
| 5′/3′ UTR | — | — | — |
| C. Number of species sharing human BRCA PLP | |||
| 1 | 68 (39.5) | 130 (41.6) | 198 (40.9) |
| 2 | 24 | 40 | 65 |
| 3 | 14 | 36 | 50 |
| 4 | 7 | 18 | 25 |
| 5 | 19 | 15 | 34 |
| 6 | 6 | 12 | 18 |
| 7 | 8 | 8 | 16 |
| 8 | 3 | 5 | 8 |
| 9 | 2 | 4 | 6 |
| 10 | 2 | 4 | 6 |
| 11 | 4 | 1 | 5 |
| 12 | 2 | 2 | 4 |
| 13 | 1 | 4 | 5 |
| 14 | 1 | 3 | 4 |
| 15 | 3 | 5 | 8 |
| 16 | 2 | 5 | 7 |
| 17 | 3 | 2 | 5 |
| 18 | 2 | 6 | 8 |
| 19 | 1 | 2 | 4 |
| 20 | — | 4 | 4 |
| 21 | — | 2 | 2 |
| 22 | — | — | — |
| 23 | — | — | — |
| 24 | — | — | — |
| 25 | — | 1 | 1 |
| 26–33 | — | — | — |
| 34 | — | 1 | 1 |
| 35 | — | — | — |
| 36 | — | 1 | 1 |
| 37–56 | — | — | — |
| 57 | — | 1 | 1 |
| 172 (100) | 312 (100) | 484 (100) |
Figure 1. Distribution of human BRCA1 PLP variants in 100 vertebrates.
Red cell: same as the human PLPs; empty cell: same base as human wild type; blue cell: different from human wild type and PLPs; gray cell: gaps; “-”: no aligned base; “=”: gap with at least one un-aligned base. The figure was generated using GraphPad Prism (version 9.0.0 for Windows, GraphPad Software). 185delAG, 5382insC: the BRCA1 founder pathogenic variants in Ashkenazi Jews population.
Figure 2. Distribution of human BRCA2 PLP variants in 100 vertebrates.
Ibid as in Fig 1. 6174delT: the BRCA2 founder pathogenic variants in Ashkenazi Jews population.
Figure S1. Examples of sequence alignment in phylogenetic and archeological analyses of human BRCA PLP variants.
(A) Phylogenetic alignment. Red box shows the alignment of the four human BRCA PLP variants in 14 species in Aves clade. (B) Archeological alignment. It shows that three BRCA PLP variants were shared in three ancient cases. Red box: the BRCA PLP variants.
Table S2 Human BRCA PLP variants used in the study. (330.9KB, xlsx)
Table S3 Distribution of the 172 shared human BRCA1 PLP variants in other species. (82.6KB, xlsx)
Table S4 Distribution of the 312 shared human BRCA2 PLP variants in other species. (135.5KB, xlsx)
Of the BRCA1 PLP variants shared in the eight clades, Aves had the highest sharing number of 14 species and Sarcopterygii the second highest of eight species than other clades (P < 0.001) (Fig S2). For example, BRCA1 c.3268C>T (p.Gln1090Ter) was shared with 19 species from Rock pigeon in Aves to Lizard in Sarcopterygii, c.2498T>A (p.Leu833Ter) with 18 species from Saker falcon in Aves to Spiny softshell turtle in Sarcopterygii, and c.2138C>A (p.Ser713Ter) with 18 species from Rock pigeon in Aves to Lizard in Sarcopterygii; all 11 PLP variants located in the BRCA1 BRCT domain were shared with the species within Aves and Sarcopterygii clades. Tasmanian devil, a species with high risk of developing facial cancer, also shared 20 human BRCA1 PLP variants. In the shared BRCA2 PLP variants, the highest one including BRCA2 c.8933C>A (p.Ser2978Ter) shared with 36 species from Lesser Egyptian jerboa in Euarchontoglires to Opossum in mammal, and c.8933C>A (p.Ser2978Ter) shared with 36 species from Lesser Egyptian jerboa in Euarchontoglires to Opossum in Mammal to Spotted gar in Fish, the most distal species sharing this PLP. As the most used animal models in biomedical study, mouse shared eight and rat shared seven human BRCA1 PLP variants, of which only two were located at the BRCT domain. Zebrafish, another important biological model, shared 25 human BRCA2 PLP variants.
Figure S2. Quantitative distribution of human BRCA PLP in 100 vertebrates.
It shows that among the eight clades, Aves and Sarcopterygii had high sharing with human BRCA PLP, no sharing of BRCA PLP between human and primates of Chimp, Gorilla, Orangutan, Gibbon, Rhesus, Crab-eating macaque, and no sharing of human BRCA1 PLPs in Fish.
Of the species sharing with human BRCA PLP variants, Spiny softshell turtle in Sarcopterygii had the highest sharing number of 34 BRCA1 PLP variants, and Lizard in Sarcopterygii had the highest sharing number of 57 BRCA2 PLP variants. There were no human BRCA PLP variant shared with primates of Chimp, Gorilla, Orangutan, Gibbon, Rhesus, Crab-eating macaque and Baboon whereas there were 10 PLP variants shared with the distal primates of Marmoset, Squirrel monkey, and Bushbaby: Marmoset shared BRCA1 c.850C>T (p.Gln284Ter) and BRCA2 c.1642C>T (p.Gln548Ter), Squirrel monkey and Bushbaby shared an BRCA1 intronic c.1058G>A (p.Trp353Ter), Squirrel monkey and Bushbaby shared BRCA2 c.4689G>A (p.Trp1563Ter), and Bushbaby shared BRCA2 c.2651C>A (p.Ser884Ter), c.2978G>A (p.Trp993Ter), c.4689G>A (p.Trp1563Ter), c.3504G>T (p.Met1168Ile) and c.5263G>T (p.Glu1755Ter). Marmoset, Squirrel monkey, and Bushbaby had the divergent time of 40, 12.5, 15 million yr from hominins, accordingly. As control, we searched BRCA1 c. 68_69del, BRCA1 c.5266dup and BRCA2 c.5946del, the three BRCA founder PLP variants in Ashkenazi Jews population (Levy-Lahad et al, 1997), in the 100 vertebrates. We found no evidence for their presence in these species: the wildtype AG at the position of 68–69 in BRCA1 was present across 51 species from Chimp to Armadillo, the wild-type C at the position of 5,266 in BRCA1 was present across 83 species from Rhesus to Coelacanth, and the wild-type T at the position of 5,946 of BRCA2 was present across 87 species from Chimp to Tetraodon.
Of the 172 BRCA1 and 312 BRCA2 PLP variants shared with non-human species, the major types were stop-gain/nonsense variants (156 [90.1%] shared BRCA1 PLP variants and 280 [89%] shared BRCA2 PLP variants). Although frameshift indels constituted the majority of human BRCA PLP variants, none of them was present in other species (Table 1). The shared PLP variants did not enrich in specific functional domains, but distributed across coding region in both BRCA1 and BRCA2 (Fig S3A and B, P > 0.1): of the 388 of 2,972 (13%) BRCA1 PLP variants located in functional domains, only 13 (3.4%) were shared with other species (11 in BRCT domain and 2 in Coiled-Coil domain); of the 1,077 of 3,652 (29.5%) BRCA2 PLP variants in functional domains, only 54 (5.0%) were shared with other species (15 in BRC repeats, 38 in DBD, 1 in NLS and 8 in Transactivation domain) (Tables S3 and S4).
Figure S3. Distribution of the shared human BRCA PLP variants.
It shows that the shared human BRCA PLP variants were not enriched at specific functional domains. (A) BRCA1. (B) BRCA2. Different colors within coding region indicate the locations of functional domains. Generated by using the ProteinPaint program (https://proteinpaint.stjude.org/).
The results from our phylogenetic analysis reported above do not support evolution conservation as the major source of human BRCA PLP.
Archeological analysis of BRCA PLP in ancient humans
As our phylogenetic analysis did not find evidence to support evolution conservation as the major source of human BRCA PLP, we then tested whether human BRCA PLP could be originated from human itself. We performed an archeological analysis by searching human BRCA PLP in ancient human genome sequence data. We first searched the three Neanderthal and two Denisovan genome sequences (Green et al, 2010; Meyer et al, 2012; Prufer et al, 2014, 2017; Mafessoni et al, 2020), which were diverged from the ancestors of modern humans 400–800 thousand year ago (kya). We did not identify any matched human BRCA PLP variants. Next, we searched human BRCA PLP variants in 2,792 ancient human genome sequences dated from 45,000 to 300 yr ago. We identified 46 BRCA PLP variants in 50 ancient individuals, including 24 BRCA1 PLP variants in 28 individuals and 22 BRCA2 PLP variants in 22 individuals (Table 2 and Fig S1B), lived in Africa, Europe, Asia, Oceania, and Central and North America (Fig 3). Each matched PLP variant had dbSNP ID, further ensured the fidelity of the matched variants. The oldest matched BRCA1 PLP variant was BRCA1 c.181T>G (p.C61G) in an individual in Voronezh Oblast, Russia dated about 37,470 yr ago (Seguin-Orlando et al, 2014), the youngest matched BRCA1 PLP variant was BRCA1 c.2599C>T (Q867X) in an individual in Fujian, China dated 307 yr ago (Yang et al, 2020); the oldest matched BRCA2 PLP variant was BRCA2 c.9573G>A (p.W3191X) in an individual in Bor District, Serbia dated about 7,874 yr ago (Mathieson et al, 2018), the youngest matched BRCA2 PLP variant was BRCA2 c.8009C>T (p.S2670L) in an individual in Shefa, Vanuatu dated 350 yr ago (Lipson et al, 2020). Of the 24 matched BRCA1 PLP variants, 14 (58.3%) were stop-gain and six (25%) were located at the BRCT domain; of the 22 matched BRCA2 PLP variants, 15 (68.2%) were stop-gain and three were located at the nucleic acid binding domain. Four of the 24 BRCA1 PLP variants were detected in two individuals.
Table 2.
BRCA PLP variants identified in ancient humans.
| Age (BP) | Fossil site | Variationa | Type | dbSNP155 | Domain | Referencesb | |
|---|---|---|---|---|---|---|---|
| cDNA | Protein | ||||||
| BRCA1 | |||||||
| 37,470 ± 1,210 | Voronezh Oblast, Russia | c.181T>G | p.C61G | missense | rs28897672 | Zinc finger | 38 |
| 9,871 ± 1,650 | Bor District, Serbia | c.5095C>T | p.R1699W | missense | rs55770810 | BRCT | 20 |
| 7,796.5 ± 75.5 | Shandong, China | c.2059C>T | p.Q687X | stopgain | rs273898674 | — | 35 |
| 4,971 ± 450 | Catalonia, Spain | c.1A>G | p.D2_M18del | startloss | rs80357287 | — | 27 |
| 4,671 ± 50 | Sistan and Baluchestan, Iran | c.4327C>T | p.R1443X | stopgain | rs41293455 | — | 24 |
| 4,121 ± 100 | Senec, Slovakia | c.3607C>T | p.R1203X | stopgain | rs62625308 | — | 31 |
| 3,350 ± 200 | Sardinia, Italy | c.1618G>T | p.E540X | stopgain | rs730881471 | — | 3 |
| 3,096 ± 20 | Hope Town, Bahamas | c.5503C>T | p.R1835X | stopgain | rs41293465 | BRCT | 4 |
| 2,110 ± 30 | Ballito Bay, South Africa | c.643C>T | p.Q215X | stopgain | rs886037979 | 37 | |
| 2,110 ± 30 | Ballito Bay, South Africa | c.949C>T | p.Q317X | stopgain | rs80357211 | 37 | |
| 2,110 ± 30 | Ballito Bay, South Africa | c.441+1G>A | — | splice site | rs397509172 | 37 | |
| 2,031 | Khovsgol, Mongolia | c.135-1G>A | — | splice site | rs80358158 | — | 2 |
| 1,980 ± 20 | Ballito Bay, South Africa | c.3967C>T | p.Q1323X | stopgain | rs80357262 | 37 | |
| 1,933 | Rostov Oblast, Russia | c.5353C>T | p.Q1785X | nonsense | rs80356969 | BRCT | 2 |
| 1,695 ± 195 | Chukotka, Russia | c.2761C>T | p.Q921X | nonsense | rs80357377 | — | 5 |
| 1,471 ± 50 | Girona, Catalonia | c.4255G>T | p.E1419X | stopgain | rs80357309 | — | 27 |
| 1,221 ± 100 | Monsenor Nouel, Dominica | c.2309C>A | p.S770X | stopgain | rs80357063 | — | 4 |
| 921 ± 500 | Monsenor Nouel, Dominica | c.3607C>T | p.R1203X | stopgain | rs62625308 | — | 4 |
| 921 ± 500 | Monsenor Nouel, Dominica | c.5096G>A | p.R1699Q | missense | rs41293459 | BRCT | 4 |
| 921 ± 500 | Monsenor Nouel, Dominica | c.4327C>T | p.R1443X | stopgain | rs41293455 | — | 4 |
| 921 ± 500 | Monsenor Nouel, Dominica | c.5216A>T | p.D1739V | missense | rs80357227 | BRCT | 4 |
| 921 ± 500 | Monsenor Nouel, Dominica | c.5095C>T | p.R1699W | missense | rs55770810 | BRCT | 4 |
| 721 ± 100 | San Pedro de Macors, Dominica | c.2389G>T | p.E797X | stopgain | rs62625306 | — | 4 |
| 721 ± 100 | San Pedro de Macors, Dominica | c.5503C>T | p.R1835X | stopgain | rs41293465 | BRCT | 4 |
| 480 ± 30 | Eland Cave, South Africa | c.5074G>A | p.D1692N | nonsynonymous | rs80187739 | BRCT | 37 |
| 480 ± 30 | Eland Cave, South Africa | c.3403C>T | p.Q1135X | stopgain | rs80357136 | 37 | |
| 419 ± 94 | Guangxi, China | c.4573C>T | p.Q1525X | stopgain | rs886040237 | SRD | 34 |
| 307.5 ± 26.5 | Fujian, China | c.2599C>T | p.Q867X | stopgain | rs886038001 | — | 35 |
| BRCA2 | |||||||
| 7,874 ± 72 | Bor District, Serbia | c.9573G>A | p.W3191X | stopgain | rs398122617 | NAB | 27 |
| 7,030 ± 50 | Castile and Leon, Spain | c.9466C>T | p.Q3156X | stopgain | rs276174925 | NAB | 27 |
| 4,950 ± 150 | Baden, Germany | c.6952C>T | p.R2318X | stopgain | rs80358920 | — | 14 |
| 4,571 ± 350 | Kladno, Central Bohemian Region | c.9382C>T | p.R3128X | stopgain | rs80359212 | — | 26 |
| 4,384 ± 200 | Scotland, UK | c.3922G>T | p.E1308X | stopgain | rs80358638 | — | 26 |
| 4,250 ± 1,200 | Vrancea County, Romania | c.7806-2A>G | — | splicing | rs81002836 | — | 9 |
| 3,858 ± 200 | Budapest, Hungary | c.8695C>T | p.Q2899X | stopgain | rs397507411 | — | 26 |
| 3,546 ± 325 | Atyrau region, Kazakhstan | c.244A>T | p.K82X | stopgain | rs397507628 | — | 24 |
| 2,921 ± 100 | NWFP, Pakistan | c.1189C>T | p.Q397X | stopgain | rs760815829 | — | 24 |
| 2,921 ± 100 | NWFP, Pakistan | c.3469G>T | p.E1157X | stopgain | rs80358595 | — | 24 |
| 2,821 ± 100 | East Kazakhstan, Kazakhstan | c.1825C>T | p.Q609X | stopgain | rs80358472 | — | 33 |
| 2,821 ± 100 | East Kazakhstan, Kazakhstan | c.2455C>T | p.Q819X | stopgain | rs397507629 | — | 33 |
| 2,330 ± 25 | St. Helena, South Africa | c.2905C>T | p.Q969X | stopgain | rs886038080 | 36 | |
| 2,321 ± 100 | NWFP, Pakistan | c.5200G>T | p.E1734X | stopgain | rs786202543 | — | 24 |
| 2,131 | Khovsgol, Mongolia | c.316+1G>A | — | splice site | rs397507303 | — | 2 |
| 2,083 | Omnogovi, Mongolia | c.7617+1G>A | — | splice site | rs397507922 | — | 2 |
| 1,980 ± 20 | Ballito Bay, South Africa | c.7480C>T | p.R2494X | stopgain | rs80358972 | 37 | |
| 1,783 | Issyk Kul, Kyrgyzstan | c.7007G>A | p.R2336H | missense | rs28897743 | — | 2 |
| 1,697 | South Kazakhstan, Kazakhstan | c.7977-1G>A | — | splice site | rs81002874 | — | 2 |
| 1,100 ± 500 | Santo Domingo, Dominica | c.171C>A | p.Y57X | stopgain | rs201523522 | — | 4 |
| 480 ± 30 | Eland Cave, South Africa | c.475G>A | p.V159M | nonsynonymous | rs80358702 | 37 | |
| 350 ± 100 | Shefa, Vanuatu | c.8009C>T | p.S2670L | missense | rs80359035 | NAB | 18 |
Bold refers to the variants detected in more than one individual.
References listed in Table S6
Figure 3. Geographic distribution of human BRCA PLPs in ancient humans.

Circle dots: matched BRCA1 PLPs; triangle dot: matched BRCA2 PLPs. The irregular dots: overlapped BRCA1 and BRCA2 PLPs. Colors in icons: time before present. The red circle dot was BRCA1 c.181T>G, dated 37,470 ± 1,210 yr ago (Table 2).
Dated BRCA founder PLP and ethnic-specific distribution of BRCA PLP
Many BRCA PLP variants are determined as BRCA founder PLP variants in different human ethnic populations, and their arisen times were determined by haplotyping analysis. For example, the arisen times for the three BRCA PLP founder variants (BRCA1 c.68_69del, BRCA1 c.5266dup, and BRCA2 c.5946del) in Ashkenazi Jews population were determined as 1,720, 1,800, and 580 yr ago, respectively (Table 3). We identified 34 BRCA founder PLP variants including 22 in BRCA1 and 12 in BRCA2. Their arisen times were from 3,225 to 140 yr ago (Table 3). For example, BRCA1 c.3228_3229del (p.G1077fs) was the oldest arisen 3,225 yr ago in Tuscany, Italy, BRCA1 c.1175_1214del (p.L345fs) was the youngest arisen 180 yr ago; BRCA2 c.9026_9030del (p.T3009fs) was the oldest arisen 2,760 yr ago in Spanish, BRCA2 c.9118-2A>G was the youngest arisen 220-144 yr ago in Finnish.
Table 3.
BRCA founder PLP variants dated by haplotype analysis.
| Age (BP) | Population | Mutation (HGVS) | Referencesa | ||
|---|---|---|---|---|---|
| cDNA | Protein | Variation type | |||
| BRCA1 | |||||
| 3,225 | Tuscany, Italy | 3347delAG (c.3228_3229del) | p.G1077fs | frameshift | 62 |
| 2,400–1,600 | Iberia | c.3331_3334delCAAG | p.Q1064fs | frameshift | 39 |
| 1,800 | Ashkenazi Jewish | 5832insC (c.5266dupC) | p.Q1756fs | frameshift | 40 |
| 1,720 | Northeastern Italy | c.676delT | p.C226fs | frameshift | 41 |
| 1,500–750 | Ashkenazi Jewish | 185delAG (c.68_69del) | p.E23fs | frameshift | 21 |
| 1,500 | Swedish | 3171ins5 (c.3052_3053ins5) | — | — | 43 |
| 1,480 | Columbia | 3450delCAAG (c.3331_3334del) | p.Q1064fs | frameshift | 44 |
| 1,440 | Mexican | c.548-?_4185 + ?del | — | exon 9–12del | 45 |
| 800 | Moroccan | c.5309G>T | p.G1770V | missense | 46 |
| 750 | Tuscany, Italy | 1499insA (c,1380dup) | p.F414fs | frameshift | 47 |
| 720–460 | Finnish | 3744delT (c.3626del) | — | nonsense | 48 |
| 600 | Norwegians | 1675delA (c.1556del) | p.K472fs | frameshift | 49 |
| 600 | Norwegians | 1135insA (c.1135_1136insA) | p.K472fs | frameshift | 49 |
| 500 | Norwegians | 816delGT (c.697_698del) | p.V186fs | frameshift | 50 |
| 500 | Norwegians | 3347delAG (c.3228_3229del) | p.G1077fs | frameshift | 50 |
| 500 | South Africans | c.2641G>T | p.E881X | nonsense | 51 |
| 380 | Spanish | c.5153-1G>A | — | splice site | 52 |
| 275 | Greek | G1738R (c.5212G>A) | p.G1738R | missense | 53 |
| 200 | Dutch | 2804delAA (c.2685_2686del) | p.P850fs | frameshift | 54 |
| 200 | Afro-Americans | 943ins10 (c.824_825ins10) | — | — | 55 |
| <200 | Finnish | 4216-2A>G (c.4097-2A>G) | — | splice site | 56 |
| 180 | Unknown | 1294del40 (c.1175_1214del) | p.L345fs | frameshift | 48 |
| BRCA2 | |||||
| 2,760 | Spanish | 9254del5 (c.9026_9030del) | p.T3009fs | frameshift | 57 |
| 2,600–2,400 | Portugal | 156-157insAlu (c.1205T>C) | p.L402P | missense | 58 |
| 1,904 | Spanish | 5344delAATA (c.5116_5119del) | p.N1706fs | frameshift | 59 |
| 1,600 | US and Canda | c.3036_3039del | p.S1013fs | frameshift | 60 |
| 1,365 | Spanish | 9538delAA (c.9310_9311delAA) | p.L3104fs | frameshift | 59 |
| 1,200 | Spanish | 5374delTATG (c.5146_5149del) | p.T1716fs | frameshift | 52 |
| 580 | Ashkenazi Jewish | 6174delT (c.5946del) | p.S1982fs | frameshift | 60 |
| 500 | Icelaner | 995del5 (c.771_775del) | p.A257fs | frameshift | 61 |
| 400–200 | Finnish | 7708C>T (c.7480C>T) | p.R2494X | nonsense | 58 |
| 360 | US and France | 982del4 (c.755_758del) | p.D252fs | frameshift | 60 |
| 220–140 | Finnish | 8555T>G (c.8327T>G) | p.L2776X | nonsense | 56 |
| 220–140 | Finnish | c.9118-2A>G | — | splice site | 56 |
references are listed in Table S6.
We also traced the ethnic origins for the BRCA PLP variants used in the study. Of the 1,054 BRCA PLP variants with available ethnic information, 548 (52%) were originated from single ethnic population, 327 (31%) were shared only between two ethnic populations. The rates were consistent in both BRCA1 and BRCA2 PLP variants (Table S5A–C).
Table S5 Ethnic origins of human BRCA PLP variants. (59.6KB, xlsx)
Discussion
Our study analyzed the evolutionary origin of BRCA PLP in modern human population. Our phylogenetic analysis across 100 vertebrates found no direct evidence for cross-species evolution conservation as the source for human BRCA PLP. Our archeological analysis in 2,792 ancient human individuals dated back to 45,000 yr ago identified 46 BRCA PLP variants of which 45 were arisen within the last 10,000 yr. Our analysis in the haplotyping-dated human BRCA founder PLP variants observed that nearly all were arisen within the last 3,000 yr. We further traced the ethnic origins of the BRCA PLP variants used in the study and observed that the majority were present in single or few ethnic populations. Based on these observations, we propose that human BRCA PLP was most likely arisen in recent human history, possibly within a few thousand years, after the latest human out-of-Africa migration and settlement at different global destinations. We consider that the positive selection on human BRCA could play a major role, and the population expansion could further increase the spectrum of human BRCA PLP in modern human population (Fig 4).
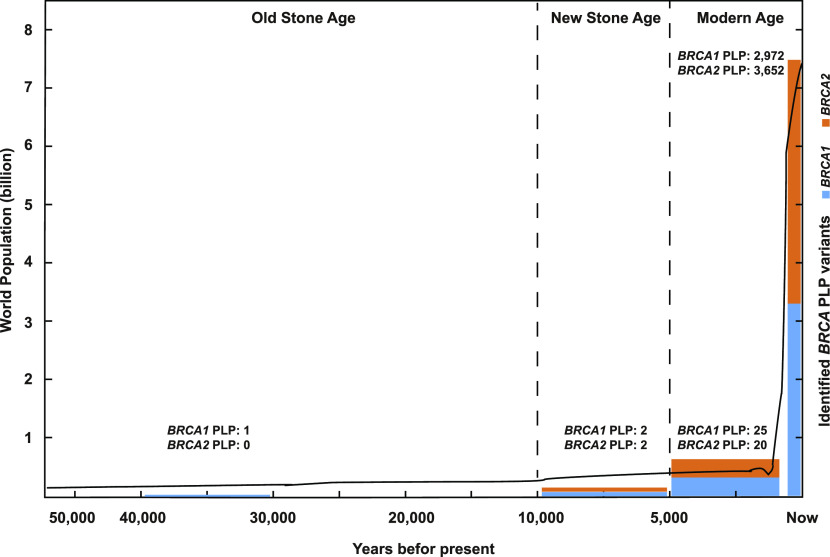
Figure 4. Model for the origin of human BRCA PLP.
It shows that the positive selection on human BRCA causes high human BRCA variation and accompanied PLP variation, and the expansion of human population further increases the spectrum of human BRCA PLP. X-axis: the timing of modern human after out-of-Africa migration; Left y-axis: size of human population; Right y-axis: number of human BRCA PLP variants. Curve: the growth of human population in the past 50,000 yr.
The prevalence of BRCA PLP is between 0.2% and 0.5% in modern human population. For example, the prevalence is 0.26% in Japanese population (Momozawa et al, 2018), 0.29% in Macau population (Qin et al, 2021), 0.38% in Chinese population (Dong et al, 2021), 0.38% in Mexican population (Fernandez-Lopez et al, 2019), 0.39% in Malaysian population (Wen et al, 2018), 0.53% in Taiwanese population (Chian et al, 2021), and 0.53% in Caucasian populations (Kurian et al, 2019). That implies that one in several hundreds of human individuals carries a BRCA PLP variant. The prevalence of BRCA PLP can be the highest in disease-causing genetic predisposition genes in human. It is interesting to understand why BRCA PLP can reach such high level in human population regardless their deleterious impact. Possible explanations can be (1). The cancer caused by BRCA PLP occurs mostly at later reproduction stage. Before reaching the stage, the PLP has already been transmitted to the next generation; (2). The loss of tumor suppressing function of BRCA due to BRCA PLP could be developmental stage-dependent. It states that the PLP could be beneficial at the reproductive stage but deleterious at later reproduction stage. Positive selection imposed on the reproductive stage can select the BRCA PLP not deleterious at the stage. However, our current study does not determine whether one or both of the explanations could contribute to the higher prevalence of BRCA PLP in modern human population.
Extensive animal model studies, especially from mouse studies, have provided rich evidence showing the pathogenic consequences by BRCA PLP in non-human species. Brca1 knockout-mice showed abnormal post-implantation development and embryonic proliferation (Liu et al, 1996; Hakem et al, 1996), embryonic lethality (Ludwig et al, 1997), neuroepithelial abnormalities (Gowen et al, 1996), irradiation hypersensitivity and genetic instability (Shen et al, 1998), abnormal T cell development (Mak et al, 2000; Xu et al, 2001), and tumorigenesis (Xu et al, 1999; Brodie et al, 2001). Without the presence of other mutated genes such as the mutated TP53, however, the mutated Brca1 alone is not sufficient to directly cause cancer (Xu et al, 2001). This can further explain that the higher prevalence of BRCA PLP in human may have less deleterious impact in the absence of synergetic effects from other genetic events.
Many human BRCA PLP variants were shared with species in Aves and Sarcopterygii clads. This could happen by chance rather than by cross-species conservation, as it is unlikely that the conservation would allow the presence of the gaps across such wide distances. Alternatively, it could also be related to species-specific pathogenicity of the same BRCA PLP variants that the BRCA PLPs in humans may not be pathogenic in non-human species (Gao & Zhang, 2003). For example, Tasmanian devil shared 20 human BRCA PLP variants. Tasmanian has a high risk of developing facial cancer, which is related with the fusion of chromosome 1 and X (Hawkins et al, 2006; Murchison et al, 2010; Taylor et al, 2017), but no evidence to show that the cancer is related to the BRCA PLP variants shared with human. A study analyzed human deleterious mutations in multiple genes including BRCA shared with mouse (Gao & Zhang, 2003), and evaluated multiple theories to explain the biological correlation including the “funder effect,” “fixations of slightly deleterious mutations,” “relaxed selection on late-onset phenotypes,” and “compensatory changes.” The study considered that the “compensatory changes,” which stated that “compensatory mutations at other sites of the same or a different protein render the deleterious mutations neutral,” can be the best to explain the conservation of human deleterious mutation with the distant species of mouse. We consider that the compensation theory can also be used to explain the sharing of human BRCA PLPs with the species in Aves and Sarcopterygii clads, although direct evidence is not available to validate the explanation.
Only limited numbers of BRCA PLP variants were identified in the ancient humans. This can be related to the limited data available from ancient humans. In addition, genomic sequences in many ancient samples had limited coverage due to the rarity and poor quality of ancient DNA. Furthermore, the size differences between ancient and modern human populations can also be a factor. It is estimated that the number of indivuduals in the latest out-of-Africa migration 65,000–50,000 yr ago was about 1,000–2,500 (Henn et al, 2012), the size of the human population in 1,800s was about 1 billion, and the size of modern human population is close to eight billion (https://ourworldindata.org/world-population-growth, https://data.worldbank.org/indicator/SP.POP.TOTL). All human BRCA PLP variants used in our current study were derived from the current human population. With the positive selection, continually increased human population size, and more powerful DNA sequencing technologies under developing, it is expected that more new human BRCA PLP variants would arise and be identified.
Human BRCA1 has 1,863 residues and BRCA2 has 3,418 residues. Although gene with large size can have more chance to generate more genetic variants, it is unlikely that the size in BRCA is a major factor for the large quantity of human BRCA PLP. For example, our study showed that mouse Brca does not change although the size of mouse Brca is very close to the human BRCA (Brca1 1,812 residues and Brca2 3,329 residues) (Wang & Wang, 2021). Another factor related with ethnic-specific PLPs can be that the same BRCA PLP could arise by chance in different ethnic populations at different times. For example, the BRCA1 68-69del founder variant in Ashkenazi Jews is also present in other ethnic populations although at very low frequency, and BRCA1 3607C>T shared between the Slovakia case dated 4,121 ± 100 ago and Dominica case dated 921 ± 500 ago (Table 2). However, this unlikely played a major role in contributing to human BRCA PLP. Pleiotropy effects may also exist that variation in non-BRCA genes may contribute to the selection of BRCA PLPs, particularly for these selected in the post-reproductive age (Williams, 1957). This is particularly interesting as BRCA PLP mainly cause high cancer risk after reproductive age. Homozygotic BRCA PLP carriers seldom survive or develop Fanconi anemia due to the embryonic lethal effect, as BRCA is essential for development (Seo et al, 2018).
A limitation of our study is that most of the BRCA founder PLP variants and ethnic distribution data used in the study were derived from European and descendants. More data from non-European populations should enhance the significance of our study.
Materials and Methods
Source of human BRCA PLPs
Human BRCA PLP variants were from ClinVar database (https://ftp.ncbi.nlm.nih.gov/pub/clinvar/vcf_GRCh38/weekly/clinvar_20201031.vcf.gz, accessed 1 November 2020). Only the variants classified as “pathogenic,” “likely pathogenic,” and “pathogenic and likely pathogenic” were included in the study. The variants with conflict classifications were excluded to ensure the reliability of the analysis.
Vertebrate genomic mapping analysis
Human BRCA PLP variants were divided into single nucleotide variants and indel groups. Their genomic positions were annotated by referring to the following reference sequences: BRCA1: genome hg38 NC_000017.11, cDNA NM_007294, NP_009225; BRCA2: genome hg38 NC_000013.11, cDNA NM_000059, NP_000050. BRCA1, and BRCA2 sequences across 100 vertebrate species were downloaded from UCSC Genome Browser (https://genome.ucsc.edu/cgi-bin/hgGateway?redirect=manual&source=genome.ucsc.edu). Sequence alignment followed the procedures in Multiz Alignments of 100 Vertebrates part in UCSC genome browser (https://genome.ucsc.edu/cgi-bin/hgc?db=hg38&c=chr17&l=43106526&r=43106527&o=43106526&t=43106527&g=multiz100way&i=multiz100way). Tree model of the 100 vertebrate species was generated using the phyloFit program in the PHAST package (Murphy et al, 2001). The phylogenetic tree for the 100 vertebrate species was from UCSC resource (http://hgdownload.soe.ucsc.edu/goldenPath/hg38/multiz100way/), and the distance between species on the tree was adjusted to ensure the readability in the figures. PhastCons and phyloP from the PHAST package were used for evolution conversion measurement. Sequence alignment between repeat-masked human hg38 and non-human genome sequences was made by using Lastz (BLASTZ) and multiz (Blanchette et al, 2004; Hubisz et al, 2011; Armstrong et al, 2019; Ramani et al, 2019). Based on the phylogenetic distance from the references, the scoring matrix and parameters for pairwise were adjusted for each species. The high-score chains were placed along genome sequences, and the low-score chains were used to fill gaps. The results of single base-level alignment were collected from the “Multiz Alignments of 100 Vertebrates” section in UCSC genome browser by entering variant position in hg38 using a Python-based tool (https://github.com/Skylette14/GetBase). For indel alignments, the base number of insertion or deletion between human and other species was the same. Each indel alignment across the matched species were manually checked to ensure reliability of the alignment. The positions corresponding to human PLP variants in the aligned non-human sequences were obtained by using GetBase.
Ancient human genomic mapping analysis
Ancient human DNA sequences and related publications were from “Allen Ancient DNA Resource (version 42.2, https://reich.hms.harvard.edu/allen-ancient-dna-resource-aadr-downloadable-genotypes-present-day-and-ancient-dna-data, accessed 1 March 2020), containing genomic sequences from a total of 2,792 ancient human individuals dated from 37,470 to 300 before present. Bam files of ancient genomic sequences were downloaded. The sequences containing BRCA1 (chr17:41,196,312-41,277,500, hg19 by Ensembl) and BRCA2 (chr13:32,889,611-32,973,805, hg19 by Ensembl) were identified and used for mapping to hg19. Mpileup command in SAMtools was used for variant calling from the mapped sequences with the minimal base quality set as 1 (Li et al, 2009). After generation of ancient human vcf files, the called variants were annotated using wANNOVAR (https://wannovar.wglab.org/) (Chang & Wang, 2012), compared with human BRCA PLP variants and manually checked in ClinVar to obtain the related information for these matched by the ancient BRCA sequences. The locations for the ancient PLP carrier’s fossil excavation and the estimated age were based on the original publications. The geographical distribution map of the ancient BRCA PLP variants were generated by using Matlab (The MathWorks, Inc.).
Statistical analysis
Kruskal–Wallis test was used for statistical comparison for the BRCA PLP variants shared between clades (GraphPad Software). Chi-Squared test was used to compare the distribution of the shared BRCA PLP variants, P < 0.01 was considered as statistical significance.
Data Availability
All data used in the study were from public domains as indicated in the text. The data generated from the study were provided as online Figs S1–S3 and Tables S1–S6.
Table S6 References of ancient human genomes and BRCA founder PLP variants. (22KB, docx)
Supplementary Material
Acknowledgements
We thank Information and Communication Technology Office (ICTO), University of Macau for providing the High-Performance Computing Cluster resources for the study. This work was supported by the grants from Macau Science and Technology Development Fund (085/2017/A2, 0077/2019/AMJ), the grants from the University of Macau (SRG2017-00097-FHS, MYRG2019-00018-FHS), the Faculty of Health Sciences, University of Macau (Startup fund, FHSIG/SW/0007/2020P, FHS Innovation grant) (SM Wang).
Author Contributions
J Li: data curation, software, formal analysis, visualization, methodology, and writing—original draft, review, and editing.
B Zhao: data curation, software, methodology, and writing—original draft, review, and editing.
T Huang: software and writing—original draft, review, and editing.
Z Qin: data curation, software, and writing—original draft, review, and editing.
SM Wang: conceptualization, resources, funding acquisition, project administration, and writing—original draft, review, and editing.
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
References
- Abkevich V, Zharkikh A, Deffenbaugh AM, Frank D, Chen Y, Shattuck D, Skolnick MH, Gutin A, Tavtigian SV (2004) Analysis of missense variation in human BRCA1 in the context of interspecific sequence variation. J Med Genet 41: 492–507. 10.1136/jmg.2003.015867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson BO, Yip CH, Smith RA, Shyyan R, Sener SF, Eniu A, Carlson RW, Azavedo E, Harford J (2008) Guideline implementation for breast healthcare in low-income and middle-income countries: Overview of the breast Health global initiative global summit 2007. Cancer 113: 2221–2243. 10.1002/cncr.23844 [DOI] [PubMed] [Google Scholar]
- Armstrong J, Fiddes IT, Diekhans M, Paten B (2019) Whole-genome alignment and comparative annotation. Annu Rev Anim Biosci 7: 41–64. 10.1146/annurev-animal-020518-115005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskaran SP, Chandratre K, Gupta H, Zhang L, Wang X, Cui J, Kim YC, Sinha S, Jiang L, Lu B, et al. (2019) Germline variation in BRCA1/2 is highly ethnic-specific: Evidence from over 30,000 Chinese hereditary breast and ovarian cancer patients. Int J Cancer 145: 962–973. 10.1002/ijc.32176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. (2004) Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res 14: 708–715. 10.1101/gr.1933104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie SG, Xu X, Qiao W, Li WM, Cao L, Deng CX (2001) Multiple genetic changes are associated with mammary tumorigenesis in Brca1 conditional knockout mice. Oncogene 20: 7514–7523. 10.1038/sj.onc.1204929 [DOI] [PubMed] [Google Scholar]
- Burk-Herrick A, Scally M, Amrine-Madsen H, Stanhope MJ, Springer MS (2006) Natural selection and mammalian BRCA1 sequences: Elucidating functionally important sites relevant to breast cancer susceptibility in humans. Mamm Genome 17: 257–270. 10.1007/s00335-005-0067-2 [DOI] [PubMed] [Google Scholar]
- Chang X, Wang K (2012) wANNOVAR: Annotating genetic variants for personal genomes via the web. J Med Genet 49: 433–436. 10.1136/jmedgenet-2012-100918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Lei JH, Bao J, Wang H, Hao W, Li L, Peng C, Masuda T, Miao K, Xu J, et al. (2020) BRCA1 deficiency impairs mitophagy and promotes inflammasome activation and mammary tumor metastasis. Adv Sci (Weinh) 7: 1903616. 10.1002/advs.201903616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chian J, Sinha S, Qin Z, Wang SM (2021) BRCA1 and BRCA2 variation in Taiwanese general population and the cancer cohort. Front Mol Biosci 8: 685174. 10.3389/fmolb.2021.685174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Liao RG, Parsons MT, Paten B, Alquaddoomi F, Antoniou A, Baxter S, Brody L, Cook-Deegan R, Coffin A, et al. (2018) BRCA challenge: BRCA exchange as a global resource for variants in BRCA1 and BRCA2. PLoS Genet 14: e1007752. 10.1371/journal.pgen.1007752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Chandratre K, Qin Y, Zhang J, Tian X, Rong C, Wang N, Guo M, Zhao G, Wang SM (2021) Prevalence of BRCA1/BRCA2 pathogenic variation in Chinese Han population. J Med Genet 58: 565–569. 10.1136/jmedgenet-2020-106970 [DOI] [PubMed] [Google Scholar]
- Fernández-Lopez JC, Romero-Córdoba S, Rebollar-Vega R, Alfaro-Ruiz LA, Jiménez-Morales S, Beltrán-Anaya F, Arellano-Llamas R, Cedro-Tanda A, Rios-Romero M, Ramirez-Florencio M, et al. (2019) Population and breast cancer patients' analysis reveals the diversity of genomic variation of the BRCA genes in the Mexican population. Hum Genomics 13: 3. 10.1186/s40246-018-0188-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming MA, Potter JD, Ramirez CJ, Ostrander GK, Ostrander EA (2003) Understanding missense mutations in the BRCA1 gene: An evolutionary approach. Proc Natl Acad Sci U S A 100: 1151–1156. 10.1073/pnas.0237285100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Zhang J (2003) Why are some human disease-associated mutations fixed in mice? Trends Genet 19: 678–681. 10.1016/j.tig.2003.10.002 [DOI] [PubMed] [Google Scholar]
- George A, Kaye S, Banerjee S (2017) Delivering widespread BRCA testing and PARP inhibition to patients with ovarian cancer. Nat Rev Clin Oncol 14: 284–296. 10.1038/nrclinonc.2016.191 [DOI] [PubMed] [Google Scholar]
- Gowen LC, Johnson BL, Latour AM, Sulik KK, Koller BH (1996) Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat Genet 12: 191–194. 10.1038/ng0296-191 [DOI] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. (2010) A draft sequence of the Neandertal genome. Science 328: 710–722. 10.1126/science.1188021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakem R, de la Pompa JL, Sirard C, Mo R, Woo M, Hakem A, Wakeham A, Potter J, Reitmair A, Billia F, et al. (1996) The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell 85: 1009–1023. 10.1016/s0092-8674(00)81302-1 [DOI] [PubMed] [Google Scholar]
- Hawkins CE, Baars C, Hesterman H, Hocking GJ, Jones ME, Lazenby B, Mann D, Mooney N, Pemberton D, Pyecroft S, et al. (2006) Emerging disease and population decline of an island endemic, the Tasmanian devil Sarcophilus harrisii. Biol Conservation 131: 307–324. 10.1016/j.biocon.2006.04.010 [DOI] [Google Scholar]
- Henn BM, Cavalli-Sforza LL, Feldman MW (2012) The great human expansion. Proc Natl Acad Sci U S A 109: 17758–17764. 10.1073/pnas.1212380109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz MJ, Pollard KS, Siepel A (2011) PHAST and RPHAST: Phylogenetic analysis with space/time models. Brief Bioinform 12: 41–51. 10.1093/bib/bbq072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Sy SM, Chen J (2010) BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol 11: 138–148. 10.1038/nrm2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttley GA, Easteal S, Southey MC, Tesoriero A, Giles GG, McCredie MR, Hopper JL, Venter DJ (2000) Adaptive evolution of the tumour suppressor BRCA1 in humans and chimpanzees. Australian breast cancer family study. Nat Genet 25: 410–413. 10.1038/78092 [DOI] [PubMed] [Google Scholar]
- Jasin M, Rothstein R (2013) Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol 5: a012740. 10.1101/cshperspect.a012740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo PA, Pearl LH, Carr AM (2016) DNA repair, genome stability and cancer: A historical perspective. Nat Rev Cancer 16: 35–42. 10.1038/nrc.2015.4 [DOI] [PubMed] [Google Scholar]
- Kojic M, Kostrub CF, Buchman AR, Holloman WK (2002) BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell 10: 683–691. 10.1016/s1097-2765(02)00632-9 [DOI] [PubMed] [Google Scholar]
- Kurian AW, Ward KC, Howlader N, Deapen D, Hamilton AS, Mariotto A, Miller D, Penberthy LS, Katz SJ (2019) Genetic testing and results in a population-based cohort of breast cancer patients and ovarian cancer patients. J Clin Oncol 37: 1305–1315. 10.1200/JCO.18.01854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Lahad E, Catane R, Eisenberg S, Kaufman B, Hornreich G, Lishinsky E, Shohat M, Weber BL, Beller U, Lahad A, et al. (1997) Founder BRCA1 and BRCA2 mutations in Ashkenazi Jews in Israel: Frequency and differential penetrance in ovarian cancer and in breast-ovarian cancer families. Am J Hum Genet 60: 1059–1067. [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson M, Spriggs M, Valentin F, Bedford S, Shing R, Zinger W, Buckley H, Petchey F, Matanik R, Cheronet O, et al. (2020) Three phases of ancient migration shaped the ancestry of human populations in Vanuatu. Curr Biol 30: 4846–4856.e6. 10.1016/j.cub.2020.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Flesken-Nikitin A, Li S, Zeng Y, Lee WH (1996) Inactivation of the mouse Brca1 gene leads to failure in the morphogenesis of the egg cylinder in early postimplantation development. Genes Dev 10: 1835–1843. 10.1101/gad.10.14.1835 [DOI] [PubMed] [Google Scholar]
- Lou DI, McBee RM, Le UQ, Stone AC, Wilkerson GK, Demogines AM, Sawyer SL (2014) Rapid evolution of BRCA1 and BRCA2 in humans and other primates. BMC Evol Biol 14: 155. 10.1186/1471-2148-14-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig T, Chapman DL, Papaioannou VE, Efstratiadis A (1997) Targeted mutations of breast cancer susceptibility gene homologs in mice: Lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev 11: 1226–1241. 10.1101/gad.11.10.1226 [DOI] [PubMed] [Google Scholar]
- Mafessoni F, Grote S, de Filippo C, Slon V, Kolobova KA, Viola B, Markin SV, Chintalapati M, Peyrégne S, Skov L, et al. (2020) A high-coverage neandertal genome from chagyrskaya cave. Proc Natl Acad Sci U S A 117: 15132–15136. 10.1073/pnas.2004944117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak TW, Hakem A, McPherson JP, Shehabeldin A, Zablocki E, Migon E, Duncan GS, Bouchard D, Wakeham A, Cheung A, et al. (2000) Brca1 required for T cell lineage development but not TCR loci rearrangement. Nat Immunol 1: 77–82. 10.1038/76950 [DOI] [PubMed] [Google Scholar]
- Mathieson I, Alpaslan-Roodenberg S, Posth C, Szécsényi-Nagy A, Rohland N, Mallick S, Olalde I, Broomandkhoshbacht N, Candilio F, Cheronet O, et al. (2018) The genomic history of southeastern Europe. Nature 555: 197–203. 10.1038/nature25778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C, et al. (2012) A high-coverage genome sequence from an archaic Denisovan individual. Science 338: 222–226. 10.1126/science.1224344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momozawa Y, Iwasaki Y, Parsons MT, Kamatani Y, Takahashi A, Tamura C, Katagiri T, Yoshida T, Nakamura S, Sugano K, et al. (2018) Germline pathogenic variants of 11 breast cancer genes in 7,051 Japanese patients and 11,241 controls. Nat Commun 9: 4083. 10.1038/s41467-018-06581-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Tovar C, Hsu A, Bender HS, Kheradpour P, Rebbeck CA, Obendorf D, Conlan C, Bahlo M, Blizzard CA, et al. (2010) The Tasmanian devil transcriptome reveals Schwann cell origins of a clonally transmissible cancer. Science 327: 84–87. 10.1126/science.1180616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, Eizirik E, O’Brien SJ, Madsen O, Scally M, Douady CJ, Teeling E, Ryder OA, Stanhope MJ, de Jong WW, et al. (2001) Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science 294: 2348–2351. 10.1126/science.1067179 [DOI] [PubMed] [Google Scholar]
- Narod SA, Foulkes WD (2004) BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer 4: 665–676. 10.1038/nrc1431 [DOI] [PubMed] [Google Scholar]
- O’Connell MJ (2010) Selection and the cell cycle: Positive darwinian selection in a well-known DNA damage response pathway. J Mol Evol 71: 444–457. 10.1007/s00239-010-9399-y [DOI] [PubMed] [Google Scholar]
- Pao GM, Zhu Q, Perez-Garcia CG, Chou SJ, Suh H, Gage FH, O’Leary DD, Verma IM (2014) Role of BRCA1 in brain development. Proc Natl Acad Sci U S A 111: E1240–E1248. 10.1073/pnas.1400783111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlicek A, Noskov VN, Kouprina N, Barrett JC, Jurka J, Larionov V (2004) Evolution of the tumor suppressor BRCA1 locus in primates: Implications for cancer predisposition. Hum Mol Genet 13: 2737–2751. 10.1093/hmg/ddh301 [DOI] [PubMed] [Google Scholar]
- Pfeffer CM, Ho BN, Singh ATK (2017) The evolution, functions and applications of the breast cancer genes BRCA1 and BRCA2. Cancer Genomics Proteomics 14: 293–298. 10.21873/cgp.20040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plon SE, Eccles DM, Easton D, Foulkes WD, Genuardi M, Greenblatt MS, Hogervorst FB, Hoogerbrugge N, Spurdle AB, Tavtigian SV, et al. (2008) Sequence variant classification and reporting: Recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat 29: 1282–1291. 10.1002/humu.20880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, de Filippo C, Grote S, Mafessoni F, Korlević P, Hajdinjak M, Vernot B, Skov L, Hsieh P, Peyrégne S, et al. (2017) A high-coverage neandertal genome from vindija cave in Croatia. Science 358: 655–658. 10.1126/science.aao1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. (2014) The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505: 43–49. 10.1038/nature12886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z, Kuok CN, Dong H, Jiang L, Zhang L, Guo M, Leong HK, Wang L, Meng G, Wang SM (2021) Can population BRCA screening be applied in non-ashkenazi Jewish populations? Experience in Macau population. J Med Genet 58: 587–591. 10.1136/jmedgenet-2020-107181 [DOI] [PubMed] [Google Scholar]
- Ramani R, Krumholz K, Huang YF, Siepel A (2019) PhastWeb: A web interface for evolutionary conservation scoring of multiple sequence alignments using phastCons and phyloP. Bioinformatics 35: 2320–2322. 10.1093/bioinformatics/bty966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen EM, Fan S, Ma Y (2006) BRCA1 regulation of transcription. Cancer Lett 236: 175–185. 10.1016/j.canlet.2005.04.037 [DOI] [PubMed] [Google Scholar]
- Seguin-Orlando A, Korneliussen TS, Sikora M, Malaspinas AS, Manica A, Moltke I, Albrechtsen A, Ko A, Margaryan A, Moiseyev V, et al. (2014) Paleogenomics. Genomic structure in Europeans dating back at least 36,200 years. Science 346: 1113–1118. 10.1126/science.aaa0114 [DOI] [PubMed] [Google Scholar]
- Seo A, Steinberg-Shemer O, Unal S, Casadei S, Walsh T, Gumruk F, Shalev S, Shimamura A, Akarsu NA, Tamary H, et al. (2018) Mechanism for survival of homozygous nonsense mutations in the tumor suppressor gene BRCA1. Proc Natl Acad Sci U S A 115: 5241–5246. 10.1073/pnas.1801796115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen SX, Weaver Z, Xu X, Li C, Weinstein M, Chen L, Guan XY, Ried T, Deng CX (1998) A targeted disruption of the murine Brca1 gene causes gamma-irradiation hypersensitivity and genetic instability. Oncogene 17: 3115–3124. 10.1038/sj.onc.1202243 [DOI] [PubMed] [Google Scholar]
- Smith KR, Hanson HA, Hollingshaus MS (2013) BRCA1 and BRCA2 mutations and female fertility. Curr Opin Obstet Gynecol 25: 207–213. 10.1097/GCO.0b013e32835f1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RL, Zhang Y, Schöning JP, Deakin JE (2017) Identification of candidate genes for devil facial tumour disease tumourigenesis. Sci Rep 7: 8761. 10.1038/s41598-017-08908-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang SM (2021) DNA damage repair system in C57BL/6 J mice is evolutionarily stable. BMC Genomics 22: 669. 10.1186/s12864-021-07983-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcsh PL, King MC (2001) BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum Mol Genet 10: 705–713. 10.1093/hmg/10.7.705 [DOI] [PubMed] [Google Scholar]
- Wen WX, Allen J, Lai KN, Mariapun S, Hasan SN, Ng PS, Lee DS, Lee SY, Yoon SY, Lim J, et al. (2018) Inherited mutations in BRCA1 and BRCA2 in an unselected multiethnic cohort of Asian patients with breast cancer and healthy controls from Malaysia. J Med Genet 55: 97–103. 10.1136/jmedgenet-2017-104947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GC (1957) PLEIOTROPY, natural selection, and the evolution of senescence. Evolution 11: 398–411. 10.1111/j.1558-5646.1957.tb02911.x [DOI] [Google Scholar]
- Xu X, Qiao W, Linke SP, Cao L, Li WM, Furth PA, Harris CC, Deng CX (2001) Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat Genet 28: 266–271. 10.1038/90108 [DOI] [PubMed] [Google Scholar]
- Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, Hennighausen L, Wynshaw-Boris A, Deng CX (1999) Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet 22: 37–43. 10.1038/8743 [DOI] [PubMed] [Google Scholar]
- Yang MA, Fan X, Sun B, Chen C, Lang J, Ko YC, Tsang CH, Chiu H, Wang T, Bao Q, et al. (2020) Ancient DNA indicates human population shifts and admixture in northern and southern China. Science 369: 282–288. 10.1126/science.aba0909 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Human BRCA variants used in previous BRCA evolution studies. (27.8KB, xlsx)
Table S2 Human BRCA PLP variants used in the study. (330.9KB, xlsx)
Table S3 Distribution of the 172 shared human BRCA1 PLP variants in other species. (82.6KB, xlsx)
Table S4 Distribution of the 312 shared human BRCA2 PLP variants in other species. (135.5KB, xlsx)
Table S5 Ethnic origins of human BRCA PLP variants. (59.6KB, xlsx)
Table S6 References of ancient human genomes and BRCA founder PLP variants. (22KB, docx)
Data Availability Statement
All data used in the study were from public domains as indicated in the text. The data generated from the study were provided as online Figs S1–S3 and Tables S1–S6.
Table S6 References of ancient human genomes and BRCA founder PLP variants. (22KB, docx)