Abstract
Cutaneous T cell lymphoma (CTCL) is characterized by a background of chronic inflammation, where malignant CTCL cells escape immune surveillance. To study how miRNAs (miRs) regulate T cell exhaustion, we performed miRseq analysis, qRT-PCR, and in situ hybridization on 45 primary CTCL samples, 3 healthy skin samples, and CTCL cell lines, identifying miR-155-5p, 130b-3p and -21-3p. Moreover, miR-155-5p, -130b-3p, and -21-3p positively correlated with immune checkpoint gene expression in lesional skin samples and were enriched in the IL6/JAK/STAT signaling pathway by gene set enrichment analysis. Further gene sequencing analysis demonstrated decreased mRNA expression of the major negative regulators of JAK/STAT signaling, SOCS, PIAS and PTPN. Transfection of MyLa and HuT78 cells with anti-miR-155-5p, -21-3p, and -130b revealed a considerable increase in SOCS proteins along with a significant decrease in the levels of activated STAT3 and IC surface protein expression, as well as decreased cell proliferation. Downregulation of miR-155, -130 and -21 in CTCL cell lines decreased CTCL cell growth and facilitated CD8+ T cell-mediated cytotoxic activity, with concordant production of IFNγ and CD107a expression. Our results describe the mechanisms of miR-induced T cell exhaustion, which provide a foundation for developing synthetic anti-miRs to therapeutically target the tumor microenvironment in CTCL.
Keywords: Cutaneous T-cell lymphoma, T cell exhaustion, miRNA, immune checkpoints, STAT signaling
Introduction
Cutaneous T-cell lymphomas (CTCL) constitute a heterogeneous group of non-Hodgkin lymphomas that derive from clonally expanded effector/central CD4+ T cells in a background of chronic inflammation. (Rubio Gonzalez et al., 2016). It is assumed that the underlying chronic inflammation sensitizes the CTCL cells to proliferate abnormally in response to stimulatory signals and predisposes them to genetic instability (Vermeer et al., 2001). Together, the immune cells that are thought to transmit these signals (e.g., tumor associated macrophages [TAMs]), the cytotoxic immune cells that become suppressed because of these signals (e.g., CD8+ T cells), and the malignant CD4+ T cells that receive the signals and proliferate abnormally collectively compose the tumor microenvironment. These signaling interactions within the tumor microenvironment create a permissive environment for CTCL growth and, in response to oncogenic signals from proliferating tumor cells, the tumor microenvironment changes during disease progression (Criscione and Weinstock, 2007). Immune checkpoints (ICs) play critical roles in immune exhaustion and disease progression. We have demonstrated that CTCL cells display a phenotypic and functional exhausted profile because of continuous tumor antigen overload (Querfeld et al., 2018).
One potential mechanism by which IC protein synthesis can be modulated is by the induction of miRNAs (miRs) (Omar et al., 2019). MiRs, negative regulators of mRNA, contribute to the pathogenesis and progression of mycosis fungoides/Sézary syndrome (MF/SS) (Ralfkiaer et al., 2011, Ralfkiaer et al., 2014). Diagnostic profiling indicated that miRs have high diagnostic potential in CTCL, with upregulated miR-155-5p and downregulation of miR-203 and miR-205 discriminating from benign inflammatory skin disorders (Ralfkiaer et al., 2014). Notably, enhanced expression of miR-155 has been demonstrated in CTCL and has been linked to the constitutive activation of the JAK/STAT, NF-kB, and PI3K/AKT survival pathways in vitro (Seto et al., 2018).
To date, no experimental studies have investigated the mechanisms of miR-directed T-cell exhaustion in CTCL. In this study, we hypothesized that miRs regulate IC receptor expression and enhance T cell exhaustion via STAT signaling, contributing to the pathogenesis of CTCL.
Results
miR expression profiles in CTCL patients
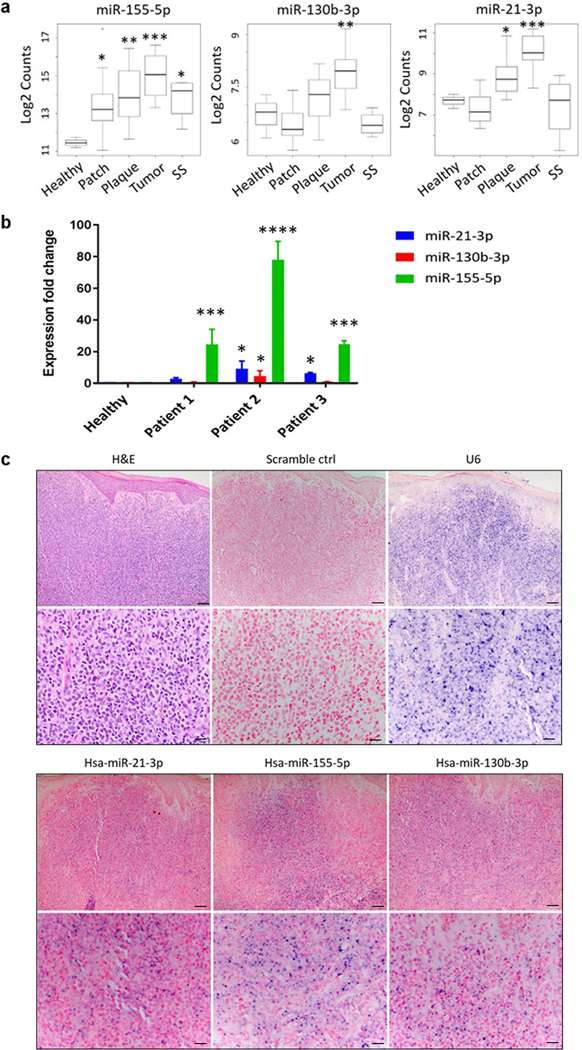
We performed miRNAseq (miRseq) analysis on 45 skin samples from CTCL patients and 3 healthy controls with Illumina Hiseq2500. Statistical analysis showed that expression of miR-155-5p, -130b-3p and -21-3p (abbreviated as miR-155, miR-130 and miR-21 herein) correlated with T-stage (patch, plaque, tumor) and was higher in advanced/tumor (T3) CTCL stages compared to healthy controls (Figure 1A). miR-155 was the only miR of those tested that was significantly increased in SS (T4) compared with normal skin tissue samples (p < 0.05). To further validate the miR expression, we performed qRT-PCR analysis using lesional CTCL skin samples and healthy controls (Figure 1B). We found significantly increased expression of miR-155, -130, and -21 in CTCL compared with healthy controls. In addition, in situ hybridization (ISH) was used to identify the location of miRs in lesional skin of CTCL. The results revealed that all three miRs were mainly expressed in dermal tumor infiltrates, although the intensity of in situ expression varied among the 3 miRs (Figure 1C). Representative hematoxylin and eosin (H&E) images highlight a predominant dermal lymphoid infiltrate composed of sheets of atypical lymphocytes. Furthermore, single cell RNAseq combined with TCR seq analysis from skin explants originating from same skin lesion provided evidence that most cells were consistent with clonal tumor cells (highlighted in blue), with small numbers of other immune cell subsets in grey color (Supplemental Figure 1A).
Figure 1.
Analysis of miR-155, -130, and -21 expression in lesional skin samples from CTCL patients. A. Expression levels of miR- 155, miR- 130, and - 21 in normal tissue, early or patch/plaque (T1, T2), tumor (T3), and erythrodermic (T4) MF/SS stages (n = 48). p- values were measured by Mann–Whitney independent t test. *p < 0.05, **p < 0.01, ***p < 0.001. B. qRT-PCR measurements of miR- 155, - 130, and - 21 expression in human lesional skin from 3 patients with CTCL were normalized to the relative expression in healthy T-cells (U6 was used as endogenous controls to normalize the miRNAs expression in CTCL tissues) and presented as mean ± SD. *p < 0.05, ***p < 0.001, ****p < 0.0001 by two-tailed Student’s t test. C. Representative H&E images and in situ hybridization images of miR- 155, miR- 130, and - 21 or scrambled (control) probes in sections of CTCL tumor lesion. The pictures were taken using optical microscope and imaging system. Upper image, scale bar = 100 μm; lower image, scale bar = 20 μm.
ICs positively correlate with miR-155, -130 and -21 in CTCL
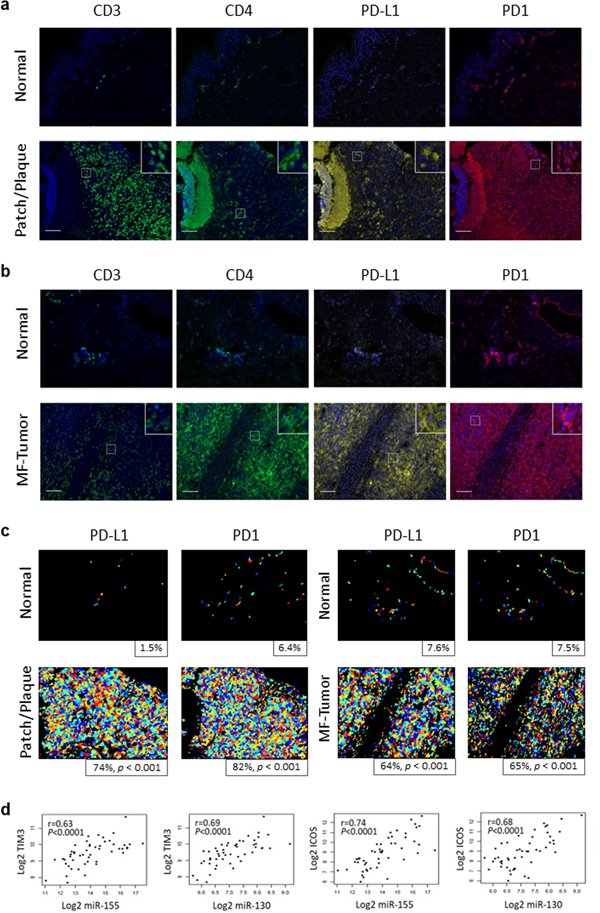
Multicolor immunohistochemistry of a representative plaque sample (T2) and a tumor lesion (T3) demonstrated overexpression of PD-L1 and PD1 compared with healthy control (Figure 2A, B, C). The Pearson’s correlation coefficient revealed a significant difference of PD1 and PD-L1 expression between plaque and tumor lesions compared to control (p <0,001 for PD1; p < 0,001 for PD-L1). Furthermore, the Pearson’s correlation coefficient between miR-155, miR-130, and miR-21 log2 expression level and the log2 expression value of various IC genes revealed that increased miR-155 expression correlated highly with CTLA4 (r = 0.59, p < 0.0001), PD1 (r = 0.42, p = 0.0021), PD-L1 (r = 0.42, p = 0.0025), TIM3 (r = 0.63, p < 0.0001), LAG3 (r = 0.57, p < 0.0001), and ICOS (r = 0.74, p < 0.0001) mRNA; increased miR-21 expression correlated highly with PD-L1 (r = 0.44, p = 0.0012), TIM3 (r = 0.52, p < 0.0001), and ICOS (r = 0.37, p < 0.0073) mRNA; and increased miR-130 expression correlated highly with CTLA4 (r = 0.5, p = 0.0002), PD-L1 (r = 0.54, p < 0.0001), TIM3 (r = 0.69, p < 0.0001), LAG3 (r = 0.49, p = 0.0003), and ICOS (r = 0.68, p < 0.0001) mRNA (Table 1 and Figure 2D). The miR profile of CTCL correlated with an exhausted T cell immunophenotype as summarized in Table 1.
Figure 2.
IC protein expression and the correlation between IC gene and miR-155, -130, -21 expression in CTCL. A-B. Immunophenotypic features of CTCL patch/plaque or tumor, with multiplex immunohistochemistry for IC expression. CD3, CD4, PD1, and PD-L1 expression on skin sections of CTCL lesion from a representative patient with patch/plaques (A) or tumor (B) are shown. Scale bar = 100 μm. C. Automated scoring of PD-L1 and PD1 staining for patch/plaque and tumor stage lesions and matched healthy controls. Positive cells are displayed in varying colors, and percentages of positive cells are shown. P-values are for Pearson’s chi-squared test of equal proportions between controls and patient samples. D. Pearson correlation scatter plots of miR-155, -130, -21 levels and PD1, PD-L1, CTLA-4, LAG3, TIM3, ICOS gene expression. p values were two-sided, and the statistical significance threshold was set at p < 0.05.
Table 1.
The miR profile of CTCL correlates with an exhausted immunophenotype.
| miRs | Log FC | P value | FDR | PD1 cor | PDL 1 cor | CTLA 4 cor | LAG3 cor | TIM3 cor | ICOS cor |
|---|---|---|---|---|---|---|---|---|---|
| hsa-miR-181a-3p | 2.44 | 8.68E-24 | 1.67E-20 | 0.37 | 0.36 | 0.49 | 0.4 | 0.53 | 0.6 |
| hsa-miR-142–3p | 2.34 | 7.53E-23 | 7.24E-20 | 0.3 | 0.47 | 0.38 | 0.41 | 0.53 | 0.54 |
| hsa-miR-21-3p | 2.14 | 3.35E-18 | 1.61E-15 | 0.24 | 0.46 | 0.27 | 0.27 | 0.53 | 0.39 |
| hsa-miR-9–5p | 2.14 | 6.31E-16 | 2.42E-13 | 0.41 | 0.21 | 0.38 | 0.37 | 0.36 | 0.36 |
| hsa-miR-625–3p | 1.95 | 1.62E-15 | 5.19E-13 | 0.43 | 0.4 | 0.58 | 0.6 | 0.59 | 0.69 |
| hsa-miR-21–5p | 1.44 | 6.70E-15 | 1.29E-12 | 0.24 | 0.52 | 0.26 | 0.34 | 0.6 | 0.33 |
| hsa-miR-130b-3p | 1.28 | 1.76E-14 | 2.70E-12 | 0.26 | 0.55 | 0.5 | 0.48 | 0.7 | 0.69 |
| hsa-miR-181b-5p | 1.52 | 2.65E-13 | 3.18E-11 | 0.41 | 0.27 | 0.49 | 0.39 | 0.43 | 0.65 |
| hsa-miR-181a-5p | 1.42 | 5.87E-13 | 5.93E-11 | 0.4 | 0.25 | 0.44 | 0.33 | 0.43 | 0.61 |
| hsa-miR-142–5p | 1.68 | 1.91E-12 | 1.67E-10 | 0.4 | 0.41 | 0.48 | 0.46 | 0.54 | 0.61 |
| hsa-miR-1246 | 2.1 | 2.25E-11 | 1.66E-09 | 0.3 | 0.4 | 0.27 | 0.45 | 0.47 | 0.13 |
| hsa-miR-146a-3p | 1.82 | 4.31E-10 | 2.12E-08 | 0.45 | 0.3 | 0.64 | 0.33 | 0.46 | 0.66 |
| hsa-miR-625–5p | 1.34 | 4.69E-10 | 2.25E-08 | 0.49 | 0.25 | 0.46 | 0.58 | 0.54 | 0.63 |
| hsa-miR-146a-5p | 1.57 | 1.95E-09 | 7.19E-08 | 0.53 | 0.24 | 0.68 | 0.39 | 0.38 | 0.76 |
| hsa-miR-766–3p | 1.02 | 3.85E-08 | 9.47E-07 | 0.13 | 0.41 | 0.37 | 0.51 | 0.45 | 0.54 |
| hsa-miR-363–3p | 1.19 | 3.69E-06 | 4.98E-05 | 0.41 | 0.13 | 0.35 | 0.53 | 0.49 | 0.33 |
| hsa-miR-20b-5p | 1.34 | 5.57E-06 | 7.18E-05 | 0.33 | 0.16 | 0.35 | 0.5 | 0.44 | 0.32 |
| hsa-miR-155-5p | 1.56 | 8.79E-06 | 0.0001068 | 0.43 | 0.42 | 0.58 | 0.55 | 0.63 | 0.74 |
| hsa-miR-150–3p | 1.29 | 3.18E-05 | 0.0003412 | 0.68 | 0 | 0.55 | 0.47 | 0.28 | 0.6 |
FDR: false discovery rate; miR: miRNA; Log FC: Log fold change
p value < 0.01 and fold-change > 1.5.
The STAT signaling pathway is activated in CTCL and is inhibited by an oligonucleotide inhibitor of miR- 155
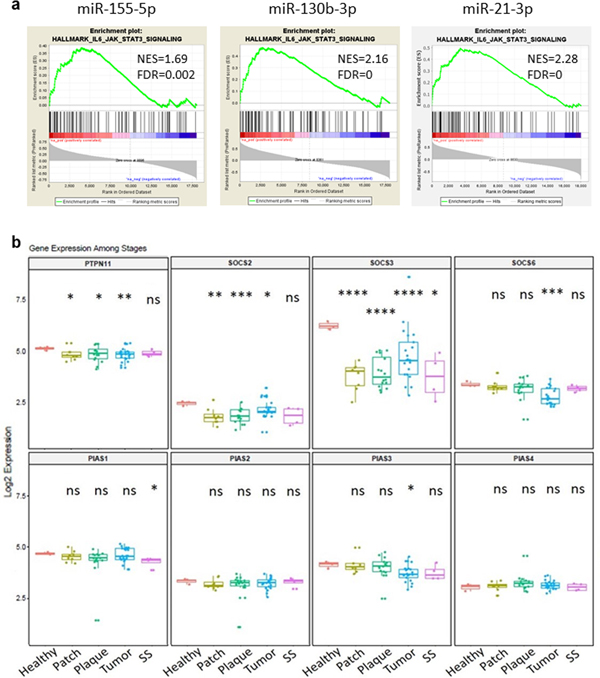
We investigated the potential signaling pathways that are associated with the increased miR-155, -130 and -21 expression profile using gene set enrichment analysis (GSEA). Our data demonstrate that the highly upregulated expression of miR-155, -130, or -21 is significantly associated with increased JAK/STAT3 signaling (Figure 3A). Furthermore, we analyzed gene expression of three major negative regulators of JAK/STAT signaling, SOCS, PIAS and PTPN. Gene expression levels of SOCS2, SOCS3, SOCS6, PIAS1, PIAS3 and PTPN11 were decreased, while there was no significant change in PIAS2 and PIAS4 expression levels in lesional CTCL samples compared to healthy controls (Figure 3B), suggesting that expression of SOCS and to a lesser extent PIAS regulators is critical in the control of STAT3 signaling in CTCL.
Figure 3.
Enrichment of miR-155, -130 and -21 in IL6/JAK/STAT3 signaling pathway and the expression of JAK/STAT signaling negative regulators. A. Enrichment plots from gene set enrichment analysis (GSEA). B. Expression level of the JAK/STAT signaling negative regulators: SOCSs, PIASs and PTPNs at different stage. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Each T- stage compared to normal control by two-tailed Student’s t test.
Cobomarsen (MRG- 106), an oligonucleotide inhibitor of miR- 155, has been evaluated in a clinical phase 1/2 trial for CTCL (Querfeld et al., 2018). We used lesional skin samples from 6 CTCL patients (baseline and after 4 weeks of treatment with cobomarsen [Supplemental Table 1]) to identify the critical pathways affected by miR- 155 inhibition in CTCL. As expected, miR-155 was the most strongly downregulated miR in the treated sample cohort when compared to baseline (Supplemental Table 2). Using the criteria of RPKM value ≥1, P value ≤0.05, and fold-change ≤ 1.5, we identified 498 up- and 370 down-regulated genes in treated vs baseline samples (Supplemental Figure 1B, Supplemental Table 3). Hierarchical clustering of all DE genes shows that treated and baseline samples are distinct except for one patient, which could be possibly explained by an overall low number of malignant T cells within the dermal inflammatory infiltrate (Supplemental Figure 1C). A multi-dimensional scaling (MDS) plot with differentially expressed (DE) genes shows a clear separation between treated and baseline samples (Supplemental Figure 1D). Furthermore, hallmark pathway analysis using GSEA identified several pathways that were significantly inhibited in treated samples vs baseline, including JAK/STAT signaling pathways (p = 0.0044, FDR= 0.019, Supplemental Table 4).
Overexpression of miR-155, -130 and -21 and T cell exhaustion signatures in CTCL cell lines
RNA seq analysis revealed the expression levels of miR-155, -130, and -21 in CTCL cell lines (H9, HH, HuT78, MyLa) and healthy control (Supplemental Figure 2A). The results highlight a distinct expression profile for miR-155, -130, and -21 in HH and MyLa (derived from skin-resident T cells in MF tumor), compared to HuT78 and H9 (derived from circulating central memory T cells in SS), indicating the biologic differences of the CTCL cell lines. qRT-PCR validation revealed that miR-155, -130 and -21 were upregulated in CTCL cell lines, but the expression level for the specific miRs varied among cell lines and could be explained based on the biologic differences of cell origin. We found that the miR expression of the MF tumor-derived CTCL cell lines MyLa and HH, and the SS-derived cell lines HuT78 and H9 are similar, respectively (Supplemental Figure 2B). Ranking of miR-155, -130, -21 expression in the CTCL cell lines is listed in Supplemental Table 5. Notably, ISH was used to validate the expression of miR-155, -130 and -21 in a CTCL cell line (HuT78 cells). The data showed that HuT78 cells express all three miRs (Supplemental Figure 2C). Analysis of RNA-seq data reveals the expression profile of STATs and the negative regulators of STAT pathway in CTCL cell lines compared with CD4+ T cells from a healthy donor (Supplemental Figure 2D). Although some differences in expression levels of the negative regulators of JAK/STAT pathway between the cell line clusters exist, they are generally downregulated. The JAK/STAT genes vary between clusters; e.g. STAT3 is higher in HuT78/H9, while STAT5 and STAT6 is higher in the MyLa/HH cell line cluster. STAT4 is only up-regulated in healthy donor CD4+ T cells. Using RT-PCR analyses, we evaluated the level of IC expression in CTCL cell lines. The ranking of the ICs expression in CTCL cell lines is listed in Supplemental Table 6.
MiR-155, -130 or -21 regulate IC expression in MyLa and HuT78 cells via the JAK/STAT signaling pathway
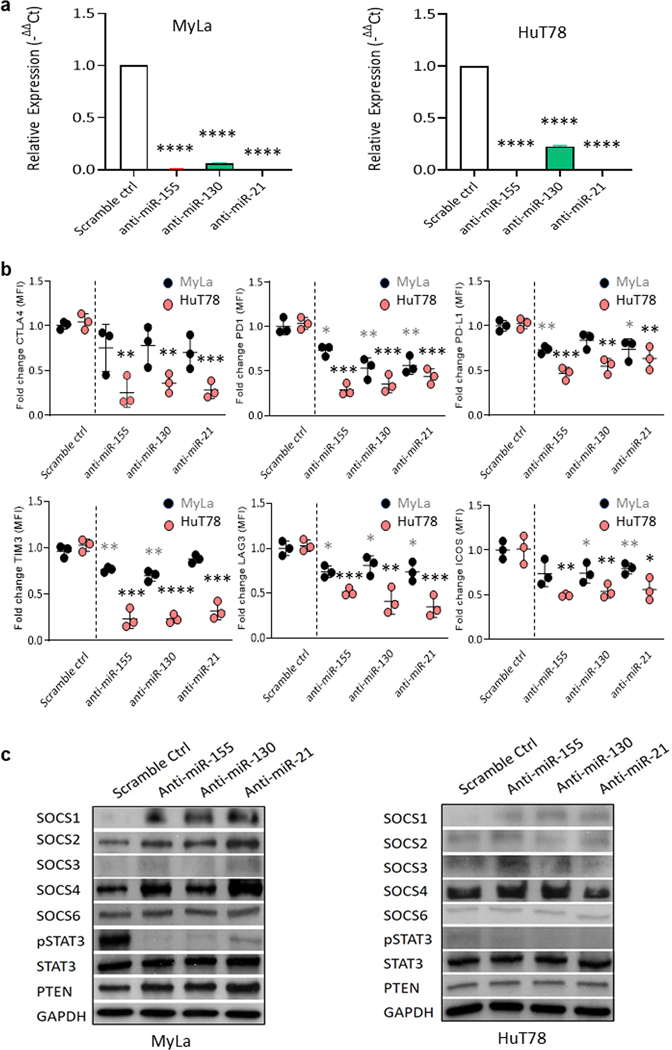
To further investigate the role of miR-155, -130 and -21 in CTCL, we transfected MyLa or Hut78 cells with anti-miR-155, -130, or -21, or scrambled control (scr). First, RT-PCR was performed to test the electroporation efficiency in MyLa and HuT78 cells (Figure 4A). We noticed that antimiR-155, -130 and -21 were significantly decreased compared to scr. After transfection, flow cytometry analysis revealed that miR-155, -130 and -21 miRs significantly decreased the IC protein surface expression of PD1 (p < 0.05, p < 0.01, p < 0.01), PD-L1 (p < 0.01, ns, p < 0.05), TIM3 (p < 0.01, p < 0.01, ns), LAG3 (p < 0.05, p < 0.05, p < 0.05), and ICOS (ns, p < 0.05, p < 0.01) in MyLa cells, and the effects were much stronger in HuT78 cells with significantly decreased expression of the extended IC panel including CTLA4 (p < 0.01, p < 0.01, p < 0.001), PD1 (p < 0.001, p < 0.001, p < 0.001), PD-L1 (p < 0.001, p < 0.001, p < 0.01), TIM3 (p < 0.001, p < 0.001, p < 0.001), LAG3 (p < 0.001, p < 0.01, p < 0.001), and ICOS (p < 0.001, p < 0.001, p < 0.05) (Figure 4B) after 72 h of transfection. The gating strategy is shown in Supplemental Figure 3A. An immunoblot assay revealed a dramatic decrease in pSTAT3 levels with a concomitant increase in SOCS1, SOCS2, and SOCS4 protein expression in MyLa, and an increase in SOCS1 expression in HuT78 cells transfected with anti-miR-155, -130 or -21, compared to cells transfected with scr (Figure 4C). There was no change in expression of PTEN (Figure 4C) or pNF-kB (Supplemental Figure 3B).
Figure 4.
Anti-miR-155, -130, and -21 reduced IC expression in MyLa and HuT78 cells. A. The relative expression levels of miRs (mean ± SD) in MyLa or HuT78 cells were analyzed by qRT-PCR, following 72 h of treatment with anti-miRs. B. ICs expression was analyzed by flow cytometry analysis in MyLa or HuT78 cells following anti-miR treatment. C. Western blot analysis was performed after MyLa or HuT78 cells were treated with anti-miR-155, -130, or -21 or scr for 48 h. Data are expressed as mean ± SEM, n = 3. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by two-tailed Student’s t test.
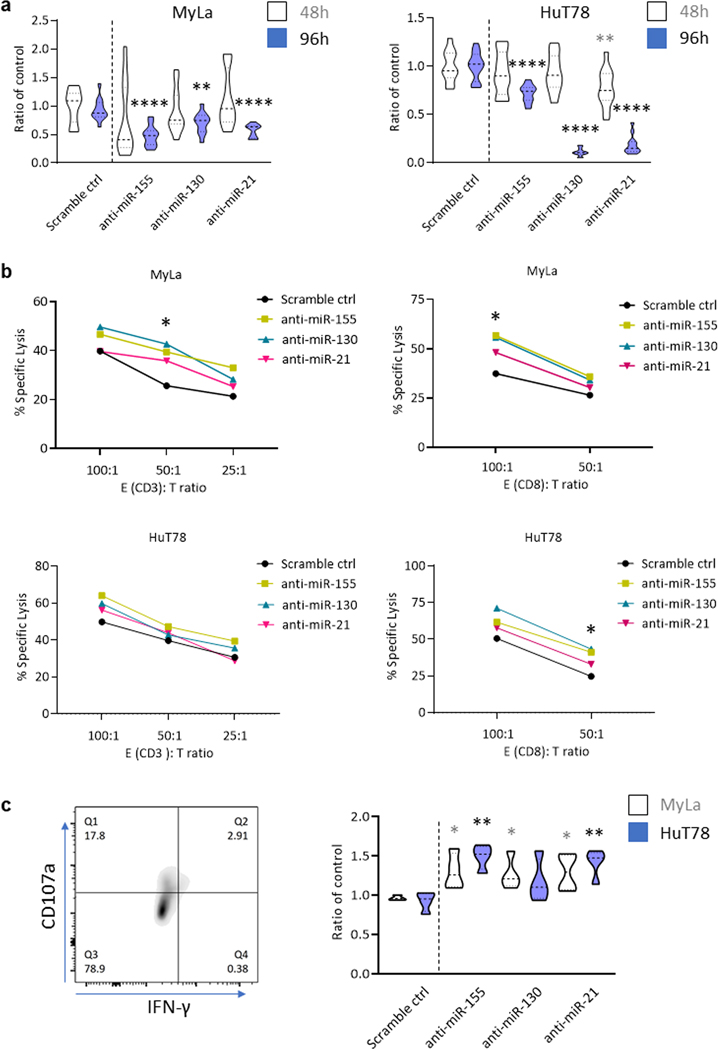
Inhibition of miR-155, -130 or -21 in CTCL cell lines decreases cell growth and promotes susceptibility to cytotoxic T-cell-mediated cytolysis
To investigate the biological role of miR-155, -130, and -21 in CTCL growth, we transfected MyLa and HuT78 cells with the corresponding anti-miRs or with scramble control. An MTT assay was utilized to assess cell growth. MyLa cells transfected with anti-miR-155, -130, or -21 showed significantly reduced cell growth compared to the control group at 96h after transfection (p < 0.0001, p < 0.01, p < 0.0001). HuT78 cells transfected with anti-miR-21 showed significantly reduced cell viability at 48h compared with scr control (Figure 5A, p < 0.01), and cell viability was significantly reduced at 96h after transfection with anti-miR-155, -130, and -21 (p < 0.0001, p < 0.0001, p < 0.0001), respectively. To evaluate the functional importance of MyLa and HuT78 cells transfected with anti-miR-155, -130, or -21, we measured T cell-induced cytolytic capacity using a 51Chromium (Cr)-release assay and cocultured effector cytotoxic CD3+ and CD8+ T cells derived from healthy donors at different ratios with the 51Cr-labelled target CTCL cell lines MyLa or Hut78 at an effector (E): target cell (T) ratio of 100:1, 50:1, or 25:1, respectively. After 24h, the percent of CD3+ or CD8+ T cell-induced cytolysis was generally higher in anti-miR-transfected MyLa and HuT78 cells compared to scr, but only reached statistical significance for anti-miR-155 and anti-miR-130-treated MyLa cells at E:T ratio of 50:1 (p < 0.05). The percent of CD8+ T cell-induced cytolysis was significantly higher for anti-miR-155 and anti-miR-130-treated MyLa cells compared to scr at E:T ratio = 100:1 (p < 0.05, p < 0.05), and for anti-miR-155 and anti-miR-130-treated HuT78 cells than the scr at E:T ratio = 50:1 (p < 0.05, p < 0.05) (Figure 5B). Significant killing of MyLa or HuT78 cells was not observed after 12h co-culture with either CD3+ or CD8+ T cells (Supplemental Figure 3C). To further validate our assay, we co-cultured CD8+ T cells with anti-miR-155, -130, or -21 treated MyLa or HuT78 cells for 48h at E:T ratio =100:1 and then measured by CD107a cell surface expression and IFN-γ production. There was a significant upregulation in CD107a expression and IFN-γ production in CD8+ T cells co-cultured with anti-miR-155, -130, and -21 treated MyLa (p<0.05, p<0.05, p<0.05), and HuT78 cells (p<0.01, ns, p<0.01) (Figure 5C).
Figure 5.
IC expression changes of MyLa and HuT78 cells are associated with decreased cell growth and enhanced cytotoxic T cell-mediated immune responses. A. MyLa or HuT78 cells treated with anti-miR-155, -130, or -21 for 48 h or 96 h were analyzed using MTT assay. Absorbance of cellular metabolic activity was measured at 590 nm and normalized to scramble control. **p < 0.01, ****p < 0.0001 by two-tailed Student’s t test. B. Anti-miRs or scramble control transfected CTCL cell lines were co-cultured with human effector cells for 24 hours at various E:T ratios. The % specific lysis of the target MyLa and HuT78 cells were plotted against multiple E:T ratios. C. CD8+ T cells, co-cultured with CTCL cells transfected with anti-miRs or scramble control, were analyzed for CD107a cell surface expression and IFN-γ production. *p < 0.05, **p < 0.01 by two-tailed Student’s t test.
Discussion
Widespread immunologic abnormalities and T cell exhaustion is a hallmark of CTCL, and ICs are known to play a key role in tumor immune escape mechanisms (Anzengruber et al., 2019, Rubio Gonzalez et al., 2016). Our previous study demonstrated that the expression of T cell exhaustion genes increases with advanced stages of disease (Querfeld et al., 2018). Here, our experimental approach was used to identify critical miRs that regulate the transcription of ICs in CTCL. Our combined miRNA and mRNA targeting screening revealed 3 critical miRs, miR-155, miR-130, and miR-21, that strongly correlated with multiple IC gene expression levels in CTCL by Pearson’s correlation coefficient analysis. These targets were validated using RT-PCR and ISH analysis in CTCL skin samples and CTCL cell lines, and their functional relevance was identified by proliferation and cytotoxic T cell assays, and in vitro analysis of CTCL cell lines transfected with antagomirs to miR-155, miR-130 and miR-21. miR-155 was previously shown to contribute to CTCL pathogenesis and disease progression (Ralfkiaer et al., 2011, Ralfkiaer et al., 2014). Recent studies also identified that miR-155 and miR-21 correlated with diagnosis and prognosis in MF and SS patients, respectively (Narducci et al., 2011, van der Fits et al., 2011). High expression of miR-155 was also shown in other lymphomas, such as DLBCL, and in solid malignancies. Notably, miR-155, miR-130, and miR-21 not only correlated with IC mRNA expression but were also all upregulated in CTCL samples compared to healthy control and correlated with tumor burden, with higher expression levels in plaques and tumors compared to those in patch and erythrodermic lesions. miR-130 and miR-21 have not been shown to be upregulated in the MF subtype of CTCL. A possible explanation could be differences in patient subgroups and control populations. Accordingly, we performed RT-PCR to further validate miR expression, and we were able to confirm and extend the findings of van Kester et al (van Kester et al., 2011) that miR-130 and miR-21 in addition to miR-155 were significantly differentially expressed in CTCL.
MiRs that modulate immune responses via regulation of IC expression have increasingly received attention (Cortez et al., 2019, Jang et al., 2017, Omar et al., 2019). Studies have shown that the expressions of ICs may directly or indirectly be repressed by miRs in multiple types of cancers (Cristino et al., 2019, Smolle et al., 2017). To our knowledge previously unreported that miRs correlated with IC expression and T cell exhaustion in CTCL. We found that miR-155, miR-130, and miR-21 strongly correlated with multiple IC gene expression levels in CTCL by Pearson’s correlation coefficient analysis. Notably, miR-155 has been linked to the PD1/PD-L1 pathway in other malignancies, but to our surprise they were also correlated with LAG3, TIM3 and ICOS. Our in vitro data show that anti-miR treatment inhibits MyLa and HuT78 cell growth and targets multiple IC mRNA expression via STAT signaling. There are several negative regulators of the JAK/STAT pathway: protein inhibitor of activated STAT (PIAS) (Chung et al., 1997); protein tyrosine phosphatase, non-receptor type (PTPN) (You et al., 1999); and suppressors of cytokine signaling (SOCS) (Hanada et al., 2003, Wormald et al., 2006). Our data showed that pSTAT3 levels decreased while levels of the negative STAT regulator SOCS increased after anti-miR transfection in both MyLa and HuT78 cells, indicating that the regulation of STAT3 by miR-155, -130, and -21 is mediated through an alteration of the expression of SOCS family members in CTCL. However, given the heterogeneity of the 2 different CTCL cell lines demonstrating different STAT gene expression levels, other pathways or tumor-intrinsic factors that play a role in IC regulation cannot be ruled out. (Kalbasi and Ribas, 2020)
The JAK/STAT pathway is known to be constitutively activated in CTCL (Netchiporouk et al., 2014, Olszewska et al., 2020, Wu et al., 2016). Our data showed that miR-155, miR-130, and miR-21 control the expression of checkpoint receptors through STAT3 signaling. STAT3-mediated regulation in PD-L1 and its role in modifying anti-tumor immunity has been recently identified (Song et al., 2018). Our findings provide additional evidence of STAT3-mediated regulation of the co-inhibitory receptors LAG3, TIM3, PD1 and CTLA4, and the co-stimulatory receptor ICOS. Our findings did not support the involvement of PTEN or NF-κB pathways in IC regulation in CTCL. The functional significance of miR-155, -130 and miR-21-mediated immune evasion was demonstrated in vitro: The downregulation of these miRs has facilitated T cell-mediated cytotoxic responses owing to decreased IC expression.
Overall, our study identified miR-155, -130, and -21 as key regulators of T cell exhaustion in CTCL that could form the basis to develop therapeutic targets in combination with immune checkpoint blockade to potentiate antitumor efficacy.
Materials and Methods
Patients and samples
Ethical approval for the study was obtained from City of Hope Comprehensive Cancer Center, Duarte, CA, USA. Skin samples from patients with CTCL attending the cutaneous lymphoma clinic at City of Hope were obtained. Sections of 45 formalin-fixed paraffin-embedded (FFPE) skin biopsies of MF/SS lesions and 3 healthy skin samples identified from City of Hope’s tissue repository were used for miR and mRNA expression analysis (Querfeld et al., 2018). All patients provided written informed consent and were staged according to the revised staging and classification of MF/SS (Olsen et al., 2007). T (skin) stages included T1 (n=7), T2 (n=16), T3 (n=18), and T4 (n=4)). Three FFPE tissue samples of healthy skin from 3 patients undergoing reconstructive plastic surgery served as healthy controls. A separate set of lesional skin sections were obtained from six early stage MF patients treated with anagomiR-155 (MRG 106, cobomarsen) at baseline and 4 weeks into treatment (Supplemental Table 1). Skin stages were T1 (n=3) and T2 (n=3), respectively. Healthy PBMC and CD3+, CD8+ T cells from City of Hope blood donor center.
Flow cytometry staining
MyLa or HuT78 cell pellets were resuspended in FACS buffer (PBS, 5% FBS) and stained with predetermined antibody volumes: anti-CD107a (Cat# 555800, BD Bioscience), anti-IFNγ (Cat# 506510, Biolegend), anti-PD1 (Cat# 560908, BD Biosciences), anti-PD-L1 (Cat# 563741, Biolegend), anti-ICOS (Cat# 564778, Biolegend), anti-CTLA4 (Cat# 561717, BD Biosciences), anti-TIM3 (Cat# 565566, BD Biosciences), anti-LAG3 (Cat# 369318, BD Biosciences) and the fixation/permeabilization solution kit (Cat# 554714, BD Bioscience). Cells were analyzed using a Fortessa flow cytometer (BD Bioscience, San Jose, CA). The FlowJo 10.6.2 software program (Tree Star, Inc.) was used for data analysis.
RNA sequencing and bioinformatics analysis
RNA sequencing was performed by the Integrative Genomics Core with Illumina Hiseq 2500 as previously described (Pal et al., 2015). Reads were aligned to human genome assembly hg19 using Tophat v2. For each sample, expression counts for ensemble genes were summarized by HTseq v0.6. 1, and reads per kilobase of transcript per million mapped reads (RPKM) were calculated. For miRNA sequencing, adapters were trimmed using cutadapt v1.9.1, and miR expression levels were counted based on miRBase v19 mature miR sequences using customized scripts as described previously (Querfeld et al., 2018).
Count normalization and differential expression analysis between groups were conducted using Bioconductor package “edgeR”. Multiple comparisons were adjusted using FDR. Heatmaps were generated using cluster v3. The Gene Ontology and pathway analysis was performed with GSEA v4.
Automated scoring of multiplex immunohistochemistry images
CellProfiler (McQuin et al., 2018) was used to quantify expression of PD-L1 and PD1 in digitized multiplex fluorescent immunohistochemistry images from representative patients with patch/plaque stage and tumor stage CTCL, and from healthy matched controls (Figure 2A, 2B, 2C). Images were de-multiplexed, and built-in object segmentation algorithms were used to detect DAPI-stained nuclei and rhodamine-stained PD-L1/PD1-expressing cells. Algorithm performance was evaluated by two pathologists (DS and CQ). The percentage of positive cells was calculated for CTCL patient samples and controls, and Pearson’s chi-squared test for equal proportions was used to derive a p-value for the null hypothesis of equal proportions.
Statistical analysis
Data analysis was performed using GraphPad Prism (Version 8.0, GraphPad Software, Inc., La Jolla, CA). The Mann–Whitney ranked sum test was used to compare the miR expression between different stages. The Pearson’s correlation coefficient used to test the linear relationship between miR expression and IC levels. The Student’s t test was used unless stated otherwise. Comparisons of > 2 groups used one-way analysis of variance (ANOVA) followed by Dunnett’s or Tukey’s post hoc analysis. The mean ± standard deviation was provided, and at least three experiments were performed. P values were two-sided, and the statistical significance threshold was set at p < 0.05.
Additional methods are discussed within supplemental material and methods.
Supplementary Material
Acknowledgments
Research reported in this publication included work performed in the Integrative Genomics Core, Pathology Core, Center for Informatics and Shared Resources Core supported by the NIH/NCI Cancer Center Support Grant (P30CA033572) to the City of Hope, NIH/NCI grants (P50CA107399, R01CA213131 to M.K.), NIH/NCI grant (R01 CA229510-01) and Leukemia Lymphoma Society Clinical Scholar Award (LLS Grant ID: 2325-19) to C. Querfeld.
Footnotes
Conflict of interest
STR: Speaker’s Bureau: Celgene, Global Education Group and Paradigm Medical Communications, LLC; Abbvie. Consulting: Novartis Pharmaceuticals Corporation, Pepromene Bio, Inc.; Exicure, Apobiologix/Apotex, Inc. MK: Scientific advisor and shareholder of Scopus Biopharma Inc.; CQ: Advisory board: Miragen, Bioniz, Trillium, Kyowa Kirin, Medivir, Helsinn/Actelion, Mallinckrodt; research funding: Celgene.
All other authors have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability Statement
RNASeq data from tissue samples of CTCL patients and healthy controls can be found under [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113113], hosted at [GEO] (Querfeld C, Leung S, Myskowski PL, Curran SA et al. Primary T Cells from Cutaneous T-cell Lymphoma Skin Explants Display an Exhausted Immune Checkpoint Profile. Cancer Immunol Res 2018 Aug;6(8):900–909. PMID: 29895574). MiRNASeq data set from CTCL patients and healthy controls, miRNA and RNASeq data from tissue samples of 6 patients treated with anagomiR-155 (MRG 106, cobomarsen), and the miRNA and RNASeq data from CTCL cell lines (HH, H9, MyLa, and HuT78) and CD4+ T cells of healthy controls are deposited in Gene Expression Omnibus (GEO) data repository and publicly available under GSE181130 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181130].
References
- Anzengruber F, Ignatova D, Schlaepfer T, Chang YT, French LE, Pascolo S, et al. Divergent LAG-3 versus BTLA, TIGIT, and FCRL3 expression in Sezary syndrome. Leukemia & lymphoma 2019;60(8):1899–907. [DOI] [PubMed] [Google Scholar]
- Betts MR, Brenchley JM, Price DA, De Rosa SC, Douek DC, Roederer M, et al. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. Journal of immunological methods 2003;281(1–2):65–78. [DOI] [PubMed] [Google Scholar]
- Chung CD, Liao J, Liu B, Rao X, Jay P, Berta P, et al. Specific inhibition of Stat3 signal transduction by PIAS3. Science (New York, NY) 1997;278(5344):1803–5. [DOI] [PubMed] [Google Scholar]
- Cortez MA, Anfossi S, Ramapriyan R, Menon H, Atalar SC, Aliru M, et al. Role of miRNAs in immune responses and immunotherapy in cancer. Genes, chromosomes & cancer 2019;58(4):244–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscione VD, Weinstock MA. Incidence of cutaneous T-cell lymphoma in the United States, 1973–2002. Arch Dermatol 2007;143(7):854–9. [DOI] [PubMed] [Google Scholar]
- Cristino AS, Nourse J, West RA, Sabdia MB, Law SC, Gunawardana J, et al. EBV microRNA-BHRF1–2-5p targets the 3’UTR of immune checkpoint ligands PD-L1 and PD-L2. Blood 2019;134(25):2261–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Kinjyo I, Inagaki-Ohara K, Yoshimura A. Negative regulation of cytokine signaling by CIS/SOCS family proteins and their roles in inflammatory diseases. Rev Physiol Biochem Pharmacol 2003;149:72–86. [DOI] [PubMed] [Google Scholar]
- Jang HJ, Lee HS, Burt BM, Lee GK, Yoon KA, Park YY, et al. Integrated genomic analysis of recurrence-associated small non-coding RNAs in oesophageal cancer. Gut 2017;66(2):215–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbasi A, Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nature reviews Immunology 2020;20(1):25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW, et al. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol 2018;16(7):e2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narducci MG, Arcelli D, Picchio MC, Lazzeri C, Pagani E, Sampogna F, et al. MicroRNA profiling reveals that miR-21, miR486 and miR-214 are upregulated and involved in cell survival in Sezary syndrome. Cell death & disease 2011;2:e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netchiporouk E, Litvinov IV, Moreau L, Gilbert M, Sasseville D, Duvic M. Deregulation in STAT signaling is important for cutaneous T-cell lymphoma (CTCL) pathogenesis and cancer progression. Cell cycle (Georgetown, Tex) 2014;13(21):3331–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen E, Vonderheid E, Pimpinelli N, Willemze R, Kim Y, Knobler R, et al. Revisions to the staging and classification of mycosis fungoides and Sezary syndrome: a proposal of the International Society for Cutaneous Lymphomas (ISCL) and the cutaneous lymphoma task force of the European Organization of Research and Treatment of Cancer (EORTC). Blood 2007;110(6):1713–22. [DOI] [PubMed] [Google Scholar]
- Olszewska B, Glen J, Zablotna M, Nowicki RJ, Sokolowska-Wojdylo M. The polymorphisms of IL-6/STAT3 signaling pathway may contribute to cutaneous T-cell lymphomas susceptibility. Archives of dermatological research 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omar HA, El-Serafi AT, Hersi F, Arafa EA, Zaher DM, Madkour M, et al. Immunomodulatory MicroRNAs in cancer: targeting immune checkpoints and the tumor microenvironment. FEBS J 2019;286(18):3540–57. [DOI] [PubMed] [Google Scholar]
- Pal SK, He M, Tong T, Wu H, Liu X, Lau C, et al. RNA-seq reveals aurora kinase-driven mTOR pathway activation in patients with sarcomatoid metastatic renal cell carcinoma. Molecular cancer research : MCR 2015;13(1):130–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Wong DJ, Ooi CC, Kurtz DM, Vermesh O, Aalipour A, et al. Molecular profiling of single circulating tumor cells from lung cancer patients. Proceedings of the National Academy of Sciences of the United States of America 2016;113(52):E8379–E86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querfeld C, Foss FM, Kim YH, Pinter-Brown L, William BM, Porcu P, et al. Phase 1 trial of cobomarsen, an inhibitor of Mir-155, in cutaneous T cell lymphoma. Blood 2018;132. [Google Scholar]
- Querfeld C, Leung S, Myskowski PL, Curran SA, Goldman DA, Heller G, et al. Primary T Cells from Cutaneous T-cell Lymphoma Skin Explants Display an Exhausted Immune Checkpoint Profile. Cancer immunology research 2018;6(8):900–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralfkiaer U, Hagedorn PH, Bangsgaard N, Lovendorf MB, Ahler CB, Svensson L, et al. Diagnostic microRNA profiling in cutaneous T-cell lymphoma (CTCL). Blood 2011;118(22):5891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralfkiaer U, Lindahl LM, Litman T, Gjerdrum LM, Ahler CB, Gniadecki R, et al. MicroRNA expression in early mycosis fungoides is distinctly different from atopic dermatitis and advanced cutaneous T-cell lymphoma. Anticancer Res 2014;34(12):7207–17. [PubMed] [Google Scholar]
- Rubio Gonzalez B, Zain J, Rosen ST, Querfeld C. Tumor microenvironment in mycosis fungoides and Sezary syndrome. Current opinion in oncology 2016;28(1):88–96. [DOI] [PubMed] [Google Scholar]
- Rubio V, Stuge TB, Singh N, Betts MR, Weber JS, Roederer M, et al. Ex vivo identification, isolation and analysis of tumor-cytolytic T cells. Nature medicine 2003;9(11):1377–82. [DOI] [PubMed] [Google Scholar]
- Seto AG, Beatty X, Lynch JM, Hermreck M, Tetzlaff M, Duvic M, et al. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br J Haematol 2018;183(3):428–44. [DOI] [PubMed] [Google Scholar]
- Smolle MA, Calin HN, Pichler M, Calin GA. Noncoding RNAs and immune checkpoints-clinical implications as cancer therapeutics. The FEBS journal 2017;284(13):1952–66. [DOI] [PubMed] [Google Scholar]
- Song TL, Nairismagi ML, Laurensia Y, Lim JQ, Tan J, Li ZM, et al. Oncogenic activation of the STAT3 pathway drives PD-L1 expression in natural killer/T-cell lymphoma. Blood 2018;132(11):1146–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits L, van Kester MS, Qin Y, Out-Luiting JJ, Smit F, Zoutman WH, et al. MicroRNA-21 expression in CD4+ T cells is regulated by STAT3 and is pathologically involved in Sezary syndrome. The Journal of investigative dermatology 2011;131(3):762–8. [DOI] [PubMed] [Google Scholar]
- van Kester MS, Ballabio E, Benner MF, Chen XH, Saunders NJ, van der Fits L, et al. miRNA expression profiling of mycosis fungoides. Mol Oncol 2011;5(3):273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer MH, van Doorn R, Dukers D, Bekkenk MW, Meijer CJ, Willemze R. CD8+ T cells in cutaneous T-cell lymphoma: expression of cytotoxic proteins, Fas Ligand, and killing inhibitory receptors and their relationship with clinical behavior. J Clin Oncol 2001;19(23):4322–9. [DOI] [PubMed] [Google Scholar]
- Wormald S, Zhang JG, Krebs DL, Mielke LA, Silver J, Alexander WS, et al. The comparative roles of suppressor of cytokine signaling-1 and -3 in the inhibition and desensitization of cytokine signaling. The Journal of biological chemistry 2006;281(16):11135–43. [DOI] [PubMed] [Google Scholar]
- Wu CS, Wei KL, Chou JL, Lu CK, Hsieh CC, Lin JM, et al. Aberrant JAK/STAT Signaling Suppresses TFF1 and TFF2 through Epigenetic Silencing of GATA6 in Gastric Cancer. International journal of molecular sciences 2016;17(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- You M, Yu DH, Feng GS. Shp-2 tyrosine phosphatase functions as a negative regulator of the interferon-stimulated Jak/STAT pathway. Molecular and cellular biology 1999;19(3):2416–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNASeq data from tissue samples of CTCL patients and healthy controls can be found under [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113113], hosted at [GEO] (Querfeld C, Leung S, Myskowski PL, Curran SA et al. Primary T Cells from Cutaneous T-cell Lymphoma Skin Explants Display an Exhausted Immune Checkpoint Profile. Cancer Immunol Res 2018 Aug;6(8):900–909. PMID: 29895574). MiRNASeq data set from CTCL patients and healthy controls, miRNA and RNASeq data from tissue samples of 6 patients treated with anagomiR-155 (MRG 106, cobomarsen), and the miRNA and RNASeq data from CTCL cell lines (HH, H9, MyLa, and HuT78) and CD4+ T cells of healthy controls are deposited in Gene Expression Omnibus (GEO) data repository and publicly available under GSE181130 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181130].