Abstract
Clustered regularly interspaced short palindromic repeats (CRISPR)-associated systems have revolutionized traditional gene-editing tools and are a significant tool for ameliorating gene defects. Characterized by high target specificity, extraordinary efficiency, and cost-effectiveness, CRISPR/Cas systems have displayed tremendous potential for genetic manipulation in almost any organism and cell type. Despite their numerous advantages, however, CRISPR/Cas systems have some inherent limitations, such as off-target effects, unsatisfactory efficiency of delivery, and unwanted adverse effects, thereby resulting in a desire to explore approaches to address these issues. Strategies for improving the efficiency of CRISPR/Cas-induced mutations, such as reducing off-target effects, improving the design and modification of sgRNA, optimizing the editing time and the temperature, choice of delivery system, and enrichment of sgRNA, are comprehensively described in this review. Additionally, several newly emerging approaches, including the use of Cas variants, anti-CRISPR proteins, and mutant enrichment, are discussed in detail. Furthermore, the authors provide a deep analysis of the current challenges in the utilization of CRISPR/Cas systems and the future applications of CRISPR/Cas systems in various scenarios. This review not only serves as a reference for improving the maturity of CRISPR/Cas systems but also supplies practical guidance for expanding the applicability of this technology.
Keywords: CRISPR/Cas system, optimized strategies, highly efficient, mutant, off-target effect
1 Introduction
In aquatic systems, host–pathogen interactions are meaningful in the ecology and evolution of all organisms. These interactions are often characterized by a strong immune defense between prokaryotic cells (archaea) and viruses, leading to their co-evolution (England and Whitaker, 2013). The strong immune defense mechanism utilized by these organisms is known as the clustered regulatory interspaced short palindromic repeats (CRISPR) system, which is used in prokaryotes to combat a viral infection. Earlier reports of CRISPR/Cas systems report three different types: I, II, and III (Makarova et al., 2011). Each type of system is characterized by a signature protein(s). The most common type, type II CRISPR/Cas9 system, mediates the immune response in three stages as follows: (1) adaption, (2) expression, and (3) interference (Makarova et al., 2020). In the adaption stage, DNA fragments of invading plasmids or phages (termed protospacers) are incorporated into the host CRISPR locus as spacers in the form of CRISPR RNA (crRNA) repeats. In the expression stage, the precursor CRISPR RNA (pre-crRNA) molecules are processed by expressed Cas proteins and cofactors into short, mature crRNA. Next, in the interference stage, the Cas9 protein recognizes and targets the crRNA, silencing the foreign sequences (Gasiunas et al., 2012; Janik et al., 2020). Single-guide RNA (sgRNA) synthesized by crRNA and tracrRNA then guides the Cas protein to generate double-strand breaks (DSBs) three base pairs upstream from the protospacer adjacent motifs (PAM) (Jinek et al., 2012). Through this mechanism, CRISPR/Cas systems can also serve as a precise gene-editing tool for genetic manipulation.
So far, the CRISPR/Cas systems have been divided into six types (types I–VI), in which type II-A (CRISPR-Cas9), type V-A (CRISPR-Cas12a or Cpf1), and (CRISPR-Cas12b or C2c1) have been most widely studied (Adli, 2018; Yao et al., 2018a). More than 10 different CRISPR/Cas proteins have been repurposed for genome editing. Among them, some of the most recently discovered Cas proteins are hotspots for research, such as the Cas12a proteins from Acidaminococcus sp. (AsCas12a) and Lachnospiraceae bacteria (LbCas12a). Beyond Cas proteins, optimization of CRISPR systems has been thoroughly studied, including sgRNA design, cell enrichment, editing conditions, etc. With the rapid development and progress of gene editing technology, CRISPR systems have been shown to be powerful and highly efficient gene-editing tools in various fields. Through numerous experiments in model and non-model organisms (Oh et al., 2010), these systems have been utilized to reveal cancer mechanisms (Sottnik et al., 2021), define gene function and phenotypes (Johansen et al., 2017), and treat human diseases (Torre et al., 2021).
As to traditional editing tools, zinc finger nucleases and transcription activator-like effector nucleases (TALENs) have overwhelmingly contributed to developments in biomedical research and application (Urnov et al., 2005). Their application is greatly limited, however, due to limitations such as high cost, low efficiency, and low throughput targeting (Batool et al., 2021). In contrast, the CRISPR technology has some unique advantages, including targeted editing of multiple genomic sites (Zhang and Showalter, 2020), fast generation of mutants (Zhang and Showalter, 2020), and accessible sgRNA design (Xu et al., 2020b). These advantages have led to a surge in CRISPR applications in various fields, such as agriculture (Zheng et al., 2019), animal husbandry (Liu et al., 2020b), chemical fields (Liu et al., 2021b), materiology (Demirer et al., 2021), etc. Although the framework of the structures and functions of CRISPR/Cas systems has been built, there are still several challenges in this system (Wang et al., 2016), including off-target effects (Coelho et al., 2020), variable efficiency (Jin et al., 2020), requirement of PAM and sgRNA (Heussler et al., 2015; Cameron et al., 2017), and inactive mutants (Ren et al., 2019). This review proposes some strategies to overcome these issues by reducing off-target effects, improving the repair efficiency of the homology-directed repair (HDR) pathway, choosing the optimal delivery system, and utilizing variants of Cas proteins. Additionally, regulation of nuclease-dead mutants of Cas9, anti-CRISPR (Acrs) protein application, and enrichment of cells and sgRNA may be effective strategies for the efficacy of CRISPR/Cas systems.
2 Strategies for Reducing Off-Target Effects
Presently, off-target effects in CRISPR/Cas systems are a major issue for gene editing. Whether the Cas protein is off- or on-target to a PAM site is mainly determined by the sgRNA, Cas proteins, ribonucleoprotein (RNP) concentration, as well as other factors, such as editing temperature and action time. The off-target cleavage of CRISPR/Cas systems often originates from the unsuccessful design or modification of gRNA, low specificity of Cas proteins, or excessive and prolonged expression of CRISPR/Cas9. Accordingly, various strategies are proposed to overcome these issues. Additionally, methods for sgRNA selection with off-target predictions have been established, such as PEM-seq (Yin et al., 2019b), CRISPR-PLANT v2 (Minkenberg et al., 2019), and CRISPR-GE (Xie et al., 2017), which avoid a waste of manpower and material resources and improve editing efficiency.
2.1 Reasonable Design and Modification of sgRNA
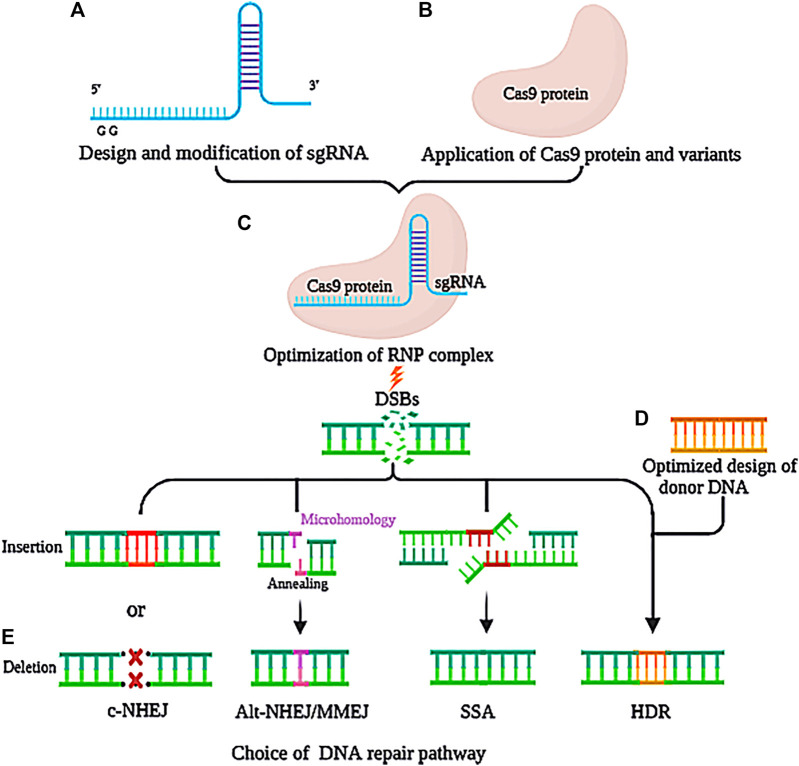
In CRISPR/Cas systems, the binding of sgRNA to the PAM site is a critical step in gene editing (Figure 1A). An unsuccessful design of sgRNA will result in lower specificity and higher miss rate (Doench et al., 2016). To avoid this, sgRNA must be accurately designed using computational tools (Liu et al., 2020a), such as CRISPR-P 2.0 (Liu et al., 2017a), E-CRISP (Heigwer et al., 2014), and CasFinder (Abby et al., 2014). On the basis of rational design, further modification of sgRNA can improve the specificity of RNA-guided Cas9 by truncation or addition of nucleotides to the 5′ or 3′ end (Pattanayak et al., 2013; Lin et al., 2014b). The 5′ end-truncated sgRNAs (2-3 bp) considerably reduce off-target mutations, but with the same on-target mutation efficiency as the full-length sequence (Fu et al., 2014). By decreasing the binding affinity of the sgRNA, the binding stringency of Cas9 to the target sequence was increased, and the off-target effect was reduced (Fu et al., 2014). Since truncated sgRNAs can reduce the off-target effect of paired Cas9 nickases without compromising the efficiency of on-target genome editing, their combination results in a much greater target specificity (Guilinger et al., 2014). Contrarily, the 3′ end-truncated sgRNA or 5′ end-added sgRNA (-GG) can decrease the on-target activity. Meanwhile, if they consist of 16 nucleotides or fewer, truncated sgRNAs exhibit lower or undetectable activity compared to matched full-length sgRNAs. Thus, at least 17-nucleotide sgRNAs are required for the CRISPR/Cas9 system to be active during gene editing. Due to the disadvantages of the traditional enzymatic preparation of sgRNAs, such as complexity, time consumption, and safety concerns, the direct chemical synthesis of sgRNAs has been widely accepted, with high sgRNA stability and low off-target effect. Recently, a potential strategy has been reported to reduce off-target editing by DNA–RNA chimera (Yin et al., 2018a). Using the Cas9–sgRNA complex as a guide, the 5′- and 3′-DNA-replaced crRNA enables more efficient genome editing—for example, replacing the crRNA with 10 DNA nucleotides could provide the same level of off-target site indel formation as the truncated sgRNA. Additionally, the synthesis cost of DNA bases is much lower (10-fold cheaper) than that of native crRNA. In light of this, the DNA–RNA chimera could provide a novel approach to reduce the cost and off-target effect of CRISPR/Cas systems.
FIGURE 1.
Optimization strategies of different steps of the CRISPR/Cas system. (A) The sgRNA sequence was optimally designed and modified by truncation or addition of 3’ or 5’ end of sgRNA, DNA-RNA chimera, etc. (B) Cas9 protein was optimized for concentration, temperature, and time, as well as application of variants. (C) Optimization of the RNP complex was conducted for proportion, function conditions, and transformation methods. (D) Donor DNA was optimized for design of the DNA template, proximity to CRISPR components, and choice of high-efficient delivery system. (E) DNA repair pathway was optimized with inhibition of the NHEJ pathway, enhancement of the HDR pathway, and modification of other pathways.
2.2 Cas Variant Application
So far, several highly specific Cas9 variants have been identified, including eSpCas9 (Slaymaker et al., 2016), SpCas9-HF1 (Kleinstiver et al., 2016), HypaCas9 (Chen et al., 2017), xCas9 (Legut et al., 2020), Sniper-Cas9 (Lee et al., 2018), evo Cas9 (evolved Cas9) (Casini et al., 2018), HiFiCas9 (Vakulskas et al., 2018), and HeFSpCas9 (Kulcsar et al., 2017). The main mechanism and characteristics of each variant are comprehensively summarized in Table 1. Among these mechanisms, the most common cause of alteration in Cas9 function is amino acid substitution of the critical domain. Due to the minimum binding energy required to introduce DSBs into the genome, non-specific interactions between Cas9 and target DNA were reduced by decreasing the excess energy of wild-type SpCas9. As shown in Table 1, although they exhibit greater target specificity, each variant has its own limitations, such as low activity (Slaymaker et al., 2016), scope limitation (Kleinstiver et al., 2016), strict dependency on a PAM site (Legut et al., 2020), etc. Future studies should be conducted to increase the efficiency of genome editing using Cas9 variants. For a given target sequence, the optimal variant should be selected based on a comparison of activity, specificity, and PAM compatibility. By comparing 13 SpCas9 variants, the results demonstrated that the overall activity order of high-fidelity variants could be ranked as SpCas9 ≥ Sniper-Cas9 > eSpCas9 (1.1) > SpCas9-HF1 > HypaCas9 ≈ xCas9 > evoCas9, whereas their overall specificity could be ranked as evoCas9 > HypaCas9 ≥ SpCas9-HF1 ≈ eSpCas9 (1.1) > xCas9 > Sniper-Cas9 > SpCas9 (Kim et al., 2020b). Using established computational models, these results provide guidance for the selection of Cas9 variants and offer a more effective exploration of variants for future research (Figure 1B).
TABLE 1.
The features of different Cas protein variants.
| Cas variants | Description | Mechanisms | Target sequence | Average indel frequency | Advantages | Limitation | Reference |
|---|---|---|---|---|---|---|---|
| xCas9 | Generation of xCas9 by “phage-assisted continuous evolution (PACE)” method | Closing to PAM or the DNA-sgRNA interface refines the DNA-RNA contact region | Refer to the three lentiviral libraries (Kim et al., 2020a) | 32% | Improve the target specificity and extend the target range, present a higher DNA specificity and lower off-target activity | Profoundly diminished of xCas9 activity at target sites with NGH PAM | Nishimasu et al. (2018) |
| SpCas9-HF1 | The quadruple substitution variant (N497A/R661A/Q695A/Q926A) of wild-type SpCas9 | Reduce the rate of DNA cleavage but have no effect on the rate of DNA reversion and release | Refer to the three lentiviral libraries (Kim et al., 2020b) | 34% | A high-fidelity variant retains on-target activities comparable to wild-type SpCas9 with >85% of sgRNAs | The unclear mechanism of target discrimination and fidelity needs to be further improved | Kleinstiver et al. (2016) |
| eSpCas9 | SpCas9 mutants consisting of individual alanine substitutions at 32 positively charged residues within the nt-groove | Neutralization of positively charged residues within this non-target strand and then weaken non-target strand binding and encourage re-hybridization between the target and non-target DNA strands | Refer to the three lentiviral libraries (Kim et al., 2020a) | 40% | Decrease the off-target activities and maintain efficient on-target editing | The unclear mechanism of target discrimination and fidelity needs to be further improved | Slaymaker et al. (2016) |
| HypaCas9 | The quadruple substitution variant (N692A/M694A/Q695A/H698A) of wild-type SpCas9 | The quadruple substitutions in the REC3 domain of wild-type SpCas9 | Refer to the three lentiviral libraries (Kim et al., 2020b) | 30% | Higher genome-wide fidelity without affecting the on-target genome editing | Not mentioned | Chen et al. (2017) |
| Cas9n | Inactivating of HNH or RuvC nuclease domains | Use dual-RNAs for site-specific DNA cleavage | Two human genes: C4BPB and CCR5 | 75 and 60% | Greater target specificity | Rational design of sgRNAs on the plus and minus strands within a limited distance | Trevino and Zhang (2014) |
| Sniper-Cas9 | The quadruple substitution variant (F539S/M763I/K890N) of wild-type SpCas9 | Weakening non-specific interactions between RNP and its substrate DNA | Refer to the three lentiviral libraries (Kim et al., 2020a) | 46% | Retain WT-level on-target activity with diminished off-target effect | Not mentioned | Lee et al. (2018) |
| evoCas9 | The quadruple substitution variant (M495V/Y515N/K526E/R661Q) of wild-type SpCas9 | Weakening non-specific interactions between RNP and its substrate DNA | Refer to the three lentiviral libraries (Kim et al., 2020b) | 15% | Retain WT level on-target activity with diminished off-target effect | Not mentioned | Casini et al. (2018) |
| HiFiCas9 | The quadruple substitution variant (R691A) of wild-type SpCas9 | Weakening non-specific interactions between RNP and its substrate DNA | Five human genes: HBB, IL2RG, CCR5, HEXB, and TRAC | Similar to WT Cas9 | Retain WT level on-target activity with diminished off-target effect | Not mentioned | Vakulskas et al. (2018) |
| HeFSpCas9 | The quadruple substitution variant (N497A/R661A/K846A/Q926A/K1003A/R1060A) of wild-type SpCas9 | Combinations of mutation domain from both eSpCas9 and SpCas9-HF1 | Not shown | Not shown | Retain WT level on-target activity with diminished off-target effect | Not mention | Kulcsar et al. (2017) |
2.3 Determination of the Optimal RNP Concentration
In general, the specificity and the activity of enzymes are often highly dependent on reaction conditions. RNP delivery produces at least twofold more colonies than plasmid transfection does (Kim et al., 2014). In the CRISPR/Cas9 system, RNP concentration plays a decisive role in both specificity and activity. After delivery to cells, RNPs almost immediately cleave chromosomal DNA and then degrade rapidly. With a high RNP concentration, the off-target effects of a CRISPR/Cas system may be amplified (Figure 1C). Meanwhile, a low RNP concentration leads to a reduction of on-target cleavage efficiency. Therefore, a suitable concentration of RNP is of paramount importance to minimize nonspecific cleavage (Figure 1E). This can be achieved by using either low concentrations of plasmids or different promoters. The former method directly reduces RNP transcription, while the latter alters the 5′-untranslated region of the target sequence, ultimately affecting translation efficiency (Hsu et al., 2013). Therefore, extensive measurements should be performed with consideration of both Cas9 activity and specificity. Compared with typical RNP concentrations, on-target activity will inevitably be inhibited to some extent. By modifying Cas9 and sgRNA instead (Figure 2A), the intrinsic specificity of Cas9 can be improved without sacrificing cleavage efficiency.
FIGURE 2.
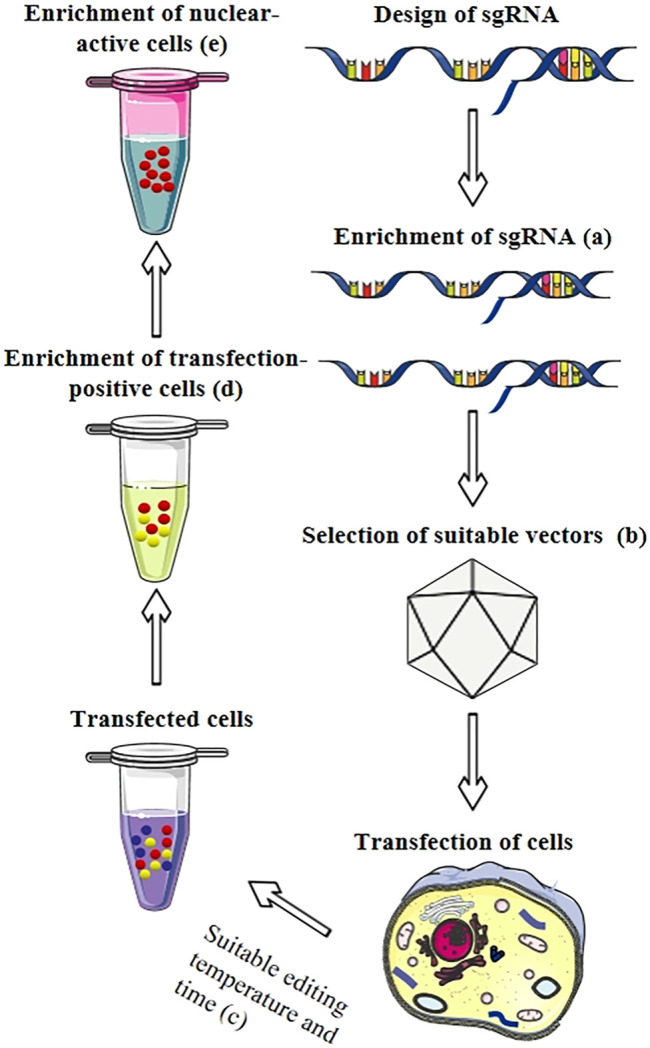
Enrichment strategies for sgRNA and mutants in CRISPR/Cas systems. (A) The sgRNA was enriched with by PCR or CRISPR. (B) The optimal vector was selected according to the different host cell and other factors. (C) Optimization of editing time and temperature was conducted through application of nuclease-dead mutants of Cas9 and anti-CRISPR proteins, heat stress method. (D) Transfection-positive cells were enriched based on fluorescent proteins, antibiotic-resistance genes, and cell-surface antigens. (E) Nuclear-active cells were enriched using NHEJ-based and SSA-based surrogate reporters.
2.4 Suitable Editing Time and Temperature
During gene editing, the efficacy, efficiency, and accuracy of CRISPR systems are often limited by temperature and time (Figure 2C). Studies have shown that a longer editing time of the Cas proteins in cells increases the off-target effects and negatively impacts outcomes (Ruan et al., 2017). Therefore, reducing the action time or overexpression of Cas proteins enhances the efficiency of gene editing. Early reports have utilized tissue culture-free systems (Manghwar et al., 2020), DNA-free systems (Kim et al., 2020a), and transient systems (Canto, 2016) to overcome these limitations. Now, however, anti-CRISPR proteins and nuclease-dead Cas proteins can be used to address these limitations. Temperature has been shown to affect Cas protein activity, but the findings are inconsistent. Hoyland-Kroghsbo et al. (2018) reported that a lower temperature is more effective than a higher one in P. aeruginosa PA14 due to the greater stability of the CRISPR/Cas complex. Additionally, low temperatures may enhance interference and adaptation by increasing the annealing efficiency of the crRNA to its target. Another report has shown that exposure to heat stress results in a greater amount of targeted mutations than with exposure to standard temperature (22°C) (LeBlanc et al., 2018). This is likely due to the fact that the activities of Cas9 and Cas12a at 37 and 34°C, respectively, are higher than at 28°C, and the expression level of sgRNA is raised at 39°C (Xiang et al., 2017). The mechanism of the effect of temperature on CRISPR/Cas systems is still unclear and should be further elucidated.
2.5 Application of Anti-CRISPR Proteins
In the course of long-term antagonism between bacteria and phages, the phages have evolved Acr proteins to evade CRISPR/Cas-mediated immunity. Up to now, a total of 44 Acr proteins have been identified and named (Zhang et al., 2019a). Within the CRISPR/Cas system subtypes, these Acr proteins are present in types I, II, and V, but not in other subtypes—for instance, FnCas9 (Green and Hu, 2017) and C2c2 (Zhang et al., 2019a) Acr proteins were not found in the subtype II-B CRISPR/Cas9 system and the type VI CRISPR/Cas13a system, respectively. In view of this, the identification and characterization of a novel Acr is a key focus for future studies. As natural inhibitors, Acrs protect the host genome from destruction by inhibiting Cas nuclease activity. This inhibition can be achieved through the following three mechanisms: (1) inhibition of Cas9 binding to DNA (Malone et al., 2020), (2) interference of Cas9 binding to gRNA (Harrington et al., 2017), and (3) blocking the activity of Cas9 (Harrington et al., 2017). Currently, only AcrIIA2 and AcrIIA4 have been utilized as tools to reduce off-target effects. Through competition with the PAM site and/or other Cas9 sequences, these Acr proteins block the cleavage activity of Cas9, preventing the excessive and prolonged expression of CRISPR/Cas9 and thus decreasing off-target effects (Hoffmann et al., 2019; Liu et al., 2019). Acrs appear to be a new agent to improve the accuracy and safety of CRISPR-based therapies. Other special functions of Acr proteins also deserve attention—for example, AcrII-C3 can precisely regulate gene expression with dCas9-based tools, which is very helpful for the development of versatile genome engineering modulators (Liu et al., 2018c). The optogenetic-controlled AcrIIA4 enables light-mediated genome and epigenome editing. By inserting the AsLOV2 domain into the most C-terminal loop of AcrIIA4, the protein can switch the CRISPR-Cas9 activity according to light/dark conditions (Bubeck et al., 2018). The mechanisms of other Acrs, such as AcrIIA5-10 and AcrVA2-3, have yet to be described (Zhang et al., 2019a).
3 Strategies to Improve the Efficiency of the HDR Repair Pathway
After Cas9 nuclease cleavage, DSBs can be repaired in a host through at least one of two different pathways: nonhomologous end joining (NHEJ)/canonical NHEJ (c-NHEJ) and HDR (Ghezraoui et al., 2014; Sander and Joung, 2014). While c-NHEJ is the predominant approach, due to its speed and high efficiency, it is also prone to error because of leading uncertain inserts or deletions (indels). Indels contribute to the generation of a targeted knockout during cell repair (Pawelczak et al., 2018). The HDR pathway enables accurate genome editing in a variety of manners, such as gene knock-in, knockout, replacement, and point mutations (Platt et al., 2014; Zuo et al., 2017; Vakulskas et al., 2018; Lu et al., 2020). However, due to competition with the NHEJ pathway, the HDR pathway tends to be less efficient (Liu et al., 2018a). Given this, different approaches have been established to improve the repair efficiency of the HDR pathway, including inhibition of the NHEJ pathway (Maruyama et al., 2015), regulation of HDR-related factors (Paulsen et al., 2017), cell cycle synchronization (Ferrari et al., 2020), optimal design of the donor DNA template (Renaud et al., 2016), and optimizing the proximity of the CRISPR component and donor DNA template (Ma et al., 2017a). These strategies are discussed in detail in the following paragraphs.
3.1 Inhibition of Nonhomologous End Joining Pathway
In theory, because of the competition between the two repair pathways, the efficiency of the HDR pathway can be boosted by inhibiting key factors of the NHEJ pathway. Among different inhibitors of the NHEJ pathway, SCR7 is a key factor that interferes with the affinity of DNA ligase IV to DSBs (Srivastava et al., 2012; Chu et al., 2015; Maruyama et al., 2015; Li et al., 2017; Shao et al., 2017; Hu et al., 2018). Maruyama et al. (2015) reported that using SCR7 increased the efficiency of HDR-mediated genome editing by up to 19-fold with the most significant enhancement effect, primarily due to co-injection of the CRISPR-Cas9 constructs with SCR7 into zygotes rather than other cells. The combination of SCR7 with other factors could significantly improve the efficiency of the HDR pathway by either downregulating KU expression (Chu et al., 2015), optimizing the donor template (Hu et al., 2018), or upregulating Rad52 expression and other small molecules (Li et al., 2017; Shao et al., 2017). Among these methods, the efficiency of the HDR pathway using Rad52 combined with SCR7 is the highest, reaching up to 40% (Shao et al., 2017). However, the effect of SCR7 in enhancing the HDR pathway remains controversial at present (Greco et al., 2016), with some reporting that embryonic stem cells tend to occur intrinsically HDR incident, suggesting that the effect of SCR7 is likely uncertain (Yang et al., 2020). Therefore, the effect of SCR7 in increasing HDR efficiency needs to be further explored. Additionally, the use of SCR7 should be extended to other fields as well instead of being limited to human- and mammalian-related diseases only, whether used alone or in combination with other medicines (Manjunath et al., 2021).
Aside from SCR7, other approaches also improved the efficiency of the HDR pathway—for instance, by downregulating KU protein expression via siRNA silencing, the frequency of the HDR pathway can be increased at least slightly (Li et al., 2018a). This method raised the affinity of downstream NHEJ enzymatic components by attaching to DNA termini upstream of the NHEJ pathway (Mateos-Gomez et al., 2017). The combination of multiple inhibitors can further increase the inhibitory effect on the NHEJ pathway—for example, M3814 combined with trichostatin A inhibits the NHEJ pathway 3-fold (Fu et al., 2021). NU7441 and KU-0060648, inhibitors of a key NHEJ pathway factor, DNA-PK, caused a 2-fold increase in HDR efficiency in HEK-293T cells (Robert et al., 2015). Due to the fact that the NHEJ pathway is crucial for the stabilization of the genome, excessive inhibition of the NHEJ pathway may eventually lead to the accumulation of unrepaired DSBs in cells, inducing cell death or embryonic lethality (Beumer et al., 2013). Therefore, it is worth noting that the safety of these inhibitors needs to be carefully evaluated in future work.
3.2 Regulation of HDR-Related Factors
Alternatively, compared with inhibition of the NHEJ pathway, direct regulation of HDR-related factors can result in more precise editing and improve the efficiency of the HDR pathway. Several HDR-related factors have been well studied, including Rad51 (Ma et al., 2020), Rad52 (Shao et al., 2017), C-terminal-binding protein interacting protein (CtIP) (Charpentier et al., 2018), and RS-1 (Song et al., 2016). All of these factors enhance a link in the HDR pathway, thereby boosting repair efficiency. Among these, the overexpression of Rad proteins and the application of CtIP result in superior outcomes. In the HDR pathway, Rad51 proteins search for a DNA donor template to perform reconstitution through the formation of filaments on the DNA (Symington, 2014). As a back-up for Rad51, Rad52 is involved in the formation of Rad51 (Lok and Powell, 2012). When Rad52 fuses with any other factor or factors, HDR efficiency can be boosted at least 2- to 7-folds and sometimes much more (Paulsen et al., 2017; Shao et al., 2017; Tran et al., 2019)—for example, Rad52 fused with Cas9 yielded an approximately 3-fold increase in the efficiency of the HDR pathway, while Rad52 combined with SCR7 increased the HDR efficiency by about 40% (Shao et al., 2017). In the early stage of the HDR pathway, a key protein, CtIP, initiates the resection process and then creates 3′ single-stranded overhangs with exonuclease 1 and bloom syndrome protein complex (Symington, 2014). When combined with Cas9 or the MS2 system, CtIP can shift the ratio of the HDR/NHEJ pathway activities by a factor of 14.9 (Tran et al., 2019). A minimal N-terminal fragment of CtIP can also be used as an HDR enhancer, which is sufficient to stimulate the HDR pathway and improve repair efficiency by at least 2-fold (Charpentier et al., 2018). Other small molecules, such as L755507 and resveratrol, can also increase HDR efficiency 2- to 3-fold (Li et al., 2017). Almost all key factors of the HDR pathway were upregulated after treatment with the small molecules mentioned above, but a high concentration of resveratrol (more than 50 μM) resulted in severe cytotoxicity, significantly reducing cell viability and slightly upregulating the expression of the NHEJ factor. Therefore, the application of these factors needs to be further explored to improve the efficiency of the HDR pathway.
The selection of the NHEJ or HDR pathway plays a pivotal role in DNA repair. As an inhibitor of the HDR pathway, the tumor suppressor p53-binding protein 1 (53BP1) blocks DNA end resection and simultaneously inhibits BRCA1 recruitment to DSB sites (Panier and Boulton, 2014; Zimmermann and de Lange, 2014). By means of fusing, inhibiting, and binding 53BP1, HDR repair frequency can be increased from 20 to 86% (Paulsen et al., 2017; Canny et al., 2018; Jayavaradhan et al., 2019). Compared with a control, the correction frequency of the HDR pathway increased by nearly 20% when RAD52 was ectopically co-expressed with dominant-negative 53BP1 (dn53BP1) (Paulsen et al., 2017). Interestingly, dn53BP1 itself did not improve HDR efficiency unless combined with Rad52, suggesting that this fragment may not effectively promote the HDR pathway. To avoid the unwanted effects of global NHEJ inhibition, the fusion of DN1S and dn53BP1 significantly blocked NHEJ events locally while improving the correction frequency of HDR up to 86% (Jayavaradhan et al., 2019). This effect was likely due to the restrictively suppressive effect of dn53BP1 around the DSB site, which makes the CRISPR-Cas9-DN1S system a far more effective and stable approach in clinical treatments with high HDR frequency. Inhibition of 53BP1 is another indispensable strategy for regulating different repair pathways—for instance, utilization of 53BP1 inhibitor (i53) resulted in up to a 5.6-fold gene conversion and demonstrated an effective impact on the conversion mediated by single-stranded oligodeoxynucleotides (ssODN) compared to canonical modality double-stranded DNA (dsDNA) donors (Canny et al., 2018). However, the unknown toxicities or adverse incidents of i53 need to be carefully considered before its use. Moreover, the direct binding of related proteins to 53BP1 is a new target for enhancing the HDR pathway. A typical example is the TIRR protein, which acts similar to i53 and provides strong suppression by binding directly to 53BP1 (Anuchina et al., 2020). Since the function of TIRR is unclear, more studies should focus on its mechanism and the potential effects on the HDR pathway. TIRR may hold promise as a new target for enhancing the HDR pathway in genome editing.
3.3 Synchronization of Cas9 Activity and HDR-Active Cell Cycle
Since HDR repair activity is restricted to the S and G2 phases of the cell cycle, synchronizing cells in these phases can significantly enhance the repair activity. In terms of a single aspect of cell cycle synchronization, chemicals that maintain the cell cycle in the S and G2 phases containing nocodazole (Yiangou et al., 2019), ABT-751 (Yang et al., 2016), and RO-3306 (Sunada et al., 2021) have been commonly used in CRISPR systems and increased the HDR efficiency by a factor of 1.38–6 (Lin et al., 2014a; Yang et al., 2016; Wienert et al., 2020). ABT-751 and nocodazole arrest the cells in the G2/M phase by inhibiting microtubule polymerization (Vasquez et al., 1997; Hande et al., 2006). Meanwhile, RO-3306 can transiently arrest cells at the transition from G2 to M phase by inhibiting the CDK1 function, enriching the number of cells in the S and G2 phases (Vassilev, 2006). Recently, Lomova et al. reported that the transient suppression of Cas9 activity and synchronization of the HDR-active cell cycle may have a prominent effect on the HDR pathway. RO-3306 combined with Cas9, which nuclease activity is reduced in the G1 phase, can improve the HDR/NHEJ ratio 20-fold, thereby limiting unnecessary NHEJ events (Lomova et al., 2019). In addition, the timed delivery of pre-assembled Cas9 RNP and chemical synchronization agents can also enhance the HDR rates by up to 38% (Lin et al., 2014a). Thus, synchronizing the cell cycle paired with controlled timing of Cas9 activity might be more practical and safer than synchronizing the cell cycle alone. In conclusion, more efficient synchronization agents for in vitro application need to be further studied. Agents with lower toxicity should also be explored for in vivo application in subsequent research. More importantly, there is no doubt that the combination of multiple technologies, such as controlled timing of Cas9 activity and cell cycle synchronization, will result in better outcomes.
3.4 Increasing the Proximity of CRISPR Components to the Donor DNA Template
At the time of DNA cleavage, if the donor DNA template is in closer proximity to the CRISPR components or has a higher concentration in the nucleus, the efficiency of the HDR pathway can be significantly increased (Devkota, 2018). Based on this hypothesis, many studies have demonstrated its feasibility and potential value in clinical applications (Liang et al., 2017). By attaching the donor DNA template to modified sgRNA, a S1mplex strategy improves the enhancement of the HDR pathway. In this system, the modified S1m-sgRNA adds an aptamer, which binds the streptavidin protein. Biotinylated ssODN linked to the streptavidin then increases proximity. Through this powerful S1mplex strategy, the ratio of HDR increased 18-fold compared with the unlinked components (Carlson-Stevermer et al., 2017). By virtue of the affinity between avidin and biotin, Ma et al. devised a Cas9 variant that was fused to avidin via a flexible linker and bridged with biotin-modified ssDNA to increase the proximity. This system achieved ∼20% HDR frequency in mouse embryos (Ma et al., 2017a). HUH endonuclease is a bridge that is also capable of forming robust covalent bindings with unmodified donor DNA templates (Lovendahl et al., 2017). Utilizing this convenient technique could create a stable Cas9 RNP–ssODN complex (Aird et al., 2018). Additionally, Natasa et al. linked ssODN to Cas9 through SNAP-tag technology, allowing O6-benzylguanine-labeled ssODN to covalently bind to SNAP-tag fusion proteins (Savic et al., 2018). Both SNAP-tag and HUH-tag enable the spatio-temporal co-localization of the donor templates at DSBs, thus achieving 24- and 30-fold enhancement of HDR efficiency, respectively. In addition to ssODN attachment to sgRNA in the S1mplex system, other experiments use a variety of ways to attach ssODN to Cas9 protein. Among them, the HUH-tag strategy displayed a more promising prospect due to the superior ease of use and lower cost associated with modifying donor DNA. As noted above, Cas9 RNP complexes can connect with donor DNA templates through a variety of chemical modifications, all of which significantly enhance the transient expression of the HDR pathway. Furthermore, Cas9 and sgRNA delivered in the RNP format also exhibited a lower frequency of unwanted mutations and off-target effects (Svitashev et al., 2016), suggesting that the RNP format may be a promising approach in the broad field of gene editing.
3.5 Selection of the Donor DNA Template
To a considerable extent, the repair efficiency of the HDR pathway depends on the selection of donor DNA, including DNA modality, length, and flanking homologous sequences (Song and Stieger, 2017; Renaud et al., 2016). Generally, the modality of a DNA template can be divided into three forms: plasmid, ssODN, and linear dsDNA (Figure 1D). As the most common form of genetic material, circular plasmids are widely used in gene editing but will not be discussed in this review (Bosch et al., 2020; Sondergaard et al., 2020; Yoshimi et al., 2021). Compared with other donor DNA templates, ssODNs have the advantages of ease of design, lower time investment, less illegitimate random integration (introducing less than 200 nucleotides), and site-specific modification for precise editing (Yang et al., 2013; Miura et al., 2018). By comparing the modification efficiency of CRISPR/mRNA and CRISPR/nuclease for a target gene, the results have shown that the 36-nt length of ssODN with the CRISPR/nuclease form achieved the optimal condition for modification of the target gene, with a slight advantage over the CRISPR/mRNA approach (Kumita et al., 2019). Currently, ssODNs have become a routine editing tool both in vitro and in vivo, especially in multigene knock-in experiments (Yoshimi et al., 2016; Miura et al., 2018; Lim et al., 2020). For large sequence DNA modifications, linear dsDNA templates (up to 11 kb) were commonly used for CRISPR systems with homology arms of 500–800 bp (Yang et al., 2013). The targeted integration of linearized dsDNA–CRISPR can increase the knock-in efficiency 12-fold by injecting PCR-amplified donor DNA, Cas9 mRNA, and sgRNA (Yao et al., 2018b). Therefore, optimal editing outcomes can be obtained by selecting the suitable donor DNA modality according to experimental requirements.
4 Selection of a Highly Efficient Delivery System
So far, numerous delivery systems to deliver drugs and genes have been developed (Figure 2B). In this section, we have selected the current delivery systems with high delivery efficiency, potential for development, high biological safety, and strong tissue specificity for overview. According to their biological characteristics, they can be classified as either bioactive or abiotic. In bioactive systems, common CRISPR delivery systems contain viral vectors (Jarrett et al., 2018; Boucher et al., 2020; Lee et al., 2021), extracellular vehicles (Yao et al., 2021), cell-penetrating peptides (CPPs) (Ramakrishna et al., 2014b), or lipid nanoparticles (Cheng et al., 2020). In abiotic systems, gold nanomaterials (Wang et al., 2018), polymers (Lv et al., 2018), and graphene oxide (Yue et al., 2018) had a better effect on CRISPR system delivery. Several prominent reviews have comprehensively described the mechanisms, efficiency, challenges, and future directions for each of these systems (Yin et al., 2017; Li et al., 2018b; Glass et al., 2018; Wilbie et al., 2019; Zhang et al., 2021). The current status of these delivery systems will be exposited in the following paragraphs. Conventional physical delivery methods, such as electroporation (Shi et al., 2018), microfluidics (DiTommaso et al., 2018), and microinjection (Xu, 2019), possess unique advantages, including high local tissue transfection efficiency and extensive cellular adaptability (Mashel et al., 2020). They are not good candidates for this review, however, as they can also cause cell damage and potentially substantial cell death. Therefore, we did not repeat their descriptions in this article.
4.1 Bioactive Delivery Systems
4.1.1 Viral Vectors
In recent years, viruses have been represented as an essential and powerful tool for CRISPR due to their efficient gene delivery and long-term stable transgenic expression (Heckl et al., 2014). The most commonly utilized viral vectors are derived from adeno-associated virus (AAV) (Jarrett et al., 2018), lentivirus (LV) (Lee et al., 2021), adenovirus (Boucher et al., 2020), and baculovirus (Yin et al., 2021). These viral vectors have been widely used to deliver CRISPR/Cas9 elements for remedying genetic defects, like hearing loss (Omichi et al., 2019), neurological disorders (Pena et al., 2020), muscular dystrophies (Crudele and Chamberlain, 2019), and cystic fibrosis lung disease (Wold and Toth, 2013; Hart and Harrison, 2017). Several excellent reviews concerning different aspects of viral vectors for CRISPR-based genome editing have been published, covering topics such as viral mechanisms (Xu et al., 2019), viral vector application (Song et al., 2021), and viral vector progress (DiCarlo et al., 2017). Although highly efficient, viral vectors are presently hindered by their inherent disadvantages of carcinogenesis, insertion size limitation, immune response, genotoxicity, cytotoxicity, and difficulties of large-scale production (Matrai et al., 2010; Kotterman et al., 2015; Chen and Goncalves, 2016; Chen et al., 2020a; Shirley et al., 2020). These viral vectors have been improved in other aspects, such as pseudotyped LV and dual-AAV systems. When delivering cargo into cells, LVs need to interact with a cellular receptor to trigger the fusion of the viral envelope with the cell membrane. The envelope glycoprotein on the LV surface is exchanged with a heterologous glycoprotein in a process known as pseudotyping. Pseudotyped LVs consist of virus particles bearing glycoproteins derived from other enveloped viruses. Thus far, a variety of viral glycoproteins, including vesicular stomatitis virus (Liu et al., 2017c; Sena-Esteves and Gao, 2018; Rust et al., 2020), baboon endogenous retrovirus (Belot et al., 2019), and feline endogenous retrovirus (Zucchelli et al., 2017), have been incorporated into LVs to improve their infectivity and confer a more selective tropism. The versatile tropism of pseudotyped LVs has been utilized in the treatment of tumors (Lee et al., 2021) and gene modification (Gutierrez-Guerrero et al., 2020). AAV vectors are hindered by their relatively low packaging capacity (Wu et al., 2010), with a packaging range of no more than 5 kb, making them inappropriate for the delivery of larger Cas9 variants (Mali et al., 2013). To address this issue, dual-AAV systems have been explored, in which one encodes Cas9 and another encodes gRNA, resulting in a large target gene transfer (Zhi et al., 2022). It needs to be pointed out, however, that the disadvantages of this system limit its clinical application, such as low probability of delivering both viral vectors to the same cell and insufficient expression efficiency.
4.1.2 Extracellular Vesicles
Whether in vitro or in vivo, extracellular vesicles (EVs) have been widely used to efficiently deliver genes or drugs (Choi et al., 2016; Montagna et al., 2018; Campbell et al., 2019; Mangeot et al., 2019; Gee et al., 2020). As natural cell-derived membrane vesicles, EVs serve the function of cell-to-cell communication with outstanding biocompatibility and immune-privileged characteristics. EVs are also hardly cleared by the immune system, avoiding the occurrence of hypersensitivity reactions (Zhang et al., 2014). Since EVs do not contain viral genomes, they have significant biosafety without the risk of endogenous virus recombination (Fuenmayor et al., 2017). Additionally, EVs transmit Cas9 with transient exposure, reducing the off-target chance triggered by Cas9 overexpression (Wu et al., 2014). All these advantages demonstrate an excellent potential for EVs as endogenous nano-vehicles in various fields. However, a major obstacle for EVs is the lack of robust tissue-specific delivery to specific cells. Targeted ligand modification on the surface of EVs is a promising avenue to ameliorate this weakness (Mathieu et al., 2019)—for instance, valency-controlled tetrahedral DNA nanostructures (TDNs) conjugated with DNA aptamers can be anchored on the EV surface via cholesterol, improving cell-specific delivery (Zhuang et al., 2020). The 3D tetrahedral steric superiority of TDN DNA aptamers can minimize lateral interactions among DNA, resulting in increased receptor–ligand binding and greatly enhancing tissue specificity. Compared with a control group, the TDN1-EVs-RNP group maximally restrained tumor growth in terms of tumor weight, volume, and percentage of tumor cells, demonstrating that the modified group accomplished a 2-fold increase in indel rate (up to 30%). Recently, EVs have been used in chimeric-antigen receptor (CAR) T-cell therapy to deliver CRISPR components to target cells precisely. By expressing chimeric-antigen receptors on vesicles derived from T cells, the anti-CD19-CAR-EVs preferentially accumulated in tumors compared to the liver, kidney, and other healthy tissues. Nevertheless, normal EVs were more evenly distributed throughout the body (Xu et al., 2020a). In addition to delivering CRISPR/Cas9 components, EVs also show great potential for drug delivery (Mateescu et al., 2017; Yang et al., 2018), anticancer therapy (Pascucci et al., 2014; Saari et al., 2015), and antigen delivery for vaccine development (Rabu et al., 2019).
4.1.3 Lipid Nanoparticles
Lipid nanoparticles (LNP) as CRISPR delivery vehicles have attracted the interest of scientists (Yin et al., 2014; Kulkarni et al., 2019). They not only help CRISPR components cross cell membranes but also protect them from enzymatic degradation and immune responses (Liu et al., 2018b; Noll et al., 2018). Due to the advantages of excellent controlled release, targeting, and high stability, LNPs have been widely used as a CRISPR delivery vector for all kinds of cargo modality, such as plasmid DNA, mRNA, and RNP complexes (Li et al., 2018c; Li et al., 2019). Theoretically, endocytosis is considered to be the key to cell internalization for almost all common LNP materials. To improve tissue specificity and delivery efficacy, several new strategies have been reported in recent years. Firstly, based on the hypothesis that charge adjustment can mediate tissue-specific delivery, a new strategy termed selective organ targeting (SORT) has been established. By adding DOTAP (a permanently cationic lipid) and constantly regulating its proportion to the original composition of LNP, we can control the charge for tissue-specific delivery (Cheng et al., 2020). The results show that this SORT strategy can achieve high organ selectivity for CRISPR cargos delivered in the lung, spleen, liver, and other organs. Among these organs, delivery to hepatocytes has the highest specificity at 93%. Secondly, ultrasound has been reported to facilitate the delivery of CRISPR components (Shen et al., 2016; Yoon et al., 2017). Ultrasound at specific locations can cause microbubbles to create local membrane deformations and pore formation in response to acoustic energy (Taniyama et al., 2011; Zhou et al., 2012). LNP released by microbubbles can then be transferred directly into the cytoplasm by diffusion. The results show that LNP incorporated with microbubbles can effectively facilitate cargo to the target site for RNP delivery, and the editing efficiency of Cas9 RNP was improved by 71.6% (Ryu et al., 2020). Thirdly, under optimized synthetic conditions, microfluidic device-designed lipid nanoparticles achieved intracellular RNP delivery with 97% gene disruption and 23% base substitution without any apparent cytotoxicity (Suzuki et al., 2020). In short, optimizing the formulation of LNP or integrating other technologies into the delivery system will be a crucial direction for achieving tissue-specific and efficient systems. Lipid-based formulations, however, do have some disadvantages. Once nanoparticles pass through the surface of cells, they are typically encased within an endosome. The encased contents then enter the lysosomal pathway directly and are eventually degraded. Therefore, coating polymers on the LNP surface or developing other unique chemical modifications to facilitate cellular uptake and disrupting endosomal membranes are promising directions that could prompt endosomal escape and avoid detection by the immune system.
4.1.4 Cell-Penetrating Peptides
As short stretches of amino acids, CPPs are polycationic, amphipathic, or non-polar in nature and possess an intrinsic ability to translocate across cell membranes (Suresh et al., 2017). Owing to the advantages of low cytotoxicity, better biological tolerance, less off-target effect, and no chemical reagent, CPPs have been exploited to deliver different cargos into cells in vitro and in vivo (Liu et al., 2014; Gagat et al., 2017). When delivering RNP complexes, CPPs conjugated with RNP to form CPPs–RNP, which can improve cellular uptake and/or fusion. However, few studies have been reported on CPP-mediated CRISPR component delivery at present. Moreover, both delivery efficiency and subsequent editing efficiency were usually at a low level of just 10–20% (Ramakrishna et al., 2014b; Yin et al., 2018b; Del’Guidice et al., 2018; Yin et al., 2019a). This likely stems from the indefinite mechanism of CPP internalization and requirement for extensive optimization for targeting each type of cargo and cell. As the major CPP cargo is trapped in endosomes, they end up being recycled or degraded in a targeted manner instead of releasing cargo to the specific destination. Thus, enhancing endosomal escape would be a potential approach to improve the efficiency of delivery and editing (LeCher et al., 2017).
4.2 Abiotic Delivery Systems
As an alternative, abiotic vectors may offer tantalizing possibilities for CRISPR/Cas9 delivery systems due to their low immunogenicity, larger delivery gene payload, ease of large-scale production (Li et al., 2015), and absence of endogenous virus recombination. Many excellent delivery systems with new properties have been established in various fields, such as gold nanomaterials (Wang et al., 2018), polymers (Lv et al., 2018), and other systems. The characteristics of each material are described in detail in the following sections.
4.2.1 Gold Nanomaterials
Due to their tunable surface functionalization, non-toxic nature, favorable size, optical properties, biocompatibility, and photothermal effect, inorganic gold nanocarriers have proved to be a promising platform for systemic gene delivery (Ghosh et al., 2008; Ma et al., 2017b). They are mainly characterized by their photothermal effect and ease of functionalization for delivering CRISPR components. As photothermal transducers, gold nanomaterials can regulate the conditional control of Cas9 activity through different optical means (Nihongaki et al., 2015). In locally specific tissues, heat converted by the second near-infrared optical window (1,000 to 1,700 nm) induces endonuclear transformation of the heat-shock factor (HSF) from an inactive monomer to an active trimer. Under the action of active HSF, the combined transfection of a cationic polymer-coated Au nanorod, Cas9 plasmid, and a heat-inducible promoter HSP70 can result in 90% GFP-positive cells, which is much higher than that of Lipofectamine 2000 or 25-kDa polyethyleneimine (Chen et al., 2020b). In the LACM system, the protective DNA-modified gold nanorod hybridizes with the target binding domain of sgRNA to protect sgRNA. Upon NIR laser irradiation, heat subsequently denatures the hybridized DNA and sgRNA, accomplishing the controlled release of sgRNA into cells (Peng et al., 2020). Thus, gold nanomaterials act as an optogenetic switch to regulate the expression and activity of Cas9 proteins with high spatial specificity.
Tunable surface functionalization is another outstanding feature of gold nanocarriers that accelerates the entry of foreign genes into cells. Various biomolecules, such as proteins, DNA, peptides, and polymers, can endow gold nanomaterials with tremendous functions for surface bioengineering (Miao et al., 2018). Protamine, as a natural protein that originates from sperm, has intrinsic cell-penetrating properties and nucleus-targeting abilities and can be used for the efficient delivery of the Cas9–sgRNA plasmid. Protamine can form a compact structure with anionic DNA and then deliver the DNA to the egg nucleus (Biju et al., 2012; Priya et al., 2014). Nanocomplexes of Cas9-gRNAEGFP and protamine-functionalized gold nanoclusters disrupt the EGFP gene effectively and convert approximately 30% of the EGFP-positive transformants to EGFP-negative cells (Tao et al., 2021). Meanwhile, AuNCs can be functionalized by electrostatic action to control the self-assembly process. In a highly pH-dependent manner, AuNCs assembled with Cas9 protein (SpCas9–AuNCs) can deliver SpCas9 into the cell and nucleus in physiological conditions (Ju et al., 2019). The self-assembled SpCas9–AuNC nanoparticles effectively transfect HPV18 E6 sgRNA into cervical cancer cells, knocking out the E6 oncogene at a rate of 34%. More importantly, self-assembled SpCas9–AuNCs had little effect on normal cells, showing a considerable potential for clinical application. However, concerning the application of gold nanomaterials, cytokine production, the extensive modification requirement, fewer in vivo experiments, and potential toxicity need to be fully considered (Dykman and Khlebtsov, 2017). Gold nanomaterials are potentially an excellent delivery system and a bright prospect for improving CRISPR systems. Additionally, they can be extensively applied to bioimaging, optical and electrochemical sensing, and medical diagnostics (Chen et al., 2016). The multifunctional integrated gold nanomaterial platform may make great contributions to biological research in the future.
4.2.2 Polymers
Polymers can also be used to deliver RNP complexes to target sites with many distinct advantages, such as ease of synthesis, structural and component flexibility, functionalization, and degradability (Chen et al., 2016). Their significant flexibility is the most fascinating feature, resulting in multifunctionality by the reasonable and convenient design of the chemical structure (Hsu and Uludag, 2012; Zhang et al., 2019b). Currently, commonly used polymers to deliver drugs or RNP include polylysine, chitosan nanoparticles, poly-(β-amino ester)s, and dendrimers. The first two kinds are commonly used for drug delivery, while the latter two are mostly used for RNP delivery. Studies of drug delivery with polymers have been described in detail in other reviews (Huo et al., 2017; Hasheminejad et al., 2019). For a wide range of unmet therapeutic needs and personalized medicine, poly-(β-amino esters), as a class of amphiphilic and pH-sensitive polymers, can efficiently bind to cargo proteins to facilitate efficient intracellular RNP delivery via hydrogen bonding as well as hydrophobic and ionic interactions (Dwivedi et al., 2012). This characteristic allows them to be customized specifically to overcome delivery barriers in varied applications (Karlsson et al., 2020). Dendrimers are a class of synthetic polymer with a spherical and hyperbranched structure, whose surface is functionalized with a high density of phenylboronic acid moieties to ensure that RNPs are efficiently bound to the dendrimer scaffold and transmit RNP to specific cells (Dixit et al., 2014). As a novel therapeutic tool for genetic disorders, dendrimers allow the efficient delivery of RNP targeting multiple genetic loci in different cell lines, proving to be a useful material for the delivery of genome-editing tools with broad biomedical applications (Taharabaru et al., 2020). Several issues exist with RNP delivery using polymers, however, such as low efficiency, high cytotoxicity, and narrow application range, which need to be overcome in the future.
5 CRISPR Regulation With Nuclease-Dead Cas Proteins
Through the same mechanism mentioned above, sgRNA-directed dCas9 binds to specific DNA sequences. When dCas9 binds specifically to a genomic locus, it can sterically block or activate RNP progression to downstream genes. These two dCas9-based strategies are called CRISPR interference (CRISPRi) (Ji et al., 2020) and CRISPR activation (CRISPRa), respectively (Larson et al., 2013). Both strategies can precisely regulate the expression of the sgRNA module or dCas9 via an inducible expression system. As of yet, several dCas9-based CRISPRa methods have been established, including dCas9-P65AD (Gilbert et al., 2013), dCas9-VPR (Chavez et al., 2015), dCas9-p300 (Hilton et al., 2015), and dCas9-TET (Xu et al., 2018). Some CRISPRi methods have also been reported, including dCas9-KRAB (Abudayyeh et al., 2017), dCas9-LSD1/KDM1A (Gilbert et al., 2013), and dCas13-YTHDF2 (Rauch et al., 2018). Several excellent reviews concerning different dCas-based CRISPRi and CRISPRa strategies describe their mechanism and principle in detail (Kampmann, 2018; Xu et al., 2020b). Currently, they are utilized to screen cellular genomes, including for cell survival/proliferation, sensitivity to drugs or toxins, fluorescent reporters, and single-cell transcriptomes (Kampmann, 2018). They are expected to precisely regulate editing time to reduce off-target effects.
6 Enrichment of Mutants
Due to off-target effects, not all genetically modified cells are equipped with positive mutants in vitro. The selection of mutants from original gene-edited cells is still a challenge at present (Ren et al., 2015). Thus, new strategies need to be investigated for enrichment and selection (Figure 2C). The most common selection markers to enrich positive cells are fluorescent proteins, antibiotic resistance genes, cell surface antigens, and so forth. Due to the merits of visualization, time saving, and decreased labor, fluorescent proteins are widely utilized in CRISPR/Cas systems (Ren et al., 2019). For a variety of cellular and environmental contexts, the variety of fluorescent genes gives scientists immense flexibility in choosing tailored reporters, such as green fluorescent protein, red fluorescent proteins (Liu et al., 2021a), and fluorescent proteins (Cao et al., 2019). Nevertheless, isolated cells are easily damaged by the solid lasers and hydrostatic pressure of flow cytometry. Compared to fluorescent proteins, the antibiotic-based method offers an alternative strategy that does not require expensive equipment but needs more time (Moriarity et al., 2014; Liesche et al., 2016). Although numerous antibiotic resistance genes have been applied in various fields, such as hygromycin (Moriarity et al., 2014), neomycin (Gu et al., 2021), zeocin (Kobayashi et al., 2019), gentamicin (Mulsant et al., 1988) and puromycin (Pandey et al., 2021), marker-free strategies are the preferred method, ameliorating public concerns for the biosafety of antibiotic resistance genes. Another non-fluorescence activated cell sorting-based enrichment method is antigen gene H-2Kk, which has a high enrichment efficiency with magnetic bead separation (Wei et al., 2001). However, when insertions or deletions are generated at the target sequences, these reporter systems express H-2Kk and hygromycin resistance protein, respectively, enabling the efficient enrichment of mutants without flow cytometry (Kim et al., 2013).
However, no matter what efficient strategies are used to select mutants, mutant enrichment alone cannot classify all stable and highly expressed mutants (Figure 2D). Thus, to select nuclear-active mutants, two surrogate reporters based on the NHEJ and single-strand annealing (SSA) have been published (Pattanayak et al., 2013). Nuclease triggers a DSB on the target sequence within the surrogate reporter construct, resulting in the formation of small random indels by the error-prone NHEJ repair pathway and leading to the correction of reporter genes with 1/3 frequency. Compared with unsorted cells, the enrichment efficiency of mutants can be increased up to 8.6- and 18-fold with the first and second generation of NHEJ-based surrogate reporters, respectively (Ramakrishna et al., 2014a). The second surrogate reporter has the capacity to identify more nuclease-positive cells via SSA. Due to its higher sensitivity, this reporter significantly increases the possibility of obtaining the desired genetically modified cell clones (Yasuda et al., 2016). Although DNA repair pathways are influenced by cell type and the nature of broken DNA ends, genomic modification within mutants may be independent of repair pathways in surrogate reporters (Ren et al., 2015). On the basis of transfection-positive cells, these two surrogate reporter strategies can produce highly efficient, nuclease-active cells.
7 Conclusion and Future Prospects
Aside from the above-mentioned approaches, other strategies can also significantly improve the editing efficiency of CRISPR/Cas systems. Firstly, owing to the fact that the nucleosome poses a strong barrier to Cas9, restoring Cas9 access to nucleosomes through the chromatin remodeling enzyme yChd1 therefore results in high efficiency editing. Nucleosome organization represents only one aspect of eukaryotic chromatin, however; thus, future research on how chromatin affects Cas9 activity needs to be done (Horlbeck et al., 2016). Secondly, cytosine base editors (CBE) and adenine base editors (ABE) have been utilized to change C/G to T/A and A/T to G/C. CBE deaminates cytosine to uracil, which is recognized by the cell replication machinery as thymine, resulting in a C/G to T/A transition. ABE-mediated DNA editing operates under a similar mechanism as that of CBE (Koblan et al., 2018; Richter et al., 2020). Despite efforts to improve DNA base editors, base editing is confined to transition mutations (incapable of transversion mutation) and is not capable of inducing indel mutations. Next, by combining reverse transcriptase with prime editors gRNA and Cas-nickase nuclease, prime editing technology can edit or “search and replace” bases in a genome (Anzalone et al., 2019). It can also be used as an alternative genome editing tool to investigate various challenges, such as editing large genes, targeting autosomal dominant diseases, and editing premature stop codons and splice-site variants (Kantor et al., 2020). When prime editors are undesirable and the base editing window is well defined, base editors are typically more efficient than prime editors. On the contrary, when prime editors are acceptable and multiple editable bases are within a defined editing window, prime editors offer unsurmountable advantages.
In the last few years, we have seen the extraordinary growth and expansion of gene editing, particularly in the field of gene therapy. Based on CRISPR technology, a series of highly efficient and targeted transcription factor components has been developed and used to construct intelligent gene circuits, making tumor gene therapy possible (Zhou et al., 2019). In cardiovascular medicine, CRISPR-based tools have multiple applications, with a primary focus on direct therapeutic interventions to treat inherited cardiac disorders (Vermersch et al., 2020). CRISPR also represents a breakthrough advance in genetically engineered immune cells (Huang et al., 2020), personalized cancer medicine (Li and Kasinski, 2020), and modification of human embryos (Tang et al., 2017). Even in the current novel coronavirus (COVID-19) outbreak, CRISPR-based technology has shown strong application value. All-in-one dual CRISPR-Cas12a is instrumental in the detection of COVID-19, offering the advantages of being instrument-free, rapid, sensitive, one-pot, and point-of-care (Ding et al., 2020). Applications in microbiology are still being newly discovered and improved, specifically in the identification and modification of industrial-related lactobacilli and streptococci as well as foodborne pathogens, including E. coli (Altenbuchner, 2016), Saccharomyces cerevisiae (Biot-Pelletier and Martin, 2016), and thermophilic fungi (Liu et al., 2017b). As a new generation of precision gene editing tools, the great success of CRISPR/Cas systems in various fields shows that these have a wide range of application and wonderful prospects.
Collectively, knowledge and technologies of genome editing are ceaselessly developing in intricately interwoven fields and are creating huge synergies. With the recent developments in CRISPR/Cas systems, they are becoming increasingly accurate, efficient, and reliable. Although massive advances have been achieved, the CRISPR/Cas systems are far from their optimal state. Among various challenges, off-target effects are still the foremost barrier in CRISPR/Cas systems. We have listed above several strategies for reducing off-target effects. Among them, special attention should be paid to optimizing time and temperature, which are often inadvertently neglected. The CRISPR/Cas systems have other limitations, including inactive mutants, variable efficiency, requirement of PAM and sgRNA, fault-prone programmed DNA repair pathways, and the lack of an efficient and safe delivery system. Apart from these, future research will involve the enhancement of Cas9 activity, application of ACR proteins, and determination of the optimal Cas9 and sgRNA ratio so as to further improve the efficiency of CRISPR systems. Simultaneously, continuous optimization of external measures, including dCas9 regulation, delivery vector development, mutant enrichment, etc., will help to further improve the efficiency. Although we are far from eliminating off-target effects completely, we are confident that CRISPR technology will continue to be perfected to meet the demands of different fields by adopting the aforementioned strategies.
Author Contributions
All authors took part in writing, reviewing, and editing the manuscript. SF, ZW, and AL wrote the manuscript. XX and SL prepared the figures. YL and BW collected and organized the literature. JL and LH created the table. LY and TG modified the paper. All authors reviewed the manuscript and approved it for publication.
Funding
This work was supported by the National Natural Science Foundation of China (nos. U1804112 and 31571289) and the Zhongjing Core Scholar’s Research Initial Fund of Henan University of Chinese Medicine (no. 00104311-2021).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Glossory
- CRISPR
clustered regulatory interspaced short palindromic repeats
- ZFNs
zinc finger nucleases
- TALENs
transcription activator-like effector nucleases crRNA
- PAM
proto-spacer adjacent motifs
- sgRNA
single guide RNA
- DSBs
double-strand breaks
- PCR
Polymerase chain reaction
- Acr
anti-CRISPR
- NHEJ
nonhomologous end joining
- c-NHEJ
canonical NHEJ
- HDR
homology-directed repair
- 53BP1
tumor suppressor p53-binding protein 1
- CtIP
CtIP C-terminal-binding protein interacting protein
- dn53BP1
dominant-negative 53BP1
- ssODN
single-stranded oligodeoxynucleotides
- dsDNA
double-stranded DNA
- EVs
extracellular vehicles
- CPPs
cell-penetrating peptides
- AAV
adeno-associated virus
- LV
lentivirus
- AdV
adenovirus
- GO
graphene oxide
- TDNs
tetrahedral DNA nanostructures
- CAR
chimeric-antigen receptor
- LNP
lipid nanoparticles
- NIR
near-infrared
- HSF
heat-shock factor
- AuNR
gold nanorods
- AuNCs
gold nanoclusters
- CRISPRi
CRISPR interference
- CRISPRa
CRISPR activation
- eGFP
enhanced green fluorescent proteins
- RFP
red fluorescent proteins
- FPs
fluorescent proteins
- CBE
cytosine base editors
- ABE
adenine base edito
References
- Abby S. S., Néron B., Ménager H., Touchon M., Rocha E. P. C. (2014). MacSyFinder: a Program to Mine Genomes for Molecular Systems with an Application to CRISPR-Cas Systems. PLoS One 9 (10), e110726. 10.1371/journal.pone.0110726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh O. O., Gootenberg J. S., Essletzbichler P., Han S., Joung J., Belanto J. J., et al. (2017). RNA Targeting with CRISPR-Cas13. Nature 550 (7675), 280–284. 10.1038/nature24049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adli M. (2018). The CRISPR Tool Kit for Genome Editing and beyond. Nat. Commun. 9 (1), 1911. 10.1038/s41467-018-04252-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aird E. J., Lovendahl K. N., St. Martin A., Harris R. S., Gordon W. R. (2018). Increasing Cas9-Mediated Homology-Directed Repair Efficiency through Covalent Tethering of DNA Repair Template. Commun. Biol. 1, 54. 10.1038/s42003-018-0054-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenbuchner J. (2016). Editing of the Bacillus Subtilis Genome by the CRISPR-Cas9 System. Appl. Environ. Microbiol. 82 (17), 5421–5427. 10.1128/aem.01453-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anuchina A. A., Lavrov A. V., Smirnikhina S. A. (2020). TIRR: a Potential Front Runner in HDR Race−hypotheses and Perspectives. Mol. Biol. Rep. 47 (3), 2371–2379. 10.1007/s12033-020-05285-x [DOI] [PubMed] [Google Scholar]
- Anzalone A. V., Randolph P. B., Davis J. R., Sousa A. A., Koblan L. W., Levy J. M., et al. (2019). Search-and-replace Genome Editing without Double-Strand Breaks or Donor DNA. Nature 576 (7785), 149–157. 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batool A., Malik F., Andrabi K. I. (2021). Expansion of the CRISPR/Cas Genome-Sculpting Toolbox: Innovations, Applications and Challenges. Mol. Diagn. Ther. 25 (1), 41–57. 10.1007/s40291-020-00500-8 [DOI] [PubMed] [Google Scholar]
- Belot L., Albertini A., Gaudin Y. (2019). Structural and Cellular Biology of Rhabdovirus Entry. Adv. Virus. Res. 104, 147–183. 10.1016/bs.aivir.2019.05.003 [DOI] [PubMed] [Google Scholar]
- Beumer K. J., Trautman J. K., Mukherjee K., Carroll D. (2013). Donor DNA Utilization during Gene Targeting with Zinc-Finger Nucleases. G3 (Bethesda) 3 (4), 657–664. 10.1534/g3.112.005439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biju V., Anas A., Akita H., Shibu E. S., Itoh T., Harashima H., et al. (2012). FRET from Quantum Dots to Photodecompose Undesired Acceptors and Report the Condensation and Decondensation of Plasmid DNA. ACS Nano 6 (5), 3776–3788. 10.1021/nn2048608 [DOI] [PubMed] [Google Scholar]
- Biot-Pelletier D., Martin V. J. J. (2016). Seamless Site-Directed Mutagenesis of the Saccharomyces cerevisiae Genome Using CRISPR-Cas9. J. Biol. Eng. 10, 6. 10.1186/s13036-016-0028-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch J. A., Colbeth R., Zirin J., Perrimon N. (2020). Gene Knock-Ins in Drosophila Using Homology-independent Insertion of Universal Donor Plasmids. Genetics 214 (1), 75–89. 10.1534/genetics.119.302819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher P., Cui X., Curiel D. T. (2020). Adenoviral Vectors for In Vivo Delivery of CRISPR-Cas Gene Editors. J. Controlled Release 327, 788–800. 10.1016/j.jconrel.2020.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubeck F., Hoffmann M. D., Harteveld Z., Aschenbrenner S., Bietz A., Waldhauer M. C., et al. (2018). Engineered Anti-CRISPR Proteins for Optogenetic Control of CRISPR-Cas9. Nat. Methods 15 (11), 924–927. 10.1038/s41592-018-0178-9 [DOI] [PubMed] [Google Scholar]
- Cameron P., Fuller C. K., Donohoue P. D., Jones B. N., Thompson M. S., Carter M. M., et al. (2017). Mapping the Genomic Landscape of CRISPR-Cas9 Cleavage. Nat. Methods 14 (6), 600–606. 10.1038/nmeth.4284 [DOI] [PubMed] [Google Scholar]
- Campbell L. A., Coke L. M., Richie C. T., Fortuno L. V., Park A. Y., Harvey B. K. (2019). Gesicle-Mediated Delivery of CRISPR/Cas9 Ribonucleoprotein Complex for Inactivating the HIV Provirus. Mol. Ther. 27 (1), 151–163. 10.1016/j.ymthe.2018.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canny M. D., Moatti N., Wan L. C. K., Fradet-Turcotte A., Krasner D., Mateos-Gomez P. A., et al. (2018). Inhibition of 53BP1 Favors Homology-dependent DNA Repair and Increases CRISPR-Cas9 Genome-Editing Efficiency. Nat. Biotechnol. 36 (1), 95–102. 10.1038/nbt.4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto T. (2016). Transient Expression Systems in Plants: Potentialities and Constraints. Adv. Exp. Med. Biol. 896, 287–301. 10.1007/978-3-319-27216-0_18 [DOI] [PubMed] [Google Scholar]
- Cao X., Zhang C., Gao Z., Liu Y., Zhao Y., Yang Y., et al. (2019). Ultrafast Internal Conversion Dynamics of Bilirubin Bound to UnaG and its N57A Mutant. Phys. Chem. Chem. Phys. 21 (5), 2365–2371. 10.1039/c8cp07553k [DOI] [PubMed] [Google Scholar]
- Carlson-Stevermer J., Abdeen A. A., Kohlenberg L., Goedland M., Molugu K., Lou M., et al. (2017). Assembly of CRISPR Ribonucleoproteins with Biotinylated Oligonucleotides via an RNA Aptamer for Precise Gene Editing. Nat. Commun. 8 (1), 1711. 10.1038/s41467-017-01875-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casini A., Olivieri M., Petris G., Montagna C., Reginato G., Maule G., et al. (2018). A Highly Specific SpCas9 Variant Is Identified by In Vivo Screening in Yeast. Nat. Biotechnol. 36 (3), 265–271. 10.1038/nbt.4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M., Khedher A. H. Y., Menoret S., Brion A., Lamribet K., Dardillac E., et al. (2018). CtIP Fusion to Cas9 Enhances Transgene Integration by Homology-dependent Repair. Nat. Commun. 9 (1), 1133. 10.1038/s41467-018-03475-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A., Scheiman J., Vora S., Pruitt B. W., Tuttle M., P R Iyer E., et al. (2015). Highly Efficient Cas9-Mediated Transcriptional Programming. Nat. Methods 12 (4), 326–328. 10.1038/nmeth.3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Alphonse M., Liu Q. (2020a). Strategies for Nonviral Nanoparticle-Based Delivery of CRISPR/Cas9 Therapeutics. Wiley Interdiscip. Rev. Nanomed Nanobiotechnol 12 (3), e1609. 10.1002/wnan.1609 [DOI] [PubMed] [Google Scholar]
- Chen J., Guo Z., Tian H., Chen X. (2016). Production and Clinical Development of Nanoparticles for Gene Delivery. Mol. Ther. - Methods Clin. Develop. 3, 16023. 10.1038/mtm.2016.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. S., Dagdas Y. S., Kleinstiver B. P., Welch M. M., Sousa A. A., Harrington L. B., et al. (2017). Enhanced Proofreading Governs CRISPR-Cas9 Targeting Accuracy. Nature 550 (7676), 407–410. 10.1038/nature24268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Chen Y., Xin H., Wan T., Ping Y. (2020b). Near-infrared Optogenetic Engineering of Photothermal nanoCRISPR for Programmable Genome Editing. Proc. Natl. Acad. Sci. USA 117 (5), 2395–2405. 10.1073/pnas.1912220117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Gonçalves M. A. F. V. (2016). Engineered Viruses as Genome Editing Devices. Mol. Ther. 24 (3), 447–457. 10.1038/mt.2015.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q., Wei T., Farbiak L., Johnson L. T., Dilliard S. A., Siegwart D. J. (2020). Selective Organ Targeting (SORT) Nanoparticles for Tissue-specific mRNA Delivery and CRISPR-Cas Gene Editing. Nat. Nanotechnol. 15 (4), 313–320. 10.1038/s41565-020-0669-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. G., Dang Y., Abraham S., Ma H., Zhang J., Guo H., et al. (2016). Lentivirus Pre-packed with Cas9 Protein for Safer Gene Editing. Gene Ther. 23 (7), 627–633. 10.1038/gt.2016.27 [DOI] [PubMed] [Google Scholar]
- Chu V. T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., et al. (2015). Increasing the Efficiency of Homology-Directed Repair for CRISPR-Cas9-Induced Precise Gene Editing in Mammalian Cells. Nat. Biotechnol. 33 (5), 543–548. 10.1038/nbt.3198 [DOI] [PubMed] [Google Scholar]
- Coelho M. A., De Braekeleer E., Firth M., Bista M., Lukasiak S., Cuomo M. E., et al. (2020). CRISPR GUARD Protects Off-Target Sites from Cas9 Nuclease Activity Using Short Guide RNAs. Nat. Commun. 11 (1), 4132. 10.1038/s41467-020-17952-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crudele J. M., Chamberlain J. S. (2019). AAV-based Gene Therapies for the Muscular Dystrophies. Hum. Mol. Genet. 28 (R1), R102–R107. 10.1093/hmg/ddz128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del'Guidice T., Lepetit-Stoffaes J. P., Bordeleau L. J., Roberge J., Théberge V., Lauvaux C., et al. (2018). Membrane Permeabilizing Amphiphilic Peptide Delivers Recombinant Transcription Factor and CRISPR-Cas9/Cpf1 Ribonucleoproteins in Hard-To-Modify Cells. PLoS One 13 (4), e0195558. 10.1371/journal.pone.0195558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirer G. S., Silva T. N., Jackson C. T., Thomas J. B., Ehrhardt D, W., Rhee S. Y., et al. (2021). Nanotechnology to advance CRISPR-Cas Genetic Engineering of Plants. Nat. Nanotechnol. 16 (3), 243–250. 10.1038/s41565-021-00854-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devkota S. (2018). The Road Less Traveled: Strategies to Enhance the Frequency of Homology-Directed Repair (HDR) for Increased Efficiency of CRISPR/Cas-mediated Transgenesis. BMB Rep. 51 (9), 437–443. 10.5483/bmbrep.2018.51.9.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo J. E., Deeconda A., Tsang S. H. (2017). Viral Vectors, Engineered Cells and the CRISPR Revolution. Adv. Exp. Med. Biol. 1016, 3–27. 10.1007/978-3-319-63904-8_1 [DOI] [PubMed] [Google Scholar]
- Ding X., Yin K., Li Z., Lalla R. V., Ballesteros E., Sfeir M. M., et al. (2020). Ultrasensitive and Visual Detection of SARS-CoV-2 Using All-In-One Dual CRISPR-Cas12a Assay. Nat. Commun. 11 (1), 4711. 10.1038/s41467-020-18575-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiTommaso T., Cole J. M., Cassereau L., Buggé J. A., Hanson J. L. S., Bridgen D. T., et al. (2018). Cell Engineering with Microfluidic Squeezing Preserves Functionality of Primary Immune Cells In Vivo . Proc. Natl. Acad. Sci. USA 115 (46), E10907–E10914. 10.1073/pnas.1809671115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit S., Singh S. R., Yilma A. N., Agee R. D., 2nd, Taha M., Dennis V. A. (2014). Poly(lactic Acid)-Poly(ethylene Glycol) Nanoparticles Provide Sustained Delivery of a Chlamydia trachomatis Recombinant MOMP Peptide and Potentiate Systemic Adaptive Immune Responses in Mice. Nanomedicine: Nanotechnology, Biol. Med. 10 (6), 1311–1321. 10.1016/j.nano.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J. G., Fusi N., Sullender M., Hegde M., Vaimberg E. W., Donovan K. F., et al. (2016). Optimized sgRNA Design to Maximize Activity and Minimize Off-Target Effects of CRISPR-Cas9. Nat. Biotechnol. 34 (2), 184–191. 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi V., Manickam C., Binjawadagi B., Joyappa D., Renukaradhya G. J. (2012). Biodegradable Nanoparticle-Entrapped Vaccine Induces Cross-Protective Immune Response against a Virulent Heterologous Respiratory Viral Infection in Pigs. PLoS One 7 (12), e51794. 10.1371/journal.pone.0051794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykman L. A., Khlebtsov N. G. (2017). Immunological Properties of Gold Nanoparticles. Chem. Sci. 8 (3), 1719–1735. 10.1039/c6sc03631g [DOI] [PMC free article] [PubMed] [Google Scholar]
- England W. E., Whitaker R. J. (2013). Evolutionary Causes and Consequences of Diversified CRISPR Immune Profiles in Natural Populations. Biochem. Soc. Trans. 41 (6), 1431–1436. 10.1042/bst20130243 [DOI] [PubMed] [Google Scholar]
- Ferrari S., Jacob A., Beretta S., Unali G., Albano L., Vavassori V., et al. (2020). Efficient Gene Editing of Human Long-Term Hematopoietic Stem Cells Validated by Clonal Tracking. Nat. Biotechnol. 38 (11), 1298–1308. 10.1038/s41587-020-0551-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.-W., Dai X.-Y., Wang W.-T., Yang Z.-X., Zhao J.-J., Zhang J.-P., et al. (2021). Dynamics and Competition of CRISPR-Cas9 Ribonucleoproteins and AAV Donor-Mediated NHEJ, MMEJ and HDR Editing. Nucleic Acids Res. 49 (2), 969–985. 10.1093/nar/gkaa1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Sander J. D., Reyon D., Cascio V. M., Joung J. K. (2014). Improving CRISPR-Cas Nuclease Specificity Using Truncated Guide RNAs. Nat. Biotechnol. 32 (3), 279–284. 10.1038/nbt.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuenmayor J., Gòdia F., Cervera L. (2017). Production of Virus-like Particles for Vaccines. New Biotechnol. 39 (Pt B), 174–180. 10.1016/j.nbt.2017.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagat M., Zielińska W., Grzanka A. (2017). Cell-penetrating Peptides and Their Utility in Genome Function Modifications (Review). Int. J. Mol. Med. 40 (6), 1615–1623. 10.3892/ijmm.2017.3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G., Barrangou R., Horvath P., Siksnys V. (2012). Cas9-crRNA Ribonucleoprotein Complex Mediates Specific DNA Cleavage for Adaptive Immunity in Bacteria. Proc. Natl. Acad. Sci. 109 (39), E2579–E2586. 10.1073/pnas.1208507109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee P., Lung M. S. Y., Okuzaki Y., Sasakawa N., Iguchi T., Makita Y., et al. (2020). Extracellular Nanovesicles for Packaging of CRISPR-Cas9 Protein and sgRNA to Induce Therapeutic Exon Skipping. Nat. Commun. 11 (1), 1334. 10.1038/s41467-020-14957-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezraoui H., Piganeau M., Renouf B., Renaud J.-B., Sallmyr A., Ruis B., et al. (2014). Chromosomal Translocations in Human Cells Are Generated by Canonical Nonhomologous End-Joining. Mol. Cel 55 (6), 829–842. 10.1016/j.molcel.2014.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P., Han G., De M., Kim C., Rotello V. (2008). Gold Nanoparticles in Delivery Applications☆. Adv. Drug Deliv. Rev. 60 (11), 1307–1315. 10.1016/j.addr.2008.03.016 [DOI] [PubMed] [Google Scholar]
- Gilbert L. A., Larson M. H., Morsut L., Liu Z., Brar G. A., Torres S. E., et al. (2013). CRISPR-mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 154 (2), 442–451. 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass Z., Lee M., Li Y., Xu Q. (2018). Engineering the Delivery System for CRISPR-Based Genome Editing. Trends Biotechnol. 36 (2), 173–185. 10.1016/j.tibtech.2017.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco G. E., Matsumoto Y., Brooks R. C., Lu Z., Lieber M. R., Tomkinson A. E. (2016). SCR7 Is Neither a Selective Nor a Potent Inhibitor of Human DNA Ligase IV. DNA Repair 43, 18–23. 10.1016/j.dnarep.2016.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J. C., Hu J. S. (2017). Editing Plants for Virus Resistance Using CRISPR-Cas. Acta Virol. 61 (2), 138–142. 10.4149/av_2017_02_02 [DOI] [PubMed] [Google Scholar]
- Gu X., Wang D., Xu Z., Wang J., Guo L., Chai R., et al. (2021). Prevention of Acquired Sensorineural Hearing Loss in Mice by In Vivo Htra2 Gene Editing. Genome Biol. 22 (1), 86. 10.1186/s13059-021-02311-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger J. P., Thompson D. B., Liu D. R. (2014). Fusion of Catalytically Inactive Cas9 to FokI Nuclease Improves the Specificity of Genome Modification. Nat. Biotechnol. 32 (6), 577–582. 10.1038/nbt.2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Guerrero A., Cosset F. L., Verhoeyen E. (2020). Lentiviral Vector Pseudotypes: Precious Tools to Improve Gene Modification of Hematopoietic Cells for Research and Gene Therapy. Viruses 12 (9), 1016. 10.3390/v12091016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hande K. R., Hagey A., Berlin J., Cai Y., Meek K., Kobayashi H., et al. (2006). The Pharmacokinetics and Safety of ABT-751, a Novel, Orally Bioavailable Sulfonamide Antimitotic Agent: Results of a Phase 1 Study. Clin. Cancer Res. 12 (9), 2834–2840. 10.1158/1078-0432.ccr-05-2159 [DOI] [PubMed] [Google Scholar]
- Harrington L. B., Doxzen K. W., Ma E., Liu J.-J., Knott G. J., Edraki A., et al. (2017). A Broad-Spectrum Inhibitor of CRISPR-Cas9. Cell 170 (6), 1224–1233. e15. 10.1016/j.cell.2017.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart S. L., Harrison P. T. (2017). Genetic Therapies for Cystic Fibrosis Lung Disease. Curr. Opin. Pharmacol. 34, 119–124. 10.1016/j.coph.2017.10.006 [DOI] [PubMed] [Google Scholar]
- Hasheminejad N., Khodaiyan F., Safari M. (2019). Improving the Antifungal Activity of Clove Essential Oil Encapsulated by Chitosan Nanoparticles. Food Chem. 275, 113–122. 10.1016/j.foodchem.2018.09.085 [DOI] [PubMed] [Google Scholar]
- Heckl D., Kowalczyk M. S., Yudovich D., Belizaire R., Puram R. V., McConkey M. E., et al. (2014). Generation of Mouse Models of Myeloid Malignancy with Combinatorial Genetic Lesions Using CRISPR-Cas9 Genome Editing. Nat. Biotechnol. 32 (9), 941–946. 10.1038/nbt.2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer F., Kerr G., Boutros M. (2014). E-CRISP: Fast CRISPR Target Site Identification. Nat. Methods 11 (2), 122–123. 10.1038/nmeth.2812 [DOI] [PubMed] [Google Scholar]
- Heussler G. E., Cady K. C., Koeppen K., Bhuju S., Stanton B. A., O'Toole G. A. (2015). Clustered Regularly Interspaced Short Palindromic Repeat-dependent, Biofilm-specific Death of Pseudomonas aeruginosa Mediated by Increased Expression of Phage-Related Genes. mBio 6 (3), e00129–15. 10.1128/mBio.00129-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton I. B., D'Ippolito A. M., Vockley C. M., Thakore P. I., Crawford G. E., Reddy T. E., et al. (2015). Epigenome Editing by a CRISPR-Cas9-Based Acetyltransferase Activates Genes from Promoters and Enhancers. Nat. Biotechnol. 33 (5), 510–517. 10.1038/nbt.3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M. D., Aschenbrenner S., Grosse S., Rapti K., Domenger C., Fakhiri J., et al. (2019). Cell-specific CRISPR-Cas9 Activation by microRNA-dependent Expression of Anti-CRISPR Proteins. Nucleic Acids Res. 47 (13), e75. 10.1093/nar/gkz271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlbeck M. A., Witkowsky L. B., Guglielmi B., Replogle J. M., Gilbert L. A., Villalta J. E., et al. (2016). Nucleosomes Impede Cas9 Access to DNA In Vivo and In Vitro . Elife 5, e12677. 10.7554/elife.12677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høyland-Kroghsbo N. M., Muñoz K. A., Bassler B. L. (2018). Temperature, by Controlling Growth Rate, Regulates CRISPR-Cas Activity in Pseudomonas aeruginosa . mBio 9 (6), e02184–18. 10.1128/mBio.02184-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C. Y. M., Uludağ H. (2012). A Simple and Rapid Nonviral Approach to Efficiently Transfect Primary Tissue-Derived Cells Using Polyethylenimine. Nat. Protoc. 7 (5), 935–945. 10.1038/nprot.2012.038 [DOI] [PubMed] [Google Scholar]
- Hsu P. D., Scott D. A., Weinstein J. A., Ran F. A., Konermann S., Agarwala V., et al. (2013). DNA Targeting Specificity of RNA-Guided Cas9 Nucleases. Nat. Biotechnol. 31 (9), 827–832. 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Shi Z., Guo X., Jiang B., Wang G., Luo D., et al. (2018). Ligase IV Inhibitor SCR7 Enhances Gene Editing Directed by CRISPR-Cas9 and ssODN in Human Cancer Cells. Cell Biosci 8, 12. 10.1186/s13578-018-0200-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D., Miller M., Ashok B., Jain S., Peppas N. A. (2020). CRISPR/Cas Systems to Overcome Challenges in Developing the Next Generation of T Cells for Cancer Therapy. Adv. Drug Deliv. Rev. 158, 17–35. 10.1016/j.addr.2020.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Q., Zhu J., Niu Y., Shi H., Gong Y., Li Y., et al. (2017). pH-Triggered Surface Charge-Switchable Polymer Micelles for the Co-delivery of Paclitaxel/disulfiram and Overcoming Multidrug Resistance in Cancer. Int. J. Nanomedicine 12, 8631–8647. 10.2147/ijn.s144452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janik E., Niemcewicz M., Ceremuga M., Krzowski L., Saluk-Bijak J., Bijak M. (2020). Various Aspects of a Gene Editing System-CRISPR-Cas9. Int. J. Mol. Sci. 21 (24), 9604. 10.3390/ijms21249604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett K. E., Lee C., De Giorgi M., Hurley A., Gillard B. K., Doerfler A. M., et al. (2018). Somatic Editing of Ldlr with Adeno-Associated Viral-CRISPR Is an Efficient Tool for Atherosclerosis Research. Arteriosclerosis, Thromb. Vasc. Biol. 38 (9), 1997–2006. 10.1161/atvbaha.118.311221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayavaradhan R., Pillis D. M., Goodman M., Zhang F., Zhang Y., Andreassen P. R., et al. (2019). CRISPR-Cas9 Fusion to Dominant-Negative 53BP1 Enhances HDR and Inhibits NHEJ Specifically at Cas9 Target Sites. Nat. Commun. 10 (1), 2866. 10.1038/s41467-019-10735-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X., Zhao H., Zhu H., Zhu K., Tang S.-Y., Lou C. (2020). CRISPRi/dCpf1-mediated Dynamic Metabolic Switch to Enhance Butenoic Acid Production in Escherichia coli . Appl. Microbiol. Biotechnol. 104 (12), 5385–5393. 10.1007/s00253-020-10610-2 [DOI] [PubMed] [Google Scholar]
- Jin J., Jia B., Yuan Y.-J. (2020). Yeast Chromosomal Engineering to Improve Industrially-Relevant Phenotypes. Curr. Opin. Biotechnol. 66, 165–170. 10.1016/j.copbio.2020.07.003 [DOI] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E. (2012). A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337 (6096), 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen A. K., Molenaar B., Versteeg D., Leitoguinho A. R., Demkes C., Spanjaard B., et al. (2017). Postnatal Cardiac Gene Editing Using CRISPR/Cas9 with AAV9-Mediated Delivery of Short Guide RNAs Results in Mosaic Gene Disruption. Circ. Res. 121 (10), 1168–1181. 10.1161/circresaha.116.310370 [DOI] [PubMed] [Google Scholar]
- Ju E., Li T., Ramos da Silva S., Gao S.-J. (2019). Gold Nanocluster-Mediated Efficient Delivery of Cas9 Protein through pH-Induced Assembly-Disassembly for Inactivation of Virus Oncogenes. ACS Appl. Mater. Inter. 11 (38), 34717–34724. 10.1021/acsami.9b12335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmann M. (2018). CRISPRi and CRISPRa Screens in Mammalian Cells for Precision Biology and Medicine. ACS Chem. Biol. 13 (2), 406–416. 10.1021/acschembio.7b00657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor A., McClements M. E., MacLaren R. E. (2020). CRISPR-Cas9 DNA Base-Editing and Prime-Editing. Int. J. Mol. Sci. 21 (17), 6240. 10.3390/ijms21176240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson J., Rhodes K. R., Green J. J., Tzeng S. Y. (2020). Poly(beta-amino Ester)s as Gene Delivery Vehicles: Challenges and Opportunities. Expert Opin. Drug Deliv. 17 (10), 1395–1410. 10.1080/17425247.2020.1796628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Choi J., Won K.-H. (2020a). A Stable DNA-free Screening System for CRISPR/RNPs-mediated Gene Editing in Hot and Sweet Cultivars of Capsicum Annuum. BMC Plant Biol. 20 (1), 449. 10.1186/s12870-020-02665-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Kim M.-S., Wee G., Lee C.-i., Kim H., Kim J.-S. (2013). Magnetic Separation and Antibiotics Selection Enable Enrichment of Cells with ZFN/TALEN-induced Mutations. PLoS One 8 (2), e56476. 10.1371/journal.pone.0056476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N., Kim H. K., Lee S., Seo J. H., Choi J. W., Park J., et al. (2020b). Prediction of the Sequence-specific Cleavage Activity of Cas9 Variants. Nat. Biotechnol. 38 (11), 1328–1336. 10.1038/s41587-020-0537-9 [DOI] [PubMed] [Google Scholar]
- Kim S., Kim D., Cho S. W., Kim J., Kim J.-S. (2014). Highly Efficient RNA-Guided Genome Editing in Human Cells via Delivery of Purified Cas9 Ribonucleoproteins. Genome Res. 24 (6), 1012–1019. 10.1101/gr.171322.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B. P., Pattanayak V., Prew M. S., Tsai S. Q., Nguyen N. T., Zheng Z., et al. (2016). High-fidelity CRISPR-Cas9 Nucleases with No Detectable Genome-wide Off-Target Effects. Nature 529 (7587), 490–495. 10.1038/nature16526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Tsukiyama T., Nakaya M., Kageyama S., Tomita K., Murai R., et al. (2019). Generation of an OCT3/4 Reporter Cynomolgus Monkey ES Cell Line Using CRISPR/Cas9. Stem Cel Res. 37, 101439. 10.1016/j.scr.2019.101439 [DOI] [PubMed] [Google Scholar]
- Koblan L. W., Doman J. L., Wilson C., Levy J. M., Tay T., Newby G. A., et al. (2018). Improving Cytidine and Adenine Base Editors by Expression Optimization and Ancestral Reconstruction. Nat. Biotechnol. 36 (9), 843–846. 10.1038/nbt.4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotterman M. A., Chalberg T. W., Schaffer D. V. (2015). Viral Vectors for Gene Therapy: Translational and Clinical Outlook. Annu. Rev. Biomed. Eng. 17, 63–89. 10.1146/annurev-bioeng-071813-104938 [DOI] [PubMed] [Google Scholar]
- Kulcsár P. I., Tálas A., Huszár K., Ligeti Z., Tóth E., Weinhardt N., et al. (2017). Crossing Enhanced and High Fidelity SpCas9 Nucleases to Optimize Specificity and Cleavage. Genome Biol. 18 (1), 190. 10.1186/s13059-017-1318-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni J. A., Witzigmann D., Chen S., Cullis P. R., van der Meel R. (2019). Lipid Nanoparticle Technology for Clinical Translation of siRNA Therapeutics. Acc. Chem. Res. 52 (9), 2435–2444. 10.1021/acs.accounts.9b00368 [DOI] [PubMed] [Google Scholar]
- Kumita W., Sato K., Suzuki Y., Kurotaki Y., Harada T., Zhou Y., et al. (2019). Efficient Generation of Knock-in/Knock-Out Marmoset Embryo via CRISPR/Cas9 Gene Editing. Sci. Rep. 9 (1), 12719. 10.1038/s41598-019-49110-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson M. H., Gilbert L. A., Wang X., Lim W. A., Weissman J. S., Qi L. S. (2013). CRISPR Interference (CRISPRi) for Sequence-specific Control of Gene Expression. Nat. Protoc. 8 (11), 2180–2196. 10.1038/nprot.2013.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc C., Zhang F., Mendez J., Lozano Y., Chatpar K., Irish V. F., et al. (2018). Increased Efficiency of Targeted Mutagenesis by CRISPR/Cas9 in Plants Using Heat Stress. Plant J. 93 (2), 377–386. 10.1111/tpj.13782 [DOI] [PubMed] [Google Scholar]
- LeCher J. C., Nowak S. J., McMurry J. L. (2017). Breaking in and Busting Out: Cell-Penetrating Peptides and the Endosomal Escape Problem. Biomol. Concepts 8 (3-4), 131–141. 10.1515/bmc-2017-0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. K., Jeong E., Lee J., Jung M., Shin E., Kim Y.-h., et al. (2018). Directed Evolution of CRISPR-Cas9 to Increase its Specificity. Nat. Commun. 9 (1), 3048. 10.1038/s41467-018-05477-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Kim Y.-Y., Ahn H. J. (2021). Systemic Delivery of CRISPR/Cas9 to Hepatic Tumors for Cancer Treatment Using Altered Tropism of Lentiviral Vector. Biomaterials 272, 120793. 10.1016/j.biomaterials.2021.120793 [DOI] [PubMed] [Google Scholar]
- Legut M., Daniloski Z., Xue X., McKenzie D., Guo X., Wessels H.-H., et al. (2020). High-Throughput Screens of PAM-Flexible Cas9 Variants for Gene Knockout and Transcriptional Modulation. Cel Rep. 30 (9), 2859–2868. 10.1016/j.celrep.2020.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Kasinski A. L. (2020). In Vivo Cancer-Based Functional Genomics. Trends Cancer 6 (12), 1002–1017. 10.1016/j.trecan.2020.07.004 [DOI] [PubMed] [Google Scholar]
- Li G., Liu D., Zhang X., Quan R., Zhong C., Mo J., et al. (2018a). Suppressing Ku70/Ku80 Expression Elevates Homology-Directed Repair Efficiency in Primary Fibroblasts. Int. J. Biochem. Cel Biol. 99, 154–160. 10.1016/j.biocel.2018.04.011 [DOI] [PubMed] [Google Scholar]
- Li G., Zhang X., Zhong C., Mo J., Quan R., Yang J., et al. (2017). Small Molecules Enhance CRISPR/Cas9-mediated Homology-Directed Genome Editing in Primary Cells. Sci. Rep. 7 (1), 8943. 10.1038/s41598-017-09306-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., He Z.-Y., Wei X.-W., Gao G.-P., Wei Y.-Q. (2015). Challenges in CRISPR/CAS9 Delivery: Potential Roles of Nonviral Vectors. Hum. Gene Ther. 26 (7), 452–462. 10.1089/hum.2015.069 [DOI] [PubMed] [Google Scholar]
- Li L., Hu S., Chen X. (2018b). Non-viral Delivery Systems for CRISPR/Cas9-based Genome Editing: Challenges and Opportunities. Biomaterials 171, 207–218. 10.1016/j.biomaterials.2018.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Bolinger J., Yu Y., Glass Z., Shi N., Yang L., et al. (2019). Intracellular Delivery and Biodistribution Study of CRISPR/Cas9 Ribonucleoprotein Loaded Bioreducible Lipidoid Nanoparticles. Biomater. Sci. 7 (2), 596–606. 10.1039/c8bm00637g [DOI] [PubMed] [Google Scholar]
- Li Y., Yang T., Yu Y., Shi N., Yang L., Glass Z., et al. (2018c). Combinatorial Library of Chalcogen-Containing Lipidoids for Intracellular Delivery of Genome-Editing Proteins. Biomaterials 178, 652–662. 10.1016/j.biomaterials.2018.03.011 [DOI] [PubMed] [Google Scholar]
- Liang X., Potter J., Kumar S., Ravinder N., Chesnut J. D. (2017). Enhanced CRISPR/Cas9-mediated Precise Genome Editing by Improved Design and Delivery of gRNA, Cas9 Nuclease, and Donor DNA. J. Biotechnol. 241, 136–146. 10.1016/j.jbiotec.2016.11.011 [DOI] [PubMed] [Google Scholar]
- Liesche C., Venkatraman L., Aschenbrenner S., Grosse S., Grimm D., Eils R., et al. (2016). Death Receptor-Based Enrichment of Cas9-Expressing Cells. BMC Biotechnol. 16, 17. 10.1186/s12896-016-0250-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D., Sreekanth V., Cox K. J., Law B. K., Wagner B. K., Karp J. M., et al. (2020). Engineering Designer Beta Cells with a CRISPR-Cas9 Conjugation Platform. Nat. Commun. 11 (1), 4043. 10.1038/s41467-020-17725-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Staahl B. T., Alla R. K., Doudna J. A. (2014a). Enhanced Homology-Directed Human Genome Engineering by Controlled Timing of CRISPR/Cas9 Delivery. Elife 3, e04766. 10.7554/eLife.04766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Cradick T. J., Brown M. T., Deshmukh H., Ranjan P., Sarode N., et al. (2014b). CRISPR/Cas9 Systems Have Off-Target Activity with Insertions or Deletions between Target DNA and Guide RNA Sequences. Nucleic Acids Res. 42 (11), 7473–7485. 10.1093/nar/gku402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A., Huang X., He W., Xue F., Yang Y., Liu J., et al. (2021a). pHmScarlet Is a pH-Sensitive Red Fluorescent Protein to Monitor Exocytosis Docking and Fusion Steps. Nat. Commun. 12 (1), 1413. 10.1038/s41467-021-21666-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Zhang Y., Zhang T. (2020a). Computational Approaches for Effective CRISPR Guide RNA Design and Evaluation. Comput. Struct. Biotechnol. J. 18, 35–44. 10.1016/j.csbj.2019.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Ding Y., Zhou Y., Jin W., Xie K., Chen L.-L. (2017a). CRISPR-P 2.0: An Improved CRISPR-Cas9 Tool for Genome Editing in Plants. Mol. Plant 10 (3), 530–532. 10.1016/j.molp.2017.01.003 [DOI] [PubMed] [Google Scholar]
- Liu J., Gaj T., Patterson J. T., Sirk S. J., Barbas III C. F., 3rd (2014). Cell-penetrating Peptide-Mediated Delivery of TALEN Proteins via Bioconjugation for Genome Engineering. PLoS One 9 (1), e85755. 10.1371/journal.pone.0085755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Yin M., Wang M., Wang Y. (2019). Phage AcrIIA2 DNA Mimicry: Structural Basis of the CRISPR and Anti-CRISPR Arms Race. Mol. Cel 73 (3), 611–620. e3. 10.1016/j.molcel.2018.11.011 [DOI] [PubMed] [Google Scholar]
- Liu M., Rehman S., Tang X., Gu K., Fan Q., Chen D., et al. (2018a). Methodologies for Improving HDR Efficiency. Front. Genet. 9, 691. 10.3389/fgene.2018.00691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P.-F., Zhao K.-R., Liu Z.-J., Wang L., Ye S.-Y., Liang G.-X. (2021b). Cas12a-based Electrochemiluminescence Biosensor for Target Amplification-free DNA Detection. Biosens. Bioelectron. 176, 112954. 10.1016/j.bios.2020.112954 [DOI] [PubMed] [Google Scholar]
- Liu Q., Chen F., Hou L., Shen L., Zhang X., Wang D., et al. (2018b). Nanocarrier-Mediated Chemo-Immunotherapy Arrested Cancer Progression and Induced Tumor Dormancy in Desmoplastic Melanoma. ACS Nano 12 (8), 7812–7825. 10.1021/acsnano.8b01890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Gao R., Li J., Lin L., Zhao J., Sun W., et al. (2017b). Development of a Genome-Editing CRISPR/Cas9 System in Thermophilic Fungal Myceliophthora Species and its Application to Hyper-Cellulase Production Strain Engineering. Biotechnol. Biofuels 10, 1. 10.1186/s13068-016-0693-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Dong Y., Wang J., Li L., Zhong Z., Li Y.-p., et al. (2017c). VSV-G Viral Envelope Glycoprotein Prepared from Pichia pastoris Enhances Transfection of DNA into Animal Cells. J. Microbiol. Biotechnol. 27 (6), 1098–1105. 10.4014/jmb.1611.11082 [DOI] [PubMed] [Google Scholar]
- Liu X., Qiu S., Mei L., Jing H., Lin X., Wang Q. (2020b). A High-Resolution Melting Analysis with an Unlabeled Probe for CRISPR/Cas9-Induced ZBED6 Knockout Pigs Detection. J. AOAC Int. 104, 541–545. 10.1093/jaoacint/qsaa161 [DOI] [PubMed] [Google Scholar]
- Liu X. S., Wu H., Krzisch M., Wu X., Graef J., Muffat J., et al. (2018c). Rescue of Fragile X Syndrome Neurons by DNA Methylation Editing of the FMR1 Gene. Cell 172 (5), 979–992. 10.1016/j.cell.2018.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok B. H., Powell S. N. (2012). Molecular Pathways: Understanding the Role of Rad52 in Homologous Recombination for Therapeutic Advancement. Clin. Cancer Res. 18 (23), 6400–6406. 10.1158/1078-0432.ccr-11-3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomova A., Clark D. N., Campo‐Fernandez B., Flores‐Bjurström C., Kaufman M. L., Fitz‐Gibbon S., et al. (2019). Improving Gene Editing Outcomes in Human Hematopoietic Stem and Progenitor Cells by Temporal Control of DNA Repair. Stem Cells 37 (2), 284–294. 10.1002/stem.2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovendahl K. N., Hayward A. N., Gordon W. R. (2017). Sequence-Directed Covalent Protein-DNA Linkages in a Single Step Using HUH-Tags. J. Am. Chem. Soc. 139 (20), 7030–7035. 10.1021/jacs.7b02572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Tian Y., Shen R., Yao Q., Wang M., Chen M., et al. (2020). Targeted, Efficient Sequence Insertion and Replacement in rice. Nat. Biotechnol. 38 (12), 1402–1407. 10.1038/s41587-020-0581-5 [DOI] [PubMed] [Google Scholar]
- Lv J., He B., Yu J., Wang Y., Wang C., Zhang S., et al. (2018). Fluoropolymers for Intracellular and In Vivo Protein Delivery. Biomaterials 182, 167–175. 10.1016/j.biomaterials.2018.08.023 [DOI] [PubMed] [Google Scholar]
- Ma L., Ruan J., Song J., Wen L., Yang D., Zhao J., et al. (2020). MiCas9 Increases Large Size Gene Knock-In Rates and Reduces Undesirable On-Target and Off-Target Indel Edits. Nat. Commun. 11 (1), 6082. 10.1038/s41467-020-19842-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M., Zhuang F., Hu X., Wang B., Wen X.-Z., Ji J.-F., et al. (2017a). Efficient Generation of Mice Carrying Homozygous Double-Floxp Alleles Using the Cas9-Avidin/Biotin-Donor DNA System. Cell Res 27 (4), 578–581. 10.1038/cr.2017.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Hartmann R., Jimenez de Aberasturi D., Yang F., Soenen S. J. H., Manshian B. B., et al. (2017b). Colloidal Gold Nanoparticles Induce Changes in Cellular and Subcellular Morphology. ACS Nano 11 (8), 7807–7820. 10.1021/acsnano.7b01760 [DOI] [PubMed] [Google Scholar]
- Makarova K. S., Haft D. H., Barrangou R., Brouns S. J. J., Charpentier E., Horvath P., et al. (2011). Evolution and Classification of the CRISPR-Cas Systems. Nat. Rev. Microbiol. 9 (6), 467–477. 10.1038/nrmicro2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K. S., Wolf Y. I., Iranzo J., Shmakov S. A., Alkhnbashi O. S., Brouns S. J. J., et al. (2020). Evolutionary Classification of CRISPR-Cas Systems: a Burst of Class 2 and Derived Variants. Nat. Rev. Microbiol. 18 (2), 67–83. 10.1038/s41579-019-0299-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Esvelt K. M., Church G. M. (2013). Cas9 as a Versatile Tool for Engineering Biology. Nat. Methods 10 (10), 957–963. 10.1038/nmeth.2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone L. M., Birkholz N., Fineran P. C. (2020). Conquering CRISPR: How Phages Overcome Bacterial Adaptive Immunity. Curr. Opin. Biotechnol. 68, 30–36. 10.1016/j.copbio.2020.09.008 [DOI] [PubMed] [Google Scholar]
- Mangeot P. E., Risson V., Fusil F., Marnef A., Laurent E., Blin J., et al. (2019). Genome Editing in Primary Cells and In Vivo Using Viral-Derived Nanoblades Loaded with Cas9-sgRNA Ribonucleoproteins. Nat. Commun. 10 (1), 45. 10.1038/s41467-018-07845-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manghwar H., Li B., Ding X., Hussain A., Lindsey K., Zhang X., et al. (2020). CRISPR/Cas Systems in Genome Editing: Methodologies and Tools for sgRNA Design, Off‐Target Evaluation, and Strategies to Mitigate Off‐Target Effects. Adv. Sci. 7 (6), 1902312. 10.1002/advs.201902312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunath M., Choudhary B., Raghavan S. C. (2021). SCR7, a Potent Cancer Therapeutic Agent and a Biochemical Inhibitor of Nonhomologous DNA End‐joining. Cancer Rep. 4, e1341. 10.1002/cnr2.1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., Dougan S. K., Truttmann M. C., Bilate A. M., Ingram J. R., Ploegh H. L. (2015). Increasing the Efficiency of Precise Genome Editing with CRISPR-Cas9 by Inhibition of Nonhomologous End Joining. Nat. Biotechnol. 33 (5), 538–542. 10.1038/nbt.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashel T. V., Tarakanchikova Y. V., Muslimov A. R., Zyuzin M. V., Timin A. S., Lepik K. V., et al. (2020). Overcoming the Delivery Problem for Therapeutic Genome Editing: Current Status and Perspective of Non-viral Methods. Biomaterials 258, 120282. 10.1016/j.biomaterials.2020.120282 [DOI] [PubMed] [Google Scholar]
- Mateescu B., Kowal E. J. K., van Balkom B. W. M., Bartel S., Bhattacharyya S. N., Buzás E. I., et al. (2017). Obstacles and Opportunities in the Functional Analysis of Extracellular Vesicle RNA - an ISEV Position Paper. J. Extracellular Vesicles 6 (1), 1286095. 10.1080/20013078.2017.1286095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Gomez P. A., Kent T., Deng S. K., McDevitt S., Kashkina E., Hoang T. M., et al. (2017). The Helicase Domain of Polθ Counteracts RPA to Promote Alt-NHEJ. Nat. Struct. Mol. Biol. 24 (12), 1116–1123. 10.1038/nsmb.3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu M., Martin-Jaular L., Lavieu G., Théry C. (2019). Specificities of Secretion and Uptake of Exosomes and Other Extracellular Vesicles for Cell-To-Cell Communication. Nat. Cel Biol 21 (1), 9–17. 10.1038/s41556-018-0250-9 [DOI] [PubMed] [Google Scholar]
- Mátrai J., Chuah M. K., VandenDriessche T. (2010). Recent Advances in Lentiviral Vector Development and Applications. Mol. Ther. 18 (3), 477–490. 10.1038/mt.2009.319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z., Gao Z., Chen R., Yu X., Su Z., Wei G. (2018). Surface-bioengineered Gold Nanoparticles for Biomedical Applications. Cmc 25 (16), 1920–1944. 10.2174/0929867325666180117111404 [DOI] [PubMed] [Google Scholar]
- Minkenberg B., Zhang J., Xie K., Yang Y. (2019). CRISPR ‐ PLANT V2: an Online Resource for Highly Specific Guide RNA Spacers Based on Improved Off‐target Analysis. Plant Biotechnol. J. 17 (1), 5–8. 10.1111/pbi.13025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura H., Quadros R. M., Gurumurthy C. B., Ohtsuka M. (2018). Easi-CRISPR for Creating Knock-In and Conditional Knockout Mouse Models Using Long ssDNA Donors. Nat. Protoc. 13 (1), 195–215. 10.1038/nprot.2017.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagna C., Petris G., Casini A., Maule G., Franceschini G. M., Zanella I., et al. (2018). VSV-G-Enveloped Vesicles for Traceless Delivery of CRISPR-Cas9. Mol. Ther. - Nucleic Acids 12, 453–462. 10.1016/j.omtn.2018.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarity B. S., Rahrmann E. P., Beckmann D. A., Conboy C. B., Watson A. L., Carlson D. F., et al. (2014). Simple and Efficient Methods for Enrichment and Isolation of Endonuclease Modified Cells. PLoS One 9 (5), e96114. 10.1371/journal.pone.0096114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulsant P., Gatignol A., Dalens M., Tiraby G. r. (1988). Phleomycin Resistance as a Dominant Selectable Marker in CHO Cells. Somat Cel Mol Genet 14 (3), 243–252. 10.1007/bf01534585 [DOI] [PubMed] [Google Scholar]
- Nishimasu S., Shi X., Ishiguro S., Gao L., Hirano S., Okazaki S., et al. (2018). Engineered CRISPR-Cas9 Nuclease with Expanded Targeting Space. Science 361, 1259–1262. 10.1126/science.aas9129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihongaki Y., Kawano F., Nakajima T., Sato M. (2015). Photoactivatable CRISPR-Cas9 for Optogenetic Genome Editing. Nat. Biotechnol. 33 (7), 755–760. 10.1038/nbt.3245 [DOI] [PubMed] [Google Scholar]
- Noll L. W., Chall R., Shridhar P. B., Liu X., Bai J., Delannoy S., et al. (2018). Validation and Application of a Real-Time PCR Assay Based on the CRISPR Array for Serotype-specific Detection and Quantification of Enterohemorrhagic Escherichia coli O157:H7 in Cattle Feces†. J. Food Prot. 81 (7), 1157–1164. 10.4315/0362-028x.jfp-18-049 [DOI] [PubMed] [Google Scholar]
- Oh J., Fung E., Price M. N., Dehal P. S., Davis R. W., Giaever G., et al. (2010). A Universal TagModule Collection for Parallel Genetic Analysis of Microorganisms. Nucleic Acids Res. 38 (14), e146. 10.1093/nar/gkq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omichi R., Shibata S. B., Morton C. C., Smith R. J. H. (2019). Gene Therapy for Hearing Loss. Hum. Mol. Genet. 28 (R1), R65–R79. 10.1093/hmg/ddz129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P., Zhang N., Curtis B. R., Newman P. J., Denomme G. A. (2021). Generation of 'designer Erythroblasts' Lacking One or More Blood Group Systems from CRISPR/Cas9 Gene‐edited Human‐induced Pluripotent Stem Cells. J. Cel Mol Med 25, 9340–9349. 10.1111/jcmm.16872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S., Boulton S. J. (2014). Double-strand Break Repair: 53BP1 Comes into Focus. Nat. Rev. Mol. Cel Biol 15 (1), 7–18. 10.1038/nrm3719 [DOI] [PubMed] [Google Scholar]
- Pascucci L., Coccè V., Bonomi A., Ami D., Ceccarelli P., Ciusani E., et al. (2014). Paclitaxel Is Incorporated by Mesenchymal Stromal Cells and Released in Exosomes that Inhibit In Vitro Tumor Growth: a New Approach for Drug Delivery. J. Controlled Release 192, 262–270. 10.1016/j.jconrel.2014.07.042 [DOI] [PubMed] [Google Scholar]
- Pattanayak V., Lin S., Guilinger J. P., Ma E., Doudna J. A., Liu D. R. (2013). High-throughput Profiling of Off-Target DNA Cleavage Reveals RNA-Programmed Cas9 Nuclease Specificity. Nat. Biotechnol. 31 (9), 839–843. 10.1038/nbt.2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen B. S., Mandal P. K., Frock R. L., Boyraz B., Yadav R., Upadhyayula S., et al. (2017). Ectopic Expression of RAD52 and dn53BP1 Improves Homology-Directed Repair during CRISPR-Cas9 Genome Editing. Nat. Biomed. Eng. 1 (11), 878–888. 10.1038/s41551-017-0145-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelczak K. S., Gavande N. S., VanderVere-Carozza P. S., Turchi J. J. (2018). Modulating DNA Repair Pathways to Improve Precision Genome Engineering. ACS Chem. Biol. 13 (2), 389–396. 10.1021/acschembio.7b00777 [DOI] [PubMed] [Google Scholar]
- Pena S. A., Iyengar R., Eshraghi R. S., Bencie N., Mittal J., Aljohani A., et al. (2020). Gene Therapy for Neurological Disorders: Challenges and Recent Advancements. J. Drug Target. 28 (2), 111–128. 10.1080/1061186x.2019.1630415 [DOI] [PubMed] [Google Scholar]
- Peng H., Le C., Wu J., Li X.-F., Zhang H., Le X. C. (2020). A Genome-Editing Nanomachine Constructed with a Clustered Regularly Interspaced Short Palindromic Repeats System and Activated by Near-Infrared Illumination. ACS Nano 14 (3), 2817–2826. 10.1021/acsnano.9b05276 [DOI] [PubMed] [Google Scholar]
- Platt R. J., Chen S., Zhou Y., Yim M. J., Swiech L., Kempton H. R., et al. (2014). CRISPR-Cas9 Knockin Mice for Genome Editing and Cancer Modeling. Cell 159 (2), 440–455. 10.1016/j.cell.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priya S. S., Rekha M. R., Sharma C. P. (2014). Pullulan-protamine as Efficient Haemocompatible Gene Delivery Vector: Synthesis and In Vitro Characterization. Carbohydr. Polym. 102, 207–215. 10.1016/j.carbpol.2013.11.024 [DOI] [PubMed] [Google Scholar]
- Rabu C., Rangan L., Florenceau L., Fortun A., Charpentier M., Dupré E., et al. (2019). Cancer Vaccines: Designing Artificial Synthetic Long Peptides to Improve Presentation of Class I and Class II T Cell Epitopes by Dendritic Cells. Oncoimmunology 8 (4), e1560919. 10.1080/2162402x.2018.1560919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna S., Cho S. W., Kim S., Song M., Gopalappa R., Kim J.-S., et al. (2014a). Surrogate Reporter-Based Enrichment of Cells Containing RNA-Guided Cas9 Nuclease-Induced Mutations. Nat. Commun. 5, 3378. 10.1038/ncomms4378 [DOI] [PubMed] [Google Scholar]
- Ramakrishna S., Kwaku Dad A.-B., Beloor J., Gopalappa R., Lee S.-K., Kim H. (2014b). Gene Disruption by Cell-Penetrating Peptide-Mediated Delivery of Cas9 Protein and Guide RNA. Genome Res. 24 (6), 1020–1027. 10.1101/gr.171264.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch S., He C., Dickinson B. C. (2018). Targeted m6A Reader Proteins to Study Epitranscriptomic Regulation of Single RNAs. J. Am. Chem. Soc. 140 (38), 11974–11981. 10.1021/jacs.8b05012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C., Xu K., Liu Z., Shen J., Han F., Chen Z., et al. (2015). Dual-reporter Surrogate Systems for Efficient Enrichment of Genetically Modified Cells. Cell. Mol. Life Sci. 72 (14), 2763–2772. 10.1007/s00018-015-1874-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C., Xu K., Segal D. J., Zhang Z. (2019). Strategies for the Enrichment and Selection of Genetically Modified Cells. Trends Biotechnol. 37 (1), 56–71. 10.1016/j.tibtech.2018.07.017 [DOI] [PubMed] [Google Scholar]
- Renaud J.-B., Boix C., Charpentier M., De Cian A., Cochennec J., Duvernois-Berthet E., et al. (2016). Improved Genome Editing Efficiency and Flexibility Using Modified Oligonucleotides with TALEN and CRISPR-Cas9 Nucleases. Cel Rep. 14 (9), 2263–2272. 10.1016/j.celrep.2016.02.018 [DOI] [PubMed] [Google Scholar]
- Richter M. F., Zhao K. T., Eton E., Lapinaite A., Newby G. A., Thuronyi B. W., et al. (2020). Phage-assisted Evolution of an Adenine Base Editor with Improved Cas Domain Compatibility and Activity. Nat. Biotechnol. 38 (7), 883–891. 10.1038/s41587-020-0453-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F., Barbeau M., Éthier S., Dostie J., Pelletier J. (2015). Pharmacological Inhibition of DNA-PK Stimulates Cas9-Mediated Genome Editing. Genome Med. 7, 93. 10.1186/s13073-015-0215-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan G.-X., Barry E., Yu D., Lukason M., Cheng S. H., Scaria A. (2017). CRISPR/Cas9-Mediated Genome Editing as a Therapeutic Approach for Leber Congenital Amaurosis 10. Mol. Ther. 25 (2), 331–341. 10.1016/j.ymthe.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust B. J., Becker P. S., Chandrasekaran D., Kubek S. P., Peterson C. W., Adair J. E., et al. (2020). Envelope-Specific Adaptive Immunity Following Transplantation of Hematopoietic Stem Cells Modified with VSV-G Lentivirus. Mol. Ther. - Methods Clin. Develop. 19, 438–446. 10.1016/j.omtm.2020.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J.-Y., Won E.-J., Lee H. A. R., Kim J. H., Hui E., Kim H. P., et al. (2020). Ultrasound-activated Particles as CRISPR/Cas9 Delivery System for Androgenic Alopecia Therapy. Biomaterials 232, 119736. 10.1016/j.biomaterials.2019.119736 [DOI] [PubMed] [Google Scholar]
- Saari H., Lázaro-Ibáñez E., Viitala T., Vuorimaa-Laukkanen E., Siljander P., Yliperttula M. (2015). Microvesicle- and Exosome-Mediated Drug Delivery Enhances the Cytotoxicity of Paclitaxel in Autologous Prostate Cancer Cells. J. Controlled Release 220 (Pt B), 727–737. 10.1016/j.jconrel.2015.09.031 [DOI] [PubMed] [Google Scholar]
- Sander J. D., Joung J. K. (2014). CRISPR-cas Systems for Editing, Regulating and Targeting Genomes. Nat. Biotechnol. 32 (4), 347–355. 10.1038/nbt.2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savic N., Ringnalda F. C., Lindsay H., Berk C., Bargsten K., Li Y., et al. (2018). Covalent Linkage of the DNA Repair Template to the CRISPR-Cas9 Nuclease Enhances Homology-Directed Repair. Elife 7, e33761. 10.7554/elife.33761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena-Esteves M., Gao G. (2018). Titration of Lentivirus Vectors. Cold Spring Harb Protoc. 2018 (4), pdb.prot095695. 10.1101/pdb.prot095695 [DOI] [PubMed] [Google Scholar]
- Shao S., Ren C., Liu Z., Bai Y., Chen Z., Wei Z., et al. (2017). Enhancing CRISPR/Cas9-mediated Homology-Directed Repair in Mammalian Cells by Expressing Saccharomyces cerevisiae Rad52. Int. J. Biochem. Cel Biol. 92, 43–52. 10.1016/j.biocel.2017.09.012 [DOI] [PubMed] [Google Scholar]
- Shen Z. Y., Xia G. L., Wu M. F., Ji L. Y., Li Y. J. (2016). The Effects of Percutaneous Ethanol Injection Followed by 20-kHz Ultrasound and Microbubbles on Rabbit Hepatic Tumors. J. Cancer Res. Clin. Oncol. 142 (2), 373–378. 10.1007/s00432-015-2034-y [DOI] [PubMed] [Google Scholar]
- Shi J., Ma Y., Zhu J., Chen Y., Sun Y., Yao Y., et al. (2018). A Review on Electroporation-Based Intracellular Delivery. Molecules 23 (11), 3044. 10.3390/molecules23113044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley J. L., de Jong Y. P., Terhorst C., Herzog R. W. (2020). Immune Responses to Viral Gene Therapy Vectors. Mol. Ther. 28 (3), 709–722. 10.1016/j.ymthe.2020.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker I. M., Gao L., Zetsche B., Scott D. A., Yan W. X., Zhang F. (2016). Rationally Engineered Cas9 Nucleases with Improved Specificity. Science 351 (6268), 84–88. 10.1126/science.aad5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søndergaard J. N., Geng K., Sommerauer C., Atanasoai I., Yin X., Kutter C. (2020). Successful Delivery of Large-Size CRISPR/Cas9 Vectors in Hard-To-Transfect Human Cells Using Small Plasmids. Commun. Biol. 3 (1), 319. 10.1038/s42003-020-1045-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F., Stieger K. (2017). Optimizing the DNA Donor Template for Homology-Directed Repair of Double-Strand Breaks. Mol. Ther. - Nucleic Acids 7, 53–60. 10.1016/j.omtn.2017.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Yang D., Xu J., Zhu T., Chen Y. E., Zhang J. (2016). RS-1 Enhances CRISPR/Cas9- and TALEN-Mediated Knock-In Efficiency. Nat. Commun. 7, 10548. 10.1038/ncomms10548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Liu C., Wang N., Huang H., He S., Gong C., et al. (2021). Delivery of CRISPR/Cas Systems for Cancer Gene Therapy and Immunotherapy. Adv. Drug Deliv. Rev. 168, 158–180. 10.1016/j.addr.2020.04.010 [DOI] [PubMed] [Google Scholar]
- Sottnik J. L., Vanderlinden L., Joshi M., Chauca-Diaz A., Owens C., Hansel D. E., et al. (2021). Androgen Receptor Regulates CD44 Expression in Bladder Cancer. Cancer Res. 81, 2833–2846. 10.1158/0008-5472.CAN-20-3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M., Nambiar M., Sharma S., Karki S. S., Goldsmith G., Hegde M., et al. (2012). An Inhibitor of Nonhomologous End-Joining Abrogates Double-Strand Break Repair and Impedes Cancer Progression. Cell 151 (7), 1474–1487. 10.1016/j.cell.2012.11.054 [DOI] [PubMed] [Google Scholar]
- Sunada S., Saito H., Zhang D., Xu Z., Miki Y. (2021). CDK1 Inhibitor Controls G2/M Phase Transition and Reverses DNA Damage Sensitivity. Biochem. Biophysical Res. Commun. 550, 56–61. 10.1016/j.bbrc.2021.02.117 [DOI] [PubMed] [Google Scholar]
- Suresh B., Ramakrishna S., Kim H. (2017). Cell-Penetrating Peptide-Mediated Delivery of Cas9 Protein and Guide RNA for Genome Editing. Methods Mol. Biol. 1507, 81–94. 10.1007/978-1-4939-6518-2_7 [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Onuma H., Sato R., Sato Y., Hashiba A., Maeki M., et al. (2021). Lipid Nanoparticles Loaded with Ribonucleoprotein-Oligonucleotide Complexes Synthesized Using a Microfluidic Device Exhibit Robust Genome Editing and Hepatitis B Virus Inhibition. J. Control. Release 330, 61–71. 10.1016/j.jconrel.2020.12.013 [DOI] [PubMed] [Google Scholar]
- Svitashev S., Schwartz C., Lenderts B., Young J. K., Mark Cigan A. (2016). Genome Editing in maize Directed by CRISPR-Cas9 Ribonucleoprotein Complexes. Nat. Commun. 7, 13274. 10.1038/ncomms13274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S. (2014). End Resection at Double-Strand Breaks: Mechanism and Regulation. Cold Spring Harb Perspect. Biol. 6 (8), a016436. 10.1101/cshperspect.a016436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taharabaru T., Yokoyama R., Higashi T., Mohammed A. F. A., Inoue M., Maeda Y., et al. (2020). Genome Editing in a Wide Area of the Brain Using Dendrimer-Based Ternary Polyplexes of Cas9 Ribonucleoprotein. ACS Appl. Mater. Inter. 12 (19), 21386–21397. 10.1021/acsami.9b21667 [DOI] [PubMed] [Google Scholar]
- Tang L., Zeng Y., Du H., Gong M., Peng J., Zhang B., et al. (2017). CRISPR/Cas9-mediated Gene Editing in Human Zygotes Using Cas9 Protein. Mol. Genet. Genomics 292 (3), 525–533. 10.1007/s00438-017-1299-z [DOI] [PubMed] [Google Scholar]
- Taniyama Y., Azuma J., Rakugi H., Morishita R. (2011). Plasmid DNA-Based Gene Transfer with Ultrasound and Microbubbles. Curr. Gene Ther. 11 (6), 485–490. 10.2174/156652311798192851 [DOI] [PubMed] [Google Scholar]
- Tao Y., Yi K., Hu H., Shao D., Li M. (2021). Coassembly of Nucleus-Targeting Gold Nanoclusters with CRISPR/Cas9 for Simultaneous Bioimaging and Therapeutic Genome Editing. J. Mater. Chem. B 9 (1), 94–100. 10.1039/d0tb01925a [DOI] [PubMed] [Google Scholar]
- Torre E. A., Arai E., Bayatpour S., Jiang C. L., Beck L. E., Emert B. L., et al. (2021). Genetic Screening for Single-Cell Variability Modulators Driving Therapy Resistance. Nat. Genet. 53 (1), 76–85. 10.1038/s41588-020-00749-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran N.-T., Bashir S., Li X., Rossius J., Chu V. T., Rajewsky K., et al. (2019). Enhancement of Precise Gene Editing by the Association of Cas9 with Homologous Recombination Factors. Front. Genet. 10, 365. 10.3389/fgene.2019.00365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov F. D., Miller J. C., Lee Y.-L., Beausejour C. M., Rock J. M., Augustus S., et al. (2005). Highly Efficient Endogenous Human Gene Correction Using Designed Zinc-finger Nucleases. Nature 435 (7042), 646–651. 10.1038/nature03556 [DOI] [PubMed] [Google Scholar]
- Vakulskas C. A., Dever D. P., Rettig G. R., Turk R., Jacobi A. M., Collingwood M. A., et al. (2018). A High-Fidelity Cas9 Mutant Delivered as a Ribonucleoprotein Complex Enables Efficient Gene Editing in Human Hematopoietic Stem and Progenitor Cells. Nat. Med. 24 (8), 1216–1224. 10.1038/s41591-018-0137-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez R. J., Howell B., Yvon A. M., Wadsworth P., Cassimeris L. (1997). Nanomolar Concentrations of Nocodazole Alter Microtubule Dynamic Instability In Vivo and In Vitro . Mol. Biol. Cel 8 (6), 973–985. 10.1091/mbc.8.6.973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L. T. (2006). Cell Cycle Synchronization at the G2/M Phase Border by Reversible Inhibition of CDK1. Cell Cycle 5 (22), 2555–2556. 10.4161/cc.5.22.3463 [DOI] [PubMed] [Google Scholar]
- Vermersch E., Jouve C., Hulot J.-S. (2020). CRISPR/Cas9 Gene-Editing Strategies in Cardiovascular Cells. Cardiovasc. Res. 116 (5), 894–907. 10.1093/cvr/cvz250 [DOI] [PubMed] [Google Scholar]
- Wang H., La Russa M., Qi L. S. (2016). CRISPR/Cas9 in Genome Editing and beyond. Annu. Rev. Biochem. 85, 227–264. 10.1146/annurev-biochem-060815-014607 [DOI] [PubMed] [Google Scholar]
- Wang P., Zhang L., Zheng W., Cong L., Guo Z., Xie Y., et al. (2018). Thermo-triggered Release of CRISPR-Cas9 System by Lipid-Encapsulated Gold Nanoparticles for Tumor Therapy. Angew. Chem. Int. Ed. 57 (6), 1491–1496. 10.1002/anie.201708689 [DOI] [PubMed] [Google Scholar]
- Wei Q., Croy B. A., Etches R. J. (2001). Selection of Genetically Modified Chicken Blastodermal Cells by Magnetic-Activated Cell Sorting. Poult. Sci. 80 (12), 1671–1678. 10.1093/ps/80.12.1671 [DOI] [PubMed] [Google Scholar]
- Wienert B., Nguyen D. N., Guenther A., Feng S. J., Locke M. N., Wyman S. K., et al. (2020). Timed Inhibition of CDC7 Increases CRISPR-Cas9 Mediated Templated Repair. Nat. Commun. 11 (1), 2109. 10.1038/s41467-020-15845-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbie D., Walther J., Mastrobattista E. (2019). Delivery Aspects of CRISPR/Cas for In Vivo Genome Editing. Acc. Chem. Res. 52 (6), 1555–1564. 10.1021/acs.accounts.9b00106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold W. S., Toth K. (2013). Adenovirus Vectors for Gene Therapy, Vaccination and Cancer Gene Therapy. Curr. Gene Ther. 13 (6), 421–433. 10.2174/1566523213666131125095046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Kriz A. J., Sharp P. A. (2014). Target Specificity of the CRISPR-Cas9 System. Quant Biol. 2 (2), 59–70. 10.1007/s40484-014-0030-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Yang H., Colosi P. (2010). Effect of Genome Size on AAV Vector Packaging. Mol. Ther. 18 (1), 80–86. 10.1038/mt.2009.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang G., Zhang X., An C., Cheng C., Wang H. (2017). Temperature Effect on CRISPR-Cas9 Mediated Genome Editing. J. Genet. Genomics 44 (4), 199–205. 10.1016/j.jgg.2017.03.004 [DOI] [PubMed] [Google Scholar]
- Xie X., Ma X., Zhu Q., Zeng D., Li G., Liu Y.-G. (2017). CRISPR-GE: A Convenient Software Toolkit for CRISPR-Based Genome Editing. Mol. Plant 10 (9), 1246–1249. 10.1016/j.molp.2017.06.004 [DOI] [PubMed] [Google Scholar]
- Xu C. L., Ruan M. Z. C., Mahajan V. B., Tsang S. H. (2019). Viral Delivery Systems for CRISPR. Viruses 11 (1). 10.3390/v11010028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Zhang Z., Zhao L., Qin Y., Cai H., Geng Z., et al. (2020a). Tropism-facilitated Delivery of CRISPR/Cas9 System with Chimeric Antigen Receptor-Extracellular Vesicles against B-Cell Malignancies. J. Controlled Release 326, 455–467. 10.1016/j.jconrel.2020.07.033 [DOI] [PubMed] [Google Scholar]
- Xu W. (2019). Microinjection and Micromanipulation: A Historical Perspective. Methods Mol. Biol. 1874, 1–16. 10.1007/978-1-4939-8831-0_1 [DOI] [PubMed] [Google Scholar]
- Xu X., Hulshoff M. S., Tan X., Zeisberg M., Zeisberg E. M. (2020b). CRISPR/Cas Derivatives as Novel Gene Modulating Tools: Possibilities and In Vivo Applications. Int. J. Mol. Sci. 21 (9), 3038. 10.3390/ijms21093038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Tan X., Tampe B., Wilhelmi T., Hulshoff M. S., Saito S., et al. (2018). High-fidelity CRISPR/Cas9- Based Gene-specific Hydroxymethylation Rescues Gene Expression and Attenuates Renal Fibrosis. Nat. Commun. 9 (1), 3509. 10.1038/s41467-018-05766-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Scavuzzo M. A., Chmielowiec J., Sharp R., Bajic A., Borowiak M. (2016). Enrichment of G2/M Cell Cycle Phase in Human Pluripotent Stem Cells Enhances HDR-Mediated Gene Repair with Customizable Endonucleases. Sci. Rep. 6, 21264. 10.1038/srep21264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Ren S., Yu S., Pan H., Li T., Ge S., et al. (2020). Methods Favoring Homology-Directed Repair Choice in Response to CRISPR/Cas9 Induced-Double Strand Breaks. Int. J. Mol. Sci. 21 (18), 6461. 10.3390/ijms21186461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Wang H., Shivalila C. S., Cheng A. W., Shi L., Jaenisch R. (2013). One-step Generation of Mice Carrying Reporter and Conditional Alleles by CRISPR/Cas-mediated Genome Engineering. Cell 154 (6), 1370–1379. 10.1016/j.cell.2013.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Shi G., Guo J., Wang C., He Y. (2018). Exosome-encapsulated Antibiotic against Intracellular Infections of Methicillin-Resistant Staphylococcus aureus . Int. J. Nanomedicine 13, 8095–8104. 10.2147/ijn.s179380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R., Liu D., Jia X., Zheng Y., Liu W., Xiao Y. (2018a). CRISPR-Cas9/Cas12a Biotechnology and Application in Bacteria. Synth. Syst. Biotechnol. 3 (3), 135–149. 10.1016/j.synbio.2018.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Lyu P., Yoo K., Yadav M. K., Singh R., Atala A., et al. (2021). Engineered Extracellular Vesicles as Versatile Ribonucleoprotein Delivery Vehicles for Efficient and Safe CRISPR Genome Editing. J. Extracell Vesicles 10 (5), e12076. 10.1002/jev2.12076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Zhang M., Wang X., Ying W., Hu X., Dai P., et al. (2018b). Tild-CRISPR Allows for Efficient and Precise Gene Knockin in Mouse and Human Cells. Develop. Cel 45 (4), 526–536. 10.1016/j.devcel.2018.04.021 [DOI] [PubMed] [Google Scholar]
- Yasuda H., Kim E., Reza A. M. M. T., Kim J.-H. (2016). A Highly Efficient Method for Enriching TALEN or CRISPR/Cas9-edited Mutant Cells. J. Genet. Genomics 43 (12), 705–708. 10.1016/j.jgg.2016.10.006 [DOI] [PubMed] [Google Scholar]
- Yiangou L., Grandy R. A., Morell C. M., Tomaz R. A., Osnato A., Kadiwala J., et al. (2019). Method to Synchronize Cell Cycle of Human Pluripotent Stem Cells without Affecting Their Fundamental Characteristics. Stem Cel Rep. 12 (1), 165–179. 10.1016/j.stemcr.2018.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin D., Ling S., Wang D., Dai Y., Jiang H., Zhou X., et al. (2021). Targeting Herpes Simplex Virus with CRISPR-Cas9 Cures Herpetic Stromal Keratitis in Mice. Nat. Biotechnol. 39, 567–577. 10.1038/s41587-020-00781-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Kanasty R. L., Eltoukhy A. A., Vegas A. J., Dorkin J. R., Anderson D. G. (2014). Non-viral Vectors for Gene-Based Therapy. Nat. Rev. Genet. 15 (8), 541–555. 10.1038/nrg3763 [DOI] [PubMed] [Google Scholar]
- Yin H., Kauffman K. J., Anderson D. G. (2017). Delivery Technologies for Genome Editing. Nat. Rev. Drug Discov. 16 (6), 387–399. 10.1038/nrd.2016.280 [DOI] [PubMed] [Google Scholar]
- Yin H., Song C.-Q., Suresh S., Kwan S.-Y., Wu Q., Walsh S., et al. (2018a). Partial DNA-Guided Cas9 Enables Genome Editing with Reduced Off-Target Activity. Nat. Chem. Biol. 14 (3), 311–316. 10.1038/nchembio.2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Hou S., Wang Q., Bao L., Liu D., Yue Y., et al. (2019a). Microenvironment-Responsive Delivery of the Cas9 RNA-Guided Endonuclease for Efficient Genome Editing. Bioconjug. Chem. 30 (3), 898–906. 10.1021/acs.bioconjchem.9b00022 [DOI] [PubMed] [Google Scholar]
- Yin J., Liu M., Liu Y., Wu J., Gan T., Zhang W., et al. (2019b). Optimizing Genome Editing Strategy by Primer-Extension-Mediated Sequencing. Cell Discov 5, 18. 10.1038/s41421-019-0088-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Wang Q., Hou S., Bao L., Yao W., Gao X. (2018b). Potent Protein Delivery into Mammalian Cells via a Supercharged Polypeptide. J. Am. Chem. Soc. 140 (49), 17234–17240. 10.1021/jacs.8b10299 [DOI] [PubMed] [Google Scholar]
- Yoon S., Wang P., Peng Q., Wang Y., Shung K. K. (2017). Acoustic-transfection for Genomic Manipulation of Single-Cells Using High Frequency Ultrasound. Sci. Rep. 7 (1), 5275. 10.1038/s41598-017-05722-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi K., Kunihiro Y., Kaneko T., Nagahora H., Voigt B., Mashimo T. (2016). ssODN-mediated Knock-In with CRISPR-Cas for Large Genomic Regions in Zygotes. Nat. Commun. 7, 10431. 10.1038/ncomms10431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi K., Oka Y., Miyasaka Y., Kotani Y., Yasumura M., Uno Y., et al. (2021). Combi-CRISPR: Combination of NHEJ and HDR Provides Efficient and Precise Plasmid-Based Knock-Ins in Mice and Rats. Hum. Genet. 140 (2), 277–287. 10.1007/s00439-020-02198-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue H., Zhou X., Cheng M., Xing D. (2018). Graphene Oxide-Mediated Cas9/sgRNA Delivery for Efficient Genome Editing. Nanoscale 10 (3), 1063–1071. 10.1039/c7nr07999k [DOI] [PubMed] [Google Scholar]
- Zhang B., Yin Y., Lai R. C., Tan S. S., Choo A. B. H., Lim S. K. (2014). Mesenchymal Stem Cells Secrete Immunologically Active Exosomes. Stem Cell Develop. 23 (11), 1233–1244. 10.1089/scd.2013.0479 [DOI] [PubMed] [Google Scholar]
- Zhang F., Song G., Tian Y. (2019a). Anti-CRISPRs: The Natural Inhibitors for CRISPR-Cas Systems. Anim. Model Exp Med 2 (2), 69–75. 10.1002/ame2.12069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Shen J., Li D., Cheng Y. (2021). Strategies in the Delivery of Cas9 Ribonucleoprotein for CRISPR/Cas9 Genome Editing. Theranostics 11 (2), 614–648. 10.7150/thno.47007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Showalter A. M. (2020). CRISPR/Cas9 Genome Editing Technology: A Valuable Tool for Understanding Plant Cell Wall Biosynthesis and Function. Front. Plant Sci. 11, 589517. 10.3389/fpls.2020.589517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Wan T., Chen Y., Chen Y., Sun H., Cao T., et al. (2019b). Cationic Polymer-Mediated CRISPR/Cas9 Plasmid Delivery for Genome Editing. Macromol Rapid Commun. 40 (5), e1800068. 10.1002/marc.201800068 [DOI] [PubMed] [Google Scholar]
- Zheng W., Li Q., Sun H., Ali M. W., Zhang H. (2019). Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated 9-mediated Mutagenesis of the Multiple Edematous Wings Gene Induces Muscle Weakness and Flightlessness in Bactrocera Dorsalis (Diptera: Tephritidae). Insect Mol. Biol. 28 (2), 222–234. 10.1111/imb.12540 [DOI] [PubMed] [Google Scholar]
- Zhi S., Chen Y., Wu G., Wen J., Wu J., Liu Q., et al. (2022). Dual-AAV Delivering Split Prime Editor System for In Vivo Genome Editing. Mol. Ther. 30 (1), 283–294. 10.1016/j.ymthe.2021.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Zhan H., Liao X., Fang L., Liu Y., Xie H., et al. (2019). A Revolutionary Tool: CRISPR Technology Plays an Important Role in Construction of Intelligentized Gene Circuits. Cell Prolif 52 (2), e12552. 10.1111/cpr.12552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S. J., Li S. W., Wang J. J., Liu Z. J., Yin G. B., Gong J. P., et al. (2012). High-intensity Focused Ultrasound Combined with Herpes Simplex Virus Thymidine Kinase Gene-Loaded Ultrasound-Targeted Microbubbles Improved the Survival of Rabbits with VX₂ Liver Tumor. J. Gene Med. 14 (9-10), 570–579. 10.1002/jgm.2668 [DOI] [PubMed] [Google Scholar]
- Zhuang J., Tan J., Wu C., Zhang J., Liu T., Fan C., et al. (2020). Extracellular Vesicles Engineered with Valency-Controlled DNA Nanostructures Deliver CRISPR/Cas9 System for Gene Therapy. Nucleic Acids Res. 48 (16), 8870–8882. 10.1093/nar/gkaa683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M., de Lange T. (2014). 53BP1: Pro Choice in DNA Repair. Trends Cel Biol. 24 (2), 108–117. 10.1016/j.tcb.2013.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchelli E., Pema M., Stornaiuolo A., Piovan C., Scavullo C., Giuliani E., et al. (2017). Codon Optimization Leads to Functional Impairment of RD114-TR Envelope Glycoprotein. Mol. Ther. - Methods Clin. Develop. 4, 102–114. 10.1016/j.omtm.2017.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo E., Cai Y.-J., Li K., Wei Y., Wang B.-A., Sun Y., et al. (2017). One-step Generation of Complete Gene Knockout Mice and Monkeys by CRISPR/Cas9-mediated Gene Editing with Multiple sgRNAs. Cel Res 27 (7), 933–945. 10.1038/cr.2017.81 [DOI] [PMC free article] [PubMed] [Google Scholar]