Abstract
Immunoglobulin G (IgG) monoclonal antibodies are a prominent and expanding class of therapeutics used for the treatment of diverse human disorders. The chemical composition of the N-glycan on the Fc region determines the effector functions through interaction with the Fc gamma receptors and complement proteins. The chemoenzymatic synthesis using endo-β-N-acetylglucosaminidases (ENGases) emerged as a strategy to obtain antibodies with customized glycoforms that modulate their therapeutic activity. We discuss the molecular mechanism by which ENGases recognize different N-glycans and protein substrates, especially those that are specific for IgG antibodies, in order to rationalize the glycoengineering of immunotherapeutic antibodies, which increase the impact on the treatment of myriad diseases.
Graphical abstract
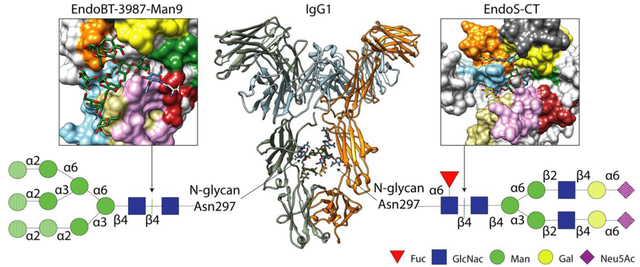
Introduction
All four IgG subclasses (IgG1–4) are glycoproteins that contain a single N-glycosylation site at the constant heavy chain 2 (CH2) domains of the Fc region. Specifically, IgG1 antibodies, by far the most prevalent antibodies in the blood, bear a conserved N-glycosylation site at residue Asn297 on the Fc region (Figure 1A). The presence of this N-glycan is critical for IgG function, contributing to both Fc γ receptor binding and activation of the complement pathway [1] (Figure 1A). Aside from IgG antibodies, other antibodies found in humans,IgA [2,3], IgM [4]and IgE [5,6], also contains N-glycosylation sites that are relevant for their function
Figure 1. IgG glycosylation and glycoengineering of IgG antibodies using ENGases and glycosynthase mutants.
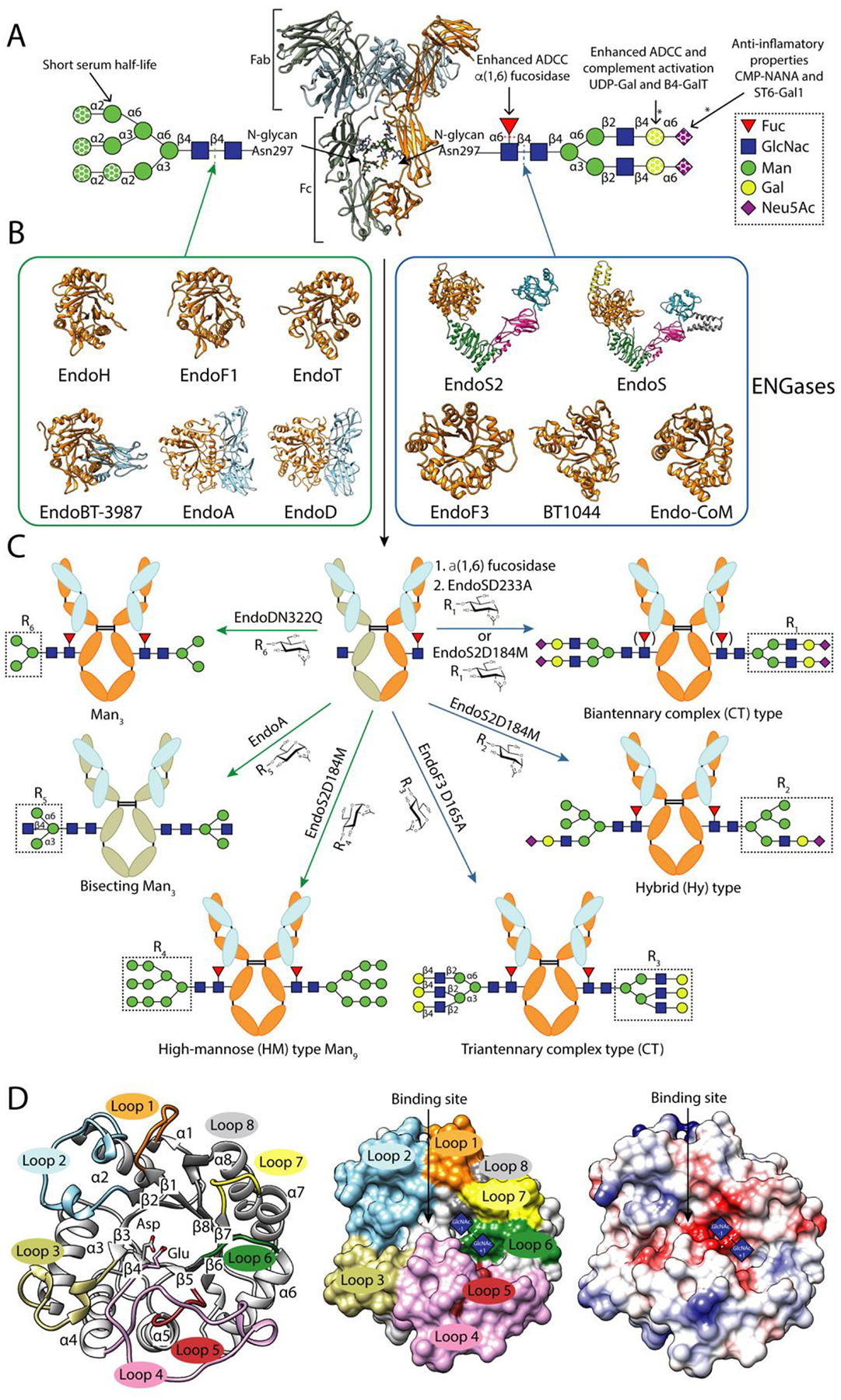
A. Cartoon representation of the overall structure of a human IgG antibody (PDB code 1HZH). Human IgG antibodies are composed of two identical heavy chains (HC) and two identical light chains (LC) organized in three globular domain structures: two identical antigen-binding fragments (Fab) and the crystallizable fragment (Fc) region. Main N-glycans found at Asn297 of Fc region in recombinant and biological IgG antibodies. In addition, 15–25% of the human serum IgG antibodies carry a second N-glycan in the LC of the Fab region [65]. This N-glycan is also highly variable, comprising over 37 different glycoforms, affecting the immunoreactivity, antigen affinity and half-life of the antibody [66]. All eukaryotic N-glycans share a common core sequence, Manα1–3(Manα1–6)Manβ1–4GlcNAcβ1–4GlcNAcβ1–Asn-X-Ser/Thr, and are classified into three major types according to the composition and structural arrangement of the α(1,3) and α(1,6) antennae: complex-type (CT-type), high-mannose-type (HM-type), and hybrid type (Hy-type) [67]. The structural repertoire of N-glycans is further expanded by (i) additional antennae (e.g., bisecting, tri-antennary and tetra-antennary CT-type N-glycans), (ii) the addition of sugars to the N-glycan core (e.g., α(1,6)-Fuc to the Asn-linked GlcNAc), and (iii) the elongation of antennae (e.g., the addition of α(2,6)-Neu5Ac to the terminal Gal in CT-type N-glycans)IgGs from human serum are composed of up to 33 different glycoforms, the most predominant N-glycan being the fucosylated biantennary CT-type [68]. Fc N-glycan structural heterogeneity markedly impacts the functionality, immunogenicity and pharmacokinetics of the IgG antibodies [69]. The dotted shapes represent that carbohydrate may or may not be present, indicating the high structural variability of the N-glycan. The lack of α(1,6)-Fuc increases the binding of IgGs to FcγRIIIa, enhancing the antibody-dependent cellular cytotoxicity (ADCC) [16,17], and in some cases improving their antitumoral activity. The addition of terminal α(2,6)-Neu5Ac to the Fc region of IgGs is associated with increased anti-inflammatory properties [70]. The presence of Gal residues is important for IgGs to interact with C1q and activate the complement pathway [71]. IgG antibodies bearing HM-type N-glycans have a shorter half-life in humans than antibodies with other glycoforms, increasing serum clearance [72].The reactions catalysed by glycosyl hydrolyses and glycosyltransferases in order to modify the N-glycan composition are marked by dotted lines and asterisks, respectively. B. Characterized ENGases from the GH18 and GH85 family with known crystal structure classified in two groups according to the N-glycans they can hydrolyze: HM-type N-glycans (green square) and CT-type N-glycans (blue square). C. In vitro chemoenzymatic remodeling of IgG antibodies. After hydrolysis of heterogenous IgG antibody glycoforms by ENGases, glycosynthase mutants can transfer a defined glycan from an oxazoline donor to the remaining GlcNAc attached to Asn297, according to the enzyme N-glycan specificity. In addition, AlfC, an α-fucosidase that only hydrolyze fucose from single GlcNAcs attached to Asn297, can first be used to remove this carbohydrate moiety that reduces the binding to FcγRIIIa. The HCs of the IgG substrates bear HM-type N-glycans (grey) or CT-type N-glycans (orange). The LC is highlighted in light blue. D. Cartoon (left), surface (center) and coulombic surface (right) representations of the X-ray crystal structure of EndoF3 in its unliganded (PDB code 1EOK), showing the overall structure of the glycosyl hydrolase domain of an ENGase. The β-strands and the α-helices that formed the (β/α)8 TIM-barrel and the loops surrounding the binding site that defined the N-glycan specificity of this enzymes are annotated. Members of both families follow a conserved substrate-assisted mechanism with retention of the anomeric configuration. In a first step, a glutamic acid acts as an acid and protonates the anomeric carbon and an aspartic acid (GH18 family) or asparagine (GH85 family) stabilizes the reaction intermediate and orients the oxygen of the 2-acetamide groupo of GlcNAc (−1) that attacks the anomeric carbon and forms an oxazolinium intermediate. In a second step, the glutamic acid acts as a base and deprotonates a water molecular provoking the second nucleophilic attack and the hydrolysis of the oxazolinium intermediate. The catalytic residues, aspartic and glutamic acids, are highlighted and the sugar binding subsites of GlcNAcs (+1) and (−1) are represented by blue squares.
IgG antibodies have emerged as one of the most powerful and effective classes of drugs to treat a wide variety of human disorders, including different types of cancer, autoimmunity, and infectious diseases. Antibody-based therapeutics come in many forms, including monoclonal antibodies [7], bispecific antibodies, antibody-drug conjugates [8] and antibody fragments [9]; those used clinically are predominantly monoclonal antibodies [10]. The N-glycan structure and heterogeneity of therapeutic IgG monoclonal antibodies have a remarkable impact on their efficacies and effector functions (Figure 1A). Several approaches have been developed to obtain homogenous glycoforms on antibodies, mostly by engineering the N-glycoprotein biosynthetic pathway in mammalian cell lines, with some antibodies produced by such methods having been recently approved for clinical use [11,12] (Figure 1).
Chemoenzymatic synthesis of glycoengineered IgG antibodies
Chemoenzymatic glycan remodeling represents an alternative strategy not only to understand the role of individual glycoforms on the effector functions, but also to generate high yields of homogeneously glycosylated IgG antibodies. This approach has the advantage of producing certain glycoforms that cannot be generated using cell lines with modified biosynthetic pathways; it also produces completely homogeneous glycoproteins, which is not achievable using cell-based methods. One chemoenzymatic synthesis method is based on the use of exoglycosidases that trim the N-glycan and/or glycosyltransferases that add monosaccharide residues to extend the N-glycan, which has been used to generate IgG antibodies with enriched glycoforms of fully galactosylated [13] and sialylated [14,15] N-glycans (Figure 1A).
The absence of core α(1,6)-fucose significantly increases antibody-dependent cellular cytotoxicity (ADCC) and, for some antibodies, therapeutic potency [16,17]. There are currently three FDA-approved afucosylated therapeutic antibodies – obinutuzumab, mogamulizumab and benralizumab [18]. AlfC, from Lactobacillus casei, is an α-L-fucosidase (EC 3.2.1.51) active on a wide range of linkages, and belongs to the 29A subfamily of glycosyl hydrolase (GH), according to the sequence-based Carbohydrate Active enZYme database (CAZy, www.cazy.org [19,20]). AlfC is approximately 10,000-fold more active on α(1,6) glycosidic bonds compared to other linkages [21,22]. This α(1,6) linkage is primarily found between the core fucose for glycans, attached to the GlcNAc adjacent to the protein. Structural and mechanistic studies of AlfC [23] indicate that it hydrolyzes fucose by a classical Koshland double-displacement reaction [24], that the active site accommodates the GlcNAc, suggesting that its enzymatic activity is mediated by distinct open and closed conformations of an active site loop.
AlfC, however, is only able to hydrolyze fucose from GlcNAc-Fuc disaccharides (Figure 1A,B). Indeed, there are currently no fucosidases known to be capable of removing fucose from fully branched glycans on antibodies. Intriguingly, transfucosylation variants of AlfC transfer fucose onto the (+1) GlcNAc of an antibody bearing a fully branched glycan, suggesting that such mutations shift the conformational equilibrium of the AlfC active site towards an open state that favors transfucosylation over hydrolysis [21,23].
The use of exoglycosidases and glycosyltransferases to remodel N-glycans is dependent on the glycan chemistry of the antibody substrate and the efficiency and sugar specificity of these enzymes. On the contrary, the chemoenzymatic synthesis approach based on the use of endoglycosidases with endo-β-N-acetylglucosaminidase (ENGases; EC 3.2.1.96) activity has aroused great interest in recent years for glycan remodeling of proteins, including antibodies. ENGases catalyze the hydrolysis of the β-1,4 linkage between the first two N-acetylglucosamine residues in the core of N-linked glycans [11] (Figure 1), and are classified into two GH families, GH18 and GH85. Although they share a common hydrolytic and glycosynthase catalytic mechanisms [25], they exhibit distinct molecular mechanisms to specifically recognize and hydrolyze complex-type (CT-type), high-mannose (HM-type) and hybrid-type (Hy-type) N-glycans [26–31]. To date, three types of enzymes can be distinguished according to N-glycan specificity: (i) enzymes that hydrolyze CT-type N-glycans but not HM-type N-glycans, (ii) enzymes that hydrolyze CT-type, HM-type and Hy-type N-glycans, and (iii) enzymes that hydrolyze HM-type and Hy-type but not CT-type N-glycans. Here we discuss recently reported structural and functional data that define the N-glycan recognition mechanisms of these three classes of ENGases.
Overall structure of ENGases
The catalytic domain of ENGases, including both GH18 and GH85 families, adopts a typical (β/α)8 barrel architecture, consisting of eight consecutive β-strand/loop/α-helix modules (Figure 1C). The loops connecting the β-strands to the α-helices display the greatest variation and form the N-glycan binding site, governing substrate specificity (Supplemental Figure 1 and 2). The electrostatic surface representation of several reported crystal structures of ENGases in complex with substrates/products reveal that the glycan is mainly located in a negatively charged cleft, of variable shape and depth, above the β-barrel (Figure 1C). Members of both families follow a conserved substrate-assisted mechanism with retention of the anomeric configuration [32,33]. In the presence of the two reaction products (i.e. a protein with a GlcNAc residue attached and a hydrolyzed N-glycan), ENGases are able to catalyze the reverse reaction and restore the glycosidic bond [34]. The reverse reaction can be enhanced by using ENGase mutants that display glycosynthase activity and a N-glycan oxazoline that acts as an activated glycosyl donor substrate. The catalytic residues, aspartic acid/asparagine and glutamic acid, are located at the tip of the β4 strand of the β-barrel and loop 4, respectively (Figure 1C). In all reported X-ray crystal structures of ENGases in complex with a N-glycan, the β-barrel mostly comprises aromatic and hydrophobic residues providing approximately 70% of the interactions with the conserved pentasaccharide core Man3GlcNAc2. The remaining contacts are distributed between the α(1,3) and α(1,6) antennae and determine the ability of the enzymes to exclusively distinguish between a diverse set of N-glycan chemistries.
Structural basis of CT-type N-glycan recognition mechanism by ENGases
EndoS and EndoS2 from Streptococcus pyogenes serotype M1 [35] and serotype M49 strains [36], respectively, are ENGases from the GH18 family that exhibit strict specificity for the Asn297-linked glycans on IgG antibodies. They are the only ENGases known to be specific to the protein part of the substrate. EndoS and EndoS2 are multidomain proteins that share 37% amino acid sequence identity. The identification of glycosynthase mutants of EndoS and EndoS2, which retain the IgG-specificity of their ENGase counterparts, was a major advancement for methods to glycoengineer IgG Fc N-glycans [37–39]. The reported X-ray crystal structures of EndoS (PDB codes 4NUY, 4NUZ and 6EN3) and EndoS2 (PDB codes 6MDV, 6MDS and 6E58) [26–28] show that both enzymes adopt “V”-shape structure with a GH domain and a β-sandwich domain located at each tip of the “V”. EndoS is the most restrictive of all known ENGases and only accepts biantennary CT-type N-glycans from IgG antibodies, while EndoS2, preferentially hydrolyzes bisected and non-bisected CT-type, but also processes HM-type and Hy-type N-glycans [36], all of which must also be linked to Asn297 of IgG antibodies. In the X-ray crystal structure of the catalytically inactive EndoSD233A/E235L variant in complex with the biantennary CT-type N-glycan product, Gal2GlcNAc2Man3GlcNAc (PDB code 6EN3, Figure 2A) [27], each antenna of the CT-type N-glycan is located in two well defined grooves of the GH domain. Groove 1 mainly comprises loops 2, 3 and 4 and recognizes the α(1,6) antenna, while groove 2 interacts with the α(1,3) antenna through loops 1, 2 and 7 (Figure 2A). Loop 2 bisects both grooves with an aromatic residue, W153 (Figure 2A), that makes a hydrogen bond with Man (−3) and CH-π interactions with GlcNAc (−7). EndoS2 also contains two grooves shaped by the equivalent loops found in EndoS, where both α(1,3) and α(1,6) antennae of the CT-type N-glycan bind (EndoS2-CT, PDB code 6MDS, Figure 2B). The length of loops 3 and 4, which accommodate the α(1,6) antenna, are shorter in EndoS2 than EndoS, creating an additional space in groove 2. The crystal structure of EndoS2 in complex with a HM-type N-glycan (PDB code 6MDV, Figure 2C) reveals that groove 1 accommodates the bulky α(1,6) antenna. The B-factor values of the sugar residues of CT-type and HM-type N-glycans in the crystal structures that comprise the α(1,6) antenna are higher than residues in other parts of the glycan structure, suggesting structural flexibility and transient interactions with the enzyme (Figure 2B–C). Interestingly, EndoS2 has a histidine residue, H109, in loop 2 instead of tryptophan, W153, as observed in EndoS. Accordingly, EndoS2, but not EndoS, can bind bisected CT-type N-glycans displaying an additional GlcNAc linked to the oxygen O4 of Man (−3). EndoS2 is also able to hydrolyze Hy-type from IgG antibodies due to the dual nature of this N-glycan. Supporting this notion, molecular docking calculations showed that the conformation of the α(1,3) and α(1,6) antennae of a Hy-type N-glycan in the active site of EndoS2, are equivalent to the α(1,3) antenna of a CT-type and the α(1,6) antenna of a HM-type N-glycans in the corresponding crystal structures [31]. Alanine scanning mutagenesis studies indicated that the recognition of CT-type N-glycans by EndoS and the recognition of CT-type, HM-type and Hy-type N-glycans by EndoS2 are mainly driven by interaction of the enzymes with the glycan core and α(1,3) antenna [27,28,31]. Altogether, these experimental data strongly support the idea that EndoS and EndoS2 share a common recognition mechanism for N-glycans driven by interaction with the α(1,3) antenna. The particular structural arrangement of loops 3 and 4 creates additional space for EndoS2 to accept different N-glycan chemistries in the α(1,6) antenna, such as those exhibited by the HM-type and Hy-type N-glycans.
Figure 2. Structural basis of CT-type N-glycan recognition by ENGases.
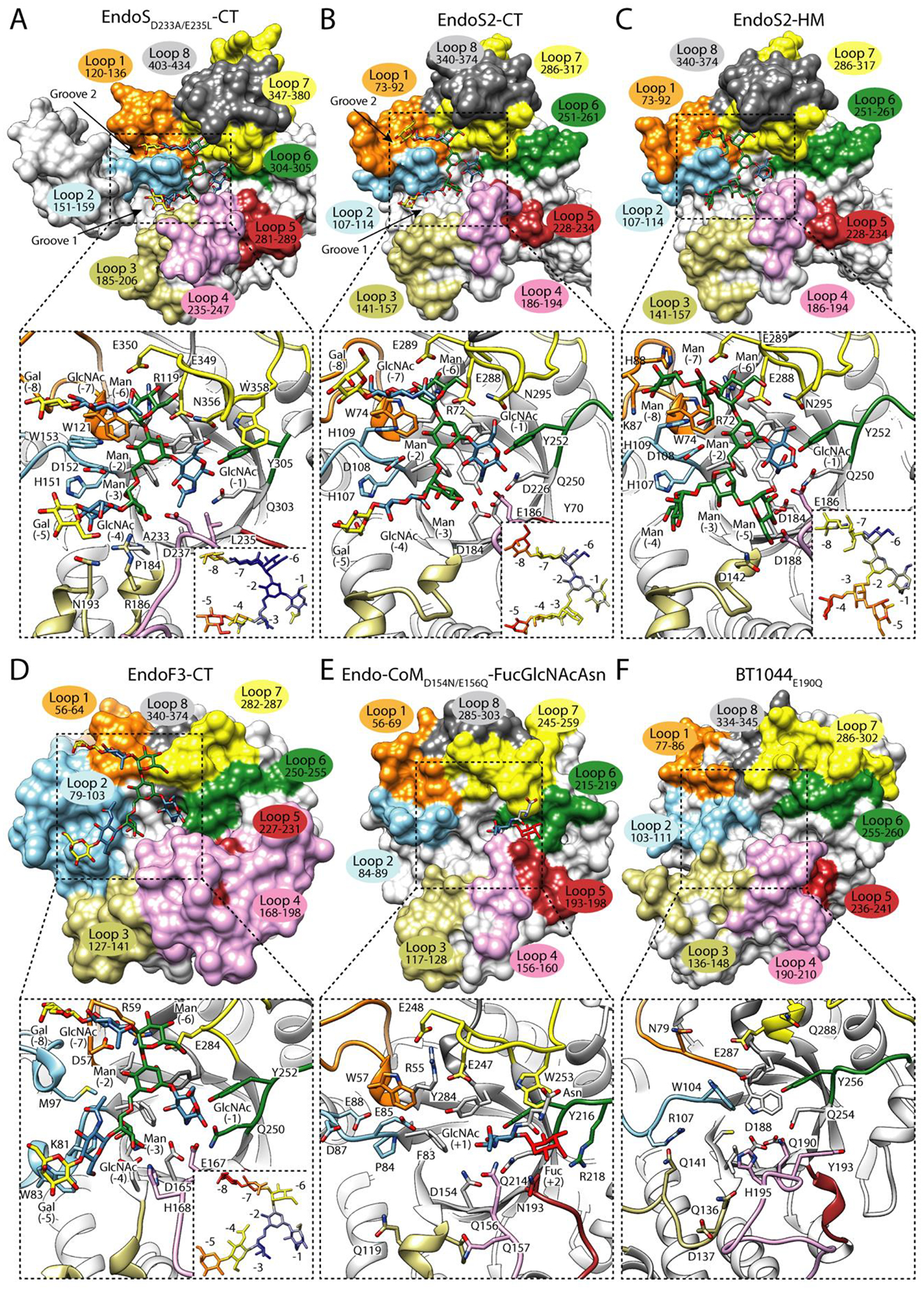
In the upper panel, surface representation of the glycosyl hydrolase domain of A. EndoSD233A/E235L in complex with the CT-type product, Gal2GlcNAc2Man3GlcNAc (PDB code 6EN3), B. EndoS2 in complex with the CT-type product, Gal2GlcNAc2Man3GlcNAc (PDB code 6MDS), C. EndoS2 in complex with the HM-type product, Man7GlcNAc (PDB code 6MDV), D. EndoF3 in complex with the CT-type product, Gal2GlcNAc2Man3GlcNAc (PDB code 1EOM), E. Endo-CoMD154N/E156Q in complex with FucGlcNAcAsn (PDB code 6KPO) and, F. BT1044E190Q in its unliganded form (PDB code 6Q64). Loops surrounding the binding site are annotated. In the lower panel, cartoon representation of the binding site of the glycosidase domain of each enzyme, showing the main residues that interact with the glycan in the active site, and relative B-factors of glycan products found in the crystal structures. The colors from blue to yellow and to red indicate B-factors from small to large.
EndoF3 from Elizabethkingia meningoseptica of the GH18 family hydrolyzes both bi- and triantennary N-glycans [40] but not HM-type N-glycans. In addition, EndoF3 glycosynthase mutants are capable of transferring both N-glycans to intact antibodies [41], whereas EndoS and EndoS2 cannot process triantennary N-glycans with a third β(1–4) antenna attached to the Man (−3). The crystal structures of EndoF3 in its unliganded form (PDB code 1EOK) and in complex with the CT-type N-glycan product,Gal2GlcNAc2Man3GlcNAc (PDB code 1EOM, Figure 2D) show that residues interacting with the pentasaccharide core are highly conserved with EndoS and EndoS2 [42]. In EndoF3, the most important differences are observed in a longer loop 2 that block the entrance of bisecting GlcNAc structures [42] and a shorter loop 7 that allows the entrance of triantennary CT-type N-glycans [27] (Figure 2A–D).
Endo-CoM from Cordyceps militaris is a GH18 ENGase that, like EndoF3 [43], also requires core fucosylation for its activity [44]. The crystal structure of catalytically inactive Endo-CoM in complex with Fuc-α1,6-GlcNAc-Asn [45] (PDB code 6KPO, Figure 2E) shows that the fucose residue interacts with the side chains of N193 of loop 5, Y216 and R218 of loop 6, and W253 of loop 7. Interestingly, the Asn residue of Fuc-α1,6-GlcNAc-Asn is well-exposed to the solvent in the complex Endo-CoM crystal structure. Alanine mutations of Y216 and R218 produce a drastically reduced hydrolytic activity of Endo-CoM against a fucosylated substrate [45]. However, the reduction caused by the Y216A mutation could be due to the fact that this residue stabilizes the reaction intermediate. In addition, Y216 and W253 are conserved in other non fucose dependent enzymes, including EndoS (Figure 2A–B,E). Structural comparison with the crystal structure of EndoF3 reveals that Y187 and Y191 from loop 4, D229 from loop 5, and R258 from α7 could also mediate fucose recognition (Figure 2D) [45].
BT1044 from Bacteroides thetaiotaomicron is a recently characterized GH18 ENGase that also has the ability to process CT-type N-glycans from IgG, among other protein substrates [46] (PDB code 6Q64, Figure 2E). Hydrolytic activity experiments against α1AGP showed that BT1044 processes predominantly bi-antennary CT-type N-glycans, including bisecting GlcNAcs, and tri-antennary N-glycans, but only if the third antenna is composed of a single GlcNAc, and do not show preference for α(1,6) fucosylation [46]. Single alanine mutations of the conserved residues in BT1044 with other GH18 ENGases resulted in inactive mutants against α1AGP [46]. Specifically, BT1044, like EndoS or EndoS2, displays an open pocket comprised by loops 4, 5 and 6, where the α(1,6)-fucose can be accommodated [46]. Moreover, the recognition of bisecting CT-type N-glycans is due to the architecture and arrangement of loop 2 and short loops 3 and 4. In summary, these data suggest that the enzymes that recognize CT-type N-glycans from the family GH18 share a common substrate recognition mechanism by which they primarily recognize the core and the α(1,3) antennae of CT-type N-glycans. In addition, GH18 ENGases show subtle structural features that provide them with the ability to process CT N-glycans with different carbohydrate substitutions (e.g., fucosylation, bisection, additional antennae).
All the structural data available for ENGases that specifically process CT-type N-glycans come from ENGases of the GH18 family. However, there are no structural data to support the molecular mechanism by which enzymes from the GH85 family recognize this type of N-glycan. Furthermore, enzymes from the GH85 family that process CT-type N-glycans have been widely used as glycosynthases [11,47]. This is the case of EndoCC from Coprinopsis cinereal and EndoM from Mucor hiemalis that have a broad N-glycan specificity, being able to process CT-type, HM- and Hy-type N-glycans.
Structural basis of HM-type N-glycan recognition mechanism by ENGases
In contrast to the lack of structural data for GH85 enzymes capable of hydrolyzing CT-type N-glycans, the crystal structures of two GH85 ENGases, EndoA [48,49] (PDB code 2VTF, 3FHA, 3FHQ) from Arthrobacter protophormiae and EndoD from Streptococcus pneumoniae (PDB code 3GDB, 2W91 and 2W92), support the mechanism by which these enzymes recognize HM-type and not CT-type N-glycans. EndoA hydrolyzes HM-type and, less efficiently, Hy-type N-glycans [50], while EndoD processes Man5 and Man3 truncated core structures [51]. Wild-type and glycosynthase mutants of these enzymes have been used for glycoengineering the Fc region of IgG antibodies (Figure 1A). The crystal structure of EndoA in complex with the intermediate mimic, Man3GlcNAc-thiazoline [49] (PDB code 3FHQ, Figure 3A) shows that loop 2 blocks the entrance of the β(1,2)GlcNAc of α(1,6) antenna of the CT-type N-glycan, while loops 1 and 2 create a narrow but deep pocket to accommodate the α(1,6) antenna of the HM-type N-glycan. In contrast, loops 1, 7 and 8 shape a solvent-exposed cavity that can recognize Hy-type N-glycans and extended HM-type N-glycans such as Man6–9. EndoA has the particularity that it can transfer bisecting Man3 to the GlcNAc-Fc [52]. EndoD displays 35% sequence identity with EndoA, however, the crystal structure of EndoD in complex with GlcNAc-thiazoline (PDB code 2W92, Figure 3B) [53] shows structural differences in the binding site that explain the specificity of EndoD for shorter N-glycans than EndoA. In EndoD, loop 7 is slightly longer than in EndoA, limiting the access to N-glycans with only one mannose residue in the α(1,3) antenna like Man5, while excluding Man9, CT-type and Hy-type N-glycans with additional carbohydrate moieties in this antenna. EndoD prefers hydrolyzing fucosylated over non-fucosylated N-glycans. In contrast, EndoA only hydrolyzes non-fucosylated HM-type N-glycans. In EndoA, W216 and W244, located in loops 4 and 5, respectively, can make clashes with the α(1,6)-fucose, inhibiting the activity of the enzyme against fucosylated N-glycans. In EndoD, the equivalent residues H384 and N413 are smaller and can make hydrogen bond and hydrophobic interactions with the α(1,6)-fucose, promoting the activity of this enzyme against fucosylated substrates.
Figure 3. Structural basis of HM-type N-glycan recognition by ENGases.
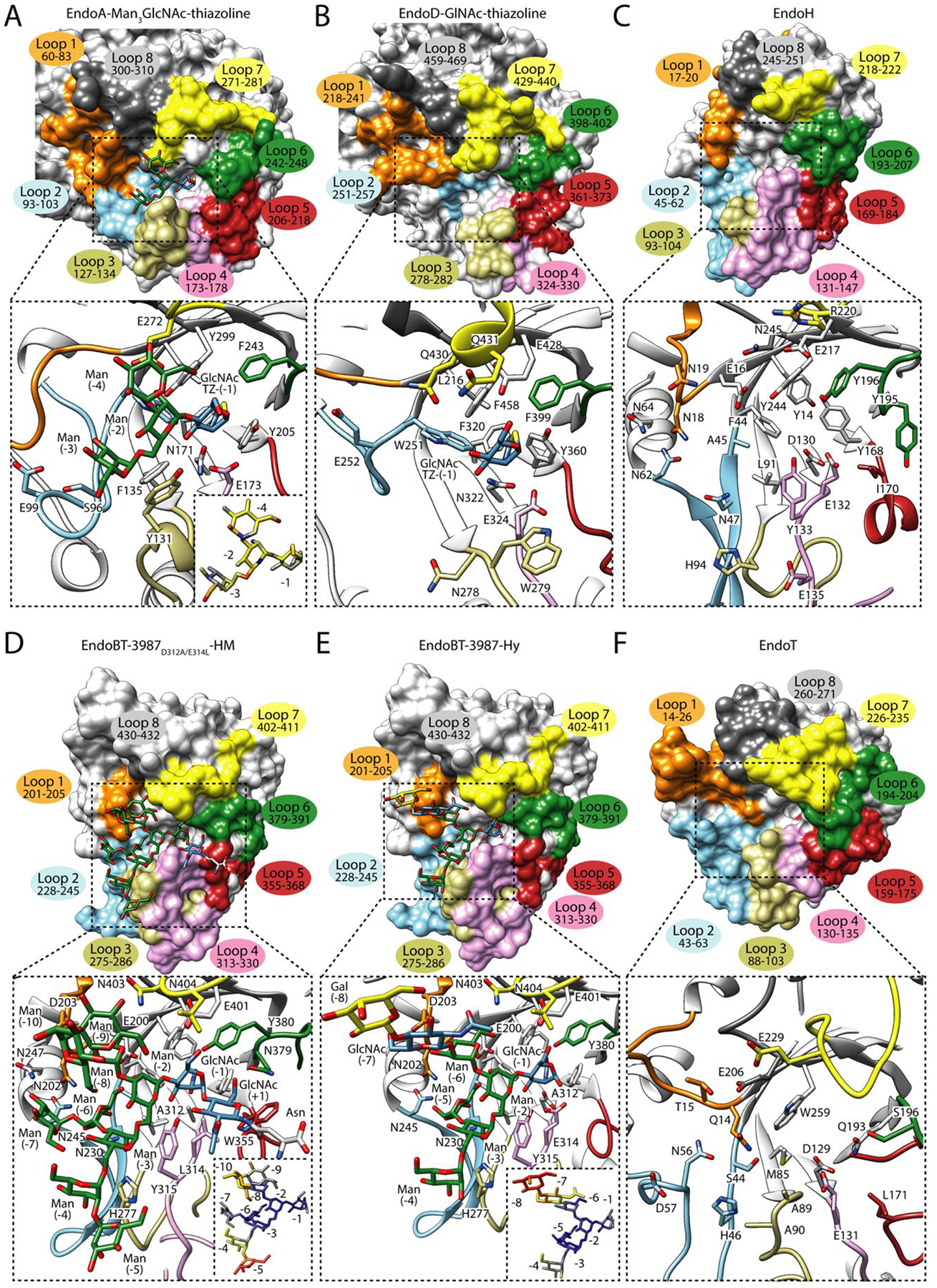
In the upper panel, surface representation of (A) the glycosyl hydrolase domain of EndoA in complex with Man3GlcNAc-thiazoline (PDB code 3FHQ), (B) EndoD in complex with GlcNAc-thiazoline (PDB code 2W92), (C) EndoH in its unliganded form (PDB code 1EDT), (D) EndoBT-3987D312A/E314L in complex with the HM-type substrate, Man9GlcNAc2Asn (PDB code 6TCV), (E) EndoBT-3987 in complex with the Hy-type product, GalGlcNAcMan5GlcNAc (PDB code 7NWF) and, (F) EndoT in its unliganded form (PDB code 4AC1). Loops surrounding the binding site are annotated. In the lower panel, cartoon representation of the binding site of the glycosidase domain of each enzyme, showing the main residues that interact with the glycan in the active site, and relative B-factors of glycan products found in the crystal structures. The colors from blue to yellow and to red indicate B-factors from small to large. The amino acid numbering used in figure is the one used in the literature for EndoH, EndoT, EndoA and starting after the amino acid position 42, 26 and, 24, respectively.
EndoH from Streptomyces plicatus belongs to the GH18 family and hydrolyzes HM-type and Hy-type N-glycans from glycoproteins [54]. This enzyme has been extensively used as tool for glycan analysis [55], to monitor protein trafficking [56] and to deglycosylate glycoproteins to obtain homogenous samples for crystallographic purposes [57]. EndoH [58] (PDB code 1EDT; Figure 3C) shares substrate specificity and displays high structural homology with other ENGases from the GH18 family, including EndoF1 from Elizabethkingia meningoseptica [59] (PDB code 2EBN) and EndoBT-3987 from B. thetaiotaomicron (PDB code 3POH). The core Manα1–3Manα1–6Manβ1–4GlcNAcβ1–4GlcNAc [Man (−6), Man (−3), Man (−2), GlcNAc (−1), and GlcNAc (+1)] is the smallest HM-type N-glycan structure that EndoH and EndoF1 are able to recognize [60]. Very recently, the crystal structures of EndoBT-3987 in complex with the substrate Man9GlcNAc2Asn (PDB code 6TCV; Figure 3D), with two HM-type products, Man5GlcNAc (PDB code 6TCW) and Man9GlcNAc (PDB codes 6T8L and 6T8K) and a Hy-type product Gal1GlcNAc1Man3GlcNAc1 (PDB code 7NWF; Figure 3E) revealed the molecular mechanisms of HM-type and Hy-type recognition and specificity for this important group of enzymes from the GH18 family, including EndoH and EndoF1 [30,31]. The Man9GlcNAc2Asn substrate is located above the (β/α)8 TIM barrel, with the asparagine residue completely exposed to the solvent (Figure 3D). The first GlcNAc (+1) interacts with residues of loop 5, 6 and 7, while GlcNAc (−1) is in a skew boat conformation (1S5) favored by interaction with hydrophobic and polar residues that are located at the tip of the β-strands of the barrel and residues of loops 5 and 7 (Figure 3D). The α(1,6) antenna interacts with loops 1, 2, 3 and 4, whereas the α(1,3) antenna is solvent exposed and only interacts with loops 1 and 7 [30] as opposed to enzymes of family GH18 that hydrolyzes CT-type N-glycans (e.g. EndoS, EndoS2 and EndoF3). The structural comparison between crystal structures of EndoBT-3987 in complex with HM-type and Hy-type products revealed that loops that interact with the common fragment of both N-glycans adopt a similar conformation, supporting the notion that EndoBT-3987 and EndoH invoke a common interaction mode (Figure 3C–D) [31]. The B-factor values of the sugar residues that comprise the α(1,3) antenna are higher than those of the α(1,6) antenna, suggesting that EndoBT-3987 and EndoH mainly recognize the α(1,6) antenna of HM-type and Hy-type N-glycans (Figure 3D–E) [31]. In addition, hydrolytic experiments of EndoBT-3987 mutants against HM-type substrates, showed that mutations in loop 3 (H277) drastically reduce the hydrolytic activity of the enzyme against both substrates, while mutations of residues in loop 2 (N230 and N235) only partially affect the activity of the enzyme. These residues are also conserved in EndoH, EndoF1 and other HM-hydrolyzing enzymes from the GH18 family [30] and interact with carbohydrates of the α(1,6) antenna that form the minimum N-glycan structure [Man (−3) and Man (−6)] that the enzyme needs to be active. However, other enzymes from the same family have other residues at these positions – a histidine, phenylalanine and alanine instead of N230, N245, and H277, respectively [30], suggesting an alternative mechanism for HM-type N-glycan recognition for these enzymes with a similar N-glycan specificity than EndoH.. This is the case for EndoT from Hypocrea jecorina [61] (PDB code 4AC1; Figure 3F), which has been used successfully to produce homogenous N-glycans on IgG antibodies [62]. Structural comparison of loops of GH18 family of ENGases (Supplementary Figure 1) [27,31] showed that loop 2 is markedly different between ENGases of the GH18 family. It adopts a β-hairpin in ENGases capable of hydrolyzing HM-type N-glycans while it is short in ENGases that hydrolyze CT N-glycans [31]. However, ENGases from the GH85 family that hydrolyze HM-type N-glycans (e.g., EndoA and EndoD) display a completely different conformation of loop 2 (Supplementary Figure 1). This, added to the fact that ENGAses from the GH85 family that hydrolyze HM-type can accept shorter N-glycans (e.g., Man3GlcNac2) than the ones from the GH18 family suggest that both families of enzymes recognize HM-type and Hy-type N-glycans following different substrate recognition mechanisms.
Accessory domains of ENGases to gain substrate specificity
In addition to the (β/α)8 GH domain responsible for catalytic activity, many GHs contain one or more carbohydrate binding module (CBM), usually adjacent to the catalytic domain. CBMs have been grouped into 84 families by the CAZy database based on sequence similarity. They have also been categorized by their 3D-folds into 7 families: β-sandwich, β-trefoil, cysteine knot, OB, hevein, hevein-like and unique fold [63]. These domains play multiple roles; binding glycans in an optimal orientation to promote hydrolysis and increasing their concentration around the catalytic site, disrupting the structure of polysaccharides such as starch to enable the GH domain to access the substrate and as an anchor point for adhesins to localize the GH domains where they are required, such as the cell surface [63,64].
The crystal structures of EndoS and EndoS2 show that the β-sandwich domain is located on the opposite end of the ‘V’-shaped protein (Figure 4A), with respect to the GH domain. Hydrogen-Deuterium Exchange Mass-Spectrometry (HDX-MS) experiments to assess the EndoS2-Fc complex show, at early time points, that there are large changes in protection in the GH and β-sandwich domains (Figure 4B). These regions correspond to the areas where the glycan binds, according to the crystal structure (loops 2, 6 and 7 in the GH domain) and where the proposed N-glycan binding site is located on the β-sandwich [28]. However, the mechanism by which these two domains cooperate together in EndoS to recognize the Fc of IgG antibodies remains unknown.
Figure 4. ENGase accessory domains that define substrate specificity.
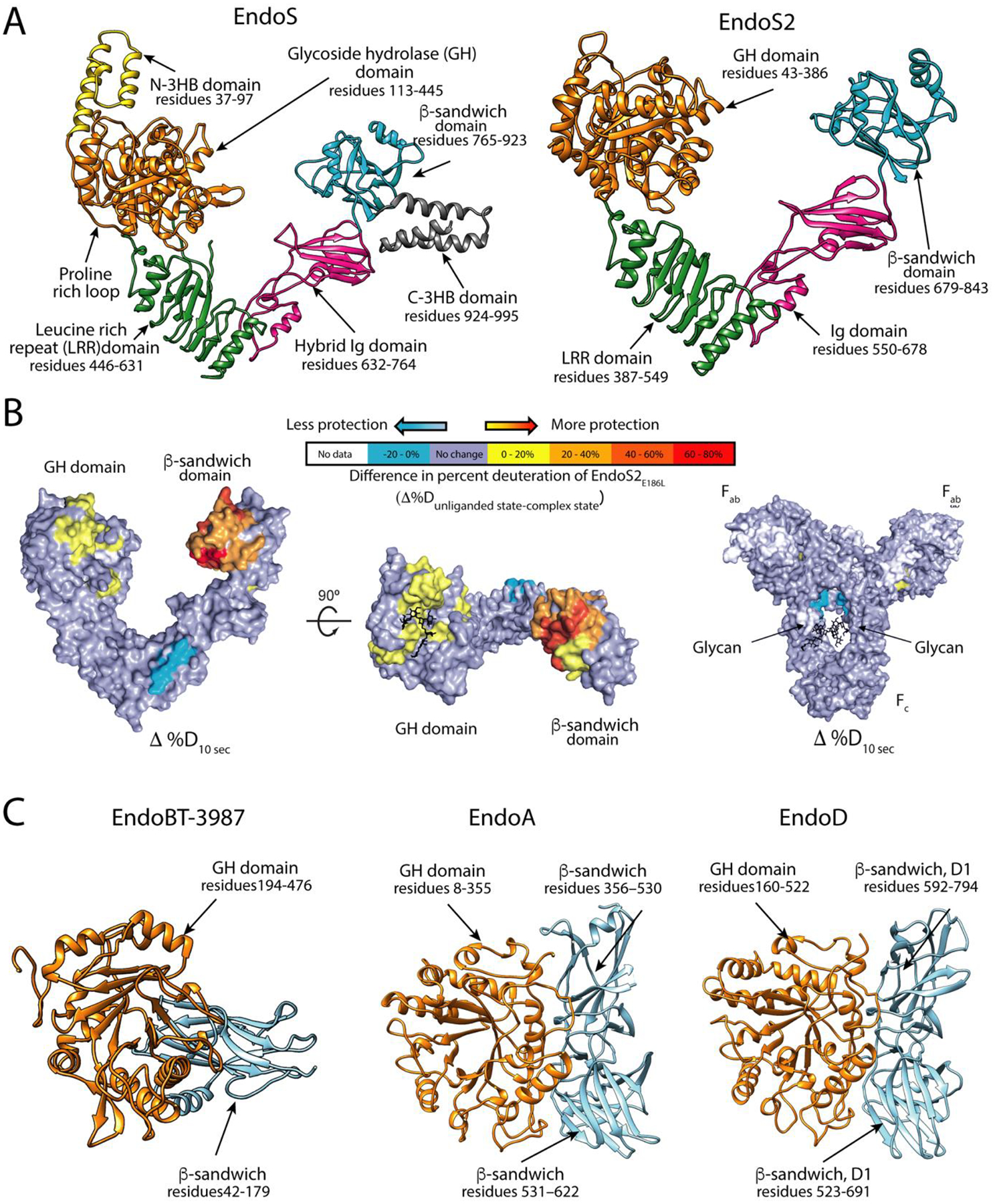
A. Overall structure of EndoS and EndoS2. B. Hydrogen-deuterium exchange mass spectrometry of EndoS2 and IgG1. Difference in deuteration between unliganded- and IgG1-bound EndoS2E186L (left and center) and IgG1 and EndoS2E186L-bound IgG1 (right), mapped onto a surface representation of EndoS2 (PDB 6MDS) and IgG1 (PDB 1HZH) at the earliest time point tested (10 s). IgG1 glycosylation sites annotated, and Fc, Fab, GH and CBM domains are labeled for orientation. C. Overall structure of EndoBT-3987, EndoA and EndoD.
EndoBT-3987 contains a β-sandwich domain at the N-terminus of the GH domain (Figure 4C). The crystal structure shows that there are two grooves located at the interface between the β-sandwich and GH domains, suggesting a potential role in carbohydrate binding. Alanine scanning mutagenesis studies, however, revealed that all alanine mutants were as enzymatically active as the wild type enzyme, suggesting that region is dispensable for hydrolytic activity [30] but could serve other mechanistic roles by binding glycans. EndoA and EndoD both contain two domains after the GH85 domain (Figure 4C). The first auxiliary domains of these ENGases exhibit structural similarities to CBM36 (PDB code 1UX7) and CBM44 (PDB code 2C4X) respectively, suggesting a potential role in glycan binding. The second domain in both cases shows some similarity to fibronectin binding domains. These domains are common in carbohydrate processing enzymes; however, their functions are unclear [48,49,53].
Conclusions and Future Challenges
Significant advances have been made over the last 2 years in understanding the molecular mechanism of substrate specificity and catalysis of ENGases, mostly due to the 3D structural characterization of several GH18 and GH85 family members. Exciting times are anticipated in the field because considerable challenges remain to be overcome to fully understand the mode of action of these enzymes. Among these challenges are:
Identifying novel enzymatic activities/specificities to process/engineer N-glycans on IgG monoclonal antibodies.
Identifying a fucosidase that defucosylated fully branched N-glycans.
Understanding the structural basis of protein, like IgG, specificity, by ENGases.
Chemoenzymatic glycan remodeling will facilitate production of custom glycoforms, enabling more comprehensive studies of antibody-mediated effector functions.
Supplementary Material
Acknowledgements
This work was supported by the MINECO/FEDER EU contracts BFU2016-77427-C2-2-R, PID2019-105649RB-I00 and Severo Ochoa Excellence Accreditation SEV-2016-0644; and the Basque Government contract KK-2019/00076 (to M.E.G.); NIH R01 AI149297 (to E.J.S.); and NIH R01 GM096973 (to L.-X.W.). This project was supported by the European Union Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 844905 (to B.T) and from La Caixa Foundation (ID 100010434) grant LCF/BQ/DR19/11740011 (to M.G.A.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• of special interest
•• of outstanding interest
- 1.de Taeye SW, Rispens T, Vidarsson G: The ligands for human IgG and their effector functions. Antibodies 2019, 8:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mantis NJ, Forbes SJ: Secretory IgA: Arresting microbial pathogens at epithelial borders. Immunol Invest 2010, 39:383–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Royle L, Roos A, Harvey DJ, Wormald MR, Van Gijlswijk-Janssen D, Redwan ERM, Wilson IA, Daha MR, Dwek RA, Rudd PM: Secretory IgA N- and O-glycans provide a link between the innate and adaptive immune systems. J Biol Chem 2003, 278:20140–20153. [DOI] [PubMed] [Google Scholar]
- •4.Arnold JN, Wormald MR, Suter DM, Radcliffe CM, Harvey DJ, Dwek RA, Rudd PM, Sim RB: Human serum IgM glycosylation: Identification of glycoforms that can bind to Mannan-binding lectin. J Biol Chem 2005, 280:29080–29087. [DOI] [PubMed] [Google Scholar]; This study reviews the role of antibody N-glycosylation, the mechanism and drivers for N-glycosylation during biotherapeutic production, the analytical approaches used to characterized glycosylation in order to lead to more rational glycan control.
- 5.Johansson SG: Raised levels of a new immunoglobulin class (IgND) in asthma. Lancet (London, England) 1967, 2:951–953. [DOI] [PubMed] [Google Scholar]
- 6.Plomp R, Hensbergen PJ, Rombouts Y, Zauner G, Dragan I, Koeleman CAM, Deelder AM, Wuhrer M: Site-specific N- glycosylation analysis of human immunoglobulin E. 2013, [DOI] [PubMed]
- 7.Breedveld FC: New drug classes therapeutic monoclonal antibodies. Lancet 2000, 355:735–740. [DOI] [PubMed] [Google Scholar]
- 8.Zolot RS, Basu S, Million RP: Antibody-drug conjugates. Nat Rev Drug Discov 2013, 12:259–260. [DOI] [PubMed] [Google Scholar]
- 9.Nelson AL, Reichert JM: Development trends for therapeutic antibody fragments. Nat Biotechnol 2009, 27:331–337. [DOI] [PubMed] [Google Scholar]
- 10.Ecker DM, Jones SD, Levine HL: The therapeutic monoclonal antibody market. MAbs 2015, 7:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang LX, Tong X, Li C, Giddens JP, Li T: Glycoengineering of antibodies for modulating functions. Annu Rev Biochem 2019, 88:433–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sjögren J, Lood R, Nägeli A: On enzymatic remodeling of IgG glycosylation; unique tools with broad applications. Glycobiology 2020, 30:254–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodoniczky J, Yuan ZZ, James DC: Control of recombinant monoclonal antibody effector functions by Fc N-glycan remodeling in vitro. Biotechnol Prog 2005, 21:1644–1652. [DOI] [PubMed] [Google Scholar]
- 14.Washburn N, Schwabb I, Ortiz D, Bhatnagar N, Lansing JC, Medeiros A, Tyler S, Mekala D, Cochran E, Sarvaiya H, et al. : Controlled tetra-Fc sialylation of IVIg results in a drug candidate with consistent enhanced anti-inflammatory activity. Proc Natl Acad Sci U S A 2015, 112:E1297–E1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pagan JD, Kitaoka M, Anthony RM: Engineered sialylation of pathogenic antibodies in vivo attenuates autoimmune disease. Cell 2018, 172:564–577.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shields RL, Lai J, Keck R, O’Connell LY, Hong K, Gloria Meng Y, Weikert SHA, Presta LG: Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human FcγRIII and antibody-dependent cellular toxicity. J Biol Chem 2002, 277:26733–26740. [DOI] [PubMed] [Google Scholar]
- 17.Ferrara C, Grau S, Jäger C, Sondermann P, Brünker P, Waldhauer I, Hennig M, Ruf A, Rufer AC, Stihle M, et al. : Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose. Proc Natl Acad Sci U S A 2011, 108:12669–12674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pereira NA, Chan KF, Lin PC, Song Z: The “less-is-more” in therapeutic antibodies: Afucosylated anti-cancer antibodies with enhanced antibody-dependent cellular cytotoxicity. MAbs 2018, 10:693–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B: The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 2014, 42:D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ardèvol A, Rovira C: Reaction mechanisms in carbohydrate-active enzymes: glycoside hydrolases and glycosyltransferases. Insights from ab Initio quantum mechanics/molecular mechanics dynamic simulations. J Am Chem Soc 2015, 137:7528–7547. [DOI] [PubMed] [Google Scholar]
- ••21.Li C, Zhu S, Ma C, Wang LX: Designer α1,6-Fucosidase mutants enable direct core fucosylation of intact N-glycopeptides and N-glycoproteins. J Am Chem Soc 2017, 139:15074–15087. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review highlights the main strategies used for glycoengineering antibodies.
- •22.Rodríguez-Díaz J, Monedero V, Yebra MJ: Utilization of natural fucosylated oligosaccharides by three novel alpha-L-fucosidases from a probiotic Lactobacillus casei strain. Appl Environ Microbiol 2011, 77:703–705. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this review, the authors focus on the importance of N-glycosylation of IgGs glycans, the enzymatic tools available to study the IgG glycans and highlight the applications for several modifications of the IgG glycans utilizing these enzymes.
- 23.Klontz EH, Li C, Kihn K, Fields JK, Beckett D, Snyder GA, Wintrode PL, Deredge D, Wang LX, Sundberg EJ: Structure and dynamics of an α-fucosidase reveal a mechanism for highly efficient IgG transfucosylation. Nat Commun 2020, 11:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lammerts van Bueren A, Ardèvol A, Fayers-Kerr J, Luo B, Zhang Y, Sollogoub M, Blériot Y, Rovira C, Davies GJ: Analysis of the reaction coordinate of alpha-L-fucosidases: a combined structural and quantum mechanical approach. J Am Chem Soc 2010, 132:1804–1806. [DOI] [PubMed] [Google Scholar]
- 25.Joosten RP, Salzemann J, Bloch V, Stockinger H, Berglund AC, Blanchet C, Bongcam-Rudloff E, Combet C, Da Costa AL, Deleage G, et al. : PDB-REDO: Automated re-refinement of X-ray structure models in the PDB. J Appl Crystallogr 2009, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trastoy B, Lomino JV, Pierce BG, Carter LG, Gunther S, Giddens JP, Snyder GA, Weiss TM, Weng Z, Wang L-XL-X, et al. : Crystal structure of Streptococcus pyogenes EndoS, an immunomodulatory endoglycosidase specific for human IgG antibodies. Proc Natl Acad Sci 2014, 111:6714–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trastoy B, Klontz E, Orwenyo J, Marina A, Wang L-X, Sundberg EJ, Guerin ME: Structural basis for the recognition of complex-type N-glycans by Endoglycosidase S. Nat Commun 2018, 9:1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klontz EH, Trastoy B, Deredge D, Fields JK, Li C, Orwenyo J, Marina A, Beadenkopf R, Günther S, Flores J, et al. : Molecular basis of broad spectrum N-glycan specificity and processing of therapeutic IgG monoclonal antibodies by Endoglycosidase S2. ACS Cent Sci 2019, 5:524–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du JJ, Klontz EH, Guerin ME, Trastoy B, Sundberg EJ: Structural insights into the mechanisms and specificities of IgG-active endoglycosidases. Glycobiology 2020, 30:268–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trastoy B, Du JJ, Klontz EH, Li C, Cifuente JO, Wang LX, Sundberg EJ, Guerin ME: Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides. Nat Commun 2020, 11:899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trastoy B, Du JJ, Li C, García-Alija M, Klontz EH, Roberts BR, Donahue TC, Wang L-X, Sundberg EJ, Guerin ME: GH18 endo-β-N-acetylglucosaminidases use distinct mechanisms to process hybrid-type N-linked glycans. J Biol Chem 2021, 297:101011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••32.White A, Rose DR: Mechanism of catalysis by retaining β-glycosyl hydrolases. Curr Opin Struct Biol 1997, 7:645–651. [DOI] [PubMed] [Google Scholar]; This study presents crystal structures of α-fucosidase AlfC from Lactobacillus casei that reveal the molecular mechanims of the hydrolsis and transfucosylation activities of the wild type and transfucosidase mutants, respectively.
- 33.Williams SJ, Mark BL, Vocadlo DJ, James MNGG, Withers SG: Aspartate 313 in the Streptomyces plicatus hexosaminidase plays a critical role in substrate-assisted catalysis by orienting the 2-acetamido group and stabilizing the transition state. J Biol Chem 2002, 277:40055–40065. [DOI] [PubMed] [Google Scholar]
- 34.Danby PM, Withers SG: Advances in enzymatic glycoside synthesis. ACS Chem Biol 2016, 11:1784–1794. [DOI] [PubMed] [Google Scholar]
- 35.Collin M, Olsén A: EndoS, a novel secreted protein from Streptococcus pyogenes with endoglycosidase activity on human IgG. EMBO J 2001, 20:3046–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••36.Sjögren J, Struwe WB, Cosgrave E, Rudd PM, Stervander M, Allhorn M, Hollands A, Nizet V, Collin M: EndoS2 is a unique and conserved enzyme of serotype M49 group A Streptococcus that hydrolyzes N-linked glycans on IgG and α1-acid glycoprotein. Biochem J 2013, 455:107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study presents crystal structures of EndoS from Streptococcus pyogenes that reveal the molecular mechanism of CT-type N-glycan recognition and specificity for and an important group of the GH18 ENGases.
- ••37.Li T, Tong X, Yang Q, Giddens JP, Wang LX: Glycosynthase mutants of endoglycosidase S2 show potent transglycosylation activity and remarkably relaxed substrate specificity for antibody glycosylation remodeling. J Biol Chem 2016, 29:16508–16518. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study presents crystal structures of EndoS2 from Streptococcus pyogenes that reveal the molecular mechanism of CT-type and HM-type N-glycan recognition and specificity.
- 38.Shivatare SS, Huang LY, Zeng YF, Liao JY, You TH, Wang SY, Cheng T, Chiu CW, Chao P, Chen LT, et al. : Development of glycosynthases with broad glycan specificity for the efficient glyco-remodeling of antibodies. Chem Commun 2018, 54:6161–6164. [DOI] [PubMed] [Google Scholar]
- ••39.Huang W, Giddens J, Fan S-QQ, Toonstra C, Wang L-XX: Chemoenzymatic glycoengineering of intact IgG antibodies for gain of functions. J Am Chem Soc 2012, 134:12308–12318. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study presents crystal structures of EndoBT-3987 from Bacteroides thetaiotaomicron that reveal the molecular mechanism of HM-type N-glycan recognition and specificity for and an important group of the GH18 ENGases, including EndoH, an enzyme extensively used in biotechnology.
- 40.O’Neill RA: Enzymatic release of oligosaccharides from glycoproteins for chromatographic and electrophoretic analysis. J Chromatogr A 1996, 720:201–215. [DOI] [PubMed] [Google Scholar]
- 41.Giddens JP, Lomino JV, Amin MN, Wang LX: Endo-F3 glycosynthase mutants enable chemoenzymatic synthesis of core-fucosylated triantennary complex type glycopeptides and glycoproteins. J Biol Chem 2016, 291:9356–9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waddling CA, Plummer THJ, Tarentino AL, Van Roey P: Structural basis for the substrate specificity of endo-beta-N-acetylglucosaminidase F(3). Biochemistry 2000, 39:7878–7885. [DOI] [PubMed] [Google Scholar]
- 43.Plummer TH, Phelan AW, Tarentino AL: Porcine fibrinogen glycopeptides: Substrates for detecting endo-β-N-acetylglucosaminidases F2 and F3 1. Anal Biochem 1996, 235:98–101. [DOI] [PubMed] [Google Scholar]
- 44.Huang Y, Higuchi Y, Kinoshita T, Mitani A, Eshima Y, Takegawa K: Characterization of novel endo-β-N-acetylglucosaminidases from Sphingobacterium species, Beauveria bassiana and Cordyceps militaris that specifically hydrolyze fucose-containing oligosaccharides and human IgG. Sci Rep 2018, 8:6–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seki H, Huang Y, Arakawa T, Yamada C, Kinoshita T, Iwamoto S, Higuchi Y, Takegawa K, Fushinobu S: Structural basis for the specific cleavage of core-fucosylated N-glycans by endo-N-acetylglucosaminidase from the fungus Cordyceps militaris. J Biol Chem 2019, 294:17143–17154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Briliūtė J, Urbanowicz PA, Luis AS, Baslé A, Paterson N, Rebello O, Hendel J, Ndeh DA, Lowe EC, Martens EC, et al. : Complex N-glycan breakdown by gut Bacteroides involves an extensive enzymatic apparatus encoded by multiple co-regulated genetic loci. Nat Microbiol 2019, 4:1571–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fairbanks AJ: The ENGases: versatile biocatalysts for the production of homogeneous N-linked glycopeptides and glycoproteins. Chem Soc Rev 2017, 46:5128–5146. [DOI] [PubMed] [Google Scholar]
- 48.Ling Z, Suits MDL, Bingham RJ, Bruce NC, Davies GJ, Fairbanks AJ, Moir JWB, Taylor EJ: The X-ray crystal structure of an Arthrobacter protophormiae endo-β-N-acetylglucosaminidase reveals a (β/α)8 catalytic domain, two ancillary domains and active site residues key for transglycosylation activity. J Mol Biol 2009, 389:1–9. [DOI] [PubMed] [Google Scholar]
- 49.Yin J, Li L, Shaw N, Li Y, Song JK, Zhang W, Xia C, Zhang R, Joachimiak A, Zhang HC, et al. : Structural basis and catalytic mechanism for the dual functional Endo-β-N-acetylglucosaminidase A. PLoS One 2009, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takegawa K, Nakoshi M, Iwahara S, Yamamoto K, Tochikura T: Induction and purification of endo-β-N-acetylglucosaminidase from Arthrobacter protophormiae grown in ovalbumin. Appl Environ Microbiol 1989, 55:3107–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muramatsu H, Tachikui H, Ushida H, Song XJ, Qiu Y, Yamamoto S, Muramatsu T: Molecular cloning and expression of endo-β-N-acetylglucosaminidase D, which acts on the core structure of complex type asparagine-linked oligosaccharides. J Biochem 2001, 129:923–928. [DOI] [PubMed] [Google Scholar]
- 52.Zou G, Ochiai H, Huang W, Yang Q, Li C, Wang LX: Chemoenzymatic synthesis and Fcγ receptor binding of homogeneous glycoforms of antibody Fc domain. Presence of a bisecting sugar moiety enhances the affinity of Fc to FcγIIIa receptor. J Am Chem Soc 2011, 133:18975–18991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abbott DW, Macauley MS, Vocadlo DJ, Boraston AB: Streptococcus pneumoniae endohexosaminidase D, structural and mechanistic insight into substrate-assisted catalysis in family 85 glycoside hydrolases. J Biol Chem 2009, 284:11676–11689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •54.Tarentino L, Maley F: Purification and properties of an Endo-β-N-acetylglucosaminidase from Streptomyces griseus. J Biol Chem 1974, 249:811–817. [PubMed] [Google Scholar]; This study presents crystal structures of catalytically inactive Endo-CoM from Cordyceps militaris in complex with Fuc-α1,6-GlcNAc-Asn that reveal the molecular mechanism by which this GH18 ENGase requires fucose for its activity.
- 55.Cao L, Diedrich JK, Ma Y, Wang N, Pauthner M, Park SKR, Delahunty CM, McLellan JS, Burton DR, Yates JR, et al. : Global site-specific analysis of glycoprotein N-glycan processing. Nat Protoc 2018, 13:1196–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Freeze HH, Kranz C: Endoglycosidase and glycoamidase release of N-linked glycans. Curr Protoc Mol Biol 2010, 62:12.4.1–12.4.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis SJ, Crispin M: Solutions to the glycosylation problem for low- and high-throughput structural glycoproteomics. In Functional and Structural Proteomics of Glycoproteins. Edited by Owens R, Nettleship JE. Springer, Dordrecht; 2011:127–158. [Google Scholar]
- 58.Rao V, Guan C, Van Roey P: Crystal structure of endo-beta-N-acetylglucosaminidase H at 1.9 A resolution: active-site geometry and substrate recognition. Structure 1995, 3:449–457. [DOI] [PubMed] [Google Scholar]
- 59.Van Roey P, Rao V, Plummer THJ, Tarentino AL: Crystal structure of endo-beta-N-acetylglucosaminidase F1, an alpha/beta-barrel enzyme adapted for a complex substrate. Biochemistry 1994, 33:13989–13996. [DOI] [PubMed] [Google Scholar]
- 60.Trimble RB, Tarentino AL: Identification of distinct endoglycosidase (endo) activities in Flavobacterium meningosepticum: Endo F1, endo F2, and endo F3: Endo F1 and endo H hydrolyze only high mannose and hybrid glycans. J Biol Chem 1991, 266:1646–1651. [PubMed] [Google Scholar]
- 61.Stals I, Karkehabadi S, Kim S, Ward M, Van Landschoot A, Devreese B, Sandgren M: High resolution crystal structure of the endo-N-Acetyl-beta-D-glucosaminidase responsible for the deglycosylation of Hypocrea jecorina cellulases. PLoS One 2012, 7:e40854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meuris L, Santens F, Elson G, Festjens N, Boone M, Dos Santos A, Devos S, Rousseau F, Plets E, Houthuys E, et al. : GlycoDelete engineering of mammalian cells simplifies N-glycosylation of recombinant proteins. Nat Biotechnol 2014, 32:485–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ: Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 2004, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guillén D, Sánchez S, Rodríguez-Sanoja R: Carbohydrate-binding domains: multiplicity of biological roles. Appl Microbiol Biotechnol 2010, 85:1241–1249. [DOI] [PubMed] [Google Scholar]
- 65.Bondt A, Rombouts Y, Selman MHJ, Hensbergen PJ, Reiding KR, Hazes JMW, Dolhain RJEM, Wuhrer M: Immunoglobulin G (IgG) fab glycosylation analysis using a new mass spectrometric high-throughput profiling method reveals pregnancy-associated changes. Mol Cell Proteomics 2014, 13:3029–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stadlmann J, Pabst M, Altmann F: Analytical and functional aspects of antibody sialylation. J Clin Immunol 2010, 30:15–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stanley P, Taniguchi N, Aebi M: N-Glycans. In Essentials of Glycobiology. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Print. Edited by Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, et al. 2015:99–111. [PubMed] [Google Scholar]
- 68.Pučić M, Knežević A, Vidič J, Adamczyk B, Novokmet M, Polašek O, Gornik O, Šupraha-Goreta S, Wormald MR, Redžić I, et al. : High throughput isolation and glycosylation analysis of IgG–variability and heritability of the IgG glycome in three isolated human populations. Mol Cell Proteomics 2011, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Majewska NI, Tejada ML, Betenbaugh MJ, Agarwal N: N-glycosylation of IgG and IgG-like recombinant therapeutic proteins: Why is it important and how can we control it? Annu Rev Chem Biomol Eng 2020, 7:311–338. [DOI] [PubMed] [Google Scholar]
- 70.Anthony RM, Ravetch JV.: A novel role for the IgG Fc glycan: The anti-inflammatory activity of sialylated IgG Fcs. J Clin Immunol 2010, 30:9–14. [DOI] [PubMed] [Google Scholar]
- 71.Nimmerjahn F, Ravetch JV.: Fc-Receptors as Regulators of Immunity. Adv Immunol 2007, 96:179–204. [DOI] [PubMed] [Google Scholar]
- 72.Goetze AM, Liu YD, Zhang Z, Shah B, Lee E, Bondarenko PV, Flynn GC: High-mannose glycans on the Fc region of therapeutic IgG antibodies increase serum clearance in humans. Glycobiology 2011, 21:949–959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


