Supplemental Digital Content is available in the text.
Keywords: arrhythmias, cardiac; epigenesis, genetic; myocytes, cardiac; sequence analysis, RNA; T-box transcription factor 5; transcription factors
Background:
The pathogenic missense variant p.G125R in TBX5 (T-box transcription factor 5) causes Holt–Oram syndrome (also known as hand–heart syndrome) and early onset of atrial fibrillation. Revealing how an altered key developmental transcription factor modulates cardiac physiology in vivo will provide unique insights into the mechanisms underlying atrial fibrillation in these patients.
Methods:
We analyzed ECGs of an extended family pedigree of Holt–Oram syndrome patients. Next, we introduced the TBX5-p.G125R variant in the mouse genome (Tbx5G125R) and performed electrophysiologic analyses (ECG, optical mapping, patch clamp, intracellular calcium measurements), transcriptomics (single-nuclei and tissue RNA sequencing), and epigenetic profiling (assay for transposase-accessible chromatin using sequencing, H3K27ac [histone H3 lysine 27 acetylation] CUT&RUN [cleavage under targets and release under nuclease sequencing]).
Results:
We discovered high incidence of atrial extra systoles and atrioventricular conduction disturbances in Holt–Oram syndrome patients. Tbx5G125R/+ mice were morphologically unaffected and displayed variable RR intervals, atrial extra systoles, and susceptibility to atrial fibrillation, reminiscent of TBX5-p.G125R patients. Atrial conduction velocity was not affected but systolic and diastolic intracellular calcium concentrations were decreased and action potentials were prolonged in isolated cardiomyocytes of Tbx5G125R/+ mice compared with controls. Transcriptional profiling of atria revealed the most profound transcriptional changes in cardiomyocytes versus other cell types, and identified over a thousand coding and noncoding transcripts that were differentially expressed. Epigenetic profiling uncovered thousands of TBX5-p.G125R-sensitive, putative regulatory elements (including enhancers) that gained accessibility in atrial cardiomyocytes. The majority of sites with increased accessibility were occupied by Tbx5. The small group of sites with reduced accessibility was enriched for DNA-binding motifs of members of the SP (specificity protein) and KLF (Krüppel-like factor) families of transcription factors. These data show that Tbx5-p.G125R induces changes in regulatory element activity, alters transcriptional regulation, and changes cardiomyocyte behavior, possibly caused by altered DNA binding and cooperativity properties.
Conclusions:
Our data reveal that a disease-causing missense variant in TBX5 induces profound changes in the atrial transcriptional regulatory network and epigenetic state in vivo, leading to arrhythmia reminiscent of those seen in human TBX5-p.G125R variant carriers.
Clinical Perspective.
What Is New?
A pathogenic variant in the fifth exon of TBX5 ([T-box transcription factor 5] p.G125R) found in a Dutch, atypical Holt–Oram syndrome family with early onset atrial fibrillation was modeled in mice.
This is the first human pathogenic variant based on a patient family in this key cardiac transcription factor explored in vivo.
We identify widespread electrophysiologic, transcriptional, and epigenetic changes (including coding and noncoding RNA, chromatin accessibility, and H3K27ac [histone H3 lysine 27 acetylation] association) in the atria of Tbx5-p.G125R heterozygous mice distinct from the changes in atria of Tbx5 insufficiency models.
What Are the Clinical Implications?
The characterization of the Tbx5 p.G125R mouse model indicates that a patient-specific pathogenic variant in TBX5 induces changes in regulatory element activity, altered balance in the regulatory network of atrial cardiomyocytes, and clinically relevant changes in cardiomyocyte function.
These findings shed light on the target and off-target genes of Tbx5-p.G125R because of its altered properties, which are at the basis of distinct electrophysiologic changes observed in a subset of Holt–Oram syndrome patients with atrial fibrillation.
This work may provide insight into the epigenetic changes and transcriptional underpinning of arrhythmia in the general population caused by small increases in TBX5-expression caused by common variants predisposing to atrial fibrillation.
Pathogenic variants in T-box transcription factor (TF) TBX5 (T-box transcription factor 5) have been linked to Holt–Oram syndrome (HOS), which is typically characterized by varying degrees of limb and cardiac malformations and conduction defects (OMIM [Online Mendelian Inheritance in Man] #142900).1–3 Tbx5 is dose-dependently required for heart development and specification and function of the conduction system, and determines the working myocardial phenotype of the atrium.3–8 Tbx5 has been put forward as key regulator of ion-handling protein-encoding gene expression and rhythm control in vivo.9–13 Genome-wide association studies uncovered several variants near TBX5 associated with atrial fibrillation (AF). Moreover, transcriptome-wide analysis of human heart tissue identified an association of increased TBX5 expression with AF risk.14
A pathogenic variant in the fifth exon of TBX5 causing a glycine to arginine substitution (c.373G>A; p.G125R) in the T-box domain was found in a Dutch, atypical HOS family.15 In this family, affected members show HOS symptoms such as mild skeletal abnormalities and septal defects, as well as atypical early onset paroxysmal AF. The function of missense variants in this key TF and the impact on gene regulation in the context of heart function in vivo are poorly understood. Therefore, we engineered the p.G125R variant in the highly conserved T-box of mouse Tbx5, and used this model to gain insight into the molecular mechanisms underlying structural and electrophysiological changes predisposing to AF in TBX5 missense variant carriers.
Methods
Detailed Methods are provided in the Supplemental Material.
Data Availability
Whole-tissue RNA-sequencing ([RNA-seq] left and right atrial), cardiomyocyte (CM) nucleus H3K27ac (histone H3 lysine 27 acetylation) CUT&RUN (cleavage under targets and release under nuclease), CM nucleus assay for transposase-accessible chromatin using sequencing (ATAC-seq), and single-nuclei RNA-seq are available on Gene Expression Omnibus (GSE167082). All other data and supporting materials have been provided with the published article.
Informed Consent Statement
The study was conducted according to the guidelines of the Declaration of Helsinki. Written informed consent was obtained from the families presented in the paper.
Animal Care
Animal care and experiments conform to the Directive 2010/63/EU of the European Parliament. All animal work was approved by the Animal Experimental Committee of the Amsterdam University Medical Centers (location Academic Medical Center, Amsterdam, the Netherlands), was performed in compliance with the Dutch government guidelines, and was approved by the Central Committee Animal Experiments.
Statistics
A detailed description of the statistics can be found in the Supplemental Material which also describes the specific statistical tests used to analyze large data sets. Results of statistical analysis are given in the text or the figure legends.
Results
Mouse Model Recapitulates Atrial Arrhythmia and ECG Abnormalities in TBX5-G125R Patients
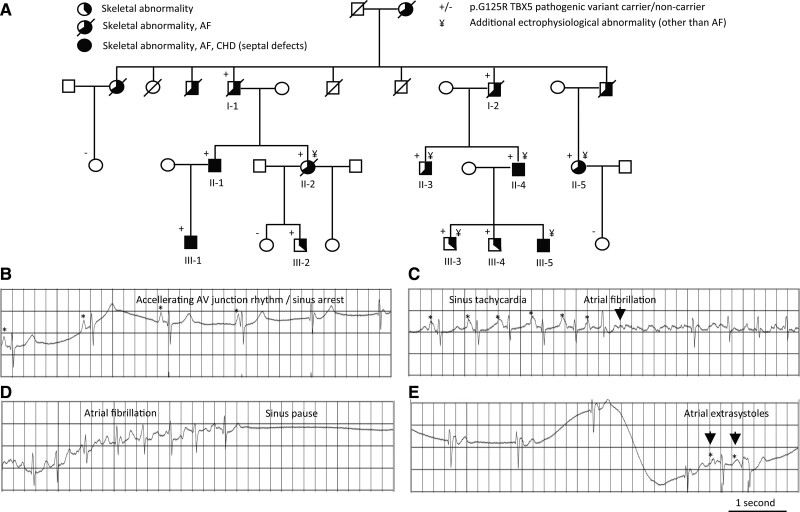
Heterozygous TBX5-p.G125R carriers show atypical HOS, including cardiac septal defects, mild skeletal abnormalities, and early onset AF (Figure 1A).15 Here, we reanalyzed in detail the available ECG traces (as obtained by 24-h Holter or 12-lead ECG) of the patients previously analyzed.15 We found a range of additional abnormalities—including irregular RR interval, right bundle branch block, atrioventricular (AV) junctional escape, AV junctional rhythm, sinus arrest, left ventricular noncompaction, atrial extras, and sick sinus syndrome—that were not checked for previously (Figure 1B–1E; Table S1). This shows that these patients suffer from other types of supraventricular arrhythmia that indicate dysfunctionality of the atrial muscle, including the sinoatrial and AV nodes.
Figure 1.
Pedigree and electrophysiological abnormalities found in a family with atypical Holt–Oram syndrome with early onset AF. A, Pedigree showing the presence of skeletal abnormalities, congenital heart disease (ie, septal defects) and AF in TBX5p.G125R carriers. B through E, Example ECG traces of electrophysiological abnormalities found within 1 individual with the TBX5p.G125R variant (Patient II-5; Table S1). AF indicates atrial fibrillation; AV, atrioventricular; CHD, congenital heart disease; and TBX5, T-box transcription factor 5.
To investigate how the p.G125R pathogenic variant in TBX5 affects heart function, we generated a mouse model with this point mutation in Tbx5 using CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats–CRISPR-associated protein 9). Interbreeding of Tbx5G125R/+ animals showed that from embryonic day (E) 12.5 to E16.5, homozygous Tbx5G125R/G125R fetuses were necrotic or dead (Table S2), revealing that homozygosity of the mutation is incompatible with life. Tbx5G125R/G125R fetuses showed ventricular and atrial septal defects (E14.5–E16.5), as well as enlarged right atria at E16.5 (Figure S1), which were also present in patients heterozygous for this pathogenic variant (Figure 1A; Table S1). We did not observe differences in CM proliferation rates in embryonic (E12.5) right atria or left ventricles between different genotypes (Figure S2), nor differences in the heart weight–tibia length ratio between control (n=6) and Tbx5G125R/+ mice (n=10) at 20 days after birth (Table S3).
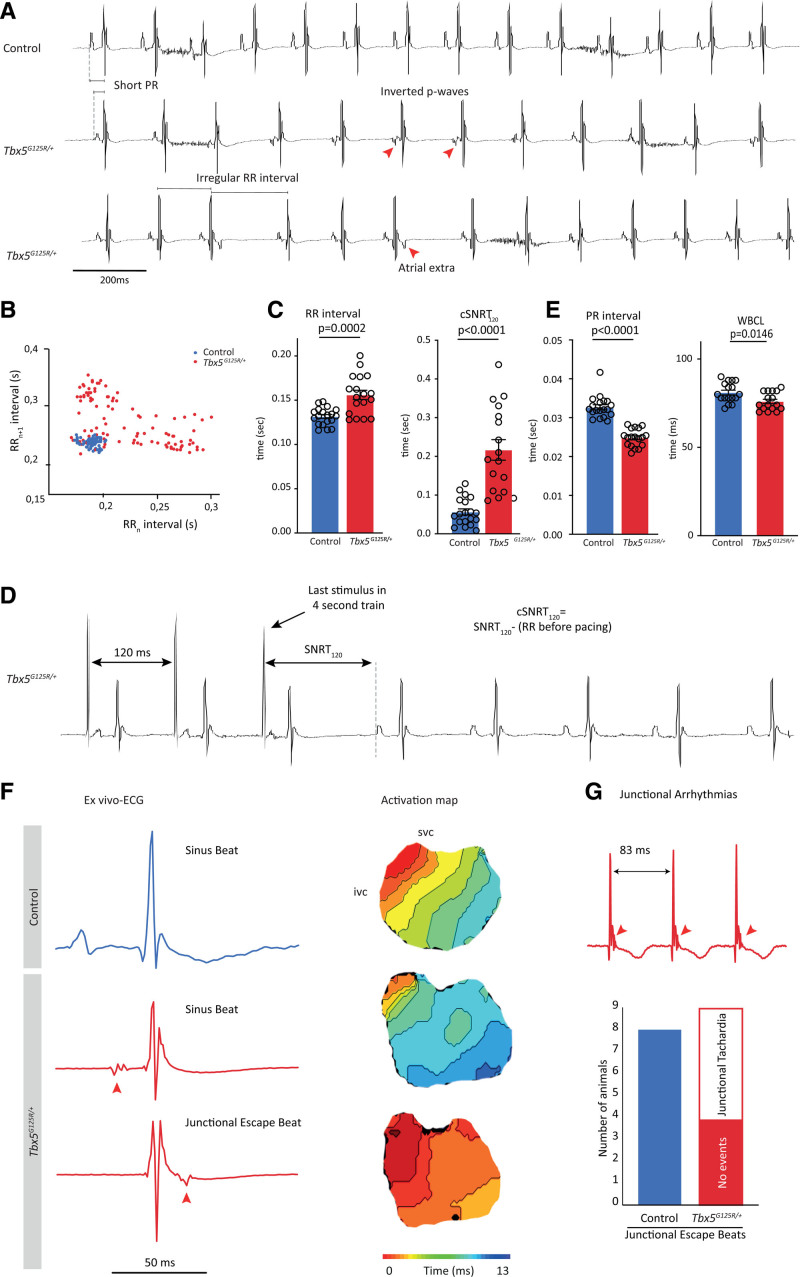
To determine the impact of Tbx5-p.G125R on electrophysiologic function, we recorded ECGs from male and female control mice (n=19) and Tbx5G125R/+ mice (n=18; Figure 2A). While QRS duration was unchanged (data not shown), the Tbx5G125R/+ mice showed longer and more variable RR intervals than controls (Figure 2B and 2C). In addition, the sinus node recovery time measured during transesophageal pacing was prolonged in Tbx5G125R/+ mice (Figure 2C and 2D). This presumably sinus nodal phenotype of the Tbx5G125R/+ mice was accompanied by dysfunction of the AV junction as evidenced by shorter PR intervals (difference between means ± SEM, 0.007 ± 0.001 s [males]; 0.011 ± 0.002 s [females]) and Wenckebach cycle lengths compared with controls (Figure 2E). Moreover, we found atrial ectopic beats with inverted P-waves in 6 of 18 animals (Figure 2A), which explains the variable RR intervals observed in Tbx5G125R/+ mice. To investigate the origin of these ectopic beats we recorded optical action potentials from isolated Langendorff-perfused hearts (Figure S3A). Hearts isolated from Tbx5G125R/+ mice also showed longer and more variable RR intervals, as well as shorter PR intervals, than controls (Figure S3B and S3C). In addition, these experiments show that the atrial ectopic beats occurred during or after the QRS complex, and occurred in all cases in response to a premature ventricular beat (Figure 2F). The morphology of the QRS complexes of these premature beats were similar, indicating that the origin was in the AV junction and that it caused retrograde activation of the atria. The junctional escape beats were present in 100% of Tbx5G125R/+ mice and turned into junctional tachycardia in 60% of the cases (Figure 2G).
Figure 2.
Transesophageal burst pacing (TEBP) of Tbx5G125R/+ mice shows atrial premature activation, shortened PR interval and ventricular premature activation. A, Typical example ECG traces of controls (n=19) and Tbx5G125R/+ mice (n=18). B, Scatterplot illustrating the beat-to-beat variability in RR interval of 1 control (blue) and 1 Tbx5G125R/+ mouse (red). C, Significant changes were observed in RR interval, RRsd (heart rate variability RRsd=SD of RRn–RRn+1), and SNRT of Tbx5G125R/+ mice (red). Genotypes were compared using Mann–Whitney U test; P values are given in the graphs. D, Typical example trace showing the analysis of cSNRT120. E, PR interval and WBCL are significantly shortened in Tbx5G125R/+ mice. Genotypes were compared using Mann–Whitney U test. F, Ex vivo typical example ECGs and corresponding activation maps (optical mapping) show atrial ectopic beats occurring during or after the QRS complex in response to a premature ventricular beat that originates in the atrioventricular junction in Tbx5G125R/+ mice. Red arrows indicate inverted premature beat (P-top). G, Junctional escape beats, not observed in 8 control mice, turn into junctional tachycardia in 55% (5/9) Tbx5G125R/+ mice. cSNRT120 indicates cyclic sinus node recovery time after 120-ms interval stimulation; ivc, inferior vena cava; SNRT, sinus node recovery time; svc, superior vena cava; TBX5, T-box transcription factor 5; and WBCL, Wenckebach cycle length.
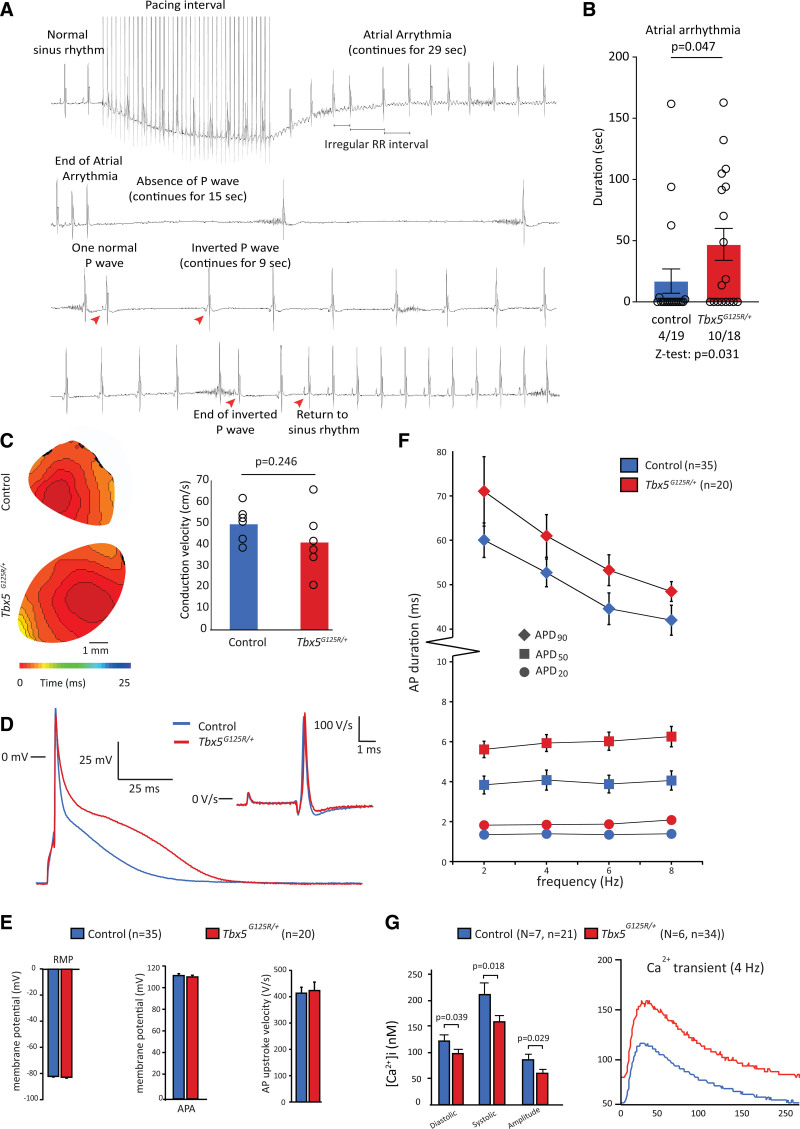
To assess whether Tbx5G125R/+ mice were susceptible to atrial arrhythmias, we performed programmed atrial stimulation in vivo using a transesophageal catheter (Figure 3A). We were able to induce atrial arrhythmias in 10 of 19 Tbx5G125R/+ mice and 4 of 18 control mice (duration of AA >10 s; Figure 3B). We optically measured conduction velocity in the atrium of isolated hearts, which was not different between genotypes (Figure 3C; Table S4). Similarly, recorded action potentials in isolated atrial CMs (Figure 3D) revealed no difference in resting membrane potential, action potential amplitude, or upstroke velocity (Figure 3E). Action potential duration, on the other hand, was longer in atrial myocytes from Tbx5G125R/+ at all frequencies tested (2, 4, 6, and 8 Hz) and at all action potential repolarization phases (ie, action potential duration [APD] measured at 20%, 50%, and 90% of repolarization; Figure 3F). Intracellular Ca2+ measurements revealed that Ca2+ transients had lower amplitudes in Tbx5G125R/+ mice, as well as decreased diastolic and systolic Ca2+ concentrations, while the decay of the Ca2+ transient was unaffected (Figure 3G; Table S5).
Figure 3.
Conduction velocity, cellular electrophysiology, and intracellular Ca2+ measurements of control and Tbx5G125R/+ mice. A, Typical ECG trace of atrial arrhythmia in Tbx5G125R/+ mice. B, Transesophageal burst pacing shows more Tbx5G125R/+ mice with atrial arrhythmia >1 s compared with controls. Atrial arrhythmia occurred significantly more often in Tbx5G125R/+ mice (Z-test) and, when present, atrial arrhythmia duration was longer in these mice (Mann–Whitney U test). C, Conduction velocity does not differ between control and Tbx5G125R/+ mice (Mann–Whitney U test). D, Typical action potentials at 6 Hz of isolated cardiomyocytes. E, No significant between-genotype differences were observed for average resting membrane potential, action potential amplitude, and action potential upstroke velocity using an independent sample t test. F, APD at 20%, 50%, and 90% of repolarization (APD20, APD50, APD90, respectively) was tested using 3-way ANOVA, showing significant interactions of genotype and frequency with APD group, followed by 2-way ANOVA per repolarization group. These 2-way ANOVAs showed that APD differs between genotypes independently of frequency in APD20 (frequency: P=0.302, genotype: P < 0.001) and APD50 (frequency: P=0.459; genotype: P < 0.001). In APD90, there is both a frequency (P < 0.001) and a genotype (P < 0.001) effect but this genotype effect is independent of the frequency (interaction: P=0.766). G, At a frequency of 8 Hz, significantly lower Ca2+ transient amplitudes, as well as a decreased diastolic and systolic Ca2+ concentrations, were observed in Tbx5G125R/+ mice (Mann–Whitney U test; performed on cells per group). On the right, typical Ca2+ transient amplitudes example measured at 4 Hz are shown. APD indicates action potential duration; and Tbx5, T-box transcription factor 5.
Deregulated Gene Expression in Atrial CMs of Tbx5G125R/+ Mice
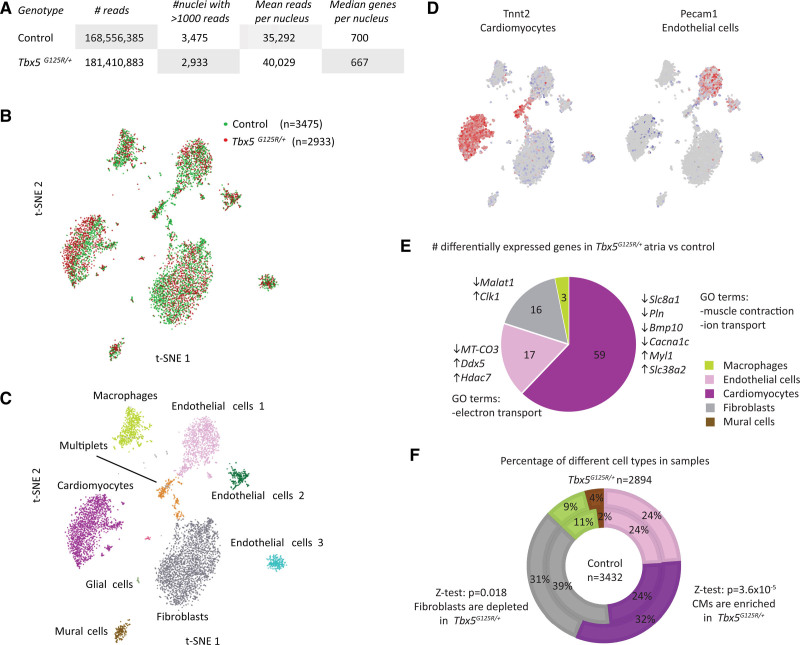
To gain insight into the cell types affected by Tbx5-p.G125R, we performed single-nucleus RNA-seq on right atrial tissue. Profiles from 3475 wild-type nuclei and 2933 Tbx5G125R/+ nuclei were obtained (Figure 4A). T-distributed stochastic neighbor embedding was used to identify 9 clusters (Figure 4B), corresponding to cardiac cell types based on marker gene expression (Figure 4C and 4D; Figure S4). Only the CM cluster showed some degree of segregation between genotypes (Figure 4B). We determined differential expression within each cluster by Kruskal–Wallis test (Bonferroni corrected P < 0.05). CMs showed 59 genes that were significantly differentially expressed; endothelial cells showed 17 genes and fibroblasts showed 16 (Figure 4E; Table S6). Analyses of the Gene Ontology terms for the differentially expressed genes in CMs showed mostly terms involving muscle contraction regulation and ion transport (Table S7). The fraction of CMs and fibroblasts differed between genotypes (CMs enriched in Tbx5G125R/+ [936/2894] vs controls [828/3432]; Z-test, P=3.68×10-5; fibroblasts depleted in Tbx5G125R/+ [255/2894] vs controls [363/3432]; Z-test, P=0.018; Figure 4F). Immunofluorescence analysis revealed ratios of Nkx2-5+ (NK2 homeobox 5) CM nuclei over non-CM nuclei were significantly higher in atria of 8-month-old Tbx5G125R/+ mice compared with controls (P=0.014; Figure S5A). Picrosirius red–staining sections of 8-month-old control (n=4) and Tbx5G125R/+ (n=5) atria revealed a trend toward a smaller fibrotic area in the atria of Tbx5G125R/+ mice compared with controls; however, it was not significant (Figure S5B and 5C). Together, these findings suggest that the Tbx5G125R/+ atria do not show elevated fibrosis even though fibrosis has been associated with AF.
Figure 4.
Single-nucleus transcriptomics of control and Tbx5G125R/+ atria. A, Number of reads and nuclei, mean reads, and mean genes per nucleus for control and Tbx5G125R/+ sample. B, tSNE map of control and Tbx5G125R/+ clusters. C, tSNE map showing different cell-type clusters. D, tSNE maps showing expression of different marker genes. E, Distribution of differential expression in different cell types (upregulated in Tbx5G125R/+ vs control ↑, and downregulated ↓). Significance of differential gene expression in single-nucleus RNA sequencing was determined by Mann–Whitney U test in the R2 genomics analysis and visualization platform (http://r2.amc.nl). F, Contribution of different cell types to control and Tbx5G125R/+ samples showing increase in CMs and decrease in fibroblast contribution in Tbx5G125R/+ (Z-test was performed to test the significance of the difference between genotypes of the fractions of cell types). Bmp10 indicates bone morphogenetic protein 10; CM, cardiomyocyte; Clk1, CDC Like kinase 1; Cacna1c, calcium voltage-gated channel subunit α1C; Ddx5, DEAD-Box Helicase 5; GO, Gene Ontology; Hdac7, histone deacetylase 7; Malat1, Metastasis Associated Lung Adenocarcinoma Transcript 1; Myl1, Myosin Light Chain 1; MT-CO3, XXX; Pecam1, Platelet And Endothelial Cell Adhesion Molecule 1; Pln, phospholamban; Slc8a1; solute carrier family 8 member A1; Slc38a2, solute carrier family 38 member A2; t-SNE, T-distributed stochastic neighbor embedding; Tbx5, T-box transcription factor 5; and Tnnt2, Troponin T2, Cardiac Type.
To determine at greater depth the transcriptional responses to heterozygosity of Tbx5-p.G125R, RNA-seq was performed on whole right atria of adult female control (n=5) and Tbx5G125R/+ (n=5) animals (Figure S6A). The point mutation may affect mRNA levels of Tbx5G125R (ie, nonsense-mediated decay). Using RNA-seq reads and Varscan,16 we quantified the number of Tbx5 transcripts tags containing the wild-type allele (G) and the number containing the mutant allele (C). In control animals, the wild-type allele had a frequency of 100%. In Tbx5G125R/+ animals, the variant allele had a frequency of 54% (202 wild-type [C] vs 242 mutant [G] tags; Fisher exact test, P=0.98), indicating both the wild-type and Tbx5G125R/+ transcripts are present at equal levels in Tbx5G125R/+ animals.
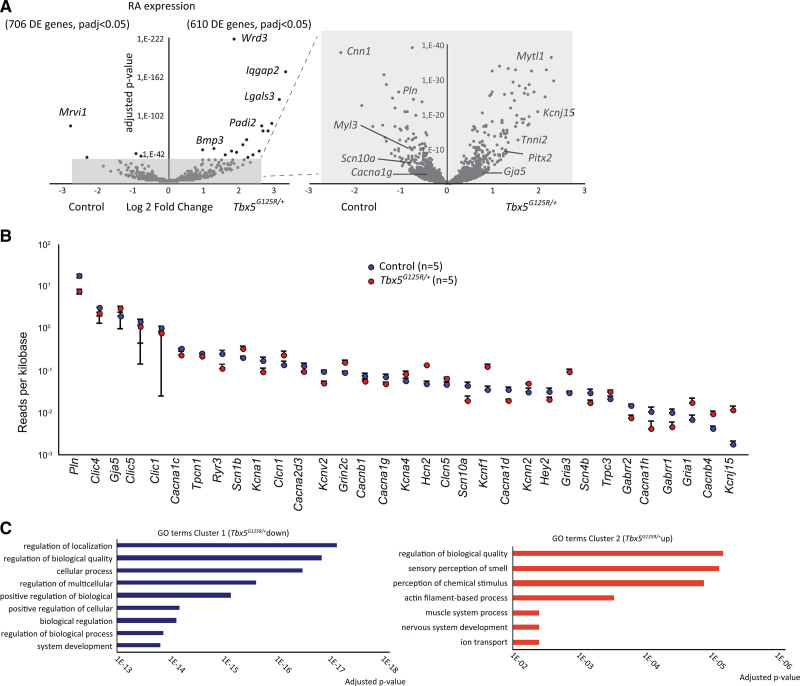
We found 1316 genes differentially expressed (Padj < 0.05; Table S8; Figure 5A); 706 were downregulated in the Tbx5G125R/+ atria, 610 were upregulated. While the cardiac function of many of the most significantly differentially expressed genes (Table S8) has not been defined, Lgals3 (galectin-3) and Padi2 (peptidyl arginine deiminase-2) have been implicated in cardiac dysfunction.17,18 We observed differential expression in 33 of 300 analyzed gene-encoding proteins involved in ion handling/electrophysiology, among which were several intracellular Ca2+-modulating and gap junction genes (eg, Cacna1c [calcium voltage-gated channel subunit α1C], Pln [phospholamban], Gja5 [gap junction protein α5], and Slc8a1 [solute carrier family 8 member A1] which could account for the low Ca2+ transient amplitudes observed in Tbx5G125R/+ mice), as well as K+ channel genes such as Kcnj15 (potassium inwardly rectifying channel subfamily J member 15) and Kcnn2 ([potassium calcium-activated channel subfamily N member 2] Figure 5B; Table S9). Reduced expression of Scn5a (sodium voltage-gated channel α subunit 5), Ryr2 (ryanodine receptor 2), and Atp2a2 (ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2) was linked to conduction slowing and altered intracellular Ca2+ concentrations in Tbx5 haploinsufficient mice.8–11 However, these and other genes that were previously found to respond to Tbx5 insufficiency (Nppa [natriuretic peptide A] Tbx3 [T-box transcription factor 3], Dsp [desmoplakin], Gja1 [gap junction protein α1]5, 9, 10 [Table S8]) were not responsive to Tbx5-p.G125R. Gene Ontology term analysis yielded general terms for genes downregulated in the Tbx5G125R/+, but pointed to actin filament and muscle process, as well as ion transport, for upregulated genes (Figure 5C; Tables S10 and S11). Transcriptional profiles of left atria of mutants and controls were in accordance with those of the right atria (Table S8).
Figure 5.
Transcription analysis of whole tissue right atrium of adult Tbx5G125R/+ mice. A, Expression analysis of the RA shows hundreds of genes differentially expressed between control (n=5) and Tbx5G125R/+ (n=5) atria. Differential expression analysis on whole-tissue RNA-sequencing was performed using the DESeq2 package. P values were corrected for multiple testing using the false discovery rate with 0.05 as control level. B, Expression analysis of ion handling genes between control and Tbx5G125R/+ mouse atria RNA-sequencing shows that 33 of 300 analyzed genes are differentially expressed (see Table S9; mean ± SEM, sorted by decreasing expression in the control group). C, GO-term analysis using PANTHER (protein analysis through evolutionary relationships). Bmp3 indicates bone morphogenetic protein 3; Cacna1c, Calcium Voltage-Gated Channel Subunit Alpha1 C; Cacna1d, Calcium Voltage-Gated Channel Subunit Alpha1 D; Cacna1g, Calcium Voltage-Gated Channel Subunit Alpha1 G; Cacna1h, Calcium Voltage-Gated Channel Subunit Alpha1 H; Cacna2d3, Calcium Voltage-Gated Channel Auxiliary Subunit Alpha2delta 3; Cacnb1, Calcium Voltage-Gated Channel Auxiliary Subunit Beta 1; Cacnb4, Calcium Voltage-Gated Channel Auxiliary Subunit Beta 4; Clcn1, Chloride Voltage-Gated Channel 1; Clcn5, Chloride Voltage-Gated Channel 5; Clic1, Chloride Intracellular Channel 1; Clic4, Chloride Intracellular Channel 4; Clic5, Chloride Intracellular Channel 5; Ccn1, Cellular Communication Network Factor 1; DE, differentially expressed; Gabrr1, Gamma-Aminobutyric Acid Type A Receptor Subunit Rho1; Gabrr2, Gamma-Aminobutyric Acid Type A Receptor Subunit Rho2; Gja5, Gap Junction Protein Alpha 5; Gria1, Glutamate Ionotropic Receptor AMPA Type Subunit 1; Gria3, Glutamate Ionotropic Receptor AMPA Type Subunit 3; Grin2c, Glutamate Ionotropic Receptor NMDA Type Subunit 2C; GO, Gene Ontology; Hcn2, Hyperpolarization Activated Cyclic Nucleotide Gated Potassium And Sodium Channel 2; Hey2, Hes Related Family BHLH Transcription Factor With YRPW Motif 2; Iqgap2, IQ Motif Containing GTPase Activating Protein 2; Kcna1, Potassium Voltage-Gated Channel Subfamily A Member 1; Kcna4, Potassium Voltage-Gated Channel Subfamily A Member 4; Kcnf1, Potassium Voltage-Gated Channel Modifier Subfamily F Member 1; Kcnj15, Potassium Inwardly Rectifying Channel Subfamily J Member 15; Kcnn2, Potassium Calcium-Activated Channel Subfamily N Member 2; Kcnv2, Potassium Voltage-Gated Channel Modifier Subfamily V Member 2; Lgals3, Galectin 3; Myl3, Myosin Light Chain 3; Mytl1, Myelin Transcription Factor 1 Like; Padi2, Peptidyl Arginine Deiminase 2; Padj, P-adjusted value; Pln, Phospholamban; Pitx2, Paired Like Homeodomain 2; RA, right atrium; Ryr3, Ryanodine Receptor 3; Scn10a, Sodium Voltage-Gated Channel Alpha Subunit 10; Scn1b, Sodium Voltage-Gated Channel Beta Subunit 1; Scn4b, Sodium Voltage-Gated Channel Beta Subunit 4; Tbx5, T-box transcription factor 5; Tnni2, Troponin I2, Fast Skeletal Type; Tpcn1, Two Pore Segment Channel 1; and Trpc3, Transient Receptor Potential Cation Channel Subfamily C Member 3.
We explored fibrotic tissue markers (Col1a1 [collagen type I α1 chain], Col3a1 [collagen type III α1 chain], Ctgf [connective tissue growth factor], Tgfb1 [transforming growth factor β1], Postn [periostin]) in whole-tissue RNA-seq of right and left atria, of which only Postn (activated fibroblast marker) was decreased in expression in right atria of Tbx5G125R/+ animals (Figure S6B; Table S8). These data further support the absence of fibrosis and activated fibroblasts in Tbx5G125R/+ atria. Furthermore, as the right atrial samples all contained sinoatrial node tissue, we explored the expression of sinoatrial node–specific genes19–23 in the whole-tissue RNA-seq data. Of 30 markers tested, 14 showed differential expression. Notably, transcripts for key TFs Tbx3, Isl1 (insulin gene enhancer protein ISL-1), and Shox2 (short stature homeobox 2), and key ion channels Hcn4 (hyperpolarization activated cyclic nucleotide gated potassium channel 4), Hcn1 (hyperpolarization activated cyclic nucleotide gated potassium channel 1), and Cacna2d2 (calcium voltage-gated channel auxiliary subunit α2 δ2) were not differentially expressed between genotypes. However, Cacna1d, (calcium voltage-gated channel subunit α1 D), Cacna2d3, calcium voltage-gated channel auxiliary subunit α2 δ3), Cacna1g (calcium voltage-gated channel subunit α1 G), Cacna1h (calcium voltage-gated channel subunit α1 H), and Ryr3 (ryanodine receptor 3), as well bone morphogenetic protein (BMP)–signaling components (Bmp2, Bmp3, Bmp10 [bone morphogenetic proteins 2, 3, and 10, respectively], Bmpr1b [bone morphogenetic protein receptor type 1B]), showed significant differential abundance in the data sets (Padj < 0.05; Table S8).
Tbx5-p.G125R Induces Changes in Epigenetic States in Atrial CMs
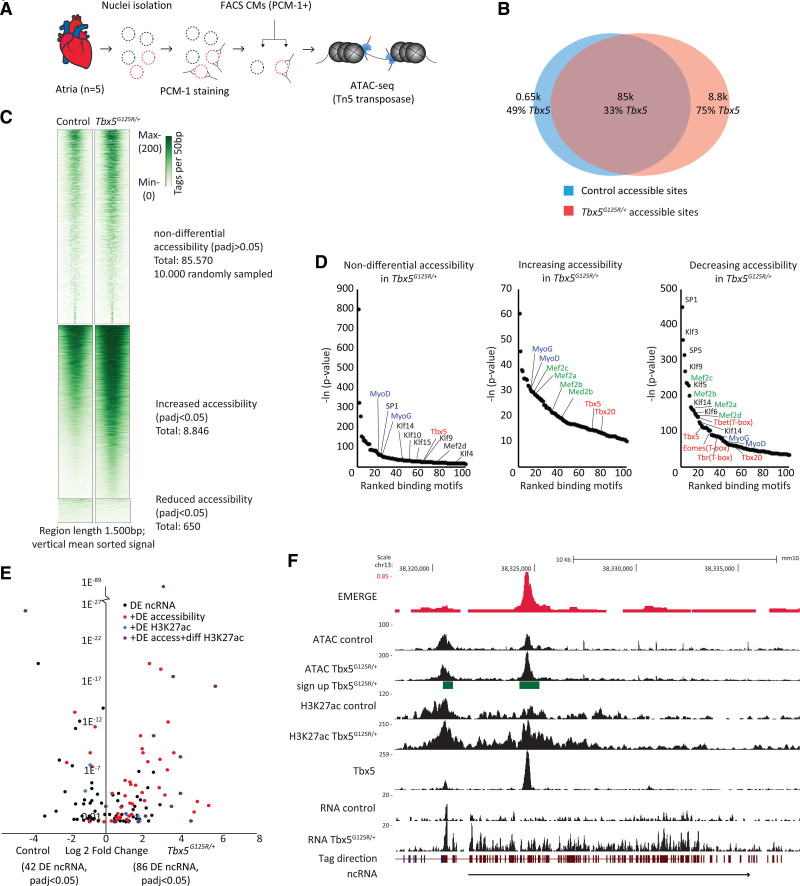
To gain insight into the changes in epigenetic state and regulatory element deployment in response to Tbx5-p.G125R, we assessed the chromatin accessibility profile of control and Tbx5G125R/+ atrial CMs (n=4 each) using ATAC-seq (Figure 6A). There were 85 570 sites that showed similar accessibility between control and Tbx5G125R/+ atrial CMs, 8846 showed increased accessibility, and 650 showed decreased accessibility (Figure 6B and 6C). To identify Tbx5-occupied accessible sites, we crossed a chromatin immunoprecipitation followed by sequencing dataset of Tbx5 occupancy in the fetal mouse heart.24 There was a striking enrichment of Tbx5 occupancy, both in sites of gained (6634/8846; Z-test P < 0.001) and reduced accessibility (318/650; Z-test P < 0.0001) compared with sites of unchanging accessibility (26 664/85 570; Figure 6B). The sites of increasing as well as decreasing accessibility were enriched in motifs for MEF2 (myocyte enhancer factor-2) TF family members as revealed by HOMER (Hypergeometric Optimization of Motif Enrichment) analysis (Tables S12–S14; Figure 6D). Indeed, heart development involves physical interaction between TBX5 and MEF2C (myocyte enhancer factor 2C).25 The sites of decreasing accessibility were strikingly enriched for motifs for KLF/SP TFs (Figure 6D; Table S14). TBX5 was reported to interact and cooperatively regulate cardiac target gene expression with KLF13 (Krüppel-like factor 13).26 These data suggest that Tbx5-p.G125R may have reduced interaction with KLF/SP factors, leading to reduced accessibility of sites normally co-occupied by Tbx5 and KLF/SP factors.
Figure 6.
ATAC-sequencing shows thousands of newly accessible chromatin overlapping Tbx5 binding sites in Tbx5G125R/+ mice. A, Method of atrial CM nucleus isolation and ATAC-seq. B, Venn diagram showing overlap of accessibility sites in control and Tbx5G125R/+ mouse atria, as well as the overlap between accessibility sites and Tbx5 binding sites (in %). C, Heatmap clustering showing increasing and decreasing accessibility. Differential accessibility was assessed using the DESeq2 package. P values were corrected for multiple testing using false discovery rate. D, HOMER motif analysis shows reduced accessibility sites in Tbx5G125R/+ atria were highly enriched for motifs of SP, MEF2, KLF, and TBX families of TFs, whereas Myo family member sites are enriched in increasingly accessible regions. False discovery rate adjusted P values are reported by HOMER. E, ncRNA expression analysis was performed using Limma to determine differential expression. F, University of California, Santa Cruz track showing a ncRNA upstream of Bmp6 (bone morphogenetic protein 6) and differential accessibility. ATAC-seq indicates assay for transposase-accessible chromatin sequencing; CM, cardiomyocyte; DE, differential expression; diff, differential; Eomes (T-box), Eomesodermin; FACS, fluorescence-activated cell sorting; H3K27Ac, histone H3 lysine 27 acetylation; HOMER, Hypergeometric Optimization of Motif Enrichment; Klf10, Krüppel-like factor 10; Klf14, Krüppel-like factor 14; Klf16, Krüppel-like factor 16; Klf3, Krüppel-like factor 3; Klf4, Krüppel-like factor 4; Klf9, Krüppel-like factor 9; Mef2a, myocyte enhancer factor-2a; Mef2b, myocyte enhancer factor-2b; Mef2c, myocyte enhancer factor-2c; Mef2d, myocyte enhancer factor-2d; MyoD, Myogenic Differentiation; MyoG, Myogenin; ncRNA, noncoding RNA; Padj, P-adjusted value; PCM-1, pericentriolar material-1; SP1, specificity protein 1; SP5, specificity protein 5; Tbet (T-box), T-Box Transcription Factor 21; Tbr (T-box), T-Box Brain Transcription Factor 1; Tbx20, T-box transcription factor 20; TF, transcription factors; and Tn5, transposon 5.
To further examine the impact of Tbx5-p.G125R on genome-wide regulatory element activity, we examined the genomic localization of H3K27ac association by CUT&RUN in isolated PCM1+ (pericentriolar material 1) CM nuclei of 8-week-old control (n=5) and Tbx5G125R/+ (n=6) pooled left and right atria (Figure S7A). We detected 127 286 regions enriched for H3K27ac association and 1653 sites with significantly differential H3K27ac regions (P < 0.05; Table S15), of which 1335 regions have increased signal in Tbx5G125R/+ and 318 regions have reduced signal (Figure S7B). Of the 127 286 H3K27ac regions, 18% (22 911/127 286) overlapped with Tbx5 binding sites in the fetal mouse heart24 (Table S16), whereas in the regions with increased H3K27ac association, 40% (347/1335; Z-test P < 0.0001) overlapped Tbx5 binding sites (Figure S7C). For example, accessibility and H3K27ac signal of Tbx5-bound regulatory elements in Padi2 and Wdr3 (WD repeat-containing protein 3) are strongly increased in Tbx5G125R/+ atria (Figure S7E and S7F). Interestingly, the H3K27ac regions that are reduced in the Tbx5G125R/+ atria all overlap Tbx5 binding sites (318/318; Z-test P < 0.0001; Figure S7C). These data are in agreement with the chromatin accessibility analysis, and together indicate that Tbx5-p.G125R induces altered epigenetic states of sites (including putative enhancers) normally occupied by Tbx5 and—to a lesser extent—of sites not occupied by Tbx5.
Previously, Tbx5-dependent enhancers and noncoding RNA (ncRNA) were identified by analyzing Tbx5-dependent enhancer transcription.12 By crossing our RNA-seq data with the ATAC-seq and CUT&RUN data sets (see Supplemental Methods; Table S17), we detected 128 enhancer-derived transcripts (ncRNA, transcriptional start site colocalizing with accessible and H3K27ac-associated site) that were differentially expressed between control and Tbx5G125R/+ animals in the right atria (n=5; Padj < 0.05), of which 42 were more abundant in the control and 86 more abundant in the Tbx5G125R/+ animals (Figure 6E; Table S17). Out of the 128 thus identified elements, 71 showed Tbx5 occupation (Table S17).24 The remaining 57 ncRNA-associated elements often showed Tbx5 occupation of accessible/H3K27ac-marked elements at a few-kbp distance, but these were considered to not colocalize. An example of a differentially expressed ncRNA with a transcriptional start site overlapping differential accessibility and H3K27ac is shown in Figure 6F near Bmp6 (bone morphogenetic protein 6). Of these differentially active regulatory elements, we identified 166 candidate target genes that were differently expressed between genotypes (Table S17). Highly interesting candidates include Tbx20 (T-box transcription factor 20), Sema3a (semaphorin 3A), Kcna4 (potassium voltage-gated channel subfamily A member 4), Slc8a1, and Tgfb2 (transforming growth factor β2), which are likely to impact atrial function in Tbx5G125R/+ animals (Table S17, column N)27–31 Differentially transcribed ncRNAs, in combination with differential accessibility and differential target gene expression, are shown for the Tgfb2, Sema3a, and Kcnj15 loci in Figure S8. Interestingly, previously identified prominent Tbx5-dependent ncRNAs (close to Ryr2 and to Atp2a2 [Serca2a (sarcoplasmic/endoplasmic reticulum CA2+ ATPase 2a])10,12 and their putative target genes (Ryr2, Atp2a2) were not affected in Tbx5G125R/+ atria (Figure S9).
Discussion
Previous characterization of a Dutch family with the pathogenic TBX5-p.G125R missense variant revealed atypical HOS, with mild skeletal abnormalities and lower penetrance of congenital heart defects but a high prevalence of AF.15 Here, we show that these patients suffer from other types of supraventricular arrhythmias that indicate dysfunctionality of the atrial muscle, including the sinoatrial node and AV node, that were not identified previously (Table S1). Typical HOS is usually associated with loss-of-function variants leading to haploinsufficiency; the atypical HOS phenotype of TBX5-p.G125R carriers suggest the pathogenic missense variant causes TBX5 to gain specific function(s). In the mouse model with this variant, the quantity of Tbx5 (wild-type) and Tbx5-p.G125R mRNA is equal in the adult Tbx5G125R/+ mouse atria, indicating that loss of expression (ie, by nonsense-mediated decay) causing haploinsufficiency does not occur. We did identify broadly changed expression profiles and epigenetic states induced by heterozygosity for Tbx5-p.G125R. However, important genes and enhancers previously found to respond to Tbx5 insufficiency (eg, Ryr2, Atp2a2, Nppa, RACER)5,9,10,12 did not respond to Tbx5-p.G125R. This indicates Tbx5-p.G125R has gained functions invoking molecular mechanisms different from those invoked by reduced Tbx5 dose. Intriguingly, expression quantitative trait loci analysis revealed that AF risk variant carriers in the human population express significantly more, not less, TBX5 in the heart.14 It will be interesting to investigate whether the Tbx5-p.G125R TF and increased TBX5 dose in risk variant carriers share transcriptional responses underlying AF propensity.
The patients displayed AV junctional escape beats, atrial extras, sinus bradycardia, and sick sinus syndrome in addition to AF. These arrhythmias were reproduced in the Tbx5-p.G125R mouse model, indicating Tbx5-p.G125R function and impact on cardiac electrophysiology in vivo are conserved between human and mice. Tbx5 is critical for the development and gene regulation in the sinoatrial and the AV nodes.3,5,6,32,33 The right atrial RNA-seq data sets indicated differential expression in 14 of 30 selected sinoatrial node markers. While these findings require validation, they suggest that the sinoatrial node may be transcriptionally affected in Tbx5G125R/+ mice. The differentially expressed genes included genes encoding calcium channels and BMP-signaling components important for sinoatrial node development and function,20,22 which may contribute to dysfunction of the sinoatrial node in Tbx5G125R/+ mice. Moreover, the balance between Tbx5 and the transcriptional repressor Tbx3 was found to determine the function of conduction system components.34,35 The altered DNA binding properties of Tbx5-p.G125R could affect both the regulation of important regulators in the nodes, and change the competitive balance with Tbx3. Further studies are required to explore the impact and underlying mechanisms of Tbx5-p.G125R on the sinoatrial and AV nodes.
Transcriptional profiling of the atria showed that heterozygosity of Tbx5-p.G125R leads to extensive dysregulation of gene expression in the atria, indicating multiple and complex mechanisms underlie the arrhythmia phenotype. While studies of Tbx5-haploinsufficient and -deficient mouse models implicate reduced expression of Ryr2 and Atp2a2 in AF propensity,9,11,36 these genes are not deregulated in atria of heterozygous Tbx5-p.G125R mice. However, we observed changes in expression of many other ion-handling protein-encoding genes, and in particular, genes associated with atrial function. Notably, the transcriptional profiling and differential regulatory element usage analysis identified Tbx5-p.G125R–mediated increased atrial expression of Tbx20, Sema3a, Kcna4, and Pitx2c (Paired Like Homeodomain 2 isoform c; the latter of which is specifically in the right atrium), and decreased atrial expression of Tgfb2. Each of these genes has been causally linked to heart function and rhythm control, further indicating that multiple complex mechanisms underlie the arrhythmia phenotype. Tbx20 is an essential cardiac TF, controlling CM function, ion channel gene expression, and atrial development.27,28 Because T-box TF function is highly dose-sensitive, increased Tbx20 expression in Tbx5+/G125R atria is likely to broadly affect cardiac gene expression and function. Sema3a, produced by CMs, controls cardiac sympathetic innervation and is associated with arrhythmia when disrupted or overexpressed in mice.29 Moreover, Sema3a was identified as a naturally occurring Kv4.3 (potassium voltage-gated channel) inhibitor,37 suggesting its induction could affect Kv4.3. An induction of Kv1.4 (encoded by Kcna4) has been observed in pathophysiologic conditions in the human heart (see Ni et al31). An increase of Kv1.4 (partly replacing Kv4.3) in atria was suggested to prolong ADP and to alter its dynamics, causing AF predisposition.31
Pitx2c is specifically expressed in the left atrium, where it imposes left identity on the atrium, initiates pulmonary vein myocardium formation, and suppresses sinoatrial node development during embryogenesis.38–40 In the formed left atrium, Pitx2c was shown to interact with Tbx5 to regulate ion channel gene expression and electrophysiology.10 Ectopic induction of Pitx2c in the right atrium could potentially affect sinoatrial node development and function, consistent with the observation that both human and mouse TBX5-pG125R carriers display sinoatrial node dysfunction. Furthermore, the atrial TF network around Tbx5 is likely affected by ectopic presence of Pitx2c in the right atrium, leading to altered ion channel gene expression.10 The decreased expression of Tgfb2 in the atria of heterozygous Tbx5-p.G125R mice may lead to reduced TGF-β signaling and diminished fibrosis,30 as is observed in the atria of these mice.
Patch clamp measurements of isolated atrial Tbx5G125R/+ CMs show a prolongation of the APD. While APD is often shortened in AF patients, mainly in the setting of sustained arrhythmias,41,42 APD prolongation is associated with increased risk of developing AF in patients.43–45 In atria of Tbx5G125R/+ mice, APD was prolonged as well, which is consistent with the patient clinical picture as well as with the increase in atrial arrhythmias we observed. The decreased Ca2+ amplitude and intracellular Ca2+ concentrations may be a result of longer Ca2+ currents because of reduced Ca2+-induced Ca2+ current inactivation, as well as decreased activation of Ca2+-activated K+ channels, and both mechanisms may contribute to the prolonged ADP that was observed. Moreover, deregulation of genes encoding channel subunits for various voltage-gated potassium currents (eg, Kcna1 [Kv1.1], Kcnj15 [Kir4.2 (inwardly rectifying potassium channel 4.2)], Kcna4 [Kv1.4]; see Figure 5B) may also contribute to prolonged ADP. Decreased expression of genes encoding intracellular Ca2+-modulating proteins such as Cacna1c and Cacna1g could be at the basis of the decreased of Ca2+ transient amplitude observed in the mutant mice. Another explanation could involve Mrvi1 (murine retrovirus integration site 1 homolog), which uniquely responds to Tbx5-p.G125R and is decreased in the Tbx5G125R/+ atria. Mrvi1 is associated with Pln and plays a key role in cardiac contractility by modulating nitric oxide–induced inhibition of Ca2+ signaling.46,47
A relatively large number of sites with increased accessibility and with increased H3K27ac association were observed in heterozygous Tbx5-p.G125R atrial CMs. These sites were enriched for Tbx5 binding in the normal heart,24 suggesting TBX5-p.G125R may display increased interaction with genuine Tbx5-occupied sites in vivo, in keeping with its increased DNA-binding affinity in vitro.15 However, sites normally not occupied by Tbx5 also showed increased accessibility, and the sites displaying decreased accessibility were enriched for Tbx5-binding and T-box motifs. Together, these observations suggest Tbx5-p.G125R has acquired properties that both quantitatively and qualitatively influence chromatin interaction. Additional analyses (eg, chromatin immunoprecipitation followed by sequencing, proteomics) are required to define these altered interaction properties of Tbx5-p.G125R. The changed chromatin interaction may involve a shift in affinity for binding partner TFs or cofactors, as was previously also reported for missense mutations in GATA4 (GATA-binding protein 4) and NKX2-5.48,49 The sites of reduced accessibility in heterozygous mutants were highly enriched for motifs of Sp/KLF, MEF2, and T-box families of TFs, pointing to candidate TFs that normally interact with Tbx5, and to a lesser extent, Tbx5-p.G125R. Mef2c and Klf13 are reported functional interaction partners of Tbx5 in the regulation of cardiac development and gene regulation.25,26 The Sp4 TF is expressed in the heart, including the atria and AV node, and Sp4-deficient mice display a high incidence of sinus bradycardia and AV block.50 Future studies could explore whether the Tbx5-p.G125R interactions with Mef2c—as well as the Sp and Klf TF family members expressed in the atria—are affected.
In conclusion, Tbx5-p.G125R displays altered properties that induce changes in the epigenetic state, such as chromatin accessibility and H3K27ac, that underlie deregulated expression of both Tbx5 target genes and off-target genes, which is at the basis of distinct electrophysiologic changes that recapitulate clinical observations in patients heterozygous for TBX5-p.G125R.
Article Information
Acknowledgments
We thank Berend de Jonge, Aho Ilgun, and Corrie de Gier-de Vries for their contributions. We thank Jan Ruijter for assisting with statistical analysis and presentations.
Sources of Funding
This work was supported by CardioVasculair Onderzoek Nederland (RACE-V Acceleration and Career Development to A.F.O.; project 2014-18 CONCOR-genes Young Talent Program to F.M.B.; and project 2014-18 CONCOR-genes to V.M.C.), Leducq Foundation (14CVD01 to V.M.C.), and Dutch CardioVascular Alliance (OUTREACH to V.M.C.).
Disclosures
None.
Supplemental Material
Supplemental Methods
Figures S1–S9
Table S1–S18
References 51–72
Supplementary Material
Nonstandard Abbreviations and Acronyms
- AF
- atrial fibrillation
- ATAC-seq
- assay for transposase-accessible chromatin using sequencing
- AV
- atrioventricular
- CM
- cardiomyocyte
- CUT&RUN
- cleavage under targets and release under nuclease
- H3K27ac
- histone H3 lysine 27 acetylation
- HOS
- Holt–Oram Syndrome
- RNA-seq
- RNA sequencing
- TBX5
- T-box transcription factor 5
- TF
- transcription factor
Circulation is available at www.ahajournals.org/journal/circ
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/circulationaha.121.054347.
For Sources of Funding and Disclosures, see page 617.
References
- 1.Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, et al. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15:30–35. doi: 10.1038/ng0197-30 [DOI] [PubMed] [Google Scholar]
- 2.Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis AR, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T, et al. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet. 1997;15:21–29. doi: 10.1038/ng0197-21 [DOI] [PubMed] [Google Scholar]
- 3.Steimle JD, Moskowitz IP. TBX5: a key regulator of heart development. Curr Top Dev Biol. 2017;122:195–221. doi: 10.1016/bs.ctdb.2016.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7 [DOI] [PubMed] [Google Scholar]
- 5.Mori AD, Zhu Y, Vahora I, Nieman B, Koshiba-Takeuchi K, Davidson L, Pizard A, Seidman JG, Seidman CE, Chen XJ, et al. Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev Biol. 2006;297:566–586. doi: 10.1016/j.ydbio.2006.05.023 [DOI] [PubMed] [Google Scholar]
- 6.Moskowitz IP, Pizard A, Patel VV, Bruneau BG, Kim JB, Kupershmidt S, Roden D, Berul CI, Seidman CE, Seidman JG. The T-Box transcription factor Tbx5 is required for the patterning and maturation of the murine cardiac conduction system. Development. 2004;131:4107–4116. doi: 10.1242/dev.01265 [DOI] [PubMed] [Google Scholar]
- 7.Moskowitz IP, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, Nobrega MA, Yokota Y, Berul C, Izumo S, et al. A molecular pathway including Id2, Tbx5, and Nkx2-5 required for cardiac conduction system development. Cell. 2007;129:1365–1376. doi: 10.1016/j.cell.2007.04.036 [DOI] [PubMed] [Google Scholar]
- 8.Arnolds DE, Liu F, Fahrenbach JP, Kim GH, Schillinger KJ, Smemo S, McNally EM, Nobrega MA, Patel VV, Moskowitz IP. TBX5 drives Scn5a expression to regulate cardiac conduction system function. J Clin Invest. 2012;122:2509–2518. doi: 10.1172/JCI62617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Y, Gramolini AO, Walsh MA, Zhou YQ, Slorach C, Friedberg MK, Takeuchi JK, Sun H, Henkelman RM, Backx PH, et al. Tbx5-dependent pathway regulating diastolic function in congenital heart disease. Proc Natl Acad Sci U S A. 2008;105:5519–5524. doi: 10.1073/pnas.0801779105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nadadur RD, Broman MT, Boukens B, Mazurek SR, Yang X, van den Boogaard M, Bekeny J, Gadek M, Ward T, Zhang M, et al. Pitx2 modulates a Tbx5-dependent gene regulatory network to maintain atrial rhythm. Sci Transl Med. 2016;8:354ra115. doi: 10.1126/scitranslmed.aaf4891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai W, Laforest B, Tyan L, Shen KM, Nadadur RD, Alvarado FJ, Mazurek SR, Lazarevic S, Gadek M, Wang Y, et al. A calcium transport mechanism for atrial fibrillation in Tbx5-mutant mice. Elife. 2019;8:e41814. doi: 10.7554/eLife.41814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang XH, Nadadur RD, Hilvering CR, Bianchi V, Werner M, Mazurek SR, Gadek M, Shen KM, Goldman JA, Tyan L, et al. Transcription-factor-dependent enhancer transcription defines a gene regulatory network for cardiac rhythm. Elife. 2017;6:e31683. doi: 10.7554/eLife.31683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burnicka-Turek O, Broman MT, Steimle JD, Boukens BJ, Petrenko NB, Ikegami K, Nadadur RD, Qiao Y, Arnolds DE, Yang XH, et al. Transcriptional patterning of the ventricular cardiac conduction system. Circ Res. 2020;127:e94–e106. doi: 10.1161/CIRCRESAHA.118.314460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roselli C, Chaffin MD, Weng LC, Aeschbacher S, Ahlberg G, Albert CM, Almgren P, Alonso A, Anderson CD, Aragam KG, et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat Genet. 2018;50:1225–1233. doi: 10.1038/s41588-018-0133-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Postma AV, van de Meerakker JB, Mathijssen IB, Barnett P, Christoffels VM, Ilgun A, Lam J, Wilde AA, Lekanne Deprez RH, Moorman AF. A gain-of-function TBX5 mutation is associated with atypical Holt-Oram syndrome and paroxysmal atrial fibrillation. Circ Res. 2008;102:1433–1442. doi: 10.1161/CIRCRESAHA.107.168294 [DOI] [PubMed] [Google Scholar]
- 16.Koboldt DC, Larson DE, Wilson RK. Using VarScan 2 for germline variant calling and somatic mutation detection. Curr Protoc Bioinformatics. 2013;44:15.4.1–15.417. doi: 10.1002/0471250953.bi1504s44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suthahar N, Meijers WC, Silljé HHW, Ho JE, Liu FT, de Boer RA. Galectin-3 activation and inhibition in heart failure and cardiovascular disease: an update. Theranostics. 2018;8:593–609. doi: 10.7150/thno.22196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fert-Bober J, Giles JT, Holewinski RJ, Kirk JA, Uhrigshardt H, Crowgey EL, Andrade F, Bingham CO, 3rd, Park JK, Halushka MK, et al. Citrullination of myofilament proteins in heart failure. Cardiovasc Res. 2015;108:232–242. doi: 10.1093/cvr/cvv185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vedantham V, Galang G, Evangelista M, Deo RC, Srivastava D. RNA sequencing of mouse sinoatrial node reveals an upstream regulatory role for Islet-1 in cardiac pacemaker cells. Circ Res. 2015;116:797–803. doi: 10.1161/CIRCRESAHA.116.305913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Eif VWW, Stefanovic S, van Duijvenboden K, Bakker M, Wakker V, de Gier-de Vries C, Zaffran S, Verkerk AO, Boukens BJ, Christoffels VM. Transcriptome analysis of mouse and human sinoatrial node cells reveals a conserved genetic program. Development. 2019;146:dev173161. doi: 10.1242/dev.173161 [DOI] [PubMed] [Google Scholar]
- 21.Goodyer WR, Beyersdorf BM, Paik DT, Tian L, Li G, Buikema JW, Chirikian O, Choi S, Venkatraman S, Adams EL, et al. Transcriptomic profiling of the developing cardiac conduction system at single-cell resolution. Circ Res. 2019;125:379–397. doi: 10.1161/CIRCRESAHA.118.314578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linscheid N, Logantha SJRJ, Poulsen PC, Zhang S, Schrölkamp M, Egerod KL, Thompson JJ, Kitmitto A, Galli G, Humphries MJ, et al. Quantitative proteomics and single-nucleus transcriptomics of the sinus node elucidates the foundation of cardiac pacemaking. Nat Commun. 2019;10:2889. doi: 10.1038/s41467-019-10709-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang D, Xue J, Geng L, Zhou L, Lv B, Zeng Q, Xiong K, Zhou H, Xie D, Zhang F, et al. Cellular and molecular landscape of mammalian sinoatrial node revealed by single-cell RNA sequencing. Nat Commun. 2021;12:287. doi: 10.1038/s41467-020-20448-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akerberg BN, Gu F, VanDusen NJ, Zhang X, Dong R, Li K, Zhang B, Zhou B, Sethi I, Ma Q, et al. A reference map of murine cardiac transcription factor chromatin occupancy identifies dynamic and conserved enhancers. Nat Commun. 2019;10:4907. doi: 10.1038/s41467-019-12812-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh TK, Song FF, Packham EA, Buxton S, Robinson TE, Ronksley J, Self T, Bonser AJ, Brook JD. Physical interaction between TBX5 and MEF2C is required for early heart development. Mol Cell Biol. 2009;29:2205–2218. doi: 10.1128/MCB.01923-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darwich R, Li W, Yamak A, Komati H, Andelfinger G, Sun K, Nemer M. KLF13 is a genetic modifier of the Holt-Oram syndrome gene TBX5. Hum Mol Genet. 2017;26:942–954. doi: 10.1093/hmg/ddx009 [DOI] [PubMed] [Google Scholar]
- 27.Shen T, Aneas I, Sakabe N, Dirschinger RJ, Wang G, Smemo S, Westlund JM, Cheng H, Dalton N, Gu Y, et al. Tbx20 regulates a genetic program essential to adult mouse cardiomyocyte function. J Clin Invest. 2011;121:4640–4654. doi: 10.1172/JCI59472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boogerd CJ, Zhu X, Aneas I, Sakabe N, Zhang L, Sobreira DR, Montefiori L, Bogomolovas J, Joslin AC, Zhou B, et al. Tbx20 is required in mid-gestation cardiomyocytes and plays a central role in atrial development. Circ Res. 2018;123:428–442. doi: 10.1161/CIRCRESAHA.118.311339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ieda M, Kanazawa H, Kimura K, Hattori F, Ieda Y, Taniguchi M, Lee JK, Matsumura K, Tomita Y, Miyoshi S, et al. Sema3a maintains normal heart rhythm through sympathetic innervation patterning. Nat Med. 2007;13:604–612. doi: 10.1038/nm1570 [DOI] [PubMed] [Google Scholar]
- 30.Nattel S, Burstein B, Dobrev D. Atrial remodeling and atrial fibrillation: mechanisms and implications. Circ Arrhythm Electrophysiol. 2008;1:62–73. doi: 10.1161/CIRCEP.107.754564 [DOI] [PubMed] [Google Scholar]
- 31.Ni H, Zhang H, Grandi E, Narayan SM, Giles WR. Transient outward K+ current can strongly modulate action potential duration and initiate alternans in the human atrium. Am J Physiol Heart Circ Physiol. 2019;316:H527–H542. doi: 10.1152/ajpheart.00251.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puskaric S, Schmitteckert S, Mori AD, Glaser A, Schneider KU, Bruneau BG, Blaschke RJ, Steinbeisser H, Rappold G. Shox2 mediates Tbx5 activity by regulating Bmp4 in the pacemaker region of the developing heart. Hum Mol Genet. 2010;19:4625–4633. doi: 10.1093/hmg/ddq393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Eif VWW, Devalla HD, Boink GJJ, Christoffels VM. Transcriptional regulation of the cardiac conduction system. Nat Rev Cardiol. 2018;15:617–630. doi: 10.1038/s41569-018-0031-y [DOI] [PubMed] [Google Scholar]
- 34.Burnicka-Turek O, Broman MT, Steimle JD, Boukens BJ, Petrenko NB, Ikegami K, Nadadur RD, Qiao Y, Arnolds DE, Yang XH, et al. Transcriptional patterning of the ventricular cardiac conduction system. Circ Res. 2020;127:e94–e106. doi: 10.1161/CIRCRESAHA.118.314460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohan RA, Bosada FM, van Weerd JH, van Duijvenboden K, Wang J, Mommersteeg MTM, Hooijkaas IB, Wakker V, de Gier-de Vries C, Coronel R, et al. T-box transcription factor 3 governs a transcriptional program for the function of the mouse atrioventricular conduction system. Proc Natl Acad Sci U S A. 2020;117:18617–18626. doi: 10.1073/pnas.1919379117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laforest B, Dai W, Tyan L, Lazarevic S, Shen KM, Gadek M, Broman MT, Weber CR, Moskowitz IP. Atrial fibrillation risk loci interact to modulate Ca2+-dependent atrial rhythm homeostasis. J Clin Invest. 2019;129:4937–4950. doi: 10.1172/JCI124231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boczek NJ, Ye D, Johnson EK, Wang W, Crotti L, Tester DJ, Dagradi F, Mizusawa Y, Torchio M, Alders M, et al. Characterization of SEMA3A-encoded semaphorin as a naturally occurring Kv4.3 protein inhibitor and its contribution to Brugada syndrome. Circ Res. 2014;115:460–469. doi: 10.1161/CIRCRESAHA.115.303657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mommersteeg MT, Hoogaars WM, Prall OW, de Gier-de Vries C, Wiese C, Clout DE, Papaioannou VE, Brown NA, Harvey RP, Moorman AF, et al. Molecular pathway for the localized formation of the sinoatrial node. Circ Res. 2007;100:354–362. doi: 10.1161/01.RES.0000258019.74591.b3 [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Klysik E, Sood S, Johnson RL, Wehrens XH, Martin JF. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci U S A. 2010;107:9753–9758. doi: 10.1073/pnas.0912585107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Bai Y, Li N, Ye W, Zhang M, Greene SB, Tao Y, Chen Y, Wehrens XH, Martin JF. Pitx2-microRNA pathway that delimits sinoatrial node development and inhibits predisposition to atrial fibrillation. Proc Natl Acad Sci U S A. 2014;111:9181–9186. doi: 10.1073/pnas.1405411111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu TJ, Kim YH, Yashima M, Athill CA, Ting CT, Karagueuzian HS, Chen PS. Progressive action potential duration shortening and the conversion from atrial flutter to atrial fibrillation in the isolated canine right atrium. J Am Coll Cardiol. 2001;38:1757–1765. doi: 10.1016/s0735-1097(01)01606-0 [DOI] [PubMed] [Google Scholar]
- 42.Kim BS, Kim YH, Hwang GS, Pak HN, Lee SC, Shim WJ, Oh DJ, Ro YM. Action potential duration restitution kinetics in human atrial fibrillation. J Am Coll Cardiol. 2002;39:1329–1336. doi: 10.1016/s0735-1097(02)01760-6 [DOI] [PubMed] [Google Scholar]
- 43.Olson TM, Alekseev AE, Liu XK, Park S, Zingman LV, Bienengraeber M, Sattiraju S, Ballew JD, Jahangir A, Terzic A. Kv1.5 channelopathy due to KCNA5 loss-of-function mutation causes human atrial fibrillation. Hum Mol Genet. 2006;15:2185–2191. doi: 10.1093/hmg/ddl143 [DOI] [PubMed] [Google Scholar]
- 44.Simpson RJ, Jr, Amara I, Foster JR, Woelfel A, Gettes LS. Thresholds, refractory periods, and conduction times of the normal and diseased human atrium. Am Heart J. 1988;116:1080–1090. doi: 10.1016/0002-8703(88)90163-9 [DOI] [PubMed] [Google Scholar]
- 45.Lee JM, Lee H, Janardhan AH, Park J, Joung B, Pak HN, Lee MH, Kim SS, Hwang HJ. Prolonged atrial refractoriness predicts the onset of atrial fibrillation: a 12-year follow-up study. Heart Rhythm. 2016;13:1575–1580. doi: 10.1016/j.hrthm.2016.03.037 [DOI] [PubMed] [Google Scholar]
- 46.Desch M, Sigl K, Hieke B, Salb K, Kees F, Bernhard D, Jochim A, Spiessberger B, Höcherl K, Feil R, et al. IRAG determines nitric oxide- and atrial natriuretic peptide-mediated smooth muscle relaxation. Cardiovasc Res. 2010;86:496–505. doi: 10.1093/cvr/cvq008 [DOI] [PubMed] [Google Scholar]
- 47.Schlossmann J, Ammendola A, Ashman K, Zong X, Huber A, Neubauer G, Wang GX, Allescher HD, Korth M, Wilm M, et al. Regulation of intracellular calcium by a signalling complex of IRAG, IP3 receptor and cGMP kinase Ibeta. Nature. 2000;404:197–201. doi: 10.1038/35004606 [DOI] [PubMed] [Google Scholar]
- 48.Ang YS, Rivas RN, Ribeiro AJS, Srivas R, Rivera J, Stone NR, Pratt K, Mohamed TMA, Fu JD, Spencer CI, et al. Disease model of GATA4 mutation reveals transcription factor cooperativity in human cardiogenesis. Cell. 2016;167:1734–1749.e22. doi: 10.1016/j.cell.2016.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouveret R, Waardenberg AJ, Schonrock N, Ramialison M, Doan T, de Jong D, Bondue A, Kaur G, Mohamed S, Fonoudi H, et al. NKX2-5 mutations causative for congenital heart disease retain functionality and are directed to hundreds of targets. Elife. 2015;4doi: 10.7554/eLife.06942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyên-Trân VT, Kubalak SW, Minamisawa S, Fiset C, Wollert KC, Brown AB, Ruiz-Lozano P, Barrere-Lemaire S, Kondo R, Norman LW, et al. A novel genetic pathway for sudden cardiac death via defects in the transition between ventricular and conduction system cell lineages. Cell. 2000;102:671–682. doi: 10.1016/s0092-8674(00)00089-1 [DOI] [PubMed] [Google Scholar]
- 51.Ruijter JM, Van Kampen AH, Baas F. Statistical evaluation of SAGE libraries: consequences for experimental design. Physiol Genomics. 2002;11:37–44. doi: 10.1152/physiolgenomics.00042.2002 [DOI] [PubMed] [Google Scholar]
- 52.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mi H, Muruganujan A, Huang X, Ebert D, Mills C, Guo X, Thomas PD. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat Protoc. 2019;14:703–721. doi: 10.1038/s41596-019-0128-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270 [DOI] [PubMed] [Google Scholar]
- 55.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verheule S, van Batenburg CA, Coenjaerts FE, Kirchhoff S, Willecke K, Jongsma HJ. Cardiac conduction abnormalities in mice lacking the gap junction protein connexin40. J Cardiovasc Electrophysiol. 1999;10:1380–1389. doi: 10.1111/j.1540-8167.1999.tb00194.x [DOI] [PubMed] [Google Scholar]
- 57.Verheule S, Sato T, Everett T, IV, Engle SK, Otten D, Rubart-von der Lohe M, Nakajima HO, Nakajima H, Field LJ, Olgin JE. Increased vulnerability to atrial fibrillation in transgenic mice with selective atrial fibrosis caused by overexpression of TGF-beta1. Circ Res. 2004;94:1458–1465. doi: 10.1161/01.RES.0000129579.59664.9d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barry PH, Lynch JW. Liquid junction potentials and small cell effects in patch-clamp analysis. J Membr Biol. 1991;121:101–117. doi: 10.1007/BF01870526 [DOI] [PubMed] [Google Scholar]
- 59.Baartscheer A, Schumacher CA, Fiolet JW. Small changes of cytosolic sodium in rat ventricular myocytes measured with SBFI in emission ratio mode. J Mol Cell Cardiol. 1997;29:3375–3383. doi: 10.1006/jmcc.1997.0567 [DOI] [PubMed] [Google Scholar]
- 60.Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 61.van Ouwerkerk AF, Bosada FM, van Duijvenboden K, Hill MC, Montefiori LE, Scholman KT, Liu J, de Vries AAF, Boukens BJ, Ellinor PT, Goumans MJTH, Efimov IR, Nobrega MA, Barnett P, Martin JF, Christoffels VM. Identification of atrial fibrillation associated genes and functional non-coding variants. Nat Commun. 2019;10:4755. doi: 10.1038/s41467-019-12721-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skene PJ, Henikoff S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife. 2017;6:e21856. doi: 10.7554/eLife.21856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hainer SJ, Fazzio TG. High-resolution chromatin profiling using CUT&RUN. Curr Protoc Mol Biol. 2019;126:e85. doi: 10.1002/cpmb.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Assefa AT, De Paepe K, Everaert C, Mestdagh P, Thas O, Vandesompele J. Differential gene expression analysis tools exhibit substandard performance for long non-coding RNA-sequencing data. Genome Biol. 2018;19:96. doi: 10.1186/s13059-018-1466-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Duijvenboden K, de Boer BA, Capon N, Ruijter JM, Christoffels VM. EMERGE: a flexible modelling framework to predict genomic regulatory elements from genomic signatures. Nucleic Acids Res. 2016;44:e42. doi: 10.1093/nar/gkv1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Doynova MD, Markworth JF, Cameron-Smith D, Vickers MH, O’Sullivan JM. Linkages between changes in the 3D organization of the genome and transcription during myotube differentiation in vitro. Skelet Muscle. 2017;7:5. doi: 10.1186/s13395-017-0122-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fernandez-Perez A, Sathe AA, Bhakta M, Leggett K, Xing C, Munshi NV. Hand2 selectively reorganizes chromatin accessibility to induce pacemaker-like transcriptional reprogramming. Cell Rep. 2019;27:2354–2369.e7. doi: 10.1016/j.celrep.2019.04.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole-tissue RNA-sequencing ([RNA-seq] left and right atrial), cardiomyocyte (CM) nucleus H3K27ac (histone H3 lysine 27 acetylation) CUT&RUN (cleavage under targets and release under nuclease), CM nucleus assay for transposase-accessible chromatin using sequencing (ATAC-seq), and single-nuclei RNA-seq are available on Gene Expression Omnibus (GSE167082). All other data and supporting materials have been provided with the published article.