Abstract
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeat) technology is a powerful tool in biology and medicine. However, the safety and application of this technology is hampered by excessive activity of CRISPR machinery. It is particularly important to develop methods for switching off CRISPR activity in human cells. The current study demonstrates the concept of supramolecular CRISPR-OFF switches by employing host-guest chemistry. We demonstrate that the CRISPR systems show considerable tolerance to adamantoylation on guide RNAs (gRNAs), whereas supramolecular complexation tremendously affects the function of adamantoyl gRNAs. Host–guest chemistry is demonstrated to be novel and effective tools to reduce unwanted excessive activities of CRISPR complexes in human cells. This work indicates considerable potential of supramolecular strategy for controlling and enhancing CRISPR systems.
INTRODUCTION
Many prokaryotes use Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) as an immune mechanism of self-defense (1). To date, CRISPR systems were adapted and widely used in different areas, especially for gene editing (2–4). For Class 2 CRISPR systems, a single CRISPR-associated effector protein (Cas) uses guide RNAs (gRNAs) to recognize and cleave target sequences at specific positions (5). CRISPR/Cas9 is an efficient and versatile tool for genome engineering in many species (6). In contrast to Cas9, the Cas13a (also known as C2c2) degrades foreign RNAs through RNA-guided RNA cleavage (7–9). However, the reliability and safety of CRISPR technology is hampered by excessive activity of CRISPR complexes (10–16). This technology can be greatly improved if the activity of CRISPR complexes can be controlled properly. Some important studies demonstrated the concept of stimuli-triggered activation of Cas9 to eliminate the adverse effects of CRISPR systems (17–19). In these studies, activation was triggered by specific stimuli to remove the protecting groups at the essential sites on CRISPR complexes. Different external triggers were studied for this purpose (20–23). However, the adverse effects are mainly caused due to sequence complementarity and can still happen once the protecting groups are removed.
An important strategy to battle excessive activity of CRISPR complexes would be to develop CRISPR-OFF switches (10). In this respect, the significance of supramolecular chemistry has been overlooked. The specific recognition of RNA elements is a fundamental challenge in the biological sciences. Synthetic supramolecular molecules are creating new opportunities for RNA recognition, which are orthogonal to classical small molecules (24). Supramolecular science examines intermolecular noncovalent bonding interactions (25–27). On account of the dynamic and noncovalent nature of supramolecular interactions, they have been introduced as temporary switches to construct stimuli-responsive tools (28,29). Cucurbit[n]uril (CBn) represents an attractive host due to its ability to encapsulate various guests (30,31). Of the known CBn homologues, CB7 molecules have drawn particular attention because of their remarkable biocompatibility (32,33). They have been widely used as structural and functional units for high-precision assemblies (34–36). However, the capability of supramolecular approach has not been extensively studied in nucleic acid studies by deficiencies in the methods currently available for supramolecular recognition of RNA elements.
The current study demonstrates the concept of supramolecular CRISPR-OFF switches by utilizing host–guest chemistry (Figure 1). This strategy relies on the use of adamantoyl RNAs (blue strands with purple moieties in Figure 1A), which are synthesized using a post-synthetic modification approach. Different adamantane imidazolides (ADIs in Supplementary Figure S1) are used to perform adamantoylation of RNAs. The noncovalent binding of CB7 to adamantoyl RNAs is investigated (complexed RNAs in Figure 1A), we further demonstrate that supramolecular complexation can be used to regulate RNA–protein interactions. The CRISPR systems show considerable tolerance to adamantoyl gRNAs, whereas supramolecular complexation tremendously affects the activity of CRISPR systems (Figure 1B). This is proved to be applicable to RNA-guided RNA cleavage (Cas13a mediated) and RNA-guided DNA cleavage (Cas9 mediated). We further show that this supramolecular strategy can be used to switch off CRISPR/Cas9-mediated genome editing in human cells. This work therefore indicates considerable potential of supramolecular strategy for controlling and enhancing CRISPR systems.
Figure 1.
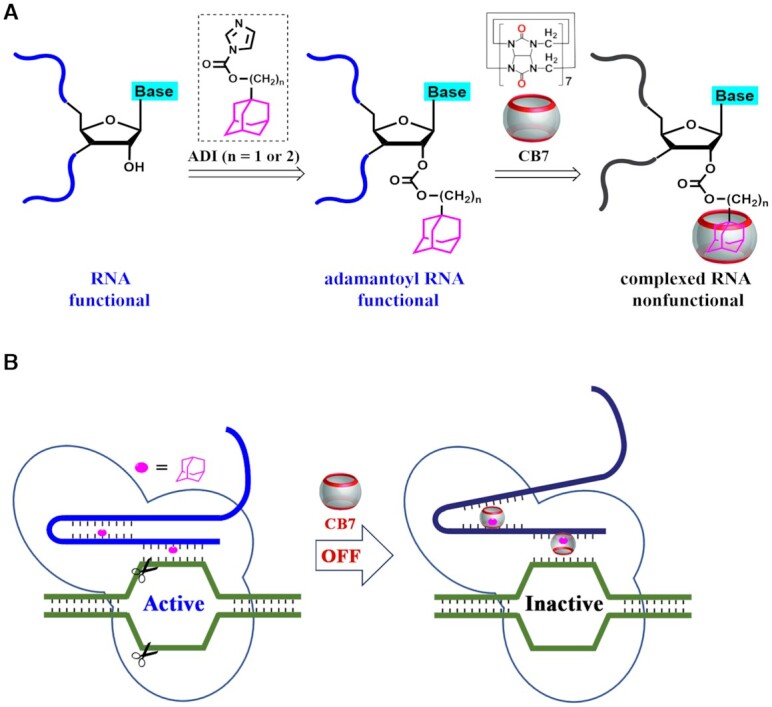
Schematic illustration of the design and workflow. (A) The illustration of supramolecular complexation on adamantoyl RNAs. The post-synthetic modification approach is used to prepare adamantoyl RNAs. Different ADI compounds were used for installing adamantane moieties to manipulate RNA functions. (B) The illustration of supramolecular CRISPR-OFF switches. Adamantoyl gRNAs are well tolerated by CRISPR systems, while supramolecular complexation tremendously affects the CRISPR functions.
MATERIALS AND METHODS
Materials
The sequences of all used oligonucleotides are demonstrated in Supplementary Table S1. These oligonucleotides were ordered from TaKaRa company (Dalian, China). Unless otherwise specified, all chemicals were purchased from Shanghai Aladdin Bio-Chem Technology Co., LTD. The M-MuLV RT (product number: M0253), the Cas9 Nuclease, Streptococcus pyogenes (product# M0646) were purchased from New England Biolabs, Inc. (USA). Transcript Aid T7 High Yield Transcription kit (product# K0441) and Glycogen (product# R0561) were purchased from Thermo Fisher Scientific Inc.
Chemical synthesis
Experimental details for synthesis of each ADI are demonstrated in Supporting Data.
The preparation of adamantoyl RNAs
For adamantoylation of RNA, ADI stock solutions (1 M) were prepared by dissolving powder in dimethyl sulfoxide (DMSO), and further dilutions were made in water. The modification reaction was performed in 1 × adamantoylation buffer, which contained 100 mM HEPES, 6 mM MgCl2, 100 mM NaCl in 60% DMSO (pH 7.5, 25°C) (37). The reaction mixture containing RNA, each ADI and DMAP was incubated at 37°C for various durations with shaking (speed, 200 r/min). The final concentration of each component for a typical 10 μl reaction is as follows: 20 μM RNA, 100 mM ADI, 100 mM DMAP. After incubation, 2.0 μl NaOAc (3 M), 75 μl ethanol and 1.0 μl glycogen (5 g/L) were added into the mixture to quench the reaction. The sample was placed at −80°C for 3.0 h and centrifuged at 12 000 g for 1.0 h subsequently. The resulting RNA pellet was resuspended in nuclease-free water.
Supramolecular complexation in unstructured RNA contexts
For supramolecular complexation assay, RNA with or without adamantoylation was incubated with 200 μM CB7 for 20 min at room temperature and subjected to native or denaturing PAGE analysis. The gel was run in 1 × TBE buffer (89 mM Tris-borate, 2 mM EDTA) in the absence or presence of 33 μM CB7. The acrylamide concentration of the separating gel was 20% (19:1 monomer to bis ratio). About 100 ng of RNA were loaded on the gel. Either denaturing or native PAGE analysis was run in a temperature-controlled vertical electrophoretic apparatus (DYCZ-22A, Liuyi Instrument Factory, Beijing, China). Electrophoresis was run at 10°C for 4.0 h at 400 V. After electrophoresis, the oligomers in the gel were visualized using a Pharos FX Molecular imager (Bio-Rad, USA) in the fluorescence mode (λex = 488 nm or 590 nm).
In vitro transcription and purification of long RNAs
In vitro transcription with the T7 RNA polymerase was performed to synthesize long RNAs, including the Spinach RNA, tracrRNA and various sgRNAs. For preparation of long RNAs, separate shorter overlapping constructs were ordered as DNA with an appended T7 promoter sequence (Supplementary Table S1) (21). A brief template-free PCR was performed to generate a template DNA containing the target-encoding sequence under the control of a T7 promoter. Transcription reactions were performed at 37 °C for 4.0 h in 1 × Transcript aid reaction buffer containing 100 mM HEPES-KOH (pH 7.9), 20 mM MgCl2, 30 mM DTT, 2 mM each NTP, 2 mM spermidine, 0.1 mg/ml T7 RNA polymerase and 300 ng PCR fragments. Following DNA degradation using DNase I at the end of the transcription, the transcribed RNA products were then purified using the NaOAc/phenol/chloroform method. The purified long RNAs were resuspended in RNase-free H2O.
Supramolecular complexation in structured RNA contexts
This study was performed by using long RNA aptamers. The adamantoylation of Spinach RNA was performed using the above general protocol. For aptamer folding and fluorescence study, the Spinach RNA at a final concentration of 0.5 μM was incubated with 10 μM DFHBI in 150 μl folding buffer (40 mM HEPES at pH 7.4, 100 mM KCl, 5 mM MgCl2). After 1 h dark incubation at room temperature, the emission spectrum was collected in wavelength range 460–600 nm. The excitation wavelength was set to 450 nm and a 1 cm path length cell is used. Fluorescence detection was performed at room temperature using a F-4600 FL Spectrophotometer (Hitachi). Slit width: excitation = 5 nm; emission = 5 nm.
Supramolecular OFF-switches for CRISPR/Cas13a
In vitro Cas13a cleavage assay was performed in 1 × Cas13a buffer, which contained 20 mM HEPES, 50 mM KCl, 5 mM MgCl2 and 5% glycerol at pH 6.8, 25°C. The gRNAs were pre-folded by heating to 65°C for 5 min and then slowly cooling to room temperature in 1 × Cas13a buffer. Briefly, gRNAs were complexed with a 2.0 molar excess of protein in 1 × Cas13a buffer at 37°C for 10 min, before adding 5’-FAM labeled RNA targets.
Adamantoyl gRNAs were prepared using the above general protocol. For supramolecular complexation assay, the gRNA (45 nM) with or without adamantoylation was mixed with the Cas13a (90 nM) and CB7 at different concentrations. The reaction was started by rapid mixing of equal volumes of the Cas13a:gRNA complex with an solution containing RNA targets (300 nM) in 1 × Cas13a buffer. The reaction mixture was incubated at 37°C for 2 h.
Collateral cleavage assay
For this study, the reactions were started by rapid mixing of equal volumes of the Cas13a/gRNA preparation with a solution containing target and reporter RNAs in 1 × Cas13a buffer. Final concentration of all components except CB7 was as following: 45 nM purified LbuCas13a, 22.5 nM gRNA, 22.5 nM target (Cas13a target1 or target2 in Supplementary Table S1), 125 nM quenched fluorescent RNA reporter (Cas13a reporter in Supplementary Table S1) and 0.5 U RiboLock RNase inhibitor. The curve describing fluorescence increase versus time is determined using the LS55 fluorescence spectrometer (Perkin–Elmer Inc., USA) with the kinetics mode at room temperature. Reactions were allowed to proceed for 1 h measured every 1 s. The excitation and emission wavelengths are set to 496 and 520 nm and a 1 cm path length cell is used. Slit width: excitation = 5 nm; emission = 5 nm.
EMSA assay using dCas13a
This assay was performed in 1 × EMSA buffer, which contained 20 mM HEPES, 50 mM KCl, 10 μg/ml BSA, 100 μg/ml yeast tRNA, 0.01% Igepal CA-630 and 5% glycerol at pH 6.8, 25°C (7). The gRNA-Cy3 and target-FAM were incubated with the dCas13a for 10 min at 37°C. Samples were then resolved by 6% native PAGE at 4°C (0.5 × TBE buffer). Electrophoresis was run at 6°C for 1 h at 300 V and 1 h at 200 V. After electrophoresis, in gel targets were analyzed using a Pharos FX Molecular imager (Bio-Rad, USA) in the fluorescence mode (λex = 488 and 590 nm).
MB fluorescence assay
The fluorescence of MB probe in the absence of gRNA was recorded to ensure that fluorescence signal does not increase. About 120 nM gRNA with or without adamantoylation was incubated with 100 nM of MB in 1 × hybridization buffer (100 mM Tris-HCl, pH 8.0, 25°C, 100 mM NaCl, 5 mM MgCl2). The CB7 was added to different concentrations and final volume of 400 μl. The fluorescence signal was recorded for 1 h and a 1 cm path length cell was used. MB fluorescence signals were recorded at λex = 496 nm and λem = 520 nm at room temperature. Slit width: excitation = 5 nm; emission = 5 nm.
EMSA assay using dCas9
The Halo domain of dCas9 was labeled by incubation with fluorescent Halo ligand (G1002 HaloTag® Alexa Fluor® 488 ligand, Promega) at ratio of 1:10 at room temperature for 30 min, and purified using 40-K MWCO Zeba spin desalting columns (Thermo Scientific). Gel mobility shift assay was carried out in 1 × EMSA buffer (38). dCas9 protein was preincubated for 10 min with 9 μl of EMSA buffer. Then 1 μL gRNA was added, and the reaction was incubated at room temperature for 15 min to examine the binary complex. Samples were then resolved by 6% native PAGE at 4°C (0.5 × TBE buffer). After electrophoresis (100 V, 1.0 h), in gel targets were visualized using a Pharos FX Molecular imager (Bio-Rad, USA) in the fluorescence mode (λex = 488 nm).
Supramolecular OFF-switches for CRISPR/Cas9
Adamantoyl gRNAs were prepared using the above general protocol for adamantoylation. Each sgRNA was designed using an online tool (39). Target DNA fragments were PCR amplified from genomic DNA (HeLa-OC cells) using the following PCR primers: t-SLX4IP-F and t-SLX4IP-R for t-SLX4IP; t-HPRT1-F and t-HPRT1-R for t-HPRT1; t-HBEGF-F and t-HBEGF-R for t-HBEGF. In vitro Cas9 cleavage assays were performed in 1 × NEBuffer™ 3.1, which contained 100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2 and 100 μg/ml BSA at pH 7.9, 25°C. Briefly, unmodified or adamantoyl sgRNAs (50 ng) were incubated with CB7 at various concentrations in the presence of Cas9 (0.2 μM), in 1 × NEBuffer™ 3.1 buffer at room temperature for 30 min. The Cas9-mediated DNA cleavage reactions were started by rapid mixing of equal volumes of the sgRNA/Cas9 preparation with a solution containing target DNA fragments (50 ng) in 1 × NEBuffer™ 3.1 buffer and subjected to an incubation at 37°C for 0.5 h. Reactions were quenched by adding SDS containing loading dye and loaded onto a 1.5% agarose gel containing 1.5 × Super GelRed for visualization (100 V, 1.5 h). The GeneRuler 100-bp DNA Ladder was used as DNA size marker for these experiments.
For the 2-part gRNA study, each crRNA (unmodified or adamantoyl) and tracrRNA (unmodified or adamantoyl) were incubated at room temperature for 10 min to form the gRNA complexes. For supramolecular complexation step, similar reactions were set up using 15 ng of each crRNA and 25 ng of tracrRNA instead of 50 ng of each sgRNA. For Cas9-mediated DNA cleavage reactions, the conditions were the same as the above sgRNA protocol except that a 11 h incubation at 37°C was used.
Cellular studies
Human HeLa-OC cell line was a kind gift from Prof. Wen-Sheng Wei, School of Life Sciences, Peking University, Beijing, China. Cellular studies were performed using HeLa-OC cells. HeLa-OC cells were cultured in Dulbecco’s Modified Eagle Medium with high glucose, sodium pyruvate and GlutaMAX supplemented with 10% fetal bovine serum and 1 × penicillin–streptomycin (Thermo Fisher Scientific). Cells were passaged to maintain confluency below 70%. In a 6-well plate, 4 × 105 cells were seeded a day prior to transfection. About 2.5 μg of each sgRNA and 5.0 μl of Lipofectamine® 3000 transfection agent (Thermo Fisher Scientific) were mixed and transfected in accordance with the manufacturer’s protocol.
For initial tests with transfection in the presence of CB7, the sgRNA:Lipofectamine complexes were incubated with CB7 at a final concentration of 500 μM. These complexes were left for 20 min at room temperature before transferring them to the well containing cells and growth medium. To determine the action time effect of supramolecular complexation, the CB7 at a final concentration of 500 μM was added to each well at different times after the addition of transfection complexes. At the end of the 4 h incubation period, the transfection medium was then replaced with fresh, complete medium supplemented with 500 μM CB7. To determine the effect of supramolecular complexation on transfected sgRNAs, the transfection of each sgRNA was performed in the absence of CB7. After a 4 h incubation, the medium was replaced with fresh, complete medium that contained CB7 at various concentrations (100, 300 and 500 μM). For cellular studies with the 2-part gRNAs, 0.6 μg of each crRNA and 2.0 μg of tracrRNA were annealed by heating to 90 °C for 2 min and slow-cooled to room temperature to form the gRNA complexes. The transfection of 2-part gRNAs was performed according to the above transfection steps. After a 4 h incubation, the medium was replaced with fresh medium that contained various concentrations of CB7.
In each of the above studies, transfected cells were further incubated at 37°C with 5% CO2 for 24 h. Genomic DNAs were extracted for mutation detection using Qiagen DNeasy Blood and Tissue Kit. Subsequently, target fragments containing target sites were amplified from genomic DNA (200 ng) using PrimeSTAR HS DNA Polymerase with the above primer sets. The amplified DNA products were purified via Zymo Research DNA Clean and Concentrator Kit. T7 Endonuclease I digestion of DNA substrates carrying the target loci (100 ng) was performed according to manufacturer’s protocol. Reaction was quenched by adding SDS containing loading dye and loaded onto a 1.5% agarose gel containing 1.5 × Super GelRed for visualization (100 V, 1.5 h).
RESULTS
The design of supramolecular CRISPR-OFF switches
The goal of this study is to establish a chemical strategy to effectively switch off CRISPR systems using small molecules. It is promising to manipulate CRISPR systems by perturbing interactions between gRNA and Cas. There are two general strategies for this purpose: the Cas engineering and gRNA engineering. In contrast to the extensive studies for the Cas engineering (40–42), very few studies have been performed for engineering gRNAs. The feature of host–guest chemistry motivated us to explore a related gRNA-engineering strategy to deactivate CRISPR complexes. Our strategy is based on the idea that the steric bulk of derivative group could be increased by adding biologically inert bulk that simply increased its molecular size (43,44). The possibility to form large steric complexes with macrocyclic host molecules (Figure 2) and thereby influence the activity of gRNAs remains an important application area of supramolecular interactions (45).
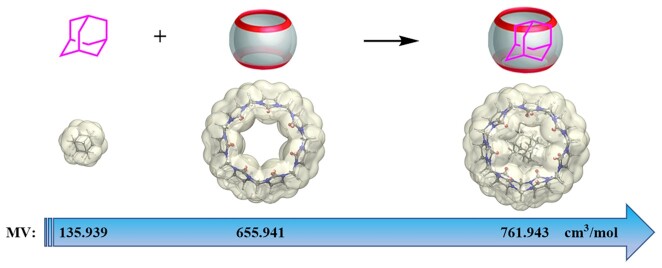
Figure 2.
The steric bulk of adamantane group is increased by forming large complexes with macrocyclic CB7 molecules The optimized molecule geometries and electron density contour surface are demonstrated with isovalue of 0.001. All optimizations were done at M062X/def2-SVP level with Grimme's D3 empirical dispersion correction (45). MV indicates molecular volume.
Two major approaches have been used for synthesizing RNA with new chemical properties. Despite the exciting applications, it is not easy to perform the solid-phase chemical RNA synthesis with phosphoramidite method. Moreover, it is difficult to introduce chemical functionality into one of the four primary RNA bases. Given the significant role of 2’-OH groups of RNA ribose, the post-synthetic modification of these groups is desired (37,46). Different ADI molecules are designed and synthesized for supramolecular functionalization of RNAs (Supplementary Figure S1). Structure determination of the synthesized compounds were performed (details in Supporting Data).
Host–guest complexation in both unstructured and structured RNA contexts
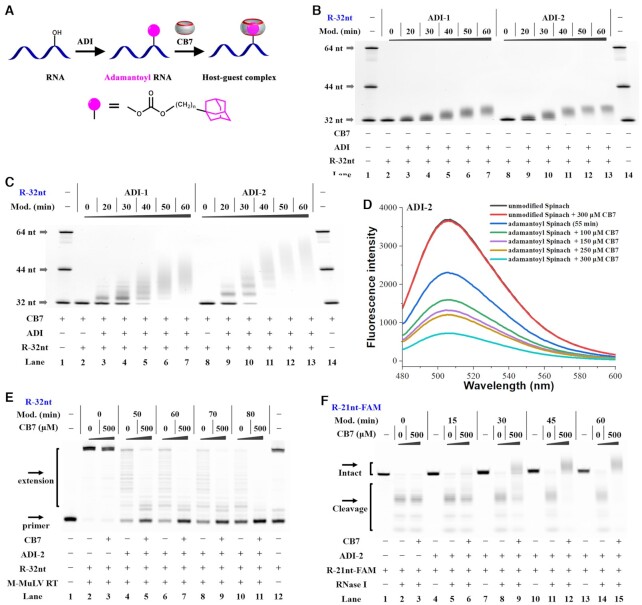
The first step is the adamantoylation of RNAs (Figure 3A). To properly evaluate the efficiency of this modification, an unstructured RNA was used (R-32nt in Supplementary Table S1). For our studies, R-32nt was incubated with each ADI at a specified concentration (100 mM) for varied periods at room temperature. After quenching, the reaction products were recovered by cold ethanol precipitation and analyzed with polyacrylamide gel electrophoresis (PAGE). As we observed, the products show indistinguishable smears with a lower mobility than unmodified RNAs (Figure 3B and Supplementary Figure S2A). This result clearly demonstrates that each ADI reacts efficiently with RNAs.
Figure 3.
Supramolecular complexation in RNA contexts. Reactions were carried out as described in the Materials and Methods Section. All samples were tested in three biological replicates. Image of representative data is shown here. (A) Schematic representation of supramolecular complexation on adamantoyl RNAs. (B) Denaturing PAGE analysis of RNA strands in the absence of CB7. (C) Denaturing PAGE analysis of RNA strands in the presence of 33 μM CB7. Supramolecular complexation significantly impedes the movement of adamantoyl RNAs on gel. For (B) and (C), lanes 1, 14: RNA marker (R-32nt, R-44nt, R-64 nt in Supplementary Table S1); lanes 2, 8: unmodified RNAs; lanes 3–7, 9–13: adamantoyl RNAs with different modification levels. (D) Supramolecular complexation in structured RNA contexts. In this assay, Spinach RNAs were modified with 100 mM ADI-2 for 55 min at room temperature. (E) The influence of supramolecular complexation on reverse transcription with M-MuLV RT. Lane 1: no enzyme control; lanes 2, 4, 6, 8, 10: no CB7 control; lanes 3, 5, 7, 9, 11: the 500-μM CB7 treatments; lane 12: RNA marker. Lanes 2–3 contain unmodified RNAs; lanes 4–5, 6–7, 8–9, 10–11 contain adamantoyl RNAs with increasing modification levels. (F) The influence of supramolecular complexation on RNase degradation. Treating adamantoyl RNAs with CB7 almost completely abolishes RNase I action. Lanes 1, 4, 7, 10, 13: no enzyme control; lanes 2, 5, 8, 11, 14: no CB7 control; lanes 3, 6, 9, 12, 15: the 500-μM CB7 treatments. Lanes 1–3 contain unmodified RNAs; lanes 4–6, 7–9, 10–12, 13–15 contain adamantoyl RNAs with increasing modification levels.
We next studied the host–guest complexation between CB7 and adamantoyl RNAs (47). For this purpose, a denaturing gel retardation assay is designed to be performed in the presence of CB7. This is based on the expectation that CB7-bound RNAs migrate more slowly than free RNA fragments when subjected to electrophoresis. By comparing the bands in each sample to the same RNA marker, we find that adamantoyl RNAs are much more retarded in the CB7-present gel than in the CB7-absent gel (Figure 3B and C). In the absence of CB7, no additional band was seen with mobility less than that of the 44 nt marker (lanes 3–7 and 9–13 in Figure 3B), even for the sample with highest modification level (lanes 7 and 13 in Figure 3B). In the presence of 200 μM CB7, very slow moving species were usually seen with mobility less than that of the 44-nt marker (lanes 6–7 and 11–13 in Figure 3C). Additionally, the CB7 treatment does not affect the migration of unmodified RNAs (lanes 2 and 8 in Figure 3C). We also analyzed supramolecular interactions between CB7 and adamantoyl RNAs with native PAGE (Supplementary Figure S2A and S2B). These results clearly show that CB7 binds specifically to adamantoyl RNAs in unstructured contexts. We also tested a number of other RNAs with different lengths and demonstrated the general applicability of our strategy (Supplementary Figures S3 and S4).
We further studied supramolecular complexation between CB7 and adamantoyl RNAs in structured contexts. In this study, we examined the Spinach RNAs that bind and activate the fluorescence of certain fluorophores (37,48). These light-up aptamers have been exploited as RNA mimics of Green Fluorescent Protein (GFP). Not surprisingly, unmodified Spinach can bind to 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI) and dramatically increase its fluorescence, which was not influenced by the addition of CB7 (Supplementary Figure S5). Although the adamantoylation step reduced the fluorescence of the Spinach-DFHBI complexes to some extent, further CB7 treatment dramatically inhibited the staining of aptamers with DFHBI. This effect became more evident with increasing modification level of RNAs (Supplementary Figure S5). We further observed that supramolecular complexation inhibits the Spinach-induced fluorescence signal in a concentration-dependent manner (Figure 3D). Thus, supramolecular complexation between CB7 and adamantoyl RNAs can be used to control the folding and function of long structured RNAs.
Encouraged by the above results, we set out to examine supramolecular interactions between CB7 and adamantoyl RNAs in more biological contexts. Here, reverse transcription of RNA templates is used as a model system to examine RNA-protein interactions. In an initial study, the Moloney Murine Leukemia Virus reverse transcriptase (M-MuLV RT) was used as the elongating enzyme. The elongation scaffold was set up by assembling M-MuLV RT with the primer/template duplex. We demonstrate that RT enzymes do not tolerate adamantoylation of template RNAs well. Moreover, supramolecular complexation leads to a substantial decrease in the processivity of M-MuLV RT as judged from product lengths (Figure 3E and Supplementary Figure S6). This effect became more evident with increasing modification level of RNAs. Additionally, the CB7 treatment does not influence M-MuLV RT elongation along unmodified templates. Similar results were also seen using a second RT (HIV-RT) with the same substrate (Supplementary Figure S7).
To further test supramolecular complexation in biological contexts, a RNase degrading assay was employed using fluorogenic RNA substrates. We show that RNase I readily degrades the majority of unmodified RNAs in 10 min. We further demonstrate that the adamantoylation of RNAs does not reduce the RNase I action (Figure 3F and Supplementary Figure S8A). Importantly, the CB7 treatment essentially stops the activity of this enzyme. Additionally, RNase I almost completely degrades unmodified RNAs in the presence of CB7 after 10 min. Further results show that supramolecular complexation can inhibit RNase I activity in a concentration-dependent manner (Supplementary Figure S8B, S8C). Similar results were also seen with a second RNase (RNase T1) (Supplementary Figure S9). Overall, these results demonstrate that supramolecular interactions can act as a strong mechanism for blocking the recognition between RNAs and proteins.
Supramolecular OFF-switches for CRISPR/Cas13a
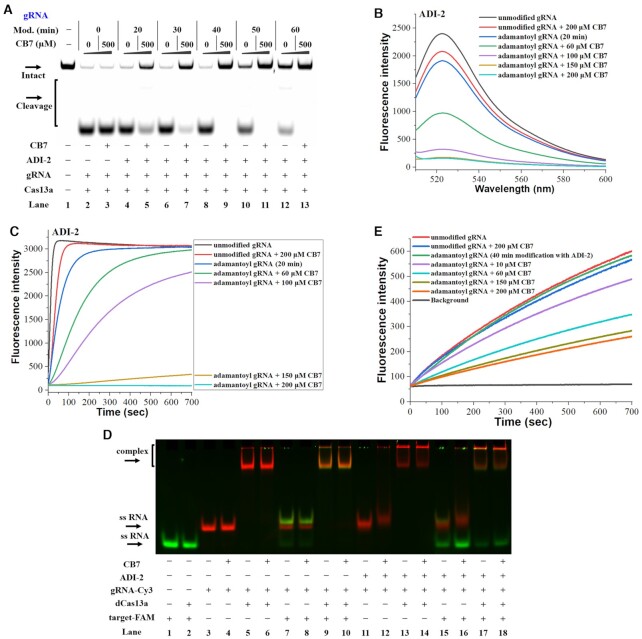
The above studies demonstrate that host–guest chemistry can be used to switch off RNA functionality. A successful CRISPR application depends greatly on the function of gRNAs (49). We tend to regulate the Cas13a activity by disrupting the function of gRNAs. The first step is the adamantoylation of gRNAs (Supplementary Figure S10). An important thing is to determine how much steric stress the gRNAs can tolerate. We demonstrate that the adamantoylation of gRNAs is well tolerated by CRISPR/Cas13a (Figure 4A). We next asked whether host–guest interaction can be used to turn off the function of CRISPR/Cas13a. The results demonstrate that supramolecular complexation reduces the potency of adamantoyl gRNAs to support Cas13a-mediated RNA cleavage. The degree of reduction depends on the modification level of adamantoyl gRNAs (Figure 4A and Supplementary Figure S11A). We further demonstrate that the addition of CB7 decreased the level of RNA cleavage in a concentration-dependent manner (Supplementary Figure S11B and S11C). Similar results were also seen with a second target RNA (Supplementary Figure S12). As a control, the cleavage reaction using unmodified gRNAs was not influenced in the presence of a relatively high concentration of CB7. These results demonstrate that the Cas13a gRNA tolerates a certain extent of adamantoylation and is much less tolerant to host–guest complexes with large steric hindrance.
Figure 4.
Supramolecular OFF-switches for CRISPR/Cas13a. Reactions were performed as described in the Materials and Methods section. All samples were tested in three biological replicates. Image of representative data was shown here. (A) Modification-dependent inhibition of CRISPR/Cas13a upon supramolecular complexation. In this study, adamantoyl gRNAs and Cas13a were employed to cleave target1 in the absence or presence of 500 μM CB7. Lane 1: no enzyme control; lanes 2, 4, 6, 8, 10, 12: no CB7 control; lanes 3, 5, 7, 9, 11, 13: the 500 μM CB7 treatments. Lanes 2–3 contain unmodified gRNA; lanes 4–5, 6–7, 8–9, 10–11, 12–13 contain adamantoyl gRNAs with increasing modification levels. (B) The influence of supramolecular complexation on the Cas13a collateral cleavage. The fluorescence spectra were measured at room temperature after 100 s incubation at 37°C. (C) Dose-dependent effect of supramolecular complexation on inhibiting CRISPR/Cas13a. (D) Effect of supramolecular complexation on the formation of ternary dCas13a-gRNA-target complexes. Lanes 1–2: target-FAM control; lanes 3–10 contain unmodified gRNA-Cy3; lanes 11–18 contain adamantoyl gRNA-Cy3 (a 60 min modification with 100 mM ADI-2). (E) Effect of supramolecular complexation on RNA hybridization. This study uses adamantoyl gRNAs (a 40 min modification with 100 mM ADI-2).
An important and well-known feature of Cas13a is that it exhibits a ‘collateral effect’ of non-target RNA cleavage (7,50). We proceeded to examine supramolecular interactions on adamantoyl gRNAs using this unique feature of Cas13a. A spectral fluorescence assay was performed to determine the cleavage ends produced by Cas13a collateral activity. In this study, the Cas13a starts cutting a quenched RNA reporter once it find a perfect complementary sequence to its gRNAs (50). The fluorescence intensity can therefore be largely increased. With adamantoyl gRNAs, the sample exhibits a comparable fluorescence compared to that of unmodified control after 100 s. Importantly, supramolecular complexation inhibited the Cas13a collateral activity in a dose-dependent manner (Figure 4B and Supplementary Figure S13A). In a kinetic fluorescence assay, the characteristic fluorescent signals for reporter RNA were plotted as a function of reaction time. In agreement with the above results, the CB7 effectively inhibited fluorescence enhancement of the reaction (Figure 4C and Supplementary Figure S13B). In addition, RNA cleavage in the control samples was not evidently influenced through the CB7 treatment. In this respect, supramolecular interactions can function as OFF-switches to terminate Cas13a-mediated RNA cleavage.
We next investigated how supramolecular complexation affects the functions of gRNAs to regulate CRISPR/Cas13a. Hence, an electrophoretic mobility shift assay (EMSA) was performed by using a dCas13a (LbuCas13a R1079A/K1080A double mutant) and a fluorescently labeled gRNA (gRNA-Cy3 in Supplementary Table S1) (7). We observed the dCas13a protein can bind with adamantoyl gRNAs very well in both absence and presence of CB7 (Supplementary Figure S14). These results demonstrate that supramolecular complexation does not evidently affect the formation of binary gRNA/Cas13a complexes. We further studied the effects of supramolecular complexation on the formation of ternary Cas13a RNP. The results demonstrated that only low interference of Cas13a RNP formation was provided by supramolecular complexation (lanes 17–18 in Figure 4D and Supplementary Figure S15A). However, supramolecular complexation significantly reduced the binding of adamantoyl gRNA to its targets (lanes 16, 18 in Figure 4D and Supplementary Figure S15A). In this respect, supramolecular complexation may play a more important role in inhibiting proper RNA hybridization and can significantly hinder the function of CRISPR/Cas13a. It is also possible that the bulky CB7 interacts with RNP to prevent cleavage but not binding between Cas and gRNA.
We proceeded to better define the role of supramolecular complexation on RNA hybridization. To this end, a fluorescence assay was performed by incubating gRNA with a complementary molecular beacon (MB in Supplementary Table S1) (37). The fluorescence of unhybridized MB is quenched due to the close proximity between quencher and fluorophore (black line in Figure 4E and Supplementary Figure S15B). When unmodified gRNA is added, we observed a large increase in fluorescence upon the separation of fluorophore and quencher in MB (red line in Figure 4E and Supplementary Figure S15B). We further show that adamantoyl gRNA leads to a fluorescent signal comparable to that of unmodified control, indicating that adamantoylation does not evidently affect RNA hybridization (at the modification levels achieved here). Importantly, treating adamantoyl gRNA with CB7 largely abolished the MB fluorescence (Figure 4E and Supplementary Figure S15B). Moreover, the CB7 treatment can inhibit the fluorescence in a concentration-dependent manner. This study further confirms that supramolecular complexation strongly impairs RNA hybridization.
Supramolecular OFF-switches for CRISPR/Cas9
The above Cas13a studies encouraged us to fabricate supramolecular OFF-switches for CRISPR/Cas9. It is known that the Cas9 protein binds its gRNA and this complex executes cleavage in the presence of target sequences (51,52). We therefore investigated whether the host-guest interactions can be used to regulate the Cas9–gRNA interactions. For CRISPR/Cas9 study, the gRNA can be provided using different formats (2). The most frequently used format is the single chimeric gRNA (sgRNA) (2,3). As a first step, in vitro transcription using the PCR-product template is used to produce sgRNAs targeting human genes (the SLX4 interacting protein, SLX4IP gene in Supplementary Figure S16A, sg-SLX4IP in Supplementary Table S1) (21,53). The adamantoylation of sg-SLX4IP is demonstrated with the electrophoresis (Supplementary Figure S17A and S17B). In the EMSA study, we used an engineered nuclease-deficient Cas9 (dCas9), which contains a Halo tag at the C-terminus (38). The fused Halo tag was fluorescently labeled by a Halo ligand conjugated to a fluorescent dye. It is observed that adamantoyl sg-SLX4IP is capable of binding to the dCas9 protein, with comparable efficiency to unmodified control (lanes 5, 7, 9, 11 and 13 in Figure 5A, Supplementary Figure S17C). Importantly, supramolecular complexation introduced significant interfering effects to the formation of dCas9/sg-SLX4IP complexes (lanes 6, 8, 10, 12 and 14 in Figure 5A, Supplementary Figure S17C). These conclusions held true for an additional study using another sgRNA targeting the GFP gene (sg-GFP in Supplementary Table S1; results in Supplementary Figures S18 and S19) (21).
Figure 5.
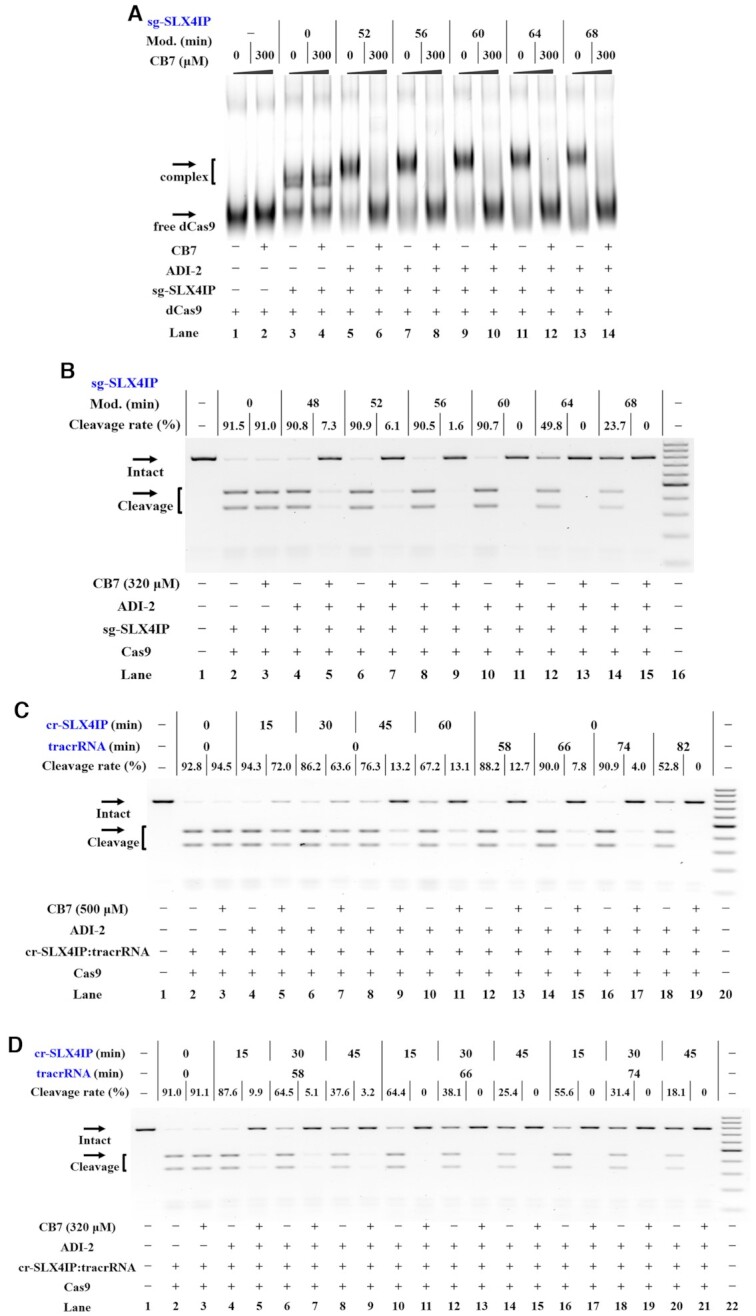
Supramolecular OFF-switches for CRISPR/Cas9. Reactions were performed as described in the Materials and Methods section. All samples were tested in three biological replicates. Image of representative data was shown here. (A) Supramolecular control of the complex formation of Cas9/sgRNA. EMSA analysis of fluorescently labeled dCas9 is demonstrated. Lanes 1–2: no sgRNA control; lanes 3–4 contain unmodified sg-SLX4IP; lanes 5–6, 7–8, 9–10, 11–12, 13–14 contain adamantoyl sg-SLX4IP with increasing modification levels. (B) Modification-dependent inhibition of CRISPR/Cas9 upon supramolecular complexation. Lane 1: target control; lanes 2–3 contain unmodified sg-SLX4IP; lanes 4–5, 6–7, 8–9, 10–11, 12–13, 14–15 contain adamantoyl sg-SLX4IP with increasing modification levels; lane 16: DNA marker (GeneRuler 100-bp DNA Ladder). (C) Modification-dependent inhibition of CRISPR/Cas9 with adamantoyl cr-SLX4IP or adamantoyl tracrRNA. Lane 1: no Cas9 control; lanes 2–3 contain unmodified cr-SLX4IP and unmodified tracrRNA; lanes 4–11 contain unmodified tracrRNA and adamantoyl cr-SLX4IP with increasing modification levels; lanes 12–19 contain unmodified cr-SLX4IP and adamantoyl tracrRNA with increasing modification levels; lane 20: DNA marker. (D) Influence of supramolecular complexation on CRISPR/Cas9 with a separate crRNA and tracrRNA. Lane 1: no Cas9 control; lanes 2–3 contain unmodified cr-SLX4IP and tracrRNA; lanes 4–21 contain adamantoyl cr-SLX4IP and tracrRNA with increasing modification levels; lane 22: DNA marker.
In view of the good efficiency of this supramolecular strategy in inhibiting Cas9/sgRNA interactions, we proceeded to investigate the effects of this strategy in controlling the activity of CRISPR/Cas9 complexes. In this study, we examined the most adopted CRISPR/Cas9 originating from Streptococcus pyogenes (2,54). For our studies, the target DNA sequences were amplified by PCR from human genomic DNA (Supplementary Figure S16A). In vitro DNA cleavage assay was performed in the absence or presence of CB7 (21). Although extensive adamantoylation of sg-SLX4IP leads to low levels of Cas9 activity, moderate adamantoylation is well tolerated for this sgRNA (Supplementary Figures S20A and S21A). We demonstrate that supramolecular complexation can inhibit the function of adamantoyl sg-SLX4IP. The observed effects of supramolecular complexation on the function of adamantoyl gRNAs are evident, with greater effects occurring at higher levels of modification (Figure 5B and Supplementary Figure S20B). The CB7 treatment leads to inhibition of Cas9-mediated DNA cleavage in a concentration-dependent manner (Supplementary Figures S20C and S21B). Further studies were performed to demonstrate the specificity of supramolecular regulation. Based on the results, the treatment with other macrocyclic hosts, such as α-cyclodextrin (α-CD), β-CD and γ-CD, produced negligible inhibition of the DNA cleavage (Supplementary Figure S21C). It is necessary to mention that other CB derivatives (CB6 and CB8) inhibited the activity of adamantoylated sgRNA, although being less potent than CB7. These conclusions held true for two additional studies using another two sgRNAs with different target sites, suggesting a general principle (Hypoxanthine Phosphoribosyltransferase 1, HPRT1 gene in Supplementary Figure S16B, results of sg-HPRT1 in Supplementary Figures S22–S24; heparin binding EGF like growth factor, HBEGF gene in Supplementary Figure S16C, results of sg-HBEGF in Supplementary Figures S25 and S26).
Although the sgRNA becomes the most popular format, the ‘2-part’ system, using a separate CRISPR-RNA (crRNA) and trans-activating crRNA (tracrRNA), has a good feature due to its facile modularity (55). We tend to explore the effect of supramolecular CRISPR-OFF switches for turning off CRISPR/Cas9 with the ‘2-part’ gRNAs. In this study, the crRNA was chemically synthesized and the tracrRNA was prepared using in vitro transcription. The adamantoylation of cr-SLX4IP was then examined (Supplementary Figure S27). Following DNA cleavage assay demonstrated that the activity of CRISPR/Cas9 was not significantly influenced by adamantoylation of crRNAs (left half in Supplementary Figures S28A and S29A). It is observed that the CB7 treatment of adamantoyl cr-SLX4IP reduces its potency to support Cas9-mediated DNA cleavage and that the degree of reduction depends on the level of adamantoylation (lanes 4–11 in Figure 5C and Supplementary Figure S28B). We further demonstrate that supramolecular complexation inhibited the function of adamantoyl crRNAs in a concentration-dependent manner (left half in Supplementary Figures S28C and S29B).
We next investigated the use of supramolecular strategy to control the function of tracrRNAs. In this study, adamantoyl tracrRNAs were complexed with unmodified crRNAs and Cas9 to cut target DNAs. We demonstrate that supramolecular complexation efficiently inhibited the activity of adamantoyl tracrRNAs to support Cas9-catalyzed DNA cleavage (lanes 12–17 in Figure 5C and Supplementary Figure S28B). In addition to the case when both crRNA and tracrRNA are modified with adamantoyl moiety, the CB7 treatment effectively inhibits the activity of Cas9 for cleaving target DNAs (Figure 5D, Supplementary Figure S30). An additional study was performed by using cr-HPRT1 in complexation with a same tracrRNA (results in Supplementary Figures S31–S35). These results are consistent with the above assay using sgRNAs.
Supramolecular control of gene editing in human cells
Having demonstrated the effectiveness of supramolecular CRISPR-OFF switches in test tubes, we became interested in exploring the application of this strategy in human cells. The Cas9-expressing stable cell lines are used for our studies (HeLa-OC cells) (56). This study includes the editing of three endogenous genes, including the HBEGF, SLX4IP and HPRT1 (Supplementary Figure S16) (53,56,57). Before testing supramolecular interactions in cells, we determined the tolerance of HeLa-OC cells to CB7. We observed that 500 μM CB7 did not have an evident effect on cell viability (Supplementary Figure S36). Next, the gRNAs with or without adamantoylation were delivered into HeLa-OC cells using Lipofectamine reagents (56). The transfection step can require a few hours for the complexes to reach the perinuclear region. For our studies, 4 h of incubation is used for transfecting gRNAs. Transfected cells were further cultured for 24 h at 37°C. Followingly, a T7E1 nuclease assay was performed to measure the percentage of indel (insertion/deletion mutation) (56). We demonstrate that the transfection of adamantoyl gRNAs could induce targeted indels in HeLa-OC cells, although with a lower activity (Supplementary Figures S37 and S38). Hence, adamantoyl gRNAs can be fully used for genome editing.
Having shown efficient genome editing using adamantoyl gRNAs in HeLa-OC cells, we tested whether host–guest interations can be used to inhibit in-cell CRISPR functions. Four hours after complex administration, the lipofectamine-containing medium is replaced to a fresh, complete medium containing CB7. A series of experiments were carried out by treating cells with CB7 at various times after the addition of gRNA:lipofectamine complexes. We observed that targeted indel frequencies decreased in a time-dependent manner (Supplementary Figures S37A and S38A). These results are expected, since the presence of CB7 in an individual transfection may interfere with the uptake of adamantoyl gRNAs by cells. Our major aim was investigating the effect of supramolecular complexation on functions of transfected gRNAs. To our delight, evident inhibition of indel generation was observed when the CB7 was added at 4 h postinitiation of transfection and incubated with cells (lanes 11, 17 in Supplementary Figures S37A and S38A). We further demonstrate that although the CB7 treatment of gRNA:lipofectamine complexes can greatly inhibit the indel formation, complete abolishment would be still difficult in the presence of 500 μM CB7 (Supplementary Figures S37B and S38B).
We next systematically investigated the effect of supramolecular complexation after the transfection step. The adamantoyl gRNAs were delivered into cells as above before increasing amounts of CB7 were added. The results demonstrate that the CB7 treatment evidently decreased the ability of adamantoyl sgRNAs to create genome lesions in HeLa-OC cells. This effect was found to be more evident upon increasing the modification levels of adamantoyl gRNAs (Figure 6A; Supplementary Figures S39A, S40A and S41A). We further demonstrate that the CB7 treatment decreases the function of adamantoyl sgRNAs in a concentration-dependent manner (Supplementary Figures S40B and S41B).
Figure 6.
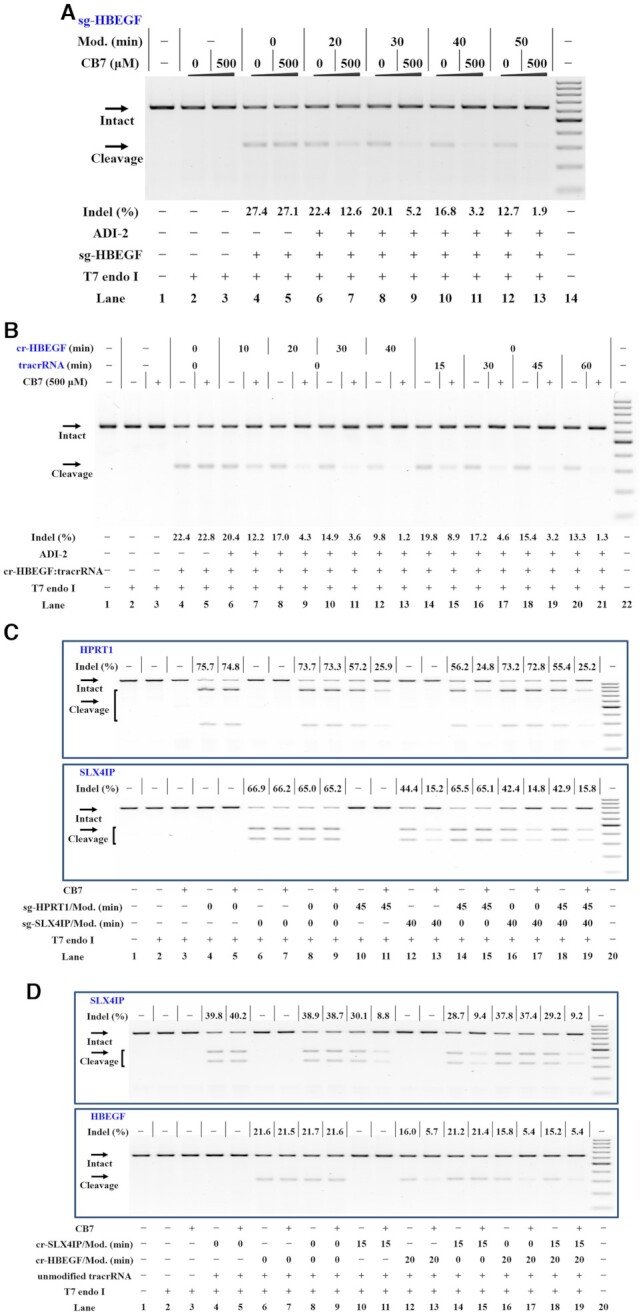
Supramolecular control of CRISPR/Cas9 in human cells. Cellular studies were performed as described in the Materials and Methods section. The gRNAs were delivered into HeLa-OC cells before CB7 was added. The treatment for each sample is indicated by the signs at the bottom of each lane. All samples were tested in three biological replicates. Image of representative data is shown here. (A) Supramolecular control of editing of a single gene using sgRNAs. Lane 1: target control; lanes 2–3: no sgRNA control; lanes 4–5 contain unmodified sg-HBEGF; lanes 6–7, 8–9, 10–11, 12–13 contain adamantoyl sg-HBEGF with increasing modification levels; lane 14: DNA marker (GeneRuler 100 bp DNA Ladder). (B) Supramolecular control of editing of a single gene using the 2-part gRNAs. Lane 1: target control; lanes 2–3: no gRNA control; lanes 4–5 contain unmodified cr-HBEGF and unmodified tracrRNA; lanes 6–13 contain unmodified tracrRNA and adamantoyl cr-HBEGF with increasing modification levels; lanes 14–21 contain unmodified cr-HBEGF and adamantoyl tracrRNA with increasing modification levels; lane 22: DNA marker. (C) Supramolecular control of sgRNA combination. Upper panel: the editing of HPRT1 gene. Lower panel: the editing of SLX4IP gene. Lane 1: target control; lanes 2–3: no sgRNA control; lanes 4–5 contain unmodified sg-HPRT1; lanes 6–7 contain unmodified sg-SLX4IP; lanes 8–9 contain unmodified sg-HPRT1 and unmodified sg-SLX4IP; lanes 10–11 contain adamantoyl sg-HPRT1; lanes 12–13 contain adamantoyl sg-SLX4IP; lanes 14–15 contain adamantoyl sg-HPRT1 and unmodified sg-SLX4IP; lanes 16–17 contain adamantoyl sg-SLX4IP and unmodified sg-HPRT1; lanes 18–19 contain adamantoyl sg-HPRT1 and adamantoyl sg-SLX4IP; lane 20: DNA marker. (D) Supramolecular control of the 2-part gRNA combination. In this study, cells were transfected with crRNA:tracrRNA complexes with different treatments. Upper panel: the editing of SLX4IP gene. Lower panel: the editing of HBEGF gene. Lane 1: target control; lanes 2–3: no gRNA control; lanes 4–5 contain unmodified cr-SLX4IP; lanes 6–7 contain unmodified cr-HBEGF; lanes 8–9 contain unmodified cr-SLX4IP and unmodified cr-HBEGF; lanes 10–11 contain adamantoyl cr-SLX4IP; lanes 12–13 contain adamantoyl cr-HBEGF; lanes 14–15 contain adamantoyl cr-SLX4IP and unmodified cr-HBEGF; lanes 16–17 contain adamantoyl cr-HBEGF and unmodified cr-SLX4IP; lanes 18–19 contain adamantoyl cr-SLX4IP and adamantoyl cr-HBEGF; lane 20: DNA marker. For (C) and (D), the separate panels come from the same experiment.
We further pursued to achieve supramolecular control of 2-part gRNAs in living cells. In this study, we determined the efficiency of genome editing by crRNA:tracrRNA co-transfection at 24 h post-transfection. We demonstrate that the transfection of adamantoyl crRNA and unmodified tracrRNA produces efficient genome editing in HeLa-OC cells with predictable banding patterns. To our delight, an evident inhibition of this process can be achieved through supramolecular complexation (lanes 6–13 in Figure 6B and Supplementary Figure S42, left half in S39B). We then transfected the cells with unmodified crRNAs and an adamantoyl tracrRNA, and took the same concentration of unmodified crRNA:tracrRNA as control. An analogous pattern of CB7-responsible inhibition was evident (lanes 14–21 in Figure 6B and Supplementary Figure S42, right half in Supplementary Figure S39B).
Many biological studies require targeting distinct genomic locations simultaneously to study complex gene regulatory networks (58). We are interested in whether supramolecular CRISPR-OFF switches could support control of specific gRNAs while other gRNAs remain unaffected. In this study, we examined the transfection of two sgRNAs targeting different genes at the same time. In addition to sgRNAs targeting HPRT1, we included sgRNAs that target the SLX4IP. We transfected each sgRNA either alone or in combination and analyzed the indel formation of target genes at 24 h post-transfection. Initial studies were performed by transfecting HeLa-OC cells with adamantoyl sg-HPRT1 and unmodified sg-SLX4IP. We demonstrate that the CB7 treatment selectively inhibited the editing of the HPRT1 gene (lanes 14–15 in Figure 6C). Next, adamantoyl sg-SLX4IP and unmodified sg-HPRT1 were transfected into HeLa-OC cells at the same time. Strong inhibition of the analyzed gene was observed when adamantoyl sgRNA was used but not with unmodified sgRNA or under CB7-free conditions (Figure 6C and Supplementary Figure S39C).
In addition to the control of multiple sgRNAs, we also tested the genome editing by transfecting tracrRNAs along with multiple crRNAs into HeLa-OC cells. We demonstrate that the CB7 treatment does not evidently influence the activity of unmodified crRNAs in HeLa-OC cells (lanes 8–9 in Figure 6D). We next transfected unmodified tracrRNA along with adamantoyl cr-SLX4IP and unmodified cr-HBEGF into HeLa-OC cells. Upon the CB7 treatment, the editing of SLX4IP gene was selectively inhibited, while the editing of HBEGF gene was not influenced (lanes 14–15 in Figure 6D). We also transfected adamantoyl cr-HBEGF and unmodified cr-SLX4IP into HeLa-OC cells at the same time. In this case, the CB7 treatment leads to selective inhibition of editing targeting the HBEGF gene, while the editing of SLX4IP gene was not influenced (Figure 6D and Supplementary Figure S39D). These results demonstrate that the manipulation of specific gRNAs offers a relatively quick and easy way to manipulate specific genes in cells.
DISCUSSION
The current study demonstrates a supramolecular strategy for the construction of CRISPR-OFF switches. The idea of the current study is simply to divide the manipulation into two steps. Step 1 is adamantoylation of gRNA by post-synthetic modifications, and step 2 is the anchoring of CB7 molecules onto the gRNA molecules through supramolecular interactions (30). To date, very little is known about the tolerance level of gRNAs to non-natural modifications. It is uncertain if the gRNA tolerates chemical modifications that are structurally unrelated to its ribonucleotide monomers, and how big these moieties can be without affecting the functionality of Cas endonuclease. We used different methods to examine the tolerance level of RNAs to adamantoylation. We demonstrate that both Cas9 and Cas13a well tolerate the ribose adamantoylations to their gRNAs. The adamantoyl groups do not seem to affect the activity of gRNAs to support Cas-mediated target cleavage, whereas larger host–guest complexes were more perturbing. We demonstrate that supramolecular manipulation of gRNAs can be used as a promising tool for controlling CRISPR systems.
A variety of side effects can occur when the CRISPR complexes are constitutively active. To date, there are currently limited means to control excessive Cas9 activity once it has been delivered or activated (13). One available method to achieve this goal is to use anti-CRISPRs (10,15,16). However, the development of anti-CRISPRs is challenging. Supramolecular control of CRISPR systems would be useful for restricting gene editing to desired times and places. Our strategy has multiple advantages. First, the adamantoylation of gRNAs is simple and easily prepared, and it is not necessarily to identify the essential sites of gRNA for CRISPR functions. Another feature of this method is that adamantoyl gRNAs exhibit an evidently higher stability compared to unmodified ones. Third, our method offers a unique and complementary approach to the current strategy to deactivate the activity of Cas9 protein using anti-CRISPRs (10).
In the current study, the control of gene editing in live cells is accomplished through supramolecular complexation on adamantoyl gRNAs. This supramolecular strategy utilizes inexpensive reagents, functions both with chemically synthesized or transcribed gRNAs and requires no Cas engineering. We believe this strategy will make CRISPR-based gene editing much safer. It is worth mentioning that supramolecular inhibition of gene editing in cells is not complete even at high CB7 concentrations. There are a number of possible reasons for this result. Our study and research by others show that CB7 molecules can cross the cell membrane (59). From a structural point of view, the CB7 molecules act as large steric hindrance to interfere with the CRISPR functions. Hence, the large size of the CB7 molecules is preferable for this purpose. However, at this point CB7 tends to encounter barrier when crossing the cell membrane than other small molecules. On the other hand, because of the noncovalent nature of CB7–adamantane interaction, we believe that some CB7 caps could be pulled away from adamantoyl gRNAs in cellular environments. The possible dissociation will leave behind a leak of the CRISPR activities. We are currently pursuing improvements of the CB7 activity through chemical engineering. Our study provides proof of principle for establishing temporal control over potentially any RNA-guided enzymatic reactions, using methods similar to those described here.
CONCLUSION
Overall, the current study establishes the use of host–guest chemistry to develop supramolecular CRISPR-OFF switches. We also demonstrates considerable potential of supramolecular strategy for controlling and enhancing CRISPR systems.
Supplementary Material
ACKNOWLEDGEMENTS
The authors also thank Prof. Wensheng Wei (Peking University), Prof. Ping Yin (Huazhong Agricultural University) and Prof. Shaoru Wang (Wuhan University) for their valuable help.
Author contributions: T.T. conceived the original idea, designed the studies and led the project. W.X.synthesized adamantane imidazolides (ADI-1 and ADI-2). X.Y.L. and Q.Q.Q. performed all biological studies. H.M.J. contributed to some of cellular studies. F.B.L. and S.M.L. contributed to the synthesis of all CB derivatives. C.Z. contributed to molecular optimization study. X.Z. provided consistent support during the running of this project. T.T. wrote the manuscript. All the authors provided feedback on the study and on the manuscript. The authors declare no competing financial interests.
Contributor Information
Wei Xiong, Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan 430072, Hubei, China.
Xingyu Liu, Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan 430072, Hubei, China.
Qianqian Qi, Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan 430072, Hubei, China.
Huimin Ji, Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan 430072, Hubei, China.
Fengbo Liu, School of Chemistry and Chemical Engineering, Wuhan University of Science and Technology, Wuhan 430081, Hubei, China.
Cheng Zhong, Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan 430072, Hubei, China.
Simin Liu, School of Chemistry and Chemical Engineering, Wuhan University of Science and Technology, Wuhan 430081, Hubei, China.
Tian Tian, Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan 430072, Hubei, China.
Xiang Zhou, Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan 430072, Hubei, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [91853119, 21877086, 21871216, 21722803, 21721005, 91753201, 51873160]; Hubei Provincial Natural Science Foundation [2019CFA064]; Fundamental Research Funds for the Central Universities [2042021kf0211]. Funding for open access charge: National Natural Science Foundation of China [91853119].
Conflict of interest statement. None declared.
REFERENCES
- 1. Garneau J.E., Dupuis M.E., Villion M., Romero D.A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadan A.H., Moineau S.. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010; 468:67–71. [DOI] [PubMed] [Google Scholar]
- 2. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A.et al.. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shmakov S., Smargon A., Scott D., Cox D., Pyzocha N., Yan W., Abudayyeh O.O., Gootenberg J.S., Makarova K.S., Wolf Y.I.et al.. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 2017; 15:169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Adli M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018; 9:1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. East-Seletsky A., O’Connell M.R., Knight S.C., Burstein D., Cate J.H., Tjian R., Doudna J.A.. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016; 538:270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B., Shmakov S., Makarova K.S., Semenova E., Minakhin L.et al.. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016; 353:aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu L., Li X., Ma J., Li Z., You L., Wang J., Wang M., Zhang X., Wang Y.. The molecular architecture for RNA-Guided RNA cleavage by cas13a. Cell. 2017; 170:714–726. [DOI] [PubMed] [Google Scholar]
- 10. Pawluk A., Amrani N., Zhang Y., Garcia B., Hidalgo-Reyes Y., Lee J., Edraki A., Shah M., Sontheimer E.J., Maxwell K.L.et al.. Naturally occurring off-switches for CRISPR-Cas9. Cell. 2016; 167:1829–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harrington L.B., Doxzen K.W., Ma E., Liu J.J., Knott G.J., Edraki A., Garcia B., Amrani N., Chen J.S., Cofsky J.C.et al.. A broad-spectrum inhibitor of CRISPR-Cas9. Cell. 2017; 170:1224–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carlson-Stevermer J., Kelso R., Kadina A., Joshi S., Rossi N., Walker J., Stoner R., Maures T.. CRISPRoff enables spatio-temporal control of CRISPR editing. Nat. Commun. 2020; 11:5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kundert K., Lucas J.E., Watters K.E., Fellmann C., Ng A.H., Heineike B.M., Fitzsimmons C.M., Oakes B.L., Qu J., Prasad N.et al.. Controlling CRISPR-Cas9 with ligand-activated and ligand-deactivated sgRNAs. Nat. Commun. 2019; 10:2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maji B., Gangopadhyay S.A., Lee M., Shi M., Wu P., Heler R., Mok B., Lim D., Siriwardena S.U., Paul B.et al.. A high-throughput platform to identify small-molecule inhibitors of CRISPR-Cas9. Cell. 2019; 177:1067–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Watters K.E., Shivram H., Fellmann C., Lew R.J., McMahon B., Doudna J.A.. Potent CRISPR-Cas9 inhibitors from staphylococcus genomes. Proc. Natl. Acad. Sci. U. S. A. 2020; 117:6531–6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shin J., Jiang F., Liu J.J., Bray N.L., Rauch B.J., Baik S.H., Nogales E., Bondy-Denomy J., Corn J.E., Doudna J.A.. Disabling cas9 by an anti-CRISPR DNA mimic. Sci. Adv. 2017; 3:e1701620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cao J., Wu L., Zhang S.M., Lu M., Cheung W.K., Cai W., Gale M., Xu Q., Yan Q.. An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res. 2016; 44:e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Davis K.M., Pattanayak V., Thompson D.B., Zuris J.A., Liu D.R.. Small molecule-triggered cas9 protein with improved genome-editing specificity. Nat. Chem. Biol. 2015; 11:316–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gonzalez F., Zhu Z., Shi Z.D., Lelli K., Verma N., Li Q.V., Huangfu D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014; 15:215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hemphill J., Borchardt E.K., Brown K., Asokan A., Deiters A.. Optical control of CRISPR/Cas9 gene editing. J. Am. Chem. Soc. 2015; 137:5642–5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jain P.K., Ramanan V., Schepers A.G., Dalvie N.S., Panda A., Fleming H.E., Bhatia S.N.. Development of light-activated CRISPR using guide RNAs with photocleavable protectors. Angew. Chem. Int. Ed Engl. 2016; 55:12440–12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luo J., Liu Q., Morihiro K., Deiters A.. Small-molecule control of protein function through staudinger reduction. Nat. Chem. 2016; 8:1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Habibian M., McKinlay C., Blake T.R., Kietrys A.M., Waymouth R.M., Wender P.A., Kool E.T.. Reversible RNA acylation for control of CRISPR-Cas9 gene editing. Chem. Sci. 2019; 11:1011–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shao Y., Jia H., Cao T., Liu D. Supramolecular hydrogels based on DNA self-assembly. Acc. Chem. Res. 2017; 50:659–668. [DOI] [PubMed] [Google Scholar]
- 25. Joyce L.A., Shabbir S.H., Anslyn E.V.. The uses of supramolecular chemistry in synthetic methodology development: examples of anion and neutral molecular recognition. Chem. Soc. Rev. 2010; 39:3621–3632. [DOI] [PubMed] [Google Scholar]
- 26. Sokkalingam P., Shraberg J., Rick S.W., Gibb B.C.. Binding hydrated anions with hydrophobic pockets. J. Am. Chem. Soc. 2016; 138:48–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cai X., Kataria R., Gibb B.C.. Intrinsic and extrinsic control of the pKa of thiol guests inside yoctoliter containers. J. Am. Chem. Soc. 2020; 142:8291–8298. [DOI] [PubMed] [Google Scholar]
- 28. Yan X., Wang F., Zheng B., Huang F.. Stimuli-responsive supramolecular polymeric materials. Chem. Soc. Rev. 2012; 41:6042–6065. [DOI] [PubMed] [Google Scholar]
- 29. Ma X., Tian H.. Stimuli-responsive supramolecular polymers in aqueous solution. Acc. Chem. Res. 2014; 47:1971–1981. [DOI] [PubMed] [Google Scholar]
- 30. Lagona J., Mukhopadhyay P., Chakrabarti S., Isaacs L.. The cucurbit[n]uril family. Angew. Chem. Int. Ed. Engl. 2005; 44:4844–4870. [DOI] [PubMed] [Google Scholar]
- 31. Barrow S.J., Kasera S., Rowland M.J., del Barrio J., Scherman O.A.. Cucurbituril-Based molecular recognition. Chem. Rev. 2015; 115:12320–12406. [DOI] [PubMed] [Google Scholar]
- 32. Bockus A.T., Smith L.C., Grice A.G., Ali O.A., Young C.C., Mobley W., Leek A., Roberts J.L., Vinciguerra B., Isaacs L.et al.. Cucurbit[7]uril-Tetramethylrhodamine conjugate for direct sensing and cellular imaging. J. Am. Chem. Soc. 2016; 138:16549–16552. [DOI] [PubMed] [Google Scholar]
- 33. Tonga G.Y., Jeong Y., Duncan B., Mizuhara T., Mout R., Das R., Kim S.T., Yeh Y.C., Yan B., Hou S.et al.. Supramolecular regulation of bioorthogonal catalysis in cells using nanoparticle-embedded transition metal catalysts. Nat. Chem. 2015; 7:597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vinciguerra B., Cao L., Cannon J.R., Zavalij P.Y., Fenselau C., Isaacs L.. Synthesis and self-assembly processes of monofunctionalized cucurbit[7]uril. J. Am. Chem. Soc. 2012; 134:13133–13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Isaacs L. Stimuli responsive systems constructed using cucurbit[n]uril-type molecular containers. Acc. Chem. Res. 2014; 47:2052–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee D.W., Park K.M., Banerjee M., Ha S.H., Lee T., Suh K., Paul S., Jung H., Kim J., Selvapalam N.et al.. Supramolecular fishing for plasma membrane proteins using an ultrastable synthetic host-guest binding pair. Nat. Chem. 2011; 3:154–159. [DOI] [PubMed] [Google Scholar]
- 37. Kadina A., Kietrys A.M., Kool E.T.. RNA cloaking by reversible acylation. Angew. Chem. Int. Ed. Engl. 2018; 57:3059–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deng W., Shi X., Tjian R., Lionnet T., Singer R.H.. CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:11870–11875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hu J.H., Miller S.M., Geurts M.H., Tang W., Chen L., Sun N., Zeina C.M., Gao X., Rees H.A., Lin Z.et al.. Evolved cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018; 556:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee J.K., Jeong E., Lee J., Jung M., Shin E., Kim Y.H., Lee K., Jung I., Kim D., Kim S.et al.. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat. Commun. 2018; 9:3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Erdogan M., Fabritius A., Basquin J., Griesbeck O.. Targeted in situ protein diversification and Intra-organelle validation in mammalian cells. Cell Chem. Biol. 2020; 27:610–621. [DOI] [PubMed] [Google Scholar]
- 43. Cao W., Qin X., Wang Y., Dai Z., Dai X., Wang H., Xuan W., Zhang Y., Liu Y., Liu T.. A general supramolecular approach to regulate protein functions by Cucurbit[7]uril and unnatural amino acid recognition. Angew. Chem. Int. Ed. Engl. 2021; 60:11196–11200. [DOI] [PubMed] [Google Scholar]
- 44. Wang S.R., Song Y.Y., Wei L., Liu C.X., Fu B.S., Wang J.Q., Yang X.R., Liu Y.N., Liu S.M., Tian T.et al.. Cucurbit[7]uril-Driven host-guest chemistry for reversible intervention of 5-Formylcytosine-Targeted biochemical reactions. J. Am. Chem. Soc. 2017; 139:16903–16912. [DOI] [PubMed] [Google Scholar]
- 45. Grimme S., Ehrlich S., Goerigk L.. Effect of the damping function in dispersion corrected density functional theory. J. Comput. Chem. 2011; 32:1456–1465. [DOI] [PubMed] [Google Scholar]
- 46. Velema W.A., Kietrys A.M., Kool E.T.. RNA control by photoreversible acylation. J. Am. Chem. Soc. 2018; 140:3491–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Persch E., Dumele O., Diederich F.. Molecular recognition in chemical and biological systems. Angew. Chem. Int. Ed. 2015; 54:3290–3327. [DOI] [PubMed] [Google Scholar]
- 48. Filonov G.S., Moon J.D., Svensen N., Jaffrey S.R.. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc. 2014; 136:16299–16308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Aman R., Ali Z., Butt H., Mahas A., Aljedaani F., Khan M.Z., Ding S., Mahfouz M.. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 2018; 19:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A.et al.. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017; 356:438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jinek M., Jiang F., Taylor D.W., Sternberg S.H., Kaya E., Ma E., Anders C., Hauer M., Zhou K., Lin S.et al.. Structures of cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014; 343:1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O.. Crystal structure of cas9 in complex with guide RNA and target DNA. Cell. 2014; 156:935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Panier S., Maric M., Hewitt G., Mason-Osann E., Gali H., Dai A., Labadorf A., Guervilly J.H., Ruis P., Segura-Bayona S.et al.. SLX4IP antagonizes promiscuous BLM activity during ALT maintenance. Mol. Cell. 2019; 76:27–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Heler R., Samai P., Modell J.W., Weiner C., Goldberg G.W., Bikard D., Marraffini L.A.. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015; 519:199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Karvelis T., Gasiunas G., Miksys A., Barrangou R., Horvath P., Siksnys V.. crRNA and tracrRNA guide Cas9-mediated DNA interference in streptococcus thermophilus. RNA Biol. 2013; 10:841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhou Y., Zhu S., Cai C., Yuan P., Li C., Huang Y., Wei W.. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014; 509:487–491. [DOI] [PubMed] [Google Scholar]
- 57. Gasperini M., Findlay G.M., McKenna A., Milbank J.H., Lee C., Zhang M.D., Cusanovich D.A., Shendure J.. CRISPR/Cas9-Mediated scanning for regulatory elements required for HPRT1 expression via thousands of large, programmed genomic deletions. Am. J. Hum. Genet. 2017; 101:192–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McCarty N.S., Graham A.E., Studena L., Ledesma-Amaro R.. Multiplexed CRISPR technologies for gene editing and transcriptional regulation. Nat. Commun. 2020; 11:1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Montes-Navajas P., Gonzalez-Bejar M., Scaiano J.C., Garcia H.. Cucurbituril complexes cross the cell membrane. Photochem. Photobiol. Sci. 2009; 8:1743–1747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.