Abstract
Six distinct clones were present among Greek multidrug-resistant Salmonella enterica serotype Typhimurium phage type DT104, since isolates belonging to resistance phenotypes including the ACSSuT (ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline) core could be distinguished with respect to their pulsed-field gel electrophoresis patterns, int1 integron structures, and presence or absence of antibiotic resistance genes ant(3")-Ia, pse-1, and tem-1.
In recent years, a marked increase in the number of multidrug-resistant Salmonella enterica serotype Typhimurium isolates belonging to definitive phage type 104 (DT104) and having a core pattern of resistance to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline (ACSSuT) has been reported (3, 4, 13, 14). Molecular studies have demonstrated that in DT104 some of the resistance genes responsible for the ACSSuT phenotype are integron associated (8, 12).
To date, all reports have suggested that multidrug-resistant DT104 isolates are clonally related (R. Prager, A. Liesegang, W. Streckel, B. Gerike, G. Seltmann, R. Helmuth, W. Rabsch, and H. Tschape, Fourth Int. Meet. Bacterial Epidemiol. Markers, abstr. P40, 1997; 7, 8). In this study, we aimed to analyze the genetic relationships of multidrug-resistant serotype Typhimurium isolates from Greece, with respect to their chromosomal fingerprints and mechanisms of resistance.
During 1989 to 1997, a total of 1,005 Salmonella isolates of human, animal feed, animal, and food origins from various parts of Greece were referred to the National Reference Center for Salmonella and Shigella. They were identified as serotype Typhimurium by the API 20E system (BioMerieux S.A., Marcy l'Etoile, France) and the Kauffman serotyping scheme (5), using commercially obtained antisera (BioMerieux). Serotype Typhimurium represented the second most frequent serotype isolated from humans, as for 1987 to 1993 (10), standing at 17% in 1997. The organism was also an important serotype in isolates of animal and food sources.
Of the 1,005 serotype Typhimurium isolates, 328 (33%) were randomly selected for antimicrobial susceptibility testing by a disk diffusion method on Mueller-Hinton agar (Oxoid Ltd., Basingstoke, United Kingdom), evaluated according to the standards of the National Committee for Clinical Laboratory Standards (6). Disks containing ampicillin, amoxicillin and clavulanic acid, cefalothin, cefamandole, ceftriaxone, ceftazidime, cefotaxime, streptomycin, kanamycin, gentamicin, chloramphenicol, doxycycline, nalidixic acid, ciprofloxacin, sulfonamides, sulfomethoxazole and trimethoprim, and nitrofurantoin were purchased from Oxoid. The isolates were grouped into 26 resistance phenotypes, the major ones being shown in Table 1. Eighty-four (26%) isolates were resistant to the ACSSuT core alone or to additional drugs as well. Thus, 11 distinct resistance phenotypes were compatible with the ACSSuT core resistance phenotype of multidrug-resistant serotype Typhimurium DT104. Fourteen isolates (17%) representing seven of these resistance phenotypes were therefore randomly selected for further analysis.
TABLE 1.
Main resistance phenotypes of serotype Typhimurium (1989–1997)
| Resistance phenotypea | No. of isolates for yr:
|
Total no. | |||||
|---|---|---|---|---|---|---|---|
| 1989 | 1991 | 1993 | 1995 | 1996 | 1997 | ||
| Susceptible | 10 | 7 | 16 | 8 | 7 | 6 | 54 |
| S | 2 | 1 | 2 | 1 | 7 | 12 | 25 |
| Su | 0 | 1 | 2 | 1 | 3 | 2 | 9 |
| T | 1 | 0 | 1 | 0 | 0 | 1 | 3 |
| ST | 2 | 3 | 1 | 3 | 6 | 6 | 21 |
| SSu | 2 | 5 | 6 | 4 | 14 | 19 | 50 |
| SSuT | 6 | 5 | 1 | 2 | 2 | 3 | 19 |
| STFu | 2 | 3 | 1 | 0 | 0 | 4 | 10 |
| SSuTFu | 0 | 1 | 3 | 1 | 1 | 2 | 8 |
| SSuTSxt | 0 | 1 | 3 | 4 | 3 | 7 | 17 |
| SSuTGKNx | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| ASSu | 0 | 1 | 1 | 4 | 0 | 0 | 6 |
| ASSuT | 0 | 4 | 1 | 2 | 2 | 4 | 13 |
| ASSuTSxt | 0 | 0 | 3 | 1 | 2 | 1 | 7 |
| ASSuTGKNx | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| ACSSuT | 1 | 1 | 2 | 5 | 0 | 0 | 9 |
| ACSSuTAmc | 0 | 1 | 1 | 4 | 1 | 0 | 7 |
| ACSSuTFu | 0 | 0 | 0 | 0 | 3 | 6 | 9 |
| ACSSuTAmcFu | 0 | 1 | 3 | 15 | 21 | 3 | 43 |
| ACSSuTAmcFu+ | 0 | 1 | 2 | 5 | 5 | 3 | 16 |
| Total | 26 | 35 | 49 | 60 | 78 | 80 | 328 |
A, ampicillin; T, tetracycline; S, streptomycin; Su, sulfonamides; Fu, nitrofurantoin; C, chloramphenicol; Sxt, cotrimoxazole; Amc, amoxicillin-clavulanic acid; Nx, nalidixic acid; G, gentamicin; K, kanamycin; +, any other antibiotic. Intermediate and high resistance are grouped together.
Phage typing was performed by the methods described by Callow (2). The scheme extended by Anderson et al. (1) uses 34 typing phages and differentiates in excess of 250 types. Of the 14 isolates, 11 belonged to phage type DT104 and 2 belonged to the related type DT104b, differing by a single phage reaction from DT104, while the remaining isolate belonged to the unrelated phage type DT193.
In conjugation experiments for the transfer of antibiotic resistance, carried out as previously described (16), only two DT104 isolates (195 and 1041) were able to transfer resistance to ampicillin, tetracycline, chloramphenicol, streptomycin, and cotrimoxazole.
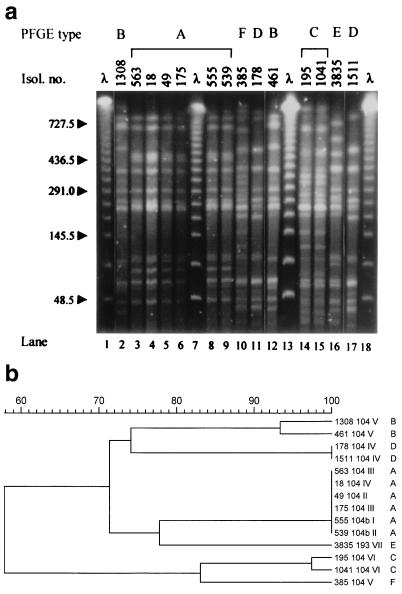
Typing by pulsed-field gel electrophoresis (PFGE) of genomic DNA digested with XbaI (New England Biolabs, Beverly, Mass.) was performed as previously described (10). DNA fragment patterns were assessed visually and compared by the GelCompar software package (Applied Maths, Kortrijk, Belgium), using the Dice coefficient, UPGMA (unweighted pair group method using arithmetic averages) clustering, and 1% tolerance in band position differences. Isolates were considered as belonging to different types if differing by four or more DNA fragments (11). Given the genetic homogeneity of Salmonella populations (10), this criterion can be considered stringent. Five distinct types, A to D and F, were observed among DT104 isolates (Fig. 1). Four isolates belonged to type A, with the remaining seven distributed among types B (two isolates), C (two), D (two), and F (one).
FIG. 1.
(a) PFGE patterns of 14 multidrug-resistant serotype Typhimurium isolates. Isol. no., isolate number. The sizes, in kilobases, of lambda phage DNA concatemers (λ) are shown to the left of the gel. All lanes are from the same gel. (b) Dendrogram of isolate similarities based on the chromosomal fingerprints shown in panel a. Isolate number, phage type, resistance phenotype code, and PFGE type are shown to the right of the dendrogram. A percent scale of similarity is shown above the dendrogram.
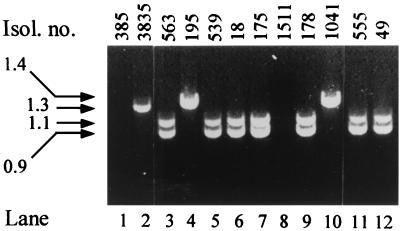
For PCR amplification of integron-related and resistance gene sequences, DNA was prepared as previously described (17). PCR was carried out in 50-μl-total-volume reactions containing 1.5 mM MgCl2, 100 μM (each) deoxynucleoside triphosphate, 0.1 U of Taq polymerase (Promega, Madison, Wis.), 1 μl of bacterial lysate, and 0.4 μM primers for int1 integron (8), ant(3")-Ia (17), pse-1 (8), and tem-1 sequences (9). Under conditions used, three distinct amplicons were obtained from the DT104 isolates with the int1 primers (Fig. 2). Two PCR products of approximately 0.9 and 1.1 kb were generated from five isolates belonging to PFGE types A and D, while a product of approximately 1.4 kb was generated from the two DT104 isolates belonging to PFGE type C. No int1 PCR product could be obtained from the remaining four DT104 isolates belonging to PFGE types B, D, and F. All DT104 isolates that were positive for int1-type integron sequences also yielded an ant(3")-I product of the expected size, 0.5 kb (Table 2). Of the four isolates (1308, 461, 385, and 1511) from which no int1 or ant(3")-I products could be generated, all were intermediately resistant to streptomycin. Amplicons of the expected size, 0.3 kb, were obtained with the pse-1 primers from five DT104 isolates, while amplicons of the expected size for tem-1, 0.7 kb, were detected in four DT104 isolates (Table 2). In contrast to ant(3")-Ia and pse-1 sequences, which were shown by PCR to be internal to int1 amplicons, the tem-1 gene was not contained in an integron gene cassette (data not shown). All strains positive for int1 sequences also contained qacE-1 and sul1 sequences (not shown).
FIG. 2.
PCR amplification products of class 1 integrons from representative multidrug-resistant serotype Typhimurium isolates. Isol. no., isolate number. The sizes, in kilobases, of the indicated amplification products are shown to the left of the gel. All lanes are from the same gel.
TABLE 2.
Genotypic and phenotypic characteristics of multidrug-resistant serotype Typhimurium isolates
| Isolate no. | Isolate yr | Origin | Resistance phenotypea | Phage type | PFGE type |
int1
|
ant(3) | pse-1 | tem-1 | Conj.c | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.9 kb | 1.1 kb | 1.3 kb | 1.4 kb | ||||||||||
| 555 | 1997 | Frozen chicken | ACSSuT [I]b | DT104b | A | + | + | + | + | ||||
| 539 | 1997 | Human feces | ACSSuTFu [II] | DT104b | A | + | + | + | + | ||||
| 49 | 1996 | Sewage filter | ACSSuTFu | DT104 | A | + | + | + | + | ||||
| 175 | 1996 | Frozen pork | ACSSuTAmcSxt [III] | DT104 | A | + | + | + | + | ||||
| 563 | 1997 | Ground chicken carcasses | ACSSuTAmcSxt [III] | DT104 | A | + | + | + | + | ||||
| 1308 | 1991 | Human feces | AC(S)SuTAmcFu [IV] | DT104 | B | + | |||||||
| 461 | 1993 | Human feces | AC(S)SuTAmcFu | DT104 | B | + | |||||||
| 385 | 1997 | Sewage filter | AC(S)SuTAmcFu | DT104 | F | ||||||||
| 18 | 1995 | Human feces | ACSSuTAmcFu [V] | DT104 | A | + | + | + | + | ||||
| 1511 | 1995 | Pigeon liver | ACSSuTAmcFu | DT104 | D | ||||||||
| 178 | 1996 | Sewage filter | ACSSuTAmcFu | DT104 | D | + | + | + | + | ||||
| 1041 | 1995 | Human feces | ACSSuTAmc(Fu)Sxt [VI] | DT104 | C | + | + | + | + | ||||
| 195 | 1997 | Human feces | ACSSuTAmc(Fu)Sxt | DT104 | C | + | + | + | + | ||||
| 3835 | 1996 | Human feces | ACSSuTAmcCFCroCtxFuGKMa [VII] | DT193 | E | + | + | ||||||
A, ampicillin; Amc, amoxicillin-clavulanic acid; T, tetracycline; S, streptomycin; C, chloramphenicol; Su, sulfonamides; Fu, nitrofurantoin; Sxt, cotrimoxazole; G, gentamicin; K, kanamycin; CF, cefalothin; Ma, cefamandole; Ctx, cefotaxime; Cro, ceftriaxone. Parentheses indicate intermediate resistance.
Roman numerals in brackets indicate the resistance phenotype.
Conj., transfer of resistance by conjugation.
Therefore, of the five PFGE types within phage type DT104, type A was prevalent. Type A isolates contained two distinct int1 integrons, detected by PCR products of 0.9 and 1.1 kb, as well as ant(3")-Ia and pse-1 sequences. Similar studies have found that the small integron harbors the aminoglycoside-resistant gene cassette ant(3")-Ia, conferring resistance to streptomycin and spectinomycin, while the large amplicon harbors the pse-1 β-lactamase gene (7, 8). The variety in integron size and presence or absence of aminoglycoside-modifying enzymes and β-lactamases, set against a background of different chromosomal types, resulting in six distinct types among Greek multidrug resistant DT104 isolates, is reminiscent of other published data on multidrug resistant serotype Typhimurium (15). In their study, Tosini et al. (15) found three distinct class 1 integrons located on two conjugative plasmids. In our study, transfer of resistance by conjugation was possible only with isolates in which the tem-1 gene was not inserted in a 1.4-kb integron. In contrast, resistance of isolates harboring the ant(3")-Ia and pse-1 gene cassettes inserted in two integrons was not transferable by conjugation, presumably due to the chromosomal location of these integrons (12).
It therefore appears that common phenotypic characteristics of antibiotic resistance and phage type may have been acquired by genotypically distinct multidrug-resistant serotype Typhimurium DT104 strains in Greece. This differentiates them from those isolated in other countries (Prager et al., Fourth Int. Meet. Bacterial Epidemiol. Markers; 7, 8), where clonality of multidrug-resistant serotype Typhimurium DT104 has been suggested. While we cannot rule out that the presence of various clones within multidrug-resistant DT104 isolates may be characteristic of the Greek situation, additional molecular studies from regions with a high rate of multidrug resistance in serotype Typhimurium, among both human and nonhuman isolates, would help to elucidate the origin and extent of this problem.
Acknowledgments
We gratefully acknowledge the expert technical assistance of Vasiliki Kontogianni, Zannina Sarandopoulou, and Yanna Pournou. We also thank Leonidas S. Tzouvelekis for critical reading of the manuscript.
This work was funded in part by the Ministry of Health.
REFERENCES
- 1.Anderson E S, Ward L R, de Saxe M J, de Sa J D H. Bacteriophage-typing designations of Salmonella Typhimurium. J Hyg. 1977;78:297–300. doi: 10.1017/s0022172400056187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Callow B R. A new phage-typing scheme for Salmonella Typhimurium. J Hyg. 1959;57:346–359. doi: 10.1017/s0022172400020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glynn M K, Bopp C, Dewitt W, Dabney P, Moktar M, Angulo F J. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N Engl J Med. 1998;338:1333–1338. doi: 10.1056/NEJM199805073381901. [DOI] [PubMed] [Google Scholar]
- 4.Gross U, Tschape H, Bednarek I, Frosch M. Antibiotic resistance in Salmonella enterica serotype Typhimurium. Eur J Clin Microbiol Infect Dis. 1998;17:385–387. doi: 10.1007/BF01691565. [DOI] [PubMed] [Google Scholar]
- 5.Kauffman F. Serological diagnosis of salmonellosis species. Copenhagen, Denmark: Muksgaard; 1972. [Google Scholar]
- 6.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility tests: tentative standards. Vol. 13 1993. , no. 24. NCCLS document M2-A5. National Committee for Clinical Laboratory Standards, Villanova, Pa. [Google Scholar]
- 7.Ridley A, Threlfall E J. Molecular epidemiology of antibiotic resistance genes in multiresistant epidemic Salmonella Typhimurium DT104. Microb Drug Res. 1998;4:113–118. doi: 10.1089/mdr.1998.4.113. [DOI] [PubMed] [Google Scholar]
- 8.Sandvang D, Aaestrup F M, Jensen L B. Characterization of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium. FEMS Microbiol Lett. 1998;160:37–41. doi: 10.1111/j.1574-6968.1998.tb12887.x. [DOI] [PubMed] [Google Scholar]
- 9.Stapleton P, Wu J-P, King A, Shannon K, French G, Phillips I. Incidence and mechanisms of resistance to the combination of amoxycillin and clavulanic acid in Escherichia coli. Antimicrob Agents Chemother. 1995;39:2778–2783. doi: 10.1128/aac.39.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tassios P T, Markogiannakis A, Vatopoulos A C, Katsanikou E, Velonakis E N, Kourea Kremastinou J, Legakis N J. Molecular epidemiology of antibiotic resistance of Salmonella enteritidis during a 7-year period in Greece. J Clin Microbiol. 1997;32:1322–1325. doi: 10.1128/jcm.35.6.1316-1321.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tenover C F, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Threlfall E J, Frost J A, Ward L R, Rowe B. Epidemic in cattle and humans of S. Typhimurium DT104 with chromosomally-integrated multiple drug resistance. Vet Rec. 1994;134:577. doi: 10.1136/vr.134.22.577. [DOI] [PubMed] [Google Scholar]
- 13.Threlfall E J, Frost J A, Ward L R, Rowe B. Increasing spectrum of resistance in multiresistant Salmonella Typhimurium. Lancet. 1996;347:1053–1054. doi: 10.1016/s0140-6736(96)90199-3. [DOI] [PubMed] [Google Scholar]
- 14.Threlfall E J, Ward L R, Skinner J A, Rowe B. Increase in multiple antibiotic resistance in non typhoidal salmonellas from humans in England and Wales: a comparison of data for 1994 and 1996. Microb Drug Resist. 1997;3:263–266. doi: 10.1089/mdr.1997.3.263. [DOI] [PubMed] [Google Scholar]
- 15.Tosini F, Visca P, Luzzi I, Dionisi A-M, Pezzella C, Petruca A, Carattoli A. Class 1 integron-borne multiple-antibiotic resistance carried by IncFI and IncL/M plasmids in Salmonella enterica serotype Typhimurium. Antimicrob Agents Chemother. 1998;42:3053–3058. doi: 10.1128/aac.42.12.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vatopoulos A C, Mainas E, Balis E, Threlfall E J, Kanelopoulou M, Kalapothaki V, Malamou-Lada H, Legakis N J. Molecular epidemiology of ampicillin-resistant clinical isolates of Salmonella enteritidis. J Clin Microbiol. 1994;32:1322–1325. doi: 10.1128/jcm.32.5.1322-1325.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Widjojoatmodjo M N, Fluit A C, Verhoef J. Molecular identification of bacteria by fluorescence-based PCR–single-strand conformation polymorphism analysis of the 16S rRNA gene. J Clin Microbiol. 1995;33:2601–2606. doi: 10.1128/jcm.33.10.2601-2606.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]