Abstract
Coronavirus Disease-19 (COVID-19) symptoms range from mild to severe illness; the cause for this differential response to infection remains unknown. Unravelling the immune mechanisms acting at different levels of the colonization process might be key to understand these differences. We carried out a multi-tissue (nasal, buccal and blood; n = 156) gene expression analysis of immune-related genes from patients affected by different COVID-19 severities, and healthy controls through the nCounter technology. Mild and asymptomatic cases showed a powerful innate antiviral response in nasal epithelium, characterized by activation of interferon (IFN) pathway and downstream cascades, successfully controlling the infection at local level. In contrast, weak macrophage/monocyte driven innate antiviral response and lack of IFN signalling activity were present in severe cases. Consequently, oral mucosa from severe patients showed signals of viral activity, cell arresting and viral dissemination to the lower respiratory tract, which ultimately could explain the exacerbated innate immune response and impaired adaptative immune responses observed at systemic level. Results from saliva transcriptome suggest that the buccal cavity might play a key role in SARS-CoV-2 infection and dissemination in patients with worse prognosis. Co-expression network analysis adds further support to these findings, by detecting modules specifically correlated with severity involved in the abovementioned biological routes; this analysis also provides new candidate genes that might be tested as biomarkers in future studies. We also found tissue specific severity-related signatures mainly represented by genes involved in the innate immune system and cytokine/chemokine signalling. Local immune response could be key to determine the course of the systemic response and thus COVID-19 severity. Our findings provide a framework to investigate severity host gene biomarkers and pathways that might be relevant to diagnosis, prognosis, and therapy.
Keywords: COVID-19 severity, Gene expression, Immune response, Pathways analysis, Multi-tissue, Differential expression analysis, Co-expression analysis, SARS-CoV-2
Abbreviations: BALF, Boncho-Alveolar Lavage Fluids; COVID-19, Coronavirus Disease 19; CRP, C-Reactive Protein; DEG, Differentially Expressed Gene; DE, Differential Expression; GEO, Gene Expression Omnibus; GO, Gene Ontology; GSEA, Gene-Set Enrichment Analysis; GS, Gene Significance; HK, House Keeping; H-B, Benjamini-Hochberg; ISGs, Interferon Stimulated Genes; IFN, Interferon; LRT, Lower Respiratory Track; MM, Module membership; NETs, Neutrophil Extracellular Traps; NP, Naso-Pharyngeal; ORA, Over-Representation; PAMPs, Pathogen-Associated Molecular Patterns; PCA, Principal Component Analysis; PRR, Pattern Recognition Receptors; QC, Quality Control; SARS-CoV-2, Evere acute Respiratory Syndrome Coronavirus 2; SD, Standard Deviation; TOM, Topological Overlap Matrix
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes the coronavirus disease 19 (COVID-19), was first identified in patients suffering from a respiratory disease in Wuhan (Hubei province, China) in December 2019 (Gómez-Carballa et al., 2020b; Pipes et al., 2020). Soon after, the virus quickly spread worldwide favored by two main catalyzers, namely, variants showing higher transmissibility (Davies et al., 2021) and superspreading events (Althouse et al., 2020; Gómez-Carballa et al., 2020a; Lemieux et al., 2020; Gómez-Carballa et al., 2021).
COVID-19 is mainly characterized by producing influenza-like illness (Jin et al., 2020; Lamers et al., 2020). A wide range of symptoms have been described, from absolutely no symptoms to severe pneumonia, lung damage, and progressive respiratory failure (Huang et al., 2020). Investigating the immunological processes underpinning SARS-CoV-2 pathogenesis is key for the development of efficient therapeutic strategies and vaccines against COVID-19. The immune system dysregulation described in severe patients is characterized by a reduce number of lymphocyte T subpopulations CD4+ and CD8+, pointing to an absence of T-cell specific response (Qin et al., 2020; Yang et al., 2020; Zheng et al., 2020b). Impairment of the adaptative immune system, together with a non-effective host immune response by innate cells, could be the cause of a higher viral replication and the activation of inflammatory processes, resulting in tissue damage and a more severe disease course (Gomez-Rial et al., 2020; Qin et al., 2020). The hyper-inflammatory response occurring during the acute phase is mainly driven by the monocyte-macrophage activation (Gómez-Rial et al., 2020), the release of pro-inflammatory cytokines (McGonagle et al., 2020; Qin et al., 2020; Zhang et al., 2020) and increase of other mediators e.g. ferritin and C-reactive protein (CRP). The infiltration of monocytes-derived macrophages from blood points to these cells as the main cause of the lung tissue damage in severe cases (Gomez-Rial et al., 2020).
Gene expression studies conducted on blood samples from COVID-19 patients indicated that genes associated with inflammatory and hypercoagulability pathways (Zhou et al., 2021) and the imbalance between innate and adaptive immune responses are the main factors responsible for severe disease course (Zheng et al., 2020a). Thair et al. (2021) found a differential transcriptomic pattern in blood that differentiated COVID-19 patients from other viral infections, suggesting that their top differentially expressed genes (DEGs) may be linked to new mechanisms of pathogen evasion from host immune response. Most recently, Aschenbrenner et al. (2021) revealed neutrophil activation signatures in severe cases, coupled with an elevated expression of genes related to coagulation and platelet function, and absence of T-cell activation.
Most of the studies to date were carried out in vitro or nasal-derived epithelial cells (Gamage et al., 2020; Sajuthi et al., 2020; Sungnak et al., 2020; Alfi et al., 2021). A gene expression study carried out on naso-pharyngeal (NP) swabs of infected patients pointed to different expression patterns depending on the viral load, age, and sex (Lieberman et al., 2020). Comparison of expression profiles of infected adults and children (Pierce et al., 2021) indicated an early and more efficient innate immune response in nasal mucosa from children, thus conditioning the final outcome of the disease. So far, only one study undertaken in three severe patients, has proposed candidate transcripts for severity and disease outcome in NP samples (Jain et al., 2021).
Saliva has also been proposed as a good alternative to NP swabs for SARS-CoV-2 detection (Pasomsub et al., 2020; To et al., 2020; Wyllie et al., 2020). The study of the specific host gene expression profile produced by the infection in oral mucosa has not raised comparable interest; only a recent study by Huang et al. (2021) highlighted the importance of the oral cavity in viral transmission and dissemination.
We carried out a multi-source gene expression study in blood, nasal, and saliva samples, collected on asymptomatic, mild, and severe patients. Our study aims at understanding the differential immune response that leads to different disease severities, and finding specific severity-related biomarkers to improve patient management and prognostic prediction.
2. Materials and methods
2.1. Patient recruitment
We prospectively collected blood, saliva, and nasal epithelium samples from 52 COVID-19 patients with confirmed PCR-positive diagnosis of SARS-CoV-2 infection at the Hospital Clínico Universitario of Santiago de Compostela (Galicia; Spain) from March to June 2020. We also recruited samples from healthy controls, defined as asymptomatic subjects with no diagnosis of COVID-19 disease at the time of sample collection. To avoid potential effects of ancestry on gene expression (Barral-Arca et al., 2019), we assured that patients and healthy controls were of (self-reported) South-European ancestry. In addition, we also carried out a maximum likelihood estimation of individual ancestries from multi-locus SNP data using whole exome sequencing data available from a significant proportion of the patients (35/52; data not shown). Procedures for exome data processing, quality control, variant annotation and estimation of ancestries were performed as done previously (Salas et al. 2017, 2018). All the analyzed samples were of European genomic ancestry (Supplementary Table 1) corroborating self-reported expectations.
Cases were classified at the time of sample collection by severity illness: severe (ICU admission), moderate (non-ICU but admitted to hospital) and mild (domiciliary lockdown patients with mild symptoms or asymptomatic; herein mild patients). Comparisons were carried out as follows: (i) whole blood from 41 patients and 13 controls, (ii) nasal epithelium from 38 patients and 11 controls, and (iii) saliva from 41 patients and 12 controls. The complete tissue sample set could be collected for 27 out of the 52 patients. Demographic and clinical data from the patients are summarized in Table 1 (see Supplementary Table 1 for more details).
Table 1.
Demographic and clinical characteristics of the COVID-19 cohort and controls. Phenotype stratification of the patients refers to the severity at the time of sample collection. Some of the clinical parameters are only available for hospitalized cases (moderate and severe). Quantitative variables are represented by the median (IQR) and qualitative variables as percentages with absolute numbers in brackets. Asterisks indicate statistically significant differences between groups.
| Controls | Mild | Moderate | Severe | ||
|---|---|---|---|---|---|
| Gender | |||||
| Female | 68.0% (17/25) | 56.5% (13/23) | 50.0% (7/14) | 26.7% (4/15) | |
| Male | 32.0% (8/25) | 43.5% (10/23) | 50.0% (7/14) | 73.3% (11/15) | |
| Age | |||||
| 41 (34–52) | 38 (29.5–46.5) | 58.5 (37.75–67.75) | 68 (58–71) | ||
| Final Severity | |||||
| Control | 100.0% (25/25) | 0.0% (0/23) | 0.0% (0/14) | 0.0% (0/15) | |
| Death | 0.0% (0/25) | 0.0% (0/23) | 0.0% (0/14) | 6.7% (1/15) | |
| Mild | 0.0% (0/25) | 100.0% (23/23) | 0.0% (0/14) | 0.0% (0/15) | |
| Moderate | 0.0% (0/25) | 0.0% (0/23) | 92.9% (13/14) | 0.0% (0/15) | |
| Severe | 0.0% (0/25) | 0.0% (0/23) | 7.1% (1/14) | 93.3% (14/15) | |
| Comorbidity | |||||
| 4.0% (1/25) | 26.1% (6/23) | 78.6% (11/14) | 86.7% (13/15) | ||
| Days of symptoms to sampling | |||||
| 8 (6.75–10.25) | 13 (12–15.75) | 12 (9.5–14.5) | |||
| Admission days | |||||
| – | – | 9.5 (6.5–11) | 34 (24.5–53) | ||
| Admission days to sampling | |||||
| – | – | 4 (3–5) | 4 (2–6) | ||
| ICU | |||||
| ICU | – | – | 7.1% (1/14) | 100% (15/15) | |
| ICU days | – | – | 11 (11–11) | 22.5 (10.75–28.75) | |
| ICU days to sampling | – | – | – | 3 (1.5–5) | |
| Symptom | |||||
| Fever | – | – | 92.9% (13/14) | 86.7% (13/15) | |
| Respiratory | – | – | 100% (14/14) | 93.3% (14/15) | |
| Gastrointestinal | – | – | 42.9% (6/14) | 28.6% (4/14) | |
| Musculoskeletal | – | – | 64.3% (9/14) | 60.0% (9/15) | |
| Sensory | – | – | 21.4% (3/14) | 26.7% (4/15) | |
| Headache | – | – | 35.7% (5/14) | 0.0% (0/15) | |
| Xray Infiltrates | |||||
| – | – | 71.4% (10/14) | 86.7% (13/15) | ||
| Xray pneumonia with consolidation | |||||
| – | – | 64.3% (9/14) | 50% (7/14) | ||
| Xray Pleural Effusion | |||||
| – | – | 14.3% (2/14) | 7.1% (1/14) | ||
| Treatments | |||||
| Oxygen | – | – | 64.3% (9/14) | 100% (14/14) | |
| Invasive Vent | – | – | 7.1% (1/14) | 78.6% (11/14) | |
| Inotropes | – | – | 0.0% (0/14) | 35.7% (5/14) | |
| Haemofiltration | – | – | 0.0% (0/14) | 14.3% (2/14) | |
| Corticoids before sample collection | – | – | 14.3% (2/14) | 66.7% (10/15) | |
| Antibiotics | – | – | 100% (14/14) | 100% (15/15) | |
| Antiviral | – | – | 78.6% (11/14) | 100% (15/15) | |
| Antimalarial | – | – | 100% (14/14) | 100% (15/15) | |
| Monoclonal antibody | – | – | 0.0% (0/14) | 46.6% (7/15) | |
| Analytical parameters | |||||
| White Cells (109/L) | – | – | 5.505 (4.82–6.3725) | 9.41 (5.275–12.36) | |
| Platelets (109/L) | – | – | 273 (227.25–400.75) | 203 (129–255.5) | |
| Neutrophils (109/L)* | – | – | 3.095 (2.445–3.78) | 6.89 (3.185–10.55) | |
| Lymphocytes (109/L) * | – | – | 1.32 (1.1875–1.52) | 0.47 (0.415–0.905) | |
| Monocytes (109/L) | – | – | 0.42 (0.29–0.58) | 0.33 (0.195–0.475) | |
| Fibrinogen (mg/dl) * | – | – | 500 (500–500) | 373.5 (318–450) | |
| Creatinine (mg/dl) | – | – | 0.75 (0.58–1.03) | 0.8 (0.765–0.94) | |
| CRP (mg/L) | – | – | 23.39 (15.5025–53.63) | 48.255 (11.15–99.6225) | |
| PCT (mg/ml) | – | – | 0.06 (0.04–0.1375) | 0.15 (0.105–0.2725) | |
| Ferritin (ng/dl) * | – | – | 398.5 (139.75–491) | 1584.5 (704–2934) | |
| D–Dimer (ng/ml) * | – | – | 518 (402–763) | 2038 (861–7832) | |
| Lactate (mmol/L) * | – | – | 0.9 (0.85–0.95) | 1.72 (1.42–2.39) | |
| Maximum CRP (mg/L) * | – | – | 49.225 (17.795–123.78) | 134.205 (71.9475–172.29) | |
| Maximum Neutrophils (109/L) * | – | – | 4.56 (3.4775–6.8) | 9.715 (5.85–11) | |
| Minimum Lymphocytes (109/L) * | – | – | 0.87 (0.7–1.1675) | 0.43 (0.345–0.4775) | |
The study was approved by the Ethical Committee of Clinical Investigation of Galicia (CEIC ref. 2020/178) and was conducted in accordance with the principles of the Declaration of Helsinki and all applicable Spanish legislation, namely Biomedical Research Act (Law 14/2007–3 of July), Act 41/2002 on the Autonomy of the Patient, Decree SAS/3470/2009 for Observational Studies, and 15/1999 Data Protection Act. Written informed consent was obtained for subjects before study inclusion.
2.2. Sample collection and processing
Blood, nasal epithelium, and saliva samples were sampled from patients and controls at the same timepoints. Blood samples (2.5 ml) were collected in PAXgene tubes (BD). RNA was isolated using PAXgene blood miRNA extraction kit (Qiagen) following manufacturer recommendations and carrying out the on-column DNase I treatment during the extraction process. Saliva and nasal epithelium samples were collected in Oragene CP-190 kit (DNA Genotek). We used sponges (SC-2 kit from DNA Genotek) to assist saliva collection in ICU patients unable to spit. RNA from saliva and nasal epithelium specimens was isolated using 500 μl of sample and the RNeasy microkit (Qiagen). We used Oragene RNA neutralizer solution (DNA Genotek) to precipitate impurities and inhibitors and slightly modified the protocol provided by the extraction kit as recommended by the Oragene tubes supplier. After isolation, RNA amount and integrity was checked using TapeStation 4200 (Agilent), and DV200 values were calculated to both ensure that >50% of the of the RNA fragments were above 200 nt and estimate the optimal sample input for the nCounter analysis. Nasopharyngeal samples were collected in Universal Transport Medium tubes (UTM) tubes to check for the presence as well as the viral load of SARS-CoV-2 at the same time as collection of blood, saliva, and nasal epithelium samples. Molecular detection of viral particles was performed through a multiplex real-time PCR using the Allplex™SARS-CoV-2 Assay (Seegene).
2.3. Nanostring nCounter expression assay
Immunological gene expression patterns of different tissues from COVID-19 patients and controls were evaluated through the SPRINT nCounter system (NanoString Technologies) and the Immunology Panel (594 genes involved in immunological processes). This panel covers the main immunological routes, including major classes of cytokines and their receptors, chemokine ligands and receptors, IFNs and their receptors, the TNF-receptor superfamily, and the KIR family genes. We used the standard gene expression protocol for 12x RNA hybridization with 5 μl of RNA as input, and a hybridization time of 18h for all samples. We mixed control and COVID-19 samples in all the same plates to avoid technical sample bias. We first carried out a quality control (QC) of the raw data checking some technical parameters following manufacturer recommendations to verify the absence of technical problems. Samples that did not pass technical QC, or with low number of genes detected, were excluded for downstream analysis.
2.4. Sample normalization and differential expression analysis
For the longitudinal multi-tissue experiment, normalization of the gene expression data was carried out for each tissue separately by selecting the optimal number and best reference candidate from the set of housekeeping (HK) genes included in the panel, as well as other endogenous genes that showed high levels of stability between samples. The GeNorm algorithm (Vandesompele et al., 2002) implemented in the R package CrtlGene (Zhong, 2019) was used to test for the most stable and optimal number of genes for normalization. We excluded HK candidate genes with less than 50 raw counts in any of the samples compared from the reference selection analysis. Data normalization was performed through a combined approach using both DESeq2 (Love et al., 2014) and RUVSeq (Risso et al., 2014) as described in (Bhattacharya et al., 2020). Genes with counts below the background (mean + 2 standard deviations [SD] of the negative control spikes in the code set) were not considered for the differential expression (DE) analysis. After data normalization and background thresholding, we eliminated lower expressed genes from the list of DEG, retaining a total of 495 genes in blood samples, 544 genes in nasal epithelium samples and 545 genes in saliva samples for downstream analysis. DEG between cohorts were obtained using DESeq2 (Love et al., 2014). We used age and sex as covariables to correct the model. Volcano plots and heatmaps were built with EnhancedVolcano (Blighe et al., 2020) and ComplexHeatmap (Gu et al., 2016) R packages, respectively. All graphics were generated using R (www.r-project.org) software and the ggplot2 package (Wickham, 2016). Wilcoxon test was used to assess statistical significance between patient groups, and Spearman test for the correlation indices. We focused on highly statistically significant expression changes by considering as thresholds an adjusted P-value < 0.05, and log2FC ≥ |1.5| for downstream analysis. For the transversal multi-tissue experiment we sub-selected 21 patients for which samples from the three tissues were available (mild: n = 11; severe: n = 10). We employed a complex model design including the tissue/severity interaction to find tissue-specific severity effects. As references for the comparisons in the transversal analysis, we used blood tissue and the mild severity category. We followed the same procedure as in the longitudinal analysis but data from the three tissues were normalized together after selecting a set of adequate reference genes.
2.5. Pathway analysis
We followed both over-representation (ORA) and gene-set enrichment (GSEA) (Subramanian et al., 2005) approaches to identify biological functions, cellular components and pathways related to the differences in gene expression. For ORA we used the DEG as input (threshold: log2FC ≥ |1.5|), and the genes included in the expression panel as pool for statistical calculations. For GSEA we used all available molecular measurements (log2FC) from the genes included in the DE analysis to detect coordinated changes in the expression of genes from same pathway. Analyses were carried out using the Clusterprofiler (Yu et al., 2012) R package. We applied the Benjamini-Hochberg (H–B) procedure for multiple test correction and a significance threshold of 0.05. We tested both the GO (Gene Ontology) and Reactome databases. Graphics were obtained with the R package enrichplot (Yu, 2021).
2.6. Validation analysis
To validate the specificity of the transcriptomic findings from our blood samples, we first used two independent datasets: (a) Thair et al., (2021) included RNA-seq transcriptomes for 62 SARS-CoV-2 positive patients and 24 healthy controls; and (b) Aschenbrenner et al., (2021) included RNA-seq transcriptomes for 39 COVID-19 patients and 10 healthy controls. We additionally checked the specificity of our COVID-19 candidate genes with respect to other viral infections by correlating our results with those from microarray data comparing mixed viral infections (n = 652) vs. healthy control samples (n = 672) reported in Thair et al. (2021).. Independently, to further test the gene expression pattern of our best candidate genes in other viral and bacterial infections, we carried out an independent validation study by meta-analyzing other datasets (n = 513 viral, n = 382 bacterial and n = 215 healthy control samples) ;downloaded from the public gene expression repository Gene Expression Omnibus (GEO) under accession numbers: GSE64456 (Mahajan et al., 2016), GSE40012 (Parnell et al., 2012), GSE42026 (Herberg et al., 2013), GSE60244 (Suarez et al., 2015), also included in Thair et al., (2021) as viral cohorts; and GSE72829 (Herberg et al., 2016), GSE69529 (DeBerg et al., 2018). To merge and integrate all these data , we first normalized and pre-processed each dataset separately using Lumi (Du et al., 2008) for Illumina® microarrays data and Oligo (Carvalho and Irizarry, 2010) for Affymetrix® microarray datasets, while RNA-seq data were pre-processed as described previously (Barral-Arca et al., 2018). Then, we merged these databases retaining only the common genes. Subsequently, we used the R package COCONUT to combine all datasets into one and reduce batch effects in the meta-analysis (Sweeney et al., 2016). Next, we used Limma package (Ritchie et al., 2015) to detect DEG and calculate log2FC values between groups. Scatterplots of log2FC values and Pearson correlation indexes were obtained for the validation analysis.
2.7. Leukocyte proportion estimation from gene expression data
We imputed immune cell subset abundances from expression data in different patient categories, including mild and healthy controls, from which real immune cell counts from blood tests were not available. We used the CIBERSORTX (Newman et al., 2019) tool and a leukocyte gene signature matrix (Newman et al., 2015) obtained from microarray data as reference. We applied a B-mode batch correction to account for cross-platform variation and 500 permutations for the analysis. We graphically represented immune cell proportions using R software and carried out a Wilcoxon test to assess statistical significance between categories.
2.8. Weighted gene co-expression network analysis
We generated a signed weighted correlation networks from gene expression data with the WGCNA R package (Langfelder and Horvath, 2008) in order to find co-expression modules related to mild-severe phenotypes in the different tissues analyzed. We used normalized expression data (and corrected for differences in gender and age) from genes showing the most variant expression values between samples (top 50% with higher variance) as input, and we chose a soft-thresholding power based on the criterion of scale-free topology after testing a set of candidate powers (powers selected were 16, 9 and 18 for blood, nasal epithelium and saliva samples, respectively). The similarity matrix between each pair of genes across all samples was calculated using correlation values and transformed into an adjacency matrix. Subsequently, the topological overlap matrix (TOM) and the corresponding dissimilarity (1–TOM) values were computed. The resulting topological overlap is a biologically meaningful measure of gene similarity based on the co-expression relationship between two genes. As co-expression module detection parameters we chose a minimum module size of 30, a medium sensitivity for cluster splitting, and a 0.2 value as dendrogram cut heigh threshold for module merging.
Correlation between module eigengenes and clinical traits were analyzed to identify modules of interest significantly associated with the mild-severe phenotype (gene significance [GS]). Module membership (MM), as a measurement of intramodular connectivity, was also calculated. We explored the correlation between GS and MM, and calculated the average absolute gene significance for all genes within a module in order to highlight the most important modules. The top hub genes within the most relevant modules were selected using both MM > 0.7 and a P-value for GS < 0.05 as thresholds.
We studied biological significance of the hub genes from the phenotype-related modules by performing an over-representation analysis and using the compare cluster function included in the Clusterprofiler R package (Yu et al., 2012).
3. Data availability
Expression data that support the findings of the present study have been deposited in Gene Expression Omnibus (GEO) database under the accession code GSE183071.
4. Results
4.1. Cohort description
Most of the clinical features were only available for hospitalized patients (Table 1). Similar median symptoms days to sampling were obtained for mild, moderate, and severe cases, as well as admission days to sampling in the case of moderate and severe patients (P-value = 0.236 and 0.704, respectively). During admission, oxygen support was required for 64.3% of the moderate cases and 100% of the severe cases (78.6% required invasive ventilation). All moderate cases were treated with a double therapy including antibiotic and antimalarial drugs (in 78.6% of the moderate cases, antiviral therapy was also administrated). Severe cases received a combined triple therapy of antibiotic, antiviral (lopinavir and ritonavir) and antimalarial (hydroxychloroquine) treatment, and 46.6% of them received tocilizumab.
No statistical differences were observed in viral loads at the time of sample collection depending on COVID-19 severity, nor statistical correlation between days of symptoms to sample collection and Ct values from the SARS-CoV-2 qPCR assay (Supplementary Fig. 1A) on nasopharyngeal swabs obtained at the point of sample collection. However, we obtained statistically significant differences in some analytical parameters between severe and moderate cases at the sampling timepoint: higher neutrophil counts (P-value = 0.012), ferritin (P-value = 0.002), D-Dimer (P-value = 0.006) and lactate (P-value = 0.019) values, but lower lymphocyte counts (P-value = 0.002) and fibrinogen (P-value = 0.004) values in severe cases. Maximum values for C-reactive protein (CRP) and neutrophils (P-values = 0.02 and 0.006, respectively) and minimum values for lymphocytes (P-value < 0.001) also showed statistically significant differences between these groups.
Immune cell fractions inferred from gene expression data also pointed to a significantly lower proportion of T lymphocyte subpopulations CD4 and CD8 in severe cases than in other severity categories and healthy controls (Supplementary Fig. 1B). Likewise, the proportion of B cells was significantly higher in mild when compared to moderate and severe cases. Cell numbers of CD4 and NK subpopulation also showed a significant decrease with disease severity. Finally, a higher proportion of monocytes was also observed in moderate cases, correlating well with real monocyte counts detected in the blood tests from moderate and severe cases.
4.2. Transcriptomic immune response in blood
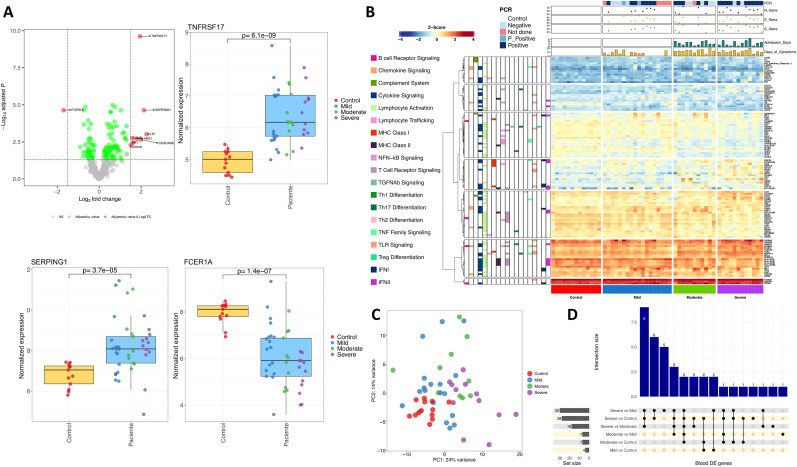
Principal component analysis (PCA) of blood gene expression revealed a moderate differentiation by severity in the first principal component (PC1; Fig. 1 A). When we compared the whole COVID-19 cohort against controls we obtained nine DEG (Supplementary Table 2, Fig. 1A), all of them over-expressed in patients except for FCER1A gene that was under-expressed (log2FC = −1.7; adjusted P-value = 2.3E-5). The most statistically significant gene was TNFRSF17 gene (log2FC = 1.9; adjusted P-value = 2.4E-10), followed by FCER1A and SERPING1 genes. Three out of the nine DEG were found to be still significant when using a model that accounted for differences in treatment with immunomodulators (steroids), administered only in some severe and moderate patients: TNFRSF17 (log2FC = 1.72; adjusted P-value = 4.1E-6), SERPING1 (log2FC = 2.2; adjusted P-value = 4.3E-4) and MX1 (log2FC = 1.97; adjusted P-value = 3.0E-3) genes. To test the robustness of this COVID-19-related immune signature we carried out a validation in two recently published COVID-19 RNA-seq datasets (Aschenbrenner et al., 2021; Thair et al., 2021) (Supplementary Fig. 3A). The 9-transcript immune signature correlated very well with the data from the two validation cohorts (R = 0.96, P-value = 5.3E-5; R = 0.92, P-value = 4.0E-4; respectively). As in our study, all genes were DE in both datasets when comparing patients vs. healthy controls.
Fig. 1.
A) Volcano plot showing the DEGs between cases and healthy controls in blood samples. Boxplots of the most relevant genes and the P-value of the statistical test are also shown. B) Heatmap and cluster analysis of the DEGs between all categories in blood samples. Only genes with a log2FC |>1| were represented. Gene clusters were generated using k-means partitioning. C) PCA of immune transcriptomic data from blood samples of the different COVID-19 categories analyzed using all genes from the panel detected above the background. D) Upsetplot of the common DEGs between categories compared in blood samples.
Next, we studied the specificity of this 9-transcript signature by examining other viral and bacterial infections. Firstly, transcriptomes of different viral infections (n = 655) and healthy controls (n = 672) from Thair et al., (2021) were contrasted with the results from our study (Supplementary Fig. 3). We found a moderate correlation (R = 0.85; P-value = 0.0037) between log2FC values from our study (COVID-19 vs. control) and their study (viral infections vs. healthy controls). Notably, TNFRSF17 gene did not show appreciable differences between non-COVID-19 viral infections and controls (log2FC = 0.16; adjusted P-value = 0.50). To further test the differential pattern of this gene in COVID-19 patients, we used a combined dataset from different microarray and RNA-seq studies that includes transcriptomes from viral and bacterial infected patients, and healthy controls. We consistently observed a moderate correlation between our results and those from the viral infections vs. healthy control dataset (R = 0.81, P-value = 0.0078), and a weaker correlation with those from bacterial infections vs. healthy control dataset (R = 0.61, P-value = 0.081). Again, TNFRSF17 gene showed a log2FC value very close to 0, and non-significant results in the viral combined dataset (log2FC = −0.30; adjusted P-value = 0.28). Furthermore, in the bacterial combined dataset, the TNFRSF17 gene (and MX1) was under-expressed in bacterial infections vs. controls, even though this contrast was also statistically significant (log2FC = −0.81, adjusted P-value = 1.9E-4) as in COVID-19 datasets.
We performed a more detailed analysis of the immunological processes underlying COVID-19 by comparing different severities (Fig. 1B; Fig. 1C; Supplementary Fig. 4). We found a similar number of DEG when comparing severe cases vs. healthy controls and severe vs. mild patients (28 and 30 respectively; Supplementary Table 2; 1D), pointing to a limited systemic immune response (Fig. 1D; Supplementary Fig. 4). We detected 14 common DEG in all comparisons involving severe cases (Fig. 1D); ARG1 gene was the most remarkable one over-expressed in severe cases (Log2FC = 3.98–4.71; adjusted P-value = 1.53E-18 – 1.78E-22) (Supplementary Table 2). Other significatively over-expressed genes in severe cases were those related to pro-inflammatory pathways IL-1 (IL1R1 and IL1R2) and IL-18 (IL18R1) and immunoregulatory functions (FKBP5 and S100A8); Supplementary Fig. 5. To further test this 14-transcript COVID-19 signature for severe patients, we compared our results to an independent cohort (Aschenbrenner et al., 2021); after excluding two genes (ZBTB16 and IL1RL1) that were not found to be DE between their severe cases vs. healthy controls , we found a notable correlation between both datasets (R = 0.87; P-value = 0.00023); Supplementary Fig. 3.
Analysis of moderate vs. healthy controls and mild vs. healthy controls returned a few DEG coinciding with the previous 14-transcript signature, e.g. TNFRSF17 and SERPING1. TNFRSF17 represents the strongest signal of COVID-19 disease of all the genes analyzed, showing an increased gene expression associated with severity (log2FC values compared to healthy controls: 1.66 in mild, 1.95 in moderate, and 2.71 in severe cases); Supplementary Table 2. Conversely, FCER1A gene was under-expressed in patients and showed a gradual decreased gene expression with severity (log2FC values compared to healthy controls: not DE in mild, −1.83 in moderate, and −3.06 in severe cases). Genes MX1 and C1QB showed an increased expression in mild (log2FC = 2.10; adjusted P-value = 1.32E-5) and moderate cases (log2FC = 1.78; adjusted P-value = 5.59E-5) with respect to controls (Supplementary Table 2). When comparing DEGs between hospitalized and non-hospitalized patients, the most relevant genes were CAMP, LTF, CEACAM8 and CEACAM6, all involved in neutrophil degranulation processes triggered by the innate immune response (Supplementary Fig. 6).
Pathway analysis by way of a GSEA of severe cases vs. healthy controls and using Reactome revealed: (i) an up-regulation of innate immune processes (mainly driven by neutrophil activation and degranulation, the IL-1 pathways, and the TLR cascade (Supplementary Table 3, Supplementary Fig. 7); and (ii) down-regulation of the adaptative immune response and related pathways such as Translocation of ZAP−70 to Immunological synapse, T cell antigen receptor (TCR) signalling, co-stimulation of CD28 family, and PD−1 signalling. Meanwhile, GSEA analysis using GO showed a comparable pattern: (i) up-regulation of the neutrophil mediated immunity and downstream processes; (ii) under-regulation of B cell proliferation/activation; and (iii) antigen receptor-mediated signalling (Supplementary Table 3, Supplementary Fig. 7). Similar findings were observed in comparisons of severe vs. moderate (Supplementary Table 3, Supplementary Fig. 8) and severe vs. mild cases (Supplementary Table 3, Supplementary Fig. 9) from Reactome, except for the IFN pathway, which was under-regulated in severe cases in comparison to moderate and mild patients. GO pathway analysis found under-expression of genes involved in antigen processing and presentation via MHC-II complex and viral defense. The only process detected by GSEA in mild cases were an up-regulation of the IFN signalling pathway with the Reactome database; whereas routes related to host response to viral infection were the most over-regulated in GO (Supplementary Table 3, Supplementary Fig. 10). Overall, there exists a significant up-regulation of the neutrophil activation and degranulation pathways, which increase with severity, and a lack of IFN response in severe cases. The differential neutrophil-driven immune response observed in moderate and severe patients was also detected when comparing hospitalized against non-hospitalized cases and healthy controls (Supplementary Fig. 11; Supplementary Table 3).
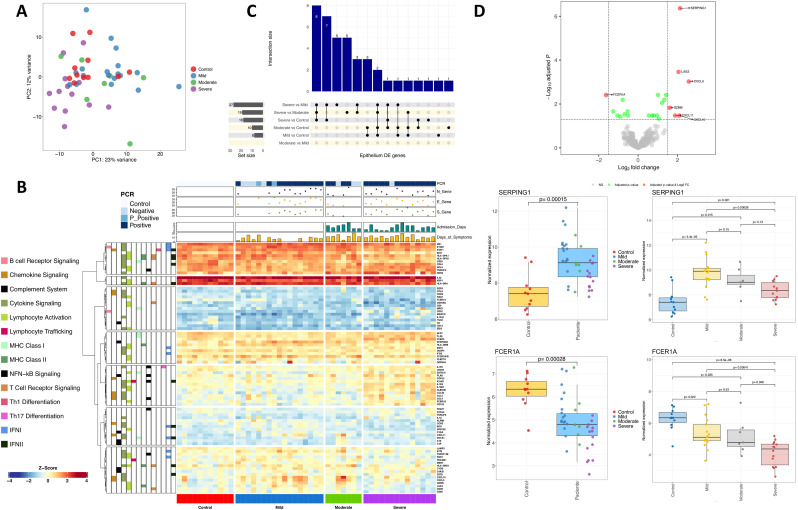
4.3. Transcriptomic immune response in nasal epithelium
PCA indicates a differentiation of severe cases against controls that is only notable in the PC2 dimension (12% of the variation); Fig. 2 A (see also heatmaps in Fig. 2B). Conversely, mild and moderate cases showed more remarkable differences with severe and controls in the PC1 dimension (23% of the variation). Of the seven DEGs observed in the comparison patients vs. healthy controls, one was under-expressed and six over-expressed (Supplementary Table 4; Fig. 2C). Thus, FCER1A was under-expressed in patients when compared against controls (log2FC = −1.63; adjusted P-value = 0.004; severe vs. controls: log2FC = −3.28; adjusted P-value = 2.75E-9; severe vs. moderate: log2FC = −2.01; adjusted P-value = 0.0004; severe vs. mild: log2FC = −1.96; adjusted P-value = 0.00097), even though it was not detected as DE when comparing moderate and mild cases with healthy controls (Supplementary Table 4; note however that the absolute expression values showed only weak statistical significance in mild and moderate patients vs. healthy controls; Fig. 2D). Of the over-expressed genes, SERPING1 gene leads the DEG list; the main signal comes from the high expression levels observed in moderate and mild patients against controls; this gene was however not DE in severe cases when compared with controls. Thus, SERPING1 over-expression decreased gradually from mild to severe disease (Supplementary Table 4; Fig. 2D). A similar pattern was observed for other genes (CXCL9, LAG3 and GZMB), all showing a gradual over-expression in moderate and mild cases against controls (Supplementary Table 4; Supplementary Fig. 12) but not in severe patients. Overall, gene expression of immune genes in nasal epithelium samples showed a very similar response to infection in mild and moderate patients, with seven overlapping DEGs (LAG3, SERPING1, CXCL9, CXCL10, CXCL11, IFITM1 and GZMB; all over-expressed) between both categories vs. controls, but no DEG when comparing mild vs. moderate cases (Fig. 2C; Supplementary Fig. 13; Supplementary Fig. 14). Despite the similarities in immune response of both phenotypic categories, these genes showed a gradual increase in expression from severe to mild disease; the differences were found to be more notable when comparing mild vs. controls than in moderate vs. controls (Supplementary Table 4).
Fig. 2.
A) PCA of immune transcriptomic data from nasal epithelium samples of the different disease categories analyzed using all genes from the panel detected above the background. B) Heatmap and cluster analysis of the DEGs between all categories in nasal epithelium samples. Only genes with a log2FC |>1| were represented. Gene clusters were generated using k-means partitioning. C) Upsetplot of the common DEGs between categories compared in nasal epithelium samples. D) Volcano plot showing the DEGs between COVID-19 samples and healthy controls in nasal epithelium samples. Boxplots of the most relevant genes and the P-value of the statistical test are also shown.
The comparison of severe vs. controls/moderate/mild yielded 18, 19 and 27 DEGs genes, respectively (Fig. 2C; Supplementary Table 4); eight genes were shared between these three comparisons (FCER1A, IL18R1, IL1R2, CCL3 CCL4, CLEC4E, CD163 and MARCO), all over-expressed in severe cases (except FCER1A) and all mainly involved in the innate immune and inflammatory response (Supplementary Fig. 15). IL18R1 and IL1R2 appeared in the top three most DEGs in severe cases compared to all categories, with a log2FC ∼2 in severe vs. moderate cases, and even higher in severe vs. controls/mild cohorts. Other under-expressed genes in severe compared to mild/moderate cases included IFIT2 (Interferon Induced Protein with Tetratricopeptide Repeats 2) and IDO1, whose encoded protein is produced in response to inflammation and has autoimmune protective features (Supplementary Table 4; Supplementary Fig. 13).
We did not find statistically significant pathways in the nasal samples from severe cases vs. controls using both GSEA and ORA. Moderate vs. controls comparisons highlighted some up-regulated pathways in GSEA mainly connected to interaction and response to other organism processes (Supplementary Table 5), and inhibition of endopeptidase activity (Supplementary Fig. 16). Comparison of mild cases vs. controls using ORA suggested two main processes: (i) antimicrobial immune humoral response (an adaptative immune response mainly driven by B lymphocytes), and (ii) chemokine related pathways (Supplementary Table 5; Supplementary Fig. 17). The comparison of severe vs. mild patient revealed up-regulated coordinated responses in various nested pathways involving NAD metabolism (Supplementary Table 5), a regulator of infection and inflammation. Molecular functions showing down-regulation in severe vs. mild cases were MHC protein binding and hydrolase activity (Supplementary Fig. 18). ORA analysis of DEGs revealed alterations in chemokine dynamics-related pathways, with over-expression of CC-type chemokines and MARCO, and under-expression of CXC-type chemokines and DEFB4A (Supplementary Fig. 18). The comparison of severe vs. moderate patients using GSEA analysis detected some similarities with that of severe vs. mild cases (Supplementary Fig. 19). Reactome and GO showed some common biological processes and/or cellular components, such as under-regulation of type I and II IFN pathway and MHC class II antigen presentation, and an over-regulation of neutrophil activation and degranulation processes. When exploring pathways associated with the comparison of moderate vs. mild cases, GSEA analysis found an upper-regulation of serine−type endopeptidase activity and a down-regulation of molecular transducer receptor activities (Supplementary Fig. 20; Supplementary Table 5).
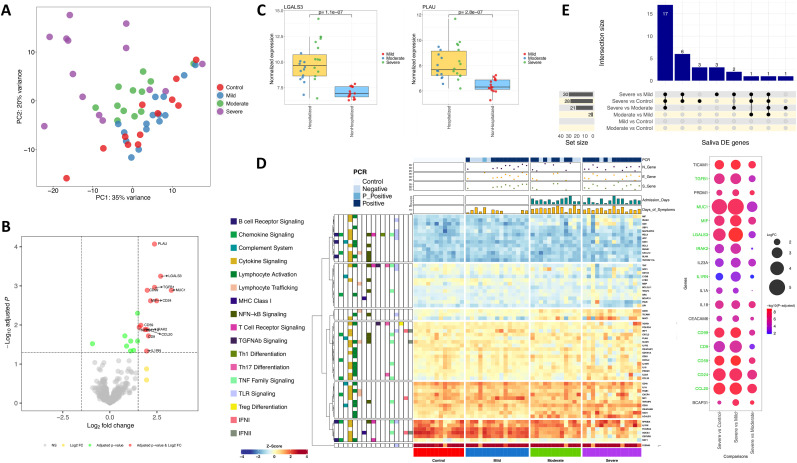
4.4. Transcriptome immune response in saliva
Among the genes detected in saliva samples, there was no DEG when comparing patients and healthy controls. This fact was reflected in a PCA (and heatmap analysis), which showed a large cluster with overlapping controls, mild and moderate, and only slightly differentiation of the severe cases in the PC1 (35% of the variation) and the PC2 (20% of the variation); Fig. 3 A,D. Therefore, the data seem to indicate very limited immune response to infection in the oral cavity of mild and moderate patients. A few DEGs were detected when comparing hospitalized with non-hospitalized patients (Fig. 3B; Supplementary Fig. 21) and against controls (Supplementary Table 6). As expected, most DEGs were the same in both comparisons, with 11 out of the 12 DEGs detected in hospitalized vs. non-hospitalized cases also appearing in hospitalized vs. controls, all of them over-expressed (Supplementary Fig. 21; Supplementary Table 6). The strongest differences in hospitalized vs. non-hospitalized patients were represented by (i) PLAU, a gene encoding a secreted serine endopeptidase that transforms plasminogen in plasmin and is implicated in the rearrangement of extracellular matrix and cell migration, and (ii) LGALS3, which plays a role in apoptosis, innate immunity (attracting macrophages and activating neutrophils), cell adhesion and T-cell regulation processes as well as in acute inflammatory responses (Fig. 3C). These two genes were the only ones detected as DE when comparing moderate vs. mild cases; they probably constitute the strongest and most specific immune signature in oral mucosa from hospitalized individuals (Supplementary Table 6; Supplementary Fig. 22). The other DEGs of the signature from hospitalized samples also exhibited expression levels that increased with severity (Supplementary Fig. 22).
Fig. 3.
A) PCA of immune transcriptomic data from saliva samples of the different disease categories analyzed using all genes from the panel detected above the background. B) Volcano plot showing the DEGs between hospitalized and non-hospitalized saliva samples. C) Boxplots of the most relevant genes from the comparison of hospitalized vs. non-hospitalized saliva samples and the P-value of the statistical test. D) Heatmap and cluster analysis of the DEGs between all categories in saliva samples. Only genes with a log2FC |>1| were represented. Gene clusters were generated using k-means partitioning. E) Upsetplot of the common DEGs between categories compared in saliva samples. Bubble plot represents log2FC and adjusted P-value from the 18 DEGs between severe and all remaining categories. Green color in gene names indicates genes also included in the hospitalized signature. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We found 18 common over-expressed DEGs that differentiate severe cases from the other phenotypic categories (Fig. 3E; Supplementary Fig. 23). CCL20 showed the highest log2FC value in all comparisons (Supplementary Table 6). As expected, higher differences were found in severe vs. mild and severe vs. controls (Fig. 3E). Eleven out of the 18 genes were also included in the signature that differentiated hospitalized from both non-hospitalized and healthy controls, respectively (Fig. 3E; Supplementary Fig. 24).
Pathway analysis in severe vs. controls using GSEA found some moderate up-regulation of processes and cell components, but only phagocytic vesicle as under-regulated cellular component (Supplementary Fig. 25). Some of these pathways were also highlighted as up-regulated in severe vs. mild patients but, in this case, a positive regulation of cell migration and motility was also detected (Supplementary Table 7; Supplementary Fig. 26). GSEA analysis of severe vs. moderate patients provided the highest number of hits from all comparisons, all showing up-regulation in severe cases, but moderate NES values (Supplementary Table 7; Supplementary Fig. 27). Among the up-regulated pathways in moderate vs. mild (Supplementary Fig. 28) cases there are pathways related to cell migration, receptor activator activity and chemoattractant activity, of which the latter two were also up-regulated in moderate cases vs. controls (Supplementary Fig. 29). GSEA also pointed to a strong under-regulation of the IFN signalling in moderate vs. mild cases in all the reference databases interrogated (Supplementary Table 7; Supplementary Fig. 28). Notably, IFN pathway was not detected in severe vs. mild cases, however it appears as highly under-regulated in hospitalized vs. non-hospitalized patients, although not when comparing to controls.
Finally, comparison of mild cases and controls resulted in several under-regulated processes and cellular components mainly involved in cell junction, cell communication and secretory vesicles (Supplementary Table 7; Supplementary Fig. 30).
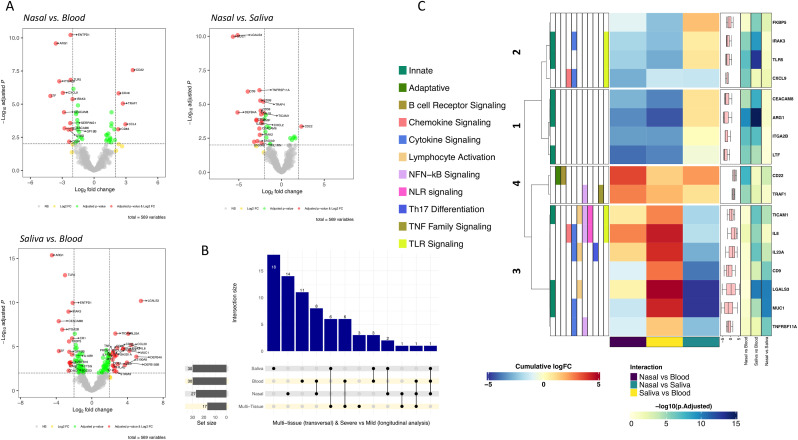
4.5. Transversal multi-tissue analysis
We selected a subset of severe and mild patients (n = 21) with samples available from all tissues collected at the same time. We explored for interactions between severity and tissue to elucidate immunological features that could illuminate the causes for the differential evolution observed in patients. Only when relaxing the thresholds for this interaction analysis (adjusted P-value < 0.05 and a cumulative log2FC > |1.4|) we found that the effect of severity for the CXCL9 gene is significant for all tissue comparisons, being much lower in nasal compared to blood tissue (P-adjusted = 1.41E-6; cumulative log2FC = −2.94) and moderately lower for both saliva vs. blood (P-adjusted = 0.03; cumulative log2FC = −1.42) and nasal vs. saliva (P-adjusted = 0.017; cumulative log2FC = −1.51) tissues (Supplementary Table 8). Looking at the CXCL9 normalized data, we found higher expression in severe than in mild cases for blood, with the opposite pattern in nasal and saliva epithelium samples (Supplementary Fig. 31). When we increased the significance thresholds (P-adjusted < 10E-6; cumulative log2FC > |2.5|) a set of 17 genes were found as DE in all possible comparisons (Supplementary Table 8; Fig. 4 B; Fig. 4C). Thirteen out of these 17 genes were also detected as DE between severe and mild cases in the longitudinal transcriptomic analysis (Fig. 4B). The transversal multi-tissue analysis generates four clusters for these 17 genes according to their cumulative Log2FC values (Fig. 4C). This analysis highlights that the main specific severity signals in blood were driven by gene cluster 1 represented by ARG1, CEACAM8, LTF and ITGA2B, and to a lesser extent by cluster 2 (specially IRAK3 and TLR5). This means that severity has a more marked effect in blood in comparison with nasal and saliva samples (Fig. 4C; Supplementary Table 8; Supplementary Fig. 31). Cluster 3 (Fig. 4C), represented by CD22 and TRAF1, indicates that severity determines an opposite behavior in nasal (lower expression in mild and higher in severe) than in blood (higher expression in mild and lower in severe (Supplementary Fig. 31). Finally, cluster 4 (Fig. 4C) represented by seven genes indicates a differential effect of severity in saliva with respect to blood and nasal epithelium (especially remarkable for MUC1 and LGALS3).
Fig. 4.
A) Volcano plots showing the DEGs between severities (severe/mild) and different tissue samples (thresholds: P-adjusted = 0.01, cumulative log2FC = |2|). B) Upsetplot of the DEGs between severe and mild categories in all tissues as well as in the multi-tissue transversal analysis. C) Heatmap and cluster analysis of the DEGs between severities (severe/mild) and different tissue samples (thresholds: P-adjusted = 10E-6, cumulative log2FC = |2.5|). Gene clusters were generated using k-means partitioning.
4.6. Co-expression analysis related to the mild-severe phenotypes
Through a co-expression network analysis, we aimed to detect modules of co-expressed genes that might be responsible for the extremely unbalance immune response to the SARS-CoV-2 infection between mild and severe phenotypes.
We detected four co-expression clusters of immune genes in blood samples (Supplementary Fig. 32A; Supplementary Table 9) constituting two meta-modules of highly related clusters (Supplementary Fig. 32B). Two of these modules were weakly significantly correlated with the mild-severe phenotype, one indicating over-expression (‘blue’ module in Supplementary Fig. 32; n = 62 genes; R = 0.41; P-value = 0.026) and the other under-expression in severe cases (‘turquoise’ module in Supplementary Fig. 32; n = 105 genes; R = −0.37; P-value = 0.043) (Supplementary Fig. 32C). Despite of not having large correlation with the phenotype, both modules showed highly significant correlation between gene significance (GS) and module membership (MM), meaning that genes associated to the phenotype were also hub genes within the module (higher number of connections) (Supplementary Figs. 33A and 33B; Supplementary Table 9). Hub genes of the most remarkable module (over-expressed in severe cases) are most likely involved in pathways related to cytokine production, IL-1 signalling, neutrophil mediated immunity, T-cell immunity, TLR pathways and NAD metabolism (Supplementary Fig. 34; Supplementary Table 9).
Stronger co-expression signals related to severity were detected in nasal epithelium and saliva samples. Two out of the four modules detected in nasal epithelium samples were statistically significant correlated with the phenotype, one composed by genes under-expressed in severe with respect to mild (see ‘turquoise’ module in Supplementary Figs. 35A, 35B, 35C; n = 129 genes; R = −0.46; P-value = 0.0099) and the other showing over-expression in severe cases (see ‘brown’ module in Supplementary Figs. 35A, 35B, 35C; n = 48 genes; R = 0.44; P-value = 0.014); see also Supplementary Table 10. The most important under-expressed genes in severe cases obtained in the DE analysis were also found among the genes placed in the ‘turquoise’ module and, by contrast, the top over-expressed genes in DE analysis from severe cases were housed in the ‘brown’ module, being many of them also hub genes in these modules (Supplementary Figs. 36A and 36B Supplementary Table 10). The core gene from the mild-related ‘turquoise’ module, defined as the significant correlated gene showing the higher MM value, was CD3D (MM = 0.95; P-value = 8.7E-17) and is part of the TCR/CD3 complex in the T-cell signal transduction with an essential role in the adaptative immune response. The core gene in the ‘brown’ module was CCRL2 (MM = 0.91; P-value = 2.2E-12), a gene that is over-expressed during the neutrophil activation and the differentiation from monocyte to macrophage. Hub genes from these modules were also involved in different pathways (most of them overlapping to those obtained from the DE analysis) pointing to a very different response in mild and severe cases. Hub genes in ‘turquoise’ module were involved in CXCR chemokine receptor binding, interferon gamma signalling and adaptative T-cell immune response, such as T-cell migration, activation and chemotaxis, TCR signalling, and MHC class II receptor activity. Nevertheless, noteworthy pathways obtained from hub genes of the ‘brown’ module were those related to CCR chemokine receptor binding, IL-1 response, NAD metabolism, neutrophil driven immune response and TLR cascades (Supplementary Fig. 37).
From the four co-expression modules detected from saliva samples only one module showed significant correlation with the phenotype (‘turquoise’ module in Supplementary Figs. 38A and 38B; Supplementary Table 11) and was also the most significant phenotype-correlated module in all tissues analyzed (n = 91 genes; R = 0.52; P-value = 0.0039; Supplementary Fig. 38C). Genes from this module were over-expressed in severe cases and displayed a good significant correlation between GS and MM (Supplementary Fig. 39A). Most genes within this module (86%) were among the top DE genes and many of them were also hub genes within the module (63%). The gene with the highest intra-modular connectivity was SRC gene (MM = 0.95; P-value = 3.48E-15; Supplementary Table 11), a non-receptor tyrosine-kinase implicated in many signalling pathways controlling different processes such as immune response, cell adhesion and migration, cell cycle and apoptosis. Among the most significant pathways we found lymphocyte proliferation and activation, interleukin signalling, apoptosis, cell-cell adhesion and cell cycle arrest (Supplementary Fig. 39A, Supplementary Fig. 40).
5. Discussion
Healthcare systems worldwide collapsed during the COVID-19 pandemic due to the high number of hospital admissions, mostly with respiratory complications that, in many cases, ended up being fatal in patients with severe disease. Usually, viral colonization begins in a local tissue environment, with the upper respiratory tract being the main gateway for the virus entrance; however, the infection does not always have a significant systemic impact. To explore differential host response to infection, we analyzed the expression of immune response genes in three different tissues from COVID-19 patients affected by different disease severities.
In the analysis of blood samples, we detected specific COVID-19 DEGs, all but one (FCER1A) over-expressed, which were subsequently validated in two independent RNA-seq datasets. A few DEGs have comparable expression levels in other viral and bacterial infections, suggesting a common response against severe infections. Moreover, ARG1 and FCER1A showed similar expression values independently of the causative pathogen origin (viral or bacterial), but with opposed log2FC values (ARG1: 1.5 to 3.5; FCER1A: −2.5 to −1.5). ARG1 encodes for the Arginase-1 protein and is expressed in neutrophils, for which a central role in innate immune and inflammatory response has been described, while FCER1A is mainly expressed in basophils and mast cells and is involved in the initiation of allergic response. We also observed that TNFRSF17 was DE in COVID-19 compared to other infections. This gene is expressed in mature B lymphocytes encoding for a member of the TNF-receptor superfamily; it is involved in MAPK8/JNK and NF-kB activation pathways and mediates the development and survival of B-cell lymphocytes to maintain humoral immunity. The number of DEGs in blood increases significantly with severity (Supplementary Fig. 4), clearly pointing to a lower immune systemic response in mild and moderate than in severe patients. Moreover, mild and moderate patients over-expressed the interferon type I (IFN–I) pathway in response to viral infection; a timely IFN-I response is critical as it represents a successful front-line protection against viral infections. IFN-I is produced by cells after pathogen-associated molecular patterns (PAMPs) recognition, inducing downstream expression of IFN stimulated genes (ISGs). Furthermore, it has an important role in immune control through the regulation of proinflammatory cytokines, activation and recruitment of different immune cells types, and shaping a successful adaptative immune response (Crouse et al., 2015). SARS-CoV-2 seems to be more susceptible to IFN-I and produces various proteins that antagonize IFN production through different mechanisms, suppressing IFN-I pathway in a more efficient manner than other coronaviruses (Lokugamage et al., 2020; Mantlo et al., 2020). Interestingly, low IFN-I alpha level has been recently related to a more severe disease outcome, and proposed as a potential therapeutic treatment (Hadjadj et al., 2020; Contoli et al., 2021). The IL-1 pathway related genes were detected as over-activated in severe cases. Conversely, moderate and severe patients showed an up-regulation of innate immune response (not observed in mild patients and healthy controls), driven by a different degree of neutrophil response pathways activation. Severe disease was associated with high neutrophil counts and increased levels of pro-inflammatory cytokines, producing microthrombus formation and organ damage. The release of neutrophil extracellular traps (NETs) could explain (at least partially) COVID-19 vascular complications through forming aggregates that produce vessel occlusion and endothelial damage (Leppkes et al., 2020; Veras et al., 2020); unbalanced NET production seems to have an important role in viral immunopathology and respiratory disease complications (Twaddell et al., 2019). Consistently, we observed a DE of S100A8/S100A9 and CAMP genes (all encoding proteins usually identified in NETs) in severe cases vs. all other categories. Likewise, the multi-tissue transversal analysis consistently indicated a higher difference in severity between systemic and local immune response in neutrophil related genes (ARG1, LTF, CEACAM8), and other blood coagulation genes that participate in the process of platelet aggregation (ITGA2B), which could explain the formation of microthrombus in severe phenotypes. Patterns of exacerbated innate response in hospitalized individuals were accompanied by a global failure of the adaptative immune response in severe cases, with under-regulation of several routes involved in T-cell signalling and maintenance of immune homeostasis, like ZAP-70 signalling. ZAP-70 kinase regulates TCR downstream signaling of antigen presentation, and an adequate regulation of its activity is key for a proper T cell proliferation; furthermore, ZAP protein can restrict SARS-CoV-2 replication (Nchioua et al., 2020). It has been described the importance of a solid T cell response in COVID-19 recovery (Zheng et al., 2020a) and, therefore, the suppression of the adaptative immune system could explain in part the characteristic lymphopenia observed in severe patients. In agreement with an over-activation of the innate and under-activation of adaptative immune response, an unbalanced activation of HLA class I and II antigen presentation was also found in severe cases. Overall, our results are in the line with previous gene expression studies carried out in blood samples (Zheng et al., 2020a; Yan et al., 2021), indicating that severe cases exhibit signals of a systemic impairment of IFN response characterized by a non-robust IFN production, a suppression of T and B responses and lymphopenia, neutrophil recruitment (with over-expression of ARG1 gene), abnormal increase of low density neutrophils (with high capacity to release NETs) and also enrichment in blood coagulation–related genes.
The most prominently over-expressed genes in the nasal epithelium of severe cases were anti-inflammatory IL-1 family receptors IL-1R2 and IL-18R1 (mainly expressed by monocytes-macrophages). Both receptors are involved in the IL-1 and IL-18 cascades, which play a role in the initiation of the inflammation process after PAMPs recognition. Nevertheless, we did not detect DE of the pro-inflammatory cytokines IL1A, IL1B, IL1RAP or IL18 in severe samples, just a moderate up-regulation of IL1R1 gene. This imbalance between agonists-antagonists/soluble receptors in severe disease with respect to mild cases was previously noted in broncho-alveolar lavage fluids (BALF) from COVID-19 patients (Xu et al., 2020), and it could lead to exacerbated inflammatory responses (Italiani et al., 2020). In addition, pathways analysis detected an alteration of NAD metabolism in severe cases with respect to those with mild disease, probably pointing to a NAD (and ATP) depletion in host cells from severe patients. It is known that some intracellular pathogens can modulate host cell NAD, impairing several innate immune routes (Mesquita et al., 2016). Nasal samples from severe cases also showed similar expression patterns of CXC-chemokines, GZMB and antiviral IFITM1/IFIT2 genes compared to controls. Unlike in mild cases, in which main signals came from cell-mediated immunity (Th1, NK and T cytotoxic cells responses) as well as from a robust IFN-II induction, severe cases showed an over-expression of genes associated to monocyte-macrophage recruitment; these cells could play a key role in the cytokine storm observed in severe patients (Merad and Martin, 2020). For instance, CC pro-inflammatory chemokines genes CCL3 and CCL4 act as chemoattractant cytokines in response to tissue injury or inflammation. Some of them have already been highlighted as severity-related biomarkers of COVID-19 in airway epithelial cells (Chua et al., 2020), and they were found highly expressed in BALF of severe compared to mild cases (Xu et al., 2020). They are ligands of chemokine receptors CCR1 (CCL3) and CCR5 (CCL3 and CCL4) and have a role in monocyte recruitment. CD163, CLEC4E and MARCO are expressed in macrophages, and the latter two can act as pattern recognition receptors (PRR) for PAMPs. The protein encoded by CLEC4E can interact with FCER1G which, unlike FCRE1A gene, is over-expressed in severe cases when compared to controls. Nasal epithelium response in moderate and mild patients was mainly characterized by an over-expression of CXCR3 receptor ligands chemokines CXCL9, CXCL10 and CXCL11. These chemokines are highly inducible by IFN gamma, and once joined to the receptor IFN is activated and triggers calcium release to the cytosol, acting as a second messenger to activate downstream signalling pathways (also highlighted in the pathway analysis of nasal samples from mild patients). These chemokines were previously described as biomarkers of respiratory viral infection in nasopharyngeal samples (Landry and Foxman, 2018). Similarly, other antiviral genes were also over-expressed in mild cases: IFITM1 has antiviral activity against several enveloped viruses, hampering the viral membrane fusion, and has demonstrated a strong inhibition to SARS-CoV-2 infection by blocking the virus entrance into cells (Buchrieser et al., 2020); and IFIT2 is involved in the inhibition of viral replication.
Co-expression network analysis yielded comparable results to those observed in DE analysis, reporting two significant modules with opposite behavior in nasal epithelium of severe vs. mild cases. One of the modules consists of genes showing over-expression in the mild phenotype, while the other includes genes over-expressed in severe patients. Hub genes from the mild-related module are mostly implicated in CXCR chemokine receptor binding processes, interferon gamma signalling and adaptative T-cell immune response. By contrast, hub genes within the severe-related module were linked to IL-1 response, CCR chemokine receptor binding, NAD metabolism and an innate response involving neutrophils and TLR cascades.
Contrary to the expression profile observed in nasal epithelium samples, there is an absence of appreciable DE in the saliva of mild cases, suggesting a lack of oral immune response against viral infection. Nevertheless, in severe, and to a lesser extent in moderate samples, some up-regulated severity related hits were detected, many of them linked to the viral dynamics. The virus has mechanisms to hijack cellular machinery to their own benefit, altering the levels of several metabolites and cellular processes; one of these molecules are galectins, which have been proposed as biomarkers to monitor viral infections as well as therapeutic targets or antagonists (Wang et al., 2020). One of the most remarkable over-expressed hits in moderate and severe saliva samples was LGALS3 (galectin-3), a contributor to pro-inflammatory response against different viral infections (Wang et al., 2020), enhancing macrophage survival, and positively regulating macrophage recruitment (Liu and Hsu, 2007), even promoting a more severe course of the infection (Chen et al., 2018). Elevated levels of galectin-3 were observed in macrophages, monocytes and dendritic cells in severe vs. mild COVID-19 patients (Caniglia et al., 2020); the regulatory role of galectin-3 in inflammatory response, fibrosis and disease progression, makes it a promising therapeutic target for severe cases (Garcia-Revilla et al., 2020). We did not observe systemic or nasal epithelium over-expression of this gene. Another important over-expressed gene in saliva was PLAU (urokinase-type plasminogen activator), which, together with CD59 (inhibitor of complement-mediated lysis) and CD9 (tetraspanin), can facilitate virus-host cell interaction, host immune system viral evasion and, thus, dissemination and viral pathogenesis through buccal epithelium. Urokinase transforms plasminogen to the active form serine protease plasmin; this airway protease (like several others e.g. trypsin, cathepsins, elastase, and TMPRSS family members) are required for coronavirus entry enhancement (Millet and Whittaker, 2015). After viral interaction with ACE2 receptor, furin site in S protein of SARS-CoV-2 is proteolytically cleaved by TMPRSS2 to enable viral internalization (Devaux et al., 2020). Plasmin has demonstrated to cleave several viral proteins of many respiratory pathogens (Dubovi et al., 1983; Goto et al., 2001; Berri et al., 2013) and also S protein in SARS-CoV in vitro (Kam et al., 2009), increasing the ability of the virus to enter host cells. Even though the plasmin protein S cleavage in SARS-CoC-2 has not been demonstrated, there is growing evidence pointing towards a probable role of the plasmin-plasminogen system and fibrinolytic pathway in the COVID-19 severity (Gralinski et al., 2013; D'Alonzo et al., 2020; Ji et al., 2020; Kwaan and Lindholm, 2021). Co-expression and upregulation of PLAU, TMPRSS2, and ACE2 was observed in airway epithelial cells from severe/moderate patients and SARS-CoV-2 infected cell lines. This, together with a plasmin-mediated cleavage, could make cells more susceptible to infection (Hou et al., 2021). Interestingly, we also observed an enhanced expression of PLAU in nasal epithelium from severe cases when compared to mild and controls, highlighting the crosslink between both tissues. In the same vein, tetraspanins are molecules associated to specific viruses and seem to play a role in different stages of viral infections (Martin et al., 2005). Tetraspanin CD9 was involved in early MERS-CoV lung cell entry (Earnest et al., 2017). Recently, it has also been proposed that extracellular vesicles may contribute to viral dissemination by transferring viral receptors, such as CD9 and ACE2, to other host cells making them more susceptible to the infection after exosomes endocytosis (Hassanpour et al., 2020). Our functional analysis revealed down-regulation of some pathways related to cell communication, adhesion and signal transduction in mild cases, probably as a mechanism to minimize viral spread. Conversely, severe cases showed up-regulation of genes involved in cell junction, adhesion and extracellular exosomes, all processes that could facilitate the viral dissemination. CD59 is a non-IFN inducible protein that acts as inhibitor of complement-mediated lysis by disruption of the membrane attack complex. Its implication in viral complement evasion of many enveloped viruses (Li and Parks, 2018), including coronaviruses (Wei et al., 2017), has been demonstrated. This escape mechanism can be mediated by incorporating CD59 to the viral envelope, controlling cellular CD59 expression or producing a counterpart protein that mimics host CD59, or even use it as cell entry receptor (Maloney et al., 2020).
We have found an important co-expression module defining the severe phenotype in saliva samples containing all these aforementioned genes. In addition, most of them were also hub genes in the module, indicating that genes highly significantly associated with the severe phenotype were also key elements within this module. These hub genes were related to lymphocyte proliferation and activation, but also to apoptosis, cell-cell adhesion and cell cycle arrest. The gene that was found to be more connected within the module was the SRC gene, a member of the Src family kinases (SFKs) that demonstrated to have an important role in the life cycle of a many viruses and have emerged as an important factor in the interplay between cell functions and viral demands (Pagano et al., 2013). It has been shown that inhibitors of SFKs can significantly reduce the replication of MERS-CoV (Shin et al., 2018) and some candidates are being proposed as treatment for COVID-19 because of its potential to effectively block the viral dissemination (Weisberg et al., 2020; Pillaiyar and Laufer, 2021).
Overall, immune response in buccal cavity, due to its anatomical location and interplay between upper and lower respiratory tract (LRT), could play a key role in viral spread and transmission not only at inter-individual level (Huang et al., 2021) but also in intra-individual viral dissemination to LRT, leading to a more severe phenotype.
Transversal analysis of different tissue samples from the same patients showed a tissue-specific behavior depending on illness severity. Most of the tissue-specific severity signals observed were represented by genes involved in innate immune response and cytokine signaling. Genes that appeared differentially expressed with severities in nasal and blood samples are related to leukocyte chemotaxis, cytokine-cytokine signaling, cell adhesion and extracellular exosomes, with higher expression of severe cases (with respect to those with mild disease) in blood and, in contrast, lower expression in nasal samples; B-cell antigen receptor signaling genes showed an opposite pattern. Specific expression signatures of severity in saliva samples correspond to genes involved in inflammatory and NK activation routes, cell adhesion, and vesicle trafficking.
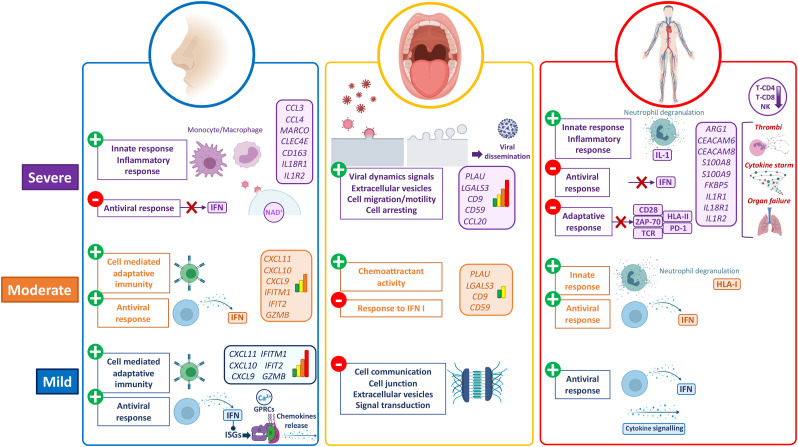
Our results point to a set of genes and pathways that are tissue-specific and correlated to severity (Fig. 5 ); many of these are involved in the innate immune system and cytokine/chemokine signaling pathways. Disease severity at systemic level might be strongly dependent on local immune response. Thus, the nasal epithelium shows a robust activation of the antiviral immune machinery in mild cases, characterized by IFN pathway involvement, thus triggering the IFN induced genes, chemokine release and transduction signaling in the cells, leading to the activation of downstream cascades related to Th1, NK and T-cytotoxic adaptative immune response. Instead, severe cases showed these local antiviral reactions switched off; they develop an exacerbated innate response represented by an over-expression of genes related to monocyte-macrophage recruitment in nasal epithelium (Fig. 5). We did not find signals of immune viral response in buccal mucosa in mild patients, which probably indicates a successful local control of the infection and, therefore, prevention of viral dissemination and systemic colonization. On the contrary, buccal mucosa in moderate and, to a greater extent, severe cases, provided evident signals of viral activity, cell arresting and viral dissemination to the LRT (Fig. 5); ultimately generating an exacerbated innate and impaired adaptative systemic immune responses. In addition, the buccal cavity might play a key role in body infection dissemination: a severe phenotype seems to be associated to limited immunological resistance to the virus in the mouth, therefore facilitating the dissemination of the virus towards the LRT. This could make saliva not only a good prognostic proxy of severity, but also a source of therapeutic targets to contain viral dissemination in early stages of the disease.
Fig. 5.
Schematic representation of the main findings in gene expression and pathway analysis of COVID-19 severity in nasal, oral and blood tissues. The figure was built with Biorender (https://biorender.com/) resources.
The present study has a few limitations. The number of genes that could be explored is limited by the technology employed (n-Counter from NanoString); in counterpart, this is the only one available that allows the analysis of RNA expression patterns from different tissues (usually involving limited and/or degraded RNA). Although we used a sample size substantially higher than usual in transcriptomic studies, results from the present study should be further validated using a similar design and other independent cohorts and, ideally, performing functional studies aimed at validating the severity related pathways detected. The growing number of high-throughput gene expression data available from COVID-19 patients could also help to validate tissue specific findings in a near future.
Overall, our study highlights the key role of the nasal epithelium in COVID-19 severity, and include transcript biomarkers and pathways that signal severity at different tissue layers. The findings might illuminate new diagnostic, prognostic, or therapeutic targets in COVID-19.
Author contribution
AS and FMT conceived, designed, and provided financial support to the study. IRC, JGR, CRV, NRN, GBC, HPF, MLC, ADU, CRT, SP, MJCT, SVL and FMT were involved in sample recruitment and analysis of the clinical data. AGC carried out the experimental lab work. AS, AGC, JPS, XB, RBA analyzed the data. AGC and AS wrote the initial draft of the article. All the authors revised and contributed to the final version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study received support from Instituto de Salud Carlos III (ISCIII): GePEM (PI16/01478/Cofinanciado FEDER; A.S.), DIAVIR (DTS19/00049/Cofinanciado FEDER, A.S.), Resvi-Omics (PI19/01039/Cofinanciado FEDER, A.S.), ReSVinext (PI16/01569/Cofinanciado FEDER, F.M.T.), Enterogen (PI19/01090/Cofinanciado FEDER, F.M.T.); Agencia Gallega para la Gestión del Conocimiento en Salud (ACIS): BI-BACVIR (PRIS-3, A.S.), and CovidPhy (SA 304 C, A.S.); Agencia Gallega de Innovación (GAIN): Grupos con Potencial de Crecimiento (IN607B 2020/08, A.S.), GEN-COVID (IN845D 2020/23, F.M.T.); Framework Partnership Agreement between the Consellería de Sanidad de la XUNTA de Galicia and GENVIP-IDIS - 2021–2024 (SERGAS-IDIS march 2021); and consorcio Centro de Investigación Biomédica en Red de Enfermedades Respiratorias (CB21/06/00103; F.M.T.). We also thank Aida Freire Valls from Nanostring for her support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envres.2022.112890.
Contributor Information
GEN-COVID study group:
Aguilera Guirao Antonio, Álvarez Escudero Julián, Antela López Antonio, Barbeito Castiñeiras Gema, Bello Paderne Xabier, Ben García Miriam, Carral García María Victoria, Cebey López Miriam, Coira Nieto Amparo, Conde Pájaro Mónica, Costa Alcalde José Javier, Currás Tuala María José, Dacosta Urbieta Ana Isabel, Díaz Esteban Blanca, Domínguez Santalla María Jesús, Fernández Pérez Cristina, Fernández Villaverde Juan, Galbán Rodríguez Cristóbal, García Allut José Luis, García Vicente Luisa, Giráldez Vázquez Elena, Gómez Carballa Alberto, Gómez Rial José, González Barcala Francisco Javier, Guerra Liñares Beatriz, Leboráns Iglesias Pilar, Lence Massa Beatriz, López Franco Montserrat, López Lago Ana, Martinón-Torres Federico, Antonio Salas, Navarro De la Cruz Daniel, Núñez Masid Eloína, Ortolá Devesa Juan Bautista, Pardo Seco Jacobo, Pazo Núñez María, Pérez del Molino Bernal Marisa, Pérez Freixo Hugo, Piñeiro Rodríguez Lidia, Pischedda Sara, Portela Romero Manuel, Pose Reino Antonio, Prada Hervella Gloria María, Queiro Verdes Teresa, Redondo Collazo Lorenzo, Regueiro Casuso Patricia, Rey García Susana, Rey Vázquez Sara, Riveiro Blanco Vanessa, Rivero Calle Irene, Rivero Velasco Carmen, Rodríguez Núñez Nuria, Rodríguez-Tenreiro Sánchez Carmen, Saborido Paz Eva, Sadiki Orayyou José Miguel, Saito Villanueva Carla, Serén Fernández Sonia, Souto Sanmartín Pablo, Taboada Muñiz Manuel, Trastoy Pena Rocío, Treviño Castellano Mercedes, Valdés Cuadrado Luis, Varela García Pablo, Vilas Iglesias María Soledad, Viz Lasheras Sandra, Ferreiro-Iglesias Rocio, Bastón-Rey iria, and Calviño-Suárez Cristina
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Alfi O., Yakirevitch A., Wald O., Wandel O., Izhar U., Oiknine-Djian E., Nevo Y., Elgavish S., Dagan E., Madgar O., et al. Human nasal and lung tissues infected ex vivo with SARS-CoV-2 provide insights into differential tissue-specific and virus-specific innate immune responses in the upper and lower respiratory tract. J. Virol. 2021 doi: 10.1128/JVI.00130-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althouse B.M., Wenger E.A., Miller J.C., Scarpino S.V., Allard A., Hebert-Dufresne L., Hu H. Superspreading events in the transmission dynamics of SARS-CoV-2: opportunities for interventions and control. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschenbrenner A.C., Mouktaroudi M., Krämer B., Oestreich M., Antonakos N., Nuesch-Germano M., Gkizeli K., Bonaguro L., Reusch N., Baßler K., et al. Disease severity-specific neutrophil signatures in blood transcriptomes stratify COVID-19 patients. Genome Med. 2021;13:7. doi: 10.1186/s13073-020-00823-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral-Arca R., Pardo-Seco J., Bello X., Martinón-Torres F., Salas A. Ancestry patterns inferred from massive RNA-seq data. RNA. 2019;25:857–868. doi: 10.1261/rna.070052.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral-Arca R., Pardo-Seco J., Martinón-Torres F., Salas A. A 2-transcript host cell signature distinguishes viral from bacterial diarrhea and it is influenced by the severity of symptoms. Sci. Rep. 2018;8:8043. doi: 10.1038/s41598-018-26239-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berri F., Rimmelzwaan G.F., Hanss M., Albina E., Foucault-Grunenwald M.L., Lê V.B., Vogelzang-van Trierum S.E., Gil P., Camerer E., Martinez D., et al. Plasminogen controls inflammation and pathogenesis of influenza virus infections via fibrinolysis. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A., Hamilton A.M., Furberg H., Pietzak E., Purdue M.P., Troester M.A., Hoadley K.A., Love M.I. An approach for normalization and quality control for NanoString RNA expression data. Briefings Bioinf. 2020 doi: 10.1093/bib/bbaa163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blighe K., Rana S., Lewis M. 2020. EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling. version 180, R package. [Google Scholar]
- Buchrieser J., Dufloo J., Hubert M., Monel B., Planas D., Rajah M.M., Planchais C., Porrot F., Guivel-Benhassine F., Van der Werf S., et al. Syncytia formation by SARS-CoV-2-infected cells. EMBO J. 2020;39 doi: 10.15252/embj.2020106267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caniglia J.L., Asuthkar S., Tsung A.J., Guda M.R., Velpula K.K. Immunopathology of galectin-3: an increasingly promising target in COVID-19. F1000Res. 2020;9:1078. doi: 10.12688/f1000research.25979.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho B.S., Irizarry R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.J., Wang S.F., Weng I.C., Hong M.H., Lo T.H., Jan J.T., Hsu L.C., Chen H.Y., Liu F.T. Galectin-3 enhances avian H5N1 influenza A virus-induced pulmonary inflammation by promoting NLRP3 inflammasome activation. Am. J. Pathol. 2018;188:1031–1042. doi: 10.1016/j.ajpath.2017.12.014. [DOI] [PubMed] [Google Scholar]
- Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., Debnath O., Thürmann L., Kurth F., Völker M.T., et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- Contoli M., Papi A., Tomassetti L., Rizzo P., Vieceli Dalla Sega F., Fortini F., Torsani F., Morandi L., Ronzoni L., Zucchetti O., et al. Blood interferon-α levels and severity, outcomes, and inflammatory profiles in hospitalized COVID-19 patients. Front. Immunol. 2021;12:648004. doi: 10.3389/fimmu.2021.648004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse J., Kalinke U., Oxenius A. Regulation of antiviral T cell responses by type I interferons. Nat. Rev. Immunol. 2015;15:231–242. doi: 10.1038/nri3806. [DOI] [PubMed] [Google Scholar]
- D'Alonzo D., De Fenza M., Pavone V. COVID-19 and pneumonia: a role for the uPA/uPAR system. Drug Discov. Today. 2020;25:1528–1534. doi: 10.1016/j.drudis.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372 doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerg H.A., Zaidi M.B., Altman M.C., Khaenam P., Gersuk V.H., Campos F.D., Perez-Martinez I., Meza-Segura M., Chaussabel D., Banchereau J., et al. Shared and organism-specific host responses to childhood diarrheal diseases revealed by whole blood transcript profiling. PLoS One. 2018;13 doi: 10.1371/journal.pone.0192082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux C.A., Rolain J.M., Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J. Microbiol. Immunol. Infect. 2020;53:425–435. doi: 10.1016/j.jmii.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P., Kibbe W.A., Lin S.M. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Dubovi E.J., Geratz J.D., Tidwell R.R. Enhancement of respiratory syncytial virus-induced cytopathology by trypsin, thrombin, and plasmin. Infect. Immun. 1983;40:351–358. doi: 10.1128/iai.40.1.351-358.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnest J.T., Hantak M.P., Li K., McCray P.B., Perlman S., Gallagher T. The tetraspanin CD9 facilitates MERS-coronavirus entry by scaffolding host cell receptors and proteases. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamage A.M., Tan K.S., Chan W.O.Y., Liu J., Tan C.W., Ong Y.K., Thong M., Andiappan A.K., Anderson D.E., Wang Y., et al. Infection of human Nasal Epithelial Cells with SARS-CoV-2 and a 382-nt deletion isolate lacking ORF8 reveals similar viral kinetics and host transcriptional profiles. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1009130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Revilla J., Deierborg T., Venero J.L., Boza-Serrano A. Hyperinflammation and fibrosis in severe COVID-19 patients: galectin-3, a target molecule to consider. Front. Immunol. 2020;11:2069. doi: 10.3389/fimmu.2020.02069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Rial J., Rivero-Calle I., Salas A., Martinon-Torres F. Role of monocytes/macrophages in covid-19 pathogenesis: implications for therapy. Infect. Drug Resist. 2020;13:2485–2493. doi: 10.2147/IDR.S258639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H., Wells K., Takada A., Kawaoka Y. Plasminogen-binding activity of neuraminidase determines the pathogenicity of influenza A virus. J. Virol. 2001;75:9297–9301. doi: 10.1128/JVI.75.19.9297-9301.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L.E., Bankhead A., Jeng S., Menachery V.D., Proll S., Belisle S.E., Matzke M., Webb-Robertson B.J., Luna M.L., Shukla A.K., et al. Mechanisms of severe acute respiratory syndrome coronavirus-induced acute lung injury. mBio. 2013;4 doi: 10.1128/mBio.00271-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Gómez-Carballa A., Bello X., Pardo-Seco J., Del Molino M.L.P., Martinón-Torres F., Salas A. Phylogeography of SARS-CoV-2 pandemic in Spain: a story of multiple introductions, micro-geographic stratification, founder effects, and super-spreaders. Zool. Res. 2020;41:605. doi: 10.24272/j.issn.2095-8137.2020.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Carballa A., Bello X., Pardo-Seco J., Martinón-Torres F., Salas A. Mapping genome variation of SARS-CoV-2 worldwide highlights the impact of COVID-19 super-spreaders. Genome Res. 2020;30:1434–1448. doi: 10.1101/gr.266221.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Carballa A., Pardo-Seco J., Bello X., Martinón-Torres F., Salas A. Superspreading in the emergence of COVID-19 variants. Trends Genet. 2021;37:1069–1080. doi: 10.1016/j.tig.2021.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Rial J., Currás-Tuala M.J., Rivero-Calle I., Gómez-Carballa A., Cebey-López M., Rodríguez-Tenreiro C., Dacosta-Urbieta A., Rivero-Velasco C., Rodríguez-Núñez N., Trastoy-Pena R. Increased serum levels of sCD14 and sCD163 indicate a preponderant role for monocytes in COVID-19 immunopathology. Front. Immunol. 2020;11:2436. doi: 10.3389/fimmu.2020.560381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanpour M., Rezaie J., Nouri M., Panahi Y. The role of extracellular vesicles in COVID-19 virus infection. Infect. Genet. Evol. 2020;85:104422. doi: 10.1016/j.meegid.2020.104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herberg J.A., Kaforou M., Gormley S., Sumner E.R., Patel S., Jones K.D., Paulus S., Fink C., Martinon-Torres F., Montana G., et al. Transcriptomic profiling in childhood H1N1/09 influenza reveals reduced expression of protein synthesis genes. J. Infect. Dis. 2013;208:1664–1668. doi: 10.1093/infdis/jit348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herberg J.A., Kaforou M., Wright V.J., Shailes H., Eleftherohorinou H., Hoggart C.J., Cebey-López M., Carter M.J., Janes V.A., Gormley S., et al. Diagnostic test accuracy of a 2-transcript host RNA signature for discriminating bacterial vs viral infection in febrile children. JAMA. 2016;316:835–845. doi: 10.1001/jama.2016.11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Ding Y., Nie H., Ji H.L. bioRxiv; 2021. Fibrinolysis Influences SARS-CoV-2 Infection in Ciliated Cells. [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N., Pérez P., Kato T., Mikami Y., Okuda K., Gilmore R.C., Conde C.D., Gasmi B., Stein S., Beach M., et al. SARS-CoV-2 infection of the oral cavity and saliva. Nat. Med. 2021 doi: 10.1038/s41591-021-01296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiani P., Mosca E., Della Camera G., Melillo D., Migliorini P., Milanesi L., Boraschi D. Profiling the course of resolving vs. Persistent inflammation in human monocytes: the role of IL-1 family molecules. Front. Immunol. 2020;11:1426. doi: 10.3389/fimmu.2020.01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R., Ramaswamy S., Harilal D., Uddin M., Loney T., Nowotny N., Alsuwaidi H., Varghese R., Deesi Z., Alkhajeh A., et al. Host transcriptomic profiling of COVID-19 patients with mild, moderate, and severe clinical outcomes. Comput. Struct. Biotechnol. J. 2021;19:153–160. doi: 10.1016/j.csbj.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H.L., Zhao R., Matalon S., Matthay M.A. Elevated plasmin(ogen) as a common risk factor for COVID-19 susceptibility. Physiol. Rev. 2020;100:1065–1075. doi: 10.1152/physrev.00013.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Lian J.S., Hu J.H., Gao J., Zheng L., Zhang Y.M., Hao S.R., Jia H.Y., Cai H., Zhang X.L., et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69:1002–1009. doi: 10.1136/gutjnl-2020-320926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam Y.W., Okumura Y., Kido H., Ng L.F., Bruzzone R., Altmeyer R. Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwaan H.C., Lindholm P.F. The central role of fibrinolytic response in COVID-19-A hematologist's perspective. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22031283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry M.L., Foxman E.F. Antiviral response in the nasopharynx identifies patients with respiratory virus infection. J. Infect. Dis. 2018;217:897–905. doi: 10.1093/infdis/jix648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux J.E., Siddle K.J., Shaw B.M., Loreth C., Schaffner S.F., Gladden-Young A., Adams G., Fink T., Tomkins-Tinch C.H., Krasilnikova L.A., et al. Phylogenetic analysis of SARS-CoV-2 in Boston highlights the impact of superspreading events. Science. 2020;371 doi: 10.1126/science.abe3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppkes M., Knopf J., Naschberger E., Lindemann A., Singh J., Herrmann I., Stürzl M., Staats L., Mahajan A., Schauer C., et al. Vascular occlusion by neutrophil extracellular traps in COVID-19. EBioMedicine. 2020;58:102925. doi: 10.1016/j.ebiom.2020.102925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Parks G.D. Relative contribution of cellular complement inhibitors CD59, CD46, and CD55 to parainfluenza virus 5 inhibition of complement-mediated neutralization. Viruses. 2018;10 doi: 10.3390/v10050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman N.A.P., Peddu V., Xie H., Shrestha L., Huang M.L., Mears M.C., Cajimat M.N., Bente D.A., Shi P.Y., Bovier F., et al. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F.T., Hsu D.K. The role of galectin-3 in promotion of the inflammatory response. Drug News Perspect. 2007;20:455–460. doi: 10.1358/dnp.2007.20.7.1149628. [DOI] [PubMed] [Google Scholar]
- Lokugamage K.G., Hage A., de Vries M., Valero-Jimenez A.M., Schindewolf C., Dittmann M., Rajsbaum R., Menachery V.D. Type I interferon susceptibility distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 2020;94 doi: 10.1128/JVI.01410-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan P., Kuppermann N., Mejias A., Suarez N., Chaussabel D., Casper T.C., Smith B., Alpern E.R., Anders J., Atabaki S.M., et al. Association of RNA biosignatures with bacterial infections in febrile infants aged 60 Days or younger. JAMA. 2016;316:846–857. doi: 10.1001/jama.2016.9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney B.E., Perera K.D., Saunders D.R.D., Shadipeni N., Fleming S.D. Interactions of viruses and the humoral innate immune response. Clin. Immunol. 2020;212:108351. doi: 10.1016/j.clim.2020.108351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantlo E., Bukreyeva N., Maruyama J., Paessler S., Huang C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antivir. Res. 2020;179:104811. doi: 10.1016/j.antiviral.2020.104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F., Roth D.M., Jans D.A., Pouton C.W., Partridge L.J., Monk P.N., Moseley G.W. Tetraspanins in viral infections: a fundamental role in viral biology? J. Virol. 2005;79:10839–10851. doi: 10.1128/JVI.79.17.10839-10851.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonagle D., Sharif K., O'Regan A., Bridgewood C. The role of cytokines including interleukin-6 in COVID-19 induced pneumonia and macrophage activation syndrome-like disease. Autoimmun. Rev. 2020;19:102537. doi: 10.1016/j.autrev.2020.102537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita I., Varela P., Belinha A., Gaifem J., Laforge M., Vergnes B., Estaquier J., Silvestre R. Exploring NAD+ metabolism in host-pathogen interactions. Cell. Mol. Life Sci. 2016;73:1225–1236. doi: 10.1007/s00018-015-2119-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nchioua R., Kmiec D., Müller J.A., Conzelmann C., Groß R., Swanson C.M., Neil S.J.D., Stenger S., Sauter D., Münch J., et al. SARS-CoV-2 is restricted by zinc finger antiviral protein despite preadaptation to the low-CpG environment in humans. mBio. 2020;11 doi: 10.1128/mBio.01930-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A.M., Steen C.B., Liu C.L., Gentles A.J., Chaudhuri A.A., Scherer F., Khodadoust M.S., Esfahani M.S., Luca B.A., Steiner D., et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano M.A., Tibaldi E., Palù G., Brunati A.M. Viral proteins and Src family kinases: mechanisms of pathogenicity from a "liaison dangereuse. World J. Virol. 2013;2:71–78. doi: 10.5501/wjv.v2.i2.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell G.P., McLean A.S., Booth D.R., Armstrong N.J., Nalos M., Huang S.J., Manak J., Tang W., Tam O.Y., Chan S., et al. A distinct influenza infection signature in the blood transcriptome of patients with severe community-acquired pneumonia. Crit. Care. 2012;16:R157. doi: 10.1186/cc11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasomsub E., Watcharananan S.P., Boonyawat K., Janchompoo P., Wongtabtim G., Suksuwan W., Sungkanuparph S., Phuphuakrat A. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce C.A., Sy S., Galen B., Goldstein D.Y., Orner E., Keller M.J., Herold K.C., Herold B.C. Natural mucosal barriers and COVID-19 in children. JCI Insight. 2021;6 doi: 10.1172/jci.insight.148694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar T., Laufer S. Kinases as potential therapeutic targets for anti-coronaviral therapy. J. Med. Chem. 2021 doi: 10.1021/acs.jmedchem.1c00335. [DOI] [PubMed] [Google Scholar]
- Pipes L., Wang H., Huelsenbeck J.P., Nielsen R. Assessing uncertainty in the rooting of the SARS-CoV-2 phylogeny. Mol. Biol. Evol. 2020 doi: 10.1093/molbev/msaa316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in wuhan, China. Clin. Infect. Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D., Ngai J., Speed T.P., Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajuthi S.P., DeFord P., Li Y., Jackson N.D., Montgomery M.T., Everman J.L., Rios C.L., Pruesse E., Nolin J.D., Plender E.G., et al. Type 2 and interferon inflammation regulate SARS-CoV-2 entry factor expression in the airway epithelium. Nat. Commun. 2020;11:5139. doi: 10.1038/s41467-020-18781-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas A., Pardo-Seco J., Barral-Arca R., Cebey-López M., Gómez-Carballa A., Rivero-Calle I., Pischedda S., Currás-Tuala M.-J., Amigo J., Gómez-Rial J. Whole exome sequencing identifies new host genomic susceptibility factors in empyema caused by streptococcus pneumoniae in children: a pilot study. Genes. 2018;9:240. doi: 10.3390/genes9050240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas A., Pardo-Seco J., Cebey-López M., Gómez-Carballa A., Obando-Pacheco P., Rivero-Calle I., Currás-Tuala M.-J., Amigo J., Gómez-Rial J., Martinón-Torres F. Whole exome sequencing reveals new candidate genes in host genomic susceptibility to respiratory syncytial virus disease. Sci. Rep. 2017;7:1–13. doi: 10.1038/s41598-017-15752-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.S., Jung E., Kim M., Baric R.S., Go Y.Y. Saracatinib inhibits Middle East respiratory syndrome-coronavirus replication in vitro. Viruses. 2018;10 doi: 10.3390/v10060283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez N.M., Bunsow E., Falsey A.R., Walsh E.E., Mejias A., Ramilo O. Superiority of transcriptional profiling over procalcitonin for distinguishing bacterial from viral lower respiratory tract infections in hospitalized adults. J. Infect. Dis. 2015;212:213–222. doi: 10.1093/infdis/jiv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney T.E., Wong H.R., Khatri P. Robust classification of bacterial and viral infections via integrated host gene expression diagnostics. Sci. Transl. Med. 2016;8:346ra391. doi: 10.1126/scitranslmed.aaf7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thair S.A., He Y.D., Hasin-Brumshtein Y., Sakaram S., Pandya R., Toh J., Rawling D., Remmel M., Coyle S., Dalekos G.N., et al. Transcriptomic similarities and differences in host response between SARS-CoV-2 and other viral infections. iScience. 2021;24:101947. doi: 10.1016/j.isci.2020.101947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K., Tsang O.T., Yip C.C., Chan K.H., Wu T.C., Chan J.M., Leung W.S., Chik T.S., Choi C.Y., Kandamby D.H., et al. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020;71:841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twaddell S.H., Baines K.J., Grainge C., Gibson P.G. The emerging role of neutrophil extracellular traps in respiratory disease. Chest. 2019;156:774–782. doi: 10.1016/j.chest.2019.06.012. [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veras F.P., Pontelli M.C., Silva C.M., Toller-Kawahisa J.E., de Lima M., Nascimento D.C., Schneider A.H., Caetité D., Tavares L.A., Paiva I.M., et al. SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J. Exp. Med. 2020;217 doi: 10.1084/jem.20201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.H., Lin C.Y., Chang M.R., Urbina A.N., Assavalapsakul W., Thitithanyanont A., Chen Y.H., Liu F.T., Wang S.F. The role of galectins in virus infection - a systemic literature review. J. Microbiol. Immunol. Infect. 2020;53:925–935. doi: 10.1016/j.jmii.2019.09.005. [DOI] [PubMed] [Google Scholar]
- Wei Y., Ji Y., Guo H., Zhi X., Han S., Zhang Y., Gao Y., Chang Y., Yan D., Li K., et al. CD59 association with infectious bronchitis virus particles protects against antibody-dependent complement-mediated lysis. J. Gen. Virol. 2017;98:2725–2730. doi: 10.1099/jgv.0.000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg E., Parent A., Yang P.L., Sattler M., Liu Q., Wang J., Meng C., Buhrlage S.J., Gray N., Griffin J.D. Repurposing of kinase inhibitors for treatment of COVID-19. Pharm. Res. (N. Y.) 2020;37:167. doi: 10.1007/s11095-020-02851-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Springer-Verlag; New York: 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., Warren J.L., Geng B., Muenker M.C., Moore A.J., et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N. Engl. J. Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Qi F., Li H., Yang Q., Wang H., Wang X., Liu X., Zhao J., Liao X., Liu Y., et al. The differential immune responses to COVID-19 in peripheral and lung revealed by single-cell RNA sequencing. Cell Discov. 2020;6:73. doi: 10.1038/s41421-020-00225-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q., Li P., Ye X., Huang X., Feng B., Ji T., Chen Z., Li F., Zhang Y., Luo K., et al. Longitudinal peripheral blood transcriptional analysis reveals molecular signatures of disease progression in COVID-19 patients. J. Immunol. 2021;206:2146–2159. doi: 10.4049/jimmunol.2001325. [DOI] [PubMed] [Google Scholar]
- Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. 2021. Enrichplot: visualization of functional enrichment result. Vol R package version 1.10.2. [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Wu Z., Li J.W., Zhao H., Wang G.Q. Cytokine release syndrome in severe COVID-19: interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int. J. Antimicrob. Agents. 2020;55:105954. doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H.Y., Xu M., Yang C.X., Tian R.R., Zhang M., Li J.J., Wang X.C., Ding Z.L., Li G.M., Li X.L., et al. Longitudinal transcriptome analyses show robust T cell immunity during recovery from COVID-19. Signal Transduct. Target Ther. 2020;5:294. doi: 10.1038/s41392-020-00457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H.Y., Zhang M., Yang C.X., Zhang N., Wang X.C., Yang X.P., Dong X.Q., Zheng Y.T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020;17:541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S. 2019. ctrlGene: Assess the Stability of Candidate Housekeeping Genes. Version 101, R Package. [Google Scholar]
- Zhou Y., Zhang J., Wang D., Guan W., Qin J., Xu X., Fang J., Fu B., Zheng X., Zhao H., et al. Profiling of the immune repertoire in COVID-19 patients with mild, severe, convalescent, or retesting-positive status. J. Autoimmun. 2021;118:102596. doi: 10.1016/j.jaut.2021.102596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Expression data that support the findings of the present study have been deposited in Gene Expression Omnibus (GEO) database under the accession code GSE183071.