Abstract
Protein–protein interaction (PPI) plays a crucial role in most biological processes, including signal transduction and cell apoptosis. Importantly, the knowledge of PPIs can be useful for identification of multimeric protein complexes and elucidation of uncharacterized protein functions. Arabidopsis thaliana , the best‐characterized dicotyledonous plant, the steadily increasing amount of information on the levels of its proteome and signaling pathways is progressively enabling more researchers to construct models for cellular processes for the plant, which in turn encourages more experimental data to be generated. In this study, we performed an overview analysis of the 10 major organelles and their associated proteins of the dicotyledonous model plant Arabidopsis thaliana via PPI network, and found that PPI may play an important role in organelle communication. Further, multilocation proteins, especially phosphorylation‐related multilocation proteins, can function as a “needle and thread” via PPIs and play an important role in organelle communication. Similar results were obtained in a monocotyledonous model crop, rice. Furthermore, we provide a research strategy for multilocation proteins by LOPIT technique, proteomics, and bioinformatics analysis and also describe their potential role in the field of plant science. The results provide a new view that the phosphorylation‐related multilocation proteins play an important role in organelle communication and provide new insight into PPIs and novel directions for proteomic research. The research of phosphorylation‐related multilocation proteins may promote the development of organelle communication and provide an important theoretical basis for plant responses to external stress.
Keywords: multilocation protein, organelle separation, organelles communication, phosphorylation, protein interaction
1. INTRODUCTION
Proteins perform the vast majority of functions in all biological domains (Subba et al., 2019). Arabidopsis thaliana, the best‐characterized dicotyledonous plant, encodes approximately 35,000 protein‐coding genes; however, the functions of the majority of these proteins remain uncharacterized, even by homology (Rhee & Mutwil, 2014). This trend is similar for Oryza sativa, a critical food crop and the best‐characterized monocotyledonous plant (Kirkwood et al., 2013). Obviously, we still know very little about the functions of proteins in plants. Hence, it is very important to strengthen the analyses and research of plants at the proteome level.
Protein interaction networks provide new opportunities for characterizing genes and proteins (McWhite et al., 2020). Determining protein–protein interactions (PPI) is a key step in discovering both gene and protein functions, which facilitates study and manipulation of critical cellular processes (Eisenberg et al., 2000; Hartwell et al., 1999; Schwikowski et al., 2000). Systematic mapping of protein complexes in model organisms such as yeast and Drosophila has led to critical functional insights, and facilitated understanding of conserved and disease‐related pathways (Vidal et al., 2011) and helped characterize proteins by association. With scientific and technological advancements, the study of PPIs in the model plant Arabidopsis thaliana has also developed rapidly. Therefore, summarizing and analyzing the PPI network in this model plant can play a guiding role for proteomics research and functional analysis of plants and crop plants.
Biological communication is ubiquitous. It exists between organic life and inorganic matter, between animals and plants, and among animals, plants, organs, cells, and organelles (Cohen et al., 2018; Frick et al., 2019; McWhite et al., 2020; Namgaladze et al., 2019; Zhang & Yang, 2018). Communication among organelles functions to regulate the size, shape, and composition of individual organelles, and coordinate transport between them (Stefan et al., 2013). The establishment of interorganellar communication, a profound consequence of the emergence of eukaryotes, enabled coordinated scaling of the biological processes necessary to meet the metabolic and energy demands of the cell (Wang & Dehesh, 2018). Recently, many studies show that the organelles of chloroplast (Chl), mitochondria (Mit), and peroxisomes (Per) actively communicate and physical interaction between peroxisomes and chloroplasts during photorespiration (Oikawa et al., 2015). Membrane contact sites (MCSs) facilitate the exchange of metabolites between organelles to support interorganellar communication (Lin et al., 2021). MCSs facilitate the exchange of metabolites between organelles to support interorganellar communication (Lin et al., 2021). Endoplasmic reticulum (ER)‐plasma membrane (PM) junctions have well‐established functions involved in the movement of small molecules, regulation of cell signaling, ER shape and architecture, and PM domain organization (Stefan et al., 2013). The nucleus–vacuole junctions establish physical contact between the perinuclear ER and the vacuole (Tosal‐Castano et al., 2021) and the contacts between the ER and lysosomes influence organelle organization and communication (Ozkan et al., 2021). The ER‐Mit encounter structure plays crucial roles in interorganelle communication, mitochondrial fission, mtDNA inheritance, lipid transfer, and autophagy (Rasul et al., 2021). A number of key molecular machinery systems participate in mediating substance exchange and signal transduction through PPI, both of which are essential processes in terms of cellular physiology and molecular biology (Lin et al., 2021), such as GRP75 interact with VDAC of mitochondrial and IP3R of ER (Kwak et al., 2020), VAPA of ER interact with KV2 of PM (Johnson et al., 2018). So, it was meaningful to study the communication between organelles based on protein interaction data.
The existence of communication between organelles raises an important question: How and by what means do organelles communicate with each other direct contact via organelle membrane contact, proteins shuttling among different organelles, or multilocation proteins interacting with other proteins to build bridges? Our previous studies showed that different pathways are involved in different organelles. In addition, we analyzed nuclear localization and its interaction proteins in Arabidopsis and found that multilocation proteins may play a key role among different pathways (Gong et al., 2021). It raises additional questions: Do multilocation proteins play a critical role in organelle communication? Recent studies have shown that multilocalized proteins play an important role in plant growth and development, such as WHY1 and BZR1, which plays a certain role in the communication of organelles (Lin et al., 2020; Wang et al., 2021; Zhang et al., 2020). Further, protein kinases in plants are divided mainly into serine/threonine, tyrosine, and histone kinases. The kinases are activated by phosphorylation, which in turn activates a cascade of events leading to the phosphorylation of different amino acids (Ardito et al., 2017). Phosphatases, which remove the phosphate group from phosphoproteins by hydrolyzing phosphoric acid monoesters into a phosphate group and a molecule with a free hydroxyl group, have the opposite function of kinases. In Arabidopsis, many kinases (such as MAPKs, CDPKs, and RPKs), phosphatases (such as PP2Cs), and PYR/PYLs (PYR1 and PYL1‐PYL13) have been identified and play key roles in the signal transduction process, including hormone signaling and biotic or abiotic response (Mayer & Yu, 2018; Raghavendra et al., 2010; Tena et al., 2011). Previously study also showed that dual location of proteins mediates diverse intercellular signaling processes, for example, MAP kinase (Chan et al., 2016) and CIPK14 (Ren et al., 2017). So, do multilocation proteins or multilocation kinase bridge organelle communication to regulating plant growth and development?
In the case of the model plant Arabidopsis, the steadily increasing amount of information on the levels of its proteome and signaling pathways is progressively enabling more researchers to construct models for cellular processes for the plant, which in turn encourages more experimental data to be generated (Holzheu & Kummer, 2020). In this study, we conducted an overview analysis of reviewed proteins in the 10 major organelles of the model plant Arabidopsis and then screened and analyzed the proteins interacting with these organelle proteins. Based on the results, we performed related bioinformatics analyses, focusing on the interactions and pathways associated with these proteins. The results indicated that multilocation proteins, especially phosphorylation‐related multilocation proteins, can function as “a needle and thread” among organelles via PPIs. Similar results have been obtained in the monocotyledonous model rice. Moreover, we provide a research strategy for multilocation proteins. Our findings will guide new directions for proteomic research based on PPIs.
2. MATERIALS AND METHODS
2.1. Data acquisition
We searched for Arabidopsis and rice proteins on UniProt on May 30, 2020, and downloaded data on the reviewed proteins. We retrieved information on their interactions from the “Subunit structure” subsection of the “Interaction” section in UniProt. This subsection contains information on protein quaternary structure and interaction with other proteins and protein complexes (PPIs, host‐pathogen PPIs, and protein‐complex interactions, with the exception of physiological receptor‐ligand interactions, which are annotated in the “Function” section). Interacting proteins from other species, such as those participating in host‐pathogen PPIs, were excluded from the analysis. Only interactions associated with “Publications” were retained for further analysis.
2.2. Protein–protein interactome network visualization
Reviewed Arabidopsis nuclear proteins that were classified as participating in protein interactions were used for protein interactome network construction with the network visualization program Cytoscape 3.8.1 (Shannon et al., 2003). Duplicate interactions, which would cause self‐looping, were removed before network construction. The network topology was analyzed using the Network Analyzer plug‐in (Assenov et al., 2008).
2.3. Biological pathway analysis of proteins in the protein–protein interactome
Proteins are the basic components of biological pathways, and PPIs play fundamental roles in these pathways. The KEGG is a database resource for the elucidation of high‐level functions and utilities of biological systems, such as the cell, organism, and ecosystem, from molecular‐level information, especially large‐scale molecular datasets generated via genome sequencing and other high‐throughput experimental technologies (Kanehisa, 2002).
3. RESULTS
3.1. Overview of reviewed and their interacted proteins in Arabidopsis thaliana
Although the functions of the majority proteins remain uncharacterized in Arabidopsis thaliana, this species remains the most intensively studied plant. To data, 11,200 proteins of Arabidopsis thaliana located mainly in 10 organelles have been reviewed in the UniProt database (May 30, 2020) (Table S1). These 10 organelles comprise the nucleus (Nuc, 3580), PM (2104), cytoplasm (Cyt, 1181), chloroplast (Chl, 1534), ER (538), Golgi apparatus (Golgi, 540), mitochondria (Mit, 329), cell wall (CW, 288), vacuole (Vac, 262), and peroxisome (Per, 144) (Table 1). As proteins located in different organelles have different molecular functions (Huber et al., 2003), we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of proteins in the 10 major organelles of Arabidopsis (Table S2). The number of KEGG pathways associated with the proteins of the different organelles varied greatly, for example, 103, 101, and 65 pathways in Cyt, Chl, and Nuc, however, 33, 47, and 14 pathways in vacuole, peroxisome and CW, respectively (Table 1).
TABLE 1.
Details of localization and interactions of reviewed proteins in Arabidopsis
| Nuc | Cyt | Mem | Chl | ER | Golgi | Mit | Per | Vac | CW | Multi | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| The reviewed proteins | ||||||||||||
| Numbers of reviewed proteins | 3580 | 1881 | 2104 | 1534 | 538 | 540 | 329 | 144 | 262 | 288 | ‐ | 11,200 |
| Numbers of involved KEGG pathways | 65 | 103 | 63 | 101 | 58 | 33 | 64 | 47 | 33 | 14 | ‐ | 130 |
| Top five involved KEGG pathways | PHST | MP | MP | MP | MP | MP | MP | MP | MP | MP | ‐ | MP |
| MP | BSM | PHST | BSM | BSM | EN | BSM | BSM | BSM | PGI | ‐ | BSM | |
| SP | CM | PLPI | BAA | PPER | ASNSM | CM | GM | PB | SSM | ‐ | PHST | |
| UMP | PLPI | EN | CM | PLPI | PGI | BAA | CM | PLPI | BSM | ‐ | CM | |
| PLPI | PHST | BSM | BC | GM | PPER | OP | PE | PH | GAM | ‐ | BAA | |
| Protein interaction of reviewed proteins | ||||||||||||
| Numbers of reviewed proteins | 1432 | 656 | 414 | 261 | 145 | 111 | 61 | 40 | 42 | 15 | 630 | 2416 |
| Numbers of interactions | 3170 | 1721 | 921 | 427 | 373 | 210 | 128 | 68 | 121 | 25 | 2211 | 4548 |
| PPI network information of single location and multilocation proteins | ||||||||||||
| Numbers of proteins with single location | 1023 | 189 | 217 | 204 | 52 | 45 | 25 | 16 | 14 | 1 | 630 | 2416 |
| Number of interactions in its own location(s) | 1234 | 46 | 131 | 149 | 23 | 35 | 11 | 2 | 2 | 0 | 451 | 3407 |
Abbreviations: ASNSM, amino sugar and nucleotide sugar metabolism; BAA, biosynthesis of amino acids; BC, biosynthesis of cofactors; BSM, biosynthesis of secondary metabolites; CM, carbon metabolism; EN, endocytosis; GAM, galactose metabolism; GM, glutathione metabolism; MP, metabolic pathways; OP, oxidative phosphorylation; PB, phenylpropanoid biosynthesis; PE, peroxisome; PGI, pentose and glucuronate interconversions; PH, phagosome; PHST, Plant hormone signal transduction; PLPI, plant‐pathogen interaction; PPER, protein processing in endoplasmic reticulum; SP, spliceosome; SSM, starch and sucrose metabolism; UMP, ubiquitin mediated proteolysis
In addition, 11,200 proteins were associated with 130 KEGG pathways, the top five being the “Metabolic pathways,” “Biosynthesis of secondary metabolites,” “Plant hormone signal transduction,” “Carbon metabolism,” and “Biosynthesis of amino acids” pathways. KEGG analysis showed that 3580 reviewed proteins of the nucleus, the largest and most important eukaryotic cell structure (Janota et al., 2020), were involved in 65 pathways, mainly the “Plant hormone signal transduction,” “Metabolic,” “Spliceosome,” “Ubiquitin mediated proteolysis,” and “Plant‐pathogen interaction” pathways. The reviewed cytoplasmic proteins involved in the “Metabolic,” “Biosynthesis of secondary metabolites,” “Carbon metabolism,” “Plant‐pathogen interaction” and “Plant hormone signal transduction” KEGG pathways. However, cell wall proteins were involved mainly involved in the “Metabolic,” “Pentose and glucuronate interconversions,” and “Starch and sucrose metabolism” KEGG pathways (Table 1). These results suggest that each organelle has unique specific functions. However, different organelle involved in some of the same pathways, and closely connected with each other.
PPI networks provide valuable information for understanding plant biological processes (Xiong, Dong, et al., 2021; Xiong, Li, et al., 2021). A PPI network provides crucial information about how biological pathways are structured and coordinated based on the functions of individual proteins. At present, many predictional analyses of PPIs have been conducted in some model plants (Ding & Kihara, 2019). To obtain the most realistic and current state of PPI research progress, we screened the interacting proteins among the 11,200 reviewed proteins of Arabidopsis. In the UniProt database, Only interactions associated with “Publications” were retained for further analysis. The results of the statistical analysis showed that 4548 pairs interacting proteins were screened (Table S3), which are involved in 3004 proteins, including 2416 reviewed proteins and 589 non‐reviewed interacting proteins. The number of proteins from different organelles and the logarithm of their participation varied greatly (Table S4). There were 1432 Nuc and 656 Cyt proteins involved in 3170 and 1721 interaction pairs, respectively, whereas only 42 Vac and 15 CW proteins were involved in 121 and 25 interaction pairs, respectively. The different organelles were ranked in terms of breadth and depth of research. There is such a sequence of different organelles at the protein interaction level as follows: Nuc > Cyt > PM > Chl > ER > Golgi > Mit > Per > Vac > CW (Table 1). Here, the research conducted on the nucleus, the central organelle of the cell, is nearly equal to the sum of the research conducted on all the other organelles, which is still the central guiding role among organelles in the cell.
3.2. Multilocation proteins play important roles in Arabidopsis PPI networks
To further understand how these proteins relate to each other, and what are the regulatory networks among them, 4548 pairs of interacting proteins were used for protein interactome network construction using the network visualization program Cytoscape 3.8.1 (Shannon et al., 2003) (Table S5). On the whole, the current research on the Arabidopsis PPIs is relatively close, to being complete, with approximately 75% of proteins (2247) participating in a large network and 25% (754) interacting in a discrete distribution (Figure 1a). However, the functions of the majority of Arabidopsis proteins remain uncharacterized, even by homology (Rhee & Mutwil, 2014). We speculate that the interactions will reveal a relatively complete protein network representing the coordinated participation of proteins in biological processes. Therefore, we believe that great potential remains for PPIs studies in Arabidopsis, which can complement and complete this network, facilitate understanding of more detailed relationships among proteins, and provide a theoretical basis for the study of crops. Further analysis revealed 630 multilocation proteins among 2416 reviewed proteins, of which 509, 113, 6, and 2 localized in two, three, four, and five locations, respectively (Table 1, Figure 1b). Based on the thickness of the lines in Figure 1c, we found that the ten organelles were closely connected via multilocation or interacting proteins (no location information). By comparison, the organelles were less closely related to each other directly (Figure 1c). Hence, whether these multilocation proteins or interacting proteins play important roles in communication among organelles is an intriguing question. Because the location information of these interacting proteins is not completely clear, there may be important interactions between multilocation or single‐location proteins and single‐location organelles. We focused mainly on the multilocation proteins in the PPI analysis and their relationships among the various organelles. For better observation and analysis, single‐ and multilocation proteins were selected to build a sub‐network (Figure 1d). From the network, it seems that almost all organelles interact with multilocation proteins. In addition, Nuc, Cyt and ER are also closely related (Figure 1d). To confirm further the role of the multilocation proteins in the various organelles, we constructed a sub‐interaction network of multilocation proteins interacting with signal‐locating proteins in different organelles (Figure 1e). The network clearly shows that the multilocation proteins are in the centre of a vortex that acts as a “bridge” among different organelles. Therefore, we believe that multilocation proteins may function mainly as bridges in the PPI network.
FIGURE 1.
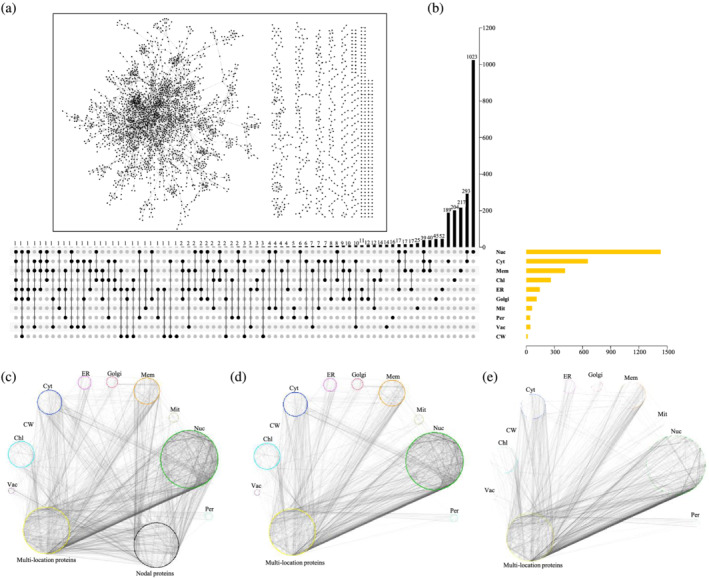
Visualization of the interactome of Arabidopsis. (a) The entire PPI network of the interactions among the reviewed proteins interaction. (b) Overview of single location and multilocation reviewed proteins in Arabidopsis. (c) Subnetwork organized by multilocation and nodal proteins. (d) Subnetwork organized by single and multilocation proteins. (e) Subnetwork of interactions between multilocation and signal‐location proteins in different organelles
To prove that multilocation proteins play an important role in organelle communication, we further analyzed the PPI network between single (1786) and multilocation proteins (630), which involved in 3407 pairs interaction. Among these, 1786 single localized proteins were involved in 1782 interactions, and total 1633 pairs (about 91.7%) were interacting proteins within an organelle. Therefore, there were only about 8.3% (149/1782) interaction pairs among different organelles. In addition, 630 multilocation proteins were involved in 451 interaction pairs. Thus, 1174 protein interaction pairs were involved in communication between the multilocation proteins and the various organelles, eight times (1174/149) as many single‐location proteins (Table 1, Figure 1d). This confirms that multilocation proteins may play a crucial role in organelle communication, which is consistent with our above results.
Thus, we evaluated how multilocation proteins function in organelles and which pathways are associated with these proteins? KEGG pathway analysis showed that “Plant hormone signal transduction” and “MAPK signaling pathway” were the top two KEGG pathways associated with the multilocation proteins. MAPK modules play key roles in the transduction of environmental and developmental signals via phosphorylation of downstream signaling targets (Jagodzik et al., 2018), and hormones such as auxin, abscisic acid, jasmonic acid, salicylic acid (SA), ethylene, brassinosteroids, and gibberellins influence signaling via MAPK cascades (Hettenhausen et al., 2015; Lu et al., 2015; Mishra et al., 2006; Rodriguez et al., 2010; Smekalova et al., 2014). To assess whether “Plant hormone signal transduction” and “MAPK signaling pathway” are important for organelle communication, we additionally performed a KEGG pathway analysis of 3004 currently reviewed proteins and their interactions, and also determined which are involved mainly in “Plant hormone signal transduction” and “MAPK signaling pathway.” The results showed that the major pathway of associated with the multilocation proteins was the same as that associated with the total proteins by KEGG analysis, indicating that multilocation proteins play an important role in biological activities.
3.3. Multilocation kinases or phosphatases in organelle communication
This raises several important questions. Which organelles are bridged by multilocation proteins, and how are they bridged? Which proteins participate in the bridging? How do they communicate with organelles and regulate biological activities? Unlike most animals, plants are immobile and cannot actively escape the effects of harsh environmental factors. Plants tend to respond to stress in the most rapid way to minimize the damage that could occur under the stress (Ashapkin et al., 2020). To address the above questions, we screened for the shortest path interaction circuits bridging multiple organelles from the network diagram of 75% of proteins (2247) participating in a large network (Figure 2a). There is only one interaction circuit connecting the 10 organelles in this network (Figure 2b), and the KEGG pathway analysis showed that the multilocation proteins connecting these 10 organelles are involved mainly in the MAPK signaling pathway, such as kinases MKK4, MPK6, MPK3, MPK4, and NDPK1. The second largest circuit involved seven organelles (Figure 2c), and KEGG analysis showed that the multilocation proteins of these organelles are mainly histidine‐related kinase such as AHP2, AHP3, AHP5, ARR9, HK2, HK3, and WOL, which are involved in “Plant hormone signal transduction.” The third interaction circuit involved six organelles in the network (Figure 2d), and KEGG analysis showed that the multilocation proteins of these organelles are mainly kinase‐ and phosphatase‐related proteins such as CPK6, CPK33, ABI1, CIPK24, KIN10, and RCAR3, which are involved in “Plant‐pathogen interaction” and “Plant hormone signal transduction.” The analysis revealed that the multilocation proteins connecting the various organelles are mainly kinases or phosphatases, suggesting that phosphorylation and dephosphorylation are key processes in organelle communication.
FIGURE 2.
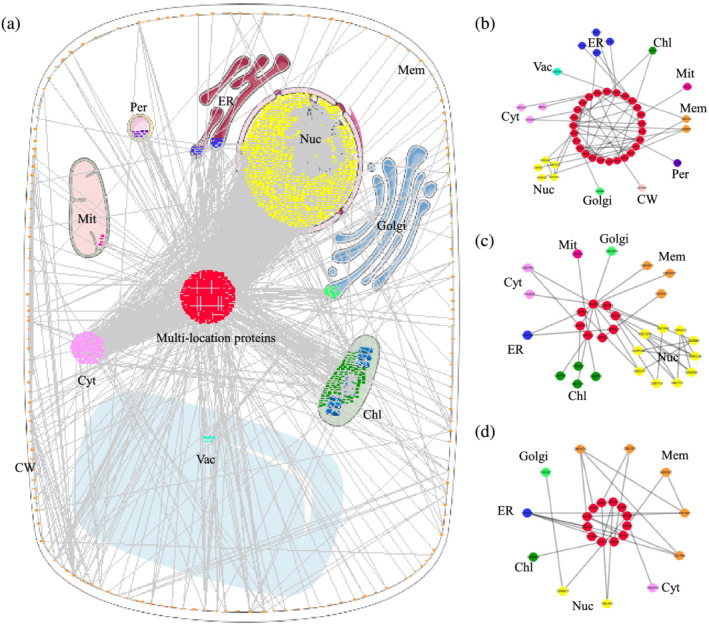
Shortest path interaction circuits bridging multiple organelles from the network diagram of 75% of proteins participating in a large network. (a) Main network. (b) Circuit of connecting the 10 organelles. (c) Circuit of connecting the seven organelles. (d) Circuit of connecting the six organelles
These directly interacting proteins elegantly link various organelles and form a path for organelle communication. On this basis, the proteins in each organelle interact with each other, and finally interact to build a PPI network that participates in the regulation of biological activities. We hypothesize that additional multilocation proteins involved in organelle communication have yet to be discovered, beyond those of the current single pathway connecting the 10 organelles. Therefore, screening of multilocation proteins has great value for understanding the growth and development of plants.
The post‐translational modification phosphorylation can alter the properties of proteins such as enzyme activity and intracellular localization. Thus, it is important to consider the localization of kinases or phosphatase, and whether they are multilocation proteins. In 630 multilocation proteins, 90 proteins (about 14.3%) were kinases or phosphatase. Moreover, these proteins involved in 529 interaction pairs (about 32.7%) of multilocation proteins. Especially pentuple location protein SNF1‐related protein kinase catalytic subunit alpha KIN10 (Q38997), involved in 51 interaction pairs. The results suggested that multilocation kinases or phosphatases may have significant roles in PPI network. Further, a total of 705 kinases‐ and 143 phosphatase‐related proteins were screened using the subcellular localization details of 11,200 Arabidopsis reviewed proteins, which were concentrated mainly in the Nuc, Cyt, PM and Chl, and among these, 129 were multilocation proteins. In addition, some widely studied kinase families, such as MAPKs and CDPKs, were selected from 11,200 reviewed proteins for further analysis. A total of 19 MAPKs, 25 CDPKs, and 212 serine/threonine‐protein related‐kinases with locational information were screened. Among the19 MAPKs, 14 (73.6%) are multilocation proteins; among the 25 CDPKs, nine (36%) are multilocation protein and 16 are membrane‐located proteins. In total, about 53% MAPKs and CDPKs were multilocation protein. The results of the analysis suggest that phosphorylation‐related proteins are not all multilocation proteins. However, the phosphorylation‐related proteins with multiple locations plays an important role in organelle communication. Compared to the results of multilocation kinases or phosphatases in PPI network, there is still a large research gap for multilocation kinases or phosphatases in 11,200 Arabidopsis reviewed proteins, and with further research, it may be found that more phosphorylation‐related proteins are multilocalized proteins. Hence, we hypothesized that although phosphorylation‐related proteins play an important role in signal transduction, not all phosphorylation‐related proteins are equally important in the plant. Some key phosphorylation‐related proteins, such as multilocation kinase proteins, may play an important role in organelle communication. Therefore, screening and obtaining more kinases or phosphatases with multilocation information may be critical for understanding cell‐to‐cell and organelle‐to‐organelle communication.
3.4. PPI network and multilocation proteins in the monocotyledonous model crop O. sativa L.
Rice is a staple food for most of the world's population. However, the PPI networks of rice have not been identified by large‐scale experiments. Here, we also summarize the recent achievements toward PPIs in the monocotyledonous model crop rice. Using the UniPort database, we reviewed 2746 proteins of rice, of which 291 were involved in 367 interaction pairs (Table 2). Similar to Arabidopsis, proteins here in Nuc and Cyt have more logarithmic interaction, with 183 proteins in Nuc involved in 249 interaction pairs and 74 proteins in Cyt involved in 147 interaction pairs. However, proteomic studies in rice are only 10%–20% of that in Arabidopsis. In addition, the PPI network with high dispersion was not a large network (Figure 3a, Table S6). Among these proteins, 63 are multilocation proteins (59 located in two and 4 in three locations), which is a lower proportion than that in Arabidopsis (Figure 3b). Although the rice genome is much larger than that of Arabidopsis, the scope and depth of research on rice are much lower, as can be seen by comparing Table 1 and Table 2. Rice protein level research is lagging, and a large gap still exists between the 40,000 annotated rice proteins and fewer than 1000 proteins that have been characterized, even with limited in vivo analyses (Rahiminejad et al., 2019). It is likely that potential interactions can be screened via comparative analyses with Arabidopsis, and made available for future reference.
TABLE 2.
Details of localization and interactions of reviewed proteins in rice
| Nuc | Cyt | Mem | Chl | ER | Golgi | Mit | Per | Vac | CW | Multi | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| The reviewed proteins | ||||||||||||
| Numbers of reviewed proteins | 951 | 463 | 398 | 459 | 133 | 114 | 37 | 30 | 69 | 62 | ‐ | 2746 |
| Protein interaction of reviewed proteins | ||||||||||||
| Numbers of reviewed proteins | 183 | 74 | 53 | 126 | 7 | 4 | 1 | 4 | 5 | 1 | 63 | 291 |
| Numbers of interactions | 249 | 147 | 68 | 26 | 10 | 5 | 1 | 7 | 6 | 1 | 142 | 367 |
| PPI network information of single location and multilocation proteins | ||||||||||||
| Numbers of proteins with single location | 131 | 18 | 41 | 26 | 3 | 2 | 1 | 2 | 3 | 1 | 63 | 228 |
| Number of interactions in its own location(s) | 59 | 2 | 25 | 12 | 0 | 1 | 0 | 0 | 0 | 0 | 27 | 99 |
FIGURE 3.
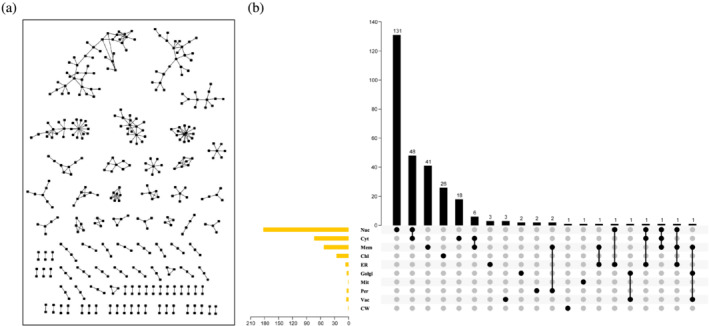
Visualization of the interactome of rice. (a) The entire PPI network of the interactions among the reviewed proteins interaction. (b) Overview of single location and multilocation reviewed proteins in rice
Recently, a genome‐wide protein labeling program for rice (RPTP) was proposed (Lu, Ronald, et al., 2020). In situ labeling of rice proteins will contribute to functional analysis of the rice proteome (Lu, Tian, et al., 2020). In this program, many proteins of interest need to be selected for an initial labeling effort, and we believe that the criterion of multilocation is important selection indicator. Here, we also conducted KEGG analysis of the multilocation proteins of rice, and “Plant hormone signal transduction” and “MAPK signaling pathway” as the top two KEGG pathways. Like Arabidopsis, the analysis showed that the multilocation proteins connecting various organelles are mainly phosphorylation related proteins, such as PP2C06, PYL8, SAPK10, SAPK8, GSK2, PYL5, PP2C53, PYL2, SAPK2, PYR1, and SAPK9. These results also suggest that multilocation proteins and phosphorylation play important roles in organelle communication in rice. Compared with Arabidopsis, there are few protein‐level interaction networks; thus, multilocation protein analysis may be a relatively rapid method to elucidate rice PPIs. We believe that this research direction will also be fruitful in the study of other plants and even animals.
4. DISCUSSION
4.1. Multilocation proteins function in PPI network and organelle communication
PPIs play a crucial role in cellular functions and biological processes in all organisms. Crosstalk among organelles can involve direct physical contact (Lin et al., 2021), especially through PPI (Johnson et al., 2018; Kwak et al., 2020). Direct physical contacts between mitochondria and the endolysosomal compartment have been reported as a rapid means of interorganelle communication, mediating lipid or other metabolite exchange (Soto‐Heredero et al., 2017). In addition, MCSs are also defined as regions where two organelles are closely apposed, and most MCSs associated with each other via protein–protein (Lin et al., 2021). For example, ER is the largest reticulum network within the cell and has extensive communication with other cellular organelles, including the PM, mitochondria, Golgi, endosomes and lipid droplets (Lin et al., 2021). GRP75 located in Mit interacts with VDAC located in mitochondrial and IP3R located in ER (Kwak et al., 2020), and VAPA located in ER interacts with KV2 located in PM (Johnson et al., 2018). In addition, proteins travel through different organelles and interact with proteins in organelles under different conditions to regulate growth and development (Isemer et al., 2012; Lin et al., 2020; Wang et al., 2021). So, PPI can be acting as a means of analyzing the degree to which organelles are linked. Here, we performed a detailed analysis using PPI data of reviewed Arabidopsis proteins, and found that 1786 single localized proteins were involved in 1782 interactions. Compared to single localized proteins, multilocation proteins are prominent in PPI networks. Among them, 630 multilocation proteins were involved in 1621 interaction. The results suggested that these multilocation proteins may play a greater role in PPI networks. So far, although many PPI networks have been analyzed, the role of multilocation proteins in PPI networks remains to be further studied. The acquisition of multilocation proteins may be more beneficial to the construction of protein interaction network.
PPIs have recently come to the fore of our understanding of organelle communications (Falz & Muller‐Schussele, 2019; Lin et al., 2021). Since multilocation proteins play an important role in PPI networks, are they equally important in organelle communication? Further analysis showed that there were only 149 interaction pairs (about 8.3%) among different organelles of signal location protein, and most interactions (about 91.7%) occur within the own organelles. If the degree of PPI connection between organelles was assumed to be an indicator of organelle information exchange. Thus, single localized proteins may play a limited role in organelle communication. Of course, it could not rule out that these few interactions may also play a decisive role in organelle communication and growth and development. Compared with single localized proteins, in the 1621 interaction pairs of 630 multilocation proteins, 1170 interaction pairs (about 72.1%) were involved in communication between the multilocation proteins and the various organelles, which is about eight times (1170/149) as many as single‐location proteins (Table 1, Figure 1d). From this point, multilocalized proteins may be more important in organelle communication than single‐localized proteins. Further, from the depth and breadth of some gene studies, some multilocalization proteins may indeed play a more important role in plant growth and development (Lin et al., 2019, 2020; Nevarez et al., 2017; Rasul et al., 2021; Swida‐Barteczka et al., 2018; Wang et al., 2016, 2021; Wang & Dehesh, 2018; Zhang et al., 2020).
Proteins will be effective only when they are in the right subcellular structure. The same protein may play different roles in different organelles and regions. Proteins with dual subcellular localization can affect transcription and display various functions in intracellular signaling. Recently, there are more and more studies on the involvement of multilocation proteins in the communication of different organelles in the regulation of plant growth and development (Li et al., 2017; Lin et al., 2020; Wang et al., 2021; Zhang et al., 2020; Zhuang et al., 2019). For example, WHY1, which is a plant‐specific DNA‐ and RNA‐binding protein that locates both in chloroplasts and nucleus (Krause et al., 2005; Prikryl et al., 2008). Due to dual location and function in the nucleus and plastids (Krause & Krupinska, 2009), it is assumed that WHY1 can move from plastids to the nucleus (Isemer et al., 2012). The plastid isoform of WHIRLY1 affects miRNA biogenesis in the nucleus (Swida‐Barteczka et al., 2018). SA accumulation could be prevented by ectopic expression of the nuclear WHY1 isoform. However, expressing the plastid WHY1 isoform greatly enhanced cellular SA levels. Altered localization of Arabidopsis WHY1, in the nucleus or chloroplast causes a perturbation in SA homeostasis, resulting in adverse plant senescence phenotypes (Lin et al., 2020). Arabidopsis why1 mutants are insensitive to ABA, it was found that WHY1 located in chloroplast enhanced the sensitivity of plants to ABA after knocking out the chloroplast WHY1 signal peptide, while WHY1 in nucleus was still not sensitive to ABA (Isemer et al., 2012). These studies also suggested that multilocalized proteins may play an important role in cell or organelle communication and participate in plant growth and development.
4.2. Multilocation kinases or phosphatases may have significant roles in organelle communication
Multilocation proteins, such as the phosphorylation‐related proteins MKK4, MPK6, MPK3, MPK4, and NDPK1, were found to be bridged with Nuc (WNK8 and BZIP1), CW (LRX3), Golgi (OFUT20), Mit (ATG11), Per (CAT3), Vac (ATG8B and ATG8D), ER (KCR1 and ATI2), Chl (NDPK2), Cyt (CRCK1 and CRCK2), and Mem (MEKK1 and ZAR1) (Figure 2a) to regulate autophagy (Li et al., 2014; Michaeli et al., 2014), phosphorylation (Hong‐Hermesdorf et al., 2006), photoperiod pathway (Wang et al., 2008), amino acid metabolism (Dietrich et al., 2011), sugar signaling (Kang et al., 2010), cell morphogenesis (Velasquez et al., 2011), reactive oxygen species (Li et al., 2015), fatty acids (Beaudoin et al., 2009), cellular redox state (Moon et al., 2003), innate immunity (Asai et al., 2002; Gao et al., 2008), and zygote development (Yu et al., 2016). In eukaryotes, cell–cell and cell–environment communication often involve cell surface receptors (Xu & Zhang, 2015). MAPK related proteins are ubiquitous signaling modules in eukaryotes. The functions of MAPK‐related proteins include immunity and stress responses. They also play essential roles in plant growth and development downstream of receptor‐like protein kinases (Wang et al., 2007; Xu & Zhang, 2015). Multiple functional pathways initiated from different receptors often involve the same MAPK components or even a complete MAPK cascade. Considering our results, in addition to their important role in cell–cell or cell‐environment communication, MAPK‐related proteins may also play an important role in organelle communication via PPIs.
Histone kinases or phosphatases are also important proteins involved in phosphorylation processes, transmitting the stress signal to a downstream MAPK cascade. Here, in the second interaction circuit, multilocation proteins such as phosphorylation‐related histidine‐containing phosphotransfer protein 1 (AHP1), AHP2, histone kinase 2 (AHK2), and PI4KB1, interact with GID1C, AHL27 and TCP10 of Nuc, GRXS15 of Mit, ETR1 of ER, DRP1A of Cyt, BETAA‐AD of Golgi, HIR1 of PM, and PNSL5 of Chl (Figure 2b). In plants, histone kinases are involved in a variety of stress responses (cold, drought, salt) by regulating hormonal signaling such as abscisic acid or cytokinin signaling, and also regulate many developmental processes including seed germination, cell division, seed size, chlorophyll retention during leaf senescence, root repression and shoot promotion (Miwa et al., 2007; Riefler et al., 2006; Tran et al., 2007). In addition, AHP has histidine phosphotransfer kinase activity, functioning as a two‐component phosphorelay mediator between cytokinin sensor histidine kinases and response regulators.
Calcium is a ubiquitous second messenger that mediates plant responses to developmental and environmental cues. In the third interaction circuit, CDPK6, CDPK33, ABI1, CIPK24, KIN10 and RCAR3 interact with MYB77 of Nuc, RBCX1 of Chl, ARF1 of Golgi, SCAR3 of Cyt, CIPK6 of ER, and CBL1 of Mem (Figure 2c). CDPKs are key factors in plant signaling that convey Ca2+ signals into physiological responses by phosphorylating various substrates, including ion channels, transcription factors and metabolic enzymes (Yip Delormel & Boudsocq, 2019). The stress‐induced Ca2+ signaling was dependent on OsPLC1, and the PLC‐mediated Ca2+ signaling was essential for controlling Na + accumulation in leaf blades. Salt stress induced the recruitment of OsPLC1 from cytoplasm to plasma membrane, where it hydrolyzed PtdIns4P, thus establishing whole plant salt tolerance (Li et al., 2017). Because of this large diversity of targets, CDPKs play pivotal roles in shoot and root development, pollen tube growth, stomatal movements, hormonal signaling, transcriptional reprogramming, and stress tolerance (Almadanim et al., 2018; Baba et al., 2018; Boudsocq & Sheen, 2013; Mori et al., 2006).
In this study, we found that multilocalized proteins may play an important role in PPI network and organelle communication both of Arabidopsis and rice, especially in multilocation kinases or phosphatases. Protein phosphorylation is an important cellular regulatory mechanism as many enzymes and receptors are activated/deactivated by phosphorylation and dephosphorylation via kinases and phosphatases (Tena et al., 2011). Signal transduction systems based on phosphorylation are central to cell–cell communication in multicellular organisms (Mayer & Yu, 2018) and reversible phosphorylation of a protein often serves as a signal modulation mechanism to regulate cellular activities (Hoang et al., 2019). In addition, phosphorylation can also affect the subcellular localization of proteins, such as BZR1. The subcellular localization and transcriptional activity are tightly regulated by reversible protein phosphorylation (Wang et al., 2021). Our study provides a new view that the phosphorylation‐related multilocation proteins plays an important role in organelle communication. The research of phosphorylation‐related multilocation proteins may promote the development of organelle communication and provide an important theoretical basis for plant responses to external stress.
4.3. Techniques, technical difficulties, and prospects of multilocation proteins
The smallest unit of life is the cell, which contains numerous protein molecules. Most of the functions critical to cell survival are performed by these proteins, which are located in different organelles, usually called “subcellular locations” (Chou, 2019). As a result of the above analyses, it speculated that multilocation proteins play key roles in the communication among organelles, which can also be used as an indicator of the closeness organelles. Thus, the method used to screen multilocation proteins is important. At present, research pertaining to subcellular localization is divided mainly into experimental evidence and software predictions. The experimental methods include immunofluorescence (Stadler et al., 2013) and expression of green fluorescent protein fusion proteins (Cui et al., 2016), and software performed predictions are based mainly on bioinformatics (Bouziane & Chouarfia, 2020; Chou, 2019; Chou et al., 2019a). The application of these experimental methods can provide more accurate information on protein subcellular localization, and the present research is based mainly on this type of method. Unfortunately, determining the subcellular locations of proteins based solely on experiments is time‐consuming and costly. In addition, external factors such as light signals induce changes in the subcellular localization of some proteins (Oikawa et al., 2008; Sakamoto & Briggs, 2002; Wan et al., 2008). Thus, inaccurate subcellular locations can be obtained by confocal microscope. In this case, many subcellular localization prediction software programs, including multilocation prediction software, have been developed (Cheng et al., 2017, 2018, 2019; Chou et al., 2011, 2019a, 2019b; Chou & Shen, 2008, 2010; Jiang et al., 2019; Sahu et al., 2019; Wu et al., 2011; Yu et al., 2014). Through comparisons with relevant proteins verified by experimental evidence, several software location information predictors, such as iLoc‐Plant and Plant‐mPLoc, have high confidence in predicting plant nuclear localization (Xiong et al., 2016). However, the prediction software results still need to be supported by experimental evidence, and a method that can ensure a certain accuracy and high‐throughput is needed.
In protein studies, one of the fundamental goals is determining the subcellular locations of proteins within an entire cell (Chou, 2019). Some proteomic techniques, such as iTRAQ, 2‐DE, and Lable‐free, are mature and can be used to analyze organelle proteins (Lv et al., 2019; Ning et al., 2016; Paik et al., 2019). If organelles are isolated effectively, the proteins of each organelle can be identified using proteomic technologies, and multilocation proteins can be obtained. However, separation and acquisition of high purity organelles are difficult. Organelle separation is usually achieved by differential centrifugation (Liao et al., 2020), but the separation efficiency and purity of this method do not meet the standards of organelle proteomic analyses. Recently, dynamic endomembrane organelle profiling has provided a novel approach, localization of organelle proteins by isotope tagging (LOPIT), for isolating and detecting protein components from multiple organelles simultaneously (Chen & Heazlewood, 2020; Geladaki et al., 2019; Mulvey et al., 2017). In addition, this method can detect and locate live proteins moving between different sub‐organelles and membrane systems, which overcomes the technical challenge of routine laboratory separation of sub‐organelles. Furthermore, dynamic endomembrane organelle profiling can be used for large‐scale qualitative and quantitative analyses of different sub‐organelle proteins by valuating localization of organelle proteins by LOPIT, free‐flow electrophoresis (FFE) and mass spectrometry. In addition, protein from different components of the Golgi, such as the cis‐, Medial‐, and trans‐Golgi proteins, can be isolated accurately (Chen & Heazlewood, 2020). By this method, it is possible to isolate the proteins of each organelle and then screen them to obtain multilocation proteins. By combining the protein location information obtained by experimental evidence, homology alignment analysis, and software prediction, more accurate location information should be obtainable. The multilocation proteins can be used for further research, such as the RPTP for rice or large‐scale gene editing, and applied to crop production, drug development, and other applications. In addition, this method (Figure 4) can be used to build databases of multilocation proteins from different species on a large scale. The databases will be important, new and comprehensive protein research resources for many researchers, allowing research to be widely extended to the protein level, which plays an important role in the regulation of biological processes. It is also expected that many new discoveries advancing animal and plant sciences will be made.
FIGURE 4.
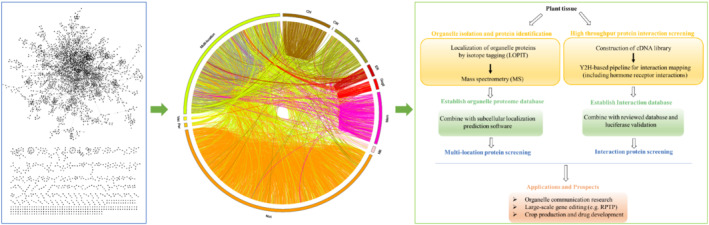
Schematic overview of multilocation protein analysis in plant tissue
4.4. Conclusion and perspectives
PPI networks often suggest protein functions and open new avenues for characterizing genes and proteins (McWhite et al., 2020). Although research on PPIs has advanced rapidly in recent years, it remains poorly developed, especially in terms of communication among organelles at the proteomic level. By summarizing and analyzing the model organisms that have been investigated thoroughly, we can find new clues and identify new research directions, which will have important impacts on research of other crops and animals. In this study, through the analysis of PPIs in the model plant Arabidopsis thaliana, we found that multilocation proteins, especially those related to phosphorylation, play an important role in organelle communication. In addition, we analyzed the research progress on PPIs and the potential role of multilocation proteins in the rice RPTP program. Considering the current research progress, we provided a research strategy and direction for multilocation proteins, which provides a theoretical basis for research pertaining to organelle communication.
CONFLICT OF INTEREST
The authors declare no competing interests.
AUTHOR CONTRIBUTIONS
XEH and GFP conceived the idea of this research. XEH, CD, GFP, ZPF, WZK, and QCX processed and analyzed the data. GFP and XEH wrote the original manuscript. YDM and ZQZ revised the manuscript. All authors read and approved the final manuscript.
Supporting information
Table S1: Details of the reviewed proteins in Arabidopsis.
Table S2: Details of the KEGG pathways associated with the reviewed proteins in Arabidopsis.
Table S3: Total interactions of reviewed proteins in Arabidopsis.
Table S4: Details of the localization of the reviewed proteins in Arabidopsis.
Table S5: Information pertaining to proteins in the organelle interactome of Arabidopsis.
Table S6: Information pertaining to proteins in the organelle interactome of rice.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (No. 32101641, 31471525, and 31971846), Henan Agricultural University (30602095), and Postdoctoral Research Foundation of China (2020M672232).
Xiong, E. , Cao, D. , Qu, C. , Zhao, P. , Wu, Z. , Yin, D. , Zhao, Q. , & Gong, F. (2022). Multilocation proteins in organelle communication: Based on protein–protein interactions. Plant Direct, 6(2), e386. 10.1002/pld3.386
Erhui Xiong and Di Cao contributed equally to this work.
DATA AVAILABILITY STATEMENT
The data that support the plots within this paper and other finding of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Almadanim, M. C. , Goncalves, N. M. , Rosa, M. T. G. , Alexandre, B. M. , Cordeiro, A. M. , Rodrigues, M. , Saibo, N. J. M. , Soares, C. M. , Romao, C. V. , Oliveira, M. M. , & Abreu, I. A. (2018). The rice cold‐responsive calcium‐dependent protein kinase OsCPK17 is regulated by alternative splicing and post‐translational modifications. Biochimica et Biophysica Acta‐Molecular Cell Research, 1865, 231–246. 10.1016/j.bbamcr.2017.10.010 [DOI] [PubMed] [Google Scholar]
- Ardito, F. , Giuliani, M. , Perrone, D. , Troiano, G. , & Lo Muzio, L. (2017). The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy. International Journal of Molecular Medicine, 40, 271–280. 10.3892/ijmm.2017.3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai, T. , Tena, G. , Plotnikova, J. , Willmann, M. R. , Chiu, W. L. , Gomez‐Gomez, L. , Boller, T. , Ausubel, F. M. , & Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature, 415, 977–983. 10.1038/415977a [DOI] [PubMed] [Google Scholar]
- Ashapkin, V. V. , Kutueva, L. I. , Aleksandrushkina, N. I. , & Vanyushin, B. F. (2020). Epigenetic mechanisms of plant adaptation to biotic and abiotic stresses. International Journal of Molecular Sciences, 21, 7457. 10.3390/ijms21207457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenov, Y. , Ramirez, F. , Schelhorn, S. E. , Lengauer, T. , & Albrecht, M. (2008). Computing topological parameters of biological networks. Bioinformatics, 24, 282–284. 10.1093/bioinformatics/btm554 [DOI] [PubMed] [Google Scholar]
- Baba, A. I. , Rigo, G. , Ayaydin, F. , Rehman, A. U. , Andrasi, N. , Zsigmond, L. , Valkai, I. , Urbancsok, J. , Vass, I. , Pasternak, T. , Palme, K. , Szabados, L. , & Cseplo, A. (2018). Functional analysis of the Arabidopsis thaliana CDPK‐related kinase family: AtCRK1 regulates responses to continuous light. International Journal of Molecular Sciences, 19, 1282. 10.3390/ijms19051282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin, F. , Wu, X. Z. , Li, F. L. , Haslam, R. P. , Markham, J. E. , Zheng, H. Q. , Napier, J. A. , & Kunst, L. (2009). Functional characterization of the Arabidopsis β‐ketoacyl‐coenzyme A reductase candidates of the fatty acid elongase. Plant Physiology, 150, 1174–1191. 10.1104/pp.109.137497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq, M. , & Sheen, J. (2013). CDPKs in immune and stress signaling. Trends in Plant Science, 18, 30–40. 10.1016/j.tplants.2012.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouziane, H. , & Chouarfia, A. (2020). Use of Chou's 5‐steps rule to predict the subcellular localization of gram‐negative and gram‐positive bacterial proteins by multi‐label learning based on gene ontology annotation and profile alignment. Journal of Integrative Bioinformatics, 18, 51–79. 10.1515/jib-2019-0091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, K. X. , Phua, S. Y. , Crisp, P. , McQuinn, R. , & Pogson, B. J. (2016). Learning the languages of the chloroplast: Retrograde signaling and beyond. Annual Review of Plant Biology, 67, 25–53. 10.1146/annurev-arplant-043015-111854 [DOI] [PubMed] [Google Scholar]
- Chen, Y. , & Heazlewood, J. L. (2020). Organellar proteomic profiling to analyze membrane trafficking pathways. Trends in Plant Science, 26, 299–300. 10.1016/j.tplants.2020.11.008 [DOI] [PubMed] [Google Scholar]
- Cheng, X. , Lin, W. Z. , Xiao, X. , & Chou, K. C. (2019). pLoc_bal‐mAnimal: Predict subcellular localization of animal proteins by balancing training dataset and PseAAC. Bioinformatics, 35, 398–406. 10.1093/bioinformatics/bty628 [DOI] [PubMed] [Google Scholar]
- Cheng, X. , Xiao, X. , & Chou, K. C. (2017). pLoc‐mPlant: Predict subcellular localization of multilocation plant proteins by incorporating the optimal GO information into general PseAAC. Molecular BioSystems, 13, 1722–1727. 10.1039/C7MB00267J [DOI] [PubMed] [Google Scholar]
- Cheng, X. , Xiao, X. , & Chou, K. C. (2018). pLoc_bal‐mPlant: Predict subcellular localization of plant proteins by general PseAAC and balancing training dataset. Current Pharmaceutical Design, 24, 4013–4022. 10.2174/1381612824666181119145030 [DOI] [PubMed] [Google Scholar]
- Chou, K. C. (2019). Advance in predicting subcellular localization of multi‐label proteins and its implication for developing multi‐target drugs. Current Medicinal Chemistry, 26, 4918–4943. 10.2174/0929867326666190507082559 [DOI] [PubMed] [Google Scholar]
- Chou, K. C. , Cheng, X. , & Xiao, X. (2019a). pLoc_bal‐mEuk: Predict subcellular localization of eukaryotic proteins by general PseAAC and quasi‐balancing training dataset. Medicinal Chemistry, 15, 472–485. 10.2174/1573406415666181218102517 [DOI] [PubMed] [Google Scholar]
- Chou, K. C. , Cheng, X. , & Xiao, X. (2019b). pLoc_bal‐mHum: Predict subcellular localization of human proteins by PseAAC and quasi‐balancing training dataset. Genomics, 111, 1274–1282. 10.1016/j.ygeno.2018.08.007 [DOI] [PubMed] [Google Scholar]
- Chou, K. C. , & Shen, H. B. (2008). Cell‐PLoc: A package of web servers for predicting subcellular localization of proteins in various organisms. Nature Protocols, 3, 153–162. 10.1038/nprot.2007.494 [DOI] [PubMed] [Google Scholar]
- Chou, K. C. , & Shen, H. B. (2010). Plant‐mPLoc: A top‐down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE, 5, e11335. 10.1371/journal.pone.0011335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, K. C. , Wu, Z. C. , & Xiao, X. (2011). iLoc‐Euk: A multi‐label classifier for predicting the subcellular localization of singleplex and multiplex eukaryotic proteins. PLoS ONE, 6, e18258. 10.1371/journal.pone.0018258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, S. , Valm, A. M. , & Lippincott‐Schwartz, J. (2018). Interacting organelles. Current Opinion in Cell Biology, 53, 84–91. 10.1016/j.ceb.2018.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, Y. , Gao, C. , Zhao, Q. , & Jiang, L. (2016). Using fluorescent protein fusions to study protein subcellular localization and dynamics in plant cells. Methods in Molecular Biology, 1474, 113–123. 10.1007/978-1-4939-6352-2_7 [DOI] [PubMed] [Google Scholar]
- Dietrich, K. , Weltmeier, F. , Ehlert, A. , Weiste, C. , Stahl, M. , Harter, K. , & Droge‐Laser, W. (2011). Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell, 23, 381–395. 10.1105/tpc.110.075390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Z. , & Kihara, D. (2019). Computational identification of protein‐protein interactions in model plant proteomes. Scientific Reports, 9, 8740. 10.1038/s41598-019-45072-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg, D. , Marcotte, E. M. , Xenarios, I. , & Yeates, T. O. (2000). Protein function in the post‐genomic era. Nature, 405, 823–826. 10.1038/35015694 [DOI] [PubMed] [Google Scholar]
- Falz, A. L. , & Muller‐Schussele, S. J. (2019). Physcomitrella as a model system for plant cell biology and organelle‐organelle communication. Current Opinion in Plant Biology, 52, 7–13. 10.1016/j.pbi.2019.05.007 [DOI] [PubMed] [Google Scholar]
- Frick, R. , Bich, L. , & Moreno, A. (2019). An organisational approach to biological communication. Acta Biotheoretica, 67, 103–128. 10.1007/s10441-019-09342-2 [DOI] [PubMed] [Google Scholar]
- Gao, M. H. , Liu, J. M. , Bi, D. L. , Zhang, Z. B. , Cheng, F. , Chen, S. F. , & Zhang, Y. L. (2008). MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen‐activated protein kinase cascade to regulate innate immunity in plants. Cell Research, 18, 1190–1198. 10.1038/cr.2008.300 [DOI] [PubMed] [Google Scholar]
- Geladaki, A. , Britovsek, N. K. , Breckels, L. M. , Smith, T. S. , Vennard, O. L. , Mulvey, C. M. , Crook, O. M. , Gatto, L. , & Lilley, K. S. (2019). Combining LOPIT with differential ultracentrifugation for high‐resolution spatial proteomics. Nature Communications, 10, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, F. P. , Cao, D. , Qu, C. X. , Yin, D. M. , Zhao, Q. Z. , & Xiong, E. H. (2021). Advances in the elucidation of nuclear proteins in the model plant Arabidopsis thaliana: Based on protein interactions and bioinformatics analysis. Journal of Plant Interactions, 16, 481–493. 10.1080/17429145.2021.1998681 [DOI] [Google Scholar]
- Hartwell, L. H. , Hopfield, J. J. , Leibler, S. , & Murray, A. W. (1999). From molecular to modular cell biology. Nature, 402, C47–C52. 10.1038/35011540 [DOI] [PubMed] [Google Scholar]
- Hettenhausen, C. , Schuman, M. C. , & Wu, J. Q. (2015). MAPK signaling: A key element in plant defense response to insects. Insect Science, 22, 157–164. 10.1111/1744-7917.12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang, Q. T. N. , Han, Y. J. , & Kim, J. I. (2019). Plant phytochromes and their phosphorylation. International Journal of Molecular Sciences, 20, 3450. 10.3390/ijms20143450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzheu, P. , & Kummer, U. (2020). Computational systems biology of cellular processes in Arabidopsis thaliana: An overview. Cellular and Molecular Life Sciences, 77, 433–440. 10.1007/s00018-019-03379-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong‐Hermesdorf, A. , Brux, A. , Gruber, A. , Gruber, G. , & Schumacher, K. (2006). A WNK kinase binds and phosphorylates V‐ATPase subunit C. FEBS Letters, 580, 932–939. 10.1016/j.febslet.2006.01.018 [DOI] [PubMed] [Google Scholar]
- Huber, L. A. , Pfaller, K. , & Vietor, I. (2003). Organelle proteomics: Implications for subcellular fractionation in proteomics. Circulation Research, 92, 962–968. 10.1161/01.RES.0000071748.48338.25 [DOI] [PubMed] [Google Scholar]
- Isemer, R. , Mulisch, M. , Schafer, A. , Kirchner, S. , Koop, H. U. , & Krupinska, K. (2012). Recombinant Whirly1 translocates from transplastomic chloroplasts to the nucleus. FEBS Letters, 586, 85–88. 10.1016/j.febslet.2011.11.029 [DOI] [PubMed] [Google Scholar]
- Jagodzik, P. , Tajdel‐Zielinska, M. , Ciesla, A. , Marczak, M. , & Ludwikow, A. (2018). Mitogen‐activated protein kinase cascades in plant hormone signaling. Frontiers in Plant Science, 9, 1387. 10.3389/fpls.2018.01387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janota, C. S. , Calero‐Cuenca, F. J. , & Gomes, E. R. (2020). The role of the cell nucleus in mechanotransduction. Current Opinion in Cell Biology, 63, 204–211. 10.1016/j.ceb.2020.03.001 [DOI] [PubMed] [Google Scholar]
- Jiang, Z. , Wang, D. , Wu, P. , Chen, Y. , Shang, H. , Wang, L. , & Xie, H. (2019). Predicting subcellular localization of multisite proteins using differently weighted multi‐label k‐nearest neighbors sets. Technology and Health Care, 27, 185–193. 10.3233/THC-199018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, B. , Leek, A. N. , Sole, L. , Maverick, E. E. , Levine, T. P. , & Tamkun, M. M. (2018). Kv2 potassium channels form endoplasmic reticulum/plasma membrane junctions via interaction with VAPA and VAPB. Proceedings of the National Academy of Sciences of the United States of America, 115, E7331–E7340. 10.1073/pnas.1805757115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M. (2002). The KEGG database. Novartis Foundation Symposium, 247, 91–101. discussion 101‐3, 119‐28, 244‐52 [PubMed] [Google Scholar]
- Kang, S. G. , Price, J. , Lin, P. C. , Hong, J. C. , & Jang, J. C. (2010). The Arabidopsis bZIP1 transcription factor is involved in sugar signaling, protein networking, and DNA binding. Molecular Plant, 3, 361–373. 10.1093/mp/ssp115 [DOI] [PubMed] [Google Scholar]
- Kirkwood, K. J. , Ahmad, Y. , Larance, M. , & Lamond, A. I. (2013). Characterization of native protein complexes and protein isoform variation using size‐fractionation‐based quantitative proteomics. Molecular & Cellular Proteomics, 12, 3851–3873. 10.1074/mcp.M113.032367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause, K. , Kilbienski, I. , Mulisch, M. , Rodiger, A. , Schafer, A. , & Krupinska, K. (2005). DNA‐binding proteins of the Whirly family in Arabidopsis thaliana are targeted to the organelles. FEBS Letters, 579, 3707–3712. 10.1016/j.febslet.2005.05.059 [DOI] [PubMed] [Google Scholar]
- Krause, K. , & Krupinska, K. (2009). Nuclear regulators with a second home in organelles. Trends in Plant Science, 14, 194–199. 10.1016/j.tplants.2009.01.005 [DOI] [PubMed] [Google Scholar]
- Kwak, C. , Shin, S. , Park, J. S. , Jung, M. , Nhung, T. T. M. , Kang, M. G. , Lee, C. , Kwon, T. H. , Park, S. K. , Mun, J. Y. , Kim, J. S. , & Rhee, H. W. (2020). Contact‐ID, a tool for profiling organelle contact sites, reveals regulatory proteins of mitochondrial‐associated membrane formation. Proceedings of the National Academy of Sciences of the United States of America, 117, 12109–12120. 10.1073/pnas.1916584117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. Q. , Chung, T. , & Vierstra, R. D. (2014). AUTOPHAGY‐RELATED11 plays a critical role in general autophagy‐ and senescence‐induced mitophagy in Arabidopsis . Plant Cell, 26, 788–807. 10.1105/tpc.113.120014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Liu, J. T. , Wang, G. Q. , Cha, J. Y. , Li, G. N. , Chen, S. , Li, Z. , Guo, J. H. , Zhang, C. G. , Yang, Y. Q. , Kim, W. Y. , Yun, D. J. , Schumaker, K. S. , Chen, Z. Z. , & Guo, Y. (2015). A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase ACTIVITY and for multiple stress responses in Arabidopsis . Plant Cell, 27, 908–925. 10.1105/tpc.114.135095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Wang, F. , Yan, P. , Jing, W. , Zhang, C. , Kudla, J. , & Zhang, W. (2017). A phosphoinositide‐specific phospholipase C pathway elicits stress‐induced Ca2+ signals and confers salt tolerance to rice. The New Phytologist, 214, 1172–1187. 10.1111/nph.14426 [DOI] [PubMed] [Google Scholar]
- Liao, P. C. , Bergamini, C. , Fato, R. , Pon, L. A. , & Pallotti, F. (2020). Isolation of mitochondria from cells and tissues. Methods in Cell Biology, 155, 3–31. 10.1016/bs.mcb.2019.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S. , Meng, T. , Huang, H. , Zhuang, H. , He, Z. , Yang, H. , & Feng, D. (2021). Molecular machineries and physiological relevance of ER‐mediated membrane contacts. Theranostics, 11, 974–995. 10.7150/thno.51871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, W. , Huang, D. , Shi, X. , Deng, B. , Ren, Y. , Lin, W. , & Miao, Y. (2019). H2O2 as a feedback signal on dual‐located WHIRLY1 associates with leaf senescence in Arabidopsis . Cell, 8, 1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, W. , Zhang, H. , Huang, D. , Schenke, D. , Cai, D. , Wu, B. , & Miao, Y. (2020). Dual‐localized WHIRLY1 affects salicylic acid biosynthesis via coordination of ISOCHORISMATE SYNTHASE1, PHENYLALANINE AMMONIA LYASE1, and S‐ADENOSYL‐L‐METHIONINE‐DEPENDENT METHYLTRANSFERASE1. Plant Physiology, 184, 1884–1899. 10.1104/pp.20.00964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, K. , Guo, W. J. , Lu, J. X. , Yu, H. , Qu, C. M. , Tang, Z. L. , Li, J. N. , Chai, Y. R. , & Liang, Y. (2015). Genome‐wide survey and expression profile analysis of the mitogen‐activated protein kinase (MAPK) gene family in Brassica rapa. PLoS ONE, 10, e0132051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y. , Ronald, P. C. , Han, B. , Li, J. , & Zhu, J. K. (2020). Rice protein tagging project: A call for international collaborations on genome‐wide in‐locus tagging of rice proteins. Molecular Plant, 13, 1663–1665. 10.1016/j.molp.2020.11.006 [DOI] [PubMed] [Google Scholar]
- Lu, Y. , Tian, Y. , Shen, R. , Yao, Q. , Wang, M. , Chen, M. , Dong, J. , Zhang, T. , Li, F. , Lei, M. , & Zhu, J. K. (2020). Targeted, efficient sequence insertion and replacement in rice. Nature Biotechnology, 38, 1402–1407. 10.1038/s41587-020-0581-5 [DOI] [PubMed] [Google Scholar]
- Lv, D. D. , Zhang, Y. Y. , Ge, H. T. , Huang, X. H. , & Wang, Y. C. (2019). Advances of the technologies in large‐scale membrane proteome identification. Yi Chuan, 41, 863–874. [DOI] [PubMed] [Google Scholar]
- Mayer, B. J. , & Yu, J. (2018). Protein clusters in phosphotyrosine signal transduction. Journal of Molecular Biology, 430, 4547–4556. 10.1016/j.jmb.2018.05.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhite, C. D. , Papoulas, O. , Drew, K. , Cox, R. M. , June, V. , Dong, O. X. , Kwon, T. , Wan, C. , Salmi, M. L. , Roux, S. J. , Browning, K. S. , Chen, Z. J. , Ronald, P. C. , & Marcotte, E. M. (2020). A pan‐plant protein complex map reveals deep conservation and novel assemblies. Cell, 181(460–474), e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaeli, S. , Honig, A. , Levanony, H. , Peled‐Zehavi, H. , & Galili, G. (2014). Arabidopsis ATG8‐INTERACTING PROTEIN1 is involved in autophagy‐dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell, 26, 4084–4101. 10.1105/tpc.114.129999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, N. S. , Tuteja, R. , & Tuteja, N. (2006). Signaling through MAP kinase networks in plants. Archives of Biochemistry and Biophysics, 452, 55–68. 10.1016/j.abb.2006.05.001 [DOI] [PubMed] [Google Scholar]
- Miwa, K. , Ishikawa, K. , Terada, K. , Yamada, H. , Suzuki, T. , Yamashino, T. , & Mizuno, T. (2007). Identification of amino acid substitutions that render the Arabidopsis cytokinin receptor histidine kinase AHK4 constitutively active. Plant & Cell Physiology, 48, 1809–1814. 10.1093/pcp/pcm145 [DOI] [PubMed] [Google Scholar]
- Moon, H. , Lee, B. , Choi, G. , Shin, S. , Prasad, D. T. , Lee, O. , Kwak, S. S. , Kim, D. H. , Nam, J. , Bahk, J. , Hong, J. C. , Lee, S. Y. , Cho, M. J. , Lim, C. O. , & Yun, D. J. (2003). NDP kinase 2 interacts with two oxidative stress‐activated MAPKs to regulate cellular redox state and enhances multiple stress tolerance in transgenic plants. Proceedings of the National Academy of Sciences of the United States of America, 100, 358–363. 10.1073/pnas.252641899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, I. C. , Murata, Y. , Yang, Y. , Munemasa, S. , Wang, Y. F. , Andreoli, S. , Tiriac, H. , Alonso, J. M. , Harper, J. F. , Ecker, J. R. , Kwak, J. M. , & Schroeder, J. I. (2006). CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S‐type anion‐ and Ca2+‐permeable channels and stomatal closure. PLoS Biology, 4, e327. 10.1371/journal.pbio.0040327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey, C. M. , Breckels, L. M. , Geladaki, A. , Britovsek, N. K. , Nightingale, D. J. H. , Christoforou, A. , Elzek, M. , Deery, M. J. , Gatto, L. , & Lilley, K. S. (2017). Using hyperLOPIT to perform high‐resolution mapping of the spatial proteome. Nature Protocols, 12, 1110–1135. 10.1038/nprot.2017.026 [DOI] [PubMed] [Google Scholar]
- Namgaladze, D. , Khodzhaeva, V. , & Brune, B. (2019). ER‐mitochondria communication in cells of the innate immune system. Cell, 8, 1088. 10.3390/cells8091088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevarez, P. A. , Qiu, Y. , Inoue, H. , Yoo, C. Y. , Benfey, P. N. , Schnell, D. J. , & Chen, M. (2017). Mechanism of dual targeting of the phytochrome signaling component HEMERA/pTAC12 to plastids and the nucleus. Plant Physiology, 173, 1953–1966. 10.1104/pp.16.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning, F. , Wu, X. , & Wang, W. (2016). Exploiting the potential of 2DE in proteomics analyses. Expert Review of Proteomics, 13, 901–903. 10.1080/14789450.2016.1230498 [DOI] [PubMed] [Google Scholar]
- Oikawa, K. , Matsunaga, S. , Mano, S. , Kondo, M. , Yamada, K. , Hayashi, M. , Kagawa, T. , Kadota, A. , Sakamoto, W. , Higashi, S. , Watanabe, M. , Mitsui, T. , Shigemasa, A. , Iino, T. , Hosokawa, Y. , & Nishimura, M. (2015). Physical interaction between peroxisomes and chloroplasts elucidated by in situ laser analysis. Nature Plants, 1, 15035. 10.1038/nplants.2015.35 [DOI] [PubMed] [Google Scholar]
- Oikawa, K. , Yamasato, A. , Kong, S. G. , Kasahara, M. , Nakai, M. , Takahashi, F. , Ogura, Y. , Kagawa, T. , & Wada, M. (2008). Chloroplast outer envelope protein CHUP1 is essential for chloroplast anchorage to the plasma membrane and chloroplast movement. Plant Physiology, 148, 829–842. 10.1104/pp.108.123075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan, N. , Koppers, M. , Van Soest, I. , Van Harten, A. , Jurriens, D. , Liv, N. , Klumperman, J. , Kapitein, L. C. , Hoogenraad, C. C. , & Farias, G. G. (2021). ER ‐ lysosome contacts at a pre‐axonal region regulate axonal lysosome availability. Nature Communications, 12, 4493. 10.1038/s41467-021-24713-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik, Y. K. , Overall, C. M. , Corrales, F. , Deutsch, E. W. , Lane, L. , & Omenn, G. S. (2019). Advances in identifying and characterizing the human proteome. Journal of Proteome Research, 18, 4079–4084. 10.1021/acs.jproteome.9b00745 [DOI] [PubMed] [Google Scholar]
- Prikryl, J. , Watkins, K. P. , Friso, G. , Van Wijk, K. J. , & Barkan, A. (2008). A member of the Whirly family is a multifunctional RNA‐ and DNA‐binding protein that is essential for chloroplast biogenesis. Nucleic Acids Research, 36, 5152–5165. 10.1093/nar/gkn492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra, A. S. , Gonugunta, V. K. , Christmann, A. , & Grill, E. (2010). ABA perception and signalling. Trends in Plant Science, 15, 395–401. 10.1016/j.tplants.2010.04.006 [DOI] [PubMed] [Google Scholar]
- Rahiminejad, M. , Ledari, M. T. , Mirzaei, M. , Ghorbanzadeh, Z. , Kavousi, K. , Ghaffari, M. R. , Haynes, P. A. , Komatsu, S. , & Salekdeh, G. H. (2019). The quest for missing proteins in rice. Molecular Plant, 12, 4–6. 10.1016/j.molp.2018.11.009 [DOI] [PubMed] [Google Scholar]
- Rasul, F. , Zheng, F. , Dong, F. , He, J. , Liu, L. , Liu, W. , Cheema, J. Y. , Wei, W. , & Fu, C. (2021). Emr1 regulates the number of foci of the endoplasmic reticulum‐mitochondria encounter structure complex. Nature Communications, 12, 521. 10.1038/s41467-020-20866-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, Y. , Li, Y. , Jiang, Y. , Wu, B. , & Miao, Y. (2017). Phosphorylation of WHIRLY1 by CIPK14 shifts its localization and dual functions in Arabidopsis . Molecular Plant, 10, 749–763. 10.1016/j.molp.2017.03.011 [DOI] [PubMed] [Google Scholar]
- Rhee, S. Y. , & Mutwil, M. (2014). Towards revealing the functions of all genes in plants. Trends in Plant Science, 19, 212–221. 10.1016/j.tplants.2013.10.006 [DOI] [PubMed] [Google Scholar]
- Riefler, M. , Novak, O. , Strnad, M. , & Schmulling, T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell, 18, 40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, M. C. S. , Petersen, M. , & Mundy, J. (2010). Mitogen‐activated protein kinase signaling in plants. Annual Review of Plant Biology, 61(61), 621–649. [DOI] [PubMed] [Google Scholar]
- Sahu, S. S. , Loaiza, C. D. , & Kaundal, R. (2019). Plant‐mSubP: A computational framework for the prediction of single‐ and multi‐target protein subcellular localization using integrated machine‐learning approaches. Aob Plants, 12, plz068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, K. , & Briggs, W. R. (2002). Cellular and subcellular localization of phototropin 1. Plant Cell, 14, 1723–1735. 10.1105/tpc.003293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwikowski, B. , Uetz, P. , & Fields, S. (2000). A network of protein‐protein interactions in yeast. Nature Biotechnology, 18, 1257–1261. 10.1038/82360 [DOI] [PubMed] [Google Scholar]
- Shannon, P. , Markiel, A. , Ozier, O. , Baliga, N. S. , Wang, J. T. , Ramage, D. , Amin, N. , Schwikowski, B. , & Ideker, T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research, 13, 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smekalova, V. , Doskocilova, A. , Komis, G. , & Samaj, J. (2014). Crosstalk between secondary messengers, hormones and MAPK modules during abiotic stress signalling in plants. Biotechnology Advances, 32, 2–11. 10.1016/j.biotechadv.2013.07.009 [DOI] [PubMed] [Google Scholar]
- Soto‐Heredero, G. , Baixauli, F. , & Mittelbrunn, M. (2017). Interorganelle communication between mitochondria and the endolysosomal system. Frontiers in Cell and Development Biology, 5, 95. 10.3389/fcell.2017.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler, C. , Rexhepaj, E. , Singan, V. R. , Murphy, R. F. , Pepperkok, R. , Uhlen, M. , Simpson, J. C. , & Lundberg, E. (2013). Immunofluorescence and fluorescent‐protein tagging show high correlation for protein localization in mammalian cells. Nature Methods, 10, 315–323. 10.1038/nmeth.2377 [DOI] [PubMed] [Google Scholar]
- Stefan, C. J. , Manford, A. G. , & Emr, S. D. (2013). ER‐PM connections: Sites of information transfer and inter‐organelle communication. Current Opinion in Cell Biology, 25, 434–442. 10.1016/j.ceb.2013.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subba, P. , Narayana Kotimoole, C. , & Prasad, T. S. K. (2019). Plant proteome databases and bioinformatic tools: An expert review and comparative insights. Omics, 23, 190–206. 10.1089/omi.2019.0024 [DOI] [PubMed] [Google Scholar]
- Swida‐Barteczka, A. , Krieger‐Liszkay, A. , Bilger, W. , Voigt, U. , Hensel, G. , Szweykowska‐Kulinska, Z. , & Krupinska, K. (2018). The plastid‐nucleus located DNA/RNA binding protein WHIRLY1 regulates microRNA‐levels during stress in barley (Hordeum vulgare L.). RNA Biology, 15, 886–891. 10.1080/15476286.2018.1481695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena, G. , Boudsocq, M. , & Sheen, J. (2011). Protein kinase signaling networks in plant innate immunity. Current Opinion in Plant Biology, 14, 519–529. 10.1016/j.pbi.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosal‐Castano, S. , Peselj, C. , Kohler, V. , Habernig, L. , Berglund, L. L. , Ebrahimi, M. , Vogtle, F. N. , Hoog, J. , Andreasson, C. , & Buttner, S. (2021). Snd3 controls nucleus–vacuole junctions in response to glucose signaling. Cell Reports, 34, 108637. 10.1016/j.celrep.2020.108637 [DOI] [PubMed] [Google Scholar]
- Tran, L. S. , Urao, T. , Qin, F. , Maruyama, K. , Kakimoto, T. , Shinozaki, K. , & Yamaguchi‐Shinozaki, K. (2007). Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 104, 20623–20628. 10.1073/pnas.0706547105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez, S. M. , Ricardi, M. M. , Dorosz, J. G. , Fernandez, P. V. , Nadra, A. D. , Pol‐Fachin, L. , Egelund, J. , Gille, S. , Harholt, J. , Ciancia, M. , Verli, H. , Pauly, M. , Bacic, A. , Olsen, C. E. , Ulvskov, P. , Petersen, B. L. , Somerville, C. , Iusem, N. D. , & Estevez, J. M. (2011). O‐glycosylated cell wall proteins are essential in root hair growth. Science, 332, 1401–1403. 10.1126/science.1206657 [DOI] [PubMed] [Google Scholar]
- Vidal, M. , Cusick, M. E. , & Barabasi, A. L. (2011). Interactome networks and human disease. Cell, 144, 986–998. 10.1016/j.cell.2011.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, Y. L. , Eisinger, W. , Ehrhardt, D. , Kubitscheck, U. , Baluska, F. , & Briggs, W. (2008). The subcellular localization and blue‐light‐induced movement of phototropin 1‐GFP in etiolated seedlings of Arabidopsis thaliana . Molecular Plant, 1, 103–117. 10.1093/mp/ssm011 [DOI] [PubMed] [Google Scholar]
- Wang, H. , Chevalier, D. , Larue, C. , Ki Cho, S. , & Walker, J. C. (2007). The protein phosphatases and protein kinases of Arabidopsis thaliana . Arabidopsis Book, 5, e0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Ding, Y. , Zhuang, X. , Hu, S. , & Jiang, L. (2016). Protein co‐localization studies: Issues and considerations. Molecular Plant, 9, 1221–1223. 10.1016/j.molp.2016.05.011 [DOI] [PubMed] [Google Scholar]
- Wang, J. Z. , & Dehesh, K. (2018). ER: The silk road of interorganellar communication. Current Opinion in Plant Biology, 45, 171–177. 10.1016/j.pbi.2018.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. , Wang, R. , Liu, M. , Yuan, W. , Zhao, Z. , Liu, X. , Peng, Y. , Yang, X. , Sun, Y. , & Tang, W. (2021). Nucleocytoplasmic trafficking and turnover mechanisms of BRASSINAZOLE RESISTANT1 in Arabidopsis thaliana . Proceedings of the National Academy of Sciences of the United States of America, 118, e2101838118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Liu, K. , Liao, H. , Zhuang, C. , Ma, H. , & Yan, X. (2008). The plant WNK gene family and regulation of flowering time in Arabidopsis . Plant Biology (Stuttgart, Germany), 10, 548–562. 10.1111/j.1438-8677.2008.00072.x [DOI] [PubMed] [Google Scholar]
- Wu, Z. C. , Xiao, X. , & Chou, K. C. (2011). iLoc‐plant: A multi‐label classifier for predicting the subcellular localization of plant proteins with both single and multiple sites. Molecular BioSystems, 7, 3287–3297. 10.1039/c1mb05232b [DOI] [PubMed] [Google Scholar]
- Xiong, E. , Dong, G. , Chen, F. , Zhang, C. , Li, S. , Zhang, Y. , Shohag, J. I. , Yang, X. , Zhou, Y. , Qian, Q. , Wu, L. , & Yu, Y. (2021). Formyl tetrahydrofolate deformylase affects hydrogen peroxide accumulation and leaf senescence by regulating the folate status and redox homeostasis in rice. Science China. Life Sciences, 64, 720–738. 10.1007/s11427-020-1773-7 [DOI] [PubMed] [Google Scholar]
- Xiong, E. , Li, Z. , Zhang, C. , Zhang, J. , Liu, Y. , Peng, T. , Chen, Z. , & Zhao, Q. (2021). A study of leaf‐senescence genes in rice based on a combination of genomics, proteomics and bioinformatics. Briefings in Bioinformatics, 22, bbaa305. 10.1093/bib/bbaa305 [DOI] [PubMed] [Google Scholar]
- Xiong, E. H. , Zheng, C. Y. , Wu, X. L. , & Wang, W. (2016). Protein subcellular location: The gap between prediction and experimentation. Plant Molecular Biology Reporter, 34, 52–61. 10.1007/s11105-015-0898-2 [DOI] [Google Scholar]
- Xu, J. , & Zhang, S. (2015). Mitogen‐activated protein kinase cascades in signaling plant growth and development. Trends in Plant Science, 20, 56–64. 10.1016/j.tplants.2014.10.001 [DOI] [PubMed] [Google Scholar]
- Yip Delormel, T. , & Boudsocq, M. (2019). Properties and functions of calcium‐dependent protein kinases and their relatives in Arabidopsis thaliana . The New Phytologist, 224, 585–604. 10.1111/nph.16088 [DOI] [PubMed] [Google Scholar]
- Yu, C. S. , Cheng, C. W. , Su, W. C. , Chang, K. C. , Huang, S. W. , Hwang, J. K. , & Lu, C. H. (2014). CELLO2GO: A web server for protein subCELlular LOcalization prediction with functional gene ontology annotation. PLoS ONE, 9, e99368. 10.1371/journal.pone.0099368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, T. Y. , Shi, D. Q. , Jia, P. F. , Tang, J. , Li, H. J. , Liu, J. , & Yang, W. C. (2016). The Arabidopsis receptor kinase ZAR1 is required for zygote asymmetric division and its daughter cell fate. PLoS Genetics, 12, e1005933. 10.1371/journal.pgen.1005933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, G. , & Yang, P. (2018). A novel cell‐cell communication mechanism in the nervous system: Exosomes. Journal of Neuroscience Research, 96, 45–52. 10.1002/jnr.24113 [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Sampathkumar, A. , Kerber, S. M. , Swart, C. , Hille, C. , Seerangan, K. , Graf, A. , Sweetlove, L. , & Fernie, A. R. (2020). A moonlighting role for enzymes of glycolysis in the co‐localization of mitochondria and chloroplasts. Nature Communications, 11, 4509. 10.1038/s41467-020-18234-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang, K. , Kong, F. , Zhang, S. , Meng, C. , Yang, M. , Liu, Z. , Wang, Y. , Ma, N. , & Meng, Q. (2019). Whirly1 enhances tolerance to chilling stress in tomato via protection of photosystem II and regulation of starch degradation. The New Phytologist, 221, 1998–2012. 10.1111/nph.15532 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Details of the reviewed proteins in Arabidopsis.
Table S2: Details of the KEGG pathways associated with the reviewed proteins in Arabidopsis.
Table S3: Total interactions of reviewed proteins in Arabidopsis.
Table S4: Details of the localization of the reviewed proteins in Arabidopsis.
Table S5: Information pertaining to proteins in the organelle interactome of Arabidopsis.
Table S6: Information pertaining to proteins in the organelle interactome of rice.
Data Availability Statement
The data that support the plots within this paper and other finding of this study are available from the corresponding author upon reasonable request.


