Abstract
Human cases of SARS-CoV-2 reinfection have been documented throughout the pandemic, but are likely under-reported. In the current study, we use the Syrian hamster SARS-CoV-2 model to assess reinfection with homologous WA1 and heterologous B.1.1.7 (Alpha) and B.1.351 (Beta) SARS-CoV-2 variants over time. Upon primary infection with SARS-CoV-2 WA1, hamsters rapidly develop a strong and long-lasting humoral immune response. After reinfection with homologous and heterologous SARS-CoV-2 variants, this immune response protects hamsters from clinical disease, virus replication in the lower respiratory tract, and acute lung pathology. However, reinfection leads to SARS-CoV-2 replication in the upper respiratory tract with the potential for virus shedding. Our findings indicate that reinfection results in restricted SARS-CoV-2 replication despite substantial levels of humoral immunity, denoting the potential for transmission through reinfected asymptomatic individuals.
Keywords: SARS-CoV-2, COVID-19, Syrian hamster, natural immunity, reinfection, humoral immunity, variants of concern, VOC, Alpha, B.1.1.7, Beta, B.1.351
Graphical abstract
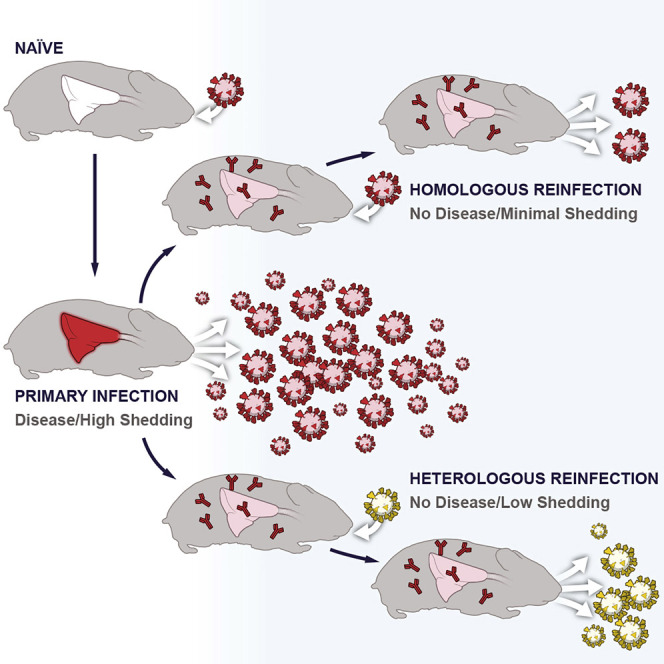
Hansen et al. show that SARS-CoV-2 infection of hamsters results in an acute pulmonary disease and that hamsters develop a strong, durable antibody response with lesions consistent with alveolar bronchiolization. SARS-CoV-2 reinfection results in low levels of viral replication in the upper respiratory tract and protection from acute respiratory disease
Introduction
In late 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19) was first reported from Wuhan, China (Chen et al., 2020; Lu et al., 2020; Wu et al., 2020). After the initial outbreak, SARS-CoV-2 was declared a pandemic on March 11, 2020 (WHO, 2020). More than 440 million cases and 5.9 million deaths have been reported globally to date (WHO, 2021). Cases of SARS-CoV-2 reinfection have been reported from many countries since the beginning of the pandemic; however, whether these cases represent real reinfection events has been difficult to verify (CDC, 2021a; Boyton and Altmann, 2021). An understanding of the dimension of reinfection resulting in viral shedding and the effect of prior immunity on acute disease development are important from a public health management perspective.
The first confirmation of reinfection by genomic sequencing was reported from Hong Kong in August 2020, with the patient experiencing an asymptomatic reinfection with a variant from a different clade 142 days after the initial symptomatic infection (To et al., 2020). Since then, several additional reinfections have been documented experiencing varying degrees of disease severity (Prado-Vivar et al., 2021; Van Elslande et al., 2021; Gupta et al., 2020; Tillett et al., 2021; Zucman et al., 2021), including an immunocompetent 25-year-old Nevada man experiencing severe COVID-19 after reinfection with a distinct 20C clade SARS-CoV-2 variant only 48 days after an initial mild disease (Tillett et al., 2021).
Reinfection with related coronaviruses is not unprecedented and has been clearly demonstrated for three of the four seasonal human coronaviruses: NL63, OC43, and 229E (Poland et al., 2020). Protective immunity gained from primary infection with these seasonal coronaviruses can wane quickly, and reinfection is possible within 6–12 months after the initial infection (Edridge et al., 2020). Rates of reinfection can be considerable, with one Kenyan study finding that, within 6 months of a primary infection, reinfection among study participants occurred at rates of 21%, 5.7%, and 4.0% for NL63, OC43, and 229E, respectively (Kiyuka et al., 2018).
Specific IgG and neutralizing antibodies have been detected in recovered COVID-19 patients for at least 5–8 months after infection, suggesting a prolonged humoral immune response and the potential for protection against reinfection and disease (Wajnberg et al., 2020; Borgonovo et al., 2021; Fotouhi et al., 2021; Chvatal-Medina et al., 2021; Gudbjartsson et al., 2020; Dan et al., 2021; Lumley et al., 2021). However, some studies have suggested that the intensity and longevity of the SARS-CoV-2 humoral responses correlate with disease severity, with IgG and neutralizing antibody titers declining more rapidly (within 1–4 months) (Ibarrondo et al., 2020; Lau et al., 2021; Long et al., 2020; Roltgen et al., 2020; Self et al., 2020).
Studies assessing the potential for reinfection with homologous SARS-CoV-2 variants in several animal models have found that primary infection induced immune responses capable of protecting against disease with evidence of low-level viral replication (Bosco-Lauth et al., 2020; Brustolin et al., 2021; Chandrashekar et al., 2020; Clark et al., 2021; Deng et al., 2020; Imai et al., 2020; Jiang et al., 2020). The relatively short time from a primary infection to reinfection in these studies (≤35 days) is a limitation and long-term studies, in which the durability of the immune response is assessed, are critical to evaluate the longevity of protection from SARS-CoV-2 reinfection. Furthermore, studies assessing the susceptibility to reinfection with heterologous SARS-CoV-2 variants of concern (VOCs) are of high importance given the continuing emergence of multiple VOCs including the Alpha, initially identified in the United Kingdom (B.1.1.7), and Beta variants, initially identified in South Africa (B.1.351). Both VOCs are associated with increases in transmission compared to prototypical variants (CDC, 2021b; Davies et al., 2021, Pearson et al., 2021). Additionally, decreases in neutralizing activity against the B.1.351 variant in vaccinated and recovered individuals has also been observed, making animal studies investigating the reinfection potential of this and other VOCs particularly important (CDC, 2021b; Wang et al., 2021; Wu et al., 2021).
The Syrian hamster is an established SARS-CoV-2 infection model that is associated with mild to moderate respiratory disease with high levels of shedding from the upper respiratory tract and virus replication in the lung (Imai et al., 2020; Rosenke et al., 2020; Sia et al., 2020). In the present study, we used the Syrian hamster model to assess the potential for homologous SARS-CoV-2 reinfection at timepoints up to 5 months after infection while concurrently tracking specific IgG and neutralizing antibody responses of primary infected hamsters. We also investigated the potential for heterologous reinfection in hamsters using the B.1.1.7 Alpha and B.1.351 Beta variants at 7 weeks after infection. Our studies showed that previous SARS-CoV-2 infection protected hamsters reinfected with homologous and heterologous SARS-CoV-2 variants from lung disease, but not from SARS-CoV-2 replication in the upper respiratory tract. This indicates that transmission through reinfected asymptomatic individuals is possible.
Results
Primary infection
Initially, we established primary infection models in the Syrian hamster with the B.1.1.7 (Alpha) and B.1.151 (Beta) VOCs in comparison with the WA1 variant. Groups of six animals were intranasally infected with 1 × 103 at 50% of the tissue culture infective dose (TCID50) of either SARS-CoV-2 isolate WA-CDC-WA1/2020 (designated WA1), SARS-CoV-2_2hCOV_19_England_204820464_2020 isolate (designated B.1.1.7, Alpha), or SARS-CoV-2 isolate nCoV-hCoV-19/USA/MD-HP01542/2021 (designated B.1.351, Beta) as described before (Rosenke et al., 2020). Hamsters were monitored for clinical signs and weight loss over 5 days. While the WA1- and B.1.1.7-infected animals lost between 5.2% and 7.0% of their body weight over those days, the B.1.151-infected hamsters only lost about 1.6% of their weight on average (Figure S1A). Additional signs of disease were rather mild, with hunched posture and ruffled fur observed in animals of all groups.
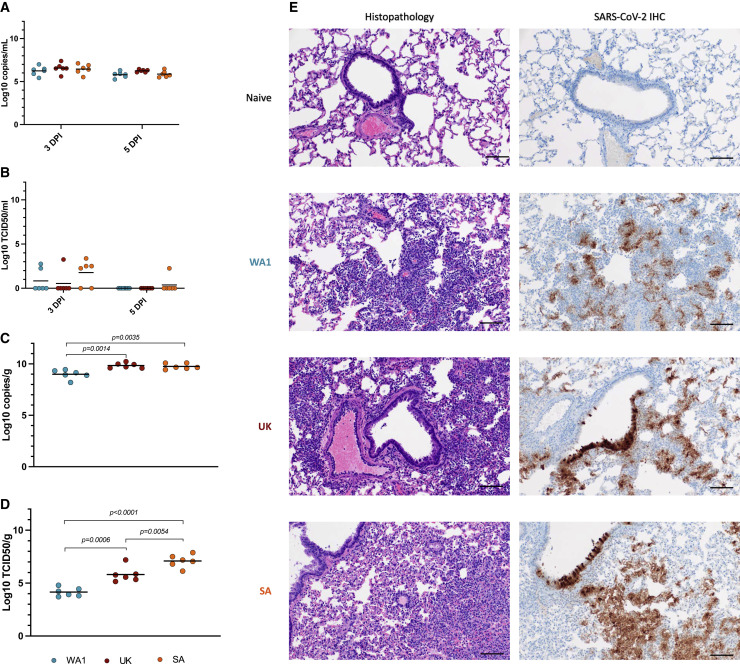
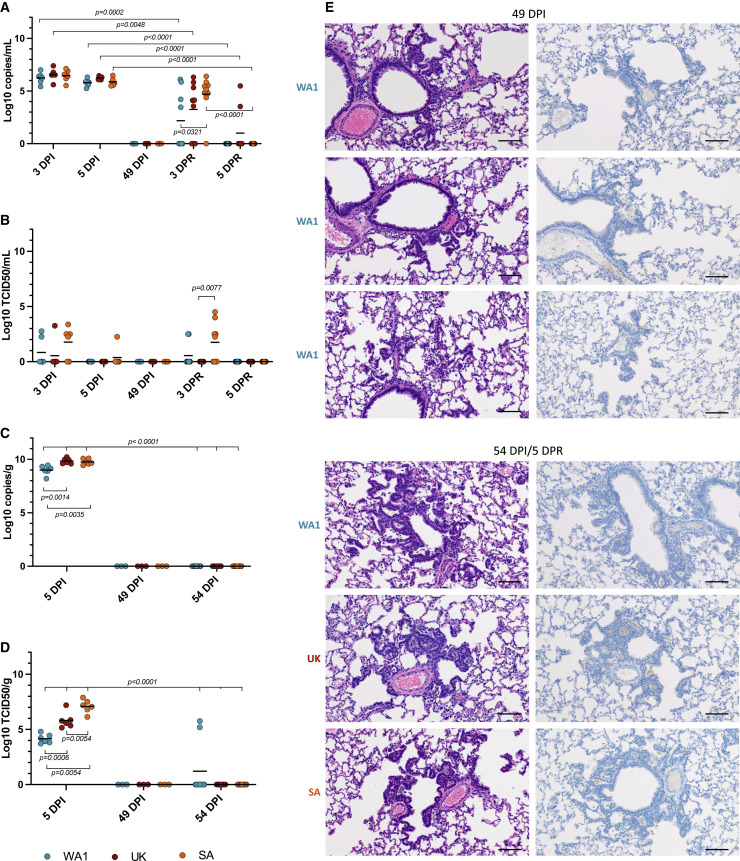
Oral swabs were collected at 3 and 5 days post-infection (DPI) and lung tissue was harvested on 5 DPI. All samples were analyzed for viral RNA and infectious virus using a previously described subgenomic (sg) E quantitative polymerase chain reaction (qPCR) (Corman et al., 2020) and TCID50 assays (Rosenke et al., 2020). Oral swabs showed comparable sgRNA levels among all three variants (Figure 1A). Infectious virus titers in the oral swabs were higher for the B.1.351-infected animals, but the difference was not statistically significant (Figure 1B). Statistically significant differences in lung viral loads, however, were noticed, with WA1-infected animals showing significantly lower sgRNA levels and infectious virus titers (Figures 1C and 1D). Although differences in lung viral loads were found between the B.1.1.7 and B.1.351 VOCs, only the infectious virus titers were statistically significant from one another (Figure 1D). During the acute stage primary infection with the B.1.1.7 and B.1.351 VOCs at 5 DPI, histopathologic lesions consisted of moderate to severe broncho-interstitial pneumonia focused on terminal airways (Figure 1E, left panels) consistent with pulmonary lesions previously described for infections with WA1 (Rosenke et al., 2020). SARS-CoV-2 antigen immunoreactivity was observed primarily in type I pneumocytes, with moderate numbers of pulmonary macrophages, type II pneumocytes, and bronchiolar epithelial cells also exhibiting the SARS-CoV-2 antigen (Figure 1E, right panels). Overall, both VOCs were associated with increased pathology and virus replication in the lower respiratory tract.
Figure 1.
Primary infection with SARS-CoV-2 variants in the Syrian hamster model
Three groups of hamsters (n = 6) were infected with either of three SARS-CoV-2 variants, WA1, B.1.1.7 (Alpha) and B.1.351 (Beta) to compare oral shedding (A, B), viral lung loads (C, D) and lung histopathology (E). A sg E qPCR assay was used to measure oral shedding of viral RNA as well as viral RNA loads in the lung tissues collected at the time of necropsy. A standard TCID50 assay was used to determine levels of infectious virus in both oral swabs and lungs. Hematoxylin and eosin (H&E) and immunohistochemistry (IHC) staining were used to determine histopathology and SARS-CoV-2 antigen distribution in lung tissue.
(A) Viral sgE RNA in oral swabs at 3 DPI and 5 DPI.
(B) Infectious virus in oral swabs at 3DPI and 5DPI.
(C) Viral sgE in lungs at 5DPI.
(D) Infectious virus in lungs at 5DPI.
(E) Histology and SARS-CoV-2 antigen distribution in lung tissue. Histopathologic lesions at 5 DPI consisted of moderate to severe broncho-interstitial pneumonia focused on terminal airways (H&E, left panels, original magnification ×200). SARS-CoV-2 antigen immunoreactivity was observed primarily in type I pneumocytes (IHC, right panels, original magnification ×200) (scale bar, 100 μM). Statistical differences were determined using nonparametric one-way AVOVA (Kruskal-Wallis) with correction for multiple comparisons in PRISM. WA1 infection (blue), B.1.1.7 infection (red), and B.1.351 infection (orange).
Homologous reinfection
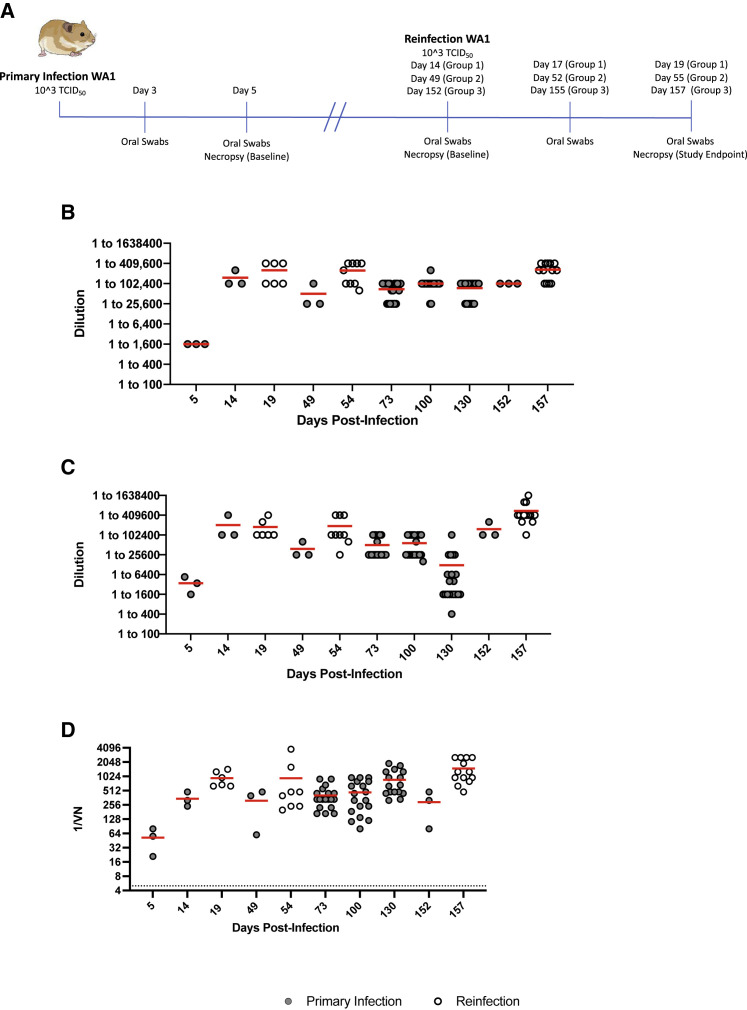
Syrian hamsters (n = 40) were infected intranasally with 200 times the median infectious dose (ID50) (1 × 103 TCID50) of WA1 as determined previously (Rosenke et al., 2020). All infected hamsters were monitored for clinical signs daily and oral swabs were taken at 3 and 5 DPI (peak virus shedding) for virology. Three animals were euthanized at 5 DPI (peak lung pathology) to establish a baseline for virology and pathology of the primary WA1 infection. The remaining infected hamsters (n = 38) were allocated to three groups that were allowed to recover for either 14 (group 1, n = 9), 49 (group 2, n = 12) or 152 (group 3, n = 17) DPI before reinfection with WA1 was performed as described above (Figure 2A). Groups 2 and 3 contained additional animals to compensate for possible attrition owing to age-related mortality. For each group, three hamsters were euthanized on the day of reinfection to establish another baseline for virology and pathology. Following homologous reinfection animals were monitored for clinical signs daily. Oral swabs were taken at 3 and 5 days post reinfection (DPR) (peak virus shedding) for virology and the remaining hamsters in each group were euthanized at 5 DPR (peak lung pathology) to collect lungs for virology and pathology. To track the humoral immune responses over time, serum samples were collected from all hamsters at the time of euthanasia. For group 3, additional serum samples were collected via retro-orbital blood collection at 73, 100, and 130 DPI.
Figure 2.
Design of homologous reinfection study and humoral immune response to infection and reinfection
(A) Homologous reinfection study. Hamsters (n = 40) were infected intranasally with 1 × 103 TCID50 of WA1 SARS-CoV-2 and swabbed at 3 DPI and 5 DPI to monitor shedding and ensure all animals were infected. Three animals were randomly selected and necropsied at 5 DPI to measure disease and infectious titers in the lung. The remaining 37 animals were divided into 3 groups for reinfection at varying timepoints. Groups 1 (n = 9), 2 (n = 12), and 3 (n = 16) were allowed to recover for 14, 49, and 152 DPI after primary infection and were reinfected with WA1 SARS-CoV-2. Three animals from each group were euthanized before reinfection as infection controls for viral replication and pathology (baseline). The humoral response was measured over the course of the study with serum collected at terminal timepoints; additionally serum was collected from group 3 at 73, 100, and 130 DPI. Oral swabs were collected at 3 DPR and 5 DPR to monitor viral shedding. Lung tissue was collected from all control and reinfected hamsters at the time of necropsy (5 DPR) to determine viral titers and lung pathology.
(B) Total IgG antibodies against the spike protein. Total IgG antibodies were determined using an in-house ELISA dilution series assay targeting the S1 region of the SARS-CoV-2 spike protein.
(C) Total IgG antibodies against the RBD. Total IgG antibodies were determined using an in-house ELISA assay targeting the RBD of the SARS-CoV-2 spike protein.
(D) Neutralizing antibodies. Neutralizing antibody titers were determined through a TCID50-based neutralization assay using two-fold serially diluted hamster serum and the homologous SARS-CoV-2 WA1-2020 strain. Solid gray circles are data points from singly infected and hollow white circles are data points generated from reinfected animals. Statistical differences were determined using nonparametric one-way AVOVA (Kruskal-Wallis) with correction for multiple comparisons.
Primary infection induced rapid, robust, and long-lasting antibody responses
Serum samples from group 1–3 animals were evaluated for the presence of neutralizing and total IgG antibodies specific to either the WA1 spike S1 subunit or spike receptor binding domain (RBD) antigens. By 5 DPI, antibody titers were 1:1,600 and between 1:1,600 and 1:6,400 against S1 and spike RBD antigens, respectively (Figures 2B and 2C). These titers correlated with a neutralizing antibody response within the range of 1:20 to 1:80 against infectious WA1 (Figure 2D). Peak IgG and neutralizing antibody responses were already observed at 14 DPI (Figures 2B–2D). High IgG and neutralizing antibody titers were maintained over the entire 152-day period, even in the absence of antigenic boost through reinfection, with no significant decrease being observed between consecutive timepoints (Figures 2B–2D). While a decrease in IgG RBD antibody titers was detected at 130 DPI (group 3), this titer decrease was not statistically significant compared with 100 and 152 DPI (Figure 2D). Although antibody titers increased after homologous reinfection in most groups in all assays, statistically significant boosting effects were not observed (Figure 2D). Together, these data show that SARS-CoV-2 primary infection in hamsters results in rapid, robust, and long-lasting IgG and neutralizing antibody responses with small but insignificant boosting effect after homologous reinfection.
Homologous reinfection was not associated with clinical disease
Hamsters lost weight after primary infection for 6 days with recovery beginning on day 7 and continuing weight gain (Figure S1B). After homologous reinfection on 14 DPI, hamsters in group 1 continued to gain weight over the 5-day period. This weight gain was attributed to the hamsters still being in their juvenile growth phase (Figure S1C). No substantial weight loss was observed for hamsters reinfected with homologous virus at 49 DPI (group 2) or 152 DPI (group 3) (Figure S1C). No signs of clinical disease, as described above for primary infected hamsters, were observed in any hamster after reinfection.
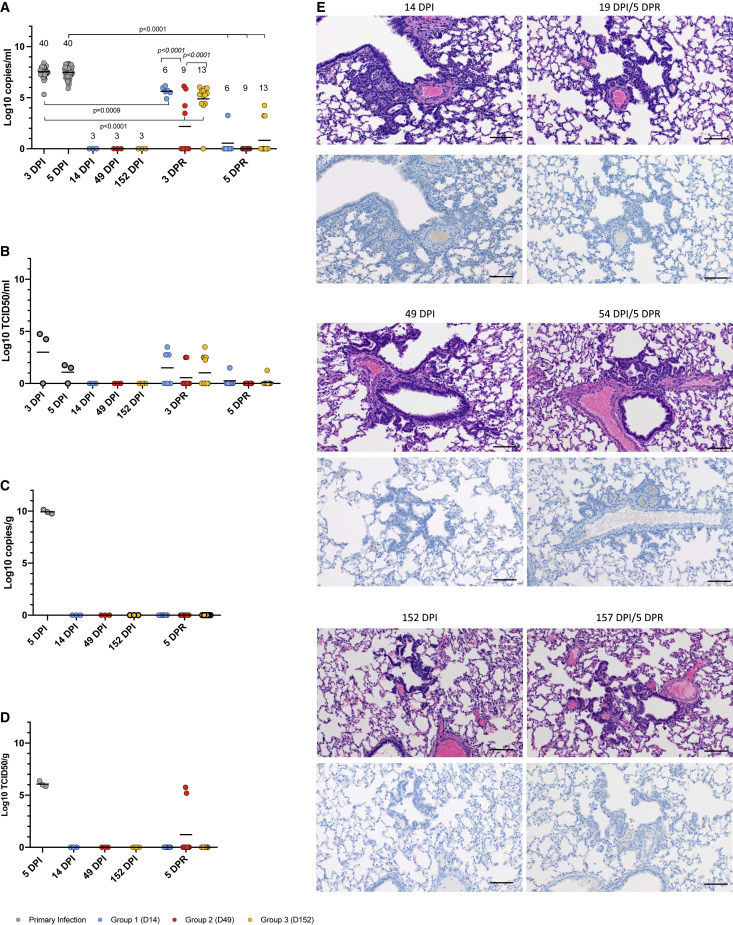
Homologous reinfection resulted in viral replication and shedding
Next, we evaluated oral swab and lung samples for infectious virus and viral RNA. Oral swabs and lung samples from animals after primary infection (3 and 5 DPI) contained high levels of viral sgRNA (Figures 3A and 3C). Infectivity assays revealed high levels of infectious virus in all lungs and low levels of infectious virus in two of three oral swabs (Figures 3B and 3D). Oral swabs and lung tissues were negative for sgRNA and infectious virus before reinfection at 14 (group 1), 49 (group 2), and 152 (group 3) DPI, indicating clearance from primary SARS-CoV-2 infection (Figures 3A–3D). After homologous reinfection, all groups had detectable sgRNA in the oral swabs at 3 DPR (Figure 3A) (group 1: 6 of 6 hamsters (100%), group 2: 4 of 9 hamsters (45%), and group 3: 12 of 13 hamsters (92%). The sgRNA levels in the reinfected hamsters were significantly lower than what was detected after the initial infection (Figure 3A). Infectious virus was isolated from oral swabs at 3 DPR in 3 of 6 animals in group 1 (50%), 2 of 9 animals in group 2 (22%), and 5 of 13 animals in group 3 (38%) (Figure 3B). While notable, these infectious titers were not statistically significant compared with the pre-reinfection baselines. At 5 DPR, oral sgRNA levels decreased for all groups, with the majority of hamsters being negative at this time point (Figure 3A). Infectious titers also decreased at 5 DPR, with only one hamster in groups 1 (16%) and 3 (8%) showing low titers of infectious virus (Figure 3B). Interestingly, no reinfected hamsters had detectable sgRNA in the lung samples at 5 DPR (Figure 3C). However, two of the nine (22%) hamsters from group 2 had infectious virus in the lungs, a result that was not statistically different from the pre-reinfection baselines (Figure 3D). Overall, homologous reinfection resulted in transient, short-lived, low-level SARS-CoV-2 replication in the upper respiratory tract with negligible spread to the lungs.
Figure 3.
Viral replication and pulmonary pathology after homologous reinfection
A sgE qPCR assay was used to detect viral RNA loads in oral swabs and lung tissue (A, C). A standard TCID50 assay was used to determine levels of infectious virus in oral swabs and lung tissues (B, D). Hematoxylin and eosin (H&E) and immunohistochemistry (IHC) staining were used to assess histopathology and SARS-CoV-2 antigen distribution in lung tissue (E).
(A) Viral RNA in oral swabs.
(B) Infectious virus in oral swabs.
(C) Viral RNA levels in lung samples.
(D) Infectious virus in lung samples.
(E) Histology and SARS-CoV-2 antigen distribution in lung tissue. H&E (original magnification ×200) and IHC (SARS-CoV-2 N protein, original magnification ×200) staining were performed immediately before (left panels; 14, 49, and 152 DPI) and 5 days after homologous reinfection (right panels; 19, 54, and 157 DPI/5DPR). H&E (upper panels) showed resolving interstitial pneumonia developing into alveolar bronchiolization. Reinfection failed to induce histopathology consistent with acute SARS-CoV-2 infection (H&E, original magnification ×200 at 19, 54, and 157 DPR/5 DPR). IHC (lower panels) showed that resolving interstitial pneumonia and alveolar bronchiolization were not associated with SARS-CoV-2 immunoreactivity. Reinfection failed to result in SARS-CoV-2-specific immunoreactivity (IHC, original magnification ×200 at 19, 54, and 157 DPI/5 DPR) (scale bar, 100 μM). A nonparametric one-way AVOVA (Kruskal-Wallis) with correction for multiple comparisons was used to determine statistical differences in PRISM. Primary infection (gray), group 1 (D14, blue), group 2 (D49, red), and group 3 (D152, orange).
Homologous reinfection did not cause acute lung pathology
Pulmonary pathology and immunohistochemistry were assessed in animals euthanized before reinfection at 14, 49, and 152 DPI (baselines) and after reinfection at 5 DPR (19, 54, and 157 DPI). Resolution of broncho-interstitial pneumonia, including terminal airway changes previously described as type II pneumocyte hyperplasia associated with resolution of disease (Rosenke et al., 2020), was observed in all animals that had been euthanized before reinfection (14, 49, and 152 DPI) (Figure 3E, left panels). Owing to the persistence of this lesion from 14 through 152 DPI as well as the presence of ciliated epithelia (Figure S2) and cells morphologically consistent with Clara cells, these foci were determined to be consistent with alveolar bronchiolization (Nettesheim and Szakal, 1972) and not the previously described type II pneumocyte hyperplasia. At 5 DPR (19, 54, and 157 DPI), there were no acute pulmonary lesions in any of the hamsters (Figure 3E, hematoxylin and eosin staining, upper panels). All reinfected animals exhibited alveolar bronchiolization with subjectively and mildly larger foci of alveolar bronchiolization noted at 54 and 157 DPI. The SARS-CoV-2 antigen was clearly observed in bronchiolar epithelium, type I and type II pneumocytes, and pulmonary macrophages during acute stage infection (5 DPI) (Figure 1E, right panel), while no evidence of viral antigen was detected in any of the groups before and after reinfection (Figure 3E, immunohistochemistry, bottom panels). Acute pulmonary damage was associated with primary SARS-CoV-2 infection and, unexpectedly, pulmonary pathology was persistent and substantial for at least 5 months into recovery. However, acute pulmonary disease consistent with primary viral interstitial pneumonia was not observed in animals reinfected with the homologous SARS-CoV-2 WA1 variant.
Heterologous reinfection
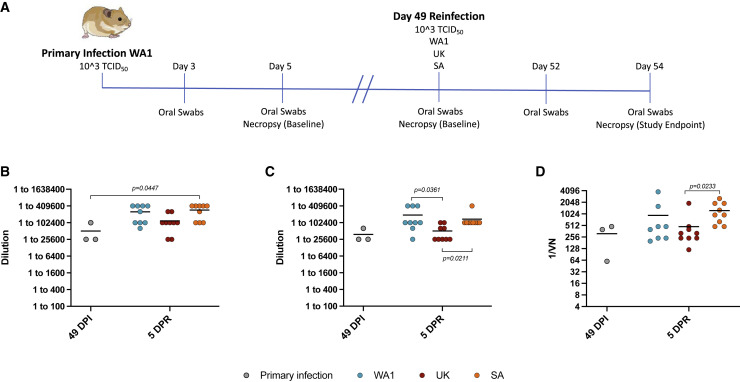
Syrian hamsters (n = 21) were intranasally infected with the WA1 SARS-CoV-2 isolate as described in the homologous reinfection study. One group (n = 9) of animals were intranasally reinfected at 49 DPI with the B.1.1.7 and the other group (n = 9) with the B.1.351 SARS-CoV-2 VOC at the same dose as for the primary WA1 infection (Figure 4A). To save animals, group 2 hamsters (n = 9) from the homologous reinfection study also served as homologous reinfection controls for the heterologous reinfection study. The monitoring of animals for disease signs, oral swab sampling (3 and 5 DPI; 3 and 5 DPR), blood sampling, and euthanasia (5 DPI, before reinfection at 49 DPI, and 5 DPR) with harvesting of lungs were performed as outlined in Figure 4A and the homologous reinfection study above (Figure 2A).
Figure 4.
Design of heterologous reinfection study and humoral immune response to infection and reinfection
(A) Heterologous reinfection study. Hamsters (n = 21) were infected intranasally with 1 × 103 TCID50 of WA1 SARS-CoV-2 and swabbed 3 DPI and 5 DPI to monitor shedding and ensure animals were infected. Three animals were randomly selected and necropsied at 5 DPI to measure disease and infectious titers in the lung. The 18 remaining animals were separated into 2 groups of 9 animals. Three animals were randomly selected from both groups and necropsied before reinfection at 49 DPI to measure lung titers and pathology (baseline). The remaining animals were reinfected with the 1 × 103 TCID50 of either the WA1, B.1.1.7 (Alpha), or B.1.351 (Beta) SARS-CoV-2 variants. Reinfected hamsters were swabbed at 3 DPR and 5 DPR to monitor shedding and necropsied at 5 DPR to determine lung viral load and pathology. To save animals, group 2 hamsters (n = 9) from the homologous reinfection study (Figures 2 and 3) also served as homologous reinfection controls for the heterologous reinfection study. Blood samples were taken at the time of necropsy (49 DPI, baseline and 54 DPI/5 DPR, reinfection).
(B) IgG antibody titers were assessed using an in-house ELISA against the WA1 spike protein.
(C) IgG antibody titers were determined using an in-house ELISA assay specific to the RBD of the WA1 spike protein.
(D) WA1-neutralizing antibody titers were determined through a dilution series using serum from either the WA1, B.1.1.7, or B.1.351 SARS-CoV-2 infected hamsters. Primary infection (gray), WA1 (blue), B.1.1.7 (red), and B.1.351 (orange). Statistical analyses were performed using nonparametric one-way AVOVA (Kruskal-Wallis) with correction for multiple comparisons in Prism.
Heterologous reinfection resulted in boosted antibody responses
Serum samples collected at 49 DPI and 5 days after reinfection (5 DPR) were used to determine IgG antibodies specific to WA1 SARS-CoV-2 spike and RBD (Figures 4B and 4C). IgG spike and RBD antibody titers were similar to what was detected in the homologous reinfection study (Figures 2B, 2C, 4B, and 4C). IgG spike and RBD antibodies were also boosted by reinfection with each variant at 5 DPR; however, this effect was only statistically significant for the B.1.351 reinfected hamsters when compared to the pre-reinfection baselines (49 DPI) (Figure 4B). The boosting effect was comparable for the animals reinfected with the WA1 and B.1.351 variants and, although the IgG spike antibody titers of animals in the B.1.1.7 reinfected group were somewhat lessoned, a boosting effect was detected as well (Figures 4B and 4C). Neutralizing activity against the WA1 was also compared at 49 DPI with the control hamsters showing equivalent neutralizing antibody titers at levels comparable with what were seen in the homologous reinfection study (Figures 2D and 4D). The neutralizing antibodies were boosted after reinfection with all variants at day 49 (5 DPR), but these increases were only significant in the animals reinfected with the B.1.351 (Figure 4D).
Heterologous reinfection was not associated with clinical disease
Weight loss in reinfected animals never exceeded 1.4% with any of the three variants (Figure S1D). While signs of clinical disease were observed in primary infected animals as described above, these signs were not apparent in any of the reinfected hamsters. These results support that primary infection with WA1 SARS-CoV-2 protected against overt clinical disease after heterologous reinfection with the B.1.1.7 and B.1.351 VOCs.
Heterologous reinfection resulted in increased virus replication and shedding
The pre-reinfection baselines (n = 3) at 49 DPI had no detectable sgRNA or infectious virus, either in the oral swabs or the lung tissues (Figures 5A and 5C). In oral swabs evaluated at 3 DPR (52 DPI), all variant groups showed evidence of viral replication in the sgE qPCR assay with four of nine (44%), six of nine (66%), and eight of nine (89%) hamsters for the WA1, B.1.1.7, and B.1.351 variant groups, respectively (Figure 5A). sgRNA was detected in all groups after reinfection, but the levels were significantly lower after reinfection (Figure 5A). In addition, the B.1.351 reinfected hamsters showed significantly higher sgRNA levels compared with the WA1, but not the B.1.1.7, reinfected animals. On average, the B.1.1.7 reinfected hamsters had higher sgRNA levels than WA1 reinfected hamsters at this time point, although this difference was not significant (Figure 5A).
Figure 5.
Viral replication and pulmonary pathology after heterologous reinfection
A sgE qPCR assay was used to detect viral RNA loads in oral swabs and lung tissue (A, C). A standard TCID50 assay was used to determine levels of infectious virus in oral swabs and lung tissues (B, D). Hematoxylin and eosin (H&E) and immunohistochemistry (IHC) staining were used to assess histopathology and SARS-CoV-2 antigen distribution in lung tissue (E).
(A) Viral RNA in oral swabs.
(B) Infectious virus in oral swabs.
(C) Viral RNA levels in lung samples.
(D) Infectious virus in lung samples.
(E) Histology and SARS-CoV-2 antigen distribution in lung tissue. H&E (original magnification ×200) and IHC (SARS-CoV-2 N protein, original magnification ×200) staining were performed immediately before (49 DPI) and 5 days after heterologous reinfection (54 DPI/5DPR). H&E (left panels) showed resolving interstitial pneumonia developing into alveolar bronchiolization. Reinfection failed to induce histopathology consistent with acute SARS-CoV-2 infection (H&E, 200 × 54 DPI/5 DPR. IHC (right panels) showed that resolving interstitial pneumonia and alveolar bronchiolization were not associated with SARS-CoV-2 immunoreactivity. Reinfection failed to result in SARS-CoV-2 specific immunoreactivity (IHC, original magnification ×200 at 54 DPI/5 DPR), (scale bar 100 μM). A nonparametric one-way AVOVA (Kruskal-Wallis) with correction for multiple comparisons was used to determine statistical differences in PRISM. WA1 reinfection (blue), B.1.1.7 reinfection (red), and B.1.351 reinfection (orange).
Higher levels of infectious virus were isolated from the 3 DPR (52 DPI) oral swabs in the majority of B.1.351 reinfected hamsters (5 of 9, 56%) compared with the WA1 group (2 of 9; 22%) and the B.1.1.7 groups (none; 0%) (Figure 5B), but these differences were not significant. At 5 DPR (54 DPI), the majority of animals had cleared the reinfection in the upper respiratory tract. At the time of necropsy (5 DPR), no hamster in any reinfection group had infectious virus present in oral swabs (Figure 5B). No sgRNA was detected in the lungs for any of the three variants at 5 DPR (54 DPI), while infectious virus was isolated in two of nine (22%) animals for the WA1 variant (Figures 5C and 5D). These results are consistent with the homologous reinfection study showing that virus replication occurred in the upper respiratory tract, but was nearly absent in the lower respiratory tract upon homologous and heterologous reinfection (Figures 3C, 3D, 5C, and 5D).
Heterologous reinfection did not cause acute lung pathology
Pulmonary pathology and immunohistochemistry were assessed in animals before (49 DPI) and after heterologous reinfection (5 DPR [54 DPI]). Alveolar bronchiolization was observed in all animals before and after reinfection; however, no evidence of acute pathology consistent with primary viral infection was observed in any reinfected hamster (Figure 5). Immunohistochemistry revealed no SARS-CoV-2 antigen detection in lung tissues from animals euthanized before (49 DPI) or after heterologous reinfection (5 DPR [54 DPI]) (Figure 5E). Thus, acute pulmonary lesions were only associated with primary SARS-CoV-2 infection with persistent pulmonary pathology observed for a prolonged period (49 DPI). Importantly, acute pulmonary pathology was not observed after reinfection with either the B.1.1.7 or B.1.351 VOC.
Discussion
Reinfection with seasonal coronaviruses has been observed in the past with cases occurring frequently within 6–12 months upon primary infection (Kiyuka et al., 2018; Edridge et al., 2020). Similarly, reinfections with SARS-CoV-2 have been described with case numbers likely being vastly under-reported (Gupta et al., 2020; Prado-Vivar et al., 2021; Tillett et al., 2021; To et al., 2020; Van Elslande et al., 2021; Zucman et al., 2021). Cases may be missed for a variety of reasons, including a general lack of testing and asymptomatic primary or secondary infections, as well as insufficient investigation and reporting of potential reinfections by overwhelmed public health agencies. Understanding the dynamics of SARS-CoV-2 reinfection will become ever more relevant as more people with previous natural or vaccine-stimulated immunity to SARS-CoV-2 may become susceptible again owing to waning immunity and the emergence of new variants that partially evade the immune response (CDC, 2021b; Wang et al., 2021; Wu et al., 2021).
The homologous reinfection study was designed to investigate the potential for reinfection of Syrian hamsters for up to 5 months while simultaneously tracking humoral immune responses. This was different to previous animal studies that have investigated SARS-CoV-2 reinfection after about 1 month (Bosco-Lauth et al., 2020; Brustolin et al., 2021; Chandrashekar et al., 2020; Deng et al., 2020; Imai et al., 2020; Jiang et al., 2020). In our study SARS-CoV-2 spike-specific IgG and neutralizing antibodies developed rapidly (within 5 days), leveled off quickly (around 14 days) and were maintained at similar levels over the entire study period (about 5 months). This rapid, strong, and long-lasting immune response protected reinfected hamsters from SARS-CoV-2-induced clinical disease and acute pulmonary pathology even after 5 months past the primary infection. Homologous reinfection at short- and long-term timepoints resulted in temporary viral replication, which seemed largely restricted to the upper respiratory tract. Localized virus replication was rapidly cleared, which might be explained by the strong humoral immune response that even got boosted after reinfection. Particularly high levels of neutralizing antibodies may quickly neutralize infectious virus, which may explain why virus was difficult to isolate. Nevertheless, infectious SARS-CoV-2 was isolated from the oral mucosa, suggesting it could be shed from the upper respiratory tract with the consequence of transmission after reinfection.
The heterologous reinfection study was designed to determine the reinfection potential with distinct variants, especially VOCs of high public health concern. First, the acute infection models were established with each variant to determine any potential differences in viral replication or disease in the hamster. Although infection with the VOCs did not change disease severity, we did observe significant increases in both viral RNA and infectious virus in the lungs of the animals infected with either the B.1.1.7 (Alpha) and the B.1.351 (Beta) variants when compared with the WA1 group. After establishing the infection model, we performed the heterologous challenge; to our knowledge, no study has thus far experimentally addressed this issue. We tested reinfection with the B.1.1.7 (Alpha) and B.1.351 (Beta) VOCs after a primary infection with the WA1 variant. In humans, both VOCs are associated with increased transmissibility and possibly pathogenicity (Pearson et al., 2021, Davies et al., 2021; CDC, 2021b). Naive Syrian hamsters were equally susceptible to both VOCs with similar or increased virus replication in the upper and lower respiratory tracts when compared with the WA1 variant. As with the homologous reinfection study, hamsters were protected from clinical disease and acute pulmonary lesions after reinfection with both VOCs. Overall, reinfection with the two VOCs resulted in temporary localized SARS-CoV-2 replication in the upper respiratory tract, but in not the lower respiratory tract, indicating a potential for shedding and thus transmission.
While infectious virus was detectable from the upper respiratory tract in both homologous and heterologous rechallenge, reinfection failed to induce acute pulmonary pathology at any evaluated time point. Interestingly, alveolar bronchiolization, a chronic pulmonary histopathologic change that can result in decreased lung function, was observed after primary SARS-CoV-2 infection of hamsters, even up to 5 months, indicating long-lasting pulmonary lesions after SARS-CoV-2 infection. It is unknown if the extent and severity of alveolar bronchiolization observed in the Syrian hamster model appropriately reflects chronic pulmonary pathology in humans, or if the lesions observed in this study are an under-representation of chronic pulmonary pathology associated with SARS-CoV-2 infection. Foci of alveolar bronchiolization seemed larger at 49 and 152 DPI relative to 14 DPI, suggesting this lesion may be even progressive in the Syrian hamster model. Importantly, it is unknown if full lesion resolution occurs in this species after SARS-CoV-2 infection. While this lesion may be subtle, it may still be associated with functional disturbances, such as activity intolerance or decreased inspiratory volume in the hamster model. So far, alveolar bronchiolization has not been documented in any SARS-CoV-2 animal model and merits further investigation.
Alveolar bronchiolization was first described after the 1957 influenza pandemic (Hers et al., 1958), and was later recapitulated in animal models of influenza infection in 1975 by Loosli et al. (1975). Interestingly, alveolar bronchiolization in the murine model of influenza infection does not resolve with time (Loosli et al., 1975). More recently, alveolar bronchiolization has been demonstrated in up to 39% of human patients diagnosed with diffuse alveolar damage (Taylor et al., 2018). Taken together, these data suggest that infectious causes of moderate to severe distal bronchiolar and alveolar disease may lead to chronic, irreparable changes in the lower respiratory tree in both humans and animal models. Moreover, as the wild-type hamster model of SARS-CoV-2 infection does not meet the clinical definition of acute respiratory distress syndrome with only mild clinical signs observed, the extent of lesions observed histologically in this model may under-represent the extent of disease in moderate-to-severe SARS-CoV-2 infection in humans.
Although significantly lower after reinfection, the presence of infectious virus in the upper respiratory tract suggests that infectious viral shedding and, therefore, transmission may occur for a limited but still undefined period, even though no respiratory disease developed. The potential for virus shedding and transmission of a reinfected host is highly relevant for epidemiologic risk assessments of SARS-CoV-2 spread after recovery from natural infection, as well as after vaccination. Viral shedding from reinfected individuals that are asymptomatic (or mildly symptomatic) could pose a serious public health concern to the unvaccinated or immunocompromised populations.
Syrian hamsters have a robust IgG and neutralizing antibody responses after SARS-CoV-2 infection, measuring magnitudes higher than what has been observed in humans (Wajnberg et al., 2020). The observed SARS-CoV-2 replication in the upper respiratory tract despite the circulation of high levels of neutralizing antibodies further illustrates the potential for transmission in this model. As COVID-19 patients have been shown to mount a much lower humoral response after SARS-CoV-2 infection (Chvatal-Medina et al., 2021; Dan et al., 2021; Gudbjartsson et al., 2020; Ibarrondo et al., 2020; Lau et al., 2021; Long et al., 2020; Roltgen et al., 2020; Self et al., 2020; Lumley et al., 2021), the potential for reinfection may be even higher in humans. This risk may be especially high for individuals who experienced only mild COVID-19, as the strength and longevity of the humoral immune response may correlate with disease severity (Ibarrondo et al., 2020; Lau et al., 2021; Long et al., 2020; Roltgen et al., 2020; Self et al., 2020), or are exposed to particular variants such as B.1.1.7 or B.1.351 VOCs that may escape certain neutralizing antibody responses generated during infection of early SARS-CoV-2 variants.
In summary, our data indicate that hamsters recovering from a primary SARS-CoV-2 infection are susceptible to homologous and heterologous SARS-CoV-2 reinfection, which seems to be localized to the upper respiratory tract and not resulting in disease and acute pulmonary pathology. Despite being restricted and short lived, virus replication may result in upper respiratory tract shedding and thus transmission. As more people recover from COVID-19 worldwide, understanding the natural immunity that SARS-CoV-2 infection confers in the short and long term will become ever more crucial. Additional longitudinal human and animal studies are needed to fully achieve this understanding.
Limitations of the study
The study design was challenging and owing to the emergence of VOCs has been adapted to include heterologous reinfection. Given the complex setup, the study has multiple limitations, of which a few are discussed below. First, study group sizes varied to decrease the number of animals required to remain practical with animal work in biocontainment. Knowing this limitation, we have carefully arranged for the different groups to gain maximum outcome with statistically significant results. Second, the dose of primary and re-infection have been chosen based on previous experience with the WA1 variant (Rosenke et al., 2020). We chose between 100 and 1,000 ID50 which is a common challenge dose. Based on our experience with the WA1 variant, we think a lower or even higher dose would not considerably alter the outcome, but can only speculate until performing such experiments. Third, owing to the fast emergence of naturally occurring VOCs, we are unable to address the current situation driven by the delta and omicron variants, as those variants had not emerged at the time of the experiment. Future studies will have to include those latest variants. Fourth, the immunology is somewhat limited by the lack of reagents for the hamster model, so the study focused on IgG antibodies as the key measure for protection against SARS-CoV-2. Further studies will address and expand on the immune response generated after reinfection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-NP1 | Genscript | U864YFA140-4/CB2093 |
| anti-rabbit IgG | Vector Laboratories | Cat #MP-6401 |
| Goat anti-hamster IgG HRP | KPL | Cat #PA1-29626 |
| Bacterial and virus strains | ||
| SARS-CoV-2 isolate SARS-CoV-2/human/USA/WA-CDC-WA1/2020 (WA1) | CDC Atlanta | Genbank Accession MN985325.1 |
| SARS-CoV-2 isolate nCoV-hCoV-19/USA/MD-HP01542/2021 (B.1.351; SA; beta) | Andy Pekosz (Johns Hopkins) | GISAID # EPI_ISL_890360 |
| SARS-CoV-2_2hCOV_19_England_204820464_ 2020 isolate (B.1.1.7; UK; alpha) | BEI Resources | GISAID # EPI_ISL_683466 |
| Critical commercial assays | ||
| Rotor-Gene probe kit | Qiagen | Cat #204374 |
| QiaAmp Viral RNA kit | Qiagen | Cat #52906 |
| RNeasy kit | Qiagen | Cat #74004 |
| ChromoMap DAB kit | Roche Tissue Diagnostics | Cat#760-159 |
| Experimental models: Cell lines | ||
| Vero E6 | Ralph Baric, UNC | N/A |
| Experimental models: Organisms/strains | ||
| Syrian hamster HsdHan:AURA | Envigo | N/A |
| Oligonucleotides | ||
| Subgenomic E primer forward 5’-ACAGGTACGTT AATAGTTAATAGCGT |
IDT | N/A |
| Subgenomic E primer reverse 5’-ATATTGCAGCA GTACGCACACA |
IDT | N/A |
| Subgenomic E probe 5’-FAM-ACACTAGCCATCC TTACTGCGCTTCG-BBQ |
IDT | N/A |
| Software and algorithms | ||
| Prism 9 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Heinz Feldmann (feldmannh@niaid.nih.gov).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animal model and sample collection
To establish the primary infection model with the B.1.1.7 and B.1.351 variants for comparison with the WA1 variant, groups of six animals (3 males, 3 females; 8 weeks of age) were intranasally infected with 1 × 103TCID50 with one of three SARS-CoV-2 variants; WA1, UK (B.1.1.7), or SA (B.1.351) as (described previously) (Rosenke et al., 2020). Briefly, hamsters were anesthetized through inhalation of vaporized isoflurane and inoculated via intranasal installation of 25 μL inoculum dropped into each naris by pipette. After infection, hamsters were weighed and monitored for signs of disease daily. Oral swabs were collected at days 3 and 5 DPI to monitor virus shedding. For this, hamsters were anesthetized, the oral cavity was swabbed using polyester flock tipped swabs (Puritan Medical Products), the swabs were immediately immersed in 1 mL of sterile tissue culture media and vortexed. At 5 DPI all hamsters were euthanized, and the lung tissue was collected for virological and histological comparisons of acute lung disease.
For the homologous reinfection study, 40 wild-type Syrian hamsters (20 males, 20 females; 8 weeks of age) were anesthetized and infected with 103 TCID50 (200 ID50) (Rosenke et al., 2020) of WA1 as described above. After infection, hamsters were weighed and monitored for signs of disease daily. Oral swabs were collected on 3 and 5 DPI as described above to monitor virus shedding. Hamsters were reinfected at 14 DPI (group 1), 49 DPI (group 2), or 152 DPI (group 3) with the same dose, strain, and route as described before. Oral swabs were collected on 3 and 5 DPR as described above to monitor virus shedding. At 5 DPR, reinfected hamsters were anesthetized, and blood was collected via cardiac puncture for serology. Animals were immediately euthanized, and lung tissue was harvested for virology and histopathology. In addition, blood was collected by peri-orbital bleeding at 73, 100, and 130 DPI from group 3 hamsters for serology.
For the heterologous reinfection study, 21 Syrian hamsters were intranasally infected with the same dose of WA1 as described above for the homologous reinfection study. Following infection, hamsters were weighed and monitored for signs of disease daily. Oral swabs were collected on 3 and 5 DPI as described above to monitor virus shedding. Subsequent reinfections were performed using either the same variant (WA 1), the B.1.1.7 (alpha), or the B.1.351 (beta) variants. The WA1-reinfected hamsters are the same animals as in the group 2 of the homologous reinfection. Oral swabs were collected on 3 and 5 DPR as described above to monitor virus shedding. At 5 DPR, reinfected hamsters were anesthetized, and blood was collected via cardiac puncture for serology. Animals were immediately euthanized, and lung tissue was harvested for virology and histopathology.
Virus and cells
SARS-CoV-2 isolate SARS-CoV-2/human/USA/WA-CDC-WA1/2020 (WA1) was kindly provided as passage 3 by the Centers for Disease Control and Prevention (Harcourt et al., 2020) and propagated one more time at Rocky Mountain Laboratories (RML) in Vero E6 cells. The virus stock used was free of contaminations and was sequenced confirmed to be identical to the initial deposited GenBank sequence (MN985325.1). SARS-CoV-2 isolate nCoV-hCoV-19/USA/MD-HP01542/2021 (B.1.351; SA; beta) was provided by Andy Pekosz (Johns Hopkins) and this passage 2 stock was propagated one more time at RML in Vero E6 cells. The virus stock was sequence confirmed and found to have amino acid changes at NSP5 (P252L: 17%) and NSP6 (L257F: 57%) when aligned to the GISAID sequence (GISAID: EPI_ISL_890360). The SARS-CoV-2_2hCOV_19_England_204820464_2020 isolate (B.1.1.7; UK; alpha) was provided by BEI Resources at passage 2 and was propagated at RML one more time in Vero E6 cells. The stock was sequence confirmed and had amino acid changes at ORF1AB (D3725G: 13%) and ORF1AB (L3826F: 18%) when aligned to the GISAID sequence (GISAID: EPI_ISL_683466). Virus propagation was performed in DMEM (Sigma) supplemented with 2% fetal bovine serum (Gibco), 1 mM L-glutamine (Gibco), 50 U/mL penicillin and 50 μg/mL streptomycin (Gibco). Vero E6 cells, kindly provided by R. Baric (University of North Carolina) were maintained in DMEM (Sigma) supplemented with 10% fetal calf serum, 1 mM L-glutamine, 50 U/mL penicillin, and 50 μg/mL streptomycin.
Biosafety and ethics
Work with infectious SARS-CoV-2 was approved by the Institutional Biosafety Committee (IBC) and performed in the high biocontainment facilities at RML, National Institute of Allergy and Infectious Diseases (NIAID), an the National Institutes of Heath (NIH). Sample removal from high biocontainment followed IBC-approved Standard Operating Protocols (Haddock et al., 2021). Animal work was approved by the Institutional Animal Care and Use Committee and performed by certified staff in an Association for Assessment and Accreditation of Laboratory Animal Care International accredited facility. Work followed the institution's guidelines for animal use, the guidelines and basic principles in the NIH Guide for the Care and Use of Laboratory Animals, the Animal Welfare Act, the United States Department of Agriculture, and the United States Public Health Service Policy on Humane Care and Use of Laboratory Animals. Syrian hamsters were group housed in HEPA-filtered cage systems enriched with nesting material and were provided with commercial chow and water ad libitum. Animals were monitored at least twice daily.
Method details
Virus titration assay
Virus endpoint titrations were performed in Vero E6 cells as previously described (Rosenke et al., 2020). Briefly, tissue was homogenized in 1 mL DMEM using a TissueLyzer (Qiagen) and clarified from cell debris by low-speed centrifugation. Cells were inoculated with 10-fold serial dilutions of homogenized lung samples or oral swabs in 100 μL DMEM (Sigma-Aldrich) supplemented with 2% fetal bovine serum, 1 mM L-glutamine, 50 U/mL penicillin, and 50 μg/mL streptomycin. Cells were incubated for 6 days and then scored for cytopathogenic effects (CPE) and the TCID50 was calculated using the Reed-Muench formula.
Viral genome detection
A qPCR was performed on RNA extracted from swabs or tissues (≤30 mg) using QiaAmp Viral RNA kit or RNeasy kit (Qiagen), respectively. A one-step real-time reverse transcriptase PCR assay was used to amplify a portion of the E gene to detect sgRNA (Corman et al., 2020). Dilutions of RNA standards counted by droplet digital PCR were run in parallel and used to calculate viral RNA genome copies. The Rotor-Gene probe kit (Qiagen) was used to run the PCRs according to the instructions of the manufacturer.
Histopathology
Tissues were fixed in 10% neutral buffered formalin for a minimum of 7 days with two changes of formalin. Tissues were placed in cassettes and processed with a Sakura VIP-6 Tissue Tek, on a 12-h automated schedule, using a graded series of ethanol, xylene, and ParaPlast Extra. Embedded tissues are sectioned at 5 μm and dried overnight at 42°C before staining.
Specific anti-CoV immunoreactivity was detected using GenScript U864YFA140-4/CB2093 NP-1 at a 1:1,000 dilution. The secondary antibody is the Vector Laboratories ImPress VR anti-rabbit IgG polymer (cat# MP-6401). The tissues were then processed for immunohistochemistry using the Discovery Ultra automated processor (Ventana Medical Systems) with a ChromoMap DAB kit (Roche Tissue Diagnostics cat#760-159).
ELISAs
Plates were coated with spike 1 or spike RBD antigen (The Native Antigen Company; 50 ng/well) in PBS for overnight adsorption at 4°C. Plates were washed in PBS/Tween (0.05) and wells blocked using 5% powdered milk in TBS/Tween (0.05%) for 1 h at RT. Hamster serum samples were added at an initial 1:100 dilution followed by 1:4 dilutions up to 1:409,600 or 1:1,638,400 in duplicate and incubated 1 h at room temperature. Plates were washed and anti-IgG goat anti-hamster HRP labelled secondary antibody (http://www.kpl.com) at 1:2,000 dilution was added to all wells for 1hr at RT. After washing ABTS substrate (seracare) was added for 15 min before a 5% SDS solution was added to stop the reaction. Optical density values for each well were measured at 405 nm. Endpoint antibody titers were based on the last positive dilution of any particular dilution series. Positives were counted as any value above the average + 3 times the standard deviation of the negative serum controls at each dilution.
Virus neutralization assay
Hamster sera were heat-inactivated (30 min, 56°C), serial diluted two-fold (prepared in 2% DMEM) and mixed with 100 TCID50 of SARS-CoV-2 WA1, B.1.1.7, or B.1.351. After 1 h incubation at 37°C and 5% CO2, virus:serum mixture was added to VeroE6 cells and incubated at 37°C and 5% CO2. At 6 DPI, the CPE was scored. The virus neutralization titer was expressed as the reciprocal value of the highest dilution of the serum which still inhibited virus replication.
Quantification and statistical analysis
Statistical analysis was performed in GraphPad PRISM version 8.2.0. Statistical differences were assessed using a nonparametric one-way AVOVA (Kruskal-Wallis) with correction for multiple comparisons to analyze viral RNA, infectious virus, and weight change data, IgG, and neutralizing antibody data.
Acknowledgments
The authors thank the animal caretakers, histopathology group, and veterinarians of the Rocky Mountain Veterinary Branch (National Institute of Allergy and Infectious Diseases [NIAID], National Institutes of Health [NIH]) for their support with animal related work. The authors acknowledge Rose Perry and Ryan Kissinger (Visual Arts, NIAID, NIH) for help with the display items and graphical abstract, respectively, and the Research Technologies Branch (NIAID, NIH) for the sequencing of stock viruses. We are grateful to Emmie de Wit and Vincent Munster for their discussions and help with virus stock preparations. The B.1.1.7 variant was obtained through BEI Resources (Bassam Hallis, Sujatha Rashid) and NIAID (Ranjan Mukul, Kimberly Stemple). The B.1.351 was obtained through Andy Pekosz (Johns Hopkins) and NIAID. This study was funded by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID) at the National Institutes of Health (NIH). The opinions, conclusions and recommendations in this report are those of the authors and do not necessarily represent the official positions of the National Institute of Allergy and Infectious Diseases at the National Institutes of Health.
Author contributions
Conceptualization, F.H., K.M.W., M.A.J., K.R., and H.F.; Investigation, F.H., K.M.W., K.R., A.O., D.H., F.F., C.C., M.A.J., B.K., and S.A.; Writing – Original Draft, F.H., K.M.W., K.R., and H.F; Writing – Review and Editing, A.O., D.H., F.F., C.C., and M.A.J.; Supervision and funding, H.F.
Declaration of interests
The authors declare no competing interests.
Published: February 22, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110515.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Borgonovo F., Passerini M., Piscaglia M., Morena V., Giacomelli A., Oreni L., Dedivitiis G., Lupo A., Falvella S., Cossu M.V., Capetti A.F. Is COVID-19 severity associated with anti-spike antibody duration? data from the ARCOVID prospective observational study. J. Infect. 2021;82:e28–e30. doi: 10.1016/j.jinf.2021.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco-Lauth A.M., Hartwig A.E., Porter S.M., Gordy P.W., Nehring M., Byas A.D., Vandewoude S., Ragan I.K., Maison R.M., Bowen R.A. Experimental infection of domestic dogs and cats with SARS-CoV-2: pathogenesis, transmission, and response to reexposure in cats. Proc. Natl. Acad. Sci. U S A. 2020;117:26382–26388. doi: 10.1073/pnas.2013102117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyton R.J., Altmann D.M. Risk of SARS-CoV-2 reinfection after natural infection. Lancet. 2021;397:1161–1163. doi: 10.1016/S0140-6736(21)00662-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brustolin M., Rodon J., Rodriguez de la Concepcion M.L., Avila-Nieto C., Cantero G., Perez M., Te N., Noguera-Julian M., Guallar V., Valencia A., et al. Protection against reinfection with D614- or G614-SARS-CoV-2 isolates in golden Syrian hamster. Emerg. Microbes Infect. 2021;10:797–809. doi: 10.1080/22221751.2021.1913974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center for Disease Control and Prevention (CDC) Common investigation Protocol for investigating suspected SARSR-CoV-2 reinfection. 2021. https://www.cdc.gov/coronavirus/2019-ncov/php/reinfection.html
- Centers for Disease Control and Prevention (CDC) SARS-CoV-2 variant Classification and definitions. 2021. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html
- Chandrashekar A., Liu J., Martinot A.J., Mcmahan K., Mercado N.B., Peter L., Tostanoski L.H., Yu J., Maliga Z., Nekorchuk M., et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chvatal-Medina M., Mendez-Cortina Y., Patino P.J., Velilla P.A., Rugeles M.T. Antibody responses in COVID-19: a review. Front. Immunol. 2021;12:633184. doi: 10.3389/fimmu.2021.633184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J.J., Sharma P., Bentley E.G., Harding A.C., Kipar A., Neary M., Box H., Hughes G.L., Patterson E.I., Sharp J., et al. Naturally-acquired immunity in Syrian Golden Hamsters provides protection from re-exposure to emerging heterosubtypic SARS-CoV-2 variants B.1.1.7 and B.1.351. bioRxiv. 2021 doi: 10.1101/2021.03.10.434447. Preprint at. [DOI] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brunink S., Schneider J., Schmidt M.L., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:eabg3055. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Bao L., Liu J., Xiao C., Liu J., Xue J., Lv Q., Qi F., Gao H., Yu P., et al. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science. 2020;369:818–823. doi: 10.1126/science.abc5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edridge A.W.D., Kaczorowska J., Hoste A.C.R., Bakker M., Klein M., Loens K., Jebbink M.F., Matser A., Kinsella C.M., Rueda P., et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- Fotouhi F., Salehi-Vaziri M., Farahmand B., Mostafavi E., Pouriayevali M.H., Jalali T., Mazaheri V., Sadat Larijani M., Tavakoli M., Eshratkhah Mohammadnejad A., et al. Prolonged viral shedding and antibody persistence in patients with COVID-19. Microbes Infect. 2021;23:104810. doi: 10.1016/j.micinf.2021.104810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson D.F., Norddahl G.L., Melsted P., Gunnarsdottir K., Holm H., Eythorsson E., Arnthorsson A.O., Helgason D., Bjarnadottir K., ingvarsson R.F., et al. Humoral immune response to SARS-CoV-2 in Iceland. N. Engl. J. Med. 2020;383:1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V., Bhoyar R.C., Jain A., Srivastava S., Upadhayay R., Imran M., Jolly B., Divakar M.K., Sharma D., Sehgal P., et al. Asymptomatic reinfection in two healthcare workers from India with genetically distinct SARS-CoV-2. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1451/5910388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddock E., Feldmann F., Shupert W.L., Feldmann H. Inactivation of SARS-CoV-2 laboratory specimens. Am. J. Trop. Med. Hyg. 2021;104:2195–2198. doi: 10.4269/ajtmh.21-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt J., Tamin A., Lu X., Kamili S., Sakthivel S.K., Murray J., Queen K., Tao Y., Paden C.R., Zhang J., et al. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg. Infect. Dis. 2020;26:1266–1273. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hers J.F., Masurel N., Mulder J. Bacteriology and histopathology of the respiratory tract and lungs in fatal Asian influenza. Lancet. 1958;2:1141–1143. doi: 10.1016/s0140-6736(58)92404-8. [DOI] [PubMed] [Google Scholar]
- Ibarrondo F.J., Fulcher J.A., Goodman-Meza D., Elliott J., Hofmann C., Hausner M.A., Ferbas K.G., Tobin N.H., Aldrovandi G.M., Yang O.O. Rapid decay of anti-SARS-CoV-2 antibodies in persons with mild covid-19. N. Engl. J. Med. 2020;383:1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. U S A. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R.D., Liu M.Q., Chen Y., Shan C., Zhou Y.W., Shen X.R., Li Q., Zhang L., Zhu Y., Si H.R., et al. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020;182:50–58 e8. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyuka P.K., Agoti C.N., Munywoki P.K., Njeru R., Bett A., Otieno J.R., Otieno G.P., Kamau E., Clark T.G., van der Hoek L., et al. Human coronavirus NL63 molecular epidemiology and evolutionary patterns in rural coastal Kenya. J. Infect. Dis. 2018;217:1728–1739. doi: 10.1093/infdis/jiy098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau E.H.Y., Tsang O.T.Y., Hui D.S.C., Kwan M.Y.W., Chan W.H., Chiu S.S., Ko R.L.W., Chan K.H., Cheng S.M.S., Perera R., et al. Neutralizing antibody titres in SARS-CoV-2 infections. Nat. Commun. 2021;12:63. doi: 10.1038/s41467-020-20247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Liao P., Qiu J.F., Lin Y., Cai X.F., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Loosli C.G., Stinson S.F., Ryan D.P., Hertweck M.S., Hardy J.D., Serebrin R. The destruction of type 2 pneumocytes by airborne influenza PR8-A virus; its effect on surfactant and lecithin content of the pneumonic lesions of mice. Chest. 1975;67:7S–14S. doi: 10.1378/chest.67.2_supplement.7s. [DOI] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumley S.F., O'Donnell D., Stoesser N.E., Matthews P.C., Howarth A., Hatch S.B., Marsden B.D., Cox S., James T., Warren F., et al. Antibody status and incidence of SARS-CoV-2 infection in health Care workers. N. Engl. J. Med. 2021;384:533–540. doi: 10.1056/NEJMoa2034545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettesheim P., Szakal A.K. Morphogenesis of alveolar bronchiolization. Lab. Invest. 1972;26:210–219. [PubMed] [Google Scholar]
- Pearson C.A.B., Russel T.W., Davies N.G., Kucharski A.J., CMMID COVID-19 Working group. Edmunds W.J., Eggo R.M. Estimates of severity and transmissibility of novel SARS-CoV-2 variant 501Y.V2 in South Africa. Preprint at CMMID Repository. 2021. https://cmmid.github.io/topics/covid19/reports/sa-novel-variant/2021_01_11_Transmissibility_and_severity_of_501Y_V2_in_SA.pdf
- Poland G.A., Ovsyannikova I.G., Kennedy R.B. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet. 2020;396:1595–1606. doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado-Vivar B., Becerra-Wong M., Guadalupe J.J., Marquez S., Gutierrez B., Rojas-Silva P., Grunauer M., Trueba G., Barragan V., Cardenas P. A case of SARS-CoV-2 reinfection in Ecuador. Lancet Infect. Dis. 2021;21:e142. doi: 10.1016/S1473-3099(20)30910-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roltgen K., Wirz O.F., Stevens B.A., Powell A.E., Hogan C.A., Najeeb J., Hunter M., Sahoo M.K., Huang C., Yamamoto F., et al. SARS-CoV-2 antibody responses correlate with resolution of RNAemia but are short-lived in patients with mild illness. medRxiv. 2020 doi: 10.1101/2020.08.15.20175794. Preprint at. [DOI] [Google Scholar]
- Rosenke K., Meade-White K., Letko M., Clancy C., Hansen F., Liu Y., Okumura A., Tang-Huau T.L., Li R., Saturday G., et al. Defining the Syrian hamster as a highly susceptible preclinical model for SARS-CoV-2 infection. Emerg. Microbes Infect. 2020;9:2673–2684. doi: 10.1080/22221751.2020.1858177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Self W.H., Tenforde M.W., Stubblefield W.B., Feldstein L.R., Steingrub J.S., Shapiro N.I., Ginde A.A., Prekker M.E., Brown S.M., Peltan I.D., et al. Decline in SARS-CoV-2 antibodies after mild infection among frontline health care personnel in a multistate hospital network - 12 States, April-August 2020. MMWR Morb Mortal Wkly Rep. 2020;69:1762–1766. doi: 10.15585/mmwr.mm6947a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia S.F., Yan L.M., Chin A.W.H., Fung K., Choy K.T., Wong A.Y.L., Kaewpreedee P., Perera R., Poon L.L.M., Nicholls J.M., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M.S., Chivukula R.R., Myers L.C., Jeck W.R., Waghray A., Tata P.R., Selig M.K., O'Donnell W.J., Farver C.F., Thompson B.T., et al. A conserved distal lung regenerative pathway in acute lung injury. Am. J. Pathol. 2018;188:1149–1160. doi: 10.1016/j.ajpath.2018.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillett R.L., Sevinsky J.R., Hartley P.D., Kerwin H., Crawford N., Gorzalski A., Laverdure C., Verma S.C., Rossetto C.C., Jackson D., et al. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect. Dis. 2021;21:52–58. doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K., Hung I.F., Ip J.D., Chu A.W., Chan W.M., Tam A.R., Fong C.H., Yuan S., Tsoi H.W., Ng A.C., et al. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin. Infect. Dis. 2020;73:e2946–e2951. doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Elslande J., Vermeersch P., Vandervoort K., Wawina-Bokalanga T., Vanmechelen B., Wollants E., Laenen L., André E., Van Ranst M., Lagrou K., Maes P. Symptomatic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reinfection by a phylogenetically distinct strain. Clin. Infect. Dis. 2021;73:354–356. doi: 10.1093/cid/ciaa1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajnberg A., Amanat F., Firpo A., Altman D.R., Bailey M.J., Mansour M., Mcmahon M., Meade P., Mendu D.R., Muellers K., et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370:1227–1230. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- WHO Director-General's opening remarks at the media briefing on COVID-19. 2020. https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020
- WHO Coronavirus (COVID-2019) dashboard. 2021. https://covid19.who.int/
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Werner A.P., Koch M., Choi A., Narayanan E., Stewart-Jones G.B.E., Colpitts T., Bennett H., Boyoglu-Barnum S., shi W., et al. Serum neutralizing activity elicited by mRNA-1273 vaccine. N. Engl. J. Med. 2021;384:1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucman N., Uhel F., Descamps D., Roux D., Ricard J.D. Severe reinfection with South African SARS-CoV-2 variant 501Y.V2: a case report. Clin. Infect. Dis. 2021;73:1945–1946. doi: 10.1093/cid/ciab129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.