Abstract
Background:
Cancer recurrence is an important predictor of survival outcomes in patients with colorectal cancer associated liver metastasis (CRLM), who undergo radical hepatectomy. Therefore, identification of patients with the greatest risk of recurrence is critical for developing a precision oncology strategy that might include frequent surveillance (in low-risk patients) or a more aggressive treatment approach (in high-risk patients). We performed genome-wide expression profiling, to identify and develop a transcriptomic signature for predicting recurrence in patients with CRLM.
Methods:
We analyzed a total of 383 CRLM patients, including 63 patients from a publicly available dataset (GSE81423) and 320 patients from whom surgical specimens were collected for independent training (n=169) and validation (n=151) of identified biomarkers. Using Cox’s proportional hazard regression analysis, we evaluated the clinical significance of the identified gene signature by comparing its performance with several key clinical factors.
Results:
We identified a 6-gene panel that robustly categorized patients with recurrence in the discovery (AUC=0.90). We showed that the panel was a significant predictor of recurrence in the clinical training (AUC=0.83) and validation cohorts (AUC=0.81). By combining our panel with key clinical factors, we established a risk-stratification model that emerged as an independent predictor of recurrence (AUC=0.85; Univariate: HR=4.34, 95% CI=2.71–6.93, P<0.001; Multivariate: HR=3.40, 95% CI=1.76–6.56, P<0.001). The stratification model revealed recurrence prediction in 89% of high-risk group and non-recurrence in 62% of low-risk group.
Conclusions:
We established a novel transcriptomic signature that robustly predicts recurrence, which has significant implications for the management of patients with CRLM.
Keywords: gene signature, genome-wide profiling, risk-stratification, prediction biomarker, tissue-based identification
INTRODUCTION
Colorectal cancer (CRC) is the second most common cause of cancer-related deaths worldwide [1, 2]. Management of patients with distant metastases is a major challenge: 20–30% of patients with CRC have distant metastatic disease at the time of diagnosis, and 5-year overall survival (OS) in these patients with metastatic CRC is less than 10% [3–5]. Approximately 50% of patients with CRC develop liver metastases after treatment of the primary tumor, and liver metastases is the most frequent cause of mortality in these patients [6–10]. Hepatectomy is the current treatment standard for colorectal liver metastases (CRLM) and can offer lead to prolonged survival, with a 5-year OS of 30–50% [11–14]. However, in spite of advances in surgical techniques, imaging modalities, and post-operative management, 50–70% of patients still experience cancer recurrence following hepatectomy, and most such recurrences occur within first 2 years of surgery [8, 15–18]. Consequently, due to these frequent recurrence events, the post-operative treatment options available to these patients are often complicated. The National Comprehensive Cancer Network guidelines suggest several selections of the treatment including surveillance with or without adjuvant chemotherapy in patients with resectable CRLM. Unfortunately, there are no randomized studies available that compared recurrence in patients of surgery or surgery with chemotherapy; underscoring the need for precise biomarkers that can enable a more personalized treatment strategy.
Several clinical trials have demonstrated that the chemotherapy improves survival outcomes after hepatectomy in patients with CRLM. For example, perioperative FOLFOX (oxaliplatin, leucovorin, and fluorouracil) chemotherapy led to a 7% improvement in progression-free survival (PFS) when followed after hepatectomy for CRLM [19, 20]. Likewise, other studies evaluating the addition of targeted therapies such as anti-EGFR antibodies in metastatic CRC also noticed improved outcomes [21, 22]. On the same lines, inclusion of anti-VEGF antibodies to a modified FOLFOX or FOLFIXIRI regimen (fluorouracil, leucovorin, oxaliplatin, and irinotecan) associated with improved response and resection rates and prolonged PFS in patients with CRLM [23–25]. These findings indicate that patients with CRLM who have a higher risk of recurrence benefit from such interventions and should be offered such intensive treatments following hepatectomy. Regarding post-surgical surveillance alone, serological carcinoembryonic antigen (CEA) level measurements, and the use of computed tomography (CT) imaging of the chest, abdomen, and pelvis every 3–6 months for the first 2 years and thereafter every 6 months for 3 more years are recommended. However, these criteria are often inadequate in high-risk patients, given that some patients need more aggressive surveillance and chemotherapy considering that more than half of patients experience tumor recurrence in 2 years of surgical treatment.
Recently, several clinical factors have been identified in patients with CRLM that predict the risk of cancer recurrence following hepatectomy [26–30]. Fong score, which combines CEA levels, tumor number, tumor size, lymph node positivity, and recurrence-free survival (RFS), is often used for predicting the recurrence after hepatectomy in patients with CRLM [31]. However, instead of or in addition to clinical factors, unravelling the molecular properties that characterize tumors may be pivotal for tailoring individualized therapies based on molecular predictors of survival outcomes. To this end, various groups have reported molecular subtypes in cancers with distinct biological properties and corresponding predictive and prognostic values [18, 26–30, 32–35]. Genetic and morphological evaluation scores, which combine clinicopathological and clinically-available biological indicators, including KRAS mutation status, have been reported to be a preoperative prognostic tool that can be used for treatment selection [36]. There is a growing consensus that molecular markers can improve the selection of patient for chemotherapy and other treatments in addition to hepatectomy or surveillance. The ability to analyze tumors at the RNA level promises to revolutionize the understanding of malignant disease, and we hope this will usher in a new biomarker.
Therefore, in this study, we conducted a comprehensive discovery effort to distinguish gene expression signatures for predicting cancer recurrence after hepatectomy in CRLM patients. We initially identified this signature in a genome-wide expression profiling dataset. Our risk-stratification model allows robust identification of patients with CRLM who are at high risk of cancer recurrence, which could improve the management of patients by recommending more intensive treatment selection in high-risk patients and sparing other low-risk patients from the unnecessary toxicity associated with adjuvant treatments.
METHODS
Gene biomarker discovery
To perform a comprehensive genome-wide biomarker discovery, gene expression profiling results from a dataset (GSE81423) were analyzed to identify and establish a biomarker panel for predicting recurrence following radical hepatectomy in patients with CRLM. The expression data was obtained from 63 patients with CRLM (33 recurrence and 30 non-recurrence), as illustrated in Fig. S1. We evaluated the prediction of recurrence by using selected biomarkers and subsequently determined area under the curve (AUC) values for each of the receiver operator characteristic (ROC) plots [37, 38].
Patient cohorts
To perform clinical training and validation of the identified gene panel, a total of 320 formalin-fixed paraffin-embedded (FFPE) specimens from two independent cohorts of resectable patients with CRLM were analyzed. This included a training cohort (n = 169; 119 with recurrence and 50 non-recurrence) of patients enrolled at the Tokushima and Kyushu University, and a validation cohort (n = 151; 102 with recurrence and 49 non-recurrence) of patients enrolled at the Kumamoto University, Japan. All patients underwent an initial radical hepatectomy between January 1995 and December 2017 in the training cohort, and between January 2001 and December 2016 in the validation cohort. The specimens were diagnosed as CRLM by pathologists. The follow-up protocol included abdominal ultrasonography, CT, and colonoscopy, evaluation of carbohydrate antigen 19-9 (CA19-9) and CEA levels, as well as a physical examination. Only regions confirmed by imaging modalities were diagnosed as tumor recurrence sites. The current study was conducted in accordance with the Declaration of Helsinki and approved by the institutional review boards of all participating institutions. A written informed consent was obtained from all patients.
RT-qPCR and RNA extraction
RNA was extracted from 10 μm sections of FFPE surgical resected tissues. Synthesis of complementary DNA (cDNA) was conducted with 500 ng of RNA. Real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR) analysis was performed as we previously reported [39]. Normalized expression values were log10-transformed [40]. The primers used in the current study are shown in Table S1.
Statistics
Clinicopathologic characteristics are shown in Table 1. OS and RFS were calculated from the date of surgery to the date of death from any cause or recurrence, or last follow-up date. Hazard ratios (HR) were calculated with 95% confidence intervals (CI) as we previously described [39]. Statistical analyses were performed using JMP Pro V 13.0 statistical software (SAS Institute Japan, Tokyo, Japan), Medcalc statistical software V.17.4.0 (Medcalc Software bvba, Ostend, Belgium).
Table 1:
Clinicopathological characteristics of clinical cohorts
| Characteristics | Training cohort (n=169), n (%) | Validation cohort (n=151), n (%) | P | |
|---|---|---|---|---|
| Age (years) | Median (range) | 65 (33–92) | 64 (25–94) | 0.113 |
| Gender | Male | 114 (67.5) | 101 (66.9) | |
| Female | 55 (32.5) | 50 (33.1) | 0.914 | |
| Recurrence status | Recurrence | 119 (70.4) | 102 (67.5) | |
| Non-recurrence | 50 (29.6) | 49 (32.5) | 0.58 | |
| Syn / Meta | Synchronous | 96 (56.8) | 98 (64.9) | |
| Metachronous | 73 (43.2) | 53 (35.1) | 0.139 | |
| Tumor number | Solitary | 83 (49.1) | 62 (41.1) | |
| Multiple | 85 (50.3) | 89 (58.9) | ||
| Missing | 1 (0.6) | 0 (0) | 0.135 | |
| Tumor size (mm) | Median (range) | 25 (5–100) | 28 (3–160) | 0.781 |
| CEA (ng/ml) | Median (range) | 9.8 (0.5–1430) | 7.1 (0.9–2061) | 0.201 |
| CA19-9 (U/ml) | Median (range) | 19.0 (0.6–6140) | 19.5 (0.1–1756) | 0.732 |
| Fong score | High score (≥3) | 44 (26.0) | 42 (27.8) | |
| Low score (<3) | 125 (74.0) | 109 (72.2) | 0.72 | |
| Neoadjuvant chemotherapy | Yes | 92 (54.5) | 83 (55.0) | |
| No | 76 (44.9) | 68 (45.0) | ||
| Missing | 1 (0.6) | 0 (0) | 0.971 | |
| Adjuvant chemotherapy | Yes | 111 (65.7) | 86 (57.0) | |
| No | 57 (33.7) | 63 (41.7) | ||
| Missing | 1 (0.6) | 2 (1.3) | 0.126 | |
| Tumor location of primary site | Right side | 33 (19.5) | 31 (20.5) | |
| Left side | 69 (40.8) | 64 (42.4) | ||
| Rectum | 67 (39.7) | 56 (37.1) | 0.895 | |
| Differentiation of primary site | Well | 72 (42.6) | 66 (43.7) | |
| Moderately | 79 (46.7) | 74 (49.0) | ||
| Poor | 6 (3.6) | 1 (0.7) | ||
| Others | 3 (1.8) | 0 (0) | ||
| Missing | 9 (5.3) | 10 (6.6) | 0.121 |
CA19-9, Carbohydrate antigen 19-9; CEA, Carcinoembryonic antigen;
RESULTS
Genome-wide expression profiling identifies a 6-gene panel that predicts cancer recurrence
To identify a gene panel that can predict cancer recurrence in CRLM patients, we first performed a genome-wide discovery in a public gene dataset from a cohort of patients with CRLM (GSE81423). By comparing gene profiles between patients with and without recurrence after radical hepatectomy, we identified a panel of 18 candidate genes differentially expressed (P < 0.05, |log2 fold-change| >1). Next, we performed lasso regression analysis with cross validation, which reduced the candidates to a panel of 6 genes: COX6A1, ERN1, IFITM2, S100P, STK24, and TMTC3. Applying the 6-gene panel to the GSE81423 data resulted in an AUC of 0.90 (95% CI = 0.79–0.96: Fig. S2A), highlighting the diagnostic potential of this panel for identifying recurrence in patients with CRLM (Fig. S2B).
Clinical cohort established the transcriptomic panel that predicts cancer recurrence
To confirm the predictive our discovered transcriptomic panel, we evaluated the performance in two clinical cohorts of patients with CRLM who underwent radical hepatectomy. The training cohort consisted of 169 patients, which included 119 patients with recurrence (70.4%), and the validation cohort consisted of 151 patients, which included 102 patients with recurrence (67.5%) with a median age of 64 years. According to the chemotherapy regimen, 92 patients in the training and 83 patients in the validation cohort received neoadjuvant chemotherapy, and 111 patients in the training and 86 patients in the validation cohort received adjuvant chemotherapy (Table 1). According to the detailed chemotherapy regimen, perioperative chemotherapy was performed in 254 of the 320 patients with CRLM. The details of these treatment are as follows: FOLFOX, 80 cases; XELOX, 51 cases; LV/UFT, 34 cases; FOLFIRI, 28 cases; oral 5FU, 18 cases; oral capecitabine, 10 cases; IRIS, 9 cases; SOX, 7 cases; LV/5FU, 7 cases and others, 10 cases.
Next, we interrogated the diagnostic performance of our panel to predict recurrence in the training cohort (119 with recurrence vs. 50 with non-recurrence). We trained the panel that robustly identified recurrence in patients with CRLM (AUC = 0.83, 95% CI = 0.76–0.88, Fig. 1A, B). Thereafter, we developed a transcriptomic panel by using parameters: Logit (P) = (COX6A1*0.1442) + (ERN1*−0.4654) + (IFITM2*−0.2096) + (S100P*−0.2334) + (STK24*−0.1346) + (TMTC3*−0.2320).
Figure 1.

The transcriptomic panel for identifying cancer recurrence. A) ROC curve for the panel in the training cohort of 169 patients (with recurrence = 119, non-recurrence = 50, AUC = 0.83). B) Risk score plot in the training cohort. C) ROC curve for the panel in the validation cohort of 151 patients (with recurrence = 102, non-recurrence = 49, AUC = 0.81). D) Risk score plot in the validation cohort.
We evaluated the accuracy of 6-gene transcriptomic panel by using same statistical model (i.e., same coefficients and cutoff values) to another validation cohort of patients with CRLM (102 with recurrence, and 49 non-recurrence). The diagnostic performance of our panel in the validation cohort was comparable in the training cohort (AUC = 0.81, 95% CI = 0.74–0.87: Fig. 1C, D).
Recurrence prediction correlates with survival outcomes
The recurrence is correlated to poor survival in CRLM patients. Thus, to determine the prognostic potential of the transcriptomic panel, we analyzed the survival outcomes (OS and PFS) in the clinical training and validation cohorts. The median follow-up period after hepatectomy were 39.7 months (95% CI = 34.5–44.4) in training and 37.1 months (95% CI = 26.9–47.8) in validation cohort. It was demonstrated that the patients with high-risk exhibited significantly poor prognosis in the training [P < 0.001]; OS [P = 0.029]; Fig. 2A, B), and validation cohorts (RFS [P < 0.001]; OS [P = 0.006]; Fig. 2C, D). We performed an univariate Cox’s proportional hazard regression analysis, which identified that our panel was an independent predictor of recurrence, compared to each clinical factors (training: HR = 3.18, 95% CI = 2.03–4.98, P < 0.001; validation: HR = 2.60, 95% CI = 1.73–3.92, P < 0.001; Table 2).
Figure 2.

The transcriptomic panel of a prognostic potential. (A) RFS and (B) OS between low- and high-risk groups of the transcriptomic panel in the training cohort. (C) RFS and (D) OS between low- and high-risk groups of the transcriptomic panel in the validation cohort.
Table 2.
Univariate and multivariate analysis for the recurrence free survival
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Factors | HR | 95% CI | p | HR | 95% CI | p |
| Training cohort (n=169) | ||||||
| Age | ||||||
| (≥65 vs. <65) | 0.85 | 0.59–1.21 | 0.365 | |||
| Gender | ||||||
| (Male vs. Female) | 0.95 | 0.64–1.39 | 0.783 | |||
| Syn / Meta | ||||||
| (Syn vs. Meta) | 1.47 | 1.01–2.12 | 0.042 | 1.13 | 0.77–1.65 | 0.554 |
| Tumor number | ||||||
| (Multi vs. Solitary) | 1.99 | 1.38–2.88 | 0.005 | 1.92 | 1.33–2.79 | 0.001 |
| Tumor size | ||||||
| (≥25mm vs. 25mm) | 1.62 | 0.97–2.71 | 0.066 | |||
| CEA | ||||||
| (≥5.0ng/mL vs. <5.0ng/mL) | 1.33 | 0.88–2.00 | 0.171 | |||
| CA19-9 | ||||||
| (≥37U/mL) vs. <37U/mL) | 1.30 | 0.89–1.91 | 0.177 | |||
| Fong score | ||||||
| (≥3 vs. <3) | 1.60 | 0.96–2.80 | 0.071 | |||
| Neoadjuvant chemotherapy | ||||||
| (Yes vs. No) | 1.18 | 0.73–1.91 | 0.492 | |||
| Adjuvant chemotherapy | ||||||
| (Yes vs. No) | 1.55 | 1.03–2.31 | 0.034 | 1.22 | 0.80–1.85 | 0.355 |
| RNA panel | ||||||
| (High risk vs. Low risk) | 3.18 | 2.03–4.98 | <0.001 | 2.87 | 1.82–4.53 | <0.001 |
| Validation cohort (n=151) | ||||||
| Age | ||||||
| (≥65 vs. <65) | 0.99 | 0.97–1.04 | 0.392 | |||
| Gender | ||||||
| (Male vs. Female) | 0.83 | 0.55–1.24 | 0.364 | |||
| Syn / Meta | ||||||
| (Syn vs. Meta) | 1.78 | 1.17–2.72 | 0.008 | 1.28 | 0.81–2.01 | 0.291 |
| Tumor number | ||||||
| (Multi vs. Solitary) | 1.96 | 1.29–2.98 | 0.002 | 1.09 | 0.70–1.72 | 0.699 |
| Tumor size | ||||||
| (≥25mm vs. 25mm) | 1.42 | 0.95–2.13 | 0.085 | |||
| CEA | ||||||
| (≥5.0ng/mL vs. <5.0ng/mL) | 1.60 | 1.05–2.42 | 0.027 | 2.24 | 1.43–3.51 | <0.001 |
| CA19-9 | ||||||
| (≥37U/mL) vs. <37U/mL) | 1.59 | 1.05–2.39 | 0.027 | 0.97 | 0.62–1.51 | 0.882 |
| Fong score | ||||||
| (≥3 vs. <3) | 1.45 | 0.96–2.32 | 0.054 | |||
| Neoadjuvant chemotherapy | ||||||
| (Yes vs. No) | 1.39 | 0.94–2.17 | 0.061 | |||
| Adjuvant chemotherapy | ||||||
| (Yes vs. No) | 0.78 | 0.52–1.16 | 0.223 | |||
| RNA panel | ||||||
| (High risk vs. Low risk) | 2.60 | 1.73–3.92 | <0.001 | 1.85 | 1.17–2.93 | 0.009 |
| Risk model | ||||||
| (High risk vs. Low risk) | 4.34 | 2.71–6.93 | <0.001 | 3.65 | 2.02–6.59 | <0.001 |
CA19-9, Carbohydrate antigen 19-9; CEA, Carcinoembryonic antigen; Meta, Metachronous; Syn, Synchronous
Risk model by combining the panel and clinical factors improves to predict recurrence
We evaluated whether the stratification model that couples known risk factors with the panel improve the accuracy in predicting recurrence. Indeed, once we incorporated four conventional clinicopathological factors (i.e., tumor number, synchronous lesions, CA19-9, and CEA,) into the stratification model, the model revealed a markedly better diagnostic performance compared with the only panel or other risk factors (AUC = 0.85, Fig. 3A).
Figure 3.
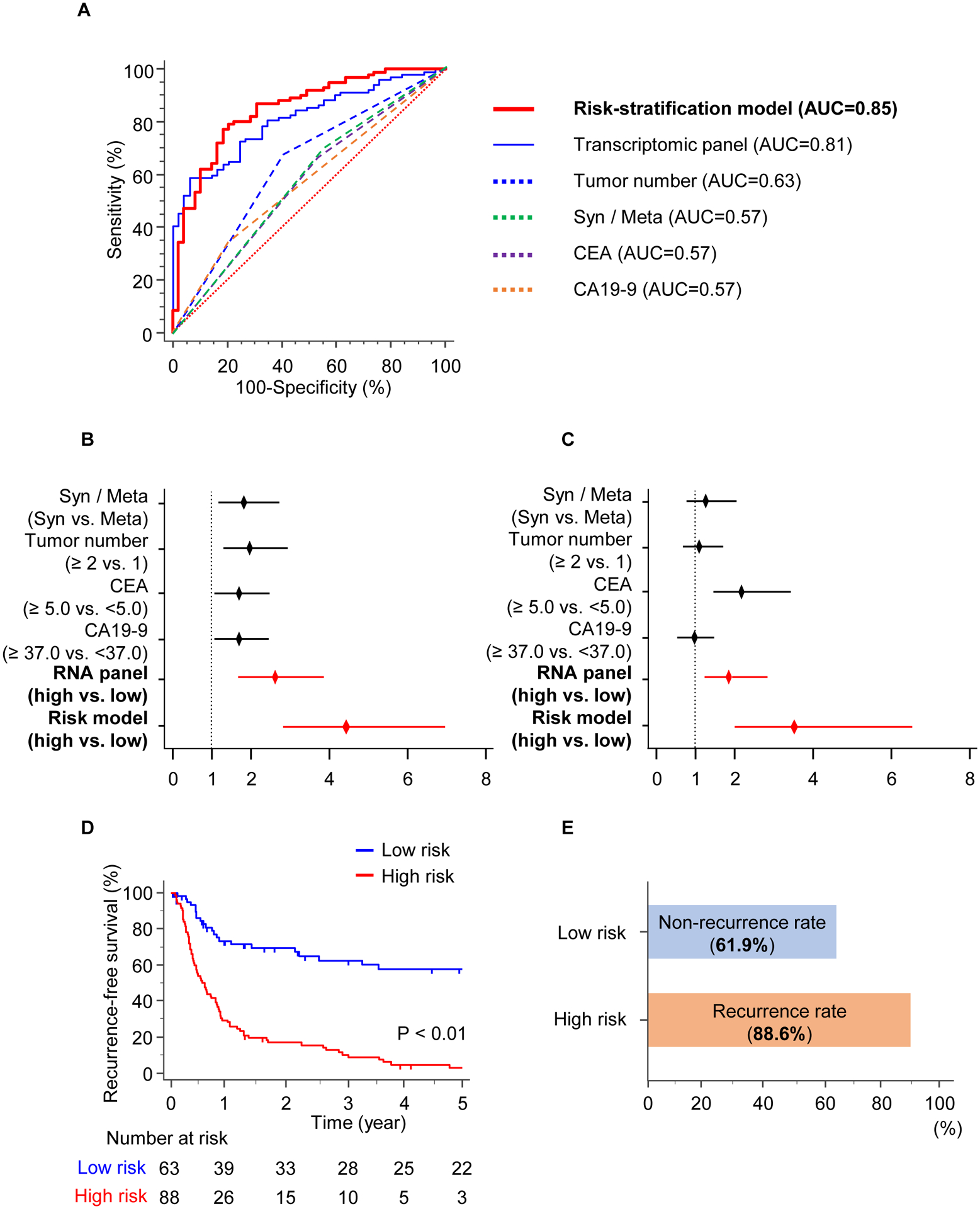
The risk-stratification model in the validation cohort. A) ROC curves for the model by combining the panel and clinical factors, vs. the transcriptomic panel or indicated risk factors in the validation cohort. B–C) Forest plot showing HRs of indicated clinicopathological factors, the panel, and the stratification model in univariate (B) and multivariate (C) analysis. D) RFS between high- (Red) and low- (Blue) risk groups by the risk-stratification model. E) Bar graphs shows 61.9% of low-risk patients with CRLM who would have experienced non-recurrence, and 88.6% of high-risk patients with CRLM who would have experienced recurrence.
We evaluated the diagnostic accuracy for our panel in validation. Therefore, we revealed that the sensitivity, specificity, positive predictive value (PPV), and negative predictive values (NPV) were 59.8%, 83.7%, 88.4%, and 50.0%, respectively (Table 3). When we performed the stratification model that also included risk factors, the model was significantly superior to the transcriptomic panel alone in predicting sensitivity, specificity, PPV, and NPV, with values of 77.2%, 81.6%, 89.7%, and 63.5%, respectively. Herein, our novel model by univariate and multivariate Cox’s regression analysis was better than the panel and various clinicopathological factors for predicting recurrence (Univariate: HR = 4.34, 95% CI = 2.71–6.93, P < 0.001, Fig. 3B; Multivariate: HR = 3.65, 95% CI = 2.02–6.59, P < 0.001, Fig. 3C) in validation (Table 2). These data highlight the potential significance of the model for identifying recurrence in CRC patients with liver metastasis.
Table 3:
Model performance for the risk of cancer recurrence
| Variable | Value (95% CI) | |||
|---|---|---|---|---|
| GSE81423 | Training cohort | Validation cohort (RNA panel) | Validation cohort (Risk model) | |
| Cutoff value | - | 1.44 | 1.44 | 2.13 |
| Sensitivity, % | 90.9 (75.7–98.1) | 79.0 (70.6–85.9) | 59.8 (49.6–69.4) | 77.2 (67.8–85.0) |
| Specificity, % | 76.7 (57.7–90.1) | 72.0 (57.5–83.8) | 83.7 (70.3–92.7) | 81.6 (68.0–91.2) |
| AUC, % | 89.5 (79.2–95.8) | 82.5 (75.9–87.9) | 81.1 (74.0–87.0) | 85.0 (78.2–90.3) |
| PPV, % | 81.1 (68.9–89.2) | 87.0 (81.0–91.4) | 88.4 (79.9–93.6) | 89.7 (82.6–94.0) |
| NPV, % | 88.5 (71.9–95.8) | 59.0 (49.4–68.0) | 50.0 (43.4–56.6) | 63.5 (54.3–71.8) |
AUC, area under the curve; CI, confidence interval;
NPV, negative predictive value; PPV, positive predictive value
Risk-stratification model robustly identifies CRLM patients at high risk for cancer recurrence
To determine the clinically usefulness of our model, we applied our risk-stratification model to dichotomize the patients within the validation cohort into high- and low-risk groups of patients for risk of cancer recurrence. We observed that the RFS was significantly lower in the low- vs. high-risk patients by our model (P < 0.001; Fig. 3D). Of the 88 cases who were classified as high-risk patients, 78 cases had cancer recurrence in 5 years (88.6%) (Fig. 3E). Of the 63 cases who were classified as low-risk patients, 39 cases did not have tumor recurrence in 5 years (61.9%), highlighting that these low-risk patients could avoid expense and toxicity from unnecessary treatment. These data highlight the clinical potential of our model to inform personalize treatment recommendations in patients with low- and high-risk CRLM.
DISCUSSION
Accumulating studies continue to emphasize the importance of identifying clinical predictive markers for recurrence in CRC patients with liver metastasis (CRLM) [8, 15–18]. However, the specific clinical risk factors that can robustly predict cancer recurrence and long-term survival outcomes in patients with CRLM remains unclear. Clinically, CRLM rarely spreads via micro-metastases adjacent to the liver metastases. Thus, the width of a negative surgical margin following hepatectomy in patients with CRLM does not affect the risk of marginal recurrence or impact survival [41–43]. Furthermore, a recent study suggested that parenchyma-sparing hepatectomy did not increase recurrence in the remnant liver and improved salvageability for recurrent patients [44]. Considering this evidence in the context of our findings about cancer outcomes, our transcriptomic panel could identify high-risk CRC patients who require close follow-up surveillance and intensive chemotherapy to prevent subsequent recurrence.
Response to chemotherapy is recognized as one of the strong predictor of survival outcome after surgery, and provides better prognosis than traditional clinicopathological features [45]. However, several issues complicate the inclusion of response to chemotherapy in a clinical scenario. The best way to assess treatment response remains unclear. Histopathological scoring of responses to chemotherapy clearly predicts treatment outcome, but such assessments can only be made after surgery on resected specimens, and different groups use different scoring systems [46]. Previous data indicated that CA19-9, CEA, synchronous lesions, and tumor number were correlated to the tumor characteristics, a higher risk of tumor recurrence, and poor survival [47–50]. In the present study, our novel model revealed better diagnostic performance to predict cancer recurrence (AUC = 0.85), compared with clinical risk factors (i.e., synchronous, CA19-9, CEA, and tumor number; AUC = 0.68 [training] and 0.68 [validation]; Fig. S3). In addition, our model robustly stratified patients into low-and high-risk patients, indicating that 88.6% cases of high-risk patients had cancer recurrence and 61.9% cases of low-risk patients did not have cancer recurrence. This highlights dramatically better performance, compared with clinicopathological risk factors and the potential importance of our findings on clinical translation for identification of recurrence and improved survival in CRLM patients.
The various genes in the panel have been shown to be genuine candidates for the pathogenesis of cancer. For instance, IFITM2 is a member of IFN-inducible transmembrane gene family, which plays key roles in modulating the immune response and thus may control tumor development [51, 52]. IFITM2 has been reported to be highly specific to human colorectal carcinogenesis and overexpressed in CRC [53]. S100P is a member of the S100 family of proteins containing 2 EF-hand calcium-binding motifs, which regulates many intracellular and extracellular activities [54, 55]. S100P is overexpressed in CRC and regulates the invasion and metastasis of CRC by promoting epithelial–mesenchymal transition [56]. Furthermore, STK24, encoding a serine/threonine protein kinase, is a member of serine/threonine kinase family that functions upstream of mitogen-activated protein kinase (MAPK) signaling and positively regulates the cell cycle, cell growth, migration, and synapse development [57]. Similarly, the STK24 activity was increased in the immunoblot analyses of CRC, which revealed an increased amount of STK24 kinase in CRC [58].
We acknowledge that our study has some potential limitations. At first, current retrospective study might have resulted in a selection bias. So, to confirm the accuracy of our stratification model, a constructed prospective clinical trial is required. Next, our research used Japanese cohort in the training and validation, who showed relatively homogenous clinicopathologic characteristics; these could vary among populations from other countries. It is important to validate the selected biomarkers and the stratification model in patient cohorts in other countries to further enhance the generalization of findings. Third, unfortunately, we did not have the mutation data for the KRAS and BRAF genes for predicting the recurrence, future studies will be needed to explore these associations.
In conclusion, we established a new stratification model that was validated in clinical cohort for the identification of high-risk CRLM patients. Our current findings reveal the clinical impact of our stratification model on accurate risk-stratification of patients, which will improve personalized management of patients with CRLM.
Supplementary Material
Supplemental Figure S1. Overview of the study.
Supplemental Figure S2. Genome-wide discovery analysis of a gene panel to predict cancer recurrence. A) ROC curve for the diagnostic accuracy of a 6-gene panel for classifying patients with cancer recurrence in GSE81423 (AUC = 0.90). B) Risk score distribution plot in GSE81423.
Supplemental Figure S3. Prediction of recurrence in training and validation cohorts without the transcriptomic panel. A) ROC curve for combined clinical risk factors for recurrence (CA19-9, CEA, synchronous, and tumor number) without the transcriptomic panel in the training cohort (AUC = 0.68). B) ROC curve for the combined clinical risk factors for recurrence without the panel in the validation cohort (AUC = 0.68).
Highlights.
Cancer recurrence is an important predictor of survival outcomes in CRLM.
Currently, there are no available biomarkers to predict the risk of CRLM.
We identified a gene signature by a genome-wide and comprehensive discovery.
Our novel signature could predict the high-risk patients of cancer recurrence.
Acknowledgements
We thank Drs. Tatsuhiko Kakisaka, Satoshi Nishiwada, Yasuyuki Okada, Huanlin Wang, Geeta Sharma, and In-Seob Lee for discussing the experiments and analysis. We thank Drs. Kensuke Yamamura, Takeo Toshima, Masaaki Nishi, Shinichiro Yamada, and Kazunori Tokuda, as well as Ms. Yumi Horikawa for collecting clinical samples and information. We also would like to extend our thanks to Dr. Sarah Wilkinson for her significant editing and useful suggestions for improving the quality of our manuscript.
Funding:
This work was supported by CA72851, CA181572, CA184792, CA202797, and CA227602 grants from the National Cancer Institute, National Institutes of Health.
Abbreviations:
- AUC
Area under the curve
- CA19-9
Carbohydrate antigen 19-9
- CEA
Carcinoembryonic antigen
- CI
Confidence intervals
- CRC
Colorectal cancer
- CRLM
Colorectal liver metastases
- CT
Computed tomography
- FFPE
Formalin-fixed paraffin-embedded
- HR
Hazard ratio
- NPV
Negative predictive value
- OS
Overall survival
- PFS
Progression-free survival
- PPV
Positive predictive value
- RFS
Recurrence-free survival
- ROC
Receiver operator characteristic
- RT-qPCR
Real-time quantitative reverse transcription polymerase chain reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: None of the authors has any potential conflicts to disclose.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES
- [1].Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145–64. [DOI] [PubMed] [Google Scholar]
- [2].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- [3].Jarnagin WR. Changing management strategies for hepatic colorectal metastasis. Oncology (Williston Park). 2009;23:1077–8, 81. [PubMed] [Google Scholar]
- [4].Van Cutsem E, Cervantes A, Nordlinger B, Arnold D. Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25 Suppl 3:iii1–9. [DOI] [PubMed] [Google Scholar]
- [5].Van Cutsem E, Nordlinger B, Adam R, Köhne CH, Pozzo C, Poston G, et al. Towards a pan-European consensus on the treatment of patients with colorectal liver metastases. Eur J Cancer. 2006;42:2212–21. [DOI] [PubMed] [Google Scholar]
- [6].Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, Bouvier AM. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg. 2006;244:254–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nordlinger B, Sorbye H, Glimelius B, Poston GJ, Schlag PM, Rougier P, et al. Perioperative FOLFOX4 chemotherapy and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC 40983): long-term results of a randomised, controlled, phase 3 trial. Lancet Oncol. 2013;14:1208–15. [DOI] [PubMed] [Google Scholar]
- [8].Tomlinson JS, Jarnagin WR, DeMatteo RP, Fong Y, Kornprat P, Gonen M, et al. Actual 10-year survival after resection of colorectal liver metastases defines cure. J Clin Oncol. 2007;25:4575–80. [DOI] [PubMed] [Google Scholar]
- [9].Van Cutsem E, Oliveira J. Advanced colorectal cancer: ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann Oncol. 2009;20 Suppl 4:61–3. [DOI] [PubMed] [Google Scholar]
- [10].Zarour LR, Anand S, Billingsley KG, Bisson WH, Cercek A, Clarke MF, et al. Colorectal Cancer Liver Metastasis: Evolving Paradigms and Future Directions. Cell Mol Gastroenterol Hepatol. 2017;3:163–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Adam R, De Gramont A, Figueras J, Guthrie A, Kokudo N, Kunstlinger F, et al. The oncosurgery approach to managing liver metastases from colorectal cancer: a multidisciplinary international consensus. Oncologist. 2012;17:1225–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kanas GP, Taylor A, Primrose JN, Langeberg WJ, Kelsh MA, Mowat FS, et al. Survival after liver resection in metastatic colorectal cancer: review and meta-analysis of prognostic factors. Clin Epidemiol. 2012;4:283–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Morris EJ, Forman D, Thomas JD, Quirke P, Taylor EF, Fairley L, et al. Surgical management and outcomes of colorectal cancer liver metastases. Br J Surg. 2010;97:1110–8. [DOI] [PubMed] [Google Scholar]
- [14].Simmonds PC, Primrose JN, Colquitt JL, Garden OJ, Poston GJ, Rees M. Surgical resection of hepatic metastases from colorectal cancer: a systematic review of published studies. Br J Cancer. 2006;94:982–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Butte JM, Gönen M, Allen PJ, Peter Kingham T, Sofocleous CT, DeMatteo RP, et al. Recurrence After Partial Hepatectomy for Metastatic Colorectal Cancer: Potentially Curative Role of Salvage Repeat Resection. Ann Surg Oncol. 2015;22:2761–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Choti MA, Sitzmann JV, Tiburi MF, Sumetchotimetha W, Rangsin R, Schulick RD, et al. Trends in long-term survival following liver resection for hepatic colorectal metastases. Ann Surg. 2002;235:759–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].D’Angelica M, Kornprat P, Gonen M, DeMatteo RP, Fong Y, Blumgart LH, et al. Effect on outcome of recurrence patterns after hepatectomy for colorectal metastases. Ann Surg Oncol. 2011;18:1096–103. [DOI] [PubMed] [Google Scholar]
- [18].de Jong MC, Pulitano C, Ribero D, Strub J, Mentha G, Schulick RD, et al. Rates and patterns of recurrence following curative intent surgery for colorectal liver metastasis: an international multi-institutional analysis of 1669 patients. Ann Surg. 2009;250:440–8. [DOI] [PubMed] [Google Scholar]
- [19].Nordlinger B, Sorbye H, Glimelius B, Poston GJ, Schlag PM, Rougier P, et al. Perioperative chemotherapy with FOLFOX4 and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC Intergroup trial 40983): a randomised controlled trial. Lancet. 2008;371:1007–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sorbye H, Mauer M, Gruenberger T, Glimelius B, Poston GJ, Schlag PM, et al. Predictive factors for the benefit of perioperative FOLFOX for resectable liver metastasis in colorectal cancer patients (EORTC Intergroup Trial 40983). Ann Surg. 2012;255:534–9. [DOI] [PubMed] [Google Scholar]
- [21].Douillard JY, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, et al. Final results from PRIME: randomized phase III study of panitumumab with FOLFOX4 for first-line treatment of metastatic colorectal cancer. Ann Oncol. 2014;25:1346–55. [DOI] [PubMed] [Google Scholar]
- [22].Van Cutsem E, Köhne CH, Láng I, Folprecht G, Nowacki MP, Cascinu S, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29:2011–9. [DOI] [PubMed] [Google Scholar]
- [23].Tang W, Ren L, Liu T, Ye Q, Wei Y, He G, et al. Bevacizumab Plus mFOLFOX6 Versus mFOLFOX6 Alone as First-Line Treatment for RAS Mutant Unresectable Colorectal Liver-Limited Metastases: The BECOME Randomized Controlled Trial. J Clin Oncol. 2020;38:3175–84. [DOI] [PubMed] [Google Scholar]
- [24].Gruenberger T, Bridgewater J, Chau I, García Alfonso P, Rivoire M, Mudan S, et al. Bevacizumab plus mFOLFOX-6 or FOLFOXIRI in patients with initially unresectable liver metastases from colorectal cancer: the OLIVIA multinational randomised phase II trial. Ann Oncol. 2015;26:702–8. [DOI] [PubMed] [Google Scholar]
- [25].Tomasello G, Petrelli F, Ghidini M, Russo A, Passalacqua R, Barni S. FOLFOXIRI Plus Bevacizumab as Conversion Therapy for Patients With Initially Unresectable Metastatic Colorectal Cancer: A Systematic Review and Pooled Analysis. JAMA Oncol. 2017;3:e170278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ito H, Mo Q, Qin LX, Viale A, Maithel SK, Maker AV, et al. Gene expression profiles accurately predict outcome following liver resection in patients with metastatic colorectal cancer. PLoS One. 2013;8:e81680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Karagkounis G, Torbenson MS, Daniel HD, Azad NS, Diaz LA Jr., Donehower RC, et al. Incidence and prognostic impact of KRAS and BRAF mutation in patients undergoing liver surgery for colorectal metastases. Cancer. 2013;119:4137–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Konopke R, Kersting S, Distler M, Dietrich J, Gastmeier J, Heller A, et al. Prognostic factors and evaluation of a clinical score for predicting survival after resection of colorectal liver metastases. Liver Int. 2009;29:89–102. [DOI] [PubMed] [Google Scholar]
- [29].Margonis GA, Kim Y, Spolverato G, Ejaz A, Gupta R, Cosgrove D, et al. Association Between Specific Mutations in KRAS Codon 12 and Colorectal Liver Metastasis. JAMA Surg. 2015;150:722–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Matias M, Casa-Nova M, Faria M, Pires R, Tato-Costa J, Ribeiro L, et al. Prognostic Factors after Liver Resection for Colorectal Liver Metastasis. Acta Med Port. 2015;28:357–69. [DOI] [PubMed] [Google Scholar]
- [31].Fong Y, Fortner J, Sun RL, Brennan MF, Blumgart LH. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg. 1999;230:309–18; discussion 18–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cremolini C, Schirripa M, Antoniotti C, Moretto R, Salvatore L, Masi G, et al. First-line chemotherapy for mCRC—a review and evidence-based algorithm. Nat Rev Clin Oncol. 2015;12:607–19. [DOI] [PubMed] [Google Scholar]
- [33].Kemeny N, Huang Y, Cohen AM, Shi W, Conti JA, Brennan MF, et al. Hepatic arterial infusion of chemotherapy after resection of hepatic metastases from colorectal cancer. N Engl J Med. 1999;341:2039–48. [DOI] [PubMed] [Google Scholar]
- [34].McMillan DC. The systemic inflammation-based Glasgow Prognostic Score: a decade of experience in patients with cancer. Cancer Treat Rev. 2013;39:534–40. [DOI] [PubMed] [Google Scholar]
- [35].Welch HG, Robertson DJ. Colorectal Cancer on the Decline--Why Screening Can’t Explain It All. N Engl J Med. 2016;374:1605–7. [DOI] [PubMed] [Google Scholar]
- [36].Margonis GA, Sasaki K, Gholami S, Kim Y, Andreatos N, Rezaee N, et al. Genetic And Morphological Evaluation (GAME) score for patients with colorectal liver metastases. Br J Surg. 2018;105:1210–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ozawa T, Kandimalla R, Gao F, Nozawa H, Hata K, Nagata H, et al. A MicroRNA Signature Associated With Metastasis of T1 Colorectal Cancers to Lymph Nodes. Gastroenterology. 2018;154:844–8.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sonohara F, Gao F, Iwata N, Kanda M, Koike M, Takahashi N, et al. Genome-wide Discovery of a Novel Gene-expression Signature for the Identification of Lymph Node Metastasis in Esophageal Squamous Cell Carcinoma. Ann Surg. 2019;269:879–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wada Y, Shimada M, Yamamura K, Toshima T, Banwait JK, Morine Y, et al. A Transcriptomic Signature for Risk-Stratification and Recurrence Prediction in Intrahepatic Cholangiocarcinoma. Hepatology. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- [41].Kokudo N, Miki Y, Sugai S, Yanagisawa A, Kato Y, Sakamoto Y, et al. Genetic and histological assessment of surgical margins in resected liver metastases from colorectal carcinoma: minimum surgical margins for successful resection. Arch Surg. 2002;137:833–40. [DOI] [PubMed] [Google Scholar]
- [42].Pawlik TM, Scoggins CR, Zorzi D, Abdalla EK, Andres A, Eng C, et al. Effect of surgical margin status on survival and site of recurrence after hepatic resection for colorectal metastases. Ann Surg. 2005;241:715–22, discussion 22–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zorzi D, Mullen JT, Abdalla EK, Pawlik TM, Andres A, Muratore A, et al. Comparison between hepatic wedge resection and anatomic resection for colorectal liver metastases. J Gastrointest Surg. 2006;10:86–94. [DOI] [PubMed] [Google Scholar]
- [44].Mise Y, Aloia TA, Brudvik KW, Schwarz L, Vauthey JN, Conrad C. Parenchymal-sparing Hepatectomy in Colorectal Liver Metastasis Improves Salvageability and Survival. Ann Surg. 2016;263:146–52. [DOI] [PubMed] [Google Scholar]
- [45].Blazer DG 3rd, Kishi Y, Maru DM, Kopetz S, Chun YS, Overman MJ, et al. Pathologic response to preoperative chemotherapy: a new outcome end point after resection of hepatic colorectal metastases. J Clin Oncol. 2008;26:5344–51. [DOI] [PubMed] [Google Scholar]
- [46].Rubbia-Brandt L, Giostra E, Brezault C, Roth AD, Andres A, Audard V, et al. Importance of histological tumor response assessment in predicting the outcome in patients with colorectal liver metastases treated with neo-adjuvant chemotherapy followed by liver surgery. Ann Oncol. 2007;18:299–304. [DOI] [PubMed] [Google Scholar]
- [47].Kawaguchi Y, Kopetz S, Lillemoe HA, Hwang H, Wang X, Tzeng CD, et al. A New Surveillance Algorithm After Resection of Colorectal Liver Metastases Based on Changes in Recurrence Risk and RAS Mutation Status. J Natl Compr Canc Netw. 2020;18:1500–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Sakamoto Y, Miyamoto Y, Beppu T, Nitta H, Imai K, Hayashi H, et al. Post-chemotherapeutic CEA and CA19-9 are prognostic factors in patients with colorectal liver metastases treated with hepatic resection after oxaliplatin-based chemotherapy. Anticancer Res. 2015;35:2359–68. [PubMed] [Google Scholar]
- [49].Kawahara H, Yoshida S, Tohyama Y, Yanagisawa S, Misawa T, Yanaga K. Serum Carcinoembryonic Antigen Levels Before the First Curative Hepatectomy for Metastatic Colorectal Cancer Is a Predictor of Recurrence. Anticancer Res. 2018;38:5351–5. [DOI] [PubMed] [Google Scholar]
- [50].Kobayashi S, Beppu T, Honda G, Yamamoto M, Takahashi K, Endo I, et al. Survival Benefit of and Indications for Adjuvant Chemotherapy for Resected Colorectal Liver Metastases-a Japanese Nationwide Survey. J Gastrointest Surg. 2020;24:1244–60. [DOI] [PubMed] [Google Scholar]
- [51].André T, Vernerey D, Mineur L, Bennouna J, Desrame J, Faroux R, et al. Three Versus 6 Months of Oxaliplatin-Based Adjuvant Chemotherapy for Patients With Stage III Colon Cancer: Disease-Free Survival Results From a Randomized, Open-Label, International Duration Evaluation of Adjuvant (IDEA) France, Phase III Trial. J Clin Oncol. 2018;36:1469–77. [DOI] [PubMed] [Google Scholar]
- [52].Kitahara O, Furukawa Y, Tanaka T, Kihara C, Ono K, Yanagawa R, et al. Alterations of gene expression during colorectal carcinogenesis revealed by cDNA microarrays after laser-capture microdissection of tumor tissues and normal epithelia. Cancer Res. 2001;61:3544–9. [PubMed] [Google Scholar]
- [53].Miyamoto C, Miyamoto N, Yamamoto H, Imai K, Shinomura Y. Detection of fecal interferon-induced transmembrane protein messenger RNA for colorectal cancer screening. Oncol Lett. 2011;2:95–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Heizmann CW, Fritz G, Schäfer BW. S100 proteins: structure, functions and pathology. Front Biosci. 2002;7:d1356–68. [DOI] [PubMed] [Google Scholar]
- [55].Donato R S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int J Biochem Cell Biol. 2001;33:637–68. [DOI] [PubMed] [Google Scholar]
- [56].Shen ZY, Fang Y, Zhen L, Zhu XJ, Chen H, Liu H, et al. Analysis of the predictive efficiency of S100P on adverse prognosis and the pathogenesis of S100P-mediated invasion and metastasis of colon adenocarcinoma. Cancer Genet. 2016;209:143–53. [DOI] [PubMed] [Google Scholar]
- [57].Madsen CD, Hooper S, Tozluoglu M, Bruckbauer A, Fletcher G, Erler JT, et al. STRIPAK components determine mode of cancer cell migration and metastasis. Nat Cell Biol. 2015;17:68–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Hennig EE, Mikula M, Rubel T, Dadlez M, Ostrowski J. Comparative kinome analysis to identify putative colon tumor biomarkers. J Mol Med (Berl). 2012;90:447–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Overview of the study.
Supplemental Figure S2. Genome-wide discovery analysis of a gene panel to predict cancer recurrence. A) ROC curve for the diagnostic accuracy of a 6-gene panel for classifying patients with cancer recurrence in GSE81423 (AUC = 0.90). B) Risk score distribution plot in GSE81423.
Supplemental Figure S3. Prediction of recurrence in training and validation cohorts without the transcriptomic panel. A) ROC curve for combined clinical risk factors for recurrence (CA19-9, CEA, synchronous, and tumor number) without the transcriptomic panel in the training cohort (AUC = 0.68). B) ROC curve for the combined clinical risk factors for recurrence without the panel in the validation cohort (AUC = 0.68).


