Abstract
Background
Apolipoprotein E (APOE) gene mediates lipoprotein clearance and is one of the most studied candidate genes for type 2 diabetes mellitus (T2DM) and coronary artery disease (CAD). This study was performed to determine the association between APOE polymorphisms and T2DM with and without CAD, and its effect on plasma lipid levels in a Chinese population.
Methods
A total of 1,414 subjects involving 869 patients and 545 health individuals were recruited. These patients were categorized into three distinct groups: 264 in T2DM group, 401 in CAD group, and 204 in T2DM+CAD group. Logistic regression analysis was used to obtain odds ratio (OR) and 95% confidence interval (CI) in predicting the risk probability of APOE. Besides, a meta-analysis was preformed to integrate an evaluation index to evaluate their associations.
Results
Genotype frequency ratio of genotype ϵ3/4 and allele ϵ4 among the CAD patients with or without T2DM was obviously increased. Compared with ϵ3/3 genotype, the ϵ3/4 genotype had a significant increased risk of CAD (adjusted OR = 1.90, 95% CI = 1.30–2.77) and T2DM+CAD (adjusted OR = 1.95, 95% CI = 1.24–3.08). In the meta-analysis, four studies were included and provided a strong evidence for the APOE ϵ4 mutation elevating the risk of CAD in patients with T2DM (ϵ3/ϵ4+ϵ4/ϵ4 vs. ϵ3/ϵ3, OR = 1.51, 95% CI = 1.13–2.02). In the T2DM group, the plasma levels of low-density lipoprotein cholesterol (LDL-C) showed significant difference among the three APOE isoforms. The high-density lipoprotein cholesterol (HDL-C) levels of CAD patients with ϵ4-bearing genotypes were lower than those with ϵ3/3 genotype.
Conclusions
Our results indicate that APOE gene polymorphisms are related to CAD with or without T2DM and have influence on lipid profiles in both T2DM and CAD patients.
Keywords: apolipoprotein E, polymorphism, type 2 diabetes mellitus, coronary artery disease, meta-analysis
Introduction
Type 2 diabetes mellitus (T2DM) is a common chronic metabolic disease characterized by high levels of sugar in the blood and is prevalent throughout the world. The incidence of T2DM is rising at an alarming rate attributed to changing dietary patterns, increasing life expectancy, and westernization of lifestyles in developing countries (1). T2DM frequently coexists with various complications such as hypertension and dyslipidemia and is also known as a major independent risk factor for coronary artery disease (CAD) (2). Cardiovascular disease including CAD is increased in T2DM subjects, which is associated with significant morbidity and mortality. Patients with T2DM have two- to fourfold greater risk of developing CAD compared to individuals without diabetes (3). The development of CAD in the setting of T2DM due to a complex combination of various risk factors plays important role in the beginning and the evolution of atherosclerosis (4). The inherited aspect of risk factors is most often a number of genes interacting with each other or with the environmental factors. Therefore, managing genetic risk factors for T2DM and CAD may improve the understanding of these disease and result in better clinical management.
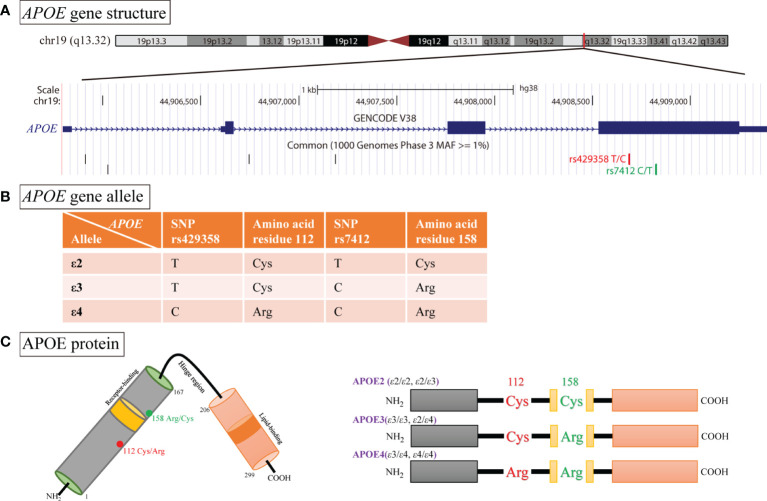
Apolipoprotein E (APOE) gene maps in the long arm of chromosome 19 at position q13.32, which encodes a multifunction glycoprotein containing 299 amino acids ( Figure 1A ). It acts as cholesterol carrier and is involved in mediating the transportation and metabolism of lipids (5). As shown in Figure 1B , two single-nucleotide polymorphisms (SNPs) in APOE, namely, rs429358 (T>C) and rs7412 (C>T), gives rise to three major alleles: ϵ2 (rs429358-T, rs7412-T), ϵ3 (rs429358-T, rs7412-C), and ϵ4 (rs429358-C, rs7412-C). Therefrom, the three alleles yield six different genotypes, of which three are homozygous, namely, ϵ2/ϵ2, ϵ3/ϵ3, and ϵ4/ϵ4, and three are heterozygous, namely, ϵ2/ϵ3, ϵ2/ϵ4, and ϵ3/ϵ4. Besides, these variants encode three different protein isoforms: APOE2 (ϵ2/ϵ2, ϵ2/ϵ3; Cys112/Cys158), APOE3 (ϵ2/ϵ4, ϵ3/ϵ3; Cys112/Arg158), and APOE4 (ϵ3/ϵ4, 4/ϵ4; Arg112/Arg158, Figure 1C ) (6). Previous studies have shown that variants of APOE could govern the metabolism of lipoproteins. Allele ϵ4 is associated with lower plasma high-density lipoprotein cholesterol (HDL-C) level but had higher plasma levels of low-density lipoprotein cholesterol (LDL-C), total cholesterol (TC), and triglycerides (TG), when compared with ϵ3 allele. Meanwhile, the presence of ϵ2 is usually coupled with lower plasma levels of LDL (7, 8). Referring to those previous studies, evidence suggests that a functional interaction between APOE polymorphisms and LDL receptor (LDL-R) influences the risk of CAD and T2DM, and ϵ4 allele has higher affinity to LDL-R than other alleles (9, 10). It is thus likely that the effects of APOE ϵ4 are due to overproduction of LDL or fewer LDL-R, overwhelming the limited ability of mediating the clearance of lipoproteins (10, 11). In contrast, Larifla et al. has reported that the lack relationship between APOE polymorphisms and CAD in Afro-Caribbean people (12). Besides, according to a recent meta-analysis including 13 eligible studies, APOE gene ϵ4 allele had a significant increased risk for CAD patients with T2DM, whereas the ϵ2 variation had null association (13). Consequently, we conducted a case–control study to investigate the association of APOE polymorphisms with T2DM and CAD in a Chinese population and its potential role in lipid metabolism.
Figure 1.
Schematic illustration of APOE genotype and APOE isoform. (A) Functional single nucleotide polymorphisms (SNPs) in the APOE gene viewed in UCSC genome browser. (B) Two SNPs (rs429358 and rs7412) in exon 4 of APOE generate three major allelic variants (ϵ2, ϵ3, and ϵ4). (C) APOE protein is a polypeptide chain with 299 amino, and the APOE2, APOE3, and APOE4 isoforms differ from one another at amino acid residues 112 or 158.
Methods
Study Population
The studied subjects were recruited from The First Affiliated Hospital of Nanjing Medical University (Nanjing, China) from January 2018 to December 2019. Our study was approved by Ethical Committee of The First Affiliated Hospital of Nanjing Medical University, and all donors signed a written informed consent before enrollment. Only Chinese subjects aged 18 years or above were recruited. Questionnaires were used to collect the information of age, sex, genetic family history, medical history, and lifestyle habits. Other clinical and biochemical data such as blood pressure, dyslipidemia, and blood glucose were obtained from clinical and laboratory examinations. Dyslipidemic or hyperlipidemic feature matches the following conditions: TC > 5.17mmol/L (200 mg/dl), TG > 1.69 mmol/L (150 mg/dl), LDL-C > 3.38 mmol/L (130 mg/dl), or HDL-C < 1.03 mmol/L (40 mg/dl). A fasting plasma glucose (FPG) level ≥7.0 mmol/L (126 mg/dl) or a 2-h plasma glucose ≥11.1 mmol/L (200 mg/dl) meets the threshold for the diagnosis of diabetes.
According to the above criteria, studied subjects were classified into four groups. First is the T2DM group that included 264 subjects that fulfilled the diabetes diagnostic criteria of FPG ≥7.0 mmol/L or were under treatment with oral antidiabetic drugs. Second is the CAD group that consisted of 401 subjects with at least 50% stenosis in a major coronary artery or one of their branches defined by coronary angiography. Third is the T2DM+CAD group included 204 subjects diagnosed to have diabetes complicated with coronary artery disease. Exclusion criteria included type 1 diabetes mellitus, malignant tumors, liver and kidney diseases, metabolic disorders, and autoimmune diseases. Fourth is the control group that included 545 healthy individuals without hyperlipidemia, hypertension, cardiovascular diseases, and diabetes.
APOE Genotyping
Genomic DNA was isolated from leukocytes of the peripheral blood using a commercial kit following the manufacturer’s protocols (Sinochip, Zhuhai, China). DNA concentration and purity were estimated by 260 and 280 nm (optical density, OD) light absorbance on a Nanodrop 2000™ spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Two APOE SNPs (rs429358 and rs7412) were genotyped using a detection kit (GeneChip Assay, Sinochip, Zhuhai, China). All samples were amplified according to the manufacturer’s instructions, and then, the amplified products were assayed by the fully automated GeneChip detection system (Sinochip, Zhuhai, China).
Lipid Profiles
After an overnight fast of at least 12 h, venous blood samples were collected from these patients. Plasma TC, TG, HDL-C, LDL-C, and FPG were quantified using the Beckman biochemical assembly line.
Systematic Review and Meta-analysis
We conducted a literature search for all studies that evaluated the association of APOE polymorphisms with T2DM and CAD in the PubMed database up to December 2021. The following key terms were used in the search: “apolipoprotein E” or “APOE,” “polymorphism,” “CAD,” and “T2DM.” References cited in each retrieved article were also manually scanned to discover additional eligible studies. Articles were recruited for this meta-analysis if they fitted the following criteria: (1) investigated the associations between APOE polymorphisms and CAD in patients with T2DM, (2) sufficient data for estimating the odds ratios (ORs) and 95% confidence intervals (CIs), and (3) published in English. Exclusion criteria were (1) duplication of previous data and (2) not using coronary angiography to confirm CAD.
Statistical Analysis
Continuous data such age and lipid profile were compared using Student’s t-test or Wilcoxon test for two groups and Kruskal–Wallis test for more than two groups. Categorical variables (sex and APOE genotypes) were expressed as frequency and compared using Pearson’s χ2 test or Fisher’s exact test. Hardy–Weinberg equilibrium was conducted to evaluate the allele and genotype difference among groups. The associations between APOE polymorphism and diseases were estimated by computing crude or adjusted ORs and 95% CIs from unconditional logistic regression. All the statistical analyses were done with R 4.0.1, and two-sided p-value <0.05 was considered statistically significant.
Results
Characteristics and Clinical Features of Subjects
The demographic and clinical features of 1,414 included individuals are summarized in Table 1 . The data from normal controls were used as a reference to compare with the data obtained from three observation groups consisted of patients with T2DM, CAD, and T2DM+CAD. A significant difference in age was found between control group (mean age, 67.2 years) and the observation groups (T2DM, 69.5 years; CAD, 65.3 years; T2DM+CAD, 70.5 years), implying a higher risk of developing T2DM with increasing age. Gender was equally distributed in CAD group with 111 female patients and 290 male patients and in T2DM+CAD group with 65 female and 139 male patients compared to control group with 206 female and 339 male subjects. There was also significant sex difference between CAD patients and the controls (p = 0.001) due to the high prevalence of male patients among CAD compared to control group (72.3% vs. 62.2%). Besides, T2DM and T2DM+CAD groups had significantly higher levels of TG than the normal control group. Patients with CAD or T2DM had lower levels of HDL-C than the controls. Therefore, the TG/HDL-C ratio was calculated, and this index was significantly higher among the observation groups.
Table 1.
Clinical characteristics and genotype distribution of APOE gene in different groups.
| Variables | Control | T2DM | CAD | T2DM+CAD | ||||
|---|---|---|---|---|---|---|---|---|
| n = 545 | n = 264 | p | n = 401 | p | n = 204 | p | ||
| Age (years) | 67.2 ± 13.4 | 69.5 ± 12.7 | 0.022 | 65.3 ± 12.3 | 0.029 | 70.5 ± 13.0 | 0.003 | |
| Sex | ||||||||
| Female | 206 (37.8%) | 85 (32.2%) | 0.120 | 111 (27.7%) | 0.001 | 65 (31.9%) | 0.132 | |
| Male | 339 (62.2%) | 179 (67.8%) | 290 (72.3%) | 139 (68.1%) | ||||
| Lipid profile (mmol/L) | ||||||||
| TC | 4.25 ± 0.96 | 4.30 ± 1.23 | 0.974 | 4.33 ± 1.13 | 0.635 | 4.29 ± 1.34 | 0.385 | |
| TG | 1.42 ± 0.91 | 1.72 ± 1.30 | <0.001 | 1.49 ± 0.89 | 0.238 | 1.86 ± 1.45 | <0.001 | |
| HDL-C | 1.15 ± 0.33 | 1.05 ± 0.28 | <0.001 | 1.09 ± 0.26 | 0.032 | 1.02 ± 0.27 | <0.001 | |
| LDL-C | 2.58 ± 0.71 | 2.65 ± 0.89 | 0.506 | 2.69 ± 0.86 | 0.173 | 2.70 ± 1.03 | 0.794 | |
| TG/HDL-C ratio | 1.38 ± 1.12 | 1.87 ± 1.88 | <0.001 | 1.49 ± 1.10 | 0.032 | 2.08 ± 2.13 | <0.001 | |
| APOE genotypes | ||||||||
| APOE2 | ϵ2/ϵ2 | 6 (1.10%) | 3 (1.14%) | 1.000 | 1 (0.25%) | 0.249 | 0 (0%) | – |
| ϵ2/ϵ3 | 87 (15.96%) | 47 (17.80%) | 0.509 | 52 (12.97%) | 0.198 | 27 (13.24%) | 0.355 | |
| APOE3 | ϵ2/ϵ4 | 6 (1.10%) | 2 (0.76%) | 1.000 | 3 (0.75%) | 0.741 | 4 (1.96%) | 0.473 |
| ϵ3/ϵ3 | 387 (71.01%) | 171 (64.77%) | 0.072 | 268 (66.83%) | 0.169 | 132 (64.71%) | 0.096 | |
| APOE4 | ϵ3/ϵ4 | 58 (10.64%) | 39 (14.77%) | 0.090 | 75 (18.70%) | <0.001 | 39 (19.12%) | 0.002 |
| ϵ4/ϵ4 | 1 (0.18%) | 2 (0.76%) | 1.000 | 2 (0.50%) | 0.577 | 2 (0.98%) | 0.182 | |
| APOE alleles | ||||||||
| ϵ2 | 105 (9.63%) | 55 (10.42%) | 0.621 | 57 (7.11%) | 0.052 | 31 (7.60%) | 0.222 | |
| ϵ3 | 919 (84.31%) | 428 (81.06%) | 0.101 | 663 (82.67%) | 0.340 | 330 (80.88%) | 0.112 | |
| ϵ4 | 66 (6.06%) | 45 (8.52%) | 0.066 | 82 (10.22%) | <0.001 | 47 (11.52%) | <0.001 | |
Data are presented as mean ± SD, or numbers (N) and percentage. p-value: comparison between T2DM/CVD/T2DM+CAD group and control group. Groups were compared using Student’s t-test or Wilcox test (for continuous variables) and Pearson’s χ2 test or Fisher’s exact test (for categorical variables). Bold values denote statistical significance at the p < 0.05 level.
APOE Genotype and Allele Frequencies of Subjects
Our extracted genomic DNA was of good quality with an OD260/OD280 ratio between 1.7 and 2.0. The genotype distributions of all included groups were in Hardy–Weinberg equilibrium (p > 0.05), exhibiting group representation. The most frequent genotype was ϵ3/ϵ3 in our subjects, followed by ϵ3/4 and ϵ2/3 genotypes, while ϵ2/2, ϵ2/4, and ϵ4/4 genotypes were the lowest, indicating that there was a significant difference in the distribution frequency of genotype ϵ3/4 among these groups. Compared with the control group, the genotype ratio of ϵ3/4 obviously increased in the CAD and T2DM+CAD groups (p < 0.001 and p = 0.002, Table 1 ). The differences in ϵ4 allele frequency distribution between the CAD and T2DM+CAD groups and the control group were considered statistically significant (both p < 0.001).
Association of APOE Polymorphism With Diseases
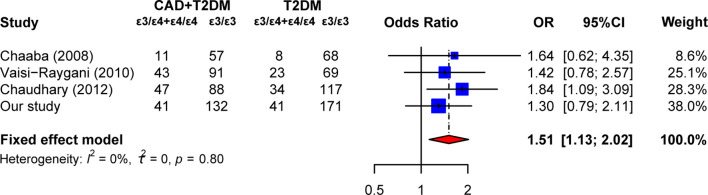
Logistic regression was performed to evaluated the correlation between APOE polymorphism and T2DM or CAD. As shown in Table 2 , when the ϵ3/3 genotype was used as the reference, the ϵ3/4 genotype had a significant increased risk of CAD and T2DM+CAD (adjusted OR = 1.90, 95% CI = 1.30–2.77 for CAD; adjusted OR = 1.95, 95% CI = 1.24–3.08 for T2DM+CAD). Furthermore, APOE allele ϵ4 appeared to increase the risk of developing CAD without or with T2DM, with adjusted OR of 1.72 (95% CI, 1.22–2.42) and 1.97 (95% CI, 1.32–2.93), respectively, compared with the allele ϵ3. However, allele ϵ4 was not found to be associated with the risk of CAD in T2DM (ϵ4 vs. ϵ3, OR = 1.36, 95% CI = 0.88–2.09). These results encouraged us to conduct a meta-analysis to explore the association between APOE allele ϵ4 and T2DM complicated with CAD. According to the above search criteria, three articles (14–16) and our study were included in the meta-analysis ( Supplementary Figure S1 ). Figure 2 displays that ϵ3/4 + ϵ4/4 genotype increased the risk of developing CAD in T2DM patients, with a pooled OR of 1.51 (95% CI, 1.13–2.02), compared to the ϵ3/3 genotype. Besides, no heterogeneity was detected, indicating that the pooled results of this meta-analysis were statistically steady and robust.
Table 2.
Comparison of APOE genotypes and alleles frequency between the control group and the case group.
| Variables | Control | T2DM | CAD | T2DM+CAD | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| APOE genotypes | n | n | OR (95% CI) | OR (95% CI) a | p a | n | OR (95% CI) | OR (95% CI) a | p a | n | OR (95% CI) | OR (95% CI) a | p a |
| ϵ2/ϵ2 | 6 | 3 | 1.13 (0.28-4.58) | 1.19 (0.29-4.83) | 0.813 | 1 | 0.24 (0.03-2.01) | 0.22 (0.03-1.82) | 0.158 | 0 | – | – | – |
| ϵ2/ϵ3 | 87 | 47 | 1.22 (0.82-1.82) | 1.22 (0.82-1.82) | 0.334 | 52 | 0.86 (0.59-1.26) | 0.86 (0.59-1.25) | 0.427 | 27 | 0.91 (0.57-1.46) | 0.90 (0.56-1.46) | 0.673 |
| ϵ2/ϵ4 | 6 | 2 | 0.75 (0.15-3.78) | 0.77 (0.15-3.86) | 0.748 | 3 | 0.72 (0.18-2.91) | 0.69 (0.17-2.81) | 0.605 | 4 | 1.96 (0.54-7.03) | 1.84 (0.5-6.72) | 0.357 |
| ϵ3/ϵ3 | 387 | 171 | Reference | Reference | – | 268 | Reference | Reference | – | 132 | Reference | Reference | – |
| ϵ3/ϵ4 | 58 | 39 | 1.52 (0.98-2.37) | 1.45 (0.93-2.27) | 0.102 | 75 | 1.87 (1.28-2.72) | 1.90 (1.30-2.77) | 0.001 | 39 | 1.97 (1.26-3.1) | 1.95 (1.24-3.08) | 0.004 |
| ϵ4/ϵ4 | 1 | 2 | 4.53 (0.41-50.25) | 5.18 (0.46-57.94) | 0.182 | 2 | 2.89 (0.26-32.01) | 2.23 (0.20-24.85) | 0.515 | 2 | 5.86 (0.53-65.19) | 6.27 (0.56-70.17) | 0.136 |
| Alleles | |||||||||||||
| ϵ2 | 105 | 55 | 1.13 (0.80-1.59) | 1.13 (0.80-1.60) | 0.484 | 57 | 0.75 (0.54-1.06) | 0.74 (0.53-1.04) | 0.081 | 31 | 0.82 (0.54-1.25) | 0.82 (0.53-1.25) | 0.346 |
| ϵ3 | 919 | 428 | Reference | Reference | – | 663 | Reference | Reference | – | 330 | Reference | Reference | – |
| ϵ4 | 66 | 45 | 1.46 (0.99-2.18) | 1.42 (0.96-2.12) | 0.082 | 82 | 1.72 (1.23-2.42) | 1.72 (1.22-2.42) | 0.002 | 47 | 1.98 (1.34-2.94) | 1.97 (1.32-2.93) | < 0.001 |
Adjusted for age and sex. Bold values denote statistical significance at the p < 0.05 level.
Figure 2.
Summary estimates of the association between the APOE ϵ4 mutation (ϵ3/ϵ4+ϵ4/ϵ4 vs. ϵ3/ϵ3) and the risk of coronary artery diseases in type 2 diabetes patients. Each study is displayed by a square whose center represents the odds ratio (OR), the area of the square is proportional to the weight of studies, and the horizontal line indicates the 95% confidence interval (CI).
Relationship Between APOE Polymorphism and Lipid Profiles
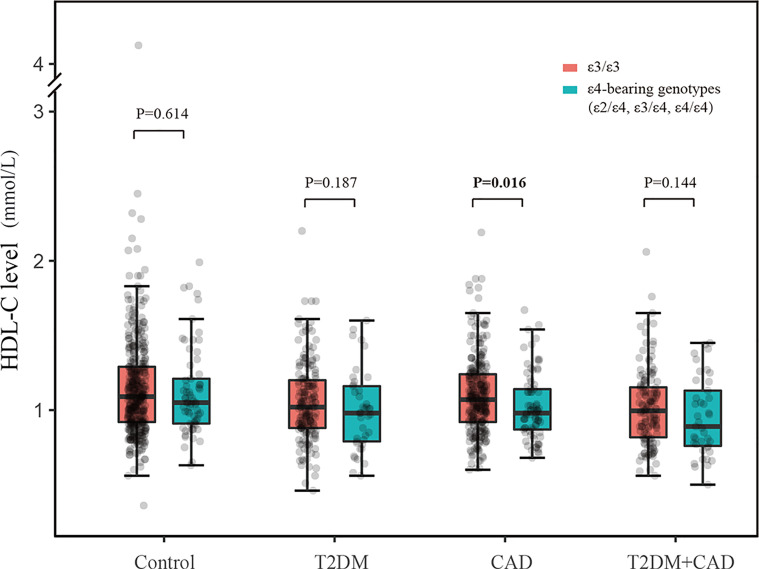
We analyzed the blood lipid profiles in subjects with different APOE genotypes. In the control and T2DM group, the levels of LDL-C showed significant difference among APOE2, APOE3, and APOE4 individuals; in the CAD group, APOE4 patients had lower levels of HDL-C than APOE3 and APOE2 patients. However, there was no significant difference in TG/HDL-C ratio across different APOE genotype groups ( Supplementary Table S1 ). The frequencies of the ϵ4 allele might contribute to the difference in APOE distribution between observation groups and control group. Therefore, we analyzed the correlation between ϵ4-bearing genotypes (ϵ2/ϵ4, ϵ3/ϵ4, and ϵ4/ϵ4) and plasma HDL-C expression. As shown in Figure 3 , ϵ3/ϵ3 genotype as a reference, ϵ4-bearing genotypes had significant decreased levels of HDL-C in CAD group (p = 0.016) but not in the other three groups. We also examined the association between ϵ4-bearing genotypes and TG/HDL-C ratio, and yet, the difference was not statistically significant ( Supplementary Figure S2 ).
Figure 3.
Correlation between plasma HDL-C level values and APOE ϵ4-bearing genotypes (ϵ2/ϵ4, ϵ3/ϵ4, and ϵ4/ϵ4).
Discussion
T2DM, a chronic condition disease, induced by a genetic predisposition together with environmental factors, is a well-established risk factor for CAD. T2DM and its related cardiovascular complications propose specific challenges at diverse stages of the life. APOE polymorphisms have been reported to significantly associate with risk for T2DM and CAD, which were considered as the most influential genetic risk factors. Here, we carried out several experiments to evaluate the association between the APOE ϵ2/ϵ3/ϵ4 polymorphisms with the risk of T2DM and CAD. When combined with the analysis the polymorphism of APOE and blood lipid levels, these results provided new understanding on the correlation between APOE gene and T2DM patients with CAD.
Our study provided evidence for the significant correlation between APOE ϵ3/ϵ4 genotype and an elevated risk of CAD without or with T2DM. After adjusting for age and sex, logistic regression analysis showed that ϵ4 allele increased the risk of CAD by 1.72 times, compared with ϵ3 allele. Besides, T2DM patients carrying ϵ4 allele had 1.97-fold higher risk of CAD as compared to the controls. Our data indicated that ϵ4 allele was an independent risk factor for CAD but not for T2DM. Previous studies have investigated the probable associations between APOE polymorphisms and patients with T2DM or CAD. Chaudhary et al. reported that the ϵ4 allele was significantly higher in both T2DM and CAD as compared with controls (16). The independent predictor of individuals carrying ϵ4 allele remained significantly associated with both CAD (adjusted OR = 2.32, 95% CI = 1.17–4.61) and T2DM (adjusted OR = 2.04, 95% CI = 1.07–3.86). El−Lebedy et al. found that the frequencies of ϵ3/ϵ4 genotype and ϵ4 allele were increased in both T2DM patients and cardiovascular disease (CVD) patients as compared with controls but were significant only in CVD patients (17). Diabetic patients who carried ϵ3/ϵ4 genotype had 2.4-fold increased risk of developing CVD (95% CI, 1.14–5.19), and the ϵ4 allele was associated with 2.23-fold higher CVD risk (95% CI, 1.09–4.59). A recent meta-analysis including 13 studies provided evidence that there were significant associations between ϵ4 allele and the risk of CAD in patients with T2DM (ϵ3/ϵ4 vs. ϵ3/ϵ3, OR = 1.69, 95% CI = 1.38–2.08; ϵ4/ϵ4 vs. ϵ3/ϵ3, OR = 2.72, 95% CI = 1.61–4.60) (13). Combined with our own data and setting of strict inclusion and exclusion criteria, our meta-analysis found that ϵ4 mutation could elevate the risk of CAD in patients with T2DM. Therefore, we may conclude that ϵ3/4 genotypes and ϵ4 allele of APOE contributed to CAD in T2DM patients.
One outcome from this work is the levels of HDL-C in CAD patients with ϵ4-bearing genotypes (ϵ2/ϵ4, ϵ3/ϵ4, and ϵ4/ϵ4) were lower than patients with ϵ3/ϵ3 genotype. It was well-known that the HDL-C levels were important factors of blood lipid levels. The relationship between ϵ4 allele and lipid profile remained controversial. Li et al. found that the APOE ϵ4 carrier had a lower HDL-C than the ϵ2 allele but not for ϵ3 allele in a Chinese population of CAD (18). In a recent study on Kashmiri population, the CAD patients carrying ϵ4 allele had significantly lower HDL-C levels (19). Chaaba et al. found that the ϵ4 allele was only associated with elevated LDL-C concentration and with CAD in type 2 diabetic men in Tunisian population, showing that gender interacted with the effects of APOE polymorphism (14). Therefore, these inconsistent findings might be complicated by considerable differences in the allele frequency distributions among different ethnic populations.
Current evidence showed that APOE is a versatile glycoprotein that plays a central role in lipoprotein metabolism (20). Although the three APOE isoforms differ in only one or two amino acids, these slight changes affect the structure and alter the affinity to lipoproteins. APOE3 binds preferentially to HDL, whereas APOE4 shows an enhanced lipid bind ability of VLDL particles, which impairs their lipolytic processing in the circulation, resulting in a more pro-atherogenic lipoprotein–cholesterol distribution. Therefore, abnormalities of lipoprotein metabolism may partly explain that the plasma HDL-C levels were lowest in APOE4 patients compared to APOE3 and APOE2 patients in CAD group. Besides, excessive amounts of circulant lipids may affect systemic inflammation and insulin resistance (IR) (21, 22). Previous studies reported the TG/HDL-C ratio as predictor of IR (22). A higher ratio indicated a large amount of circulant lipids. Our study showed that the T2DM+CAD group had the highest TG/HDL-C ratio than the control, T2DM, and CAD groups. Then, we assessed whether the APOE isoforms were associated with IR, but no association was observed. Confirming results of previous studies, the relationship between APOE allele frequencies and IR was controversial (23–25).
There are several limitations that should be pointed out as follows. First, this case–control study was hospital based, and the selection bias was inevitable. Therefore, we adjusted for potential confounding factors such as age and sex to minimize the bias in logistic regression. Second, adult populations showed specific mutations not only owing to genetic background. Life habits, diets, climate, pollution, and even pandemic might influence it. As this study recruited subjects aged 18 years or above, subgroup analysis for age (children and adults) could not be performed. Meanwhile, we did not have the detailed information of some risk factors (smoking, diet, and physical activity) in the development of CAD and T2DM, and the gene–environment interactions analysis was not conducted. Third, there was no significant association between APOE alleles and T2DM in our study, which was not consistent with previous study (26, 27), and the results from our single-center study required external validation in other training programs. A final comment on the low number of individuals in some subgroups (control, T2DM, CAD, and T2DM+CAD for ϵ2/ϵ2, ϵ2/ϵ4 and ϵ4/ϵ4) should be taken up with care and caution.
In conclusion, although the APOE ϵ4 allele was not found to be associated with T2DM, it increased the risk of CAD and related to the development of T2DM with CAD. Different APOE isoforms were also linked to variations in lipid and lipoprotein levels in circulation.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethical Committee of The First Affiliated Hospital of Nanjing Medical University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
TX and LW designed the research. LW, YZ, HZ, GR, PH, FW, and TX conducted research. LW and YZ analyzed data. LW wrote the initial draft of the manuscript. FW and TX revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.838547/full#supplementary-material
References
- 1. Zheng Y, Ley SH, Hu FB. Global Aetiology and Epidemiology of Type 2 Diabetes Mellitus and Its Complications. Nat Rev Endocrinol (2018) 14(2):88–98. doi: 10.1038/nrendo.2017.151 [DOI] [PubMed] [Google Scholar]
- 2. Emamalipour M, Seidi K, Jahanban-Esfahlan A, Jahanban-Esfahlan R. Implications of Resistin in Type 2 Diabetes Mellitus and Coronary Artery Disease: Impairing Insulin Function and Inducing Pro-Inflammatory Cytokines. J Cell Physiol (2019) 234(12):21758–69. doi: 10.1002/jcp.28913 [DOI] [PubMed] [Google Scholar]
- 3. Dunlay SM, Givertz MM, Aguilar D, Allen LA, Chan M, Desai AS, et al. Type 2 Diabetes Mellitus and Heart Failure: A Scientific Statement From the American Heart Association and the Heart Failure Society of America: This Statement Does Not Represent an Update of the 2017 ACC/AHA/HFSA Heart Failure Guideline Update. Circulation (2019) 140(7):e294–324. doi: 10.1161/CIR.0000000000000691 [DOI] [PubMed] [Google Scholar]
- 4. Huth C, Bauer A, Zierer A, Sudduth-Klinger J, Meisinger C, Roden M, et al. Biomarker-Defined Pathways for Incident Type 2 Diabetes and Coronary Heart Disease-A Comparison in the MONICA/KORA Study. Cardiovasc Diabetol (2020) 19(1):32. doi: 10.1186/s12933-020-01003-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marais AD. Apolipoprotein E in Lipoprotein Metabolism, Health and Cardiovascular Disease. Pathology (2019) 51(2):165–76. doi: 10.1016/j.pathol.2018.11.002 [DOI] [PubMed] [Google Scholar]
- 6. Singh PP, Singh M, Mastana SS. APOE Distribution in World Populations With New Data From India and the UK. Ann Hum Biol (2006) 33(3):279–308. doi: 10.1080/03014460600594513 [DOI] [PubMed] [Google Scholar]
- 7. Eichner JE, Dunn ST, Perveen G, Thompson DM, Stewart KE, Stroehla BC. Apolipoprotein E Polymorphism and Cardiovascular Disease: A HuGE Review. Am J Epidemiol (2002) 155(6):487–95. doi: 10.1093/aje/155.6.487 [DOI] [PubMed] [Google Scholar]
- 8. Vaisi-Raygani A, Rahimi Z, Nomani H, Tavilani H, Pourmotabbed T. The Presence of Apolipoprotein Epsilon4 and Epsilon2 Alleles Augments the Risk of Coronary Artery Disease in Type 2 Diabetic Patients. Clin Biochem (2007) 40(15):1150–6. doi: 10.1016/j.clinbiochem.2007.06.010 [DOI] [PubMed] [Google Scholar]
- 9. Knouff C, Hinsdale ME, Mezdour H, Altenburg MK, Watanabe M, Quarfordt SH, et al. Apo E Structure Determines VLDL Clearance and Atherosclerosis Risk in Mice. J Clin Invest (1999) 103(11):1579–86. doi: 10.1172/JCI6172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mahley RW. Apolipoprotein E: From Cardiovascular Disease to Neurodegenerative Disorders. J Mol Med (Berlin Germany) (2016) 94(7):739–46. doi: 10.1007/s00109-016-1427-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tachibana M, Holm M-L, Liu C-C, Shinohara M, Aikawa T, Oue H, et al. APOE4-Mediated Amyloid-β Pathology Depends on Its Neuronal Receptor LRP1. J Clin Invest (2019) 129(3):1272–7. doi: 10.1172/JCI124853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Larifla L, Armand C, Bangou J, Blanchet-Deverly A, Numeric P, Fonteau C, et al. Association of APOE Gene Polymorphism With Lipid Profile and Coronary Artery Disease in Afro-Caribbeans. PloS One (2017) 12(7):e0181620. doi: 10.1371/journal.pone.0181620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Luo J-Q, Ren H, Banh HL, Liu M-Z, Xu P, Fang P-F, et al. The Associations Between Apolipoprotein E Gene Epsilon2/Epsilon3/Epsilon4 Polymorphisms and the Risk of Coronary Artery Disease in Patients With Type 2 Diabetes Mellitus. Front Physiol (2017) 8:1031:1031. doi: 10.3389/fphys.2017.01031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chaaba R, Attia N, Hammami S, Smaoui M, Ben Hamda K, Mahjoub S, et al. Association Between Apolipoprotein E Polymorphism, Lipids, and Coronary Artery Disease in Tunisian Type 2 Diabetes. J Clin Lipidol (2008) 2(5):360–4. doi: 10.1016/j.jacl.2008.08.441 [DOI] [PubMed] [Google Scholar]
- 15. Vaisi-Raygani A, Rahimi Z, Tavilani H, Pourmotabbed T. Butyrylcholinesterase K Variant and the APOE-Epsilon 4 Allele Work in Synergy to Increase the Risk of Coronary Artery Disease Especially in Diabetic Patients. Mol Biol Rep (2010) 37(4):2083–91. doi: 10.1007/s11033-009-9666-4 [DOI] [PubMed] [Google Scholar]
- 16. Chaudhary R, Likidlilid A, Peerapatdit T, Tresukosol D, Srisuma S, Ratanamaneechat S, et al. Apolipoprotein E Gene Polymorphism: Effects on Plasma Lipids and Risk of Type 2 Diabetes and Coronary Artery Disease. Cardiovasc Diabetol (2012) 11:36. doi: 10.1186/1475-2840-11-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. El-Lebedy D, Raslan HM, Mohammed AM. Apolipoprotein E Gene Polymorphism and Risk of Type 2 Diabetes and Cardiovascular Disease. Cardiovasc Diabetol (2016) 15:12. doi: 10.1186/s12933-016-0329-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li S-S, Yang J, Li L-S, Wang H-C. Apolipoprotein E Polymorphism and the Characteristics of Diseased Vessels in Male Chinese Patients With Angiographic Coronary Artery Disease: A Case-Case Study. Clin Cardiol (2010) 33(6):E30–4. doi: 10.1002/clc.20703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Afroze D, Yousuf A, Tramboo NA, Shah ZA, Ahmad A. ApoE Gene Polymorphism and Its Relationship With Coronary Artery Disease in Ethnic Kashmiri Population. Clin Exp Med (2016) 16(4):551–6. doi: 10.1007/s10238-015-0389-7 [DOI] [PubMed] [Google Scholar]
- 20. Phillips MC. Apolipoprotein E Isoforms and Lipoprotein Metabolism. IUBMB Life (2014) 66(9):616–23. doi: 10.1002/iub.1314 [DOI] [PubMed] [Google Scholar]
- 21. Wang KY, Tanimoto A, Guo X, Yamada S, Shimajiri S, Murata Y, et al. Histamine Deficiency Decreases Atherosclerosis and Inflammatory Response in Apolipoprotein E Knockout Mice Independently of Serum Cholesterol Level. Arterioscler Thromb Vasc Biol (2011) 31(4):800–7. doi: 10.1161/atvbaha.110.215228 [DOI] [PubMed] [Google Scholar]
- 22. Khedr D, Hafez M, Lumpuy-Castillo J, Emam S, Abdel-Massih A, Elmougy F, et al. Lipid Biomarkers as Predictors of Diastolic Dysfunction in Diabetes With Poor Glycemic Control. Int J Mol Sci (2020) 21(14):5079. doi: 10.3390/ijms21145079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ragogna F, Lattuada G, Ruotolo G, Luzi L, Perseghin G. Lack of Association of apoE ϵ4 Allele With Insulin Resistance. Acta Diabetol (2012) 49(1):25–32. doi: 10.1007/s00592-011-0255-3 [DOI] [PubMed] [Google Scholar]
- 24. Griffin BA, Walker CG, Jebb SA, Moore C, Frost GS, Goff L, et al. APOE4 Genotype Exerts Greater Benefit in Lowering Plasma Cholesterol and Apolipoprotein B Than Wild Type (E3/E3), After Replacement of Dietary Saturated Fats With Low Glycaemic Index Carbohydrates. Nutrients (2018) 10(10):1524. doi: 10.3390/nu10101524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gonzalez-Aldaco K, Roman S, Torres-Reyes LA, Panduro A. Association of Apolipoprotein E2 Allele With Insulin Resistance and Risk of Type 2 Diabetes Mellitus Among an Admixed Population of Mexico. Diabetes Metab Syndr Obes (2020) 13:3527–34. doi: 10.2147/dmso.S268329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alharbi KK, Khan IA, Syed R. Association of Apolipoprotein E Polymorphism With Type 2 Diabetes Mellitus in a Saudi Population. DNA Cell Biol (2014) 33(9):637–41. doi: 10.1089/dna.2014.2461 [DOI] [PubMed] [Google Scholar]
- 27. Chen DW, Shi JK, Li Y, Yang Y, Ren SP. Association Between ApoE Polymorphism and Type 2 Diabetes: A Meta-Analysis of 59 Studies. Biomed Environ Sci BES (2019) 32(11):823–38. doi: 10.3967/bes2019.104 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.