Supplemental Digital Content is available in the text.
Keywords: endothelial cells, gene ontology, mice, microRNA, transcriptome
Background:
Endothelial cells (ECs) play a critical role in angiogenesis and vascular remodeling. The heterogeneity of ECs has been reported at adult stages, yet it has not been fully investigated. This study aims to assess the transcriptional heterogeneity of developmental ECs at spatiotemporal level and to reveal the changes of embryonic ECs clustering when endothelium-enriched microRNA-126 (miR-126) was specifically knocked out.
Methods:
C57BL/6J mice embryos at day 14.5 were harvested and digested, followed by fluorescence-activated cell sorting to enrich ECs. Then, single-cell RNA sequencing was applied to enriched embryonic ECs. Tie2 (Tek receptor tyrosine kinase)-cre–mediated ECs-specific miR-126 knockout mice were constructed, and ECs from Tie2-cre–mediated ECs-specific miR-126 knockout embryos were subjected to single-cell RNA sequencing.
Results:
Embryonic ECs were clustered into 11 groups corresponding to anatomic characteristics. The vascular bed (arteries, capillaries, veins, lymphatics) exhibited transcriptomic similarity across the developmental stage. Embryonic ECs had higher proliferative potential than adult ECs. Integrating analysis showed that 3 ECs populations (hepatic, mesenchymal transition, and pulmonary ECs) were apparently disorganized after miR-126 being knocked out. Gene ontology analysis revealed that disrupted ECs were mainly related to hypoxia, glycometabolism, and vascular calcification. Additionally, in vivo experiment showed that Tie2-cre–mediated ECs-specific miR-126 knockout mice exhibited excessive intussusceptive angiogenesis; reductive glucose and pyruvate tolerance; and excessive accumulation of calcium. Agonist miR-126-3p agomir significantly rescued the phenotype of glucose metabolic dysfunction in Tie2-cre–mediated ECs-specific miR-126 knockout mice.
Conclusions:
The heterogeneity of ECs is established as early as the embryonic stage. The deficiency of miR-126 disrupts the differentiation and diversification of embryonic ECs, suggesting that miR-126 plays an essential role in the maintenance of ECs heterogeneity.
Highlights.
The heterogeneity of endothelial cells (ECs) is established as early as the embryonic stage.
Embryonic ECs (eECs) has spatiotemporal heterogeneity and higher proliferation potential than adult ECs.
During development, regular embryonic ECs clustering can be disrupted in ECs-specific microRNA-126 knockout mice. And the emerged abnormal embryonic ECs clusters eventually lead to disordered glycometabolism, excessive intussusceptive angiogenesis, and increased endothelial-to-mesenchymal transition.
MicroRNA-126-HIF1A (hypoxia-inducible factor 1-alpha)-GLUT1 (glucose transporter 1) pathway is involved in glycometabolism, and microRNA-126-3p agomir restored the abnormal blood glucose tolerance in microRNA-126–deficient mice.
See accompanying editorial on page 343
Vascular endothelial cells (ECs) are the inner layer of blood vessels, which directly respond to physical and chemical signals related to vascular tension, cell adhesion, antithrombosis, smooth muscle cell proliferation, and vascular wall inflammation.1 ECs in different tissues have heterogeneous phenotypes, presumably to satisfy different physical and chemical signals.2 Using fluorescence-activated cell sorting (FACS) with common marker genes PECAM1 (platelet endothelial cell adhesion molecule-1) and CDH5 (Cadherin 5), ECs can be enriched for further analysis and application.3
With the development of single-cell RNA sequencing (scRNA-seq) technology, the heterogeneity of ECs was further characterized at the level of transcriptomics and epigenomics.4–10 Recently, EC atlases of 11 adult mouse tissues explored the endothelial diversity between and within tissues, which provided a powerful discovery tool and resource value.10 Whether endothelial heterogeneity exists in embryonic stage and how early it exists need further investigation.
The formation of vasculature starts with vasculogenesis and is followed by angiogenesis. Vasculogenesis initiates the vascular system during embryonic development by the differentiation and migration of mesoderm-derived angioblasts or ECs.11 Angiogenesis expands the vascular system by sprouting or intussusception.12,13 Sprouting angiogenesis is characterized by sprouts composed of ECs. Intussusceptive angiogenesis (IA) involves formation of blood vessels by splitting process in which elements of interstitial tissues invade existing vessels, forming transvascular tissue pillars that expand.14,15
ECs exist in another special characteristic, namely, endothelial-to-mesenchymal transition (EndoMT). EndoMT, which parallels the epithelia-to-mesenchymal transition, describes a process in which ECs lose their endothelial specification gradually and gain features of mesenchymal cell.16 Mesenchyme transited ECs obtain characteristics of endothelial and mesenchymal cells simultaneously in both morphology and transcriptome. Although accounting for a minority in normal conditions, EndoMT is essential in embryonic development, such as the formation of cardiac valve.17 In addition, EndoMT participates in multiple pathology processes, including fibrosis.18–20 EndoMT also serves as evidence for endothelial plasticity.
As ECs-exclusively enriched microRNA, microRNA-126 (miR-126) regulates angiogenesis and vascular integrity.17,21 Its dysfunction is related to various vascular diseases such as hypertension, atherosclerosis, and tumorigenesis.22–25
To illuminate the transcriptional heterogeneity of embryonic ECs (eECs) and functions of miR-126 at single-cell resolution, ECs from wild-type and Tie2 (Tek receptor tyrosine kinase)-cre–mediated ECs-specific miR-126 knockout (miR-126-ecKO) mice were enriched by FACS at embryonic day 14.5 and were followed by scRNA-seq separately. With reported organ or tissue-specific genes, the whole-embryo hierarchical landscape of eECs was described. The spatiotemporal heterogeneity of ECs was explored by aggregating public single-cell datasets. In addition, more vascular abnormal phenotypes caused by deficiency of miR-126 were identified.
Materials and Methods
Data Availability
The authors declare that all data (including scRNA-seq data and R scripts for single-cell data analysis) and methods supporting the findings of this study are available in the Supplemental Methods or from the corresponding authors on reasonable request.
An expanded Methods section is available in the Supplemental Methods. All animal experiments were performed in accordance with the institutional guidelines of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. We used male mice because this minimized estrogen-dependent fluctuations in response to angiogenesis and glucose metabolism.26,27
Statistical Analysis
For experimental studies, all quantitative data were evaluated whether they followed the normal distribution by the Shapiro-Wilk test and equal variance by F test. For data that passed both tests, data are expressed as means±SEM, Student t test was used for the comparison between 2 groups, and for comparison among multiple groups, the data were analyzed by 1-way ANOVA with Tukey post hoc test. For the data that were not normally distributed, nonparametric test (Mann-Whitney U test) was performed and presented as median±SD. All P values are 2-sided, and P<0.05 was considered a statistically significant difference.
Results
Embryonic ECs Can Be Divided Into 11 Hierarchical Populations
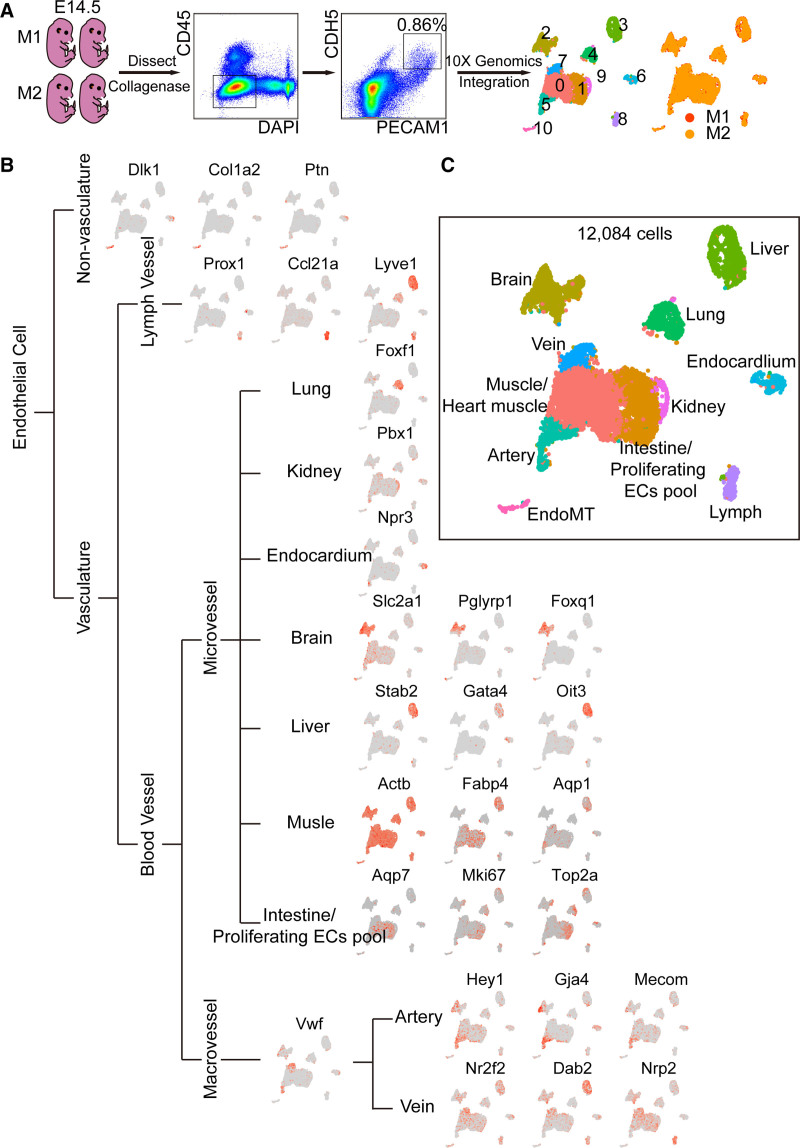
Four wild-type embryos at stage embryos at day 14.5 (E14.5) were harvested and dissociated into single-cell suspension by collagenase IV. FACS was used to purify live (DAPI−), nonmonocyte ECs (PECAM1 [CD31+], CDH5+, and CD45−), followed by scRNA-seq based on 10X Genomics strategy. False-positive cells including blood mononuclear cells and mesenchymal cells which may also be picked due to autofluorescence were excluded by unique molecular identifiers (Pecam1>0, Cdh5>0, Ptprc=0, Alas2<15). Finally, 5806 and 6278 eECs were obtained from 2 repetitions, respectively, and then integrated together. Eleven clusters were identified by clustering analysis, and cells isolated from the M1 (embryos from mouse 1) and M2 (embryos from mouse 2) were well integrated (Figure 1A). Established endothelial markers in various organs or tissues were used to annotate each cluster (Figure 1B). ECs in the vasculature are composed of blood vascular ECs and lymphatic ECs. Genes preferentially expressed in lymphatic ECs include Lyve1, Prox1, and Ccl21a28–30 (cluster 8 in Figure 1B). Blood vascular ECs differ from the microvessels with the initiative expression of Vwf and Vcam1.31 Then, the ECs in macrovessels are further classified into arterial ECs which were expressed with genes like Hey1, Gja4, Mecom specifically32–34 (cluster 5 in Figure 1B) and venous ECs which were expressed with Nr2f2, Dab2, Nrp2 specifically35–37 (cluster 7 in Figure 1B). The expression profile of ECs in microvessels is related to the function of tissues and organs where located. For example, Foxf1 is highly expressed in pulmonary ECs38 (cluster 4 in Figure 1B). The renal ECs expressed higher genes, such as Pbx1 (cluster 9 in Figure 1B). Npr3 is a marker gene of endocardium39 (cluster 6 in Figure 1B). As we know, many ECs of the brain participated in the formation of the blood-brain barrier and expressed specific genes (eg, Slc2a1, Pglyrp1, Foxq1)40 (cluster 2 in Figure 1B). However, Stab 2, Gata4, and Oit3 are highly expressed in hepatic ECs41,42 (cluster 3 in Figure 1B). Specifically, mesenchymal genes like Dlk1, Col1a2, and Ptn19,43,44 are highly expressed in ECs transited to Mesenchymal cells (EndoMT, cluster 10 in Figure 1B).
Figure 1.
Endothelial cells (ECs) at embryonic day 14.5 were divided into 11 populations. A, ECs were enriched for single-cell RNA sequencing analysis. Collagenase I was used to dissect embryos at day 14.5 (E14.5). Fluorescence-activated cell sorting (FACS) was used to isolate live (DAPI−), nonmonocyte (CD45−) ECs (PECAM1 [CD31+] and CDH5+). Seurat 3.0 was used for single-cell analysis. Colors denoted different clusters or different origins of mouse. B, Known endothelial mark genes in different tissues were used to define clusters of embryonic ECs (eECs). C, eECs were classified into 11 populations. Colors denoted different populations. DAPI indicates 4',6-diamidino-2-phenylindole; EndoMT, endothelial-to-mesenchymal transition; M1, embryos from mouse 1; M2, embryos from mouse 2; and PECAM1, platelet endothelial cell adhesion molecule-1.
Collectively, 11 heterogeneous clusters of eECs were hierarchically organized, just as the anatomic structure of vasculature (Figures 1C and 2A). That is, eECs include lymphatic ECs (cluster 8) and blood vascular ECs. Blood vascular ECs include macrovascular ECs (cluster 5 and cluster 7) and microvascular ECs (cluster 2–4, cluster 6, and cluster 9). We also identified one cluster as mesenchyme transited ECs (cluster 11). Besides, there were 2 undefined clusters (cluster 0 and cluster 1), which will be defined below.
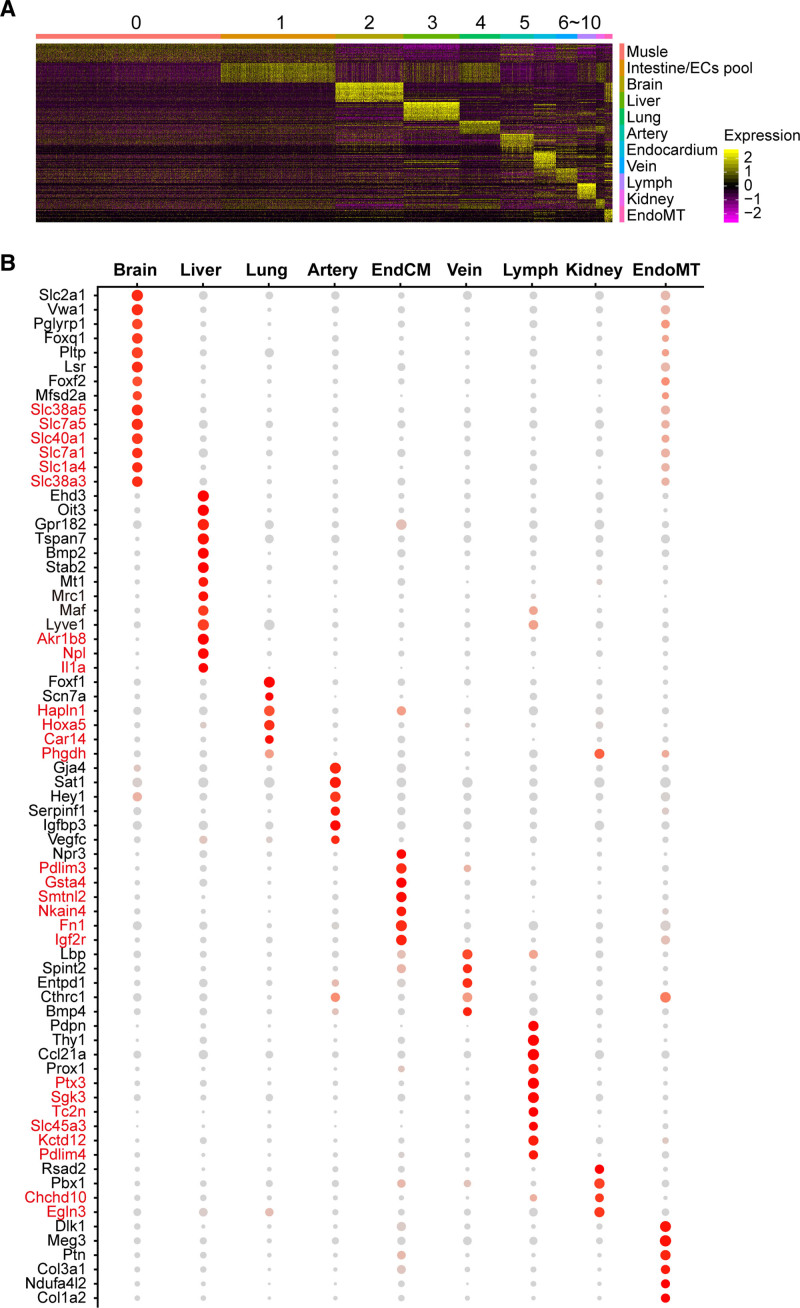
Figure 2.
Genes highly and exclusively expressed in each population of embryonic endothelial cells (eECs). A, Heat map of the top 20 enriched genes in each population reveals the heterogeneities of eECs. B, Genes highly expressed in each population of eECs were shown. The expression of genes in red font is higher in eECs than in adult endothelial cells. Dot size corresponds to percentage of cells within the cluster, and dot color corresponds to expression level. EndoMT indicates endothelial-to-mesenchymal transition.
To evaluate the reliability of the results defined in Figure 1C and clarify the origin of cluster 0 and 1, the publicly available single-cell datasets of E-MTAB-807710 was integrated with our dataset, which sequenced the ECs of 11 adult mouse tissues, respectively (Figure S1A and S1B in the Supplemental Material). As expected, most adult ECs (aECs) and embryonic ECs (eECs) from the same tissue sources were clustered together (Figure S1C in the Supplemental Material). More specifically, the majority of aECs from the brain, liver, and kidney nicely overlapped with their corresponding eECs (Figure S1D in the Supplemental Material). Part of aECs from the heart overlapped with eECs defined as endocardium, and this was consistent with anatomic distribution of ECs in the heart. Most pulmonary aECs were closed but did not overlap with eECs of the lung. This was reasonable since pulmonary circulation underwent fundamental changes after birth compared to the embryonic stage. Interestingly, we observed that clusters 0 and 1 from eECs well overlapped with muscle and intestinal aECs subclusters separately (Figure S1D in the Supplemental Material). We also observed that proliferation-related genes were mainly expressed in cluster 1 (cluster 1 in Figure 1B). Furthermore, certain genes (Actb, Fabp4, Aqp1 in muscle and Apq710 in intestinal ECs) were enriched in cluster 0 and cluster 1, respectively (cluster 0 and 1 in Figure 1B). Thus, we speculated cluster 0 as muscle eECs (including heart muscle ECs) and cluster 1 as Intestine/Proliferating ECs Pool.
We further used another public scRNA-seq dataset (ArrayExpress: GSE109774), which sequenced organs or tissues of adult mouse with different strategies (10X genomics and Droplet).45 Up to 6736 aECs of Pecam1+Cdh5+CD45− were obtained (Figure S2A and S2B in the Supplemental Material), followed by aggregation analysis with eECs (Figure S2C in the Supplemental Material). The aggregation of eECs and aECs reconfirmed the classification of eECs (Figures 1 and 2 and Figure S2 in the Supplemental Material).
Furthermore, genes highly and exclusively expressed in each subpopulation of eECs were listed (red font in Figure 2B). Taking advantage of the published datasets of adult mouse mentioned above, we highlighted some genes with red font in Figure 2B, which were relatively higher expressed in eECs compared with aECs.
Embryonic ECs Are Heterogeneous in Tissues and Have High Proliferation Potential
To study the intratissue EC heterogeneity, we performed the graph-based subclustering of ECs (aECs and eECs integrated dataset (published dataset E-MTAB-8077 for aECs and our scRNA-seq dataset for eECs) in Figure S1 in the Supplemental Material) for each tissue separately. Uniform manifold approximation and projection visualization analysis obtained comparable results (Figure 3). The expressions of the top 10 marker genes for each tissue-specific EC subclusters were shown by heatmap (Figure S3 in the Supplemental Material). According to established (canonical) marker genes of ECs subtypes, the ECs subclusters were annotated. For instance, the large vessel markers, Vwf and Vcam1, which are related to the diameter blood vessel, were enriched in some arterial and venous EC clusters. Therefore, we classified them as artery/vein. Some reports showed that capillary ECs express both large vessel and capillary markers. We, therefore, classified them as capillary arterial/venous (Figure 3A and 3B). Some EC subclusters express unexpected genes, such as Serpina1e and Alb (subcluster 6 in Figure 3C), which are highly expressed in hepatocytes.46,47 We, therefore, define them as organ-infiltrating ECs.
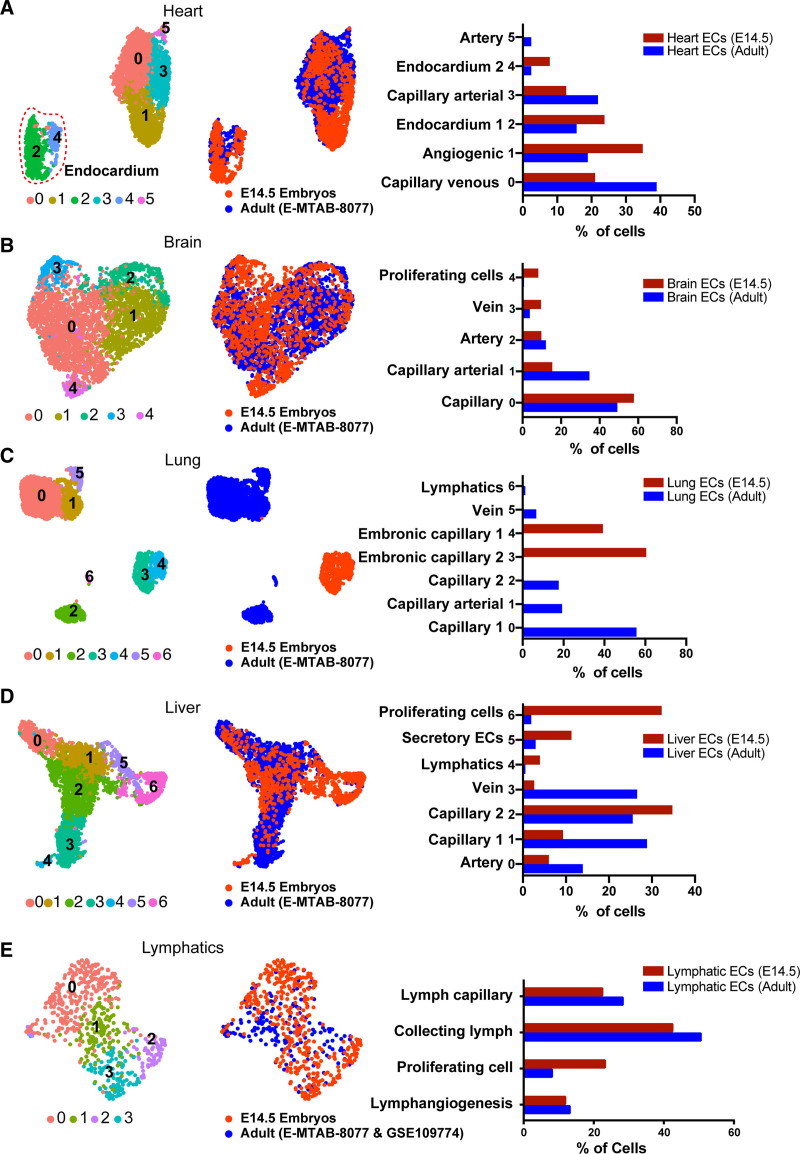
Figure 3.
The vascular heterogeneity still existed within embryonic endothelial cells (eECs) tissues which had higher proliferation potential than adult ECs (aECs). A, Five clusters subclustered from integrated heart subset and the fraction of each subcluster calculated separately from cardiac aECs and eECs. B, Five clusters subclustered from integrated brain subset and the fraction of each subcluster calculated separately from cerebral aECs and eECs. C, Seven clusters subclustered from integrated lung subset and the fraction of each subcluster calculated separately from pulmonary aECs and eECs. D, Seven clusters subclustered from integrated liver subset and the fraction of each subcluster calculated separately from hepatic aECs and eECs. E, Four clusters subclustered from integrated lymphatic subset and the fraction of each subcluster calculated separately from lymphatic aECs and eECs. E14.5 indicates embryos at day 14.5. E-MTAB-8077: published dataset E-MTAB-8077 from Kalucka et al.10 GSE109774: published dataset GSE109774 from Schaum et al.45
By integrating eECs with aECs, we reveal the heterogeneity that existed within tissues at embryonic stage, as well as between eECs and aECs. We then calculate the subcluster ECs percentage on embryonic tissues, respectively, and compared them with aECs (Figure 3). Most tissues showed the same proportion in most types of ECs (Figure 3A through 3D and 3E). However, the eECs showed higher ratio on proliferating ECs in cardiac eECs (Figure 3SA), cerebral eECs (Figure 3SB), hepatic eECs (Figure 3SD), and lymphatic eECs (Figure 3SE). We confirmed that the proliferation-related genes, such as Mki67, Top2a, and Cdk1, were highly expressed in eECs (Figure S3 in the Supplemental Material). Gene Ontology (GO) analysis on differentially expressed genes (DEG) between embryonic ECs and aECs showed that DEG mainly participated in mitotic cell cycle, tube morphogenesis, and cell population proliferation (Figure S4A in the Supplemental Material). Additionally, the proportion of proliferating cells (Mki67 positive) displayed over half ratios across all tissues (Figure S4B in the Supplemental Material). To verify the bioinformatics results, dual immunofluorescence staining of MKI67 (marker of proliferation Ki-67) and PECAM1 was applied to the frozen section of liver tissue. The result showed that in the same region, there were more double-positive (MKI67+ and PECAM1+) ECs in embryonic liver than in adult liver (Figure S4C and S4D in the Supplemental Material).
Intriguingly, we identified 3 subclusters of endocardium (Figure S5A in the Supplemental Material). According to the expression profile of marker genes, 3 endocardial subclusters were conventional endocardium (Npr3, cluster 0), cardiac valve (Postn, Hapln1, and Tbx20, cluster1), and coronary ECs (Aplnr and Fabp4, cluster 2; Figure S5C in the Supplemental Material). Furthermore, the degree of shear stress in the cardiac valve at each side differs, with the upstream surface suffering more and expressing more elastic genes compared with the downstream surface, and 4 subclusters were obtained by further clustering analysis of the cardiac valve (Figure S5A in the Supplemental Material). Then, the proliferating cell (cluster 1–0), unbiased valve cells (cluster 1–1), upstream valve cells (cluster 1–2), and downstream valve cells (cluster 1–3) were defined by considering the anatomic structure of cardiac valve and expression profiles of genes related to proliferation (Mki67, Top2a, Cdk1), cushion mesenchymal cells (Msx1, Hapln1, Postn), and elasticity (Fbln5, Fbln2, Ptn, Fbn1, Eln; Figure S5B and S5C in the Supplemental Material). The percentage of proliferating cells in eECs was extremely higher than in aECs (Figure S5D in the Supplemental Material), indicating that endocardial eECs have higher plasticity.
Classic mesenchymal cells include but are not limited to fibroblasts, mural cells (pericytes and smooth muscle cells), osteocytes, chondrocytes, and adipocytes. Wondering if EndoMT varies with the diverse mesenchymal cells, we extracted aECs and eECs transited to mesenchymal cells (cluster 12 and cluster 14 in Figure S2C in the Supplemental Material) and found EndoMT can be further classified into 3 subclusters (Figure S5E in the Supplemental Material). Then, the endothelial-to-pericyte transition, endothelial-to-smooth muscle cell transition (EndoSMT), and endothelial-to-fibroblast transition were classified with the expression profiles of gene markers in pericytes (Pdgfrb,32 Abcc948), smooth muscle cells (Acta2,49 Tagln50), and fibroblasts (Col1a1 and Ptn43,51), respectively (Figure S5E and S5G in the Supplemental Material). EndoSMT and endothelial-to-pericyte transition can be named EndoMT. When considering the developmental stages, the proportion of endothelial-to-pericyte transition was significantly higher in embryos, whereas EndoSMT was in reverse (Figure S5F in the Supplemental Material). The potential reason may be that the shear stress in embryos is relatively low. Besides, the cycling cells of EndoMT were found distributed predominantly in the embryonic stage through exploring the ratio of cycling cells in each subcluster (Figure S5E in the Supplemental Material). Additionally, the ratio of cycling cells in each subcluster was different, and it is the highest in the EndoSMT. More than half of EndoSMT in embryos were cycling cells, which indicated its augmented potential during development (Figure S5F in the Supplemental Material).
Knockout of miR-126 Affects eECs Clustering and the Abnormality of eECs Is Mainly Related to Hypoxia and Apoptosis
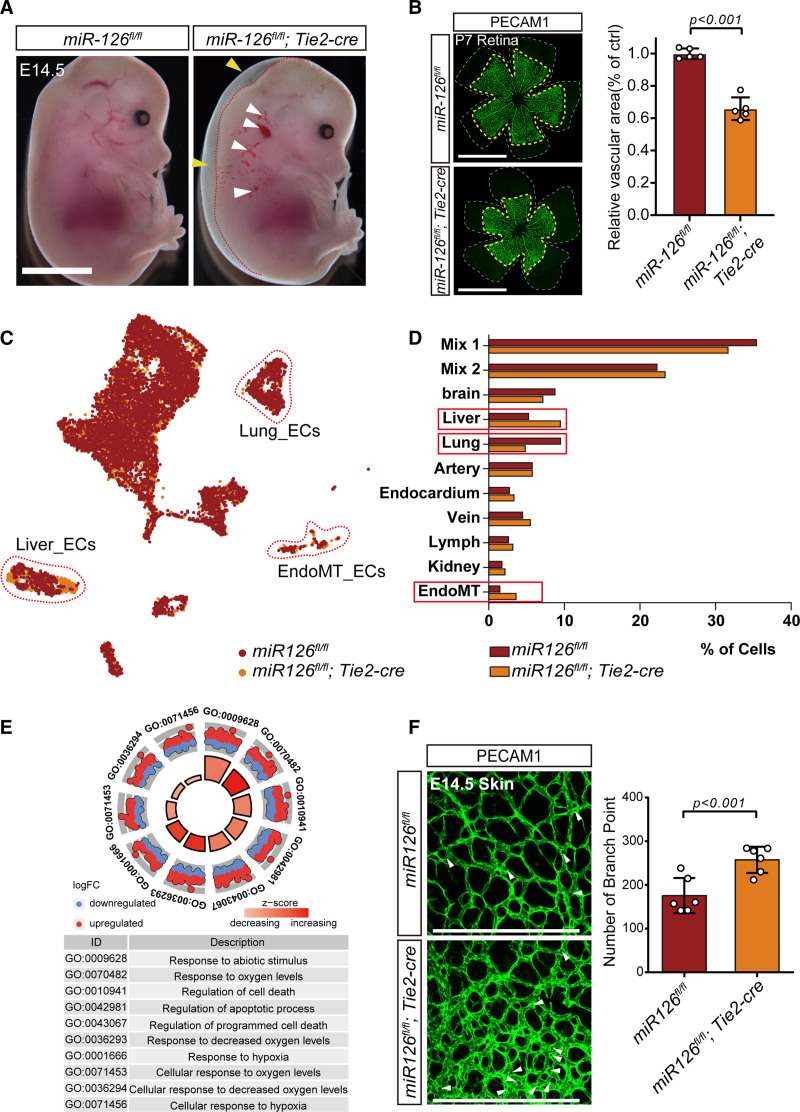
MicroRNA-126 (miR-126) is an endothelium-enriched miR and functions in vascular integrity and angiogenesis.21,52–54 To reinforce the phenotype, EC-specific miR-126 knockout mice (miR-126-ecKO) were constructed (Figure S6A and S6B in the Supplemental Material). As expected, miR-126-ecKO embryos showed vascular leakage and subcutaneous embryonic edema (Figure 4A) and a higher lethality (Figure S6C in the Supplemental Material) compared with littermate control (miR-126fl/fl). Besides, impaired migration and sprouting of ECs in embryonic lymph and postnatal retinal was observed in miR-126-ecKO, as indicated by decreased vascular area (Figure 4B, Figure S6E and S6F in the Supplemental Material) and blunted terminals (Figure S6D and S6E in the Supplemental Material). All these vascular abnormalities indicated that the angiogenesis in the vasculature of miR-126-ecKO mouse was reduced, which was consistent with reported studies.55,56
Figure 4.
Knock out of microRNA-126 (miR-126) disturbed the embryonic endothelial cells clustering during development. A, Embryos of Tie2 (Tek receptor tyrosine kinase)-cre–mediated ECs-specific miR-126 knockout (miR-126-ecKO) mouse demonstrated vascular leakage (white arrowheads) and hemorrhaging (yellow arrowheads), scale bar=2000 μm. B, The migration of ECs in the retina at p7 was reduced in miR-126-ecKO mouse. The retina vessels were stained by ECs marker gene PECAM1 (platelet endothelial cell adhesion molecule-1) in miR-126–deficient mice and wild types at P7. And the quantification of vascular area changes after miR-126 was knocked out in endothelial cells. The white dash lines indicate terminals of retina and the yellow dash lines indicate terminals of retina vessels. Scale bar, 1000 μm. C, The ECs of miR-126-ecKO embryos were aggregated with the littermate controls (miR-126fl/fl), and dashed lines demarcated the different groups. D, Quantitatively compare and analyze the proportion of each cell population in the 2 samples. E, Gene Ontology (GO) biological function analysis of differentially expressed gene in miR-126-ecKO and the littermate controls. F, Vascularization in the skin of miR-126-ecKO mouse was more complex than their littermate controls. Pillars were indicated with white arrowheads. Number of branches points was counted on the right (n=6), scale bar=200 μm. E14.5 indicates embryos at day 14.5; EndoMT, endothelial-to-mesenchymal transition; FC, fold change; and P7, posnatal day 7.
To elucidate the effect of miR-126 on the development of ECs from a single-cell perspective, 5203 ECs of miR-126-ecKO embryos were enriched and sequenced, followed by aggregation with eECs from the littermate control. Eleven populations were obtained and defined. And the fraction of most populations accounted comparable between miR-126-ecKO and littermate control embryos, except hepatic, pulmonary, and mesenchymal-transited ECs (Figure 4C and 4D), which will be further discussed below. GO analysis of genes enriched only in miR-126-ecKO eECs showed that 5 out of top 10 GO terms were related to hypoxia (Figure 4E). To confirm the analysis result, the eECs were enriched by FACS from both miR-126-eKO and littermate control mice, and the quantitative polymerase chain reaction results showed that genes related with hypoxia including Hif1a (hypoxia-inducible factor 1-alpha) were upregulated apparently after miR-126 was knocked out (Figure S7B in the Supplemental Material). In addition, feature plot analysis and coimmunofluorescence of HIF1A and PECAM1 indicated the expression profile was higher in miR-126–deficient cardiac ECs than in littermate control (Figure S7C and S7D in the Supplemental Material). Previous study reported that hypoxia stimulates excessive IA.57 Intriguingly, we did find excessive IA in the embryonic skin and the terminal region of postnatal retina in miR-126-ecKO mouse by immunofluorescence staining of PECAM1 (white arrow in Figure 4F and Figure S7A in the Supplemental Material), which means that miR-126 is essential to homeostasis of vascular network at embryonic stage. Therefore, we preliminarily concluded that knockout of miR-126 in ECs would affect angiogenesis both in sprouting angiogenesis and IA, but in opposite directions. GO analysis of genes enriched only in miR-126–deficient eECs showed that 4 out of 5 top terms were related to cell apoptosis and cell death. Genes related to apoptosis (Mex3 family, caspase family, genes related to autophagy activation, and genes related to lysosomal biogenesis) had been identified in the affected ECs clusters. As the results showed, 3 members of Mex3 family were upregulated after miR-126 knocked out (Figure S8A, S8D, and S8E in the Supplemental Material). Considering that Mex3a could form a complex with miR-126-5p to facilitate antiapoptotic effect,58 the upregulation of Mex3a at transcripts level may be due to some compensatory mechanisms in miR-126–deficient eECs. Most members of Caspase family were upregulated in miR-126–deficient eECs (Figure S8B in the Supplemental Material). Moreover, a toolbox for deducing autophagy status which was built by Bordi et al,59 had been detected in our integrated single-cell data. Only sixty percentage of genes related to autophagy activation (Induction List) were upregulated after miR-126 knocked out. Although over 80% of genes related to lysosomal biogenesis were upregulated in miR-126–deficient eECs (Figure S8C in the Supplemental Material). This indicates that the activation of autophagy in miR-126–deficient eECs might be earlier than E14.5 days. Considering the higher embryonic mortality observed in miR-126–deficient mice, the apoptosis on eECs at E14.5 was analysis by flow cytometry. The result showed that the percentage of PI (propidium iodide)-positive cells were apparently increased after miR-126 was knocked out in eECs (Figure S8F and S8G in the Supplemental Material). The increased cell apoptosis contributed to the higher embryonic lethality observed in miR-126-ecKO mice (Figure S5C in the Supplemental Material).
Next, we analyzed the expression profiles of the miR-126 target genes (predicted targets by TargetScan7.2 and validated targets by published research papers) in the integrated scRNA-seq data. The results showed that most of target genes were upregulated after miR-126 was knocked out (Figure S9A through S9C in the Supplemental Material). Interestingly, many miR-126 target genes showed tissue heterogeneity: some targets were predominantly upregulated in hepatic eECs, whereas others were mainly upregulated in EndoMT eECs (Figure S9D in the Supplemental Material). Furthermore, GO analysis was conducted on the screened miR-126-3p targets, miR-126-5p targets, and total miR-126 targets, respectively, and the results showed that the upregulated biological progresses were mainly related to cell migration, response to oxygen-containing compound (Figure S9E through S9H in the Supplemental Material). Notably, among miR-126 targets, glucose transport–related genes denoted as glucose import were also upregulated. This indicated that it may act downstream of miR-126 targets.
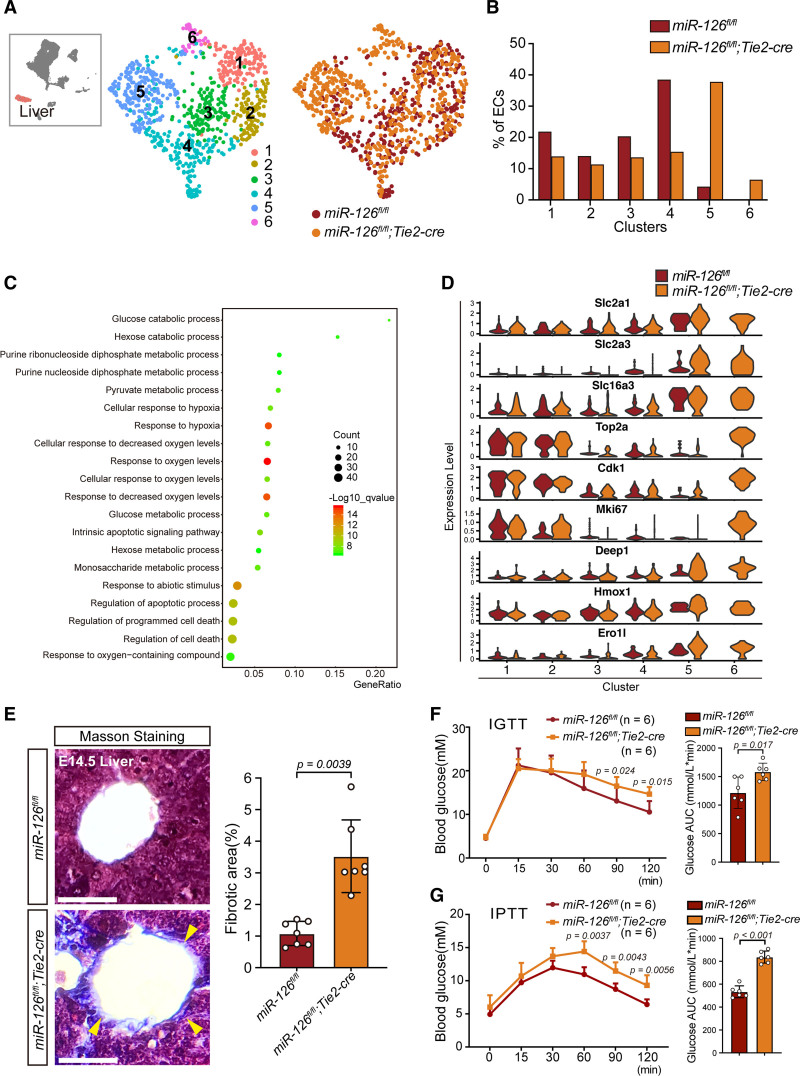
Knockout of miR-126 Disturbs Hepatic eECs Clustering to Affect Glucose Metabolism and Fibrosis
To compare the hepatic eECs with or without miR-126 in detail, we performed reclustering analysis of eECs from the liver (cluster 3 in Figure 4C) and obtained5 subclusters. Of which clusters 5 and 6 varied most in miR-126-ecKO ECs (Figure 5A and B). Most DEG in clusters 5 and 6 were similar, except for proliferative genes (Figure S10A in the Supplemental Material). GO biological progress analysis of DEG in clusters 5 and 6 revealed multiple statistically significant GO terms associated with glucose regulation and hypoxia (Figure 5C). Some glucose transmembrane genes including Slc2a1, Slc2a3, and Slc16a3 were highly expressed in clusters 5 and 6 (Figure 5D and Figure S10A in the Supplemental Material). Glucose tolerance test and pyruvate tolerance test were conducted to verify whether knocking out miR-126 in ECs affected glycose metabolism. The miR-126-ecKO mouse showed reduced tolerance to glucose and pyruvate (Figure 5F and 5G). In fact, previous studies have declared a negative correlation between miR-126 and blood glucose level, but the causality was unknown.60–62 The results suggested a potential function of miR-126 in glucose regulation. The pathways and metabolisms involved warrant further investigation. Considering that hepatic hypoxia may lead to fibrosis, Masson staining was performed and showed increased collagenous fibers in miR-126-ecKO mouse (Figure 5E). Collectively, the disorganized hepatic eECs caused by miR-126 deficiency contributes to liver fibrosis and abnormal glucose metabolism at adult stage.
Figure 5.
Knock out of microRNA-126 (miR-126) in endothelial cells (ECs) disturbed blood glucose metabolism and induced fibrosis in hepatic tissue. A, 5 subclusters were obtained by further clustering of hepatic ECs (cluster 3 in Figure 6A). Colors denote different subclusters or mice line. B, Fractions of hepatic ECs from Tie2 (Tek receptor tyrosine kinase)-cre–mediated ECs-specific miR-126 knockout (miR-126-ecKO) or littermate controls in each subcluster. C, Gene Ontology biological function analysis of differentially expressed genes in cluster 2 and 3. D, Vnplot of genes whose expression profiles were significantly different between miR-126-ecKO and the littermate controls. E, Masson staining of embryonic liver at embryos at day 14.5 (E14.5), scale bar=200μm. F, Postprandial blood glucose was elevated in miR-126-ecKO mouse compared with wild types verified by Intraperitoneal glucose tolerance test (IGTT). Sample size: n=6. G, Gluconeogenesis was elevated in miR-126-ecKO mouse compared with miR-126fl/fl mouse verified by Intraperitoneal pyruvate tolerance test (IPTT). Sample size: n=6. AUC indicates area under curves.
Knockout of miR-126 in eECs Leads to Excessive EndoMT and Vascular Calcification
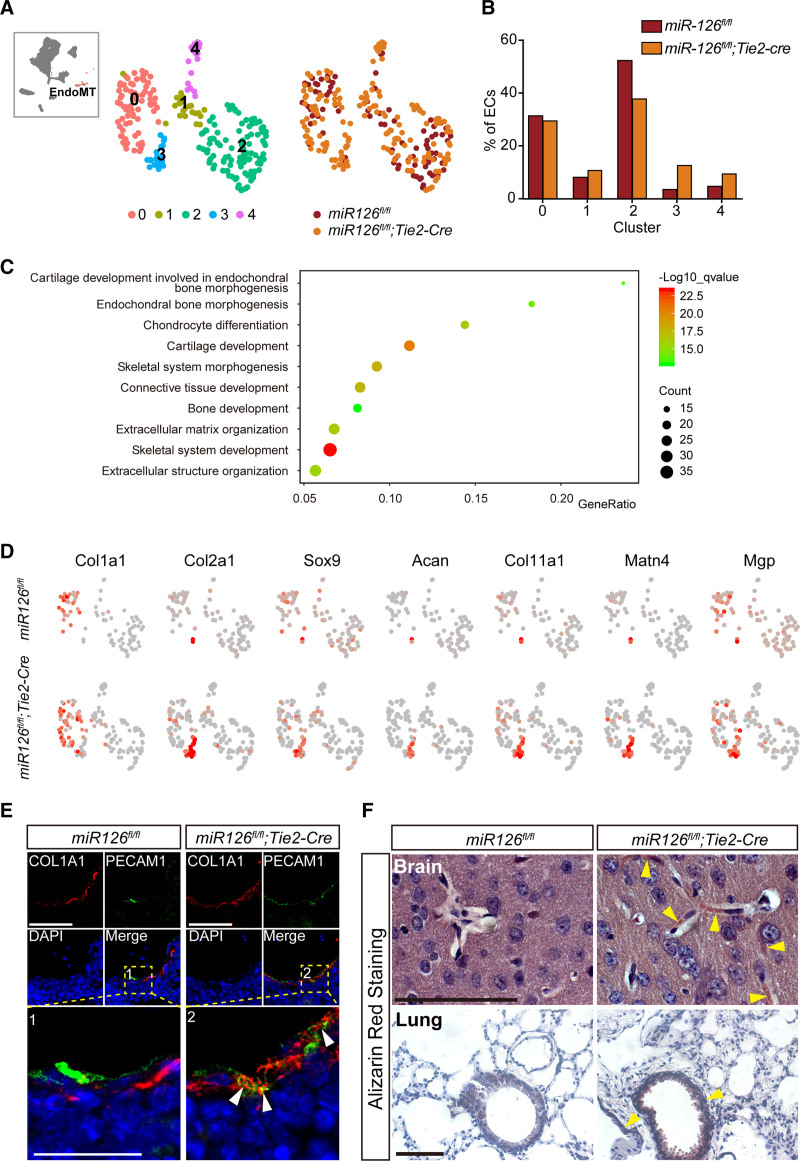
To determine the effect of miR-126 knockout on EndoMT, reclustering of mesenchymal-transited ECs (cluster 10 in Figure 4C) was performed and 5 subclusters were identified. Of which cluster 3 and cluster 4 varied most in miR-126-ecKO eECs (Figure 6A and 6B). GO analysis of DEG in cluster 3 revealed most terms related to extracellular matrix organization and skeletal system development (Figure 6C). Genes enriched in the extracellular matrix and cartilage were also highly expressed in cluster 3 (Figure 6D). Additionally, quantitative polymerase chain reaction analysis confirmed a higher expression of ECM (extracellular matrix) enriched genes in miR-126–deficient eECs (Figure S10B in the Supplemental Material). Dual immunofluorescence for an EC marker (PECAM1) and markers of extracellular matrix (COL1A1 [COL1A1 (alpha-1 type I collagen)] and LUM [lumican]) confirmed the excessive EndoMT in miR-126 knocked out eECs (Figure 6E and Figure S10C in the Supplemental Material). Previous reports demonstrated that the endochondral and intramembranous bone formation in vessels may lead to ectopic vascular calcification and eventually lead to various vascular diseases, such as atherosclerosis or hypertrophy.63,64 Multiple cells were the potential sources for calcification, including smooth muscle cells,65 pericytes,66 and ECs.67 Alizarin red staining demonstrated that vascular calcification in the brain and lung of miR-126-ecKO mouse increased (Figure 6F). Our analysis indicated knockout of miR-126 resulted in vascular calcification by endothelial-to-cartilage transition.
Figure 6.
Endothelial-to-mesenchymal transition (EndoMT) and vascular calcification were increased in Tie2 (Tek receptor tyrosine kinase)-cre–mediated ECs-specific microRNA-126 (miR-126) knockout (miR-126-ecKO) embryos. A, Five subclusters were obtained by further clustering of EndoMT (cluster 10 in Figure 6A). Colors denote different subclusters or mice line. B, Fractions of EndoMT from miR-126-ecKO or miR-126fl/fl in each subcluster. C, Gene Ontology biological function analysis of differentially expressed genes in cluster 3. D, Feather plot of representative genes in chondrogenesis and endothelial-to-mesenchymal which were enriched in cluster 3. E, Double immunofluorescent staining of COL1A1 (alpha-1 type I collagen) and PECAM1 (platelet endothelial cell adhesion molecule-1) proteins in embryonic hepatic tissue to identify the EndMT (Endothelial-to-Mesenchymal Transition). F, Alizarin Red S Staining to identify the calcium in the adult’s brain and lung slides. Scale bar=100 µm. DAPI indicates 4',6-diamidino-2-phenylindole.
Knockout of miR-126 in eECs Leads to Upregulation of Cell Adhesion-Related Genes in Pulmonary eECs
To study the abnormity of pulmonary ECs, further clustering (cluster 4 in Figure 4C) identified 2 subclusters (Figure S11A in the Supplemental Material). The ratio of ECs in each group did not change much (Figure S11B in the Supplemental Material). Expression profiles and GO analysis showed that the different genes are mainly related to 3 molecular functions: cell adhesion, glucose catabolic, and cell apoptotic process (Figure S11C and S11D in the Supplemental Material). The biological process of glucose catabolic and programmed cell death has been observed in other tissues or whole body eECs as well. However, the abnormal cell adhesion process was found only in pulmonary subclusters (Figure S11D in the Supplemental Material). This indicated that there might be a defect with junctions between the ECs.
MiR-126-HIF1A-GLUT1 Pathway Is Involved in Glycometabolism, and Treatment With miR-126 Agomir Rescues Blood Glucose Imbalance in miR-126-ecKO Mice
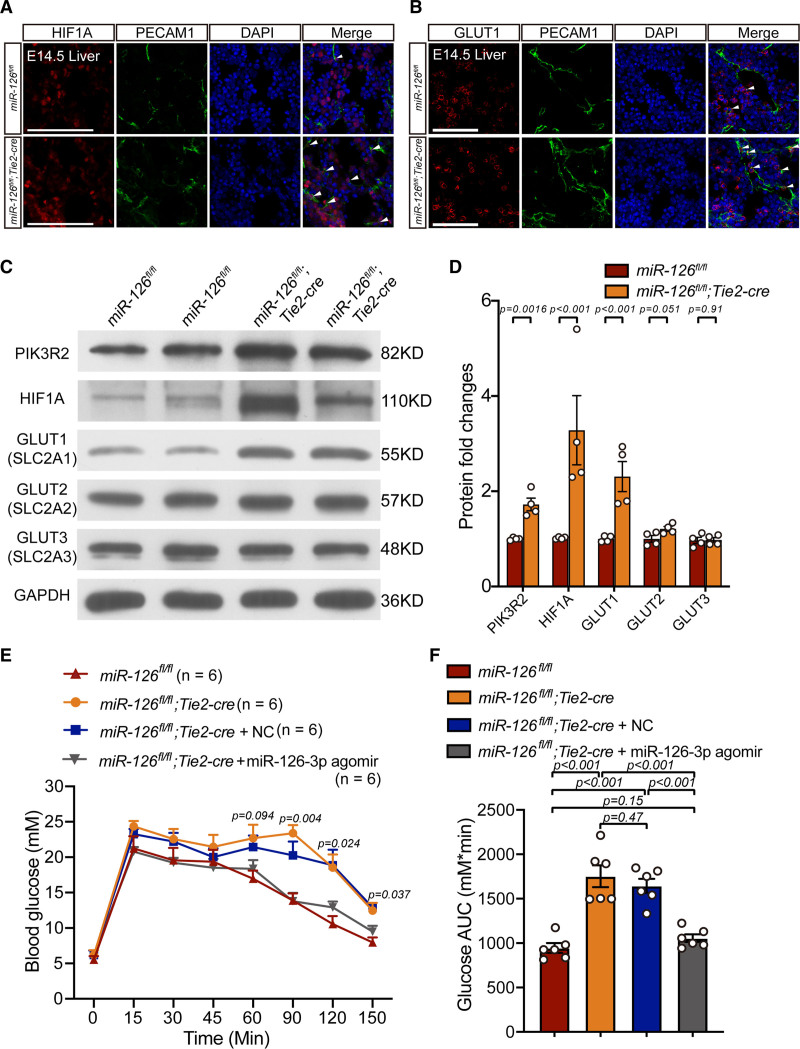
The glucose transport proteins family (including GLUT1, GLUT2, GLUT3, and GLUT4) was reported to play critical roles in blood glucose metabolism.68–71 Heilig et al72 found that overexpression of GLUT1 (Slc2a1) in rat mesangial cells cultured in a normal glucose milieu mimicked the diabetic phenotype. Additionally, published articles claimed that overexpression of miR-126 directly inhibited Hif1a in ECs.73 This means that knocking out miR-126 in ECs may result in upregulation of HIF1A. We thus proposed that when miR-126 was knocked out in ECs, hepatic ECs partially upregulated the genes related to hypoxia. The upregulated hypoxia-related genes eventually induced the upregulation of Slc2a1 (GLUT1) and Slc2a3 (GLUT3). Finally, the upregulated GLUT1 led to a disorder in blood glucose metabolism.
To verify this speculation and confirm bioinformatic analysis results in Figure 5, dual immunofluorescence was performed on hepatic tissues of miR-126–deficient and littermate control. The results showed that HIF1A and GLUT1 were both expressed more highly in miR-126–deficient eECs (Figure 7A and 7B). To figure out whether the effects of miR-126 on hepatic eECs are conservative in embryonic and adult stage. The adult hepatic ECs were enriched by collagenase perfusion which was followed by Percoll-gradient separation. The results of Immunoblotting and quantification data showed that HIF1A and GLUT1 were both upregulated in hepatic ECs when miR-126 was knocked out (Figure 7C and 7D). Conversely, miR-126 knockout had little influence on GLUT2 (SLC2A2) and GLUT3 (SLC2A3) expression profile in hepatic ECs.
Figure 7.
HIF1A-GLUT1 pathway was upregulated in microRNA-126 (miR-126)–deficient endothelial cells (ECs), and miR-126-3p agomir restored the disrupted glycometabolism in miR-126 knocked out mice. A, Dual immunofluorescence of GLUT1 (glucose transporter 1) and PECAM1 (platelet endothelial cell adhesion molecule-1) in miR-126–deficient and littermate control hepatic tissues at embryos at day 14.5 (E14.5). Scale bar=100 µm. B, Dual immunofluorescence of HIF1A (hypoxia-inducible factor 1-alpha) and PECAM1 in miR-126–deficient and littermate control hepatic tissues at E14.5. Scale bar=100 µm. C, Western Blotting of glucose transport protein family (GLUT1, GLUT2 and GLUT3) and HIF1A in adult miR-126–deficient and wild-type hepatic ECs. D, Quantification of immunoblotting results in C. E, Intraperitoneal glucose tolerance test (IGTT) on wild types, miR-126–deficient mice treated with miR-126-3p agomir, agomir-negative control (NC) and no treats. Sample size: n=6. F, Area under curves (AUC) analysis IGTT results. Sample size: n=6. PIK3R2 indicates phosphoinositide-3-kinase regulatory subunit 2; and SLC2A, solute carrier family 2.
The blood glucose metabolism was found disrupted in miR-126 knocked out mice (Figure 5E). It is important to know whether restoring miR-126 recovers the defect. Hence, we treated miR-126–deficient mice with miR-126-3p agomir via tail vein injection. The intraperitoneal glucose tolerance test results revealed that miR-126-3p treatments displayed a significant restoration of impaired blood glucose tolerance in miR-126 knocked out mice (Figure 7E and 7F). The results indicated that miR-126-Hif1a-Glut1 pathway participated in murine blood glucose metabolism in vivo.
Discussion
The present study provided a novel insight into the heterogeneity of ECs at embryonic stage and investigated the effect of miR-126 on ECs development at single-cell levels. The heterogeneous transcriptome of ECs in adult stage was investigated previously4–10 but rarely studied on developmental eECs. Single-cell analysis of the whole-embryo ECs revealed 11 clusters, in agreement with the hierarchical structure of vasculature in anatomy. Using public single-cell datasets (E-MTAB-8077 and GSE109774), the definition of each population was confirmed by aggregation of aECs and eECs. The cardiac, cerebral, pulmonary, hepatic, and lymphatic ECs were clustered in accordance with their anatomic structures. The heart ECs were further divided into endocardium, cycling cells, and cardiac valve ECs. In addition, the cardiac valves ECs were even subclustered into upstream valves, downstream valves, and coronary ECs. When considering the developmental stage (adult and embryo), eECs differed with aECs mostly by the high potential of proliferation and less shear stress. Thus, ECs are spatiotemporally characterized as heterogeneous.
Tie2 was first reported by Dumont et al74 and Tie2 transcripts were firstly detected in the extraembryonic on embryonic day 7 during development.75 Both Tie2-cre and Cdh5-cre have been used widely as constitutive active pan-endothelial Cre mouse models. Payne et al76 had given a comprehensive comparison among endothelial-specific Cre mouse models including Tie2-cre and Cdh5-cre. They reported that the earliest pan-endothelial cre activity in Tie2-cre model was around E7.5, whereas the earliest pan-endothelial cre activity in Cdh5-cre model is around E9.5. Hence, we applied Tie2-cre mouse model to investigate what happens to ECs when genes are knocked out in their initial development stage.
To clarify whether the heterogeneity in embryonic ECs is stable, we focused on a key regulator, miR-126, which is expressed predominately in ECs and is critical for angiogenesis.21,52–54 In this study, we demonstrate that miR-126 is essential for regular eECs clustering. GO analysis identified that top 50 differently expressed genes are mainly related to hypoxia and cell apoptosis. And genes related to apoptosis, including Mex3 family,58 Caspase family, and a toolbox for deducing autophagy status which was built by Bordi et al,59 had also been analyzed in single-cell data. As expected, most apoptosis-related genes were upregulated in miR-126–deficient eECs. In addition, three eECs subgroups, including hepatic, pulmonary, and EndoMT ECs, showed significant changes in the clustering via integrated analysis. Unexpectedly, the upregulated genes are different among 3 subgroups. This indicated that the influence of miR-126 on the development of eECs may be heterogeneous across tissues.
The previous studies described that circulating miR-126 is closely associated with type 2 diabetes.60,61 However, the detailed mechanisms were unclear. Here, we characterized hepatic eECs clusters which were mostly affected when miR-126 was knocked out and found that genes related to glycometabolism, such as Slc2a1 and Slc2a3,68–70 were greatly upregulated. The experimental results verified this influence could last until adulthood, suggesting that loss of endothelial miR-126 is not only a feature of type 2 diabetes but also a cause. Previous works claimed that overexpression of miR-126 directly inhibits Hif1a in porcine endothelial cell,73 and hypoxia was reported to induce GLUT1 (Slc2a1) expression in ECs.77 Our results validated that miR-126 targets Hif1a which is a potent stimulus for induction of Slc2a1 mRNA in ECs. Tail vein administration miR-126 agomir apparently rescued impaired glucose tolerance in miR-126-ecKO mice. Hence, we demonstrate that miR-126-Hif1a-Slc2a1 pathway plays a protection role in murine glycometabolism in vivo. Additional investigation is required to identify downstream genes that mediate this effect.
EndoMT is a common cellular transdifferentiation during development. It also contributes to fibrosis under pathological condition.78 In the present work, we classify EndoMT subclusters in eECs. Specifically, EndoMT was divided into 3 subpopulations (endothelial-to-fibroblast transition, EndoSMT, and endothelial-to-pericyte transition) according to various mesenchymal cell types. In addition, comparative analysis of scRNA-seq data demonstrated that EndoMT can be disrupted when miR-126 is knocked out. Moreover, excessive extracellular matrix in ECs and fibrosis in hepatic tissue were observed in miR-126-ecKO mice, which confirmed the analysis results. Therefore, these findings suggest that miR-126 may function as a guidance cue to EndoMT.
Hypoxia is reported to induce excessive IA.57 Excessive IA phenotypes in miR-126-ecKO embryos indicated the potential role of miR-126 in the homeostasis of mouse embryonic vascular network. However, the mechanism of miR-126 on IA deserves further exploration. Some new abnormal phenotypes were observed from miR-126-ecKO mice including aberrant glycometabolism, fibrosis, and vascular calcification. In addition, we demonstrated that pathway including miR-126, hypoxia-induced factors (HIF1A), and GLUT1 may play critical roles in blood glucose metabolism. Although the comprehensive interactions and causal relationships between all these new phenotypes were elusive, our findings may facilitate further study on miR-126.
Article Information
Acknowledgments
We thank all members of the laboratory for critical discussion and comments on the article. We would like to thank Lin Qiu and people from Institutional Center for Shared Technologies and Facilities of Shanghai Institute of Nutrition and Health (SINH), Chinese Academy of Sciences (CAS) for technical assistance. We thank M.F. Xie, Z.Y. Cao, and staff for their technical assistance on primary cell culture and agomir tail vein injection.
Sources of Funding
This work was supported in part by the National Key Research and Development Program of China (2017YFA0103700, 2019YFA0802700, 2021YFA0804803) and the National Natural Science Foundation of China (92168206, 81930012, 81730013).
Disclosures
None.
Supplemental Materials
Supplemental Methods
Figures S1–S11
Major Resources Tables
References 79–82
Supplementary Material
Nonstandard Abbreviations and Acronyms
- aECs
- adult ECs
- ECs
- endothelial cells
- eECs
- embryonic ECs
- EndoMT
- endothelial-to-mesenchymal transition
- EndoSMT
- endothelial-to-smooth muscle cells transition
- FACS
- fluorescence-activated cell sorting
- IA
- intussusceptive angiogenesis
- miR-126-ecKO
- Tie2-cre–mediated ECs-specific miR-126 knockout
- Tie2
- Tek receptor tyrosine kinase
F.-H. Guo and Y.-N. Guan contributed equally.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/ATVBAHA.121.317093.
For Sources of Funding and Disclosures, see page 340.
Contributor Information
Fang-Hao Guo, Email: fhguo@sibs.ac.cn.
Ya-Na Guan, Email: darlingna@yeah.net.
Jun-Jun Guo, Email: guojunjun2017@sibs.ac.cn.
Jing-Jing Qiu, Email: jjqiu@sibs.ac.cn.
Yong Ji, Email: yongji@njmu.edu.cn.
Alex F. Chen, Email: chenfengyuan@xinhuamed.com.cn.
References
- 1.Garlanda C, Dejana E. Heterogeneity of endothelial cells. Specific markers. Arterioscler Thromb Vasc Biol. 1997;17:1193–1202. doi: 10.1161/01.atv.17.7.1193 [DOI] [PubMed] [Google Scholar]
- 2.Augustin HG, Koh GY. Organotypic vasculature: from descriptive heterogeneity to functional pathophysiology. Science. 2017;357:eaal2379. doi: 10.1126/science.aal2379 [DOI] [PubMed] [Google Scholar]
- 3.Zhou F, Li X, Wang W, Zhu P, Zhou J, He W, Ding M, Xiong F, Zheng X, Li Z, et al. Tracing haematopoietic stem cell formation at single-cell resolution. Nature. 2016;533:487–492. doi: 10.1038/nature17997 [DOI] [PubMed] [Google Scholar]
- 4.Park J, Shrestha R, Qiu C, Kondo A, Huang S, Werth M, Li M, Barasch J, Suszták K. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 2018;360:758–763. doi: 10.1126/science.aar2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vanlandewijck M, He L, Mäe MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Laviña B, Gouveia L, et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018;554:475–480. doi: 10.1038/nature25739 [DOI] [PubMed] [Google Scholar]
- 6.Baryawno N, Przybylski D, Kowalczyk MS, Kfoury Y, Severe N, Gustafsson K, Kokkaliaris KD, Mercier F, Tabaka M, Hofree M, et al. A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell. 2019;177:1915–1932 e1916.. doi: 10.1016/j.cell.2019.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lukowski SW, Patel J, Andersen SB, Sim SL, Wong HY, Tay J, Winkler I, Powell JE, Khosrotehrani K. Single-cell transcriptional profiling of aortic endothelium identifies a hierarchy from endovascular progenitors to differentiated cells. Cell Rep. 2019;27:2748–2758.e3. doi: 10.1016/j.celrep.2019.04.102 [DOI] [PubMed] [Google Scholar]
- 8.Paik DT, Tian L, Williams IM, Rhee S, Zhang H, Liu C, Mishra R, Wu SM, Red-Horse K, Wu JC. Single-cell RNA sequencing unveils unique transcriptomic signatures of organ-specific endothelial cells. Circulation. 2020;142:1848–1862. doi: 10.1161/CIRCULATIONAHA.119.041433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paik DT, Tian L, Lee J, Sayed N, Chen IY, Rhee S, Rhee JW, Kim Y, Wirka RC, Buikema JW, et al. Large-scale single-cell rna-seq reveals molecular signatures of heterogeneous populations of human induced pluripotent stem cell-derived endothelial cells. Circ Res. 2018;123:443–450. doi: 10.1161/CIRCRESAHA.118.312913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalucka J, de Rooij L, Goveia J, Rohlenova K, Dumas SJ, Meta E, Conchinha NV, Taverna F, Teuwen L-A, Veys K, et al. Single-cell transcriptome atlas of murine endothelial cells. Cell. 2020;180:764–779 e720. doi: 10.1016/j.cell.2020.01.015 [DOI] [PubMed] [Google Scholar]
- 11.Schmidt A, Brixius K, Bloch W. Endothelial precursor cell migration during vasculogenesis. Circ Res. 2007;101:125–136. doi: 10.1161/CIRCRESAHA.107.148932 [DOI] [PubMed] [Google Scholar]
- 12.Potente M, Gerhardt H, Carmeliet P. Basic and therapeutic aspects of angiogenesis. Cell. 2011;146:873–887. doi: 10.1016/j.cell.2011.08.039 [DOI] [PubMed] [Google Scholar]
- 13.Djonov V, Schmid M, Tschanz SA, Burri PH. Intussusceptive angiogenesis: its role in embryonic vascular network formation. Circ Res. 2000;86:286–292. doi: 10.1161/01.res.86.3.286 [DOI] [PubMed] [Google Scholar]
- 14.Hillen F, Griffioen AW. Tumour vascularization: sprouting angiogenesis and beyond. Cancer Metastasis Rev. 2007;26:489–502. doi: 10.1007/s10555-007-9094-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mentzer SJ, Konerding MA. Intussusceptive angiogenesis: expansion and remodeling of microvascular networks. Angiogenesis. 2014;17:499–509. doi: 10.1007/s10456-014-9428-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Q, Ouyang C, Xie L, Ling Y, Huang T. The ROCK inhibitor, thiazovivin, inhibits human corneal endothelial-to-mesenchymal transition/epithelial-to-mesenchymal transition and increases ionic transporter expression. Int J Mol Med. 2017;40:1009–1018. doi: 10.3892/ijmm.2017.3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arciniegas E, Frid MG, Douglas IS, Stenmark KR. Perspectives on endothelial-to-mesenchymal transition: potential contribution to vascular remodeling in chronic pulmonary hypertension. Am J Physiol Lung Cell Mol Physiol. 2007;293:L1–L8. doi: 10.1152/ajplung.00378.2006 [DOI] [PubMed] [Google Scholar]
- 19.Díez M, Musri MM, Ferrer E, Barberà JA, Peinado VI. Endothelial progenitor cells undergo an endothelial-to-mesenchymal transition-like process mediated by TGFbetaRI. Cardiovasc Res. 2010;88:502–511. doi: 10.1093/cvr/cvq236 [DOI] [PubMed] [Google Scholar]
- 20.Maddaluno L, Rudini N, Cuttano R, Bravi L, Giampietro C, Corada M, Ferrarini L, Orsenigo F, Papa E, Boulday G, et al. EndMT contributes to the onset and progression of cerebral cavernous malformations. Nature. 2013;498:492–496. doi: 10.1038/nature12207 [DOI] [PubMed] [Google Scholar]
- 21.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2011;481:190–194. doi: 10.1038/nature10661 [DOI] [PubMed] [Google Scholar]
- 23.Potus F, Ruffenach G, Dahou A, Thebault C, Breuils-Bonnet S, Tremblay È, Nadeau V, Paradis R, Graydon C, Wong R, et al. Downregulation of MicroRNA-126 contributes to the failing right ventricle in pulmonary arterial hypertension. Circulation. 2015;132:932–943. doi: 10.1161/CIRCULATIONAHA.115.016382 [DOI] [PubMed] [Google Scholar]
- 24.Schober A, Nazari-Jahantigh M, Wei Y, Bidzhekov K, Gremse F, Grommes J, Megens RT, Heyll K, Noels H, Hristov M, et al. MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nat Med. 2014;20:368–376. doi: 10.1038/nm.3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang B, Nguyen LXT, Li L, Zhao D, Kumar B, Wu H, Lin A, Pellicano F, Hopcroft L, Su Y-L, et al. Bone marrow niche trafficking of mir-126 controls the self-renewal of leukemia stem cells in chronic myelogenous leukemia. Nat Med. 2018;24:450–462. doi: 10.1038/nm.4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan H, Yang W, Zhou F, Li X, Pan Q, Shen Z, Han G, Newell-Fugate A, Tian Y, Majeti R, et al. Estrogen improves insulin sensitivity and suppresses gluconeogenesis via the transcription factor foxo1. Diabetes. 2019;68:291–304. doi: 10.2337/db18-0638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capone C, Anrather J, Milner TA, Iadecola C. Estrous cycle-dependent neurovascular dysfunction induced by angiotensin II in the mouse neocortex. Hypertension. 2009;54:302–307. doi: 10.1161/HYPERTENSIONAHA.109.133249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aspelund A, Antila S, Proulx ST, Karlsen TV, Karaman S, Detmar M, Wiig H, Alitalo K. A dural lymphatic vascular system that drains brain interstitial fluid and macromolecules. J Exp Med. 2015;212:991–999. doi: 10.1084/jem.20142290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nurmi H, Saharinen P, Zarkada G, Zheng W, Robciuc MR, Alitalo K. VEGF-C is required for intestinal lymphatic vessel maintenance and lipid absorption. EMBO Mol Med. 2015;7:1418–1425. doi: 10.15252/emmm.201505731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y, García-Verdugo JM, Soriano-Navarro M, Srinivasan RS, Scallan JP, Singh MK, Epstein JA, Oliver G. Lymphatic endothelial progenitors bud from the cardinal vein and intersomitic vessels in mammalian embryos. Blood. 2012;120:2340–2348. doi: 10.1182/blood-2012-05-428607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gustavsson C, Agardh CD, Zetterqvist AV, Nilsson J, Agardh E, Gomez MF. Vascular cellular adhesion molecule-1 (VCAM-1) expression in mice retinal vessels is affected by both hyperglycemia and hyperlipidemia. PLoS One. 2010;5:e12699. doi: 10.1371/journal.pone.0012699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gale NW, Baluk P, Pan L, Kwan M, Holash J, DeChiara TM, McDonald DM, Yancopoulos GD. Ephrin-B2 selectively marks arterial vessels and neovascularization sites in the adult, with expression in both endothelial and smooth-muscle cells. Dev Biol. 2001;230:151–160. doi: 10.1006/dbio.2000.0112 [DOI] [PubMed] [Google Scholar]
- 33.Krebs LT, Xue Y, Norton CR, Shutter JR, Maguire M, Sundberg JP, Gallahan D, Closson V, Kitajewski J, Callahan R, et al. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000;14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 34.Fang JS, Coon BG, Gillis N, Chen Z, Qiu J, Chittenden TW, Burt JM, Schwartz MA, Hirschi KK. Shear-induced Notch-Cx37-p27 axis arrests endothelial cell cycle to enable arterial specification. Nat Commun. 2017;8:2149. doi: 10.1038/s41467-017-01742-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerety SS, Wang HU, Chen ZF, Anderson DJ. Symmetrical mutant phenotypes of the receptor EphB4 and its specific transmembrane ligand ephrin-B2 in cardiovascular development. Mol Cell. 1999;4:403–414. doi: 10.1016/s1097-2765(00)80342-1 [DOI] [PubMed] [Google Scholar]
- 36.You LR, Lin FJ, Lee CT, DeMayo FJ, Tsai MJ, Tsai SY. Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature. 2005;435:98–104. doi: 10.1038/nature03511 [DOI] [PubMed] [Google Scholar]
- 37.Su T, Stanley G, Sinha R, D’Amato G, Das S, Rhee S, Chang AH, Poduri A, Raftrey B, Dinh TT, et al. Single-cell analysis of early progenitor cells that build coronary arteries. Nature. 2018;559:356–362. doi: 10.1038/s41586-018-0288-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren X, Ustiyan V, Pradhan A, Cai Y, Havrilak JA, Bolte CS, Shannon JM, Kalin TV, Kalinichenko VV. FOXF1 transcription factor is required for formation of embryonic vasculature by regulating VEGF signaling in endothelial cells. Circ Res. 2014;115:709–720. doi: 10.1161/CIRCRESAHA.115.304382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, Pu W, Li G, Huang X, He L, Tian X, Liu Q, Zhang L, Wu SM, Sucov HM, et al. Endocardium minimally contributes to coronary endothelium in the embryonic ventricular free walls. Circ Res. 2016;118:1880–1893. doi: 10.1161/CIRCRESAHA.116.308749 [DOI] [PubMed] [Google Scholar]
- 40.Cornford EM, Hyman S, Swartz BE. The human brain glut1 glucose-transporter - ultrastructural-localization to the blood-brain-barrier endothelia. J Cereb Blood Flow Metab. 1994;14:106–112. doi: 10.1038/jcbfm.1994.15 [DOI] [PubMed] [Google Scholar]
- 41.Géraud C, Koch PS, Zierow J, Klapproth K, Busch K, Olsavszky V, Leibing T, Demory A, Ulbrich F, Diett M, et al. GATA4-dependent organ-specific endothelial differentiation controls liver development and embryonic hematopoiesis. J Clin Invest. 2017;127:1099–1114. doi: 10.1172/JCI90086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Politz O, Gratchev A, McCourt PA, Schledzewski K, Guillot P, Johansson S, Svineng G, Franke P, Kannicht C, Kzhyshkowska J, et al. Stabilin-1 and -2 constitute a novel family of fasciclin-like hyaluronan receptor homologues. Biochem J. 2002;362(Pt 1):155–164. doi: 10.1042/0264-6021:3620155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perez-Pinera P, Alcantara S, Dimitrov T, Vega JA, Deuel TF. Pleiotrophin disrupts calcium-dependent homophilic cell-cell adhesion and initiates an epithelial-mesenchymal transition. Proc Natl Acad Sci USA. 2006;103:17795–17800. doi: 10.1073/pnas.0607299103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L, Uezumi A, Kaji T, Tsujikawa K, Andersen DC, Jensen CH, Fukada SI. Expression and functional analyses of Dlk1 in muscle stem cells and mesenchymal progenitors during muscle regeneration. Int J Mol Sci. 2019;20:E3269. doi: 10.3390/ijms20133269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaum N, Karkanias J, Neff NF, May AP, Quake SR, Wyss-Coray T, Darmanis S, Batson J, Botvinnik O, Chen MB, et al. Single-cell transcriptomics of 20 mouse organs creates a tabula muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weisend CM, Kundert JA, Suvorova ES, Prigge JR, Schmidt EE. Cre activity in fetal albCre mouse hepatocytes: utility for developmental studies. Genesis. 2009;47:789–792. doi: 10.1002/dvg.20568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lomas DA, Mahadeva R. Alpha1-antitrypsin polymerization and the serpinopathies: pathobiology and prospects for therapy. J Clin Invest. 2002;110:1585–1590. doi: 10.1172/JCI16782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bondjers C, He L, Takemoto M, Norlin J, Asker N, Hellström M, Lindahl P, Betsholtz C. Microarray analysis of blood microvessels from PDGF-B and PDGF-Rbeta mutant mice identifies novel markers for brain pericytes. FASEB J. 2006;20:1703–1705. doi: 10.1096/fj.05-4944fje [DOI] [PubMed] [Google Scholar]
- 49.Nehls V, Drenckhahn D. The versatility of microvascular pericytes: from mesenchyme to smooth muscle? Histochemistry. 1993;99:1–12. doi: 10.1007/BF00268014 [DOI] [PubMed] [Google Scholar]
- 50.Williams MJ, Sugatani T, Agapova OA, Fang Y, Gaut JP, Faugere MC, Malluche HH, Hruska KA. The activin receptor is stimulated in the skeleton, vasculature, heart, and kidney during chronic kidney disease. Kidney Int. 2018;93:147–158. doi: 10.1016/j.kint.2017.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friedrich J, Leonard E, Blazer-Yose B, Basile D. Endothelial to mesenchymal transition and circulating cells contribute to fibroblast formation following renal ischemia reperfusion. Faseb J. 2008;22:22–23. doi:10.1096/fasebj.22.1supplement.942.6 [Google Scholar]
- 52.Zou J, Li WQ, Li Q, Li XQ, Zhang JT, Liu GQ, Chen J, Qiu XX, Tian FJ, Wang ZZ, et al. Two functional microRNA-126s repress a novel target gene p21-activated kinase 1 to regulate vascular integrity in zebrafish. Circ Res. 2011;108:201–209. doi: 10.1161/CIRCRESAHA.110.225045 [DOI] [PubMed] [Google Scholar]
- 53.Chen J, Zhu RF, Li FF, Liang YL, Wang C, Qin YW, Huang S, Zhao XX, Jing Q. MicroRNA-126a directs lymphangiogenesis through interacting with chemokine and Flt4 signaling in zebrafish. Arterioscler Thromb Vasc Biol. 2016;36:2381–2393. doi: 10.1161/ATVBAHA.116.308120 [DOI] [PubMed] [Google Scholar]
- 54.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci U S A. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kontarakis Z, Rossi A, Ramas S, Dellinger MT, Stainier DYR. Mir-126 is a conserved modulator of lymphatic development. Dev Biol. 2018;437:120–130. doi: 10.1016/j.ydbio.2018.03.006 [DOI] [PubMed] [Google Scholar]
- 56.Kuhnert F, Mancuso MR, Hampton J, Stankunas K, Asano T, Chen CZ, Kuo CJ. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development. 2008;135:3989–3993. doi: 10.1242/dev.029736 [DOI] [PubMed] [Google Scholar]
- 57.Taylor AC, Seltz LM, Yates PA, Peirce SM. Chronic whole-body hypoxia induces intussusceptive angiogenesis and microvascular remodeling in the mouse retina. Microvasc Res. 2010;79:93–101. doi: 10.1016/j.mvr.2010.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santovito D, Egea V, Bidzhekov K, Natarelli L, Mourão A, Blanchet X, Wichapong K, Aslani M, Brunßen C, Horckmans M, et al. Noncanonical inhibition of caspase-3 by a nuclear microRNA confers endothelial protection by autophagy in atherosclerosis. Sci Transl Med. 2020;12:eaaz2294. doi: 10.1126/scitranslmed.aaz2294 [DOI] [PubMed] [Google Scholar]
- 59.Bordi M, De Cegli R, Testa B, Nixon RA, Ballabio A, Cecconi F. A gene toolbox for monitoring autophagy transcription. Cell Death Dis. 2021;12:1044. doi: 10.1038/s41419-021-04121-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107:810–817. doi: 10.1161/CIRCRESAHA.110.226357 [DOI] [PubMed] [Google Scholar]
- 61.Zhang T, Lv C, Li L, Chen S, Liu S, Wang C, Su B. Plasma miR-126 is a potential biomarker for early prediction of type 2 diabetes mellitus in susceptible individuals. Biomed Res Int. 2013;2013:761617. doi: 10.1155/2013/761617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pordzik J, Jakubik D, Jarosz-Popek J, Wicik Z, Eyileten C, De Rosa S, Indolfi C, Siller-Matula JM, Czajka P, Postula M. Significance of circulating microRNAs in diabetes mellitus type 2 and platelet reactivity: bioinformatic analysis and review. Cardiovasc Diabetol. 2019;18:113. doi: 10.1186/s12933-019-0918-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banerjee S, Bagheri M, Sandfort V, Ahlman MA, Malayeri AA, Bluemke DA, Yao J, Grayson PC. Vascular calcification in patients with large-vessel vasculitis compared to patients with hyperlipidemia. Semin Arthritis Rheum. 2019;48:1068–1073. doi: 10.1016/j.semarthrit.2018.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fu Y, Gao C, Liang Y, Wang M, Huang Y, Ma W, Li T, Jia Y, Yu F, Zhu W, et al. Shift of macrophage phenotype due to cartilage oligomeric matrix protein deficiency drives atherosclerotic calcification. Circ Res. 2016;119:261–276. doi: 10.1161/CIRCRESAHA.115.308021 [DOI] [PubMed] [Google Scholar]
- 65.Speer MY, Yang HY, Brabb T, Leaf E, Look A, Lin WL, Frutkin A, Dichek D, Giachelli CM. Smooth muscle cells give rise to osteochondrogenic precursors and chondrocytes in calcifying arteries. Circ Res. 2009;104:733–741. doi: 10.1161/CIRCRESAHA.108.183053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Collett GD, Canfield AE. Angiogenesis and pericytes in the initiation of ectopic calcification. Circ Res. 2005;96:930–938. doi: 10.1161/01.RES.0000163634.51301.0d [DOI] [PubMed] [Google Scholar]
- 67.Wirrig EE, Yutzey KE. Conserved transcriptional regulatory mechanisms in aortic valve development and disease. Arterioscler Thromb Vasc Biol. 2014;34:737–741. doi: 10.1161/ATVBAHA.113.302071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.An Y, Varma VR, Varma S, Casanova R, Dammer E, Pletnikova O, Chia CW, Egan JM, Ferrucci L, Troncoso J, et al. Evidence for brain glucose dysregulation in Alzheimer’s disease. Alzheimers Dement. 2018;14:318–329. doi: 10.1016/j.jalz.2017.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Olson AL, Pessin JE. Structure, function, and regulation of the mammalian facilitative glucose transporter gene family. Annu Rev Nutr. 1996;16:235–256. doi: 10.1146/annurev.nu.16.070196.001315 [DOI] [PubMed] [Google Scholar]
- 70.Schmidt S, Richter M, Montag D, Sartorius T, Gawlik V, Hennige AM, Scherneck S, Himmelbauer H, Lutz SZ, Augustin R, et al. Neuronal functions, feeding behavior, and energy balance in Slc2a3+/- mice. Am J Physiol Endocrinol Metab. 2008;295:E1084–E1094. doi: 10.1152/ajpendo.90491.2008 [DOI] [PubMed] [Google Scholar]
- 71.Tang M, Gao G, Rueda CB, Yu H, Thibodeaux DN, Awano T, Engelstad KM, Sanchez-Quintero MJ, Yang H, Li F, et al. Brain microvasculature defects and Glut1 deficiency syndrome averted by early repletion of the glucose transporter-1 protein. Nat Commun. 2017;8:14152. doi: 10.1038/ncomms14152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heilig CW, Concepcion LA, Riser BL, Freytag SO, Zhu M, Cortes P. Overexpression of glucose transporters in rat mesangial cells cultured in a normal glucose milieu mimics the diabetic phenotype. J Clin Invest. 1995;96:1802–1814. doi: 10.1172/JCI118226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ben-Aicha S, Escate R, Casaní L, Padró T, Peña E, Arderiu G, Mendieta G, Badimón L, Vilahur G. High-density lipoprotein remodelled in hypercholesterolaemic blood induce epigenetically driven down-regulation of endothelial HIF-1α expression in a preclinical animal model. Cardiovasc Res. 2020;116:1288–1299. doi: 10.1093/cvr/cvz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dumont DJ, Yamaguchi TP, Conlon RA, Rossant J, Breitman ML. tek, a novel tyrosine kinase gene located on mouse chromosome 4, is expressed in endothelial cells and their presumptive precursors. Oncogene. 1992;7:1471–1480. [PubMed] [Google Scholar]
- 75.Dumont DJ, Fong GH, Puri MC, Gradwohl G, Alitalo K, Breitman ML. Vascularization of the mouse embryo: a study of flk-1, tek, tie, and vascular endothelial growth factor expression during development. Dev Dyn. 1995;203:80–92. doi: 10.1002/aja.1002030109 [DOI] [PubMed] [Google Scholar]
- 76.Payne S, De Val S, Neal A. Endothelial-specific cre mouse models. Arterioscler Thromb Vasc Biol. 2018;38:2550–2561. doi: 10.1161/ATVBAHA.118.309669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen C, Pore N, Behrooz A, Ismail-Beigi F, Maity A. Regulation of glut1 mRNA by hypoxia-inducible factor-1. Interaction between H-ras and hypoxia. J Biol Chem. 2001;276:9519–9525. doi: 10.1074/jbc.M010144200 [DOI] [PubMed] [Google Scholar]
- 78.Zeisberg EM, Tarnavski O, Zeisberg M, Dorfman AL, McMullen JR, Gustafsson E, Chandraker A, Yuan X, Pu WT, Roberts AB, et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat Med. 2007;13:952–961. doi: 10.1038/nm1613 [DOI] [PubMed] [Google Scholar]
- 79.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zeng A, Li H, Guo L, Gao X, McKinney S, Wang Y, Yu Z, Park Z, Semerad C, Ross E, Cheng L-C, et al. Prospectively isolated tetraspanin(+) neoblasts are adult pluripotent stem cells underlying planaria regeneration. Cell. 2018;173:1593–1608 e1520. doi: 10.1016/j.cell.2018.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106 [DOI] [PubMed] [Google Scholar]
- 82.Tual-Chalot S, Allinson KR, Fruttiger M, Arthur HM. Whole mount immunofluorescent staining of the neonatal mouse retina to investigate angiogenesis in vivo. J Vis Exp. 2013;9:e50546. doi: 10.3791/50546 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data (including scRNA-seq data and R scripts for single-cell data analysis) and methods supporting the findings of this study are available in the Supplemental Methods or from the corresponding authors on reasonable request.
An expanded Methods section is available in the Supplemental Methods. All animal experiments were performed in accordance with the institutional guidelines of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. We used male mice because this minimized estrogen-dependent fluctuations in response to angiogenesis and glucose metabolism.26,27