Abstract
Circular RNAs (circRNAs) belong to an abundant category of non-coding RNAs that are stable and specific, and thus have great potential in cancer treatment. However, little is known about the role of circRNAs during radiotherapy in lung adenocarcinoma (LUAD). Here, we established the expression profiles of 1,875 dysregulated circRNAs in non-irradiated and irradiated A549 cells and identified circNEIL3 as a significantly downregulated circRNA in A549 cells treated with 0, 2, or 4 Gy of radiation, respectively. Functional assays demonstrated that circNEIL3 knockdown promoted radiation-induced cell pyroptosis, whereas circNEIL3 overexpression had the opposite effects. Importantly, the effects of circNEIL3 overexpression on inhibiting pyroptosis were reversed by PIF1 knockdown. Mechanistically, circNEIL3-mediated pyroptosis was achieved through directly binding to miR-1184 as a sponge, thereby releasing the inhibition of miR-1184 on PIF1, which ultimately induces DNA damage and triggers AIM2 inflammasome activation. In vivo, circNEIL3 knockdown significantly enhanced the efficacy of radiotherapy as evidenced by decreases in tumor volume and weight. Collectively, the circNEIL3/miR-1184/PIF1 axis that mediate pyroptosis induction may be a novel, promising therapeutic strategy for the clinical treatment of lung cancer.
Subject terms: Radiotherapy, Non-small-cell lung cancer
Introduction
Lung cancer remains the leading cause of cancer-related mortality accounting for 18% globally and 19% of all cancer-related deaths in China [1]. It is estimated that 85% of human lung cancers are non-small cell lung cancer, with lung adenocarcinoma (LUAD) being the predominant subtype, accounting for 50% of all cases [2]. Radiotherapy is widely used in the treatment of clinical tumors, related to lung cancer, prostate cancer, and kidney cancer, among others [3–6]. However, ionizing radiation can cause a range of opportunistic infections and systemic inflammation, with life-threatening tumor recurrence and metastasis still prevalent in patients [7, 8]. Therefore, further investigation is needed to gain a deeper understanding of lung cancer to provide new therapeutic targets for lung cancer radiotherapy.
Pyroptosis is a recently discovered form of programmed cell death, characterized by cell swelling and large bubbles emerging from the plasma membrane [9, 10]. Pyroptosis is divided into the caspase-1-activated classical and caspase-11/4/5-activated non-classical pathways. In the classical pathway, the assembled NLRP1, NAIP, NLRC4, AIM2, NLRP3, and pyrin proteins activate and cleave pro-caspase-1 to form active caspase-1, which then cleaves gasdermin D (GSDMD) to the N-terminal form (GSDMD-N) with its pore-forming activity, and ultimately releases inflammatory factors IL-1β and IL-18, causing inflammatory reactions [11, 12]. Conversely, in the non-classical pathway, lipopolysaccharide (LPS) can combine directly and activate caspase-11/4/5, which can then cleave GSDMD into GSDMD-N [11–13]. Recent studies have shown that pyroptosis plays an important role in the radiation process [14–16]. For example, NLRP3 inflammasome activation can mediate radiation-induced pyroptosis in bone marrow-derived macrophages [17]. Furthermore, mesenchymal stem cells can attenuate radiation-induced brain injury by inhibiting microglia pyroptosis [18]. However, little is known regarding the role of pyroptosis in lung cancer radiotherapy.
Circular RNAs (circRNAs), a category of non-coding RNAs characterized by covalently closed-loop structures without 5′−3′ polarity or a poly adenosine tail [19, 20], are abundant, conserved, stable in organisms, and resistant to RNase R treatment [21–23]. Since their first discovery in viruses in 1976, the functions of circRNAs in various human diseases have been reported, including cardiovascular diseases [24, 25], lupus nephritis [26], systemic neural diseases [27], and most cancers [22, 28]. Moreover, the radiosensitivity of bone marrow mesenchymal stem cells has been shown to be affected by circRNA_014511 binding to miR‑29b‑2‑5p [29]. Functionally, circRNAs are commonly used as competitive endogenous RNAs (ceRNAs) to eliminate the inhibitory effects of microRNA (miRNA) on target genes by acting as miRNA sponges.
Some important regulatory roles of circRNAs in terms of radiation have been reported, such as radiation-induced esophageal injury [30], the radiation sensitivity of esophageal cancer cells [31], and the radioresistance of HeLa cells [32]. However, the specific role and mechanism of circRNAs in lung cancer radiotherapy remain unclear. Therefore, we investigated the potential roles of circRNAs in the radiotherapy of lung cancer by establishing the expression profile of circRNAs and verified thousands of distinct circRNAs in non-irradiated and irradiated LUAD cells using high-throughput RNA sequencing (RNA-seq). Subsequently, we identified specific differentially expressed circRNAs and elucidated their roles in pyroptosis as potential therapeutic targets in lung cancer.
Results
circNEIL3 is significantly downregulated in irradiated LUAD cells
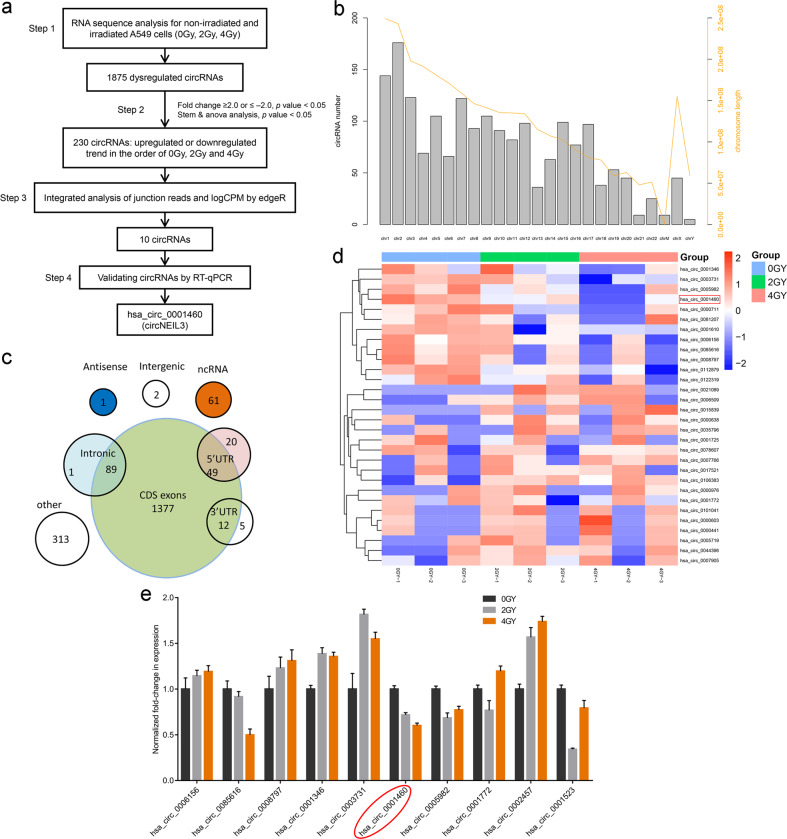
To investigate the roles of circRNAs in the radiotherapy of lung cancer, we performed RNA-seq and analyzed the expression profiles of circRNAs in non-irradiated and irradiated A549 lung cancer cells (sequences were deposited in the Gene Expression Omnibus under dataset code GSE124396; www.ncbi.nlm.nih.gov/geo). A total of 1875 dysregulated circRNAs were identified in irradiated A549 cells (Fig. 1a). The hierarchical clustering heatmap suggested that the expression levels of circRNAs were clustered and distinguishable (Supplementary Fig. S1a). The clonogenic survival assay showed that the 4 Gy dose of radiation treatment significantly reduced the proliferation ability of A549 cells (Supplementary Fig. S1b). In addition, the category and chromosomal localization of circRNAs were also identified (Fig. 1b, c). Based on the STEM and ANOVA analysis, 230 circRNAs showed an upregulated or downregulated trend in A549 cells treated with different doses (0, 2, or 4 Gy) of radiation (|fold change | ≥ 2.0, P < 0.05). Of these, the first 15 downregulated and 15 upregulated circRNAs are represented in Fig. 1d and listed in Table S1. Through integrated analysis of junction reads and logCPM by edgeR, we selected and validated 10 circRNAs using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) (Fig. 1e and Table S2). The results showed that hsa_circ_0001460 (termed circNEIL3) was the most significantly downregulated circRNA in the LUAD cells treated with either 0, 2, or 4 Gy of radiation, respectively (Fig. 1a, e).
Fig. 1. The circRNA expression profile workflow in the irradiated LUAD cells.
a Flow chart for identifying and validating circRNAs in A549 cells. b Distribution map of circRNAs based on chromosome location. c Distribution map of circRNAs based on category. d Heatmap showing the top 15 most upregulated and top 15 most downregulated circRNAs in A549 cells treated with either 0, 2, or 4 Gy radiation, respectively. e Ten most representative circRNAs verified using RT-qPCR.
Characterization of circNEIL3 in LUAD cells
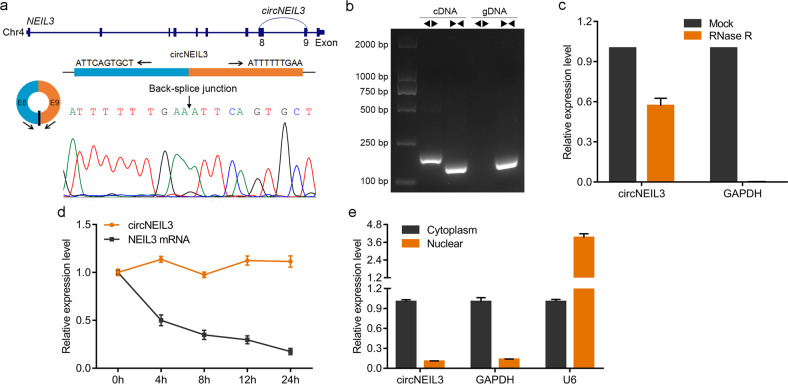
We next evaluated the exon structure of circNEIL3, which is derived from exons 8 to 9 of the NEIL3 gene with a length of 596 nucleotides located at chr4:178274462-178281831+. We also amplified the back-spliced junction of circNEIL3 using divergent primers, which was confirmed by Sanger sequencing (Fig. 2a). This sequence is consistent with the circBase database annotation (http://www.circbase.org/). Subsequently, PCR analysis showed that divergent primers only amplified a band in complementary DNA and not in genomic DNA (Fig. 2b). RT-qPCR after RNase R treatment showed that circNEIL3 was resistant to RNase R digestion compared to GAPDH (Fig. 2c). The stability of circNEIL3 was further analyzed in A549 cells treated with actinomycin D, an inhibitor of transcription, suggesting that circNEIL3 was more stable than NEIL3 (Fig. 2d). To observe the cellular localization of circNEIL3, the nuclear and cytoplasmic circNEIL3 RNA was detected by RT-qPCR. As shown in Fig. 2e, circNEIL3 transcripts were preferentially located in the cytoplasm. These results demonstrated the characteristics of circNEIL3 as a circRNA, implying that its biological stability may be beneficial to its function.
Fig. 2. Characterization of circNEIL3 in LUAD cells.
a Genomic location of circNEIL3. The back-spliced junction of circNEIL3 identified using Sanger sequencing. b PCR analysis for circNEIL3 in complementary DNA (cDNA) and genomic DNA (gDNA). c Relative levels of circNEIL3 and GAPDH detected using RT-qPCR after RNase R digestion, normalized to the value detected in the mock group. d RT-qPCR for the abundance of circNEIL3 and NEIL3 in A549 cells treated with actinomycin D at the indicated time points. e Levels of circNEIL3 in the cytoplasmic and nuclear fractions of A549 cells.
circNEIL3 deficiency is important for irradiation-induced pyroptosis in LUAD
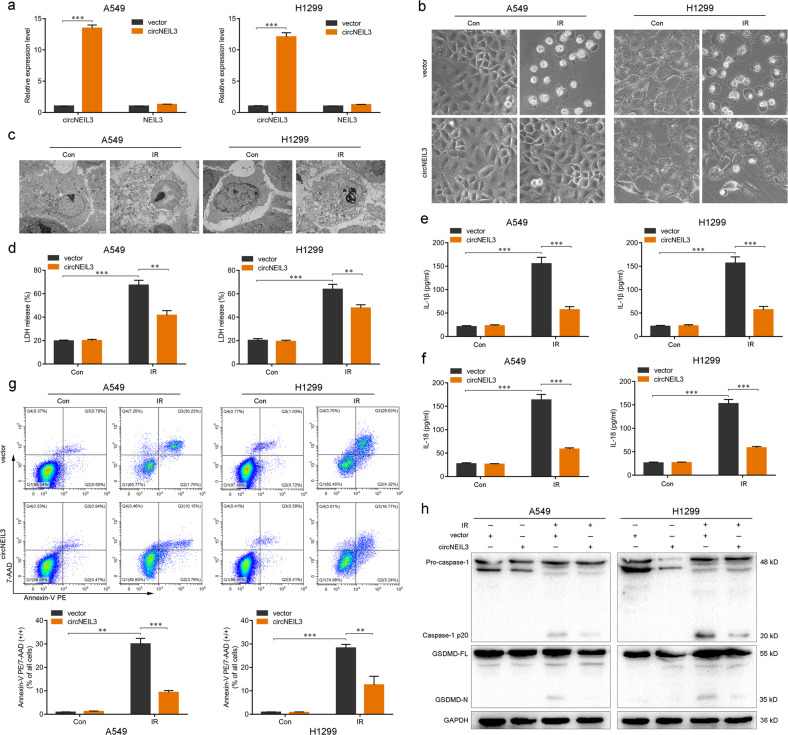
To investigate the effect of circNEIL3 on LUAD radiotherapy, we utilized a small hairpin RNA lentiviral vector for stable knockdown of endogenous circNEIL3 in the A549 and H1299 cell lines. For ectopic overexpression of circNEIL3, exons 8–9 of NEIL3 were cloned into the lentiviral vector. CircNEIL3 overexpression successfully upregulated circNEIL3 expression in both A549 and H1299 cells but had no effect on NEIL3 mRNA expression (Fig. 3a). Similarly, circNEIL3 knockdown successfully downregulated circNEIL3 expression in LUAD cells, whereas NEIL3 mRNA expression again showing no obvious change (Supplementary Fig. S2a). Morphologically, irradiation-treated A549 and H1299 cells exhibited large bubbles emerging from the plasma membrane and cell swelling, which highly resembled the characteristics of pyroptosis induced by the N-terminus of GSDMD [12], and circNEIL3 overexpression visibly inhibited this phenomenon (Fig. 3b). Conversely, stable knockdown of circNEIL3 remarkably increased the numbers of cell swelling and the formation of large bubbles (Supplementary Fig. S2b). The transmission electron microscopy also revealed multiple pores formed in the membranes of irradiation-treated A549 and H1299 cells (Fig. 3c).
Fig. 3. The importance of circNEIL3 expression in the irradiation-induced pyroptosis of LUAD cells.
a Expression levels of circNEIL3 and NEIL3 in LUAD cells. b Representative images of LUAD cells treated with a 4-Gy dose of radiation after transfection for circNEIL3 overexpression. c Transmission electron microscopy images of LUAD cells 48 h after treated with 4 Gy dose of radiation. Scale bar 2 μm. d–f Enzyme-linked immunosorbent assays for quantifying the LDH, IL-1β, and IL-18 release of LUAD cells. g Percentage of Annexin V PE and 7-AAD double-positive cells in LUAD cells as detected using flow cytometry. h Analysis of pyroptosis markers (caspase-1, GSDMD-N) by western blotting in the indicated groups. **P < 0.01, ***P < 0.001, two-tailed Student’s t-test. Con control, IR irradiated.
Moreover, circNEIL3 overexpression significantly inhibited the release of lactate dehydrogenase (LDH) and the secretion of interleukin (IL)−1β and IL-18 in irradiated A549 and H1299 cells (Fig. 3d–f), whereas circNEIL3 knockdown led to a marked increase in the release of LDH and secretion of IL-1β and IL-18 (Supplementary Fig. S2c–e). Flow cytometry analysis showed fewer annexin V-PE and 7-AAD double-positive cells in circNEIL3-overexpressed A549 and H1299 cells treated with irradiation (Fig. 3g), whereas the opposite effects were observed for the circNEIL3 knockdown group (Supplementary Fig. S2f). Furthermore, western blotting showed that circNEIL3 overexpression suppressed the activity of caspase-1 to cleave GSDMD into GSDMD-N fragments with pore-forming activity (Fig. 3h), whereas, as expected, circNEIL3 knockdown had the opposite effects (Supplementary Fig. S2g). Taken together, these findings indicated that circNEIL3 deficiency plays a critical role in irradiation-induced pyroptosis in LUAD cells.
circNEIL3 knockdown facilitates pyroptosis by targeting PIF1
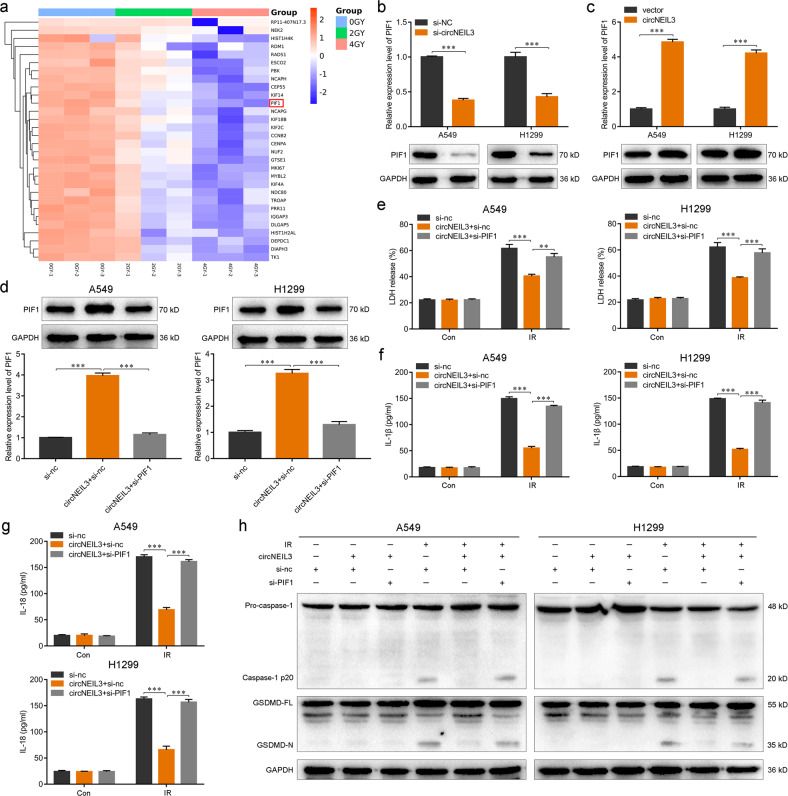
To explore the molecular mechanism of circNEIL3 in regulating the pyroptosis of LUAD cells, we constructed and analyzed the ceRNA network for each circRNA using the miRanda and TargetScan tools (Supplementary Fig. S1c). Six predicted co-expressed mRNAs were found. Based on the hierarchical clustering heatmap of the first 30 mRNAs that showed a downward trend in the A549 cells treated with different doses (0, 2, or 4 Gy) of radiation, PIF1 was the most obvious gene exhibiting a declining trend (Fig. 4a). Indeed, RT-qPCR and western blotting revealed that circNEIL3 knockdown significantly reduced the mRNA and protein levels of PIF1, and the opposite results were observed when circNEIL3 was overexpressed (Fig. 4b, c). Therefore, PIF1 was considered as a major candidate target of circNEIL3.
Fig. 4. circNEIL3 facilitates pyroptosis by targeting PIF1.
a Heatmap of the top 30 downregulated mRNAs in A549 cells treated with different doses (0, 2, or 4 Gy) of radiation. b, c Relative expression levels of PIF1 after circNEIL3 knockdown b or overexpression c. d Upregulation of PIF1 induced by circNEIL3 overexpression decreased by si-PIF1 treatment detected using both RT-qPCR and western blotting. e–g Enzyme-linked immunosorbent assays for LDH, IL-1β, and IL-18 release of cells in the indicated groups. h Upregulated expression of active-caspase-1 and GSDMD-N induced by circNEIL3 overexpression decreased by si-PIF1 treatment, detected using western blotting. **P < 0.01, ***P < 0.001, two-tailed Student’s t-test. Con, control; IR, irradiated.
Cell pyroptosis after knockdown of PIF1 was examined to further study the function of circNEIL3 on PIF1. Among the small interfering RNAs (siRNAs) tested, mRNA and protein levels of PIF1 were reduced by at least 60% with si-h-PIF1_001 (Supplementary Fig. S3); therefore, we utilized this siRNA for subsequent experiments. The PIF1 upregulation effect of circNEIL3 overexpression was clearly decreased following si-PIF1 treatment (Fig. 4d). Notably, PIF1 knockdown significantly attenuated the effects of circNEIL3 on reducing LDH, IL-1β, and IL-18 levels in irradiation-treated A549 and H1299 cells (Fig. 4e–g). Furthermore, the effects of circNEIL3 overexpression on the levels of active caspase-1 and GSDMD-N involved in pyroptosis were reversed by PIF1 knockdown (Fig. 4h). These results suggested that circNEIL3 potentially modulates LUAD pyroptosis by targeting PIF1.
circNEIL3 regulates PIF1 expression via direct binding to miR-1184
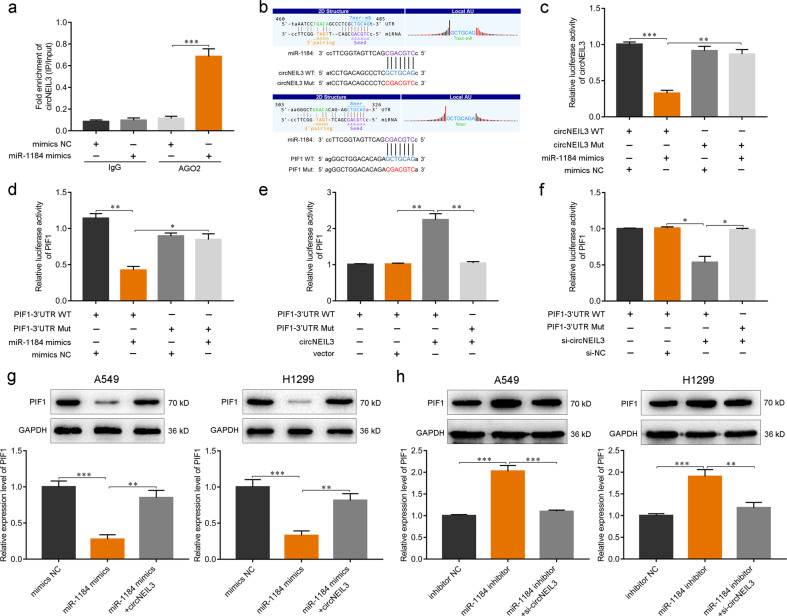
circRNAs can eliminate the inhibitory effects of miRNAs on downstream targets by acting as miRNA sponges [22, 26, 33]. Given that circNEIL3 was found to be mainly located in the cytoplasm (Fig. 2e) and its interaction is established by the ceRNA network (Supplementary Fig. S1c), we next explored whether circNEIL3 could function as an miR-1184 sponge to regulate the expression of PIF1. AGO2 immunoprecipitation was performed to determine whether circNEIL3 served as a platform for AGO2 and miR-1184. Compared with IgG, circNEIL3 and miR-1184 were significantly enriched in the AGO2 pulldown complex (Fig. 5a). Subsequently, we constructed dual luciferase reporter assays containing wild-type and mutated putative binding sites for either circNEIL3 or PIF1 transcripts to confirm that circNEIL3 and PIF1 could be regulated by miR-1184 (Fig. 5b). The miR-1184 mimics significantly reduced the luciferase activity of the circNEIL3 wild-type reporter compared with that of either the control or mutated luciferase reporter (Fig. 5c). Similarly, the luciferase activity of the PIF1 wild-type reporter was markedly decreased when transfected with miR-1184 mimics (Fig. 5d).
Fig. 5. circNEIL3 regulates PIF1 expression via directly binding miR-1184.
a RNA immunoprecipitation performed using AGO2 antibody in A549 cells transfected with miR-1184 mimics or negative control (NC) mimics, followed by the detection of circNEIL3 enrichment. b Wild-type (WT) and mutant (Mut) transcripts of circNEIL3 or the PIF1 3′-untranslated region (UTR) for luciferase reporters. c, d Dual luciferase reporter activity of circNEIL3 c and the PIF1 3′-UTR d in A549 cells co-transfected with either miR-1184 mimics or NC mimics. e, f Dual luciferase reporter activity of the PIF1 3′-UTR in A549 cells with circNEIL3 overexpression e or knockdown f. g Expression of PIF1 in LUAD cells transfected with miR-1184 mimics alone or co-transfected with circNEIL3. h Expression of PIF1 in LUAD cells transfected with miR-1184 inhibitor alone or co-transfected with si-circNEIL3. *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed Student’s t-test.
Moreover, circNEIL3 overexpression could further increase the luciferase activity of the PIF1 wild-type reporter, whereas circNEIL3 knockdown reduced the activity level (Fig. 5e, f). Consistently, the miR-1184 mimics significantly reduced the mRNA and protein levels of PIF1 in A549 and H1299 cells, whereas circNEIL3 overexpression reversed these effects of miR-1184 on PIF1 expression (Fig. 5g). Conversely, circNEIL3 knockdown significantly eliminated the elevation of PIF1 expression caused by miR-1184 inhibitors (Fig. 5h). Taken together, these results demonstrated that circNEIL3 could directly bind to miR-1184 as a sponge to regulate the expression of PIF1.
The circNEIL3/miR-1184/PIF1 axis mediates pyroptosis via the DNA damage pathway
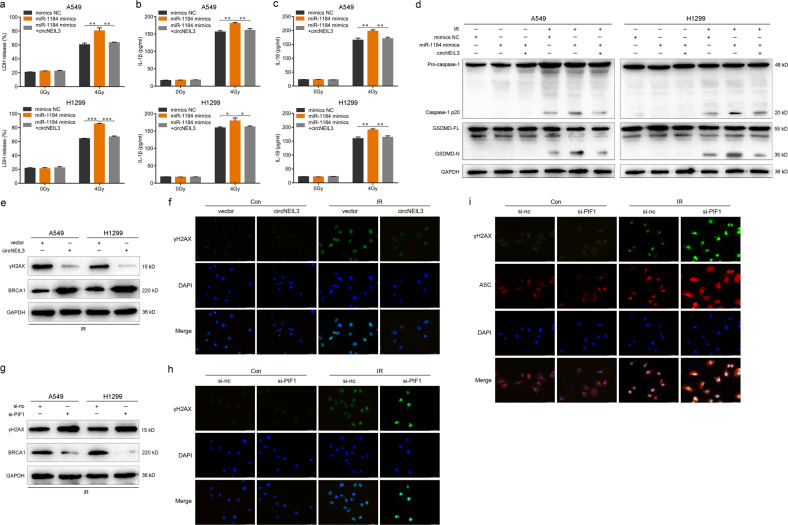
Based on the results described above, we assessed the regulatory role of the circNEIL3/miR-1184/PIF1 axis on the occurrence of cell pyroptosis. The increase in the release of LDH, IL-1β, and IL-18 induced by miR-1184 mimics was remarkably suppressed by circNEIL3 overexpression in irradiation-treated A549 and H1299 cells (Fig. 6a–c). The levels of active caspase-1 and GSDMD-N were evidently increased by miR-1184 mimics, whereas circNEIL3 overexpression abolished this effect (Fig. 6d). These results suggested that pyroptosis mediated by the circNEIL3/miR-1184/PIF1 axis is important for the radiotherapy of LUAD.
Fig. 6. The circNEIL3/miR-1184/PIF1 axis mediates pyroptosis via the DNA damage repair pathway.
a–c Enzyme-linked immunosorbent assays detecting the release of LDH, IL-1β, and IL-18 in LUAD cells transfected with miR-1184 mimics alone or co-transfected with circNEIL3. d Western blot of the expression of active-caspase-1 and GSDMD-N in the indicated groups. e Expression levels of γH2AX and BRCA1 in irradiated LUAD cells with circNEIL3 overexpression. f Immunofluorescence staining of γH2AX in A549 cells with or without circNEIL3 overexpression. g Expression levels of γH2AX and BRCA1 in irradiated LUAD cells with PIF1 knockdown. h Immunofluorescence staining of γH2AX in A549 cells treated with si-nc or si-PIF1. i Immunofluorescence co-staining of γH2AX and ASC in A549 cells treated with si-nc or si-PIF1. Scale bar, 50 μm. *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed Student’s t-test. Con, control; IR, irradiated.
Previous studies have shown that the host NEIL3 gene of circNEIL3 is a DNA glycosylase, which functions to remove DNA oxidative damage and modifies base excision repair [34, 35]. PIF1 helicase can affect the DNA replication of telomeres, ribosomes, and mitochondria; prevents the inappropriate addition of telomeres during DNA double-strand breaks; and reduces DNA damage [36–38]. Therefore, we next analyzed the γH2AX, a key marker of DNA damage.
After irradiation treatment, circNEIL3 overexpression significantly decreased γH2AX but increased BRCA1 protein levels (Fig. 6e). Whereas PIF1 knockdown promoted γH2AX, it inhibited BRCA1 protein levels compared to the control group (Fig. 6g). In parallel, immunofluorescence assay showed that the expression of γH2AX was reduced by circNEIL3 overexpression (Fig. 6f) but was increased by PIF1 knockdown (Fig. 6h). As DNA damage can activate the AIM2 inflammasome, thereby inducing cytokine production and GSDMD-mediated pyroptosis [39–41], we further detected the localization of ASC specks that co-stain with γH2AX. In A549 cells exposed to radiation following PIF1 knockdown, many of the ASC specks were detected in the nucleus of the DNA-damaged cells co-stained with γH2AX (Fig. 6i). Overall, these findings suggested that the circNEIL3/miR-1184/PIF1 axis mediates cell pyroptosis through the DNA damage pathway.
circNEIL3 knockdown promotes radiation efficiency in vivo
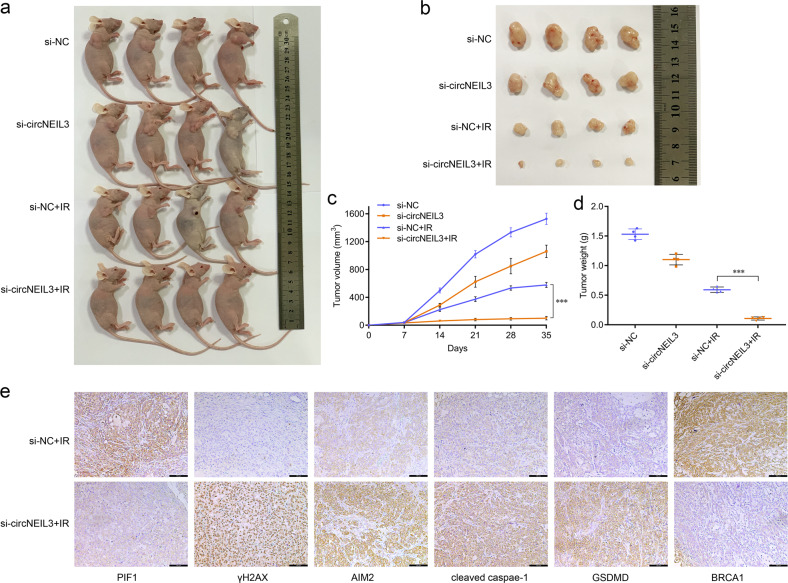
To further explore the therapeutic potential of circNEIL3 in vivo, we established a subcutaneous tumor model in nude mice using A549 cells with or without circNEIL3 knockdown (Fig. 7a). Tumor xenograft analysis indicated that circNEIL3 knockdown efficiently enhanced the sensitivity of xenograft tumors to radiation treatment, and reduced the size and weight of the xenograft tumors (Fig. 7b–d). Immunohistochemistry analysis of tumor xenograft samples suggested that in the radiation-treated group, circNEIL3 knockdown elevated the expression of γH2AX, AIM2, cleaved caspase-1, and GSDMD, but reduced the levels of PIF1 and BRCA1 (Fig. 7e).
Fig. 7. circNEIL3 knockdown promotes the radiation efficiency in vivo.
a Images of nude mice with subcutaneous xenografts in the indicated groups. b Xenograft tumors isolated from sacrificed mice with or without radiation treatment (0 or 4 Gy/time/week) at the end of the experiment. c Volumes of subcutaneous xenograft tumors. d Average weights of xenograft tumors in the indicated groups. e Immunohistochemistry staining of DNA damage and pyroptosis biomarkers in subcutaneous tumors of mice injected with si-NC+IR vs si-circNEIL3+IR (scale bar, 100 μm).
Discussion
The circRNA expression profile influences the occurrence and progression of cancer. Compared with linear parental genes, circRNAs are expressed more widely, stably, and specifically in cells, tissues, and body fluids [21, 42, 43], and thus have greater potential as targets for cancer treatment. However, the role of circRNAs in the process of pyroptotic cell death during LUAD radiotherapy remains unclear. In this study, we established, the first (to the best of our knowledge) expression profile of circRNAs in non‑irradiated and irradiated LUAD cells through RNA-seq. Subsequently, we identified circNEIL3 as a significantly downregulated circRNA in the LUAD cells treated with 0, 2, or 4 Gy of radiation. We then performed gain-of-function and loss-of-function experiments of circNEIL3 in LUAD cells, showing that circNEIL3 knockdown promoted cell pyroptosis, while circNEIL3 overexpression had the opposite effects, indicating that circNEIL3 was associated with the pyroptosis of LUAD cells treated with radiation.
The ceRNA hypothesis proposes that circRNAs eliminate the inhibitory effects of miRNAs on mRNA by acting as miRNA sponges, constructing a complex post-transcriptional regulatory network [19, 22, 44]. Comprehensive analysis of the ceRNA network and heatmap of the top 30 downregulated mRNAs revealed that PIF1 might be the downstream target of circNEIL3. The experimental findings showed that circNEIL3 knockdown significantly reduced mRNA and protein levels of PIF1, whereas circNEIL3 overexpression had the opposite effects. The effects of circNEIL3 overexpression on inhibiting pyroptosis were also reversed by PIF1 knockdown in irradiation-treated LUAD cells.
Previous studies have reported that circRNAs and mRNAs could share miRNA response elements, competing for the binding to miRNAs, and regulating each other’s expression [21–23]. We found that miR-1184 markedly reduced the luciferase activity of the circNEIL3 luciferase reporter, and this miRNA was subsequently verified as the binding target of circNEIL3. In addition, PIF1 was identified as the direct target of miR-1184, and circNEIL3 significantly attenuated the effects of miR-1184 on PIF1. These results indicate that circNEIL3 may function as a ceRNA to regulate PIF1 by binding to miR-1184 as a sponge in LUAD.
The extent of cell pyroptosis was increased by miR-1184 mimics, whereas circNEIL3 overexpression distinctly abolished the effects of miR-1184. The host gene NEIL3 of circNEIL3 plays an important role in eliminating DNA oxidative damage and base excision repair [34, 35, 45]. PIF1 helicase functions also affect DNA replication and DNA damage [36]. Therefore, we speculated that circNEIL3 could also affect DNA damage repair. Interestingly, the radiation-induced expression of γH2AX, a marker of DNA damage, was significantly weakened by circNEIL3 overexpression but was enhanced by PIF1 knockdown. In this study, we demonstrated that ASC specks co-stained with γH2AX in cells exposed to radiation after PIF1 knockdown, which suggests that the circNEIL3/miR-1184/PIF1 axis mediates cell pyroptosis induced by radiation through regulation of the DNA damage repair pathway.
To determine the effect of circNEIL3 on lung cancer radiotherapy in vivo, we established an A549 subcutaneous tumor model and showed that circNEIL3 knockdown can enhance radiation efficiency, resulting in a reduced size and weight of xenograft tumors accompanied by a higher expression of DNA damage and pyroptosis markers. Our study lacks validation with clinical specimens since it is difficult to obtain lung cancer tissue specimens following radiotherapy. Nonetheless, our results confirm the role of the circNEIL3/miR-1184/PIF1 axis in both tumor cells and subcutaneous tumors in mediating pyroptosis to regulate LUAD radiotherapy by regulating the DNA damage repair pathway.
Collectively, this study demonstrates that circNEIL3 exerts its function as a ceRNA by directly binding to miR-1184, thereby abrogating the endogenous inhibitory effect of miR-1184 on the target gene PIF1, and inducing significant DNA damage, which in turn triggers AIM2 inflammasome activation to induce pyroptosis, ultimately affecting survival in lung cancer radiotherapy (Fig. 8). These results suggest that the circNEIL3/miR-1184/PIF1 axis mediates cell pyroptosis by regulating the DNA damage repair pathway. Therefore, circNEIL3-related pyroptosis induction may serve as a potential therapeutic target to enhance radiotherapy in lung cancer.
Fig. 8. Diagram of the proposed mechanism and function of circNEIL3 in lung adenocarcinoma radiotherapy.
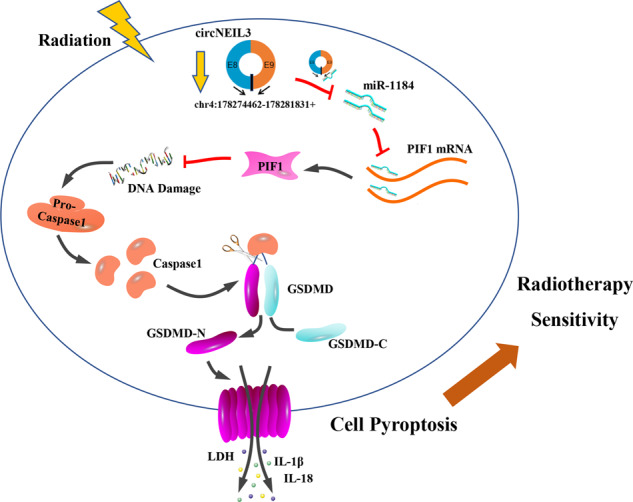
circNEIL3 exerts its function as a ceRNA by directly binding to miR-1184, thereby abrogating the endogenous inhibitory effect of miR-1184 on the target gene PIF1, and inducing significant DNA damage, which in turn triggers AIM2 inflammasome activation to induce pyroptosis, ultimately affecting survival in lung cancer radiotherapy.
Materials and methods
Cell lines and cultures
The human LUAD cell lines A549 and NCI‑H1299 were purchased from The American Type Culture Collection and were authenticated by STR DNA profiling analysis. The LUAD cell lines were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum, 10 mM l-glutamine, and 5 mg/ml penicillin/streptomycin at 37 °C with 5% CO2. All media and supplements were purchased from Invitrogen (Carlsbad, CA, USA).
Irradiation treatment
A549 cells (1 × 104 cells/well) were seeded in 96‑well plates. After incubation for 24 h, the cells were exposed to various doses (0, 2, or 4 Gy) of radiation using the X‑RAD 160‑225 instrument (42 cm; 225 kV/s; 12.4 mA; 2.0 Gy/min; filter, 2 mm aluminum; Precision X‑Ray, North Branford, CT, USA) [46]. Based on our previous study [47], we performed the optimal radiosensitivity assessment after 48 h of radiation treatment. The irradiated LUAD cell lines refer to the adherent cells in the petri dish 48 h after 4 Gy dose of radiation treatment.
Total RNA extraction and RT-qPCR
Total RNA was extracted using Total RNA Extraction Kit (Solarbio, Beijing, China), according to the manufacturer’s instructions, which was reverse-transcribed using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). qPCR was then performed using a CFX96 Real-time System (Bio-Rad) with SYBR Green Supermix (Bio-Rad). Both procedures were performed according to the manufacturer’s instructions. The β-actin was used to be the internal controls of RT-qPCR analysis. The sequences of the primers used in this study are listed in Table S3.
circRNA sequencing analysis
RNA libraries were constructed by CloudSeq Pte Ltd (Shanghai, China). circRNA-seq analysis was also performed by CloudSeq Pte Ltd. with a Bioanalyzer 2100 (Agilent, Santa Clara, CA) and sequenced by HiSeq 2000 (Illumina, San Diego, CA) on a 100 bp paired-end run [48]. The RNA-seq data were deposited in the Gene Expression Omnibus database (accession code: GSE124396).
Cell viability assay
Cell viability was assessed by clonogenic survival assay. When the cell density reached 70%, cells were exposed to various doses (0, 2, or 4 Gy) of radiation. After 48 h, trypsin digestion was performed, cells were counted and re-seeded at 500 cells per well in a 6-well plate (Corning, Corning, NY, USA). Cells were cultured for 10 days with medium changes every 3 days. Colonies were washed with PBS, fixed in methanol, and stained with crystal violet. The data were expressed as percent survival relative to the control: ((treated count)/(average ctrl count)) × 100%.
RNase R treatment
Total RNA (2 μg) was incubated at 37 °C for 30 min with or without 3 U/μg RNase R (Epicentre Technologies, Thane, Maharashtra, India), and subsequently purified using RNeasy MinElute Cleaning Kit (Qiagen, Hilden, Germany). All of the products obtained were used as substrates for complementary DNA synthesis and were analyzed by RT-qPCR.
Actinomycin D assay
A549 cells were exposed to 2 μg/ml actinomycin D (Sigma-Aldrich, St. Louis, MO, USA) at various time points (0, 4, 8, 12, and 24 h). Cells were harvested and total RNA was extracted. The stability of circNEIL3 and NEIL3 mRNA was analyzed using RT-qPCR.
Cytoplasm and nuclear localization
The cytoplasm and nuclear fractions were separated and isolated from cells using NE-PER Extraction Kit (Thermo Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. Cytoplasmic and nuclear RNAs were then converted to complementary DNAs and detected by RT-qPCR.
Vector construction and cell transfection
To overexpress circNEIL3 in LUAD cells, GM-7183: GPLVX-circRNA _Mini-GFP-Puro was used as the lentiviral vector and 1E8TU was used as the control virus. For circNEIL3 knockdown, siRNAs targeting the si-h-coding sequences were designed and inserted into a 2494-PGMLV-SC5 RNAi lentiviral vector. Both of the constructions of circNEIL3 overexpression and knockdown were performed by Genomeditech (Shanghai, China). Overexpression and knockdown efficiencies were evaluated by RT-qPCR. Cells were transfected using Lipofectamine 3000 (Invitrogen) and harvested after 48 h for further experiments.
PIF1 knockdown
PIF1 was silenced in LUAD cells with an siRNA Kit (RiboBio, Guangzhou, China), according to the manufacturer’s instructions. The target sequences were as follows: si-h-PIF1_001, 5′-GGTTGGTCATTGACGAGAT-3′; si-h-PIF1_002, 5′-GAGGCAGACCTGTTTGACA-3′; and si-h-PIF1_003, 5′-GGGAAGTCATATCTGCTAA-3′. The corresponding negative control was also purchased from RiboBio. Knockdown efficiency was assessed by RT-qPCR and western blotting.
LDH, IL-1β, and IL-18 release assay
LDH release was detected using LDH Cytotoxicity Assay Kit (Beyotime). The levels of IL-1β and IL-18 were measured with enzyme-linked immunosorbent assays using QuantiCyto IL-1β and IL-18 ELISA Kit, respectively (Enzyme-linked Biotechnology, Shanghai, China), according to the manufacturer’s instructions. The absorbance value was measured at 450 nm. Each experiment was repeated three times.
Flow cytometry
Flow cytometry was used to measure pyroptosis using Annexin V-PE/7-AAD Detection Kit (KeyGEN, Jiangsu, China), according to the manufacturer’s instructions. The cells were analyzed using a flow cytometer (FACSCalibur, Becton-Dickinson, Franklin Lakes, NJ, USA).
Western blot
The specific steps of the western blot assay were performed, as previously described [22, 33] with the following antibodies: anti-Caspase-1 (ab179515), anti-GSDMD (ab219800), anti-GSDMD-N (ab215203), anti-ASC (ab180799) and anti-AIM2 (ab93015) (Abcam, Cambridge, UK); anti-γH2AX (9718) (CST, Danvers, MA, USA); anti-PIF1 (19006-1-AP), anti-BRCA1 (22362-1-AP), anti-GAPDH (60004-1-Ig), and the secondary antibodies (Proteintech, Wuhan, China). Blots were conjugated with the chemiluminescent horseradish peroxidase substrate (Millipore), visualized, and quantified by Quantity 5.2 (Bio-Rad) according to the manufacturers’ instructions.
RNA immunoprecipitation
RNA immunoprecipitation experiments were performed using Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) according to the manufacturer’s protocols. AGO2 antibody was used for the RNA immunoprecipitation assay (Cell Signaling Technology). Co-precipitated RNA was detected by RT-qPCR.
Dual-luciferase reporter assay
For the luciferase reporter vector, the sequence of circNEIL3 and the PIF1 3′ untranslated region was cloned downstream of the pGL3-promoter. For the mutant version, the sequence fragment of circNEIL3 (478–484: GCTGCAG), the binding site for miR-1184, was changed to CGACGTC with pmiR-RB-ReportTM vector (RiboBio). Similarly, the sequence fragment of PIF1 (319–325: GCTGCAG) was changed to CGACGTC with pmiR-RB-ReportTM vector. The constructed and identified wild-type and mutant circNEIL3 and PIF1 forms were verified by RiboBio. The specific steps of the dual luciferase reporter assay were performed as described previously [22]. The luciferase activity was measured using a dual luciferase reporter assay system (Promega, Madison, WI, USA). The luciferase values were normalized to the corresponding Renilla luciferase values, and then the fold changes were calculated.
Immunofluorescence
The immunofluorescence assay was performed according to previous reports [7, 23]. Immunofluorescence images were obtained using an Olympus BX51 microscope (Olympus, Tokyo, Japan) equipped with various objectives and a DP50 camera. Images were processed using DPC controller software (Olympus).
Mouse xenograft model
All xenograft experiments were performed in accordance with the guidelines of the Laboratory Animal Ethical Committee at Chengdu Medical College. All experimental protocols were approved by the Laboratory Animal Ethical Committee at Chengdu Medical College. A subcutaneous tumorigenic model was established to study the role of circNEIL3 on the efficacy of LUAD radiotherapy in vivo. BALB/c-Female nude mice (4–5 weeks of age, 14–16 g) were purchased from Dossy Experimental Animals Co., Ltd (Chengdu, China). Before performing the experiment, mice were randomly assigned to four treatment groups (si-NC, si-circNEIL3, si-NC + IR and si-circNEIL3+IR), with four mice in each group. A549 cells with or without circNEIL3 knockdown (1 × 106) were suspended in 100 μl serum-free RPMI-1640 medium and injected subcutaneously into the axilla of each nude mouse. Seven days after cell implantation, when the tumor volume was approximately 30–40 mm3, each nude mice in the IR groups was exposed to radiation treatment under anesthesia using an X-RAD 160-225 instrument (Precision XRay, Inc., Branford, CT, USA; filter: 2 mm AI; 42 cm, 225 kV/s, 12.4 mA, 2.0 Gy/min) (4 Gy dose/each time per mice per week). Individual cylindrical lead cover shields were used so that only the subcutaneous tumor in the axilla was exposed to radiation. Vernier calipers were used to measure the tumor growth every 7 days, according to the formula V = (a × b2)/2 to detect the tumor volume, where a and b are the maximum and minimum diameter in millimeters, respectively. After four weeks, the mice were sacrificed and weighed immediately after dissection.
Immunohistochemistry
Immunohistochemistry was performed as previously reported [23]. The images were obtained using an Olympus BX51 microscope (Olympus, Tokyo, Japan) equipped with various objectives and a DP50 camera. Images were processed using DPC controller software (Olympus).
Statistical analysis
All experiments in this study were independently performed with at least three biological replicates. Data analysis was performed with the statistical program GraphPad Prism 7.0 (GraphPad Software, San Diego, CA, USA). The results are presented as the mean ± standard deviation unless otherwise indicated. Statistical analyses were performed using two-tailed Student’s t-tests to compare the significance of differences between two experimental groups. A value of P < 0.05 was considered statistically significant.
Supplementary information
Table S1. The top 30 dysregulated circRNAs in A549 cells treated with different doses of radiation.
Table S2. The top 10 meaningful circRNAs based on the junction reads and logCPM values by edgeR.
Table S3. Primer sequences for RT-qPCR used in this study.
Figure S1. Bioinformatics analysis of circRNAs in LUAD cells.
Figure S2. circNEIL3 knockdown can promote irradiation-induced pyroptosis in LUAD cells.
Figure S3. Knockdown efficiency of PIF1 detected by RT-qPCR and western blot.
Acknowledgements
We thank NovelBioinformatics Ltd., Co for their support with the bioinformatics analysis using their NovelBio Cloud Analysis Platform. This work was supported by the National Natural Science Foundation of China (Nos. 81972977 and 81802955), the Foundation of Chengdu Medical College (CYZ19-01), the Foundation of Health Commission of Sichuan Province (20ZD016), the Foundation of Sichuan Science and Technology Agency (2018JY0648 and 2019YJ0589), the Foundation of The First Affiliated Hospital of Chengdu Medical College (CYFY2017ZD03, CYFY2018ZD02, CYFY2019ZD06, and CYFY2020YB09) and the Foundation of Sichuan Medical Association (S20011).
Author contributions
Conducted the study design: YX, TZ, and D-MW; Carried out experiments and data analysis: TZ and D-MW; Provided technical support and material: P-WL, TL, RH, S-HD, MH, and Y-YZ; Wrote the manuscript: TZ and D-MW; Read and approved the final manuscript: All authors.
Data availability
The RNA-seq data were deposited in the GEO database (accession code: GSE124396).
Competing interests
The authors declare no competing interests.
Ethics statement
The study was approved by the Ethics Committee of Chengdu Medical College.
Footnotes
Edited by Dr. George Calin.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ting Zhang, Dong-Ming Wu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-022-04561-x.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyasaka Y, Komatsu S, Abe T, Kubo N, Okano N, Shibuya K, et al. Comparison of oncologic outcomes between carbon ion radiotherapy and stereotactic body radiotherapy for early-stage non-small cell lung cancer. Cancers (Basel) 2021;13:176. doi: 10.3390/cancers13020176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desideri I, Francolini G, Scotti V, Pezzulla D, Becherini C, Terziani F, et al. Benefit of ablative versus palliative-only radiotherapy in combination with nivolumab in patients affected by metastatic kidney and lung cancer. Clin Transl Oncol. 2019;21:933–8. doi: 10.1007/s12094-018-02005-7. [DOI] [PubMed] [Google Scholar]
- 5.Kyroudi A, Petersson K, Ozsahin E, Bourhis J, Bochud F, Moeckli R. Exploration of clinical preferences in treatment planning of radiotherapy for prostate cancer using Pareto fronts and clinical grading analysis. Phys Imaging Radiat Oncol. 2020;14:82–6. doi: 10.1016/j.phro.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park JS, Yu JI, Lim DH, Nam H, Kim YI, Lee J, et al. Impact of radiotherapy on kidney function among patients who received adjuvant treatment for gastric cancer: logistic and linear regression analyses. Cancers (Basel)13, 59 (2020). [DOI] [PMC free article] [PubMed]
- 7.Gao J, Peng S, Shan X, Deng G, Shen L, Sun J, et al. Inhibition of AIM2 inflammasome-mediated pyroptosis by Andrographolide contributes to amelioration of radiation-induced lung inflammation and fibrosis. Cell Death Dis. 2019;10:957. doi: 10.1038/s41419-019-2195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helm JS, Rudel RA. Adverse outcome pathways for ionizing radiation and breast cancer involve direct and indirect DNA damage, oxidative stress, inflammation, genomic instability, and interaction with hormonal regulation of the breast. Arch Toxicol. 2020;94:1511–49. doi: 10.1007/s00204-020-02752-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–22. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Man SM, Karki R, Kanneganti TD. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol Rev. 2017;277:61–75. doi: 10.1111/imr.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu F, Lan Z, Xin Z, He C, Guo Z, Xia X, et al. Emerging insights into molecular mechanisms underlying pyroptosis and functions of inflammasomes in diseases. J Cell Physiol. 2020;235:3207–21. doi: 10.1002/jcp.29268. [DOI] [PubMed] [Google Scholar]
- 12.Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, et al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 2016;535:111–6. doi: 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- 13.Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science. 2013;341:1250–3. doi: 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmad I, Muneer KM, Chang ME, Nasr HM, Clay JM, Huang CC, et al. Ultraviolet radiation-induced downregulation of SERCA2 mediates activation of NLRP3 inflammasome in basal cell carcinoma. Photochem Photobio. 2017;93:1025–33. doi: 10.1111/php.12725. [DOI] [PubMed] [Google Scholar]
- 15.Liu D, Kong F, Yuan Y, Seth P, Xu W, Wang H, et al. Decorin-modified umbilical cord mesenchymal stem cells (MSCs) attenuate radiation-induced lung injuries via regulating inflammation, fibrotic factors, and immune responses. Int J Radiat Oncol Biol Phys. 2018;101:945–56. doi: 10.1016/j.ijrobp.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Tao X, Sun M, Chen M, Ying R, Su W, Zhang J, et al. HMGB1-modified mesenchymal stem cells attenuate radiation-induced vascular injury possibly via their high motility and facilitation of endothelial differentiation. Stem Cell Res Ther. 2019;10:92. doi: 10.1186/s13287-019-1197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu YG, Chen JK, Zhang ZT, Ma XJ, Chen YC, Du XM, et al. NLRP3 inflammasome activation mediates radiation-induced pyroptosis in bone marrow-derived macrophages. Cell Death Dis. 2017;8:e2579. doi: 10.1038/cddis.2016.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao H, Wang H, Rong X, Li E, Xu RH, Peng Y. Mesenchymal stem cells attenuate radiation-induced brain injury by inhibiting microglia pyroptosis. Biomed Res Int. 2017;2017:1948985. doi: 10.1155/2017/1948985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 20.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–8. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang S, et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19:218. doi: 10.1186/s13059-018-1594-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng Z, Yu C, Cui S, Wang H, Jin H, Wang C, et al. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat Commun. 2019;10:3200. doi: 10.1038/s41467-019-11162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang X, Li Z, Zhang Q, Wang W, Li B, Wang L, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. 2019;18:71. doi: 10.1186/s12943-019-0969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan WLW, Lim BTS, Anene-Nzelu CGO, Ackers-Johnson M, Dashi A, See K, et al. A landscape of circular RNA expression in the human heart. Cardiovasc Res. 2017;113:298–309. doi: 10.1093/cvr/cvw250. [DOI] [PubMed] [Google Scholar]
- 25.Yu F, Tie Y, Zhang Y, Wang Z, Yu L, Zhong L, et al. Circular RNA expression profiles and bioinformatic analysis in coronary heart disease. Epigenomics. 2020;12:439–54. doi: 10.2217/epi-2019-0369. [DOI] [PubMed] [Google Scholar]
- 26.Luan J, Jiao C, Kong W, Fu J, Qu W, Chen Y, et al. circHLA-C plays an important role in lupus nephritis by sponging miR-150. Mol Ther Nucleic Acids. 2018;10:245–53. doi: 10.1016/j.omtn.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shao Y, Chen Y. Roles of circular RNAs in neurologic disease. Front Mol Neurosci. 2016;9:25. doi: 10.3389/fnmol.2016.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68:1214–27. doi: 10.1016/j.jhep.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Zhang J, Li J, Gui R, Nie X, Huang R. CircRNA_014511 affects the radiosensitivity of bone marrow mesenchymal stem cells by binding to miR-29b-2-5p. Bosn J Basic Med Sci. 2019;19:155–63. doi: 10.17305/bjbms.2019.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo J, Zhang C, Zhan Q, An F, Zhu W, Jiang H, et al. Profiling circRNA and miRNA of radiation-induced esophageal injury in a rat model. Sci Rep. 2018;8:14605. doi: 10.1038/s41598-018-33038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, Xue N, Guo Y, Niu K, Gao L, Zhang S, et al. CircRNA_100367 regulated the radiation sensitivity of esophageal squamous cell carcinomas through miR-217/Wnt3 pathway. Aging (Albany NY) 2019;11:12412–27. doi: 10.18632/aging.102580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu D, Li Y, Ming Z, Wang H, Dong Z, Qiu L, et al. Comprehensive circular RNA expression profile in radiation-treated HeLa cells and analysis of radioresistance-related circRNAs. PeerJ. 2018;6:e5011. doi: 10.7717/peerj.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang JF, Zhao XH, Wang YB, Ren FH, Sun DW, YB Y, et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11:32. doi: 10.1038/s41419-020-2230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krokeide SZ, Laerdahl JK, Salah M, Luna L, Cederkvist FH, Fleming AM, et al. Human NEIL3 is mainly a monofunctional DNA glycosylase removing spiroimindiohydantoin and guanidinohydantoin. DNA Repair (Amst) 2013;12:1159–64. doi: 10.1016/j.dnarep.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Semlow DR, Zhang J, Budzowska M, Drohat AC, Walter JC. Replication-dependent unhooking of DNA interstrand cross-links by the NEIL3 glycosylase. Cell. 2016;167:498–511. doi: 10.1016/j.cell.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deegan TD, Baxter J, Ortiz Bazan MA, Yeeles JTP, Labib KPM. Pif1-family helicases support fork convergence during DNA replication termination in eukaryotes. Mol Cell. 2019;74:231–44. doi: 10.1016/j.molcel.2019.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S, Wang H, Jehi S, Li J, Liu S, Wang Z, et al. PIF1 helicase promotes break-induced replication in mammalian cells. EMBO J. 2021;40:e104509. doi: 10.15252/embj.2020104509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pohl TJ, Zakian VA. Pif1 family DNA helicases: a helpmate to RNase H? DNA Repair (Amst) 2019;84:102633. doi: 10.1016/j.dnarep.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–13. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–8. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lammert CR, Frost EL, Bellinger CE, Bolte AC, McKee CA, Hurt ME, et al. AIM2 inflammasome surveillance of DNA damage shapes neurodevelopment. Nature. 2020;580:647–52. doi: 10.1038/s41586-020-2174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu X, Zhang J, Tian Y, Gao Y, Dong X, Chen W, et al. CircRNA inhibits DNA damage repair by interacting with host gene. Mol Cancer. 2020;19:128. doi: 10.1186/s12943-020-01246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu M, et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer. 2020;19:101. doi: 10.1186/s12943-020-01221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei Y, Chen X, Liang C, Ling Y, Yang X, Ye X, et al. A noncoding regulatory RNAs network driven by circ-CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatology. 2020;71:130–47. doi: 10.1002/hep.30795. [DOI] [PubMed] [Google Scholar]
- 45.Imani NM, Housh K, Rodriguez AA, Haldar T, Kathe S, Wallace SS, et al. Unhooking of an interstrand cross-link at DNA fork structures by the DNA glycosylase NEIL3. DNA Repair (Amst) 2020;86:102752. doi: 10.1016/j.dnarep.2019.102752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu D, Han R, Deng S, Liu T, Zhang T, Xie H, et al. Protective effects of flagellin A N/C against radiation-induced NLR pyrin domain containing 3 inflammasome-dependent pyroptosis in intestinal cells. Int J Radiat Oncol Biol Phys. 2018;101:107–17. doi: 10.1016/j.ijrobp.2018.01.035. [DOI] [PubMed] [Google Scholar]
- 47.Zhang T, Wu DM, Deng SH, Han R, Liu T, Li J, et al. RNAseq profiling of circRNA expression in radiation-treated A549 cells and bioinformatics analysis of radiation-related circRNA-miRNA networks. Oncol Lett. 2020;20:1557–66. doi: 10.3892/ol.2020.11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The top 30 dysregulated circRNAs in A549 cells treated with different doses of radiation.
Table S2. The top 10 meaningful circRNAs based on the junction reads and logCPM values by edgeR.
Table S3. Primer sequences for RT-qPCR used in this study.
Figure S1. Bioinformatics analysis of circRNAs in LUAD cells.
Figure S2. circNEIL3 knockdown can promote irradiation-induced pyroptosis in LUAD cells.
Figure S3. Knockdown efficiency of PIF1 detected by RT-qPCR and western blot.
Data Availability Statement
The RNA-seq data were deposited in the GEO database (accession code: GSE124396).