Abstract
Introduction
Studies have shown that genetic variation and environmental factors are associated with individual differences in therapeutic efficacy and side effects of opioids. However, the focus of these studies has been on a single factor of single-nucleotide polymorphisms (SNPs) or haplotypes, for which results have rarely been validated. For complex traits, such as cancer pain and opioid response, interactions between multiple genetic variation and environmental factors need to be considered to explain the opioid individual differences.
Methods
We conducted an exploratory two-stage cross-sectional study with 1027 Chinese patients who were taking strong opioid medications for their cancer pain, and genotyped 110 SNPs to explore the association of SNPs, haplotypes, gene–gene and gene–environment interactions with opioid dose, pain relief, and opioid-induced constipation.
Results
Due to the failure to meet Benjamini–Hochberg criteria in the discovery stage or to be validated in replication stage, no association was found between SNPs, haplotypes, paired SNP–SNP interactions or multi-dimensional gene–gene interactions and opioid response. However, for gene–environment interactions, optimal models have been constructed in all phenotypes of opioid response.
Conclusions
This study reveals for the first time that construction of multidimensional gene–environment interactions enables better interpretations of the effect of genetic variation and environmental factors on the opioid response in patients with cancer pain.
Trial registration
Chictr.org.cn, identifier, ChiCTR2000033576.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40122-022-00353-5.
Keywords: Cancer pain, Opioid response, Individual difference, Gene–gene interaction, Gene–environment interaction, GMDR
Key Summary Points
| This two-stage cross-sectional study explored the association of SNPs, haplotypes, gene–gene and gene–environment interactions with opioid dose, pain relief, and opioid-induced constipation. |
| No association was found between SNPs, haplotypes, paired SNP–SNP interactions or multi-dimensional gene–gene interactions and opioid response. However, optimal gene–environment interaction models have been constructed in all phenotypes of opioid response. |
| Construction of multidimensional gene–environment interactions enables better interpretations of the effect of genetic variation and environmental factors on the opioid response in patients with cancer pain. |
Introduction
Strong opioids are the mainstay of analgesic therapy in moderate-to-severe cancer-related pain management [1, 2]. However, the efficacy and side effects of opioids vary considerably among individuals, which results in inadequate pain control in 10–30% of patients with cancer pain [3, 4]. Both genetic and environmental factors contribute to individual differences in opioid response. Regarding genetic factors, numerous studies have identified that genetic polymorphisms of drug metabolizing enzymes (cytochrome P450 family 3 subfamily A member 4 [CYP3A4], CYP2D6, UDP glucuronosyltransferase family 2 member B7 [UGT2B7]), membrane drug transport proteins (ATP binding cassette B1 [ABCB1]), and molecules involved in opioid receptor signaling (opioid receptor mu 1 [OPRM1], opioid receptor kappa 1 [OPRK1], opioid receptor delta 1 [OPRD1], potassium inwardly rectifying channel subfamily J member 6 [KCNJ6]) and pain regulation (catachol-omethyltransferase [COMT]), which are associated with the efficacy, dose, and toxicity of opioids [5–8]. However, results from different studies are inconsistent. For example, the European Pharmacogenetic Opioid Study (EPOS), the largest study to date, which included 2294 European patients with cancer pain, did not find any association between opioid dose and 112 candidate SNPs in 25 genes [9]. In two genome-wide association studies (GWAS), although three SNPs were identified to be associated with pain relief, opioid efficacy, or opioid sensitivity in patients with cancer pain, these results have not been confirmed in subsequent studies [10, 11]. Even for the most extensively studied OPRM1 rs1799971, a recent meta-analysis revealed that there was only a statistically significant association between the OPRM1 A118G polymorphism and pain relief after opioid analgesia in Asian patients but not in Caucasian populations [12].
Given the complexity of pain biology and the size of the human genome, conflicting results on the association of genetic polymorphisms with opioid response may be explained by variable study designs, sample heterogeneity, small sample sizes, phenotype complexity, and alternative statistical approaches [13]. Pain perception and opioid response are complex traits, which are likely to be the product of numerous gene–gene and gene–environment interactions. Therefore, it is necessary to detect gene–gene and gene–environment interactions between different genes for complex traits (e.g., pain or opioid response). Most previous studies had explored the relationship between SNPs and opioid analgesia [14–20]. Few studies have investigated the interactions between genetic variants from more than one gene. However, these studies have been limited to two candidate SNPs at a time [18–20], and no further studies have analyzed gene–environment interactions. Even for the GWAS, there are some limitations, such as only a limited number of SNPs to reach the statistical threshold. Genetic variants that do reach the statistical threshold explain only a small proportion of phenotypic variation, and the function of these variants may be minor or isolated. GWAS cannot identify every genetic determinant or their interactions [21].
Moreover, genetic studies for cancer pain and opioid response in China are conducted with sample size of ten to over 100 cases, focusing on only a few SNPs and lagging behind the United States, Europe, Japan, and other countries or regions. There is evidence showing significant differences in pain sensitivity and analgesic effects between races and ethnicities [22].
To address these issues, the present study was designed to analyze polymorphisms of 136 SNPs in 54 candidate genes in the Chinese population and investigate the effects of genetic and environmental factors on the efficacy and adverse drug reaction (ADR) of opioids in Chinese cancer pain patients by combining clinical characteristics from multiple perspectives, including SNPs, haplotypes, gene–gene interactions, and gene–environment interactions to provide a reference for individualized treatment of cancer pain.
Methods
Patient Enrollment Criteria
This exploratory cross-sectional study was divided into two stages, namely discovery and replication stages. Subjects in the discovery sample were enrolled from March 1, 2018 to January 31, 2019 at a cancer center in a Grade III Level A hospital in central China. Patients were enrolled according to the following criteria: (1) age > 18 years; (2) had a histologically or cytologically diagnosed malignant tumor; (3) experienced cancer pain symptoms and received regular treatment with strong opioids (categorized as the third step for cancer pain treatment by the WHO’s pain ladder for adults [23]) for at least 72 h; and (4) voluntarily provided informed consent. Subjects in the replication sample were recruited at the same medical center from April 1, 2019 to October 31, 2019, using the same enrollment criteria as the replication sample. Each patient was only enrolled in either the discovery or the replication sample. After informed consent was obtained, blood samples that were remaining following the hematology tests were collected for DNA analysis. The clinical characteristics of all patients are summarized in Table 1.
Table 1.
Patient demographics and symptoms
| Variables | All patients | Discovery sample | Replication sample | p |
|---|---|---|---|---|
| (N = 858) | (N = 572) | (N = 286) | ||
| Age | 56.241 ± 11.449 | 56.360 ± 11.108 | 56.003 ± 12.118 | 0.667 |
| Ethnic group | ||||
| Han | 858 (100.000) | 572 (100.000) | 286 (100.000) | 1.000 |
| Others | 0 (0.000) | 0 (0.000) | 0 (0.000) | |
| Gender | 0.268 | |||
| Male | 517 (60.256) | 337 (58.916) | 180 (62.937) | |
| Female | 341 (39.744) | 235 (41.084) | 106 (37.063) | |
| BMI | 21.230 ± 3.601 | 21.340 ± 3.913 | 21.010 ± 2.872 | 0.209 |
| Tumor diagnosis | ||||
| Lung | 348 (40.559) | 220 (38.462) | 128 (44.755) | 0.227 |
| Gastrointestinal | 256 (29.837) | 176 (30.769) | 80 (27.972) | |
| Breast | 69 (8.042) | 49 (8.566) | 20 (6.993) | |
| Genitourinary | 57 (6.643) | 40 (6.993) | 17 (5.944) | |
| Hematological | 15 (1.748) | 12 (2.098) | 3 (1.049) | |
| Unknown origin | 7 (0.816) | 5 (0.874) | 2 (0.699) | |
| Others | 106 (12.354) | 70 (12.238) | 36 (12.587) | |
| PS | ||||
| 0–1 | 734 (85.548) | 489 (85.490) | 245 (85.664) | 1.000 |
| > 1 | 124 (14.452) | 83 (14.510) | 41 (14.336) | |
| Tumor metastasis status | ||||
| Bone | 404 (47.086) | 261 (45.629) | 143 (50.000) | 0.220 |
| Liver | 194 (22.611) | 125 (21.853) | 69 (24.126) | |
| CNS | 100 (11.655) | 65 (11.364) | 35 (12.238) | |
| Lung | 148 (17.249) | 100 (17.483) | 48 (16.783) | |
| Other | 400 (46.620) | 273 (47.727) | 127 (44.406) | |
| Opioid tolerance | 230 (26.807) | 153 (26.748) | 77 (26.923) | 1.000 |
| Pain category | ||||
| Bone soft tissue pain | 588 (68.531) | 379 (66.259) | 209 (73.077) | 0.213 |
| Visceral pain | 85 (9.907) | 59 (10.315) | 26 (9.091) | |
| Neuropathic pain | 15 (1.748) | 10 (1.748) | 5 (1.748) | |
| Mixed pain | 170 (19.813) | 124 (21.678) | 46 (16.084) | |
| NRS2002 score | 3.144 ± 1.169 | 3.147 ± 1.173 | 3.138 ± 1.162 | 0.918 |
| Number of adjuvant analgesics | 0.782 ± 0.749 | 0.762 ± 0.741 | 0.822 ± 0.763 | 0.273 |
| SDS score | 42.668 ± 8.891 | 42.710 ± 8.874 | 42.584 ± 8.939 | 0.845 |
| ALT (U/l) | 32.457 ± 45.268 | 32.280 ± 47.483 | 32.811 ± 40.553 | 0.871 |
| AST (U/l) | 39.152 ± 49.740 | 40.224 ± 54.887 | 37.007 ± 37.375 | 0.372 |
| LDH (U/l) | 344.867 ± 480.500 (n = 850) | 348.863 ± 447.911 (n = 569) | 336.776 ± 541.280 (n = 281) | 0.643 |
| Creatinine serum concentration (μmol/l) | 67.707 ± 26.548 | 67.713 ± 23.202 | 67.695 ± 32.261 | 0.993 |
| Opioid | ||||
| Oxycodone | 704 (82.051) | 471 (82.343) | 233 (81.468) | 0.199 |
| Morphine | 15 (1.748) | 9 (1.573) | 6 (2.098) | |
| Fentanyl | 161 (18.765) | 106 (18.531) | 55 (19.231) | |
| Time since onset of pain symptoms (months) | 2.568 ± 4.023 | 2.518 ± 3.968 | 2.669 ± 4.136 | 0.604 |
| Time since start of opioid treatment (months) | 1.469 ± 3.015 | 1.424 ± 3.098 | 1.558 ± 2.844 | 0.542 |
| Opioid dose (OEDD, mg) | 40.026 ± 36.312 | 40.162 ± 36.303 | 39.755 ± 36.391 | 0.877 |
| Pain relief (%) | 77.528 ± 15.158 | 77.124 ± 15.218 | 78.336 ± 15.031 | 0.270 |
| Constipation | 521 (60.723) | 355 (62.063) | 166 (58.042) | 0.267 |
P was calculated by SPSS (version 17.00); For continuous variables (marked mean ± SD) using independent sample t test; Chi-square test was used for categorical variables (marked as number (%) and Fisher’s exact test was used for values less than 5
BMI body mass index, PS performance status, CNS central nervous system, NRS2002 nutritional risk screening scoring, SDS Self-Rating Depression Scale, ALT alanine aminotransferase, AST aspartate transaminase, LDH lactate dehydrogenase, OEDD oral oxycodone equivalent daily dose
This study protocol was approved by the medical ethics committee of Tongji Medical College, Huazhong University of Science and Technology (approval number: 2018-S016) and registration was submitted to the China Clinical Trials Registry (registration number: ChiCTR2000033576). This study was performed in accordance with the Helsinki Declaration of 1964 and its later amendments.
Patient Enrollment Procedure
The patient’s age, ethnicity, gender, body mass index (BMI), tumor diagnosis, and known localization of metastases, were collected on the day of enrollment. The duration and dose of opioids and number of adjuvant analgesics were recorded. Opioid dose included fixed dose administered in the past 24 h and rescue dose. In this study, 81.91% of patients were on oxycodone hydrochloride sustained-release tablets. Thus, all opioid doses were converted into oral oxycodone equivalent daily dose (OEDD) according to the conversion factors in the NCCN adult cancer pain guideline [2]. According to the definition of opioid tolerance by FDA, the patient was considered to be opioid tolerant if for at least 1 week he or she had been receiving oral morphine 60 mg/day; transdermal fentanyl 25 mcg/h; oral oxycodone 30 mg/day; or an equianalgesic dose of any other opioid [24]. Characteristics of pain were assessed using the modified Edmonton Staging System [25] and performance status (PS) was assessed using the Zubrod-ECOG-WHO scoring system [26]. Patients’ nutritional status was assessed using the nutritional risk screening (NRS 2002) scoring system [27] and mental status was assessed using the Zung Self Rating Depression Scale (SDS) [28].
Phenotypes
In this study, three phenotypes that reflect opioid drug response in the past 24 h, namely opioid dose, pain relief, and constipation, were collected on the day of enrollment to examine the relevance of genetic polymorphisms in opioid drug efficacy and safety. The pain relief phenotype in the study was semi-quantitative and determined based on the Brief Pain Inventory (BPI), one of the most widely used measurement tools for assessing clinical pain [29]. Pain relief was scored on an 11-point numerical rating scale that ranged from 0% (no relief) to 100%. Constipation is one of the most common adverse effects of opioids and was used as an indicator for safety in this study. The relevance of constipation for opioids was determined by combining patients’ current therapeutic agents and medical history and using the five principles of adverse event analysis, developed by the Chinese State Food and Drug Administration [30]. Severity was graded according to the National Cancer Institute Common Toxicity Criteria (NCI-CTCAE) Version 4.0, which classifies ADR as grade 0 or other.
Genotyping Procedures
In order to make the candidate genotypes more clinically significant, only SNP loci with clinical research support for the association with cancer pain or opioid drug response will be selected. First, by using available information in the Human Pain Genetics Database (http://hpgl.ca/hpgdb), we had screened 82 SNP loci related to cancer pain or analgesic. Then we conducted a systematic search for genetic loci affecting cancer pain or opioid response to check the SNP loci screened in the first step and fill in the gaps. Finally, 136 candidate SNP loci were identified. Genomic DNA was extracted from all subjects using a blood genomic DNA extraction kit (DNP-348, Beijing Tiangen Biochemical Technology Co., Ltd.) and stored at − 80 °C until further analysis. The quality of all DNA samples was verified using agarose gel electrophoresis. Genotyping analysis was performed by CapitalBio Technology using the Mass ARRAY® DNA mass spectrometry system (Agena Bioscience, San Diego, CA).
Of the 136 candidate SNPs, seven failed for establishment of the detection method, five had a typing missing rate greater than 10%, 13 SNPs had a minor allele frequency (MAF) < 1%, and one SNP was not in Hardy–Weinberg equilibrium. After excluding these 26 SNPs, 110 were included in the final analysis. Chromosomal location, genotype frequencies, and gene function of the 136 SNPs are listed in the Supplementary Material Table 1. The MAFs in this study were similar to those reported in previous studies.
Statistical Analysis
Continuous variables are expressed as means ± SDs, and categorical variables are expressed as percentages (%). Before analysis, quality control was performed for the raw genetic data to exclude DNA samples with > 10% typing missing rate and candidate SNP loci that had > 10% typing missing rate, had MAF < 1%, or were not in the Hardy–Weinberg equilibrium (χ2 test, p < 0.01). Because OEDD was not normally distributed, it was log-transformed (log OEDD) for analysis when used as a phenotypic variable. To better explore the effects of non-genetic factors on different phenotypes, age, BMI, and time since start of opioid treatment were used as both continuous and categorical variables (Supplementary Material Table 2). Significant (p < 0.05) non-genetic factors for each phenotype were identified using the lm function in R, which were included as covariates in the subsequent SNP association and paired gene–gene interaction analyses. Collinearity diagnostics were also performed.
Single SNP Association Analysis
PLINK1.90 software was used for the association analysis of SNPs [31]. Spearman test and Wilcoxon test were used for continuous variables and dichotomous variables, respectively. The dominant model was prespecified for the primary analyses (additive and recessive models were used for secondary and exploratory) to investigate which SNPs were associated with the three phenotypes. Opioid dose and pain relief were used as both continuous and dichotomous variables (bound by the mean, opioid dose [logarithm] ≤ 1.5 vs. > 1.5; pain relief ≤ 78% vs. > 78%) for the analyses. The analysis was also repeated without the inclusion of covariates as a sensitivity check.
To explore whether the lack of significant relationship between candidate SNPs and each phenotype may have been due to case mixing, we also performed subgroup analyses. Subgroups were grouped by age: young (≤ 57 years) and old (> 57 years) (grouped by the mean value); by opioids: oxycodone, morphine and fentanyl, and by time since start of opioid treatment: patients receiving short-term (≤ 1 month) opioid treatment, and patients receiving long-term (> 1 month) opioid treatment.
Construction of Haplotypes and Association Analysis
The Haploview software (https://www.broadinstitute.org/haploview/haploview) [32] was employed and Lewontin’s D value was used as criteria for the haplotype prediction and association analysis of 110 SNPs that were screened at quality control. A total of 12 haplotype blocks were successfully constructed in both the discovery and replication samples; the frequency of each haplotype is detailed in the Supplementary Material Table 4. χ2 test was used for the haplotype association analysis, all phenotypes were analyzed as dichotomous variables (continuous phenotypes were converted into dichotomous variables according to their mean values; opioid dose [logarithm] ≤ 1.5 vs. > 1.5, pain relief ≤ 78% vs. > 78%).
Gene–gene and Gene–environment Interaction Analyses
Similar to haplotype association analysis, all phenotypes were analyzed as dichotomous variables in the gene–gene and gene–environment interaction. PLINK1.90 and generalized multifactor dimensionality reduction (GMDR) software [33] were used to explore the effects of gene–gene and gene–environment interactions on the three phenotypes. False-positive and true-positive rates were used to evaluate model prediction accuracy. Cross-validation consistency was used to assess the quality of the model, and the sign test was used to evaluate statistical significance of the model. Models with the highest testing accuracy and cross-validation consistency were regarded as the best interaction models.
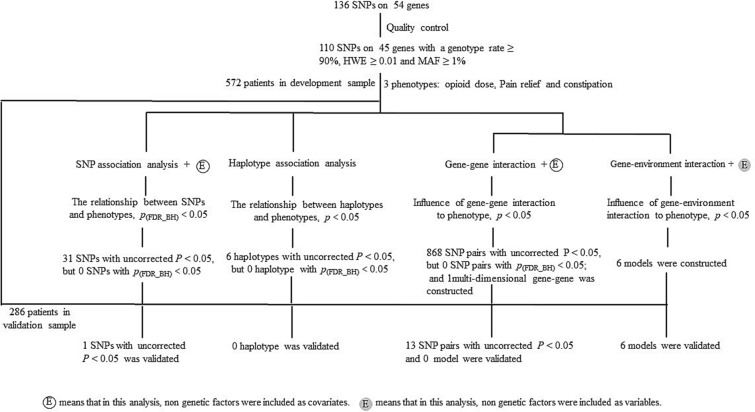
All analyses were two-tailed, and p < 0.05 was considered statistically significant. For the different genetic models, the following approaches were implemented to alleviate the problem of multiplicity. First, a two-step analysis was performed for all three phenotypes. The discovery samples were used for the initial screening of SNPs, haplotypes, and models, and replication samples were then used to validate the statistically significant results that were obtained from the discovery samples. Second, for the association analyses of SNPs and haplotypes and for the paired gene–gene interaction analysis, the Benjamini–Hochberg (BH) method [34], based on the false discovery rate (FDR) criterion, was used for multiple comparison correction to reduce the falsity of the association analysis. The flow diagram of the analysis is shown in Fig. 1.
Fig. 1.
Flow diagram of the data analysis in the study. We firstly explored the association between 45 candidate genes and opioid response in SNP or haplotype association analysis. In SNP association analysis, only one SNP was significantly associated with constipation. Although this result failed to meet the Benjamini–Hochberg FDR criterion, this association was verified in the validation sample. Then we studied the association of gene–gene with opioid response. In haplotype association analysis, six haplotypes were significantly associated with opioid response in the development stage (p < 0.05). However, none of them met Benjamini–Hochberg FDR criteria, or were verified in the validation sample. There were 280–304 SNP–SNP pairs that were significantly associated with opioid dose, pain relief, or constipation. However, only seven SNP–SNP pairs associated with opioid dose, and three with pain relief or constipation passed validation in the validation stage. At last, we explored the influence of multi-dimensional gene–gene and gene–environment interactions on opioid response. We found one multi-dimensional gene–gene model and six multi-dimensional gene–environment models in the development stage, and 0 gene–gene models and six gene–environment models were verified in the validation sample, respectively
Results
Clinical Information of Enrolled Patients
A total of 695 patients were included in this study in the discovery stage. However, blood samples of 114 patients were not collected and thus could not be used for further genetic analysis. In addition, seven patients had incomplete information, and genotyping results of two patients failed the quality control test, which resulted in 572 patients in the final analysis. Details are shown in Table 1. To examine the credibility of the results obtained in the discovery stage, we established a replication sample comprising 332 patients, of which 286 had complete clinical information and blood samples. There were no significant differences between the discovery and replication samples in demographic characteristics (Table 1).
Univariate analysis showed that 15 factors such as age, tumor diagnosis, PS score, and tumor metastasis status were associated with opioid response (Supplementary Material Table 2).
Single SNP Association Analysis
The association analysis of the 110 SNPs with the three phenotypes showed that variation at rs5275 in the prostaglandin-endoperoxide synthase 2 (PTGS2) gene was significantly associated with constipation in the dominant model and that carrying the minor allele G significantly reduced the incidence of constipation. Although this result failed to meet the Benjamini–Hochberg FDR criteria, this association was verified in the replication sample. As shown in Table 2, the risk of constipation in patients carrying the GG/GA genotype was significantly lower in both the discovery and replication groups (50.838 and 49.485%, respectively) than that in patients carrying the AA genotype (66.492 and 62.567%, respectively; odds ratio [OR] = 0.528 [0.363-0.768], p = 0.001), and the additive model showed that for each additional mutation in allele G, the risk of constipation decreased by 43.76% (p = 0.001). When we tested for sensitivity by not adding the covariates in the association analysis, variation at rs5275 remained significantly associated with constipation following multiple comparison correction (pFDR_BH = 0.032). Similar results were obtained when analyses were performed using the additive or recessive models.
Table 2.
Association between variation at rs5257 and opioid induced constipation
| Model | Genotypes | Discovery sample (n = 572) | Replication sample (n = 286) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Patients No. (%) | Constipation (%) | OR value | P1 | P2 | Patients No. (%) | Constipation (%) | OR value | P1 | ||
| DOM | GG + GA | 179 (31.294) | 91 (50.838) |
0.528 (0.363–0.768) |
0.001 | 0.093 | 97 (33.916) | 48 (49.485) |
0.570 (0.343–0.948) |
0.030 |
| AA | 382 (66.783) | 254 (66.492) | 1.000 (ref) | 187 (65.384) | 117 (62.567) | 1.000 (ref) | ||||
| ADD | GG | 19 (3.387) | 8 (42.105) |
0.562 (0.406–0.780) |
0.001 | 0.062 | 10 (3.521) | 6 (60.000) |
0.657 (0.425–1.017) |
0.060 |
| GA | 160 (28.520) | 83 (51.875) | 87 (30.634) | 42 (48.276) | ||||||
| AA | 382 (68.093) | 254 (66.492) | 1.000 (ref) | 187 (65.845) | 117 (62.567) | 1.000 (ref) | ||||
DOM dominant model, ADD additive model, OR odd ratio, CI confidence interval, P1 the uncorrected p value, P2 p value corrected by the Benjamini and Hochberg method
Haplotype Association Analysis
Twelve distinct haplotype blocks were identified in the linkage disequilibrium analysis of all 110 SNPs based on gene location (Supplementary Material Table 4). Three haplotypes were significantly associated with opioid response in the discovery stage (p < 0.05). However, none of them met Benjamini–Hochberg FDR criteria, or were verified in the replication sample. Notably, different haplotypes of OPRM1 (rs6912029, rs1799971, rs589046, and rs563649) showed significant differences in opioid dose and constipation. Patients with GATC haplotype were more likely to use lower doses of opioids (χ2 = 5.083, p = 0.024) and patients with GACC haplotype had a relatively low risk of constipation (χ2 = 3.883, p = 0.049). In contrast, patients with the GGCC haplotype had a higher risk of constipation (χ2 = 4.291, p = 0.038). However, these associations did not pass validation (Table 3). Supplementary Material Fig. 1 shows the linkage disequilibrium (LD) of the five haplotypes and locuszoom plot of association results of SNPs in different phenotypes.
Table 3.
Haplotypes with an initial significant association with the opioid response
| Phenotype | Gene | SNPs | Haplotype | Discovery sample | Replication sample | ||||
|---|---|---|---|---|---|---|---|---|---|
| Case, control frequencies | χ2 | p | Case, control frequencies | χ2 | p | ||||
| Opioid dose | OPRD1 | rs529520, rs581111 | AA | 0.049, 0.086 | 4.192 | 0.041 | 0.071, 0.085 | 0.236 | 0.627 |
| OPRM1 | rs6912029, rs1799971, rs589046, rs563649 | GATC | 0.031, 0.067 | 5.083 | 0.024 | 0.048, 0.052 | 0.036 | 0.850 | |
| ARRB2 | rs3786047, rs16954146, rs1045280, rs2271167, rs2036657 | GGTGA | 0.761, 0.822 | 5.243 | 0.022 | 0.802, 0.811 | 0.054 | 0.816 | |
| Constipation | UGT2B7 | rs4296738, rs4235108, rs4587017, rs7438135, rs7668282, rs7439366, rs10028494, rs6851533, rs12645107 | GAGATCACG | 0.054, 0.029 | 3.969 | 0.046 | 0.025, 0.016 | 0.381 | 0.537 |
| OPRM1 | rs6912029, rs1799971, rs589046, rs563649 | GACC | 0.487, 0.547 | 3.883 | 0.049 | 0.467, 0.487 | 0.234 | 0.628 | |
| GGCC | 0.362, 0.303 | 4.291 | 0.038 | 0.380, 0.374 | 0.018 | 0.892 | |||
Influence of Gene–gene and Gene–environment Interactions on Opioid Response and Side Effects
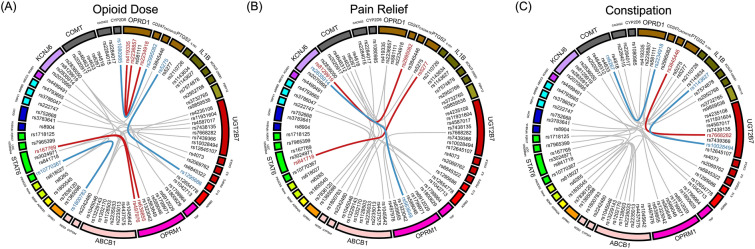
We first analyzed the association between paired gene–gene interactions and phenotypes. All 110 SNPs were found to be involved in the interaction in the discovery samples, which resulted in the presence of a large number of SNP–SNP interaction pairs. There were 280–304 SNP–SNP interaction pairs that were significantly associated with opioid dose, pain relief, or constipation. However, all of them failed to meet the Benjamini–Hochberg FDR criteria, and only seven SNP–SNP pairs associated with opioid dose, and three with pain relief or constipation passed validation in the replication stage. As shown in Fig. 2, polymorphic loci (rs419335, rs2234918, rs2236857, and rs581111) located on the OPRD1 gene were present in multiple pairs of the gene–gene interaction models, which affected phenotypes, such as opioid dose and constipation. We suggest that the action mode of these mutated loci on the OPRD1 gene on opioid effects are predominantly coactivation with other mutations.
Fig. 2.
The paired gene–gene interactions associated with opioid response. The paired gene–gene interactions were identified in opioid utilized patients of three different phenotypes associated with opioid response: A opioid dose, B pain relief, and C constipation. There were 280–304 SNP–SNP pairs that were significantly associated with opioid dose, pain relief or constipation. In order to show the interactions between different genes more clearly, only SNP–SNP pairs with p < 0.05 in the discovery stage and p < 0.1 in the replication stage were shown. The red line in the circle indicates that the OR value of this gene–gene interaction pair is over 1 in both of the discovery stage and the replication stage, while the blue line indicates that the OR value of this gene–gene interaction pair is below 1 in two stages. The gray lines indicate that these gene–gene interactions were only found to be significant in the discovery stage, or the OR values were contradictory in the replication stage. The color of these genes indicates the chromosome location of these genes, and the same color of two genes indicates that these two genes locate in the same chromosome
Subsequently, we predicted multidimensional SNP–SNP interactions using GMDR. Because of the limitation of sample size, the number of dimensions was set to 3–5. Of all the phenotypes, only one statistically significant three-dimensional model was constructed, which was for the pain relief phenotype. This model contained rs2020917, rs2952768, and rs8904 and had a test accuracy of 0.592, a CV consistency of 9/10, and a significance test of p = 0.011. However, we failed in replicating this in the replication stage (p = 0.989) (Table 4).
Table 4.
Multi-dimensional gene–gene interaction in discovery and replication stage
| Study component |
Phenotype | Model | Training Bal. Acc | Testing Bal. Acc | Sign test (p) | CV consistency | OR value (95% CI) |
|---|---|---|---|---|---|---|---|
| Discovery | Opioid dose | ||||||
| rs2110726-rs2952768-rs1045046 | 0.644 | 0.437 | 0 (1.000) | 1/10 | 3.071 (1.476, 6.389) | ||
| rs2110726-rs9524885-rs2952768-rs1045046 | 0.710 | 0.483 | 5 (0.623) | 2/10 | 6.097 (2.820, 13.186) | ||
| rs2284017-rs1202170-rs1143627-rs1042713-rs2835914 | 0.796 | 0.498 | 5 (0.623) | 2/10 | 23.737 (9.144, 61.678) | ||
| Pain relief | |||||||
| rs2020917-rs2952768-rs8904 | 0.655 | 0.592 | 9 (0.011) | 9/10 | 3.921 (2.244, 6.852) | ||
| rs2834167-rs2952768-rs1202170-rs10770367 | 0.722 | 0.550 | 7 (0.172) | 3/10 | 6.775 (3.837, 11.961) | ||
| rs2952768-rs8129919-rs8904-rs618027-rs841718 | 0.809 | 0.527 | 7 (0.172) | 2/10 | 18.744 (9.711, 36.177) | ||
| Constipation | |||||||
| rs1799971-rs222747-rs2284015 | 0.654 | 0.480 | 5 (0.623) | 3/10 | 3.723 (2.169, 6.392) | ||
| rs1799971-rs222747-rs2835914-rs2284015 | 0.717 | 0.477 | 3 (0.945) | 3/10 | 6.523 (3.713, 11.458) | ||
| rs2834167-rs2952768-rs6269-rs1143627-rs13229143 | 0.807 | 0.486 | 6 (0.377) | 2/10 | 18.297 (9.543, 35.082) | ||
| Replication | Pain relief | ||||||
| rs2020917-rs2952768-rs8904 | 0.612 | 0.465 | 2 (0.989) | 10/10 | N.A | ||
CV cross validation, OR odd ratio, CI confidence interval, N.A. not applicable
Next, we investigated whether environmental factors were significant in the univariate analysis and were involved in modulating the effect of gene–gene interactions on opioid drug efficacy or adverse effects. Because of the limitation of sample size, the number of dimensions was set to 2–5. As shown in Table 5, the two best models related to opioid dose were the opioid tolerance-opioid two-dimensional model (testing accuracy = 0.707, CV of consistency = 10/10, p = 0.001, OR = 11.248), and the opioid tolerance-rs2110726-rs9524885-rs2952768 four-dimensional model (testing accuracy = 0.684, CV of consistency = 7/10, p = 0.001, OR = 10.159). In the pain relief phenotype, only the NRS2002 score-rs2834167-rs2952768-rs6269-rs1202170 five-dimensional model had a critical statistical significance (testing accuracy = 0.539, CV of consistency = 4/10, p = 0.054, OR = 27.961). Only one three-dimensional model, diagnosis-rs5275-rs2835914, was significantly associated with the constipation phenotype (testing accuracy = 0.596, CV of consistency = 8/10, p = 0.001, OR = 4.364). All of the above models were validated in the replication stage.
Table 5.
Multi-dimensional gene–environment interaction in development sample and validation sample
| Study component | Phenotype | Model | Training Bal. Acc | Testing Bal. Acc | Sign test (p) | CV consistency | OR value (95% CI) |
|---|---|---|---|---|---|---|---|
| Discovery | Opioid dose | Opioid tolerance-Opioid | 0.711 | 0.707 | 10 (0.001) | 10/10 | 11.248 (5.506, 22.978) |
| Opioid tolerance-Opioid-rs841718 | 0.727 | 0.680 | 10 (0.001) | 5/10 | 14.732 (6.931, 31.314) | ||
| Opioid tolerance-rs2110726-rs9524885-rs2952768 | 0.756 | 0.684 | 10 (0.001) | 7/10 | 10.159 (5.658, 18.242) | ||
| Opioid tolerance-rs2110726-rs9524885-rs2952768-rs8904 | 0.806 | 0.602 | 10 (0.001) | 2/10 | 18.195 (9.666, 34.253) | ||
| Pain relief | NRS2002 score-rs8904 | 0.607 | 0.502 | 6 (0.377) | 5/10 | 2.537 (1.484, 4.337) | |
| NRS2002 score-rs8904-rs1202170 | 0.662 | 0.477 | 3 (0.945) | 3/10 | 3.929 (2.285, 6.756) | ||
| NRS2002 score-rs2952768-rs6269-rs1143627 | 0.741 | 0.514 | 5 (0.623) | 6/10 | 8.347 (4.652, 14.977) | ||
| NRS2002 score-rs2834167-rs2952768-rs6269-rs1202170 | 0.836 | 0.539 | 8 (0.054) | 4/10 | 27.961 (13.765, 56.799) | ||
| Constipation | Opioid dose -Tumor diagnosis | 0.623 | 0.538 | 7 (0.172) | 7/10 | 3.318 (1.862, 5.912) | |
| Tumor diagnosis-rs5275-rs2835914 | 0.675 | 0.596 | 10 (0.001) | 8/10 | 4.364 (2.541, 7.495) | ||
| Tumor diagnosis-rs1295686-rs6473799-rs7574878 | 0.745 | 0.502 | 5 (0.623) | 2/10 | 8.752 (4.881, 15.692) | ||
| Tumor diagnosis-rs6269-rs8904-rs1042713-rs2835914 | 0.835 | 0.501 | 4 (0.828) | 2/10 | 27.717 (13.752, 55.861) | ||
| Replication | Opioid dose | Opioid tolerance-Opioid | 0.687 | 0.662 | 9 (0.011) | 10/10 | N.A |
| Opioid tolerance-Opioid-rs841718 | 0.699 | 0.673 | 9 (0.011) | 10/10 | N.A | ||
| Opioid tolerance-rs2110726-rs9524885-rs2952768 | 0.730 | 0.635 | 10 (0.001) | 10/10 | N.A | ||
| Opioid tolerance-rs2110726-rs9524885-rs2952768-rs8904 | 0.793 | 0.626 | 9 (0.011) | 10/10 | N.A | ||
| Pain relief | NRS2002 score-rs2834167-rs2952768-rs6269-rs1202170 | 0.866 | 0.582 | 8 (0.055) | 10/10 | N.A | |
| Constipation | Tumor diagnosis-rs5275-rs2835914 | 0.641 | 0.558 | 8 (0.055) | 10/10 | N.A |
CV cross validation, OR odd ratio, CI confidence interval, N.A. not applicable
Discussion
In general, most candidate gene association studies assessing the association between genetic polymorphisms and pain perception and/or analgesic efficacy of opioids examined the association of SNPs or were limited to exploring the interaction between two specific genes, with limited data on haplotypes of multiple genes and especially interactions between different genes. No further studies have analyzed gene–environment interactions [14–20].
In this two-stage study, we analyzed the effects of individual genetic loci using different genetic models (dominant, additive, and recessive models) of opioid treatment in patients with cancer pain and comprehensively explored association between 110 SNPs in 45 genes and opioid response from the perspectives of haplotypes, gene–gene interactions, and gene–environment interactions. Similar to other researchers, we were unable to demonstrate the association between 45 candidate genes and opioid response in Chinese Han adult cancer pain in SNP or haplotype association analysis [12, 35, 36]. However, in the follow-up analysis of gene–gene and gene–environment interactions analysis, we have found novel results. The results highlight the necessity to detect gene–gene and gene–environment interactions between different genes for complex traits (e.g., pain and opioid response). The current findings, if they can be further confirmed in larger and ethnically diverse cohorts, may provide additional possibilities for the advancement of individualized treatments for cancer pain.
In the SNP association analysis, we found that variation at rs5275 in the PTGS2 gene was associated with opioid constipation. Although it did not meet Benjamini–Hochberg FDR criteria, this result was validated in the replication stage (Table 2). Rs5275 is located in the 3'UTR of the PTGS2 gene that encodes the cyclooxygenase-2 (COX-2) protein. It has previously been reported that the G allele can significantly increase COX-2 protein levels and increases the risk of constipation [37–39], which is inconsistent with the 43.76% decrease in risk of constipation for each additional mutant allele G in this study (p = 0.001). We first consider whether this inconsistency was related to the use of selective COX-2 inhibitors. However, there was no significant difference between the GG/GA and AA genotype groups in the use of selective COX-2 inhibitors (11.518 vs. 11.732%). The possible explanation for this is that the gene may not be the primary affecting factor [40], and the effect of variation at rs5275 is susceptible to interference by other factors. This is supported by other studies that showed that preoperative administration of a selective COX-2 inhibitor significantly improves postoperative ileus [41]. However, the combination of opioids with COX-2 inhibitors did not affect the incidence of constipation in patients with cancer pain [42].
Pain perception and response to analgesics is a complex process that involves multiple biochemical pathways. There may be genetic variants that have a small or insignificant effect in the single SNP association analysis. However, such effects may become important in the presence of another genetic or environmental variant [35]. Association studies focusing on individual variants may yield inconclusive results and miss clinically relevant genetic variants [43]. On the other hand, individual SNP associations are not necessarily responsible for phenotype; causal SNPs could be in linkage disequilibrium with those studied [15]. There have been several studies on SNP–SNP interactions in patients with cancer pain. However, apart from the haplotype studies, they have been limited to investigating interactions of up to two SNPs at a time [17, 18, 36]. In the present study, we analyzed haplotypes, paired SNPs, and multidimensional gene–gene interactions of all genes using PLINK and GMDR software to explore the interactions between SNPs in the same and different genes. In this part of the analysis, we found that some SNPs that were identified as ‘negative’ in the previous SNP association analyses were in fact, significantly associated with the phenotype in the form of SNP–SNP interactions.
In the haplotype association analysis, we identified six haplotypes that were associated with opioid dose or constipation (p < 0.05). However, none of these met Benjamini–Hochberg FDR criteria, or were verified in the replication sample (Table 3). It is worth noting that although no association with opioid dose was found for individual SNP in the OPRM1 gene, the OPRM1 haplotypes showed weak associations (0.01 < p < 0.05) with opioid dose and constipation phenotypes, although these were not validated in the replication group. Moreover, all patients with the rs1799971 haplotype carrying the A allele required lower opioid doses, whereas only patients with the rs1799971 haplotype carrying the G allele had a higher frequency in the high dose group (not significant). The same finding was reported in the study by Manuela et al. [16]. This result confirmed the data in a recent meta-analysis by Yu et al., which demonstrated that carriers of the G allele (AG + GG) of the OPRM1 rs1799971 (A118G) polymorphism require more opioid analgesics for cancer pain treatment [12].
Among the paired SNP–SNP interactions, 280–304 pairs of SNP–SNP interaction were significantly associated with each phenotype in the discovery stage, but all of them failed to meet the Benjamini–Hochberg FDR criteria, and only 3–7 pairs passed validation in the replication stage (Fig. 2). These SNP–SNP pairs were derived from 16 different genes and were involved in pathways and links, such as opioid receptors, drug transporters, signaling pathways, and inflammatory factors. This demonstrated the complexity of pain and opioid drug responses and supported the necessity for extensive analysis of interactions between different genes, with the most notable being OPRD1 rs2234918-COMT rs6267. The interaction between these two SNPs significantly increased the risk of constipation, with an OR of 4.953 (p = 0.017) in the discovery group and 6.441 (p = 0.034) in the replication group. This is the first study to identify such an interaction. When dimensionality of the analysis was increased to examine SNP–SNP interactions in 3–5 dimensions, statistically significant SNP interactions became obviously fewer. Of all the phenotypes, only one three-dimensional model showed an association with pain relief, and this model contained COMT rs2020917-cyclic adenosine monophosphate (cAMP) responsive element binding protein 1 (CREB1) rs2952768-NFKBIA rs8904 (OR = 3.921, p = 0.011). However, the result was not replicated in the replication group (p = 0.989) (Table 4).
For such outcomes, we believe that in addition to genes, environmental factors also play an important role. As previously mentioned, pain perception and analgesic response is a complex clinical feature that may be influenced by numerous different clinical and environmental factors [43]. The extent to which genes explain phenotypic variation may be limited, with genes explaining only 5–24.2% of the variation in morphine dose, as reported in the literature [15, 16]. Therefore, we used environmental factors as markers to explore whether the combination of environmental factors and genes improve predictive results. Results showed that statistically significant optimal gene–environment interaction models were generated successfully for each of the three phenotypes (Table 5).
It should be noted that variation at rs5257 was negatively correlated with constipation risk in the SNPs association analysis (Table 2), while the gene–environment interaction model suggested that there was an interaction among PTGS2 rs5275, diagnosis and KCNJ6 rs2835914. This interaction significantly increased the risk of constipation, which verifies our inference in SNPs association analysis. Stronger associations with phenotypes were observed from the OR values of these gene–environment models than the associations of the gene–gene or paired SNP–SNP interaction models, which suggested that environmental factors play an important role in opioid response. Opioid tolerance is defined as a predictable physiological decrease in the analgesic effect of opioids over time that requires an increase in opioid dose to achieve the same analgesic effect [44]. It is not surprising that opioid tolerance is a factor in the optimal model of the opioid dose phenotype. The model constructed above showed that the NRS2002 score was associated with pain relief. The NRS2002 score is a tool used to assess the nutritional risk of patients, and is positively correlated with the pain intensity in patients with cancer pain (r = 0.273, p = 0.001) [45]. Poor nutritional status can decrease the efficacy of fentanyl transdermal patches and increase pain scores [46], and may increase the incidence of adverse effects [47]. Cancer diagnosis is a factor in constipation, and one study examining genetic and non-genetic factors affecting opioid constipation found that tumors in female genitals and gastrointestinal tracts increase the risk of constipation [40], which was similar to our findings. In this study, the incidence of constipation in patients with genitourinary was 78.947%, which was higher than that in patients with other tumors.
In terms of genetic factors, several genes were identified as having a negative effect on the individual SNP and haplotype association, and the gene–gene interactions. CREB1 interacted with other genes for opioid dose and pain relief. The CREB1 and cAMP pathways are involved in the analgesic and rewarding effects of opioids, and mRNA expression levels of the CREB1 gene have been found to be higher in subjects with the rs2952768 CC genotype in a GWAS, with increased postoperative opioid analgesic requirements and reduced dependence on other drugs [48]. In the present study, opioid dose was also higher in patients with the rs2952768 CC genotype than those with the CT + TT type (44.410 vs. 39.072 mg), whereas pain relief was slightly lower than that of the latter genotype (76.000 vs. 77.333%).
In this study, independent discovery and replication samples were analyzed in a stepwise manner to reduce the FDR of multiple testing. However, such a necessary precaution somewhat reduced statistical power. The CaTS V0.0.2 software (http://csg.sph.umich.edu//abecasis/CaTS/index.html) [49] was used to calculate the statistical power. Due to the high proportion of case groups of opioid dose, pain relief and constipation (46.153, 34.732, and 68.687%, respectively), even under the condition of small sample size, when the MAF = 0.05, all phenotypes have more than 80% confidence in detecting an association with an OR of 1.5. In this study, only four SNPs (rs2069845, rs6853, rs1143634 and rs11931604) have MAF less than 0.05. Thus, the sample size of this study can meet the statistical requirements of most SNPs to detect the effect of genetic variation on opioid drug response.
We acknowledge that there are some limitations in this study. (1) Opioid pharmacology and pain perception involve multiple complex biological systems, and more than 400 genes have been identified in the NCBI Gene Database. The 110 candidate SNPs included in this study did not cover every relevant gene. (2) The present study was a cross-sectional study that was conducted at only one time point, which did not allow for a comprehensive assessment of changes in pain and opioid response. (3) In the analysis, the other concomitant medications received by patients were not accounted for, which may have had an impact on pain intensity and constipation. (4) This study was a single-center study, and the subjects were all from a Chinese Han population. It remains to be verified whether the findings are applicable to other ethnic groups.
Conclusions
In conclusion, this exploratory cross-sectional study demonstrated that genetic variability affects the efficacy and adverse effects of opioids in patients with cancer pain. By expanding on the analysis of individual SNPs to the construction gene–gene interactions and gene–environment interactions, this study enhances our understanding of how genetic variation impacts the response of opioids in patients with cancer pain. Understanding the complexity of opioid responses will be an ongoing challenge, and continued advances in data mining and modeling techniques will promote the integration of generated information toward achieving the ultimate goal of individualized opioid therapy.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the participants of the study.
Funding
This work was supported by the National Key Research and Development Plan of China (2017YFC0909900). The Rapid Service Fee was funded by the National Key Research and Development Plan of China (2017YFC0909900).
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Author Contributions
YZ, CS, HW, and JLH conceived and designed the experiments. XC, QY, and HXZ performed the experiments. JML, LYW, and WJG analyzed the data. XC, CW, JYX, JL, HXW, and HZ contributed reagents/materials/analysis tools. CS and JML wrote the paper. All authors reviewed the manuscript.
Disclosures
Chen Shi, Jinmei Liu, Jianli Hu, Xu Chen, Jiyi Xie, Juan Luo, Cong Wang, Hanxiang Wang, Qi Yuan, Haixia Zhu, Weijing Gong, Shijun Li, Hong Zhou, Leiyun Wang, Hui Wang, and Yu Zhang have nothing to disclose.
Compliance with Ethics Guidelines
This study was performed in accordance with the Helsinki Declaration of 1964 and its later amendments. All participants understood the purpose of the study, and signed a consent form prior to any study-related activities. The protocols were approved by the medical ethics committee of Tongji Medical College, Huazhong University of Science and Technology (approval number: 2018-S016).
Data Availability
All data generated or analyzed during this study are included in this published article/as supplementary information files.
Footnotes
Chen Shi and Jinmei Liu contributed equally to this work and share first authorship.
References
- 1.Fallon M, Giusti R, Aielli F, et al. Management of cancer pain in adult patients: ESMO Clinical Practice Guidelines. Ann Oncol. 2018;29(Suppl 4):iv166–iv191. doi: 10.1093/annonc/mdy152. [DOI] [PubMed] [Google Scholar]
- 2.Swarm RA, Paice JA, Anghelescu DL, et al. Adult cancer pain, version 3.2019, NCCN clinical practice guidelines in oncology. J Natl Compr Cancer Netw. 2019 doi: 10.6004/jnccn.2019.0038. [DOI] [PubMed] [Google Scholar]
- 3.Cherny N, Ripamonti C, Pereira J, et al. Strategies to manage the adverse effects of oral morphine: an evidence-based report. J Clin Oncol. 2001;19(9):2542–54. https://pubmed.ncbi.nlm.nih.gov/11331334 [DOI] [PubMed]
- 4.Riley J, Ross JR, Rutter D, et al. No pain relief from morphine? Individual variation in sensitivity to morphine and the need to switch to an alternative opioid in cancer patients. Support Care Cancer. 2006;14(1):56–64. https://pubmed.ncbi.nlm.nih.gov/15952009 [DOI] [PubMed]
- 5.Nielsen LM, Olesen AE, Branford R, Christrup LL, Sato H, Drewes AM. Association between human pain-related genotypes and variability in opioid analgesia: an updated review. Pain Pract. 2015;15(6):580–594. doi: 10.1111/papr.12232. [DOI] [PubMed] [Google Scholar]
- 6.Sadhasivam S, Chidambaran V. Pharmacogenomics of opioids and perioperative pain management. Pharmacogenomics. 2012;13(15):1719–1740. doi: 10.2217/pgs.12.152. [DOI] [PubMed] [Google Scholar]
- 7.Skorpen F, von Hofacker S, Bjørngaard M, et al. The rare Arg181Cys mutation in the μ opioid receptor can abolish opioid responses. Acta Anaesthesiol Scand. 2016;60(8):1084–1091. doi: 10.1111/aas.12739. [DOI] [PubMed] [Google Scholar]
- 8.Gasche Y, Daali Y, Fathi M, et al. Codeine intoxication associated with ultrarapid CYP2D6 metabolism. N Engl J Med. 2004;351(27):2827–31. https://pubmed.ncbi.nlm.nih.gov/15625333 [DOI] [PubMed]
- 9.Klepstad P, Fladvad T, Skorpen F, et al. Influence from genetic variability on opioid use for cancer pain: a European genetic association study of 2294 cancer pain patients. Pain. 2011;152(5):1139–1145. doi: 10.1016/j.pain.2011.01.040. [DOI] [PubMed] [Google Scholar]
- 10.Galvan A, Skorpen F, Klepstad P, et al. Multiple Loci modulate opioid therapy response for cancer pain. Clin Cancer Res. 2011;17(13):4581–4587. doi: 10.1158/1078-0432.CCR-10-3028. [DOI] [PubMed] [Google Scholar]
- 11.Yokoshima Y, Sumitani M, Nishizawa D, et al. Gamma-aminobutyric acid transaminase genetic polymorphism is a candidate locus for responsiveness to opioid analgesics in patients with cancer pain: an exploratory study. Neuropsychopharmacol Rep. 2018;38(4):175–181. doi: 10.1002/npr2.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu Z, Wen L, Shen X, Zhang H. Effects of the OPRM1 A118G polymorphism (rs1799971) on opioid analgesia in cancer pain: a systematic review and meta-analysis. Clin J Pain. 2019;35(1):77–86. doi: 10.1097/AJP.0000000000000636. [DOI] [PubMed] [Google Scholar]
- 13.Kim H, Clark D, Dionne RA. Genetic contributions to clinical pain and analgesia: avoiding pitfalls in genetic research. J Pain. 2009;10(7):663–693. doi: 10.1016/j.jpain.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith SB, Reenilä I, Männistö PT, et al. Epistasis between polymorphisms in COMT, ESR1, and GCH1 influences COMT enzyme activity and pain. Pain. 2014;155(11):2390–2399. doi: 10.1016/j.pain.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Wei Z, Zhang J, Hakonarson H, Cook-Sather SD. Candidate gene analyses for acute pain and morphine analgesia after pediatric day surgery: African American versus European Caucasian ancestry and dose prediction limits. Pharmacogenomics J. 2019;19(6):570–581. doi: 10.1038/s41397-019-0074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Gregori M, Diatchenko L, Ingelmo PM, et al. Human genetic variability contributes to postoperative morphine consumption. J Pain. 2016;17(5):628–636. doi: 10.1016/j.jpain.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 17.Reyes-Gibby CC, Shete S, Rakvåg T, et al. Exploring joint effects of genes and the clinical efficacy of morphine for cancer pain: OPRM1 and COMT gene. Pain. 2007;130(1–2):25–30. https://pubmed.ncbi.nlm.nih.gov/17156920 [DOI] [PMC free article] [PubMed]
- 18.Matic M, Jongen JL, Elens L, et al. Advanced cancer pain: the search for genetic factors correlated with interindividual variability in opioid requirement. Pharmacogenomics. 2017;18(12):1133–1142. doi: 10.2217/pgs-2017-0060. [DOI] [PubMed] [Google Scholar]
- 19.Tour J, Löfgren M, Mannerkorpi K, et al. Gene-to-gene interactions regulate endogenous pain modulation in fibromyalgia patients and healthy controls-antagonistic effects between opioid and serotonin-related genes. Pain. 2017;158(7):1194–1203. doi: 10.1097/j.pain.0000000000000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khalil H, Sereika SM, Dai F, et al. OPRM1 and COMT gene–gene interaction is associated with postoperative pain and opioid consumption after orthopedic trauma. Biol Res Nurs. 2017;19(2):170–179. doi: 10.1177/1099800416680474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cannon ME, Mohlke KL. Deciphering the emerging complexities of molecular mechanisms at GWAS Loci. Am J Hum Genet. 2018;103(5):637–653. doi: 10.1016/j.ajhg.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HJ, Yang GS, Greenspan JD, et al. Racial and ethnic differences in experimental pain sensitivity: systematic review and meta-analysis. Pain. 2017;158(2):194–211. doi: 10.1097/j.pain.0000000000000731. [DOI] [PubMed] [Google Scholar]
- 23.Organisation. WH. Cancer pain relief : with a guide to opioid availability 1996 [2nd ed:] Available from: http://www.who.int/cancer/palliative/painladder/en/.
- 24.FDA. Extended-release (ER) and long-acting (LA) opioid analgesics Risk Evaluation and Mitigation Strategy (REMS) 2013. Available from: www.fda.gov/downloads/drugs/drugsafety/postmarketdrugsafetyinformationforpatientsandproviders/ucm311290.pdf.
- 25.Fainsinger RL, Nekolaichuk CL, Lawlor PG, Neumann CM, Hanson J, Vigano A. A multicenter study of the revised Edmonton Staging System for classifying cancer pain in advanced cancer patients. J Pain Symptom Manage. 2005;29(3):224–37. https://pubmed.ncbi.nlm.nih.gov/15781173 [DOI] [PubMed]
- 26.Oken MM, Creech RH, Tormey DC, et al. Toxicity and response criteria of the Eastern Cooperative Oncology Group. Am J Clin Oncol. 1982;5(6):649–55. https://pubmed.ncbi.nlm.nih.gov/7165009 [PubMed]
- 27.Kondrup J, Allison SP, Elia M, Vellas B, Plauth M. ESPEN guidelines for nutrition screening 2002. Clin Nutr. 2003;22(4):415–21. https://pubmed.ncbi.nlm.nih.gov/12880610 [DOI] [PubMed]
- 28.Zung WW. A Self-rating depression scale. Arch Gen Psychiatry. 1965;12:63–70. https://pubmed.ncbi.nlm.nih.gov/14221692 [DOI] [PubMed]
- 29.Daut RL, Cleeland CS, Flanery RC. Development of the Wisconsin Brief Pain Questionnaire to assess pain in cancer and other diseases. Pain. 1983;17(2):197–210. doi: 10.1016/0304-3959(83)90143-4. [DOI] [PubMed] [Google Scholar]
- 30.Department of drug safety supervision SFada, national adverse drug reaction monitoring center. Adverse drug reaction report and monitoring manual. 2nd ed. 2012. https://www.docin.com/p-2296263456.html.
- 31.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. https://pubmed.ncbi.nlm.nih.gov/15297300 [DOI] [PubMed]
- 33.Lou X-Y, Chen G-B, Yan L, et al. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am J Hum Genet. 2007;80(6):1125–37. https://pubmed.ncbi.nlm.nih.gov/17503330 [DOI] [PMC free article] [PubMed]
- 34.Yoav Benjamini YH. On the adaptive control of the false discovery rate in multiple testing with independent statistics. J Educ Behav Stat. 2000;25:60–83. doi: 10.2307/1165312. [DOI] [Google Scholar]
- 35.Thornton-Wells TA, Moore JH, Haines JL. Genetics, statistics and human disease: analytical retooling for complexity. Trends Genet. 2004;20(12):640–7. https://pubmed.ncbi.nlm.nih.gov/15522460 [DOI] [PubMed]
- 36.Scarpi E, Calistri D, Klepstad P, et al. Clinical and genetic factors related to cancer-induced bone pain and bone pain relief. Oncologist. 2014;19(12):1276–1283. doi: 10.1634/theoncologist.2014-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore AE, Young LE, Dixon DA. A common single-nucleotide polymorphism in cyclooxygenase-2 disrupts microRNA-mediated regulation. Oncogene. 2012;31(12):1592–1598. doi: 10.1038/onc.2011.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwarz NT, Kalff JC, Türler A, et al. Prostanoid production via COX-2 as a causative mechanism of rodent postoperative ileus. Gastroenterology. 2001;121(6):1354–71. https://pubmed.ncbi.nlm.nih.gov/11729115 [DOI] [PubMed]
- 39.Shi X-Z, Lin Y-M, Powell DW, Sarna SK. Pathophysiology of motility dysfunction in bowel obstruction: role of stretch-induced COX-2. Am J Physiol Gastrointest Liver Physiol. 2011 doi: 10.1152/ajpgi.00379.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laugsand EA, Skorpen F, Kaasa S, et al. Genetic and non-genetic factors associated with constipation in cancer patients receiving opioids. Clin Transl Gastroenterol. 2015;6:e90. doi: 10.1038/ctg.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Josephs MD, Cheng G, Ksontini R, Moldawer LL, Hocking MP. Products of cyclooxygenase-2 catalysis regulate postoperative bowel motility. J Surg Res. 1999;86(1):50–4. https://pubmed.ncbi.nlm.nih.gov/10452868 [DOI] [PubMed]
- 42.Dalpiaz AS, Peterson D. Parecoxib: a shift in pain management? Expert Rev Neurother. 2004;4(2):165–77. https://pubmed.ncbi.nlm.nih.gov/15853557 [DOI] [PubMed]
- 43.Droney J, Riley J, Ross JR. Evolving knowledge of opioid genetics in cancer pain. Clin Oncol (R Coll Radiol) 2011;23(6):418–428. doi: 10.1016/j.clon.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 44.Hayhurst CJ, Durieux ME. Differential opioid tolerance and opioid-induced hyperalgesia: a clinical reality. Anesthesiology. 2016;124(2):483–488. doi: 10.1097/ALN.0000000000000963. [DOI] [PubMed] [Google Scholar]
- 45.Chen Y, Xiang Q, Li C, et al. Nutritional risk and assessment for patients with cancer pain. Nutr Cancer. 2021 doi: 10.1080/01635581.2021.1882510. [DOI] [PubMed] [Google Scholar]
- 46.Takahashi H, Chiba T, Tairabune T, et al. A retrospective study on the influence of nutritional status on pain management in cancer patients using the transdermal fentanyl patch. Biol Pharm Bull. 2014;37(5):853–7. https://pubmed.ncbi.nlm.nih.gov/24790008 [DOI] [PubMed]
- 47.Zopf Y, Rabe C, Neubert A, Hahn EG, Dormann H. Risk factors associated with adverse drug reactions following hospital admission: a prospective analysis of 907 patients in two German university hospitals. Drug Saf. 2008;31(9):789–98. https://pubmed.ncbi.nlm.nih.gov/18707193 [DOI] [PubMed]
- 48.Nishizawa D, Fukuda K, Kasai S, et al. Genome-wide association study identifies a potent locus associated with human opioid sensitivity. Mol Psychiatry. 2014;19(1):55–62. doi: 10.1038/mp.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38(2):209–13. https://pubmed.ncbi.nlm.nih.gov/16415888 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article/as supplementary information files.