Abstract
Background
Patients with a severe COVID-19 infection often require admission at an intensive care unit (ICU) when they develop acute respiratory distress syndrome (ARDS). Hyperinflammation plays an important role in the development of ARDS in COVID-19. Procalcitonin (PCT) is a biomarker which may be a predictor of hyperinflammation. When patients with COVID-19 are in the emergency department (ED), elevated PCT levels could be associated with severe COVID-19 infections. The goal of this study is to investigate the association between PCT levels and severe COVID-19 infections in the ED.
Methods
This was a retrospective cohort study including patients with a confirmed COVID-19 infection who visited the ED of Erasmus Medical Center in Rotterdam, the Netherlands, between March and December 2020. The primary outcome was a severe COVID-19 infection, which was defined as patients who required ICU admission, all cause in-hospital mortality and mortality within 30 days after hospital discharge. PCT levels were measured during the ED visit. We used logistic regression to calculate the odds ratio (OR) with 95% confidence interval (95% CI) and corresponding area under the curve (AUC) of PCT on a severe COVID-19 infection, adjusting for bacterial coinfections, age, sex, comorbidities, C-reactive protein (CRP) and D-dimer.
Results
A total of 332 patients were included in the final analysis of this study, of which 105 patients reached the composite outcome of a severe COVID-19 infection. PCT showed an unadjusted OR of 4.19 (95%CI: 2.52–7.69) on a severe COVID-19 infection with an AUC of 0.82 (95% CI: 0.76–0.87). Corrected for bacterial coinfection, the OR of PCT was 4.05 (95% CI: 2.45–7.41). Adjusted for sex, bacterial coinfection, age any comorbidity, CRP and D-dimer, elevated PCT levels were still significantly associated with a severe COVID-19 infection with an adjusted OR of 2.11 (95% CI: 1.36–3.61). The AUC of this multivariable model was 0.85 (95%CI: 0.81–0.90).
Conclusion
High PCT levels are associated with high rates of severe COVID-19 infections in patients with a COVID-19 infection in the ED. The routine measurement of PCT in patients with a COVID-19 infection in the ED may assist physicians in the clinical decision making process regarding ICU disposition.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12879-022-07144-5.
Keywords: COVID-19, Biomarkers, Emergency department, Procalcitonin
Background
Coronavirus disease (COVID-19) caused by the novel Coronavirus (SARS-CoV-2) was declared a pandemic on the 11th of March 2020 by the World Health Organization [1]. Ever since, COVID-19 caused a high burden on hospital and intensive care unit capacity [2]. Patients with COVID-19 often require hospitalization and treatment at an intensive care unit (ICU) when they develop acute respiratory distress syndrome (ARDS). The exact underlying pathophysiology of ARDS in these severe COVID-19 infections is under research with an exponentially growing amount of studies on this topic [3, 4]. Many studies have found that hyperinflammation, caused by an overwhelming upregulation of the immune system, and immune thrombosis play a major role in the development of ARDS in COVID-19 [5].
When patients with a COVID-19 infection present at the emergency department (ED), it is important to be able to identify patients who are at high risk of developing a severe COVID-19 infection. These patients may benefit from more extensive monitoring or early ICU admission.
Different biomarkers have been identified as predictors of disease severity in COVID-19 [6], including procalcitonin (PCT). PCT is a biomarker previously used for distinguishing viral infections from bacterial infections [7], although its use in the ED for this purpose is controversial [8, 9]. Elevated PCT levels are often seen in patients with a bacterial infection. PCT is the prohormone of calcitonin and in a physiological state produced by C-cells of the thyroid gland. During inflammation, PCT is synthesized in all tissues. Bacterial toxins are well known triggers of synthesis of PCT. Other triggers of synthesis include interleukin-6 and tumor necrosis factor alpha (TNF-alpha) [10, 11]. High levels of these cytokines have been reported in severe COVID-19 infections [12]. For this reason, PCT may also be elevated in a hyperinflammatory state in the absence of a bacterial pathogen. ARDS is clinically the most important severe complication of COVID-19, therefore PCT may be predictive of hyperinflammation and the development of ARDS [13]. This may aid physicians in the ED to identify patients that are at risk of developing ARDS early.
The goal of this study is to investigate the association between PCT levels and severe COVID-19 infections in the ED.
Methods
In this retrospective single center cohort study we included patients with a confirmed COVID-19 infection who visited the ED of Erasmus University Medical Center, in Rotterdam, the Netherlands, between 1 March 2020 and 31 December 2020. Erasmus University Medical Center is an academic hospital with 40.000 ED visits annually. This study was conducted in accordance with the Declaration of Helsinki (64th WMA General Assembly, Fortaleza, Brazil, October 2013). The institutional review board waivered informed consent for the retrospective use of clinical data of COVID-19 patients. The study was performed following the STROBE guidelines (Additional file 1).
Inclusion and exclusion criteria
Patients were included if they tested positive on COVID-19 by nasal pharyngeal PCR test on the day of the ED visit, the reason for ED visit was related to the COVID-19 infection and PCT levels were measured in the ED. Patients were excluded if the PCT levels were measured but invalid (e.g. hemolytic blood samples), if the COVID-19 infection was a secondary finding and not the primary reason for ED visit or when no follow-up data were available due to transfer to another hospital.
Data collection
Patient data including demographics, comorbidities, vital signs and laboratory tests were collected from the electronic patient records. Vital signs including heart rate, arterial oxygen saturation, respiratory rate, temperature and blood pressure were recorded from the time of ED visit. Laboratory testing during the ED visit included hemoglobin, red cell distribution width (RDW), leucocyte count, thrombocytes, total bilirubin, alanine aminotransferase (ALAT), aspartate aminotransferase (ALAT), lactate, D-dimer, C-reactive protein (CRP) and PCT. Patients were followed up for 30 days after hospital discharge. Patient disposition from the ED was categorized into discharge home from the ED, admission to general ward and admission to the ICU. Mortality data was classified as all cause in-hospital mortality and mortality at home within 30 days after hospital discharge. To correct for potential bacterial coinfections at ED visit, culture within 48 h of ED visit were reviewed. If blood, urine or sputum cultures or pneumococcal or legionella antigen tests were positive, patients were classified as having a bacterial coinfection.
Primary outcome
The primary outcome was a severe COVID-19 infection, defined as patients that were admitted to the ICU or patients that died by any in-hospital or within 30 days after hospital discharge. We calculated odds ratios (ORs) for PCT as continuous variable and commonly used cut-off values of PCT of 0.25 ng/mL, 0.5 ng/mL and 1.0 ng/mL. These cut-off values are derived from recommendations in previous literature [14].
Secondary outcomes
The secondary outcomes of this study were hospital admission, ICU admission and all-cause mortality, either in-hospital or within 30 days after hospital discharge. For the secondary outcomes, PCT was analyzed as both continuous variable and at the different cut-off values as previously described.
PCT measurement
PCT analysis was available as a standard laboratory test in patients who visited the ED with a suspected COVID-19 infection. Blood was collected in a lithium heparin tube and analyzed directly upon arrival in the clinical chemistry laboratory. PCT was measured using E801 Elecsys BRAHMS PCT reagent on a COBAS 8000 (Roche Diagnostics, Switzerland). The PCT values were available to the treating physician during the ED visit.
Statistical analysis
Normally distributed variables were reported as mean with standard deviation (SD), non-normally distributed variables as median with interquartile range (IQR). Multiple imputation was used for handling missing data.
Differences in dichotomous variables between the severe and non-severe COVID-19 infection patients were analyzed with a chi-square test. Differences in continuous variables were analyzed using an independent sample T-test for normally distributed data and a Mann–Whitney-U test for non-normally distributed data. Following, we did an univariate logistic regression analysis calculating the ORs of the variables that significantly differed between the groups.
For the primary outcome, we calculated unadjusted odds ratios of PCT using univariate logistic regression analysis. We calculated adjusted ORs of PCT correcting for only bacterial coinfections and in a multivariable logistic regression model correcting for the following predefined variables: age, sex, bacterial coinfection, the presence of any comorbidity, CRP and D-dimer. We calculated receiver operating characteristic (ROC) curves and the corresponding area under the curve (AUC). The calibration statistics of the univariate analysis of PCT and the multivariable regression model were analyzed using the Hosmer-Lemshow test to investigate the goodness-of-fit and graphically reported by calibration plot with intercept and slope statistics.
For the secondary outcomes, we used predefined cut-off values of 0.25 ng/mL, 0.5 ng/mL, 1.0 ng/mL of PCT and calculated the unadjusted ORs, the sensitivity, specificity, negative predictive value and positive predictive value on the primary and secondary outcomes of the study.
The correct for the skewed distribution of PCT, we transformed PCT using the natural logarithm and performed the univariate analysis and calibration analysis in Additional file 3. Additionally, we transformed PCT into a nominal variable with the ranges of 0–0.25 ng/mL (reference), 0.25–0.50 ng/mL, 0.50–1.00 ng/mL and larger than 1.00 ng/mL and used this in the multivariable logistic regression model in Additional file 4.
Statistical analyses were performed using ‘R’ version 4.1.1. We used the MICE package for multiple imputation of missing data and the RMS package for calibration statistics.
Results
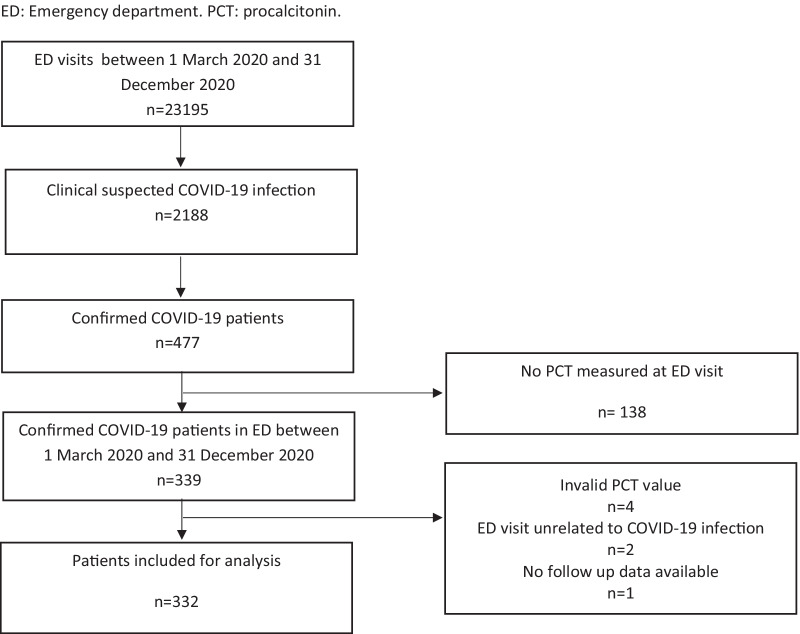
Between 1 March 2020 and 31 December 2020, a total of 23,195 patients visited the ED of which 2188 were clinically suspected of having a COVID-19 infection. Of these patients, 477 patients had a positive COVID-19 PCR-test at the day of the ED visit. Procalcitonin was measured in 339 of these patients. Four patients were excluded due to an invalid procalcitonin measurement, two patients were excluded because the ED visit was not related to the COVID-19 infection and one patient was excluded because there was no follow up data available. A total of 332 patients were included in the analysis of this study (Fig. 1). There were missing data in 3% of the data These missing data were imputated using multiple imputation. A specification of missing data per variable is shown in Additional file 2.
Fig. 1.
Flow diagram of included patients. ED emergency department, PCT procalcitonin
A total of 105 (32%) patients reached the composite outcome of a severe COVID-19 infection. Of these 105 patients, 44 (13%) were admitted to the ICU and 61 (18%) patients died. The non-severe COVID-19 infection group consisted of 227 (68%) patients, of which 67 (20%) were discharged directly from the ED and 158 (48%) were discharged from the general ward after an uncomplicated hospital admission. Baseline characteristics and differences between both groups including unadjusted ORs are shown in Table 1. The severe COVID-19 group was significantly older, more often male and had more frequently diabetes mellitus and any form of immunodeficiency as comorbidity. In vital parameters there were significant differences in heartrate, respiratory rate, oxygen saturation and diastolic blood pressure. In laboratory testing there were significant differences in PCT, CRP, leukocyte count, RDW, ASAT, ALAT, lactate, D-dimer and creatinine. Patients with severe COVID-19 infections had a bacterial coinfection more often.
Table 1.
Baseline characteristics and univariate analysis
| Patient characteristics | All patients | Non-severe COVID-19 infection | Severe COVID-19 infection | p-value | Unadjusted OR (95% CI) | |
|---|---|---|---|---|---|---|
| Demograpic data | n = 227 | n = 105 | ||||
| Sex: male | n (%) | 191 (57.5) | 115 (50.7) | 76 (72.4) | < 0.001 | 2.55 (1.56—4.26) |
| Age | Mean (SD) | 60 (16.4) | 58.1 (16.1) | 64.9 (16.3) | < 0.001 | 1.03 (1.01–1.05) |
| Comorbidity: pulmonary disease | n (%) | 83 (25) | 53 (23.8) | 29 (27.6) | 0.54 | |
| Comorbidity: cardiovascular disease | n (%) | 165 (49.7) | 115 (50.7) | 50 (47.6) | 0.691 | |
| Comorbidity: diabetes mellitus | n (%) | 89 (26.8) | 49 (21.6) | 40 (38.1) | 0.002 | 2.23 (1.35—3.71) |
| Comorbidity: malignancy | n (%) | 52 (15.7) | 35 (15.4) | 17 (16.1) | 0.986 | |
| Comorbidity: renal disease | n (%) | 55 (16.6) | 43 (18.9) | 12 (11.4) | 0.12 | |
| Comorbidity: auto-immune diseases | n (%) | 37 (11.1) | 27 (11.9) | 10 (9.5) | 0.652 | |
| Comorbidity: immunodeficiency | n (%) | 63 (19) | 53 (23.3) | 10 (9.5) | 0.005 | |
| Vital parameters | ||||||
| Heartrate | Mean (SD) | 94.1 (18.4) | 91 (17.9) | 101 (19.6) | < 0.001 | 1.03 (1.02–1.04) |
| Respiratory rate | Mean (SD) | 25.2 (9) | 24 (8.9) | 27 (8.9) | 0.001 | 1.03 (1.02–1.07) |
| Oxygen saturation | Median (IQR) | 95 (94–97) | 96 (94–97) | 94 (88–96) | < 0.001 | 0.85 (0.82–0.92) |
| Diastolic blood pressure | Mean (SD) | 79.4 (16) | 81 (15.6) | 77 (16.2) | 0.005 | 0.98 (0.96–0.99) |
| Systolic blood pressure | Mean (SD) | 134.6 (22.2) | 136 (21.8) | 135 (23.2) | 0.156 | |
| Temperature (Celcius) | Mean (SD) | 37.6 (1) | 37.5 (0.9) | 37.8 (1.2) | 0.288 | |
| Laboratory testing | ||||||
| Procalcitonin (ng/mL) | Median (IQR) | 0.14 (0.07–0.38) | 0.1 (0.06–0.18) | 0.47 (0.17–1.46) | < 0.001 | 4.19 (2.52–7.69) |
| CRP (mg/L) | Median (IQR) | 62 (26–136) | 46 (19–93) | 141 (67–213) | < 0.001 | 1.01 (1.01–1.02) |
| Leucocyte count | Median (IQR) | 6.4 (4.8–9) | 5.8 (4.4–7.7) | 7.7 (6.1–11.1) | < 0.001 | 1.16 (1.1–1.24) |
| Hemoglobin (mmol/L) | Mean (SD) | 8 (1.3) | 8 (1.2) | 7.9 (1.5) | 0.504 | |
| RDW (%) | Median (IQR) | 13.3 (12.6–14.6) | 13.2 (12.5–14.2) | 13.9 (12.9–14.6) | 0.014 | 1.04 (0.95–1.15) |
| Thrombocytes | Median (IQR) | 206 (164–265) | 205 (167–264) | 206 (160–266) | 0.851 | |
| ASAT (U/L) | Median (IQR) | 41 (28–61) | 36 (27–53) | 53 (36–81) | < 0.001 | 1.02 (1.01–1.02) |
| ALAT (U/L) | Median (IQR) | 28 (19–48) | 27 (18–43) | 31 (22–53) | 0.012 | 1.01 (1–1.01) |
| Total bilirubin (umol/L) | Median (IQR) | 8 (6–11) | 8 (6–11) | 8 (6–12) | 0.976 | |
| D-dimer (mg/L) | Median (IQR) | 1.07 (0.5–2.18) | 1 (0.47–1.48) | 1.72 (0.71–4.51) | < 0.001 | 1.22 (1.13–1.4) |
| Creatinin (umol/L) | Median (IQR) | 88 (70–122) | 82 (67–110) | 111 (83–163) | < 0.001 | 1.01 (1–1.01) |
| Lactate (mmol/L) | Median (IQR) | 1.3 (1–1.8) | 1.2 (1–1.7) | 1.6 (1.2–2.5) | < 0.001 | 2.15 (1.56–2.94) |
| Bacterial coinfection | n (%) | 37 (11) | 19 (8) | 18 (17) | 0.030 | 2.26 (1.13–4.54) |
| Outcome data | ||||||
| Admission length | Median (IQR) | 7 (214) | 6 (09) | 16 (827) | < 0.001 | |
| Mortality | n (%) | 61 (18.4) | 0 (0) | 66 (58.1) | < 0.001 |
SD standard deviation, IQR interquartile range, OR odds ratio, CRP C-reactive protein, RDW red blood cell distribution width, ASAT aspartate aminotransferase, ALAT alanine-aminotransferase
PCT showed an unadjusted OR of 4.19 (95%CI: 2.52–7.69) on a severe COVID-19 infection with an AUC of 0.82 (CI: 0.76–0.87). Corrected for bacterial coinfection, high PCT remained associated with an increase in severe COVID-19 infections with an adjusted OR of 4.05 (95% CI: 2.45–7.41). In the multivariable model adjusted for sex, bacterial coinfection, age, any comorbidity, CRP and d-dimer, PCT was still significantly associated with a severe COVID-19 infection with an adjusted OR of 2.11 (95% CI: 1.36–3.61) (Table 2). The AUC of the multivariable model was 0.85 (95% CI: 0.81–0.90).
Table 2.
Multivariable logistic regression model
| Predictor | Odds ratio | Confidence interval |
|---|---|---|
| Procalcitonin (ng/mL) | 2.11 | 1.36–3.61 |
| Bacterial coinfection | 1.95 | 0.81–4.61 |
| Sex: male | 1.32 | 0.72–2.44 |
| Age | 1.03 | 1.01–1.05 |
| Comorbidity: any | 0.97 | 0.44–2.19 |
| CRP | 1.01 | 1.00–1.01 |
| D-dimer | 1.24 | 1.12–1.41 |
CRP C-reactive protein
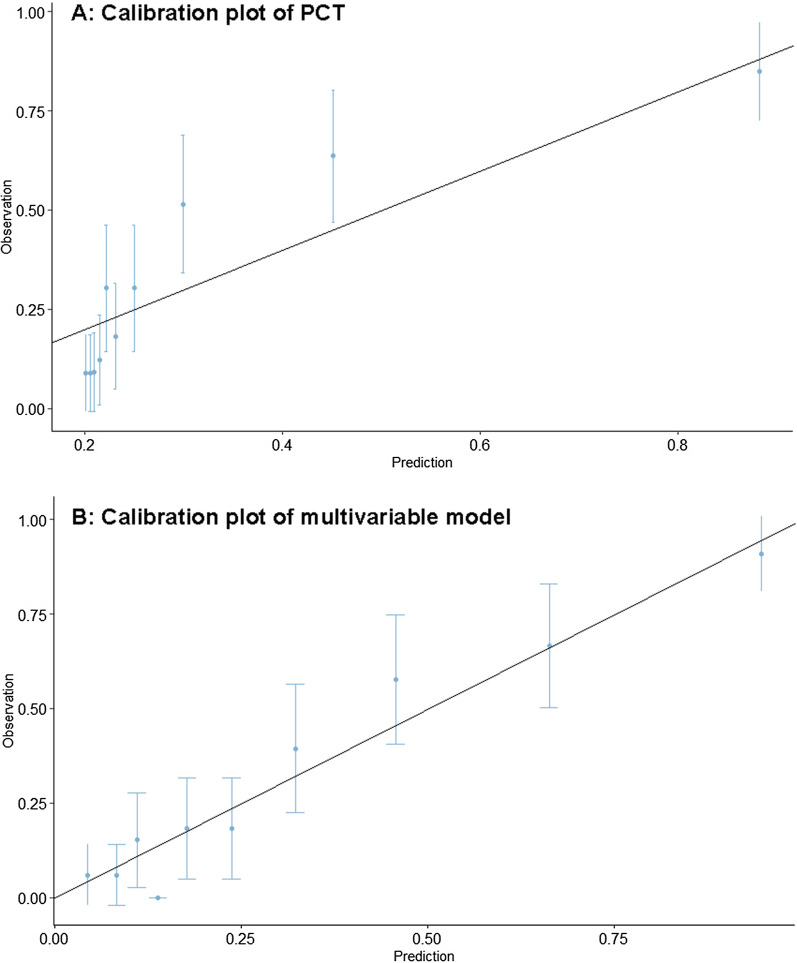
On the univariate analysis of PCT on a severe COVID-19 infection, the Hosmer-Lemshow test showed a bad fit (p = 0.003) and the calibration plot showed a slope of 1.00 (95% CI: 0.64–1.42) and intercept of 0.00 (95% CI: − 0.42–0.48) (Fig. 2A). The Hosmer-Lewshow test of the multivariable logistic regression model showed a good fit (p = 0.16) and the calibration plot showed a slope of 1.00 (CI: 0.76–1.27) with an intercept of 0.00 (CI: − 0.35–0.36) (Fig. 2B).
Fig. 2.
A Calibration plot of PCT. B Calibration plot of multivariable model. PCT Procalcitonin
For the primary outcome, the OR of PCT at these cutoff points were respectively 9.76 (95% CI: 5.79–16.83), 12.12 (95% CI: 6.61–23.2) and 14.40 (95% CI: 6.45–36.77). The ORs of PCT at the different cut-off points of 0.25 ng/mL, 0.5 ng/mL and 1.0 ng/mL for the secondary outcomes are shown in Table 3. On hospital admission, the OR of PCT was 14.68 (95% CI: 5.27–61.11) at a cutoff point of 0.25 ng/mL and 22.78 (95% CI: 4.88–406.11) at a cutoff point of 0.5 ng/mL. Since there were no patients who were discharged home with a PCT of higher than 1.0, we could not calculate the OR of PCT at a cutoff of 1.0. For ICU admission, the OR of PCT at the previously mentioned cutoff points were 5.94 (95% CI: 3.36–10.79), 7.7 (95% CI: 4.24–14.2) and 8.93 (95% CI: 4.42–18.51) respectively. For mortality the ORs were 9.72 (95% CI: 5.2–19.08), 9.38 (95% CI: 5.08–17.68) and 7.93 (95% CI: 3.92–16.29) respectively.
Table 3.
Procalcitonin at different cutoff points on primary and secondary outcomes
| Outcome | Cut-off value PCT | Odds ratio | Confidence interval | Sensitivity | Specificity | Negative predictive value | Positive predictive value |
|---|---|---|---|---|---|---|---|
| Severe outcome | Continuous scale | 4.19 | (2.52–7.69) | ||||
| 0.25 (ng/mL) | 9.76 | (5.79–16.83) | 0.67 | 0.82 | 0.84 | 0.63 | |
| 0.5 (ng/mL) | 12.12 | (6.61–23.2) | 0.49 | 0.92 | 0.79 | 0.75 | |
| 1 (ng/mL) | 14.40 | (6.45–36.77) | 0.31 | 0.96 | 0.75 | 0.82 | |
| Hospital admission | Continuous scale | 1667.00 | (67.91–98,005.22) | ||||
| 0.25 (ng/mL) | 14.68 | (5.27–61.11) | 0.40 | 0.95 | 0.28 | 0.97 | |
| 0.5 (ng/mL) | 22.78 | (4.88–406.11) | 0.25 | 0.98 | 0.25 | 0.98 | |
| 1 (ng/mL) | NA | NA | 0.15 | 1 | 0.22 | 1 | |
| ICU admission | Continuous scale | 1.15 | (1.05–1.3) | ||||
| 0.25 (ng/mL) | 5.94 | (3.36–10.79) | 0.66 | 0.74 | 0.90 | 0.39 | |
| 0.5 (ng/mL) | 7.70 | (4.24–14.2) | 0.53 | 0.87 | 0.88 | 0.50 | |
| 1 (ng/mL) | 8.93 | (4.42–18.51) | 0.36 | 0.93 | 0.85 | 0.6 | |
| Mortality | Continuous scale | 1.22 | (1.08–1.43) | ||||
| 0.25 (ng/mL) | 9.72 | (5.2–19.08) | 0.75 | 0.76 | 0.93 | 0.41 | |
| 0.5 (ng/mL) | 9.38 | (5.08–17.68) | 0.57 | 0.87 | 0.90 | 0.50 | |
| 1 (ng/mL) | 7.93 | (3.92–16.29) | 0.36 | 0.93 | 0.86 | 0.55 |
ICU intensive care unit, NA not available
Discussion
In this study we found that elevated PCT levels are associated with an increase in the combined outcome of ICU admission and mortality. When treating a COVID-19 patient in the ED with an elevated PCT, the treating physician should not only look for possible bacterial coinfections but also be cautious of disease progression. Based on these findings, physicians should beware that patients with elevated PCT levels in the ED may have an increased risk of becoming severely ill. Although these findings need to be validated before PCT levels can be used to guide clinical decision-making on disposition of COVID-19 patients, PCT levels may serve as an early warning sign of possible clinical deterioration. Due to a low sensitivity of PCT on all outcomes, normal PCT levels should not be used in disposition decisions in COVID-19 patients in the ED.
Our findings are supported by other studies, that showed that hospitalized COVID-19 patients with severe infections had higher PCT levels [15–17]. Only a few studies focused on the use of PCT in patients with a COVID-19 infection in an ED setting. Nazerian et al. found that PCT was not useful in diagnosing COVID-19 in the ED. However, the association PCT and of disease severity was not analysed [18]. Surme et al. investigated predictors of ICU admission and mortality and also found that high PCT were associated with an increase of the composite outcome of ICU admission and mortality [19]. Kaal et al. also showed similar results in a cohort of 142 patients, and showed that patients with PCT levels above 0.1 ng/mL have an elevated risk of a severe infection [20].
PCT is primarily used as biomarker for bacterial infections. The findings of this study show a new use for PCT in the specific group of COVID-19 patients as marker of disease severity. Even after correcting for bacterial coinfections and inflammatory markers CRP and d-dimer, elevated PCT levels remained associated with a severe COVID-19 infection. Our finding that PCT is highly elevated in severe COVID-19 infections can be explained when looking at the pathways of synthesis of PCT [10]. PCT synthesis is upregulated by different cytokines such as interleukine-6 and TNF-alpha. Because hyperinflammation is shown to be an important factor in progression of COVID-19 infections, the dysregulated immune response may also trigger PCT production [4]. Similar results of elevated PCT levels were found in patients with isolated severe influenza virus infections, findings that support the hypothesis that PCT is a marker of hyperinflammation [21].
At different cut-off values, PCT levels showed a high specificity for the hospital admission, ICU admission and mortality. On hospital admission, the lowest cut-off value of 0.25 ng/mL resulted in a specificity of 95%. This finding could be used as an argument to use PCT as support tool for hospital disposition decisions, because the majority of patients with a PCT above 0.25 ng/mL were admitted to the hospital. The sensitivity of PCT on all outcomes was low. Therefore, PCT is less suitable for being incorporated it in a rule out decision support tool of severe COVID-19 infections in the ED. Different cut-off values of PCT are recommended in distinguishing bacterial from viral infections, also ranging from 0.25 to 1.0 ng/mL [14]. The OR of PCT on hospital admission was 1667 (95% CI: 68–98,005). This high OR can be explained by the exponential increase in PCT levels in patients with severe infections. In our cohort, only 67 (20%) patients were discharged from the ED and the highest PCT level measured in these patients was 0.82 mg/mL. The patients that were admitted to the hospital also included the group that required ICU admission or died and several patients had a PCT levels above 10 ng/mL. This large difference in PCT levels between these groups resulted in the high OR.
Limitations
A limitation of this study is its retrospective design. Patients were included if PCT was measured in the ED. PCT was only measured in patients where the primary reason of the ED visit was suspected COVID-19 infection. Patients that were initially not suspected of having COVID-19, but were diagnosed with a COVID-19 infection nonetheless, were not included in this study.
In the secondary outcomes, the subgroups have a relatively small size, which influences the generalizability of the findings.
The patient population consisted solely of confirmed COVID-19 patients. Therefore, the results are not biased by patients who visited the ED with a suspected COVID-19 infection but were diagnosed with an alternative diagnosis. However, the results of this study cannot be used for distinguishing a COVID-19 infection from other viral or bacterial infections [18].
This study was performed in a cohort of patients who visited the ED in 2020, when no vaccinations against SARS-CoV-2 were available. It is unclear if these results are generalizable to the current ED setting, where the majority of patients is vaccinated [22].These results require validation in vaccinated patients.
Using only PCT as single predictor of the primary outcome showed a poor goodness-of-fit and the results may not be valid in other cohorts. Therefore, these results have to be validated in other cohorts. Because PCT has a skewed distribution of < 0.50 ng/mL in non-severe patients and from 0.50 up to 58 ng/mL in severely infected patients, the results may be biased by the patients with high PCT values. This can be corrected by using the natural logarithm of PCT, thereby reducing the range of PCT. The results were similar, but the goodness-of-fit was high (p = 0.91) (Additional file 3).
PCT was prospectively measured in the ED and available to the treating physician, along with other routinely measured laboratory tests. PCT levels may have been used in clinical decision making and influenced disposition decisions.
Conclusion
High PCT levels are associated with high rates of severe COVID-19 infections in patients with a COVID-19 infection in the ED. The routine measurement of PCT in patients with a COVID-19 infection in the ED may assist physicians in the clinical decision-making process regarding ICU disposition. These results should be validated in a prospective multicenter cohort.
Supplementary Information
Additional file 1. STROBE checklist (Strengthening the reporting of observational studies in epidemiology).
Additional file 2. Amount of missing data.
Additional file 3. Univariate analysis of natural logarithm of procalcitonin on a severe COVID-19 infection.
Additional file 4. Multivariable logistic regression model with different cut-off values of procalcitonin.
Acknowledgements
Not applicable.
Abbreviations
- COVID-19
Coronavirus disease
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- ICU
Intensive care unit
- ARDS
Acute respiratory distress syndrome
- ED
Emergency department
- PCT
Procalcitonin
- TNF-alpha
Tumor necrosis factor alpha
- RDW
Red cell distribution width
- ALAT
Alanine aminotransferase
- ASAT
Aspartate aminotransferase
- CRP
C-reactive protein
- OR
Odds ratio
- SD
Standard deviation
- IQR
Interquartile range
- AUC
Area under the curve
Authors' contributions
KTM, SE and HE were involved in the conception or design of the manuscript. KTM, YD and HE performed the analysis and interpretation of the data. KTM drafted the manuscript. KTM, SE, HE, EJ, CR, DG, EG and YD were involved in the critical revision of the manuscript. All authors read and approved the final manuscript.
Funding
This manuscript was not funded by any funding body.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The institutional review board of Erasmus University Medical Center granted permission to the retrospective use of data of COVID-19 patients. The data was anonymized before its use in this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health O . WHO Director-General's opening remarks at the media briefing on COVID-19-11 March 2020. Geneva: Switzerland; 2020. [Google Scholar]
- 2.Reese H, Iuliano AD, Patel NN, Garg S, Kim L, Silk BJ, et al. Estimated incidence of COVID-19 illness and hospitalization—United States, February–September, 2020. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grasselli G, Tonetti T, Protti A, Langer T, Girardis M, Bellani G, et al. Pathophysiology of COVID-19-associated acute respiratory distress syndrome: a multicentre prospective observational study. Lancet Respir Med. 2020;8(12):1201–1208. doi: 10.1016/S2213-2600(20)30370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torres Acosta MA, Singer BD. Pathogenesis of COVID-19-induced ARDS: implications for an ageing population. Eur Respir J. 2020 doi: 10.1183/13993003.02049-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manson JJ, Crooks C, Naja M, Ledlie A, Goulden B, Liddle T, et al. COVID-19-associated hyperinflammation and escalation of patient care: a retrospective longitudinal cohort study. Lancet Rheumatol. 2020;2(10):e594–e602. doi: 10.1016/S2665-9913(20)30275-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danwang C, Endomba FT, Nkeck JR, Wouna DLA, Robert A, Noubiap JJ. A meta-analysis of potential biomarkers associated with severity of coronavirus disease 2019 (COVID-19) Biomark Res. 2020;8:37. doi: 10.1186/s40364-020-00217-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuetz P, Wirz Y, Sager R, Christ-Crain M, Stolz D, Tamm M, et al. Effect of procalcitonin-guided antibiotic treatment on mortality in acute respiratory infections: a patient level meta-analysis. Lancet Infect Dis. 2018;18(1):95–107. doi: 10.1016/S1473-3099(17)30592-3. [DOI] [PubMed] [Google Scholar]
- 8.van der Does Y, Limper M, Jie KE, Schuit SCE, Jansen H, Pernot N, et al. Procalcitonin-guided antibiotic therapy in patients with fever in a general emergency department population: a multicentre non-inferiority randomized clinical trial (HiTEMP study) Clin Microbiol Infect. 2018;24(12):1282–1289. doi: 10.1016/j.cmi.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 9.Huang DT, Yealy DM, Filbin MR, Brown AM, Chang CH, Doi Y, et al. Procalcitonin-guided use of antibiotics for lower respiratory tract infection. N Engl J Med. 2018;379(3):236–249. doi: 10.1056/NEJMoa1802670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maruna P, Nedelnikova K, Gurlich R. Physiology and genetics of procalcitonin. Physiol Res. 2000;49(Suppl 1):S57–61. [PubMed] [Google Scholar]
- 11.Linscheid P, Seboek D, Nylen ES, Langer I, Schlatter M, Becker KL, et al. In vitro and in vivo calcitonin I gene expression in parenchymal cells: a novel product of human adipose tissue. Endocrinology. 2003;144(12):5578–5584. doi: 10.1210/en.2003-0854. [DOI] [PubMed] [Google Scholar]
- 12.Leisman DE, Ronner L, Pinotti R, Taylor MD, Sinha P, Calfee CS, et al. Cytokine elevation in severe and critical COVID-19: a rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. Lancet Respir Med. 2020;8(12):1233–1244. doi: 10.1016/S2213-2600(20)30404-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camporota L, Chiumello D, Busana M, Gattinoni L, Marini JJ. Pathophysiology of COVID-19-associated acute respiratory distress syndrome. Lancet Respir Med. 2021;9(1):e1. doi: 10.1016/S2213-2600(20)30505-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoeboer SH, van der Geest PJ, Nieboer D, Groeneveld AB. The diagnostic accuracy of procalcitonin for bacteraemia: a systematic review and meta-analysis. Clin Microbiol Infect. 2015;21(5):474–481. doi: 10.1016/j.cmi.2014.12.026. [DOI] [PubMed] [Google Scholar]
- 15.Hu R, Han C, Pei S, Yin M, Chen X. Procalcitonin levels in COVID-19 patients. Int J Antimicrob Agents. 2020;56(2):106051. doi: 10.1016/j.ijantimicag.2020.106051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heidari-Beni F, Vahedian-Azimi A, Shojaei S, Rahimi-Bashar F, Shahriary A, Johnston TP, et al. The level of procalcitonin in severe COVID-19 patients: a systematic review and meta-analysis. Adv Exp Med Biol. 2021;1321:277–286. doi: 10.1007/978-3-030-59261-5_25. [DOI] [PubMed] [Google Scholar]
- 17.Huang I, Pranata R, Lim MA, Oehadian A, Alisjahbana B. C-reactive protein, procalcitonin, D-dimer, and ferritin in severe coronavirus disease-2019: a meta-analysis. Ther Adv Respir Dis. 2020;14:1753466620937175. doi: 10.1177/1753466620937175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nazerian P, Gagliano M, Suardi LR, Fanelli A, Rossolini GM, Grifoni S, et al. Procalcitonin for the differential diagnosis of COVID-19 in the emergency department. Prospective monocentric study. Intern Emerg Med. 2021. [DOI] [PMC free article] [PubMed]
- 19.Surme S, Buyukyazgan A, Bayramlar OF, Cinar AK, Copur B, Zerdali E, et al. Predictors of intensive care unit admission or death in patients with coronavirus disease 2019 pneumonia in Istanbul, Turkey. Jpn J Infect Dis. 2021. [DOI] [PubMed]
- 20.Kaal A, Snel L, Dane M, van Burgel N, Ottens T, Broekman W, et al. Diagnostic yield of bacteriological tests and predictors of severe outcome in adult patients with COVID-19 presenting to the emergency department. Emerg Med J. 2021;38(9):685–691. doi: 10.1136/emermed-2020-211027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gautam S, Cohen AJ, Stahl Y, Valda Toro P, Young GM, Datta R, et al. Severe respiratory viral infection induces procalcitonin in the absence of bacterial pneumonia. Thorax. 2020;75(11):974–981. doi: 10.1136/thoraxjnl-2020-214896. [DOI] [PubMed] [Google Scholar]
- 22.Thompson MG, Stenehjem E, Grannis S, Ball SW, Naleway AL, Ong TC, et al. Effectiveness of COVID-19 vaccines in ambulatory and inpatient care settings. N Engl J Med. 2021;385(15):1355–1371. doi: 10.1056/NEJMoa2110362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. STROBE checklist (Strengthening the reporting of observational studies in epidemiology).
Additional file 2. Amount of missing data.
Additional file 3. Univariate analysis of natural logarithm of procalcitonin on a severe COVID-19 infection.
Additional file 4. Multivariable logistic regression model with different cut-off values of procalcitonin.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.