Abstract
The ciliary epithelium plays a central role in ocular homeostasis but cells of the pigmented and non-pigmented layers are difficult to isolate physically and study. Here we used single-cell RNA-sequencing (scRNA-seq) to analyze the transcriptional signatures of cells harvested from the ciliary body and contiguous tissues. Microdissected tissue was dissociated by collagenase digestion and the transcriptomes of individual cells were obtained using a droplet-based scRNA-seq approach. In situ hybridization was used to verify the expression patterns of selected differentially-expressed genes. High quality transcriptomes were obtained from 10,024 cells and unsupervised clustering distinguished 22 cell types. Although efforts were made to specifically isolate the ciliary body, approximately half of the sequenced cells were derived from the adjacent retina. Cluster identities were assigned using expression of canonical markers or cluster-specific genes. The transcriptional signature of cells in the PCE and NPCE were distinct from each other and from cells in contiguous tissues. PCE cell transcriptomes were characterized by genes involved in melanin synthesis and transport proteins such as Slc4a4. Among the most differentially expressed genes in NPCE cells were those encoding members of the Zic family of transcription factors (Zic1, 2, 4), collagen XVIII (Col18a1), and corticotrophin-releasing hormone-binding protein (Crhbp). The ocular melanocyte population was distinguished by expression of the gap junction genes Gjb2 and Gjb6. Two fibroblast signatures were detected in the ciliary body preparation and shown by in situ hybridization to correspond to uveal and scleral populations. This cell atlas for the ciliary body and contiguous layers represents a useful resource that may facilitate studies into the development of the ciliary epithelium, the production of the aqueous and vitreous humors, and the synthesis of the ciliary zonule.
Keywords: Ciliary epithelium, Transcriptomics, Single-cell RNA-seq, Fibroblast, Aqueous humor, Vitreous humor, In situ hybridization, Mouse, Melanocyte, Cell atlas
1. Introduction
The bilayered ciliary epithelium has a critical role in ocular homeostasis. Aqueous humor (AH) secreted by the epithelium nourishes the lens and other avascular structures of the anterior segment and maintains the intraocular pressure (Civan and Macknight, 2004). Cells of the ciliary epithelium also synthesize the major structural components of the zonule (Bassnett, 2021; Jones et al., 2019) and are the source of collagen, opticin, and other proteins of the vitreous humor (Bishop et al., 2002). In lower vertebrates, ciliary epithelial cells retain limited neurogenic capacity and facilitate the lifelong growth of the eye (Reh and Levine, 1998), although this appears not to be the case in humans (Fernandez-Nogales et al., 2019).
The ciliary epithelium lines the inner surface of the ciliary body; in mice, a wedge-shaped structure, approximately 250 μm wide, located between the root of the iris and the anterior lip of the retina (Fig. 1A). Much of the ciliary body surface has a fluted appearance (the pars plicata), though a narrow unfolded region (the pars plana) is also present, interposed between the pars plicata and the retina. The inner of the two ciliary epithelial layers, the non-pigmented ciliary epithelium (NPCE), faces the AH, while the outer layer (the pigmented ciliary epithelium, PCE) borders the vascularized stroma of the ciliary body (Fig. 1B). The stroma is overlaid successively by a thin layer of smooth muscle, the choroid, and the sclera. The scleral surface is in turn covered by loose connective tissue (the episclera). Posteriorly, the NPCE and the PCE are continuous with the neural retina and the retinal pigment epithelium (RPE), respectively. Anteriorly, the NPCE and PCE are continuous with the posterior and anterior layers of the iris pigment epithelium, while the stromal tissue of the ciliary body merges imperceptibly with the iris stroma. Cells of the immune system (including leukocytes, mast cells, macrophages, and dendritic cells) populate many, if not all, of these tissue compartments (Smith, 2002).
Fig. 1.
The ciliary body and contiguous tissues in the mouse eye. A. The ciliary body of the mouse eye as viewed from the interior aspect. The folded pars plicata and narrow pars plana regions are indicated. The neural retina is visible as a pale, translucent layer adjacent to the pars plana. B. Diagrammatic cross-sectional view of the ciliary body and contiguous tissues. NPCE, non-pigmented ciliary epithelium; PCE, pigmented ciliary epithelium; RPE, retinal pigment epithelium; SC, Schlemm’s canal; TM, trabecular meshwork. The dashed lines in B delineate the approximate region excised for scRNA-seq analysis.
The ciliary epithelium plays a central role in ocular homeostasis and, consequently, dysfunction of the epithelium can lead to serious ophthalmic diseases, including ectopia lentis and vitreous degeneration. To understand the etiology of such conditions and devise effective treatment strategies, a detailed knowledge of the transcriptional activity of the two layers of the ciliary epithelium is required. Investigators have used a variety of techniques to analyze gene expression patterns in this region of the eye, including expressed sequence tag (EST) libraries (Escribano and Coca-Prados, 2002; Wistow et al., 2008), microarrays (Diehn et al., 2005; Janssen et al., 2012; Ohta et al., 2005; Wagner et al., 2013), and PCR arrays (Dahlin et al., 2013). A key limitation in studying the transcriptional signatures of specific ocular cell types is the difficulty in physically isolating the cells of interest. In the case of the ciliary epithelium, this difficulty is compounded by the fact that the PCE and NPCE cells constitute collectively only a small fraction of the ciliary body tissue volume.
The recent development of single-cell RNA-sequencing (scRNA-seq) technology (Zheng et al., 2017) obviates the need for precise tissue dissection and may have particular utility in anatomically complex settings such as the ciliary body. The scRNA-seq approach, in which transcriptomes are obtained from individual cells collected from enzymatically-dissociated tissue, has been used recently to identify subtypes of ganglion cells (Macosko et al., 2015; Tran et al., 2019), bipolar cells (Shekhar et al., 2016), and amacrine cells (Yan et al., 2020) in the retina. In the anterior segment, scRNA-seq has provided insights into the stem cell niche in the corneal limbus (Collin et al., 2021; Kaplan et al., 2019), the identity of cells in the trabecular meshwork and Schlemm’s canal of human and animal eyes (Patel et al., 2020; van Zyl et al., 2020) and an initial characterization of the mouse ciliary epithelium (van Zyl et al., 2020). Here, as part of our investigations into the cellular origin of the ciliary zonule, we used scRNA-seq to characterize the transcriptional signatures of cells in the ciliary body and adjacent tissues of the mouse eye. The resulting cell atlas should prove useful when studying the role of the ciliary epithelium in ocular homeostasis.
2. Methods
2.1. Animals
All animal experiments were reviewed and approved by the Washington University animal studies committee and adhered to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. C57/BL6J or BALB/cJ mice were obtained from The Jackson Laboratory (Bar Harbor, ME). Animals were euthanized by CO2 inhalation.
2.2. Dissection and cell dissociation
Three one-month-old male C57/BL6J mice (six eyes) were used for each scRNA-seq experiment. Dissection was carried out under a stereomicroscope in pre-warmed Dulbecco’s Modified Eagle Medium (DMEM, Thermo Fisher Scientific, Waltham, MA). Enucleated globes were hemisected along a line parallel and 0.5 mm posterior to the limbus. In each case, the posterior half of the eye was removed. The cornea and iris were dissected from the anterior hemisphere and discarded. The zonular fibers were severed, freeing the lens and yielding a ring of tissue, comprising the ciliary body and contiguous tissues (see Fig. 1). Rings of tissue were then cut into four pieces and processed for cell dissociation.
2.3. Cell dissociation
Samples (each comprising tissue from six eyes, see above) were incubated in collagenase A (7 mg/ml; Sigma Aldrich, St. Louis, MO) for 30 min (37 °C) in 5 ml of TESCA buffer (50 mM TES, 0.3 mM CaCl2). For the final 10 min of the incubation period, DNase 1 (New England Bio-labs, Ipswich, MA) was added to the medium, at a final concentration of 0.5 U/μl.
The enzyme reaction was quenched by addition of 1.5 ml of DMEM/10% heat-inactivated fetal bovine serum (FBS). The sample was triturated gently for 5 min using a wide-bore pipette tip. The supernatant containing suspended cells was decanted and centrifuged for 10 min at 100×g. The cell pellet was resuspended in 220 μl of PBS containing 0.04% bovine serum albumin. Finally, cells were strained through a 40 μm Flowmi cell strainer (Sigma Aldrich). The number of cells was quantified using a hemocytometer and cell viability was measured by trypan blue exclusion. Typical cell yields from this procedure were 100,000–150,000 cells per preparation with >80% viability.
2.4. Droplet-based single-cell RNA sequencing
Two independent cell dissociation experiments were performed. In each case, suspensions of approximately 10,000 single cells were loaded into Chromium Single Cell chips (10x Genomics, Pleasanton, CA) (Zheng et al., 2017) and 5000 to 6000 cells were recovered. Single-cell libraries were generated using a Chromium Next GEM Single Cell 3' kit (v3.1). Normalized libraries were sequenced on a NovaSeq6000 S4 Flow Cell (Illumina, San Diego, CA) using the XP workflow. A median sequencing depth of 50,000 reads/cell was targeted for each Gene Expression Library. Reads were demultiplexed and aligned to the mouse reference genome (mm10–2222-A) using CellRanger software (version 6.0.0; 10x Genomics). Preprocessing of scRNA-seq data was performed using Seurat Version 4.0 (Hao et al., 2021; Satija et al., 2015), a bioinformatics package running in the R programming environment. Low quality cells were identified and filtered based on the percentage of reads mapping to the mitochondrial genome, the number of unique genes per cell, and the total number of molecules per cell. Following data normalization, a set of 2000 highly variable features (i.e., genes that were expressed strongly in some cells but only weakly in others) was identified (Macosko et al., 2015). Data were scaled and the number of principal components (PCs) was determined using the PC ElbowPlot function in Seurat. The number of PCs (in this case, 25) was used as an input variable in subsequent cell clustering operations using the FindNeighbors and FindClusters functions. The Findclusters resolution parameter, determining the granularity of the clustering operation, was set to 0.3. Data were visualized after non-linear dimension reduction using Uniform Manifold Approximation and Projection (UMAP). The identity of the resulting clusters was determined by expression of canonical markers or in situ hybridization analysis of differentially-expressed genes.
2.5. In situ hybridization
One-month-old mice (C57/BL6J or BALB/cJ) were euthanized by CO2 inhalation. Eye tissue was fixed overnight in 4% paraformaldehyde/PBS and dehydrated through a graded series of ethanols and xylene. Tissue was embedded in paraffin and sectioned in the midsagittal plane at a thickness of 4 μm. Expression of selected genes was visualized by in situ hybridization using the RNAscope technique (RNAscope 2.5 HD assay; Advanced Cell Diagnostics, Hayward, CA), as described (Shi et al., 2013; Wang et al., 2012). Target probe sets are listed in Table 1. Probes consisted of 20 pairs of oligonucleotides spanning a ≈1 kb region of the target mRNA transcript. Following proprietary preamplification and amplification steps, binding was visualized using an alkaline-phosphatase-conjugated probe with Fast Red as a substrate. As a negative control, adjacent tissue sections were incubated with a probe against DapB, a bacterial gene. The ubiquitously expressed PolR2A (DNA-directed RNA polymerase II polypeptide A) was used as a positive control. Results are representative of at least three independent experiments in each case. All in situ hybridization experiments were performed on both BALB/cJ and C57/BL6J mice with comparable results.
Table 1.
RNAscope probes used to visualize the expression of cluster-specific genes by in situ hybridization.
| Cell Type | Gene/Cat. No. | ||
|---|---|---|---|
|
| |||
| NPCE | Col18a1/483801 | Zic1/493121 | Crhbp/498971 |
| PCE | Gsta3/487251 | Slc4a4/452981 | |
| Melanocyte | Gjb2/518881 | Ednrb/473801 | |
| Fibroblast | Mmp3/480961 | Serpine2/435241 | |
2.6. Data availability
The data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE178667 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE178667).
3. Results
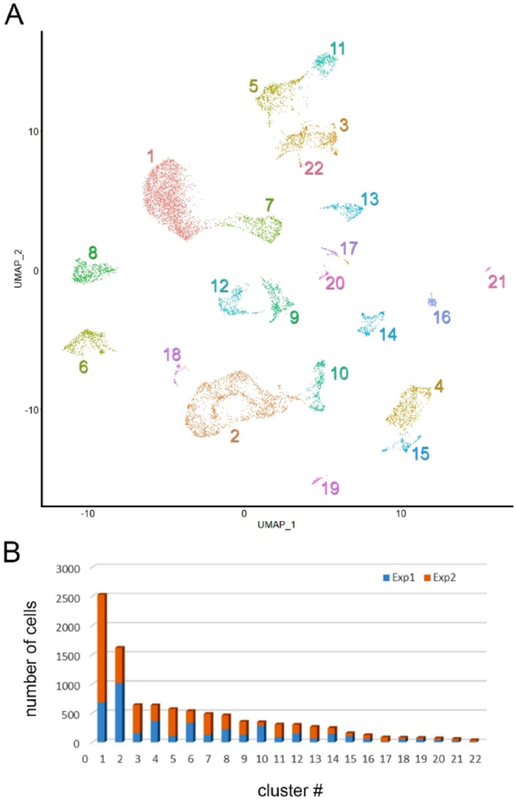
A Droplet-based single-cell sequencing approach was used to obtain transcriptomes from 10,024 individual cells dissociated from the ciliary body and adjacent tissues of the mouse by enzymatic treatment. The median gene count per cell was 1484 and the total number of genes detected was 20,804. Seurat was used to sort the cells into 22 individual clusters (Fig. 2A). Two independent experiments were performed. All clusters were identified in both experiments (Fig. 2B) and, consequently, data were combined for subsequent analyses.
Fig. 2.
Unsupervised clustering of single-cell transcriptomes obtained from dissociated mouse ciliary body and contiguous tissues. A. Clustering of 10,024 individual transcriptomes obtained from two independent experiments, visualized by uniform manifold approximation and projection (UMAP). Cell clusters are numbered according to cluster size, from the largest (cluster 1, containing 2531 cells) to the smallest (cluster 22, containing 38 cells). B. The number of cells per cluster obtained from experiment 1 (blue) and 2 (orange).
The identity of each cluster was determined using canonical markers and cluster-specific genes (Fig. 3). The marker genes used for the cell type assignment are listed in Table S1.
Fig. 3.
Cell type assignment using dot plots of differentially-expressed genes in the ciliary body and contiguous tissues (see also Table S2). Circle size is proportional to the fraction of cells within a cluster that express a particular gene. The color intensity indicates the average scaled expression of cells in that cluster. RGC, retinal ganglion cells; N/K T-cell, natural killer T-cells; rM-Macro-phage, resolution phase-macrophage; Vasc Endo, vascular endothelial cells; NPCE, non-pigmented ciliary epithelial cells; S-Fibroblasts, scleral fibroblasts, PCE, pigmented ciliary epithelial cells; RPE, retinal pigment epithelial cells; U-Fibroblasts, uveal fibro blasts.
Although we endeavored to cleanly dissect the ciliary body, several clusters corresponded to cells derived from the adjacent neural retina. These included, rods (C1 in Fig. 2a), cones (C13), amacrine cells (C3), bipolar cells (C5, C11), Müller glia (C7), and retinal ganglion cells (C22). Melanocytes (C6) and cells of the retinal pigment epithelium (RPE, C8) were also present. Immune cells, including neutrophils (C19), natural killer T-cells (C16), B-cells (C21), and a substantial population of macrophages (C4, C15) were recovered from the tissue, along with smooth muscle cells (C18), vascular endothelial cells (C14) and Schwann cells. Fibroblasts were numerous, forming two closely-related clusters (C2 and C10).
A spreadsheet showing the differentially-expressed genes that best defined the transcriptional signatures of each of the 22 cell clusters is included in the Supplemental Data (Table S2).
3.1. Comparison of PCE and NPCE
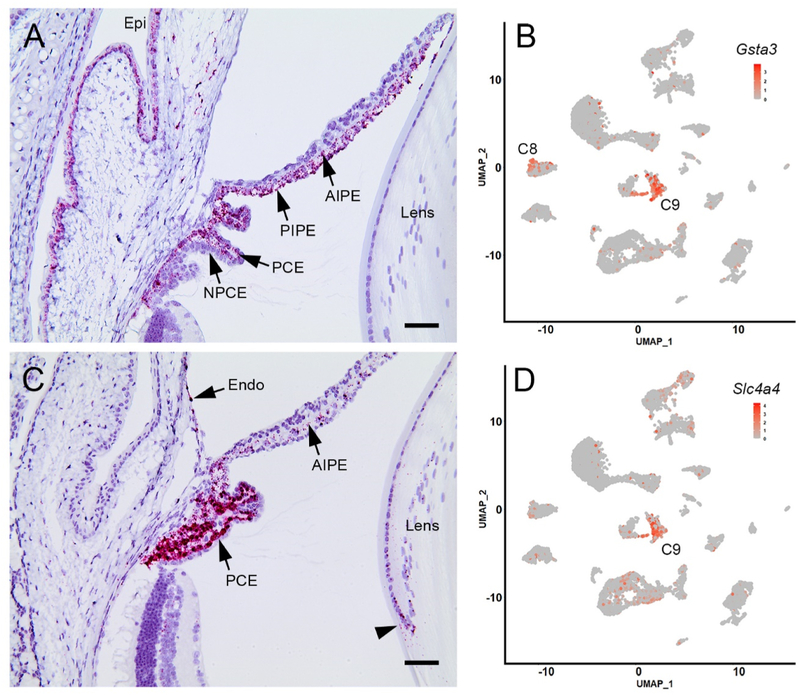
Ciliary epithelial cells formed two distinct clusters, C9 and C12 (Fig. 2A). Cells in cluster C12 expressed the canonical NPCE marker Best2 (Bakall et al., 2008). The identity of the cells was verified by in situ hybridization utilizing probes for three cluster-specific markers (Crhbp, Col18a1, Zic1, see below). Cells in cluster C9 expressed the canonical PCE marker Slc4a4 (Bok et al., 2001). PCE cell identity was verified by in situ hybridization, using probes against Slc4a4 and Gsta3, a cluster-specific gene (Fig. 4 and Table S2).
Fig. 4.
Heat map visualization of differences in gene expression between pigmented ciliary epithelial (PCE) cells and non-pigmented ciliary epithelial cells (NPCE). Genes involved in melanin synthesis are indicated by *.
Transcriptomes from 304 NPCE cells and 355 PCE cells were sequenced in the two experiments, representing 3.03% and 3.54%, respectively of the total cell population.
Heat map comparisons (Fig. 4) of the two layers showed, unsurprisingly, that with respect to the unpigmented NPCE cells, many of the most differentially expressed genes in the PCE cluster were involved in melanin synthesis (indicated by * in the heat map shown in Fig. 4). Among the differentially expressed genes that were not directly involved in the melanin synthesis pathway were Gsta3 (glutathione S-transferase A3) and Slc4a4 (encoding the electrogenic sodium-bicarbonate cotransporter). The expression of those two genes in the anterior segment of BALB/cJ mice was analyzed by in situ hybridization (Fig. 5). The absence of pigment from the BALB/cJ strain allowed the colored in situ hybridization reaction product to be visualized in the otherwise heavily pigmented cells of the PCE, RPE, and iris. Parallel experiments in C57Bl/6J mice yielded identical results, although with substantial masking of signal in pigmented layers (data not shown). In situ hybridization confirmed that Gsta3 (Fig. 5A) and Slc4a4 (Fig. 5C) transcripts were present in the PCE and absent from the NPCE, consistent with the scRNA-seq data. Scatter plot analysis (Fig. 5B,D) confirmed that the majority of Gsta3-and Slc4a4-expressing cells were located in C9. Expression of Slc4a4 ceased abruptly at the PCE/RPE border although Gsta3 expression was noted in some RPE cells (consistent with the results of scatter plot analysis showing the presence of a contingent of Gsta3-expressing cells in C8; Fig. 5B). Modest expression of Gsta3 was also noted in the corneal epithelium and both the anterior and posterior iris pigment epithelia (tissues that were excluded from the scRNA-seq analysis). Likewise, low levels of Slc4a4 expression were noted in equatorial lens cells, the corneal endothelium, and the anterior iris epithelium. PolR2A, the positive control gene, was broadly expressed in the eye, while no signal was detected for DapB, the negative control (data not shown).
Fig. 5.
Expression of the cluster 9-enriched markers Gsta3 and Slc4a4 in anterior segment tissues of BALB/cJ mice. A. In situ hybridization with a Gsta3 probe reveals strong labeling (red) in the pigmented ciliary epithelial (PCE) layer but not in the non-pigmented ciliary epithelium (NPCE). The anterior and posterior iris pigment epithelium (AIPE and PIPE) and the corneal (Epi) and conjunctival epithelia are also labeled. B. Cluster analysis indicates that Gsta3 expression is largely restricted to cells in cluster 9 (C9) although some expressing cells are located in C8. C. Slc4a4 is strongly expressed in the PCE, although transcripts are also detected at a low level in the anterior iris pigment epithelium (AIPE) corneal endothelium (endo) and equatorial lens cells (arrowhead). D. Scatter plot showing that Slc4a4-expressing cells are largely restricted to C9. Scale bar A,C = 50 μm.
Compared to the PCE, the transcriptome of the NPCE was characterized by differential expression of collagens (e.g., col9a1, col23a1, col18a1), members of the Zinc finger protein of cerebellum (Zic) family of transcription factors, opticin (optc), Na+/K+-ATPase isoforms (particularly Atp1a2 and Atp1b3), and transport proteins of the Slc6 family (e.g., Slc6a13, Slc6a6, slc6a9). One gene with unexpectedly strong expression in the NPCE was Crhbp (encoding corticotropin-releasing hormone binding protein).
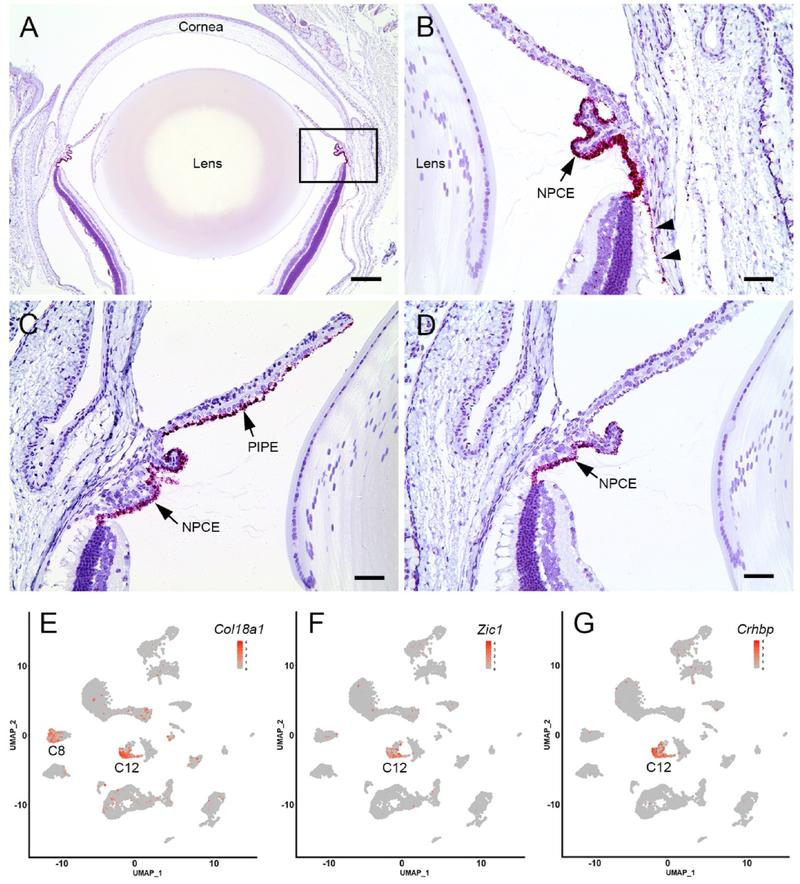
The expression of three NPCE-enriched transcripts was evaluated further using in situ hybridization (Fig. 6). Col18a1 encodes collagen XVIII, a heparin sulfate proteoglycan and a component of basal laminae. In humans, mutations in COL18A1 underlie Knobloch syndrome, characterized by multiple ocular symptoms including ectopia lentis. Marked expression of Col18a1 was detected in the NPCE with lower level expression in the RPE (Fig. 6A and B). Scatter plot analysis (Fig. 6E) indicated that Col18a1-expressing cells were located largely in cluster 12 (C12) with modest expression in C8, the RPE cell cluster (see Fig. 3). Zic1, 2, and 4 were among the most differentially expressed genes in NPCE cells (Fig. 4 and Table S2). We used in situ hybridization against one family member, Zic1, to localize its expression to the NPCE layer (Fig. 6C). Expression was also noted in the posterior iris pigment epithelium. Scatter plot analysis confirmed that Zic1-expressing cells were restricted to C12 (Fig. 6F). In situ hybridization indicated that expression of Crhbp was restricted to the NPCE (Fig. 6D) and scatter plot visualization (Fig. 6G) confirmed that Crhbp-expressing cells were confined almost exclusively to the NPCE cell cluster (C12), making Crhbp one of the most specific markers for NPCE cells identified in the present study.
Fig. 6.
Expression of three cluster 12-enriched markers in the anterior segment of the mouse eye. A. Expression of Col18a1 is particularly strong in the ciliary body. B. Higher magnification images (from the boxed region in A) indicate that the in situ hybridization reaction product (red) is present in cells of the non-pigmented ciliary epithelium (NPCE) and, to a lesser extent, the retinal pigment epithelium (arrow heads). C. Zic1 is strongly expressed by NPCE cells and cells in the posterior iris pigment epithelium (PIPE). D. In the anterior segment, Crhbp expression is limited to NPCE cells. E-G, Scatter plot analysis of Col18a1, Zic1 and Crhbp expression suggests that, in each case, expressing cells are largely restricted to cluster 12 (C12), with the exception of Col18a1, where a proportion of cells in C8 also express the gene. Scale bar A = 250 μm, B-D = 50 μm.
3.2. Melanocytes
The melanocytes (537 cells in total) recovered from the ciliary body preparation constituted about 5% of the total cell population. The transcriptional signatures of cells in the melanocyte cluster (C6) were characterized by the expression of canonical markers such as Cited1 (Shioda et al., 1996) and Mlph (Kuroda et al., 2003) (see Table S2). Among the most strongly expressed cluster-specific genes were Gjb2 and Gjb6, encoding two connexin family members (connexin26 and connexin30, respectively) and Ednrb, encoding an endothelin receptor. To visualize the distribution of the melanocyte population, in situ hybridization analysis was used to detect expression of Gjb2 and Ednrb in anterior chamber tissues of albino mice (Fig. 7). Melanocytes were distributed throughout the choroid and iris stroma but were excluded from the ciliary body. Corneal tissue was not included in the scRNA-seq analysis but in situ hybridization suggested that Ednrb and Gjb2 were expressed by corneal endothelial and epithelial cells, respectively. Scatter plots (Fig. 7B,D) confirmed that cells expressing either genes were confined to cluster 6 (C6).
Fig. 7.
Expression of two cluster 6-enriched genes, Gjb2 and Ednrb, in tissues of the anterior segment. In situ hybridization indicates that Gjb2- (A) and Ednrb-expressing cells (C) are distributed throughout the choroid (Ch) and iris stroma. Additionally, Gjb2 is expressed by corneal epithelial cells (Epi) and Ednrb is expressed by corneal endothelial cells (Endo). B,D. Scatter plots indicate that Ednrb- and Gjb2-expressing cells are largely confined to C6 (see also Fig. 2A). Scale bar A,C = 50 μm.
3.3. Fibroblasts
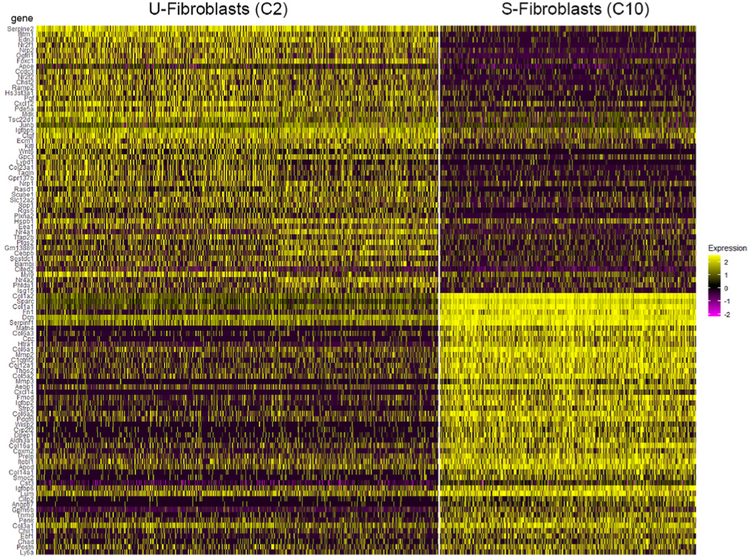
Fibroblasts were among the most abundant of the cell types identified in this study, collectively accounting for nearly 20% of all captured transcriptomes. The fibroblasts segregated into two distinct clusters, C2 and C10 (Fig. 2A) containing 1622 and 346 cells respectively. Differential expression analysis was used to compare transcriptional signatures of the two fibroblast types. The most significant differences were plotted as a heat map to show the expression across all cells in the dataset (Fig. 8).
Fig. 8.
Heat map comparison of gene expression in two fibroblast clusters (C2 and C10).
The smaller cluster, C10, was characterized by expression of genes encoding fibrillar (e.g., Col1a1, Col1a2, Col3a1, Col5a2) and nonfibrillar (e.g., Col6a3, Col14a1, Col16a1) collagens, and other ECM components, such as fibronectin (Fn1) and decorin (Dcn). A strong and relatively specific marker for this group was Mmp3, encoding matrix metalloproteinase-3. The larger cluster, C2, was characterized by comparatively weak collagen expression but strong expression of genes encoding signaling molecules such as connective tissue growth factor (Ctgf) and endothelin-3 (Edn3). The most differentially expressed gene and a good marker for this group was Serpine2, encoding a serine protease inhibitor.
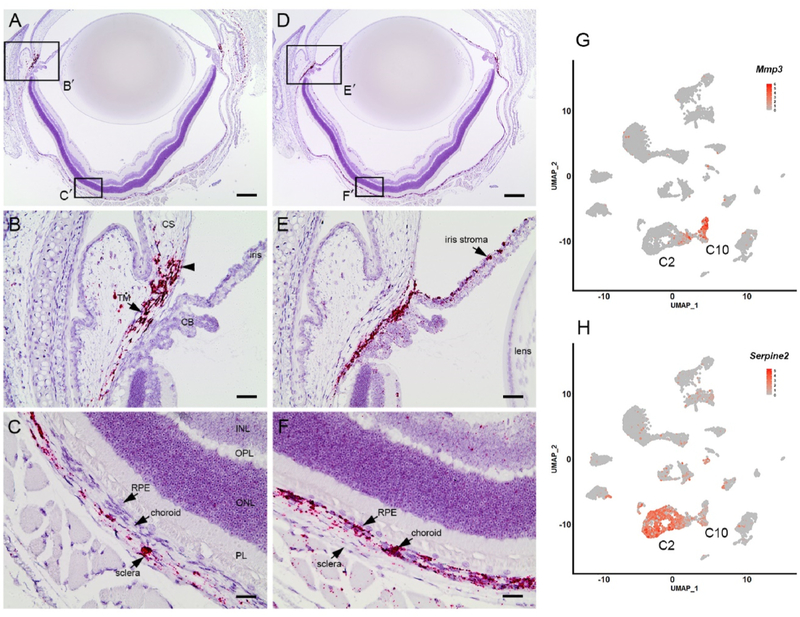
To determine whether the C10 and C2 fibroblasts had distinct anatomical distributions, we used in situ hybridization with probes against Mmp3 or Serpine2 (Fig. 9). Mmp3-expressing fibroblasts were present throughout the sclera and were consequently designated as scleral fibroblasts (S-Fibroblasts in Fig. 3A). Anteriorly, the Mmp3-expressing fibroblasts extended as far as the corneoscleral junction and the trabecular meshwork (Fig. 9A and B). Posteriorly, Mmp3-expressing cells were present throughout the sclera but were absent from the underlying choroid (Fig. 9C). By contrast, Serpine2 (a C2 marker)expressing cells were not detected in the sclera. Instead, positively-labeled cells were distributed throughout the uvea, including the choroid and the iris stroma (Fig. 9D–F). Consequently, those cells were designated as uveal fibroblasts (U-Fibroblasts in Fig. 3A).
Fig. 9.
Non-overlapping distribution of two putative fibroblast populations in the eye wall visualized using the cluster-specific markers, Mmp3 and Serpine2. The boxed regions (B', C', E', and F') in A and D are shown at higher magnification in B,C,E, and F. B. In situ hybridization identifies a population of Mmp3-expressing cells (red) at the corneoscleral junction (arrowhead) and the trabecular meshwork (TM). C. In the posterior segment, Mmp3-expressing cells are restricted to the sclera. E. In the anterior segment, Serpine2-expressing cells are present in the iris stroma. F. Posteriorly, Serpine2 expression is largely restricted to choroidal cells. G,H. Scatter plots confirm that Mmp3- and Serpine2-expressing cells are largely confined to clusters C10 and C2, respectively. CS, corneal stroma; CB, ciliary body; RPE, retinal pigment epithelium; PL, photoreceptor layer; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer. Scale bars A,D = 250 μm B,C,E, and F = 50 μm.
4. Discussion
It is impracticable to dissect specific cell types from anatomically complex tissues such as those that form the anterior segment of the eye. In such systems, gene expression measurements by bulk profiling techniques necessarily reflect population averages. With the introduction of scRNA-seq technology, however, it has become feasible to analyze the transcriptomes of previously inaccessible cell populations. We used this approach to generate a cell atlas for the ciliary body and contiguous tissues in the mouse eye.
Cluster analysis identified 22 discrete cell types, originating from the epithelial, vascular, neural, and connective tissues of the eye. A particular focus of our analysis were the NPCE and PCE layers of the ciliary body. Together, these cell types function to secrete the AH, contribute to the maintenance of intraocular pressure, synthesize the ciliary zonule, and produce key proteins of the vitreous humor. With regard to synthesis of the zonule, genes encoding the fibrillar proteins fibrillin-1 and −2, latent TGFβ-binding protein-2 (LTBP-2), and microfibril associated protein-2 (MFAP2) were expressed strongly by cells in the NPCE layer. Proteomic analysis has shown these proteins to be major components of the zonule (De Maria et al., 2017; Eckersley et al., 2018), contributing to the tensile properties of the zonular fibers (Jones et al., 2019; Shi et al., 2021). The current study showed that other microfibril-associated genes are expressed strongly by NPCE cells (e.g., Mfap4), suggesting that zonule composition is more complex than believed previously. Thus, the cell atlas can provide novel insights into NPCE function.
It has been suggested that cells in the pars plana region of the NPCE are primarily responsible for the production of zonule components such as fibrillin-1 and LTBP-2 (Shi et al., 2013, 2021). Cells in the pars plana are also thought to synthesize opticin and other components of the vitreous humor (Bishop et al., 2002). Given these observations, it was notable that the NPCE cells formed a single cluster, rather than two (or more) sub-clusters corresponding to cells derived from the pars plana and pars plicata regions. Our inability to resolve sub-clusters of NPCE cells may be a consequence of the limited number of cells (304 NPCE cells) available in this investigation. Studies in which multiple subtypes of amacrine cells or retinal ganglion cells were distinguishable utilized >30,000 cells in each case (Tran et al., 2019; Yan et al., 2020). NPCE cells constituted only 3% of the total number of cells recovered from the dissociated ciliary body. To test the hypothesis that the NPCE population includes two or more sub-clusters will likely require an enrichment step. Immunopanning or cell sorting using some of the cluster-specific cell surface markers identified in the current study may offer a practicable means of accumulating sufficient cells.
Among the most differentially expressed NPCE genes were members of the Zic family of transcription factors. Expression of Zic genes has been noted previously in the embryonic ciliary marginal zone in Xenopus and mice (Nagai et al., 1997; Nakata et al., 1998) and loss of function mutations in Zic2, Zic3, or Zic5 in mice can result in microphthalmia or anophthalmia (Diamand et al., 2018). The roles that this family of transcription factors play in the ciliary epithelium of adult animals is currently unclear but warrants further study. In addition to functioning as classical transcription factors in the regulation of gene expression, Zic proteins also modulate signaling through wnt, nodal, and sonic hedgehog, three pathways of critical importance in the eye (Diamand et al., 2018; Miesfeld and Brown, 2019).
One unexpected transcript identified in NPCE cells was Crhbp. Corticotropin-releasing hormone-binding protein (CRHBP) is an important component of the hypothalamus-pituitary-adrenal stress response axis, serving ultimately to antagonize cortisol production (Ketchesin et al., 2017). We have recently detected CRHBP in samples of human AH (De Maria et al., 2021) but its role in the eye is unclear. It is notable that some retinal amacrine cells synthesize corticotropin-releasing hormone (CRH) (Park et al., 2018; Skofitsch and Jacobowitz, 1984; Yan et al., 2020) and that CRH receptors are present on retinal cell membranes (Olianas and Onali, 1995). Therefore, intraocular production of CRHBP could potentially modulate signaling through retinal CRH receptors.
The two layers of the ciliary epithelium operate in tandem to produce AH. Fluid secretion requires the uptake of NaCl from stromal interstitial fluid by PCE cells, solute transfer between the two epithelial layers via gap junctions, and secretion of NaCl into the posterior chamber by the NPCE cells. The transepithelial movement of ions promotes the osmotic flux of water (Civan and Macknight, 2004). The physiology of AH secretion is still not completely understood and the cell atlas offers clues to the molecular identity of key transport proteins. Solute uptake into PCE cells involves the coupled movement of Na+, Cl−, and HCO3− via electroneutral symport and antiport systems. Physiological studies imply a role for the Ae2 anion exchanger (encoded by Slc4a2, and differentially expressed by PCE cells) and the Na+/H+ antiporter (encoded by Slc9a1 and broadly, if weakly, expressed by both PCE and NPCE cells) in solute uptake into PCE cells. The electroneutral Na-K-Cl symporter has also been implicated in this process (Dunn et al., 2001) and Slc12a2, the gene that encodes it, is expressed in the PCE layer. However, the expression level of Slc12a2 is weak compared to that of two electrogenic Na+/HCO3− cotransporters, Slc4a4 (encoding NBCe1) and slc4a5 (NBCe2), which are strongly and differentially expressed by PCE cells. The contribution of NBCe1/NBCe2 to AH production has not been tested directly but the notion that they have a role in AH formation is supported by the observation that mutations in SLC4A4 underlie a form of renal tubular acidosis (MIM#604278), in which glaucoma is part of the phenotype (Igarashi et al., 1999).
Both the PCE and NPCE cell layers express Gja1, the gene encoding the gap junction protein connexin43, and experiments in vitro suggest that this connexin likely constitutes the main conduit by which solutes pass from the PCE to the NPCE (Wang et al., 2010).
Extrusion of Na+ and Cl− into the posterior chamber is driven by Na+/K+-Atpase activity in NPCE cells. The catalytic subunit of the enzyme appears to be encoded by Atp1a2, which is strongly and specifically expressed by NPCE cells. The regulatory subunit, Atp1b3, is expressed more broadly in the eye, including by NPCE cells, consistent with earlier immunocytochemical studies on the distribution of Na+/K+-Atpase isoforms (Wetzel and Sweadner, 2001). Chloride is believed to enter the posterior chamber via chloride channels located in the basolateral membranes of the NPCE cells. Best2, encoding a calcium-activated chloride channel implicated previously in AH production (Bakall et al., 2008), was among the most differentially expressed genes in NPCE cells. Other Cl− channels, such as members of the epithelial chloride channel (E-ClC) and Tmem16 families, which might be expected a priori to contribute to AH production, were not expressed in NPCE cells. In many cases, the single cell data help verify the existence of transport processes identified previously using traditional physiological or pharmacological strategies. However, in some instances, the cell atlas may help identify genes with unsuspected roles in AH production. One example may be Slc26a4, a gene that is strongly and specifically expressed by NPCE cells. Slc26a4 encodes pendrin, a protein with a well-studied role in renal physiology and blood pressure regulation (Wall et al., 2020). Pendrin is an apically-located Na+-independent Cl−/HCO3− /OH− transporter which, to our knowledge, has not previously been studied in regard to AH production. The final step in AH production, water extrusion into the posterior chamber, is mediated by water channels. The scRNA-seq data indicate that Aqp4, encoding aquaporin-4, is strongly expressed by NPCE cells (Table S2).
Two distinct populations of fibroblasts were detected. One group (S-fibroblasts), characterized by expression of Mmp3, was present throughout the sclera. Anteriorly, Mmp3-expressing cells were also prominent in the chamber angle and corneoscleral junction. The second group (U-fibroblasts), marked by expression of Serpine2, was distributed throughout the uvea, including the iris. Fibroblasts are among the most heterogeneous and plastic of cell types and several fibroblast lineages can coexist within a single tissue or organ. In the healthy lung, for example, six types of pulmonary fibroblasts have been distinguished (Xie et al., 2018) and three distinct fibroblast lineages are present in endometrial tissue (Kirkwood et al., 2021). Recent scRNA-seq studies on anterior segment tissues imply that ocular fibroblasts may be similarly diverse. For example, scRNA-seq studies on human cornea have identified four distinct populations of fibroblast-like cells: limbal fibroblasts, limbal stromal keratocytes, corneal stromal stem cells, and central stromal keratocytes (Collin et al., 2021). Additionally, fibroblast-like and myofibroblast-like cell populations have been identified in the trabecular meshwork (Patel et al., 2020). Furthermore, studies on the conventional outflow pathway of humans and animal models have shown the presence of discrete populations of fibroblast-like cells associated with the trabecular beams and juxtacannalicular tissue (JCT) (van Zyl et al., 2020). The transcriptional signatures of our “U-fibroblasts ” and “S-fibroblasts ” appear to most closely resemble those of JCT and BeamY cells, respectively. To date, scRNA-seq studies have utilized a variety of ocular tissues from both human sources and animal models. Consequently, a consensus on the taxonomy of the ocular fibroblast populations has yet to emerge. This is an important issue because remodeling of ocular connective tissue, for example during myopia progression or in response to trauma or surgery, is likely facilitated by cells of a fibroblast lineage. An analysis of the interrelationships between fibroblast populations in the eye will have implications for the understanding and treatment of a range of ocular conditions.
It should soon be possible to catalog each of the cell types present in the eyes of humans and model organisms, in line with the broad aims of the human cell atlas organization (Rozenblatt-Rosen et al., 2017). This knowledge base will provide the foundation for future studies on ocular pathology and development. Single cell techniques lend themselves to studies of organogenesis (Haniffa et al., 2021) and scRNA-seq has already been successfully deployed to analyze the processes of competence, specification, and differentiation in the developing retina (Shiau et al., 2021). It is to be expected that the inductive interactions that control the developmental trajectories of cells in the anterior segment will prove similarly amenable to analysis at the single cell level.
Supplementary Material
Acknowledgements
Supported by National Institutes of Health Grants R01 EY029130, R01 EY009852, and P30 EY002687. Additional funding was provided by The Marfan Foundation, the Grace Nelson Lacy Glaucoma Research fund, and an unrestricted grant to the Department of Ophthalmology & Visual Sciences from Research to Prevent Blindness. We thank Phil Ruzycki, Brian Clark, and Fion Shiau for their helpful advice during the course of the project, Seta Dikranian and Qing Tan for their assistance with the in situ hybridization experiments, and the Genome Technology Access Center (GTAC) at Washington University School of Medicine for help with genomic analysis. GTAC is partially supported by P30 CA91842 and by ICTS/CTSA grant UL1 TR000448 from the National Center for Research Resources (NCRR).
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.exer.2021.108811.
References
- Bakall B, McLaughlin P, Stanton JB, Zhang Y, Hartzell HC, Marmorstein LY, Marmorstein AD, 2008. Bestrophin-2 is involved in the generation of intraocular pressure. Invest. Ophthalmol. Vis. Sci. 49, 1563–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassnett S, 2021. Zinn’s zonule. Prog. Retin. Eye Res. 82, 100902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop PN, Takanosu M, Le Goff M, Mayne R, 2002. The role of the posterior ciliary body in the biosynthesis of vitreous humour. Eye 16, 454–460. [DOI] [PubMed] [Google Scholar]
- Bok D, Schibler MJ, Pushkin A, Sassani P, Abuladze N, Naser Z, Kurtz I, 2001. Immunolocalization of electrogenic sodium-bicarbonate cotransporters pNBC1 and kNBC1 in the rat eye. Am. J. Physiol. Ren. Physiol 281, F920–F935. [DOI] [PubMed] [Google Scholar]
- Civan MM, Macknight AD, 2004. The ins and outs of aqueous humour secretion. Exp. Eye Res. 78, 625–631. [DOI] [PubMed] [Google Scholar]
- Collin J, Queen R, Zerti D, Bojic S, Dorgau B, Moyse N, Molina MM, Yang C, Dey S, Reynolds G, Hussain R, Coxhead JM, Lisgo S, Henderson D, Joseph A, Rooney P, Ghosh S, Clarke L, Connon C, Haniffa M, Figueiredo F, Armstrong L, Lako M, 2021. A single cell atlas of human cornea that defines its development, limbal progenitor cells and their interactions with the immune cells. Ocul. Surf 21, 279–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlin A, Geier E, Stocker SL, Cropp CD, Grigorenko E, Bloomer M, Siegenthaler J, Xu L, Basile AS, Tang-Liu DD, Giacomini KM, 2013. Gene expression profiling of transporters in the solute carrier and ATP-binding cassette superfamilies in human eye substructures. Mol. Pharm. 10, 650–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maria A, Wilmarth PA, David LL, Bassnett S, 2017. Proteomic analysis of the bovine and human ciliary zonule. Invest. Ophthalmol. Vis. Sci. 58, 573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maria A, Zientek KD, David LL, Wilmarth PA, Bhorade AM, Harocopos GJ, Huang AJW, Hong AR, Siegfried CJ, Tsai LM, Sheybani A, Bassnett S, 2021. Compositional analysis of extracellular aggregates in the eyes of patients with exfoliation syndrome and exfoliation glaucoma. Invest. Ophthalmol. Vis. Sci. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamand KEM, Barratt KS, Arkell RM, 2018. Overview of rodent zic genes. Adv. Exp. Med. Biol. 1046, 179–207. [DOI] [PubMed] [Google Scholar]
- Diehn JJ, Diehn M, Marmor MF, Brown PO, 2005. Differential gene expression in anatomical compartments of the human eye. Genome Biol. 6, R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JJ, Lytle C, Crook RB, 2001. Immunolocalization of the Na-K-Cl cotransporter in bovine ciliary epithelium. Invest. Ophthalmol. Vis. Sci. 42, 343–353. [PubMed] [Google Scholar]
- Eckersley A, Mellody KT, Pilkington S, Griffiths CEM, Watson REB, O’Cualain R, Baldock C, Knight D, Sherratt MJ, 2018. Structural and compositional diversity of fibrillin microfibrils in human tissues. J. Biol. Chem. 293, 5117–5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE, 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano J, Coca-Prados M, 2002. Bioinformatics and reanalysis of subtracted expressed sequence tags from the human ciliary body: identification of novel biological functions. Mol. Vis. 8, 315–332. [PubMed] [Google Scholar]
- Fernandez-Nogales M, Murcia-Belmonte V, Chen HY, Herrera E, 2019. The peripheral eye: a neurogenic area with potential to treat retinal pathologies? Prog. Retin. Eye Res. 68, 110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniffa M, Taylor D, Linnarsson S, Aronow BJ, Bader GD, Barker RA, Camara PG, Camp JG, Chedotal A, Copp A, Etchevers HC, Giacobini P, Gottgens B, Guo G, Hupalowska A, James KR, Kirby E, Kriegstein A, Lundeberg J, Marioni JC, Meyer KB, Niakan KK, Nilsson M, Olabi B, Pe’er D, Regev A, Rood J, Rozenblatt-Rosen O, Satija R, Teichmann SA, Treutlein B, Vento-Tormo R, Webb S, Human Cell Atlas Developmental Biological, N, 2021. A roadmap for the human developmental cell atlas. Nature 597, 196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R, 2021. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi T, Inatomi J, Sekine T, Cha SH, Kanai Y, Kunimi M, Tsukamoto K, Satoh H, Shimadzu M, Tozawa F, Mori T, Shiobara M, Seki G, Endou H, 1999. Mutations in SLC4A4 cause permanent isolated proximal renal tubular acidosis with ocular abnormalities. Nat. Genet. 23, 264–266. [DOI] [PubMed] [Google Scholar]
- Janssen SF, Gorgels TG, Bossers K, Ten Brink JB, Essing AH, Nagtegaal M, van der Spek PJ, Jansonius NM, Bergen AA, 2012. Gene expression and functional annotation of the human ciliary body epithelia. PLoS One 7, e44973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones W, Rodriguez J, Bassnett S, 2019. Targeted deletion of fibrillin-1 in the mouse eye results in ectopia lentis and other ocular phenotypes associated with Marfan syndrome. Dis. Model Mech 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Wang J, Wray B, Patel P, Yang W, Peng H, Lavker RM, 2019. Single-cell RNA transcriptome helps define the limbal/corneal epithelial stem/early transit amplifying cells and how autophagy affects this population. Invest. Ophthalmol. Vis. Sci. 60, 3570–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketchesin KD, Stinnett GS, Seasholtz AF, 2017. Corticotropin-releasing hormone-binding protein and stress: from invertebrates to humans. Stress 20, 449–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood PM, Gibson DA, Smith JR, Wilson-Kanamori JR, Kelepouri O, Esnal-Zufiaurre A, Dobie R, Henderson NC, Saunders PTK, 2021. Single-cell RNA sequencing redefines the mesenchymal cell landscape of mouse endometrium. Faseb. J. 35, e21285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda TS, Ariga H, Fukuda M, 2003. The actin-binding domain of Slac2-a/melanophilin is required for melanosome distribution in melanocytes. Mol. Cell Biol. 23, 5245–5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA, 2015. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesfeld JB, Brown NL, 2019. Eye organogenesis: a hierarchical view of ocular development. Curr. Top. Dev. Biol. 132, 351–393. [DOI] [PubMed] [Google Scholar]
- Nagai T, Aruga J, Takada S, Gunther T, Sporle R, Schughart K, Mikoshiba K, 1997. The expression of the mouse Zic1, Zic2, and Zic3 gene suggests an essential role for Zic genes in body pattern formation. Dev. Biol. 182, 299–313. [DOI] [PubMed] [Google Scholar]
- Nakata K, Nagai T, Aruga J, Mikoshiba K, 1998. Xenopus Zic family and its role in neural and neural crest development. Mech. Dev. 75, 43–51. [DOI] [PubMed] [Google Scholar]
- Ohta K, Kikuchi T, Miyahara T, Yoshimura N, 2005. DNA microarray analysis of gene expression in iris and ciliary body of rat eyes with endotoxin-induced uveitis. Exp. Eye Res. 80, 401–412. [DOI] [PubMed] [Google Scholar]
- Olianas MC, Onali P, 1995. G protein-coupled corticotropin-releasing hormone receptors in rat retina. Regul. Pept. 56, 61–70. [DOI] [PubMed] [Google Scholar]
- Park SJH, Pottackal J, Ke JB, Jun NY, Rahmani P, Kim IJ, Singer JH, Demb JB, 2018. Convergence and divergence of CRH amacrine cells in mouse retinal circuitry. J. Neurosci. 38, 3753–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel G, Fury W, Yang H, Gomez-Caraballo M, Bai Y, Yang T, Adler C, Wei Y, Ni M, Schmitt H, Hu Y, Yancopoulos G, Stamer WD, Romano C, 2020. Molecular taxonomy of human ocular outflow tissues defined by single-cell transcriptomics. Proc. Natl. Acad. Sci. U. S. A. 117, 12856–12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reh TA, Levine EM, 1998. Multipotential stem cells and progenitors in the vertebrate retina. J. Neurobiol. 36, 206–220. [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Stubbington MJT, Regev A, Teichmann SA, 2017. The human cell atlas: from vision to reality. Nature 550, 451–453. [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A, 2015. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, McCarroll SA, Cepko CL, Regev A, Sanes JR, 2016. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166, 1308–1323 e1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Jones W, Beatty W, Tan Q, Mecham RP, Kumra H, Reinhardt DP, Gibson MA, Reilly MA, Rodriguez J, Bassnett S, 2021. Latent-transforming growth factor beta-binding protein-2 (LTBP-2) is required for longevity but not for development of zonular fibers. Matrix Biol. 95, 15–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Tu Y, De Maria A, Mecham RP, Bassnett S, 2013. Development, composition, and structural arrangements of the ciliary zonule of the mouse. Invest. Ophthalmol. Vis. Sci. 54, 2504–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiau F, Ruzycki PA, Clark BS, 2021. A single-cell guide to retinal development: cell fate decisions of multipotent retinal progenitors in scRNA-seq. Dev. Biol. 478, 41–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioda T, Fenner MH, Isselbacher KJ, 1996. msg1, a novel melanocyte-specific gene, encodes a nuclear protein and is associated with pigmentation. Proc. Natl. Acad. Sci. U. S. A. 93, 12298–12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skofitsch G, Jacobowitz DM, 1984. Corticotropin releasing factor-like immunoreactive neurons in the rat retina. Brain Res. Bull. 12, 539–542. [DOI] [PubMed] [Google Scholar]
- Smith RS, 2002. Systematic Evaluation of the Mouse Eye: Anatomy, Pathology, and Biomethods. CRC Press, Boca Raton. [Google Scholar]
- Tran NM, Shekhar K, Whitney IE, Jacobi A, Benhar I, Hong G, Yan W, Adiconis X, Arnold ME, Lee JM, Levin JZ, Lin D, Wang C, Lieber CM, Regev A, He Z, Sanes JR, 2019. Single-cell profiles of retinal ganglion cells differing in resilience to injury reveal neuroprotective genes. Neuron 104, 1039–1055 e1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zyl T, Yan W, McAdams A, Peng YR, Shekhar K, Regev A, Juric D, Sanes JR, 2020. Cell atlas of aqueous humor outflow pathways in eyes of humans and four model species provides insight into glaucoma pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 117, 10339–10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AH, Anand VN, Wang WH, Chatterton JE, Sun D, Shepard AR, Jacobson N, Pang IH, Deluca AP, Casavant TL, Scheetz TE, Mullins RF, Braun TA, Clark AF, 2013. Exon-level expression profiling of ocular tissues. Exp. Eye Res. 111, 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall SM, Verlander JW, Romero CA, 2020. The renal physiology of pendrin-positive intercalated cells. Physiol. Rev. 100, 1119–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y, 2012. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Do CW, Valiunas V, Leung CT, Cheng AK, Clark AF, Wax MB, Chatterton JE, Civan MM, 2010. Regulation of gap junction coupling in bovine ciliary epithelium. Am. J. Physiol. Cell Physiol. 298, C798–C806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel RK, Sweadner KJ, 2001. Immunocytochemical localization of NaK-ATPase isoforms in the rat and mouse ocular ciliary epithelium. Invest. Ophthalmol. Vis. Sci. 42, 763–769. [PubMed] [Google Scholar]
- Wistow G, Peterson K, Gao J, Buchoff P, Jaworski C, Bowes-Rickman C, Ebright JN, Hauser MA, Hoover D, 2008. NEIBank: genomics and bioinformatics resources for vision research. Mol. Vis. 14, 1327–1337. [PMC free article] [PubMed] [Google Scholar]
- Xie T, Wang Y, Deng N, Huang G, Taghavifar F, Geng Y, Liu N, Kulur V, Yao C, Chen P, Liu Z, Stripp B, Tang J, Liang J, Noble PW, Jiang D, 2018. Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep. 22, 3625–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Laboulaye MA, Tran NM, Whitney IE, Benhar I, Sanes JR, 2020. Mouse retinal cell atlas: molecular identification of over sixty amacrine cell types. J. Neurosci. 40, 5177–5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH, 2017. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE178667 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE178667).