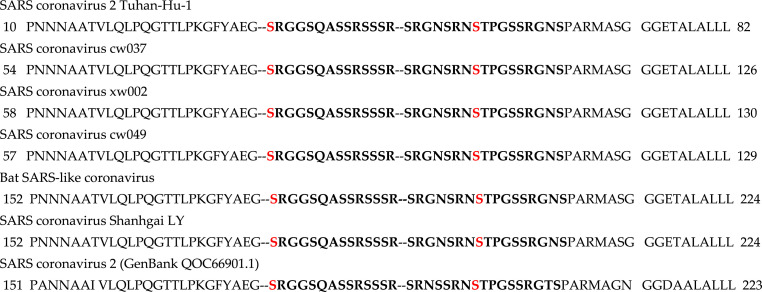Abstract
The Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) nucleocapsid protein (N-protein) is responsible for viral replication by assisting in viral RNA synthesis and attaching the viral genome to the replicase-transcriptase complex (RTC). Numerous studies suggested the N-protein as a drug target. However, the specific N-protein active sites for SARS-CoV-2 drug treatments are yet to be discovered. The purpose of this study was to determine active sites of the SARS-CoV-2 N-protein by identifying torsion angle classifiers for N-protein structural changes that correlated with the respective angle differences between the active and inactive N-protein. In the study, classifiers with a minimum accuracy of 80% determined from molecular simulation data were analyzed by Principal Component Analysis and cross-validated by Logistic Regression, Support Vector Machine, and Random Forest Classification. The ability of torsion angles ψ252 and φ375 to differentiate between phosphorylated and unphosphorylated structures suggested that residues 252 and 375 in the RNA binding domain might be important in N-protein activation. Furthermore, the φ and ψ angles of residue S189 correlated to a 90.7% structural determination accuracy. The key residues involved in the structural changes identified here might suggest possible important functional sites on the N-protein that could be the focus of further study to understand their potential as drug targets.
Keywords: Covid-19, N-Protein, Machine learning, Molecular simulation
1. Introduction
The Coronavirus (COVID-19) pandemic, which is caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), has resulted in over ∼250 million cases and over 5 million deaths worldwide currently [[1], [2], [3]]. Although isolation measures have been implemented, COVID-19 cases continue to increase exponentially [4]. Vaccines provide a limited protection solution to COVID-19 because their distribution has logistic issues, they have waning immunity, and they might still allow for transmission to others from asymptomatic infected individuals [5,6]. Although scientists have quantified binding patterns and looked at viral genetic codes, they have yet to find specific active sites for the SARS-CoV-2 N-Protein. Therefore, proposed antiviral drug-targeted treatments to suppress the viral protein through these active sites have been ineffective, with no FDA-approved antiviral therapy to date [[7], [8], [9]]. Current COVID-19 treatments include antiviral agents (remdesivir), steroids (dexamethasone), and anticoagulant medications (heparin) [10,11].
Numerous studies have suggested that coronavirus nucleocapsid protein (N-protein) phosphorylation is essential for viral replication. The protein consists of 5 domains, the N-arm (amino acid residues 1-43), the N-terminal domain (residues 44-174), the central Ser/Arg-rich flexible linker region (residues 175-254), the C-terminal domain (residues 255-364), and the C-arm (residues 365-419) [12,13]. The N-terminal domain is associated with RNA binding and the C-terminal domain is associated with RNA binding and oligomerization. When activated, with RNA binding, the N-protein undergoes a phase transition from crystal to liquid [[12], [13], [14]]. The N-arm, linker, and C-arm are considered to be disordered [13]. The region between residues 177 and 207 in the linker contains many phosphorylation sites (summarized in Refs. [[15], [16], [17], [18], [19]]). Phosphorylation is thought to modulate the phase transition [20].
The SARS-CoV-2 N-protein is activated by phosphorylation to facilitate viral transcription and assembly [7]. Moreover, mutating serine to alanine at position 177 (S177A) in bat severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) produces a virus that does not replicate. Residue S189 is also thought to be the priming site for hyperphosphorylation of this region [15]. In this role, the viral N-protein must interact with RNA, other viral proteins, and oligomerize with itself [21]. A study of 300,000 patients found that patients taking lithium which is an inhibitor of GSK3, had a 50% reduction in risk of SARS-CoV-2 infection [22]. This study uses a computational approach to provide additional information on the consequences of phosphorylation on N-protein structure.
Previous studies have analyzed variations in RNA binding sites implicating residues 248-280 in human coronavirus OC43 (HCoV-OC43) and SARS-CoV-2; and R122, G68, F57, P61, Y63, Y102, Y124, and Y126 in SARS-CoV-2 [23,24]. Other studies have implicated residues Q290, R294, and S328 as part of the oligomerization interface [21]. These “active” sites determine activity through substrate binding, which results in a significant structural and functional change. This study is an attempt to identify potential SARS-CoV-2 N-protein active sites by (i) finding potential phosphorylation sites, (ii) determining significant N-protein torsion angles, (iii) establishing significant N-protein amino acids with molecular dynamics, (iv) identifying active sites through significant angle/residue correspondence, and (v) testing accuracy through computational cross-validation.
Protein tertiary structure is characterized by torsion angles, also known as dihedral angles, representing protein molecular bond orientations [25]. Two out of the three possible torsion angles (φ for the N-Ca bond and ψ for the Ca-C bond) are visualized with the Ramachandran Plot, which shows the statistical distribution of having both angles in an amino acid [26]. Structural changes in proteins are visualized through 3-D protein representations rendered from torsion angle variations and atomic position plots [25]. Molecular Dynamics (MD), a computational system using force-fields under Newtonian mechanics, determines structural changes [26]. Specifically, MD simulation is used to provide spatial and temporal images of protein conformations and transitions throughout a trajectory, thereby resulting in an extensive sampling of conformation ensembles [27]. This study utilizes a novel approach to identify potential SARS-CoV-2 N-protein active sites that, in theory, can be suppressed in efficient drug-targeted treatments by using machine learning and computational cross-validation to discover significant structural torsion angle variations, functional amino acid variations, and the correspondence between the two.
2. Materials and methods
This was achieved through (i) finding N-protein phosphorylation sites, (ii) determining significant N-protein torsion angle variations from MD simulation, (iii) establishing significant N-protein amino acid residues with MD, (iv) identifying active sites through significant angle/residue correspondence, and (v) testing accuracy through computational cross-validation. For the first objective of identifying significant torsion angles, Nanoscale Molecular Dynamics 2 (NAMD version 3.0, Linux multicore GPU NVIDIA CUDA acceleration) was used to obtain the torsion angles [28]. Structures were viewed using Visual Molecular Dynamics viewer (VMD V1.9.3) [29].
Additionally, a meta-analysis has been conducted across relevant research studies (4, 5, 7, 8, and 13) to gather suspected relevant torsion angles in general SARS-CoV-2 N-protein. To identify feasible torsion angles, Ramachandran Plots, and the University of Tampere Institute of Biomedical Technology's RMSD (root-mean-square deviation) and RMSF (root-mean-square fluctuation) Visualizer Tool (V 1.0) were utilized. This tool is a VMD extension available at https://www.ks.uiuc.edu/Research/vmd/plugins/rmsdvt/(retrieved on January 8, 2021). Furthermore, The RMSD and RMSF were calculated for all trajectory structures using the Bio3D R package (v 2.4–1) [30,31]. To determine torsion angle classifiers, Orange Statistical Software Package (V 3.27.1) was used to build classification trees. For the second objective of identifying significant functional residues through residue removals, GROMACS Molecular Dynamics (V 4.6) has been used. After identifying significant corresponding residues, Orange was used to build the statistical analysis models (Logistic Regression, Support Vector Machine (SVM) [32], and Random Forest [33]) to determine the dihedral angles required to classify the various functional classifications of active and inactive. The default parameters were used. Briefly, Logistic regression uses a linear combination of features to build a classification model. SVM, a linear binary classification task, finds an optimal hyperplane with the maximum margin such that the distance between the closest data points of the two classes is maximized. Random Forest is an ensemble method that builds many decision trees by choosing random samples with replacement to create each tree and randomly generates a subset of features to decide the candidate split by highest Gini impurity or entropy [34]. The reason behind selecting these classifiers is to assess both linear (logistic regression) and nonlinear (random forest and SVM) classification methods to predict functionally significant residues.
2.1. Sequence retrieval of N-protein and modeling
We have retrieved the N-protein sequence from the GenBank sequence database (GenBank Accession: QIC50514.1). In order to develop a theoretical model for N-Protein, the sequence was submitted to the Iterative Threading ASSEmbly Refinement (I-TASSER) webserver (https://zhanglab.dcmb.med.umich.edu/I-TASSER/; last accessed January 8, 2021) [35].
2.2. Phosphorylation site prediction
To investigate the phosphorylation of N-protein, the FASTA-formatted sequence for SARS-CoV-2 (GenBank Accession: QOC66901.1) was submitted to GPS 3.0 (http://gps.biocuckoo.org/online.php; retrieved on April 4, 2020), NetPhos 3.1 (http://www.cbs.dtu.dk/services/NetPhos/; retrieved on April 4, 2020), and Scansite 3.0 (https://scansite4.mit.edu/; retrieved on April 4, 2020) phosphorylation prediction servers (phospho-algorithms). These tools were chosen because many researchers consider them to be among the most reliable phospho-algorithms [[36], [37], [38], [39]]. The candidates were ranked based on a combination of the highest scores and the presence of a site found by more than one tool using a prediction score cutoff (GPS >9, NetPhos >0.4, and Scansite >0.4). Similar sequences to the SARS-CoV-2 N-protein (GenBank Accession: QOC66901.1) were found using NCBI BLAST with the default parameters [40]. The top 5 sequences based on the highest scores were compared using Multiple Sequence Alignment (MSA), carried out using Clustal Omega (V 1.2.4) under the default setting, and visualized using Jalview (V 2.11.1.3) [41,42].
2.3. Process of molecular dynamics simulations
Four N-protein systems (the unphosphorylated wild-type (WT), the phosphorylated at residue 177 (S177-P), the phosphorylated at residue 198 (S198-P), alanine substitution at residue 177 (S177A) were simulated to understand the possible mechanisms that regulate phosphorylation.
2.4. Solvation
The four structures were solvated in a cubic periodic box using a three-site transferrable intermolecular potential (TIP3P) water model with a minimum distance spanning 15 Å from the system via VMD. Ions were added to neutralize the net charge of the systems and set the ion concentration to b 0.15 Mol/L.
2.5. Preparing MD simulation
The MD simulation was performed using Nanoscale Molecular Dynamics (NAMD2) with CHARMM36 all-force field parameters parallel programming model. Periodic boundary conditions were used to simulate the structures. A 12 Å cutoff for Van Der Waals interaction with a switching function distance of 8 Å, and the smooth particle-mesh Ewald (PME) method was enabled accordingly. In order to perform the MD simulation, the following procedures were applied to all four systems. Each system was energetically minimized to adjust the structure force field and relaxed possible steric clashes to obtain a low energy starting conformation. To avoid distress, a total of 1000 steps of minimization was performed. The system was heated from 0 to normal physiological conditions up to 310 K, with the Langevin thermostat applied. After heating, systems equilibration was performed to adjust the system density and equilibrate kinetic and potential energies. The 12 systems were simulated to sample the structural characteristics and dynamics at 300 K using NVE ensemble for 100 ns under NPT and time step 1 fs. The long-range electrostatics was handled with the particle-mesh Ewald (PME) methods. The atom coordinate was recorded every 1 ps throughout the simulation process. Additionally, 1 fs integration step was used for all simulations using 100 ns. NAMD version 3.0, Linux multicore GPU NVIDIA CUDA acceleration capability, The HPC node that was used to perform all simulations had the following configuration: CPU: Intel Xeon gold 6126 (12 cores) @ 2.60 GHz, Memory (RAM): 196 GB, GPU: Tesla V100 (VRAM 32 GB). The configurations files to fun the NAMD simulations and the R script to generate the analysis figures are available in the George Mason University Dataverse (https://doi.org/10.13021/orc2020/Z2WMAU retrieved on June 2, 2022).
2.6. Feature extraction
Using VMD, the last 500 trajectories of the simulation (DCD format) were loaded into the OpenGL viewer and used for the torsion angle extraction [43]. This assured that the number of frames could capture the molecular changes during the trajectory and meet or exceed the needs of effective machine learning analyses [44]. The dihedral angles of the protein backbone were obtained using a modest TCL code executed on the DCD trajectory for each frame, and the resulting matrix of φ and ψ angles was used for the structural investigations. The TCL code is available as the file getphi.tcl in the George Mason University Dataverse at https://doi.org/10.13021/orc2020/Z2WMAU (retrieved on June 2, 2022).
2.7. Determining significant N-protein torsion angles
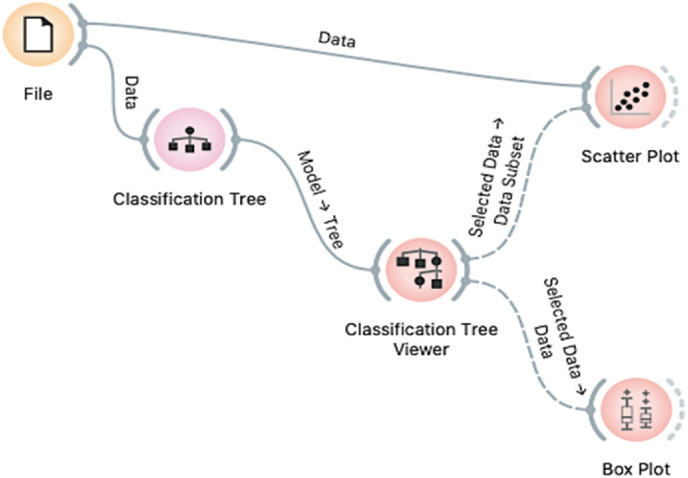
The φ and ψ angles obtained from the molecular simulation data were visualized using Ramachandran Plots and Atomic Position Visualization Software to explore the protein's torsion angles. Specifically, the data consisting of all simulated φ and ψ torsional angles in the N-protein was used in a machine learning model [25]. In addition, a meta-analysis across multiple SARS-CoV-2 peer-reviewed scientific articles yielded common torsion angles suspected to be involved in N-protein function domains were identified. Ramachandran Plots were used to identify permitted torsion angles by plotting torsion angles (φ and ψ) combinations for each residue in a protein that is feasible without steric hindrance are visible. Moreover, RMSD and RMSF visualizations of the N-protein were gathered for torsion angles insight. Specifically, normal distributions, multi-histograms, and trajectories of the RMSD and RMSF for residues were devised. RMSD and RMSF visualizations of amino acids provided insight into torsion angle calculations as RMSF indicates protein bond flexibility and RMSD indicates the degree of separation, which is used to calculate the angle between adjacent atoms [25]. After identifying applicable torsion angles that would be sterically feasible and comparing their relative angle measurements using RMSD/RMSF visualizations, The Orange data analysis package was used to build novel classification networks. Specifically, by training φ/ψ angle classifiers to a forward pruning tree algorithm, classification trees were utilized to accurately determine individual angle classifiers' success in predicting N-protein inactivity and activity. This prediction was modeled by building a network that determined that a torsion angle resulted in significant structural change if its variations resulted in significant phosphorylation changes that affected protein activity. Using the Orange software module for classification (Fig. 1 ), classifiers up to at least 80% classification accuracy were determined and distinctions between classifiers were identified by checking if there was separation between the data values in scatter and box plots.
Fig. 1.
Orange Modeling Network Built for Primary Classification. Utilizes applicable torsion angles from NAMD Molecular Simulations and meta-analysis of relevant articles. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
After identifying accurate torsion angle classifiers that resulted in a significant structural change that predicted the N-protein activity, a Principal Component Analysis (PCA) was conducted to streamline the major classifiers. The Orange *.odt files used to perform the analysis are available in the George Mason University Dataverse at https://doi.org/10.13021/orc2020/Z2WMAU (retrieved on June 2, 2022).
2.8. Establishing significant N-protein residues
After determining the streamlined major classifiers, the GROMACS Molecular Dynamics Visualization Software (V2020.2) was used to visualize the N-protein. Through the molecular dynamic simulations, spatial and temporal realistic models of N-protein dynamics and flexibility were rendered. By manipulating the N-protein, specific amino acid removals, or even alterations, were found to have resulted in N- Protein inactivity. Although this did not immediately signify that the amino acid residue was a significant active site, it could have been an intermediate amino acid needed for an activation process, such as phosphorylation transduction signaling, significant amino acid removals, and alterations resulted in N-Protein inactivity were documented. This was because these residues were characterized by having significant functional relevance to the N-protein.
2.9. Identifying SARS-CoV-2 N-protein active sites
Active sites for proteins are locations where proteins bind and conform to substrates. These sites determine the active region of the protein and, if suppressed, would render an inactive protein. Therefore, these sites are indicators of significant structural and functional changes. In Objective 1, torsion angle variations in residues that resulted in significant structural changes were identified from NAMD data and meta-analysis articles. Residues with functional value, discovered when said residues were removed, were discovered using GROMACS. A list of potential active site candidates was devised by identifying residue numbers with a torsion angle variation resulting in significant structural change and functional value. Furthermore, active site candidate residue numbers were compared to see if they fall within the numbered N-protein domains for RNA-binding, phosphorylation cascades, and oligomerization (4, 5, 7, 8, and 13). From these comparisons, N-protein active site residue(s) that were responsible for each domain showed a significant functional change in terms of N-protein activity when removed and showed N-protein activity prediction capability and significant structural change in the N-protein when altered were identified.
2.10. Testing accuracy through computational cross-validation
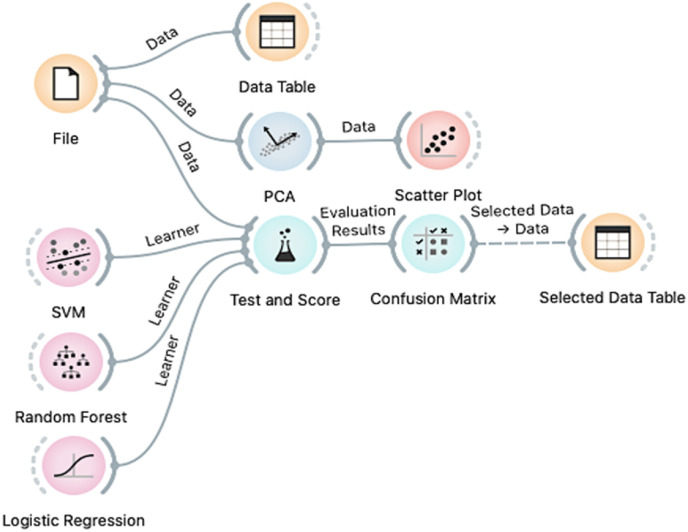
After determining the SARS-CoV-2 N-protein active sites, computational cross-validations using Logistic Regression, Random Forest, and SVM models were conducted (Fig. 2 ). These computational mathematical models were the most effective for a study that includes a large-scale protein activity [45]. By generating a confusion matrix, accuracy data were gathered for select active sites with the highest classification. Accuracies and functionally significant residues. Expressly, this accuracy referred to the predictions of determining inactivity and activity of the SARS-CoV-2 N-Protein. The error rate, accuracy, sensitivity, and specificity were calculated using the confusion matrices’ data.
Fig. 2.
Orange modeling network built for computational cross-validation. Utilizes support vector machine (SVM), random forest, and logistic regression to build accuracy confusion matrices. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3. Results
3.1. Phosphorylation site prediction
The online tools GPS 5.0, NetPhos3.1, and Scansite3 were used to predict phosphorylation sites on the SARS-CoV-2 N-protein. Many putative phosphorylation sites were found, and the residues with more than one tool predicting the site with high confidence are highlighted in bold (Table 1 ). The residues S177 and S198 chosen for the simulation are shown in red. Residues T91 and S207 are predicted with high confidence in all three tools. In particular, it is essential to note that there is a region with a large number of phosphorylation sites from residues 177-207 (bold italics). This corresponds to the region from residues 177-210 that has been suggested to be rich in phosphorylation sites [16]. In fact, Wu and coworkers have mutated this site (S177 in bat SARS) to an alanine and observed that the virus does not replicate. The residues in this region have been observed to be important for N-protein oligomerization which is critical for viral replication [46]. NCBI BLAST was used to find the five most similar sequences. Multiple sequence alignment was used to compare them. The N-protein in SARS in this region is highly conserved across coronaviruses and virtually identical between SARS-CoV-2 and SARS-CoV, the region in Fig. 3 being identical. Two phosphorylation sites, S177 and S189, were chosen as sites whose phosphorylation is vital for viral replication based upon previous experimental studies, phosphorylation site prediction, and evolutionary conservation.
Table 1.
Prediction of phosphorylation sites.
| Residue | Score |
Kinase |
||||
|---|---|---|---|---|---|---|
| GPS5.0 | Scansite3 | NetPhos3.1 | GPS5.0 | Scansite3 | Netphos3.1 | |
| S38 | 0.973 | PKC | ||||
| S80 | 0.998 | unspecified | ||||
| T92 | 0.498 | 0.988 | NEK4 | PKC,PKA | ||
| S106 | 0.996 | P38MAPK,GSK3 | ||||
| S142 | 0.97 | CKI,P38MAPK | ||||
| S177 | 0.507 | |||||
| S181 | x | 0.984 | DNAPK,ATM,PKA | |||
| S184 | x | 0.956 | PKC,cdc2 | |||
| S185 | x | 0.987 | cdc2 | |||
| S187 | x | 0.98 | cdc2 | |||
| S188 | x | 0.991 | PKC | |||
| S189 | x | 0.997 | PKC,RSK | |||
| S191 | x | 0.992 | unspecified | |||
| S194 | x | 0.997 | PKC | |||
| S195 | x | 0.995 | PKB,RSK,PKC | |||
| S198 | x | 0.986 | PKC,cdc2 | |||
| T199 | x | |||||
| S202 | 0.964 | PKC,cdc2 | ||||
| S203 | x | |||||
| T206 | 0.818 | PKC | ||||
| S207 | 0.234 | x | 0.995 | cdk5,RSK,GSK3 | ||
| S233 | 0.662 | PKC,cdc2 | ||||
| S236 | 0.987 | PKC | ||||
| S246 | 0.905 | PKC | ||||
| S251 | 0.99 | PKG | ||||
| S283 | 0.906 | DNAPK | ||||
| T319 | 0.592 | 0.493 | NEK10 | none | ||
| S326 | 0.955 | unspecified | ||||
| T392 | 0.27 | 0.647 | CAMKII | PKA | ||
| S411 | 0.976 | unspecified | ||||
| S413 | 0.956 | unspecified | ||||
Notes: Scansite2 did not specify kinases. Kinase definitions (in alphabetical order): ATM ataxia telangiectasia mutated protein kinase; CAMKII - calmodulin kinase II; cdc2-cyclin-dependent kinase 2; CKI - cyclin-dependent kinase; DNAPK - DNA-dependent protein kinase; GSK3 - glycogen synthase kinase; NEK4 -NIMA related kinase 4; NEK10 - NIMA related kinase 10; p38MAPK - mitogen-activated protein kinase; PKA - protein kinase A; PKB - protein kinase B; PKC - protein kinase C;; PKG - protein kinase G; RSK-ribosomal S6 kinase.
Fig. 3.
Coronavirus multiple sequence alignment. Residues 177-207 are shown in bold. The residues chosen for the simulations are shown in red. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.1.1. Molecular dynamics simulation
Molecular N-protein simulations were performed for four different cases: unphosphorylated, phosphorylated at S177 (S177-P), alanine substituted at position S177 (S177A), and phosphorylated at S198 (S189-P). The average RMSF for the entire molecule is 4.2 Å, 8.6 Å, 4.7 Å, and 12.2 Å for WT, S198-P, S177-P, and S177A, respectively. Fig. S1A shows that the RMSF is similar for WT and S177-P. The RMSF for S189-P is increased and the RMSD for 177A is greatly increased. Fig. S1B demonstrates that the simulations have reached a steady-state RMSD as evidence that the simulations have converged. The RMSD for WT and S177-P are similar (13.8 Å and 15.8 Å, respectively), with S177-P slightly increased. The RMSD for S198-P is increased (22.7 Å), and the RMSD for S177A is greatly increased (28.7 Å). The histogram for the RMSD in Fig. S1C shows that S177-P has a broader distribution of RMSD, suggesting some structural changes compared to WT. The distribution for 189-P is shifted to the right showing a significant structural change. The S177A shows the largest deviation from WT as it is shifted further to the right. The radius of gyration (Fig. S1D) is similar for WT and 177-P, significantly smaller for S177A, and slightly increased for S189-P. The unphosphorylated protein 177S and unphosphorylated state have been shown not to promote viral replication, and the protein phosphorylated at S177 promotes viral replication [16]. Analyses of the molecular structures will explore the differences between the “active” state capable of viral replication and the “inactive state” without viral replication.
3.2. Determining significant N-protein torsion angles
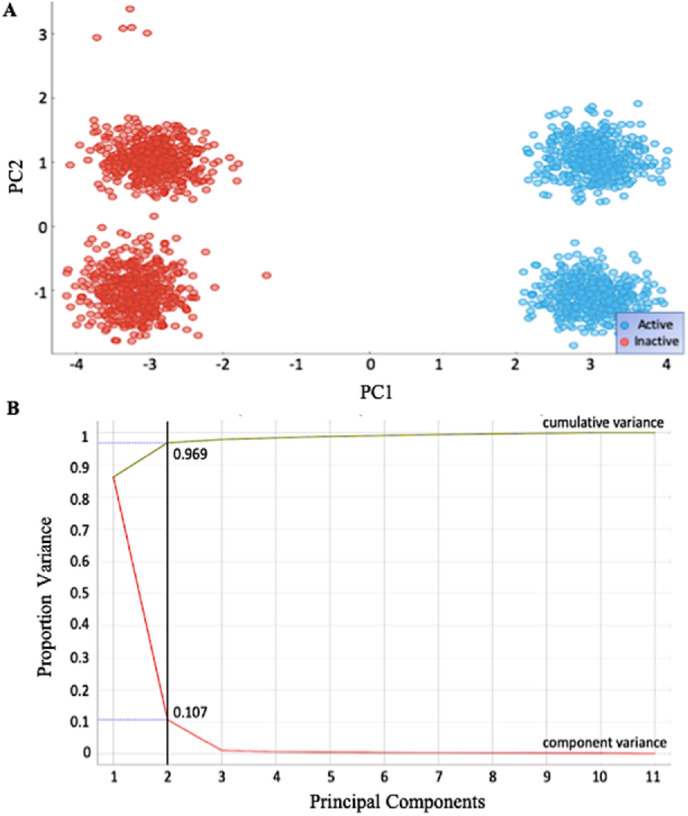
The torsional angles define a protein's structure. By understanding the differences in the phi (φ) and psi (ψ) torsional angles between the active and inactive forms of the N-protein, it is possible to identify which amino acid residues are suspected of playing a significant role in SARS-CoV-2 N-protein function. The Orange data mining software package (using the default parameters in the Principal Component Analysis example workflow) was used to perform the analysis of these torsional angles for the active (S177-P and S189-P) and inactive (WT and S177A) forms of the N-protein. First, Principal Component Analysis was applied to determine if the information contained in the torsional angles could separate the structures into the active and inactive classes (Fig. 4 ).
Fig. 4.
Principal Component Analysis of Major Angle Classifiers. Principal Component Analysis (PCA) streamlined major angle classifiers of 100% accuracy and 90.4% accuracy A. Scatter Plot Graph after PCA division showing clearly defined groups. B. PCs 1 and 2 are sufficient to explain 96.9% variance with most of the separation coming from PC1.
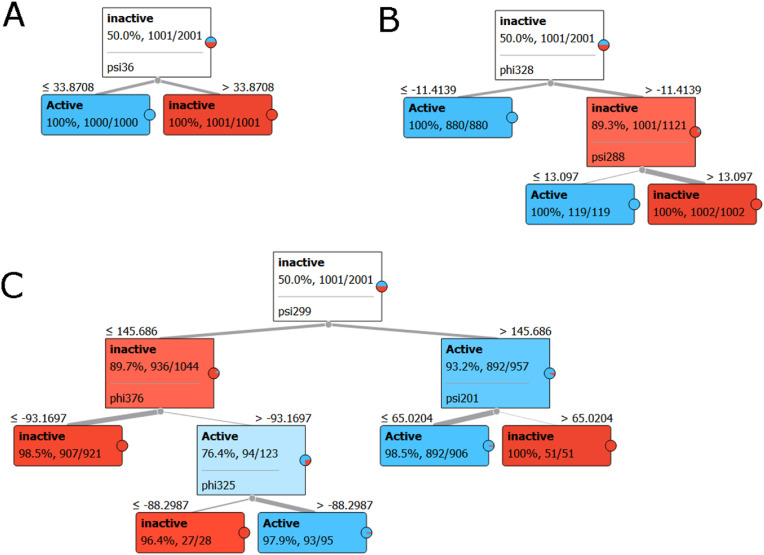
Classification trees were used to identify the angle's accuracy in predicting the N-protein's active or inactive state (Fig. 5 ). From this analysis, the critical residues that change with activation by phosphorylation could be used to infer those important for structural and functional changes. The torsion angles determined from these classifiers were differentiated using the classification tree workflow (using default parameters). Once the tree was classified, these classifying torsional angles were removed from the analysis to find the following angles that could classify the data. This process was repeated starting with those angles that achieved 100% classification and continued until 80% classification was obtained. Fig. 5A shows how one torsional angle can completely separate the data into active and inactive classes. Fig. 5, Fig. 6 C demonstrate how more torsional angles might be needed for classification. It is important to note that torsional angles that separate the data can be shown clearly to cluster the data in a scatter plot.
Fig. 5.
Classification Tree Visualization for Torsion Angles. Orange Classification Trees used phosphorylation changes attributed to structural changes. A. Single-parent trees with the greatest percentage are used for accuracy. ψ36 separates structures into active and inactive classes with 100% accuracy B. Limited branching (2 layers) tree shows greater than 80% classifiers consideration. φ328 divides a subset of data into active and inactive classes with 89.3% accuracy and needs a second torsional angle to classify the data accurately. C. More torsional angles (4 angles) are needed to classify the data with high accuracy. However, even with the additional angles, complete classification is not reached. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
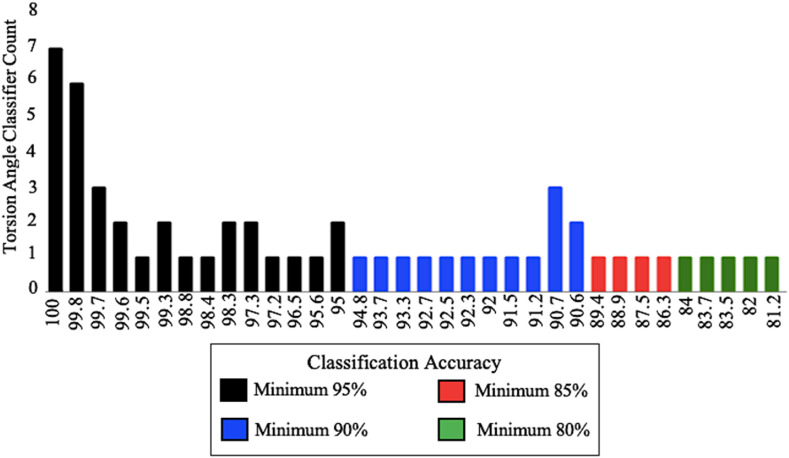
Fig. 6.
Determination of Classification Accuracy Ranges for φ and ψ Torsion Angles. Orange Classification Trees use decision-based machine learning to identify the minimum 95%, 90%, 85%, and 80% accuracy torsion angles classifiers. Classification trees determine accuracy based on prediction of inactive and active states for the respective φ/ψ angle. Classifiers shown do not include branched classifiers. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The dihedral angles ψ36, ψ50, φ76, ψ83, φ99, ψ 214, φ375, ψ252, φ406, φ416 were able to classify active from inactive with 100% accuracy. The torsional angles associated with 99.8–100% classification are φ36, ψ76, φ177, ψ177, ψ252, and φ416. The torsional angles associated with a 95–99.8% classification are φ36, ψ36, ψ50, φ76, ψ76, ψ83, φ99, φ176, ψ176, φ177, ψ177. φ178, ψ178, φ179, ψ179. φ180, ψ180, φ181, ψ181, φ182, ψ182, φ183, ψ183, φ184, ψ184, φ185, ψ185, φ186, ψ186, φ187, ψ187, φ188, ψ188, φ189. ψ189, φ190, ψ190, φ191, ψ191, φ192, ψ192, φ193, ψ193, φ194, ψ194, φ195, ψ195, φ196, ψ196, φ197, ψ197, φ198, ψ198, φ199, ψ199, φ200, ψ200, φ201, ψ201, φ202, ψ202, φ203, ψ203, φ204, v204, φ205, ψ205, φ206, ψ206, φ214, ψ252, φ375, φ406, and φ416. Fig. S1 in the Supplementary Material demonstrates that the ψ36 and ψ50 angles can completely separate the structures in the ensemble into the active and inactive classes.
To summarize this data, a histogram was created to show the number of residues able to classify the structures in the ensemble into the active and inactive classes with an accuracy of 80% or greater. Other classifiers were noted but were not considered in determining primary/supplementary active sites. These classifiers were deemed significant based on the significant predicted functional change associated with their protein structures. Classifiers that had at least 80% accuracy in determining the activity of the SARS-CoV-2 N-protein were determined (Fig. 6).
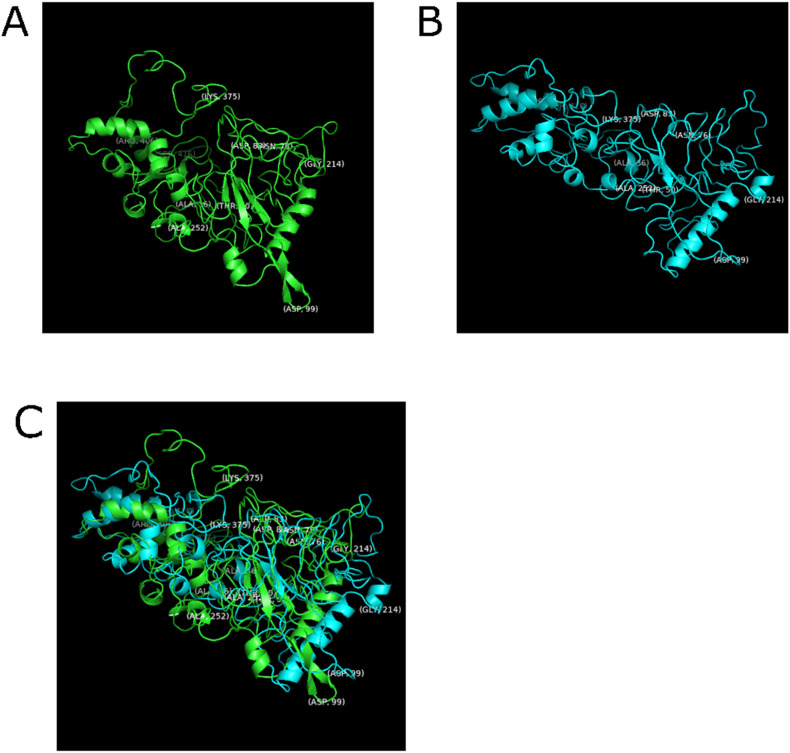
The structure of the WT and S177-P N-Protein is shown in Fig. 7 . The RMSD deviation of these two structures is 12.0 Å. The torsion angles identified as classifying the structures 100% are shown labelled. Fig. 7C shows the aligned structures for comparison. Three primary residue angles were identified as active site candidates following the determination of significant residues in correspondence with the angle classifiers previously identified. These were ψ252, φ375, and ψ189. ψ252 and φ375 have a 100% classification accuracy, and ψ189 has a 90.4% classification accuracy. The S177-P (activated structure) is elongated compared to the WT with the labelled angles more clustered than it WT. Based on comparing domain residue numbers of the N-Protein (4, 5, 7, 8, and 13), the primary active site residues for RNA-binding are predicted to be residues 252 and 375. The supplementary active site responsible for the phosphorylation cascade to activate these functions is residue 189. Cross-validation was used to ascertain the residues’ accuracy under statistical models (Fig. 8 ).
Fig. 7.
Structures from Molecular Dynamics Simulation. A. Wild-type N-protein. B. N-protein phosphorylated at S177 (S177-P). C. Wild-type and S177-P aligned structures. Residues that were able to classify structures with 100% accuracy are labelled.
Fig. 8.
Confusion matrices for logistic regression, random forest classification, and SVM models. Confusion Matrices show the cross-validation results for statistical models regarding major classifiers with left activity states (actual) and upper activity states (predicted). A. Confusion Matrices for ψ252 for Logistic Regression, Random Forest, and SVM (L–R). B. Confusion Matrices for φ375 for Logistic Regression, Random Forest, and SVM (L–R). C. Confusion Matrices for ψ189 for Logistic Regression, Random Forest, and SVM (L–R).
Machine learning was applied as a final test of the importance of identifying the torsional angles as essential to distinguish between active and inactive proteins. Random forests, logistic regression and SVM were used using 10-fold cross-validation. Fig. 8 shows the confusion matrices for ψ252, φ375, and ψ189 showing.
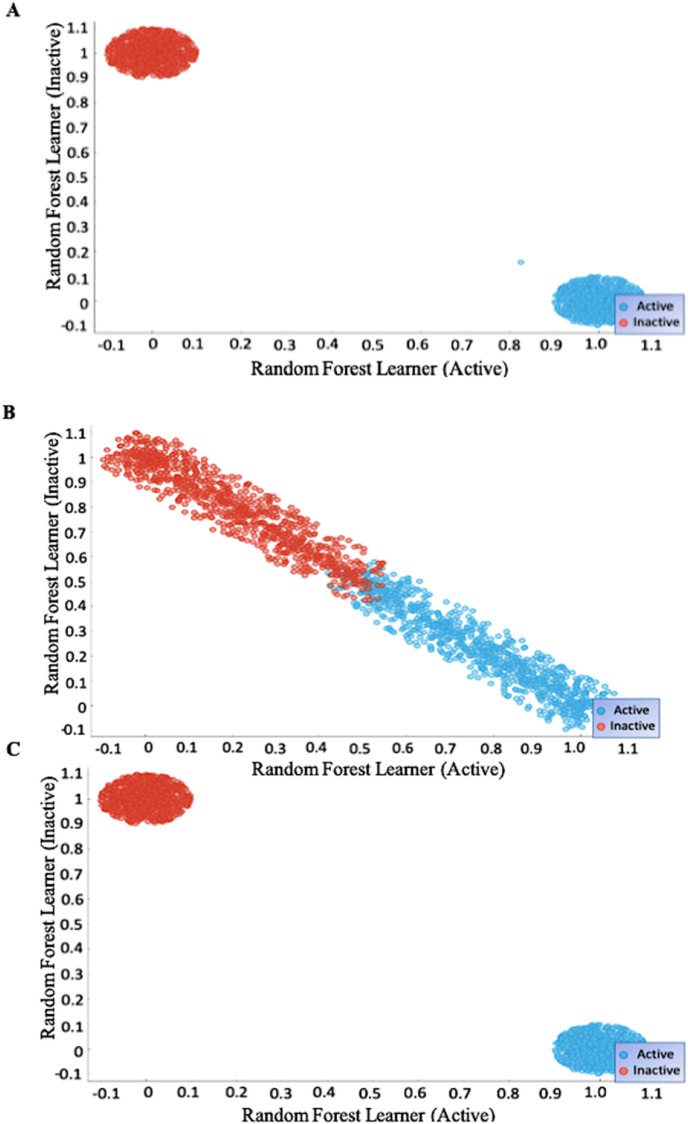
The confusion matrices support the identified active sites as ψ252 and φ375 are highly accurate in determining activity as they are primary sites, while the 50% accuracy of ψ189 is a supplementary site. Random Forest, the most accurate algorithm in our model, was visualized. Scatter plots based upon these machine learning measures are shown in Fig. 9 , supporting the full separation by ψ252 and φ375 and less accurate validation by ψ189.
Fig. 9.
Scatter plot graphs for logistic regression, random forest classification, and SVM models. A. ψ252 Random Forest Classification Scatter Plot. B. ψ189 Random Forest Classification Scatter Plot. C. φ375 Random Forest Classification Scatter Plot.
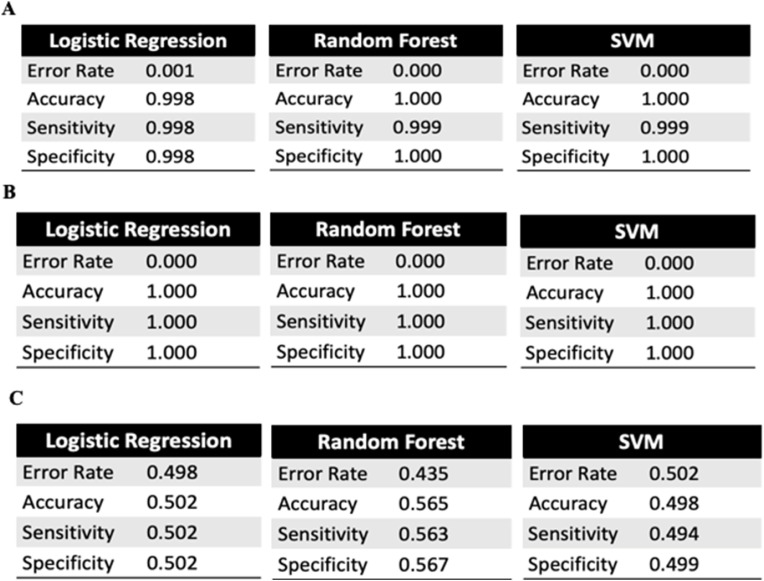
These scatter plots further support the assertion of the primary active site residues 252 and 375 and the supplementary active site 189 by showing distinctions to lack of distinctions, respectively, under the same model. To further quantify the accuracy of determining active sites, confusion matrices provided data on the true positives, true negatives, false positives, and false negatives for calculating sensitivity, specificity, error rate, and accuracy of residues (Fig. 10 ). In addition to research defining the N-protein domain characteristics, this computational cross-validation supports the determination of the active site residues 189, 252, and 375 for the SARS-CoV-2 N-protein.
Fig. 10.
Sensitivity, Specificity, Error Rate, and Accuracy Analysis of Classifiers. Data gathered from confusion matrices of statistical models to determine classifiers' sensitivity, specificity, error rate, and accuracy analysis. A. Statistical Model Analysis for ψ252. B. Statistical Model Analysis for φ375. C. Statistical Model Analysis for ψ189.
4. Discussion
The SARS-CoC-2 N-protein has been considered to be an important drug target [47]. The N-Protein has essential roles in viral replication, including interacting with the viral genome to create the ribonucleoprotein [48]. Recent studies have targeted drugs toward N-protein with ceftriaxone, cefotaxime, and cefuroxime having higher binding affinities to the N-terminal domain of N-protein, C-terminal domain of N-protein, and receptor-binding domain of S protein [49]. A molecular understanding of N-protein functional domains is vital to better understand how to target drugs against N-protein.
The goal of this study was to identify the amino acids involved in the activation of the N-protein. Machine learning applied to molecular dynamics simulation identified the φ and ψ angles that displayed differences between the active and inactive forms of the N-protein. Experimental studies by Wu and coworkers demonstrated that phosphorylation at S177 of the bat SARS virus (equivalent to S177 in our N-protein) was necessary for viral replication [16]. Furthermore, they showed that conversion of this residue to alanine (S177A) resulted in no viral activation suggesting an essential role of phosphorylation at this residue. Experimental post-translation modification analysis using mass spectroscopy by Supekar et al. found phosphorylation at T394 and S177 in SARS-Cov-2 in N-protein expressed in HEK cells [50].
Fig. 1 shows that S177 residue is highly conserved across coronaviruses. We also explored phosphorylation at S198 in SARS to determine if this had a role in viral activation. Lin and co-workers used mass spectroscopy and concluded that two residues were phosphorylated in the dense serine-rich S177-S207 span and suggested that it was likely S207 and possibly S189 but could not exclude the phosphorylation of other residues [51].
Using the observation that unphosphorylated N-protein was inactive and N-protein phosphorylated at S177 was active, machine learning was applied to the set of φ and ψ angles from molecular simulations. The study's findings suggest that there are changes in SARS-CoV-2 significant changes at amino acid residues 252 and 375, and notable changes at residue 189. Active sites are known to characterize protein activity and result in significant structural and functional change when interacted with, either through substrates or through external manipulation (https://www.britannica.com/science/active-site retrieved on January 8, 2021). These residues fulfill all three characteristics required to be an active site. To begin with, all three residues correspond with torsion angles that have high classification accuracy when determining N-protein structural change through activity, as seen above. Residues 252 and 375 have a 100% classification accuracy, while residue 189 has a 90.4% accuracy. In addition to being significant indicators of structural change, they also demonstrate significant functional change. Specifically, when removed in molecular dynamic simulations, all three residues resulted in N-protein inactivity.
Moreover, as seen in the computational cross-validation, primary sites 252 and 375 had nearly 100% accuracy, sensitivity, specificity, and a nearly negligible error rate when determining the activity of N-proteins through models, with Random Forest surprisingly being the most successful. Supplementary active site 189 had approximately 50% accuracy in determining the activity of N-proteins through statistical models. However, this supports that residue 189 is a supplementary active site as it alone does not determine activity but rather is a causation factor for the other primary sites’ functioning. This is because, as supported through previous research on N-protein domains (4, 5, 7, and 8), residues 252 and 375 fall within the general RNA-binding and oligomerization ranges. In contrast, residue 189 falls within the phosphorylation cascade domain, a transduction signaling pathway that activates other functions. Hence, as supported by these findings, residues 252, 375, and 189 serve as the SARS-CoV-2 N-protein active sites for its viral functions.
Identifying structure changes between active N-protein and inactive N-protein might be possible to infer whether phosphorylation affects N-Protein oligomerization or its nuclease activity. Experimental studies have suggested that RNA binding is needed before oligomerization occurs. Furthermore, the activation of GSK3 alone did not promote oligomerization. On the other hand, the activation of CDK1 alone had a negligible effect. Furthermore, activation of both GSK3 and CKD1 are needed together for complete oligomerization response [52]. A more recent study by Liu et al. found that GSK3 inhibitors blocked N-protein phosphorylation and impaired SARS-Cov-2 replication in lung epithelial cells [22]. Chang and coworkers observed that the C-terminal structural domain acts as an oligomerization domain in solution using a disulfide tracking technique. Furthermore, the amino acid residues Gln290, Arg294, and Ser328 are at the oligomerization interface [21]. Another study utilizing the N-Protein crystal structure identified a binding pocket for RNA binding to N-protein with the interaction residues: Arg123 and Gly69, Phe58, Pro62, Tyr64 Tyr103, Tyr125, and Tyr127 (with our protein in our case) [23]. In our study, we have found that phosphorylation causes significant changes in residues ψ69, φ69 ψ290, and ψ238 as they have found. The residues Gln290, Arg294, and Ser328, which are thought to be part of the oligomerization interface, have increased RMSF in S177-P compared to WT (4.08 vs 3.05, 9.74 vs 2.62, and 10.10 vs 5.78, respectively) [21].
Tung and Limtung recently studied mutation in the phosphorylation-rich regions (residues 177-206) of SARS-CoV-2 using a structural model to understand the importance of viral replication, transcription, and packaging [17]. Among SARS-CoV-2 strains, they observed mutations in serine residues at several positions) S187, S198, and S203; S186, S197, and S202) to Phenylalanine, Leucine, and Asparagine, respectively. The phosphorylation site S187 is a phosphorylation site for casein kinase I CKI which controls a wide variety of cellular processes, including the control of the cell cycle. The Aurora A and B kinases, which have a role in the cell cycle, act at S198. Phosphorylation sites 187, 189, 198, 199, 203, and 205 form a phosphorylation-dependent binding domain from 14-3-3 protein which is vital for controlling cell cycle, cell survival, and cell death [17]. GSK3, which is also involved in the cell cycle, phosphorylates S203. Wu and coworkers showed that phosphorylation sites critical for viral replication of SARC-CoV were located between residues 177-206 were phosphorylated by GSK3 using mass spectrometric analysis and deletion mapping [15]. The binding of 14-3-3 protein to the hyperphosphorylated region of the N-protein controls the cytoplasmic/nuclear localization of N-protein [7,53].
Sequence alignment showed in Fig. 3 that the phosphorylation-rich residues between 177 and 206 are highly conserved among SARS-CoV-2 viruses. Tung and Limtung also showed that this region is highly conserved in SARS-CoV-2 viruses [17]. Previous work by Chang and co-workers have shown that this region is also conserved across coronaviruses [54]. Specifically, S184, S191, R192, S195, and R196 were conserved across SARS-CoV, infectious bronchitis virus, mouse hepatitis virus, human coronavirus, and Middle East respiratory virus.
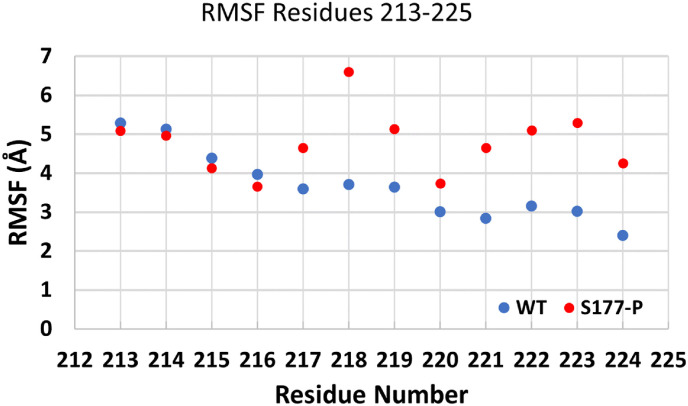
Experimental studies by Carlson et al. show that the phosphorylation of N-protein forms a more liquid-like droplet [52]. They suggest that the unphosphorylated N-protein help form a structured oligomer suitable for nucleocapsid assembly, and the phosphorylated N-protein forms a liquid-like compartment that helps with viral processing. Other studies using truncation analysis by Lu et al., have suggested that an L/Q rich region (residues 210-246) plays a crucial role in RNA-mediated phase separation [14]. Molecular simulation studies have shown that a hydrophobic α-helix spanning in residues ∼213–225 exist in this region that could be involved in this process [13]. Similarly, Savastano et al. found that phosphorylation-induced phase separation was critical for RNA polymerase activity in viral replication [55]. Fig. 11 shows that the RMSF values of WT are increased compared to S177-P for residues 217-224. Furthermore, the structure of S177P seems to be more elongated while WT is more compact. This is consistent with the suggestion that these residues are involved in the transition seen in experimental studies. Additionally, residue 214 shows a significant change in the torsional angles compared to WT as it can be used to separate the ensemble of structures into two classes. However, while there is a correlation, this study does not prove or disprove this hypothesis.
Fig. 11.
The RMSF for residues 213-225. There is an increase in RMSF for S177-P compared to WT.
Recently, Gao et al. predicted N-protein phosphorylation sites were identified using NetPhos3.1 Server, and significant sites were predicted using mutational analysis using PROVEAN PROTEIN [56]. Significant sites were predicted with the following results: Deletion of the sites T58, Y88, T116, S256, T264, G266, T272, T333 negatively affected protein function. Single amino acid insertion at Y88, T116, T264, T266, T272 also affected protein function. Substitution at Y88, T116, S177, T266, T272, T333 was also predicted to have an essential role in N-protein function. These sites did not appear as the consensus sites in our computational screen using the three software implementations: GPS5.0, Scansite3, and NetPhos3.1.
Additionally, mutational studies need to be considered carefully as they can change structure and function. In our study, we found that S177A has significant structural changes from WT. It is not clear that using an alanine mutation to show the importance of a phosphorylation site as done by Wu and coworkers is a valid test [15]. In fact, Gray and coworkers studied databases of structures with mutations and found that methionine was the best-tolerated mutation while proline was the least tolerated. They found that alanine substitution produced structural changes that were approximately the average of all changes.
5. Conclusions
This study aimed to identify SARS-CoV-2 N-protein active sites as possible areas of focus for future antiviral drug treatments. Specifically, identifying these active sites that have remained unknown to date is crucial in stopping the global COVID-19 pandemic as it allows for immunosuppression of the virus through drug treatments that suppress the active sites identified. This study found that the SARS-CoV-2 N-protein residues 252 and 375 display significant changes with N-protein activation and might play a role in RNA-binding and packaging, one the N-protein's viral functions. Also, it found the supplementary active site in charge of facilitating phosphorylation cascades to activate the functions, as mentioned earlier, to be residue 189.
Author contributions
Conceptualization, S.S. and M.S.J.; methodology, S.S., L.A., J.H., and M.S.J.; validation, M.S.J.; formal analysis, S.S..; investigation, S.S, A.A, Y.B, and M.A; resources, M.S.J. and L.A.; data curation, J.H..; writing—original draft preparation, S.S.; writing—review and editing, S.S., L.A., J.H., F.A., A.A, Y.B, M.A and M.S.J.; visualization, M.S.J. and F.A.; supervision, M.S.J.; project administration, M.S.J.; funding acquisition, L.A. and M.S.J. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by King Abdullah International Medical Research Center (KAIMRC) (LA), King Fahad Specialist Hospital-Dammam (KFSH-D) (FA). Article publishing charges were paid by KAIMRC.
Institutional review board statement
Not applicable.
Informed consent statement
Not applicable.
Data availability statement
Data is available at the George Mason University Dataverse) https://doi.org/10.13021/orc2020/Z2WMAU (retrieved on June 2, 2022).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by King Abdullah International Medical Research Center (KAIMRC) (LA), King Fahad Specialist Hospital-Dammam (KFSH-D) (FA). Article publishing charges were paid by KAIMRC.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2022.100889.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Keni R., Alexander A., Nayak P.G., Mudgal J., Nandakumar K. COVID-19: emergence, spread, possible treatments, and global burden. Front Public Health. 2020;8:216. doi: 10.3389/fpubh.2020.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johns hopkins corona virus resource. 2021. [Google Scholar]
- 3.Worldometer Coronavirus. 2021. [Google Scholar]
- 4.Pawar M. The global impact of and responses to the COVID-19. Pandemic Int J Commun Soc Develop. 2020;2:111–120. [Google Scholar]
- 5.Wouters O.J., Shadlen K.C., Salcher-Konrad M., Pollard A.J., Larson H.J., Teerawattananon Y., Jit M. Lancet; London, England): 2021. Challenges in ensuring global access to COVID-19 vaccines: production, affordability, allocation, and deployment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dolgin E. COVID vaccine immunity is waning - how much does that matter? Nature. 2021;597:606–607. doi: 10.1038/d41586-021-02532-4. [DOI] [PubMed] [Google Scholar]
- 7.McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Españo E., Kim D., Kim J., Park S.K., Kim J.K. vol. 21. Immune network; 2021. p. e7. (COVID-19 antiviral and treatment candidates: current status). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perveen R.A., Nasir M., Talha K.A., Selina F., Islam M.A. Systematic review on current antiviral therapy in COVID-19 pandemic. Med J Malaysia. 2020;75:710–716. [PubMed] [Google Scholar]
- 10.Siordia J.A., Jr., Bernaba M., Yoshino K., Ulhaque A., Kumar S., Bernaba M., Bergin E. 2020. Systematic and statistical review of coronavirus disease 19 treatment trials, SN comprehensive clinical medicine; pp. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The ATTACC, ACTIV-4a, and REMAP-CAP Investigators Therapeutic anticoagulation with heparin in noncritically ill patients with covid-19. N Engl J Med. 2021;385:790–802. doi: 10.1056/NEJMoa2105911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai Z., Cao Y., Liu W., Li J. The SARS-CoV-2 nucleocapsid protein and its role in viral structure, biological functions, and a potential target for drug or vaccine mitigation. Viruses. 2021;13:1115. doi: 10.3390/v13061115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cubuk J., Alston J.J., Incicco J.J., Singh S., Stuchell-Brereton M.D., Ward M.D., Zimmerman M.I., Vithani N., Griffith D., Wagoner J.A., Bowman G.R., Hall K.B., Soranno A., Holehouse A.S. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat Commun. 2021;12:1936. doi: 10.1038/s41467-021-21953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu S., Ye Q., Singh D., Cao Y., Diedrich J.K., Yates J.R., Villa E., Cleveland D.W., Corbett K.D. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat Commun. 2021;12:502. doi: 10.1038/s41467-020-20768-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu C.H., Yeh S.H., Tsay Y.G., Shieh Y.H., Kao C.L., Chen Y.S., Wang S.H., Kuo T.J., Chen D.S., Chen P.J. Glycogen synthase kinase-3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. J Biol Chem. 2009;284:5229–5239. doi: 10.1074/jbc.M805747200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu K., Li W., Peng G., Li F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19970–19974. doi: 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tung H.Y.L., Limtung P. Mutations in the phosphorylation sites of SARS-CoV-2 encoded nucleocapsid protein and structure model of sequestration by protein 14-3-3. Biochem Biophys Res commun. 2020;532:134–138. doi: 10.1016/j.bbrc.2020.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikolakaki E., Giannakouros T. SR/RS motifs as critical determinants of coronavirus life cycle. Front Mol Biosci. 2020;7 doi: 10.3389/fmolb.2020.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fung T.S., Liu D.X. Post-translational modifications of coronavirus proteins: roles and function. Future Virol. 2018;13:405–430. doi: 10.2217/fvl-2018-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlson C.R., Asfaha J.B., Ghent C.M., Howard C.J., Hartooni N., Morgan D.O. 2020. Phosphorylation modulates liquid-liquid phase separation of the SARS-CoV-2 N protein, bioRxiv : the preprint server for biology. [Google Scholar]
- 21.Chang C.K., Chen C.M., Chiang M.H., Hsu Y.L., Huang T.H. Transient oligomerization of the SARS-CoV N protein--implication for virus ribonucleoprotein packaging. PLoS One. 2013;8 doi: 10.1371/journal.pone.0065045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X., Verma A., Garcia G., Ramage H., Myers R.L., Lucas A., Michaelson J.J., Coryell W., Kumar A., Charney A.W., Kazanietz M.G., Rader D.J., Ritchie M.D., Berrettini W.H., Schultz D.C., Cherry S., Damoiseaux R., Arumugaswami V., Klein P.S. 2021. Targeting the Coronavirus Nucleocapsid Protein through GSK-3 Inhibition, medRxiv : the preprint server for health sciences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q., Yan Y., Zhang C., Shan H., Chen S. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm Sin B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C.Y., Chang C.K., Chang Y.W., Sue S.C., Bai H.I., Riang L., Hsiao C.D., Huang T.H. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J Mol Biol. 2007;368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung S., Bae S.E., Ahn I., Son H.S. Protein backbone torsion angle-based structure comparison and secondary structure database web server. Genom Inform. 2013;11:155–160. doi: 10.5808/GI.2013.11.3.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Childers M.C., Daggett V. Insights from molecular dynamics simulations for computational protein design. Mol Sys Des Eng. 2017;2:9–33. doi: 10.1039/C6ME00083E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dror R.O., Jensen M.Ø., Borhani D.W., Shaw D.E. Exploring atomic resolution physiology on a femtosecond to millisecond timescale using molecular dynamics simulations. J Gen Physiol. 2010;135:555–562. doi: 10.1085/jgp.200910373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phillips J.C., Hardy D.J., Maia J.D.C., Stone J.E., Ribeiro J.V., Bernardi R.C., Buch R., Fiorin G., Hénin J., Jiang W., McGreevy R., Melo M.C.R., Radak B.K., Skeel R.D., Singharoy A., Wang Y., Roux B., Aksimentiev A., Luthey-Schulten Z., Kalé L.V., Schulten K., Chipot C., Tajkhorshid E. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J Chem Phys. 2020;153 doi: 10.1063/5.0014475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 30.Grant B.J., Rodrigues A.P., ElSawy K.M., McCammon J.A., Caves L.S. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 31.Skjærven L., Yao X.Q., Scarabelli G., Grant B.J. Integrating protein structural dynamics and evolutionary analysis with Bio3D. BMC Bioinf. 2014;15:399. doi: 10.1186/s12859-014-0399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cortes C., Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–297. [Google Scholar]
- 33.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 34.Aljouie A., Roshan U. 2019 IEEE international conference on bioinformatics and biomedicine. BIBM; 2019. Multi-path convolutional neural network for glioblastoma survival group prediction with point mutations and demographic features; pp. 1274–1279. [Google Scholar]
- 35.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue Y., Liu Z., Cao J., Ma Q., Gao X., Wang Q., Jin C., Zhou Y., Wen L., Ren J. GPS 2.1: enhanced prediction of kinase-specific phosphorylation sites with an algorithm of motif length selection. Protein Eng Des Sel. 2011;24:255–260. doi: 10.1093/protein/gzq094. [DOI] [PubMed] [Google Scholar]
- 37.Blom N., Sicheritz-Ponten T., Gupta R., Gammeltoft S., Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 38.Obenauer J.C., Cantley L.C., Yaffe M.B. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vlachakis D., Bencurova E., Papageorgiou L., Bhide M., Kossida S. Protein phosphorylation prediction: limitations, merits and pitfalls. J Mol Biochem. 2015;4(2) 145- [Google Scholar]
- 40.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 41.Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–w641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(33–38):27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 44.McCoy M.D., Hamre J., Klimov D.K., Jafri M.S. Predicting genetic variation severity using machine learning to interpret molecular simulations. Biophys J. 2021;120:189–204. doi: 10.1016/j.bpj.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srivastava A., Mahmood A., Srivastava R. 2017 international conference on current trends in computer, electrical, electronics and communication. CTCEEC; 2017. A comparative analysis of SVM random forest methods for protein function prediction; pp. 1008–1010. [Google Scholar]
- 46.He R., Dobie F., Ballantine M., Leeson A., Li Y., Bastien N., Cutts T., Andonov A., Cao J., Booth T.F., Plummer F.A., Tyler S., Baker L., Li X. Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochem Biophys Res commun. 2004;316:476–483. doi: 10.1016/j.bbrc.2004.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.-P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.-Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwarteng A., Asiedu E., Sakyi S.A., Asiedu S.O. Targeting the SARS-CoV2 nucleocapsid protein for potential therapeutics using immuno-informatics and structure-based drug discovery techniques. Biomed Pharmacother. 2020;132:110914. doi: 10.1016/j.biopha.2020.110914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu X., Zhou Z., Li F., Xiao Y., Wang Z., Xu J., Dong F., Zheng H., Yu R. The study of antiviral drugs targeting SARS-CoV-2 nucleocapsid and spike proteins through large-scale compound repurposing. Heliyon. 2021;7 doi: 10.1016/j.heliyon.2021.e06387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Supekar N.T., Shajahan A., Gleinich A.S., Rouhani D.S., Heiss C., Chapla D.G., et al. Variable posttranslational modifications of severe acute respiratory syndrome coronavirus 2 nucleocapsid protein. Glycobiology. 2021;31(9):1080–1092. doi: 10.1093/glycob/cwab044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin L., Shao J., Sun M., Liu J., Xu G., Zhang X., Xu N., Wang R., Liu S. Identification of phosphorylation sites in the nucleocapsid protein (N protein) of SARS-coronavirus. Int J Mass Spectrom. 2007;268:296–303. doi: 10.1016/j.ijms.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carlson C.R., Asfaha J.B., Ghent C.M., Howard C.J., Hartooni N., Morgan D.O. Phosphorylation modulates liquid-liquid phase separation of the SARS-CoV-2 N protein. Mol Cell. 2020;80:P1092–P1103. doi: 10.1016/j.molcel.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tugaeva K.V., Hawkins D.E.D.P., Smith J.L.R., Bayfield O.W., Ker D.-S., Sysoev A.A., Klychnikov O.I., Antson A.A., Sluchanko N.N. The mechanism of SARS-CoV-2 nucleocapsid protein recognition by the human 14-3-3 proteins. J Mol Biol. 2021;433:166875. doi: 10.1016/j.jmb.2021.166875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang C.K., Hou M.H., Chang C.F., Hsiao C.D., Huang T.H. The SARS coronavirus nucleocapsid protein--forms and functions. Antivir Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Savastano A., Ibáñez de Opakua A., Rankovic M., Zweckstetter M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat Commun. 2020;11:6041. doi: 10.1038/s41467-020-19843-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao T., Gao Y., Liu X., Nie Z., Sun H., Lin K., Peng H., Wang S. Identification and functional analysis of the SARS-COV-2 nucleocapsid protein. BMC Microbiol. 2021;21:58. doi: 10.1186/s12866-021-02107-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is available at the George Mason University Dataverse) https://doi.org/10.13021/orc2020/Z2WMAU (retrieved on June 2, 2022).