Abstract
Background
Rift Valley fever virus (RVFV) is an arbovirus that causes epizootics and epidemics among livestock population and humans. Our surveillance system has revealed multiple emergences and re-emergences of RVFV in West Africa over the last decade.
Methods
The Sentinel Syndromic Surveillance Network in Senegal (4S) has been implemented since 2011. Samples from human suspected arbovirus infection in 4S sentinel sites were sent to Institut Pasteur de Dakar (IPD), where arbovirus diagnosis by enzyme-linked immunosorbent assay (ELISA), real-time reverse transcription polymerase chain reaction (RT-PCR), and virus isolation was performed. Overall, IPD has received a total of 1149 samples from arboviral suspected patients through the 4S network from January to December 2020. These samples were screened for 7 arboviruses including RVFV. Whole-genome sequencing of positive RVFV samples by RT-PCR was performed using the Illumina Miseq platform followed by genome assembly. Phylogenetic analyses were performed using MEGA X.
Results
Out of the 1149 arbovirus suspected cases, 4 RVFV-positive samples were detected with RT-PCR while 5 RVFV-positive samples were detected by ELISA. Complete genome sequences were obtained for 3 strains among the 4 positive samples by RT-PCR. Phylogenetic analyses indicated an emergence of a virus first described in South Africa during a major outbreak.
Conclusions
This strong surveillance system allowed the detection of an RVFV outbreak in Senegal in 2020. The obtained genomes clustered with strains from South Africa belonging to lineage H. This calls for implementation of a strong surveillance system for wild animals, humans, and livestock simultaneously in all African countries.
Keywords: genetic diversity, Rift Valley fever virus, Senegal, surveillance
D/C/F/TAF is the reference for combination therapy based on protease inhibitors but has not been compared with regimens containing integrase inhibitors as initial ART. We could not demonstrate D/C/F/TAF noninferiority relative to DTG/ABC/3TC, although both regimens were similarly well tolerated.
Rift Valley fever virus (RVFV) is a zoonotic arbovirus of the Phenuiviridae family [1]. The virus has a genome segmented with 3 single-stranded RNA segments: L (large), M (medium), and S (small). The RVFV genome is characterized by low genetic diversity (between 1% and 5%) and by the existence of a single RVFV serotype. Molecular classification of RVFV strains isolated from 16 countries showed that these viruses clustered into 15 lineages (A–O) [2].
RVFV has caused substantial epidemics throughout Africa and in the Arabian Peninsula. RVFV infection leads to abortions in pregnant animals and high mortality in newborn sheep and goats. Humans can be infected by being in contact with infected tissues, blood, body fluids or abortion products, or through the bite of an infected mosquito [3, 4]. Most human cases are moderate, but rare severe complications such as hepatitis, meningoencephalitis, or hemorrhagic fever could occur and lead to death. Recently, RVFV infection has also been associated with spontaneous abortion among pregnant women in Sudan [5]. Transmission between ruminants occurs via mosquito bites, involving mainly species belonging to the Aedes, Culex, and Mansonia genera, and through direct contact with infectious animal tissues and fluids [4].
Rift Valley fever (RVF) is listed by the World Health Organization for Animal Health and the World Health Organization (WHO) among the 10 priority diseases for research and development due to its potential to cause major epidemics in humans [6]. Regarding the new plan established by the WHO after the 2013–2016 Ebola epidemic in West Africa for urgent research and development toward new diagnostic tools, vaccines, and medicines for RVFV, there are currently ongoing efforts to produce safe and efficient vaccines against RVFV to prevent infections in humans.
The most recent RVFV outbreaks occurred in Mauritania (2015), Niger (2016), Angola and China (2016), Uganda (since 2016), South Africa (2018), Mayotte (2018–2019), and Sudan (2019) [7–13].
In West Africa, particularly in Senegal, RVF is endemic and has been repeatedly reported among humans, livestock, and mosquitoes, especially in the Ferlo (Northern Senegal) [14–17]. RVFV, originally described as Zinga, was first isolated in Senegal in 1974 from an Aedes dalzieli mosquito pool collected in Kedougou. The first reported outbreak took place at the Senegalese-Mauritanian border in 1987 with 1500 human infections and more than 200 deaths [18] and was due to lineage N2. Since then, several outbreaks or sporadic cases of RVF (1993–1994, 1998–1999, 2002–2003, 2010, 2012, and 2015) have been reported in Senegal and Mauritania, with serious consequences in pastoral areas where livestock is the basis of the production system [18–20].
In 2012 in Kedougou, Senegal, RVFV was isolated from Ae. dalzieli mosquitoes and from a person with mild febrile symptoms [21]. Phylogenetic analysis of the partial nonstructural protein gene on the small RNA segment showed that the Kedougou human isolate was closely related to a strain from the West African lineage (lineage C) that had circulated in Mauritania in 2010 and 2012 and in South Africa 2008–2009 [19, 20].
During 2013–2014, 11 confirmed human cases of RVF including severe cases with encephalitis and retinitis, 1 isolation from a mosquito pool (Aedes ochraceus), and 52 confirmed animal cases were reported in Mbour, Linguere, and Kedougou [22]. In addition, phylogenetic analyses showed that the strains from humans and mosquitoes in Linguere were more closely related to the ArD38661 strain previously identified in Senegal in 1984 [22] and belonged to the Eastern African clade (lineage A).
The factors that can explain the emergence or re-emergence of RVF in Senegal remain unexplained. However, heavy rainfall and high temperatures occurring during the rainy season are known to be important factors in promoting the development of the main RVFV vectors [23].
Senegal has a longstanding yellow fever surveillance system as part of the WHO network of yellow fever laboratories. However, this yellow fever surveillance based on symptoms such as fever and jaundice is not optimal for the detection of other viruses. In addition, the Institut Pasteur de Dakar (IPD), in collaboration with the Senegalese Ministry of Health, has implemented a countrywide surveillance network named “4S” (Syndromic Sentinel Surveillance in Senegal) since 2011. This principal objective of this surveillance network is to strengthen national detection capabilities of epidemics [24].
Up to 2015, the 4S surveillance system focused only on testing influenza and other respiratory viruses. Since 2015, because of an increasing number of febrile cases not related to respiratory viruses, arboviruses and bacteria have been included in this surveillance system. As a result, 9 RVFV-positive samples by real-time reverse transcription polymerase chain reaction (RT-PCR) and serological detection were obtained in 2020 through this surveillance network from suspected arbovirus infections. Our study describes the molecular epidemiology of the RVFV strains isolated in Senegal during 2020 using whole-genome sequencing, genome assembly, and phylogenetic analyses.
METHODS
The 4S network allows for detection of unexpected or unusual occurrence of specific symptoms in order to monitor the evolution of diseases under surveillance, investigate outbreaks, and implement appropriate response actions.
Presentation of the Syndromic Sentinel Surveillance System Network in Senegal (4S Network)
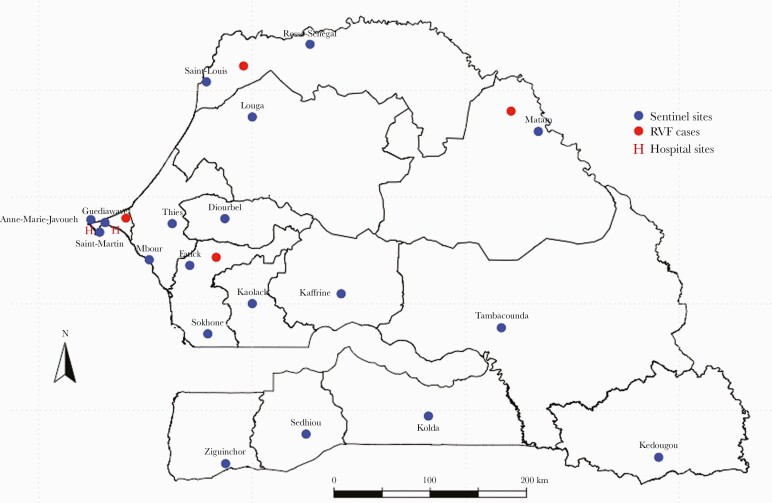
This surveillance network is based on a syndromic approach for febrile infectious diseases. Sentinel sites were selected on the basis of criteria used by the MoH. A checklist of criteria was developed based on the WHO-recommended attributes for sentinel site selection including feasibility, representativeness, and the availability of data to enable disease burden estimate [25]. These sites are composed of community health care centers in decentralized areas in each Senegalese region and 3 reference hospitals in Dakar. Overall, there currently are 20 surveillance sentinel sites in the 4S network in Senegal (Figure 1).
Figure 1.
Location of sentinel surveillance centers for the 4S network (blue circle) and distribution of Rift Valley fever cases (red circle) in Senegal 2020.
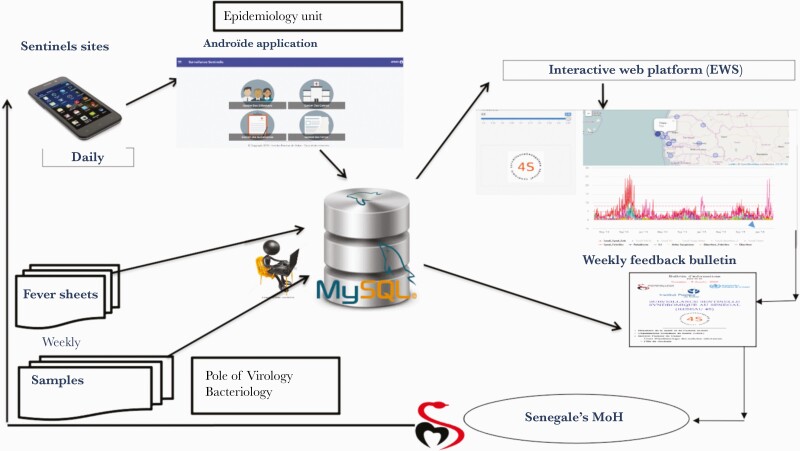
The 4S network is based on data collected by sentinel general practitioners (PGSs) through a standard data collection form available in all sentinel surveillance centers. Each PGS transmits encrypted data in the form of messages at least once a day via a smartphone connected to a real-time web platform (original feature of this surveillance system). Individual sheets containing sociodemographic, clinical, para-clinical (malaria Rapid Diagnostic Test [RDT] results), and therapeutic information are completed for all febrile patients. In addition, biological samples are taken (nasopharyngeal swabs for influenza, sera for arbovirus diagnosis, stool or rectal swabs in case of outbreak of diarrhea). All samples and the individual sheets are sent to IPD on a weekly basis (Figure 2).
Figure 2.
Syndromic Sentinel Surveillance Network in Senegel (4S).
Sample Collections for Arbovirus Diagnosis
The epidemiological survey is based on an individual questionnaire completed by practitioners for suspected cases. For each suspected case of arboviral disease, a dry tube venous sample is taken and sent to the arbovirus reference laboratory at IPD.
Differential Diagnostic for Arboviruses
The blood samples received at the Virology Department at IPD were tested for arbovirus detection using enzyme-linked immunosorbent assay (ELISA) and RT-PCR. As part of routine surveillance for acute febrile illnesses, these samples were tested for Chikungunya (CHIK), Dengue (DEN), West Nile (WN), yellow fever (YF), Zika (ZIK), RVFV, and Crimean Congo hemorrhagic fever (CCHF). For RVFV RT-PCR, primers and probes used have been previously described [26]. Immunoglobulin M (IgM) ELISA was performed using an IPD in-house method with antigens and immune ascites produced in mice. The laboratory turnaround time is ~7 days.
RT-PCR samples positive for RVFV were used for whole-genome sequencing on an Illumina Miseq platform.
Viral Genome Sequencing
We used 4 RT-PCR-positive samples for sequencing. RNA extraction using the Qiagen kit was performed before RNA depletion.
Briefly, host ribosomal RNA was depleted using specific probes and Oligo-dT. The mixture was heat-denatured at 95°C for 2 minutes, followed by slow cooling to 45°C at 0.1°C/s. RNA hybrids were degraded with RNase H (NEB) as previously described [27]. After that, samples were treated with TURBO DNase (ThermoFisher) before purification using RNAClean XP beads (Beckman Coulter). Depleted RNA was used as a template for first-stranded cDNA synthesis using the SuperScript IV Reverse Transcriptase kit (Invitrogen, Thermo Fisher, USA), and the double-stranded cDNA and sequencing libraries were produced using the Nextera XT DNA Library Preparation kit (Illumina, San Diego, CA, USA) [28]. RNA- and DNA-free water was also processed in the samples batch as a control.
Sequencing was done on an Illumina MiSeq with 150 cycles using paired-end reads.
Bioinformatics Analyses
The sequencing reads were demultiplexed with the default Illumina platform demultiplexer (bcl2fastq). Then an in-house pipeline was used to (i) trim the adapters, (ii) align reads, and (iii) call consensus sequences using cutadapt, bowtie2, and samtools, respectively. The consensus genomes were constructed with the RVFV reference genome. Once the consensus genomes were generated, their homologous sequences in public databases were identified using BLAST. Sequences were aligned using the multiple sequence comparison by log-expectation (MUSCLE) algorithm [29], available in MEGA, version X [30]. The phylogenetic relationships of the newly characterized Senegalese RVFV genome segments were analyzed using complete L, M, and S sequences from all homologous sequences available in GenBank. The best fitted nucleotide substitution model was determined for each segment data set using MEGA X and then applied for all the subsequent phylogenetic analyses. The aligned nucleotide sequences were also used to calculate the mean pairwise distances. Maximum likelihood phylogenetic trees were inferred using MEGA for 1000 replications (version X). The maximum likelihood trees were rooted on midpoints, and nodes were supported by bootstrap values. We investigated recombination with RDP4 beta 4.8 [31], and reassortment events were also analyzed using the 3 segments.
RESULTS
From January to December 2020, IPD received a total of 1149 samples from arboviral suspected patients through the 4S network. Each sentinel site has collected at least 1 sample since the implementation of the surveillance. As of the time of writing, Diamaguene-Kaffrine (71 samples), Mbour Toucouleur-Thies (73 samples), Abattoirs-Kaolack (74 samples), Ndiaye-Ndiaye-Fatick (94 samples), Pont-Tambacounda (136 samples), and Bokidiawe-Matam (226 samples) are the most active sites. Matam, Fatick, and Richard Toll have the highest number of RVF detections through the 4S network, representing, respectively, 20.7%, 8.6%, and 3.6% of all cases.
Among all collected samples, 9 were RVFV-positive by RT-PCR and/or IgM (1 RT-PCR+/IgM+, 3 RT-PCR+/IgM-, and 5 RT-PCR-/IgM+), and 1 was IgM-positive for both RVF and yellow fever. RVF-confirmed patients had a mean age (range) of 26 (17–40) years and M/F sex ratio of 3.5. Among these patients, 2 were from the Ndiaye-Ndiaye sentinel site (Fatick) located in the center of the country 148.9 km from Dakar, 4 were from Matam located in the north of the country 532.1 km from Dakar, 2 were from Richard Toll (Saint Louis) located in the north of the country 393.6 km from Dakar, and 1 was from Dantec hospital (Dakar) (Figure 1). The symptoms most commonly present in these patients were headache (89%), myalgia (78%), arthralgia (55%), and fever (100%), although the 4 patients positive by RT-PCR did not present severe clinical symptoms.
Sequencing was performed on Miseq (Illumina San Diego, CA, USA; https://www.illumina.com), and good coverage was obtained from 3 out of 4 samples (>98%). Thus, the complete genome was obtained for those 3 samples.
The nucleotide sequences of all the segments of the RVF isolates analyzed in the current study have been deposited in GenBank, with accession numbers indicated in Table 1. We compute pairwise distances of the newly described samples (2020 in Senegal) to previously published samples (2010 samples in South Africa) using MEGA X. The diversity at the nucleotide level was around 1%.
Table 1.
GenBank Accession Number
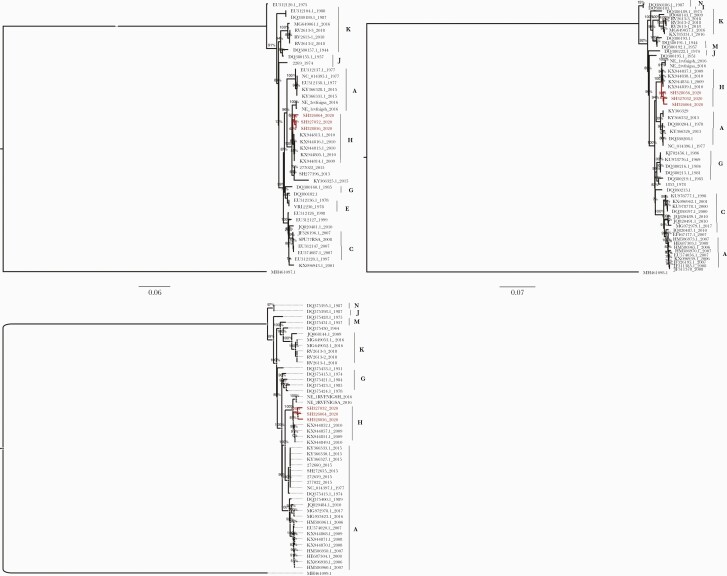
The phylogenetic analyses of the segment sequences generated in this study and the available RVFV L, M, and S sequences in Genbank are displayed in Figure 3. The generated trees show that the 2020 isolates from Senegal clustered with isolates previously identified during the 2009 and 2010 RVFV outbreaks in South Africa, belonging to lineage H (Figure 3 segments S, M, and L). This phylogenetic relationship was supported by nodes with high bootstrap values (>90%).
Figure 3.
Maximum likelihood (ML) tree based on complete sequences of Rift Valley fever virus (RVFV) isolates circulating in Senegal in 2020 (segments S, M, and L). The tree is midpoint-rooted, and nodes are labeled with local support values, computed using the Shimodaira-Hasegawa (SH) test for 5000 bootstrap replications and the generalized time-reversible model with a proportion of gamma rate. The newly characterized RVFV isolates from Senegal are color-coded in red in this tree and clustered with isolates from South Africa belonging to lineage H.
In addition, general coherence among the tree topologies from individual segment data sets suggests that recombination and re-assortment events did not occur in the newly characterized Senegalese sequences of RVFV.
DISCUSSION
Since the outbreak of 1987, several sporadic RVF cases have been reported in Senegal, with circulation of 3 main lineages, which are A, C, and N. Among these sporadic human cases (from 2012 to 2014), only 2 were detected during the early phase of the disease with RT-PCR-positive results, while most of the cases were positive by IgM or immunoglobulin G [21, 22].
However, since the implementation of the arbovirus testing in the 4S network in 2015, better detection of many arboviruses including RVFV, Crimean Congo hemorrhagic fever virus [32], Dengue virus [33], and West Nile virus (unpublished data) has been observed. Indeed, in 2020, 9 RVFV human cases were detected in different areas in Senegal, such as the Fatick, Matam, Saint-Louis, and Dakar regions. Interestingly, 4 of these cases were detected at an early phase of disease with an RT-PCR-positive result. Therefore, our data confirm the studies conducted by the Ministry of Health and show that most of the viral hemorrhagic fever cases detected through the 4S system are in the early phase of disease, while in other health facilities these cases are mainly detected during the hemorrhagic phase of disease with a limited chance of survival.
This early detection of arboviruses and hemorrhagic fever viruses through the 4S network allows authorities to implement rapid responses to manage patients as well as prevent virus propagation in the country.
In addition, this early detection with the availability of genome sequences from different cases and geographical areas leads to more efficient virus characterization. This genomic surveillance could also serve to establish links between different cases, particularly in the context of an outbreak.
Indeed, viral sequence analyses showed no evidence of genetic re-assortment in the 3 different RVFV isolates from Senegal. Similarly, RDP4 analysis showed no recombination event. Phylogenetic analyses based on the entire genome showed that the isolates belong to a monophyletic group and clustered with RVFV strains collected in South Africa during the 2009 and 2010 outbreaks, which belong to the lineage H [2]. Indeed, in 2009, an RVF outbreak with unusual clinical manifestation in animals was observed in South Africa. This outbreak occurred atypically in the absence of abnormally high rainfall and in addition to causing abortion storms; it has also been associated with high mortality among pregnant adult cattle. In 2010, an RVF outbreak caused by the same strain was also declared in South Africa, and 232 confirmed infections have been reported in humans, with 26 confirmed deaths [34]. In 2018, an outbreak was declared in Free State Province in South Africa, and the strains clustered in the distant lineage K despite extensive transmission of lineage H during 2010–2011 in the same province [35]. During 2010–2011 in Namibia, an RVFV outbreak was also reported where analysis of animal specimens confirmed virus circulation on 7 farms [36, 37]. Molecular characterization, based on analysis of partial sequences of the M segment of RVFV strains, suggested that the Namibian outbreak was probably caused by the RVFV strains first described in South Africa [36]. The RVFV strains from South Africa in the 2009 and 2010 outbreaks constitute the lineage H described by Grobelaar and collaborators [2]. This would be the first time that lineage H was detected in Senegal. These data suggest an introduction of RVFV lineage H from South Africa to Namibia in 2010 and 2011 or a reemergence of an old strain. Indeed, strains from lineage H are related to a 2004 focal outbreak in Namibia [2]. However, we observed the occurrence of lineage H in Senegal 10 years after its detection in South Africa. This introduction in Senegal could be direct from South Africa or indirect via another African country. But no circulation of RVFV lineage H has been reported elsewhere in Africa. Indeed, circulation of lineage H in Senegal could be a new detection of an old introduction. Lineage H may have emerged from strains already present in Senegal but not previously identified. These results are in line with the multiple introductions of RVFV in West Africa from South Africa described by Soumare and collaborators [38] and highlight the ability of RVFV to rapidly move across regions. This underlines the need for a strong surveillance system for RVFV, such as the 4S network, in Africa. Information on how these introductions occurred will better support the importance of improving surveillance and control strategies for RVFV. Indeed, the implementation of good surveillance systems in all African countries is crucial for better prevention and response to outbreaks caused by arboviruses.
In Senegal, human cases of RVFV were detected in several parts of the country, especially in the north, west, and center, demonstrating an introduction and rapid spread of lineage H through the country. In addition, the Ministry of Livestock and Animal Production notified the World Organization for Animal Health (OIE) on October 15, 2020, of a case of Rift Valley fever in an equine antelope (Hippotragus equinus) discovered on October 3 in a private reserve with permanent water points in the Bango district of Saint Louis [39]. However, no phylogenetics information was available for this case of animal infection.
Further phylogenetics and phylodynamics studies could give information on how the virus was introduced and rapidly spread in the country.
In addition, only sporadic human cases have been detected, and they all recovered from the RVFV infection. This suggests that lineage H did not show high virulence in Senegal compared with Namibia and South Africa. However, the Senegal virus might be just as virulent as the strains in Namibia and South Africa but less prevalent. As the difference at the nucleotide level is very low between these strains, other factors such as mosquito vector competence, host susceptibility, and environment could also be involved. The same situation occurred with lineage A, which caused a major outbreak in Mauritania in 2015, while only sporadic cases were detected in Senegal in 2013 [7, 22]. A better adaptation of these virulent lineages to the Senegalese environment might lead to major outbreaks in the future. It is therefore important to understand the factors influencing the presence of viruses in nature, as well as their emergence and spread in endemic areas, in order to implement control strategies.
The ability to generate full-length sequences of viral genomes allows us to monitor virus variation between epizootic and outbreak events and rapid progress to be made in understanding the epidemiology of RVF. Implementing genomic surveillance in wild animals, humans, and livestock (one-health concept) is crucial to enable rapid responses to the disease. Indeed, data from a one-health surveillance system will allow prevention of future widespread epidemics but also provide health authorities with an opportunity to anticipate and prepare for RVF outbreaks. Moreover, for rapid and efficient detection and monitoring of arboviruses such as RVFV across the country, the 4S network in Senegal could serve as an example to be implemented at the African continent level.
Acknowledgments
We express our gratitude to Cherif Sylla, Oumar Ndiaye, Diogop Camara, and Makhfouz Traore for excellent technical assistance in laboratory diagnosis. We also thank the authorities and the field agents of the 4S sentinel sites.
Financial support. This work was supported by the Department of Health and Human Services, Washington DC, USA (grant number: 6 IDSEP190051-02-01).
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Patient consent. This is a national surveillance study. Our study does not include factors necessitating patient consent.
References
- 1. Gaudreault NN, Indran SV, Balaraman V, Wilson WC, Richt JA.. Molecular aspects of Rift Valley fever virus and the emergence of reassortants. Virus Genes 2019; 55:1–11. [DOI] [PubMed] [Google Scholar]
- 2. Grobbelaar AA, Weyer J, Leman PA, Kemp A, Paweska JT, Swanepoel R.. Molecular epidemiology of Rift Valley fever virus. Emerg Infect Dis 2011; 17:2270–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gear J, De Meillon B, Measroch V, Davis DH, Harwin H.. Rift Valley fever in South Africa. 2. The occurrence of human cases in the Orange Free State, the North-Western Cape Province, the Western and Southern Transvaal. B. Field and laboratory investigation. S Afr Med J 1951; 25:908–12. [PubMed] [Google Scholar]
- 4. Linthicum KJ, Britch SC, Anyamba A.. Rift Valley fever: an emerging mosquito-borne disease. Annu Rev Entomol 2016; 61:395–415. [DOI] [PubMed] [Google Scholar]
- 5. Baudin M, Jumaa AM, Jomma HJE, et al. Association of Rift Valley fever virus infection with miscarriage in Sudanese women: a cross-sectional study. Lancet Glob Health 2016; 4:e864–71. [DOI] [PubMed] [Google Scholar]
- 6. World Health Organization. WHO publishes list of top emerging diseases likely to cause major epidemics. 2017. Available at: http://www.emro.who.int/pandemic-epidemic-diseases/news/list-of-blueprint-priority-diseases.html. Accessed 10 December 2015.
- 7. Bob NS, Bâ H, Fall G, et al. Detection of the Northeastern African Rift Valley fever virus lineage during the 2015 outbreak in Mauritania. Open Forum Infect Dis 2017; 4:XXX–XX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu W, Sun FJ, Tong YG, Zhang SQ, Cao WC.. Rift Valley fever virus imported into China from Angola. Lancet Infect Dis 2016; 16:1226. [DOI] [PubMed] [Google Scholar]
- 9. Lagare A, Fall G, Ibrahim A, et al. First occurrence of Rift Valley fever outbreak in Niger, 2016. Vet Med Sci 2019; 5:70–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shoemaker TR, Nyakarahuka L, Balinandi S, et al. First laboratory-confirmed outbreak of human and animal Rift Valley fever virus in Uganda in 48 Years. Am J Trop Med Hyg 2019; 100:659–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Schalkwyk A, Romito M.. Genomic characterization of Rift Valley fever virus, South Africa, 2018. Emerg Infect Dis 2019; 25:1979–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Youssouf H, Subiros M, Dennetiere G, et al. Rift Valley fever outbreak, Mayotte, France, 2018-2019. Emerg Infect Dis 2020;26:769–72. doi: 10.3201/eid2604.191147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ahmed A, Ali Y, Elduma A, et al. Unique outbreak of Rift Valley fever in Sudan, 2019. Emerg Infect Dis 2020; 26:3030–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chevalier V, Lancelot R, Thiongane Y, Sall B, Diaité A, Mondet B.. Rift Valley fever in small ruminants, Senegal, 2003. Emerg Infect Dis 2005; 11:1693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diallo M, Lochouarn L, Ba K, et al. First isolation of the Rift Valley fever virus from Culex poicilipes (Diptera: Culicidae) in nature. Am J Trop Med Hyg 2000; 62:702–4. [DOI] [PubMed] [Google Scholar]
- 16. Marrama L, Spiegel A, Ndiaye K, et al. Domestic transmission of Rift Valley fever virus in Diawara (Senegal) in 1998. Southeast Asian J Trop Med Public Health 2005; 36:1487–95. [PubMed] [Google Scholar]
- 17. Ba Y, Sall AA, Diallo D, et al. Re-emergence of Rift Valley fever virus in Barkedji (Senegal, West Africa) in 2002-2003: identification of new vectors and epidemiological implications. J Am Mosq Control Assoc 2012; 28:170–8. [DOI] [PubMed] [Google Scholar]
- 18. Jouan A, Le Guenno B, Digoutte JP, Philippe B, Riou O, Adam F.. An RVF epidemic in Southern Mauritania. Ann Inst Pasteur Virol 1988; 139:307–8. [DOI] [PubMed] [Google Scholar]
- 19. Faye O, Ba H, Ba Y, et al. Reemergence of Rift Valley fever, Mauritania, 2010. Emerg Infect Dis 2014; 20:300–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sow A, Faye O, Ba Y, et al. Rift Valley fever outbreak, Southern Mauritania, 2012. Emerg Infect Dis 2014; 20:296–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sow A, Faye O, Faye O, et al. Rift Valley fever in Kedougou, Southeastern Senegal, 2012. Emerg Infect Dis 2014; 20:504–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sow A, Faye O, Ba Y, et al. Widespread Rift Valley fever emergence in Senegal in 2013-2014. Open Forum Infect Dis 2016; 3:XXX–XX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Caminade C, Ndione JA, Diallo M, et al. Rift Valley fever outbreaks in Mauritania and related environmental conditions. Int J Environ Res Public Health 2014; 11:903–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thiam D, Niang M, Dia N, et al. Influenza sentinel surveillance network improvement in Senegal and results [in French]. Bull Soc Pathol Exot 2015; 108:21–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barry MA, Arinal F, Talla C, et al. Performance of case definitions and clinical predictors for influenza surveillance among patients followed in a rural cohort in Senegal. BMC Infect Dis 2021; 21:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weidmann M, Sanchez-Seco MP, Sall AA, et al. Rapid detection of important human pathogenic phleboviruses. J Clin Virol 2008; 41:138–42. [DOI] [PubMed] [Google Scholar]
- 27. Matranga CB, Gladden-Young A, Qu J, et al. Unbiased deep sequencing of RNA viruses from clinical samples. J Vis Exp 2016; 54117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adey A, Morrison HG, Asan , et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol 2010; 11:R119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004; 32:1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018; 35:1547–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martin DP, Murrell B, Golden M, Khoosal A, Muhire B.. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 2015; 1:vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dieng I, Barry MA, Diagne MM, et al. Detection of Crimean Congo haemorrhagic fever virus in North-eastern Senegal, Bokidiawé 2019. Emerg Microbes Infect 2020; 9:2485–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dieng I, Cunha M, Diagne MM, et al. Origin and spread of the Dengue virus type 1, genotype V in Senegal, 2015-2019. Viruses 2021; 13:57. doi: 10.3390/v13010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kegakilwe PS. An atypical outbreak of Rift Valley fever in the Northern Cape in October 2009. Paper presented at: Proceedings of the 9th Annual Congress of the Southern African Society for Veterinary Epidemiology and Preventive Medicine; 2010; Farm Inn, Republic of South Africa. [Google Scholar]
- 35. Jansen van VP, Kgaladi J, Msimang V, Paweska JT.. Rift Valley fever reemergence after 7 years of Quiescence, South Africa, May 2018. Emerg Infect Dis 2019; 25:338–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Monaco F, Pinoni C, Cosseddu GM, et al. Rift Valley fever in Namibia, 2010. Emerg Infect Dis 2013; 19:2025–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cosseddu GM, Magwedere K, Molini U, et al. Genetic diversity of Rift Valley fever strains circulating in Namibia in 2010 and 2011. Viruses 2020; 12:1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Soumaré PO, Freire CC, Faye O, et al. Phylogeography of Rift Valley fever virus in Africa reveals multiple introductions in Senegal and Mauritania. PLoS One 2012; 7:e35216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. World Organization for Animal Heath (OIE). 2020. https://www.vidal.fr/actualites/26002-cas-de-fievre-de-la-vallee-du-rift-chez-une-antilope-au-senegal.html.