Abstract
Background and Aims
Intestinal fibrosis is a common complication of Crohn’s disease [CD]. It is characterised by an accumulation of fibroblasts differentiating into myofibroblasts secreting excessive extracellular matrix. The potential role of the intestinal epithelium in this fibrotic process remains poorly defined.
Methods
We performed a pilot proteomic study comparing the proteome of surface epithelium, isolated by laser-capture microdissection, in normal and fibrotic zones of resected ileal CD strictures [13 zones collected in five patients]. Proteins of interests were validated by immunohistochemistry [IHC] in ileal and colonic samples of stricturing CD [n = 44], pure inflammatory CD [n = 29], and control [n = 40] subjects. The pro-fibrotic role of one selected epithelial protein was investigated through in-vitro experiments using HT-29 epithelial cells and a CCD-18Co fibroblast to myofibroblast differentiation model.
Results
Proteomic study revealed an endoplasmic reticulum [ER] stress proteins increase in the epithelium of CD ileal fibrotic strictures, including anterior gradient protein 2 homologue [AGR2] and binding-immunoglobulin protein [BiP]. This was confirmed by IHC. In HT-29 cells, tunicamycin-induced ER stress triggered AGR2 intracellular expression and its secretion. Supernatant of these HT-29 cells, pre-conditioned by tunicamycin, led to a myofibroblastic differentiation when applied on CCD-18Co fibroblasts. By using recombinant protein and blocking agent for AGR2, we demonstrated that the secretion of this protein by epithelial cells can play a role in the myofibroblastic differentiation.
Conclusions
The development of CD fibrotic strictures could involve epithelial ER stress and particularly the secretion of AGR2.
Keywords: CD fibrosis, ER stress, anterior gradient protein 2 homologue
1. Introduction
Intestinal fibrosis is a common and potentially serious complication of Crohn’s disease [CD], arising in a large proportion of patients.1 This is a consequence of local chronic inflammation, which is required to initiate the fibrotic process, but subsequently plays a minor role in its perpetuation and worsening.2,3 To date, no CD anti-inflammatory drugs nor any other treatments exist to reverse or prevent CD-associated strictures, and alternative therapeutic approaches [such as balloon dilatation, strictureplasty, or bowel resection] are frequently required.3–6 Thus, there is an unmet medical need for anti-fibrotic therapies targeting pathways involved in the pathophysiology of intestinal fibrosis.
Intestinal fibrosis is characterised by an excessive deposition of extracellular matrix [ECM] by activated myofibroblasts in the submucosa and muscle layers, progressively leading to bowel strictures. One of the main actors of this pathophysiological process is the fibroblast which accumulates and undergoes differentiation into these ECM-producing activated myofibroblasts, which are alpha-smooth muscle actin [α-SMA]-positive cells.7,8 This myofibroblastic differentiation is elicited by a whole series of factors, among which transforming growth factor-β1 [TGF-β1] seems to play a key role.9 Whereas in other organs, the source of ECM-producing myofibroblasts is limited to a few cell types, in the gut, multiple cells may become activated myofibroblasts,10–14 including epithelial cells. These cells can trans-differentiate into fibroblasts via epithelial-to-mesenchymal transition [EMT]15,16 characterised by a progressive loss of typical epithelial hallmarks [including E-cadherin, cytokeratins] together with the acquisition of typical mesenchymal cell markers [such as vimentin, α-SMA]. This newly acquired mesenchymal phenotype is characterised by the acquisition of a fibroblast-like elongated morphology, loss of cell polarity, and enhanced invasive capacity allowing them to migrate into the submucosa and initiate secretion of ECM.17
Next to this EMT, the role played by the surface epithelium in CD intestinal fibrosis has been insufficiently studied. The aim of our study was to highlight potential epithelial proteins involved in CD fibrosis. In this context, we performed a pilot proteomic study to identify the surface epithelium proteome changes in CD strictures. From this experiment, proteins of interest were selected and evaluated by immunohistochemistry [IHC]. Finally, the role of these proteins in intestinal fibrosis was evaluated through functional experiments performed in an in-vitro model.
2. Materials and Methods
2.1. Patient enrolment and selection
This project was approved by the Ethics Committee of the University Hospital of Liège in 2014 [reference: B707201421402]. Selected patients were searched retrospectively through our pathology database from 2009 to 2017, and their formalin-fixed paraffin-embedded [FFPE] tissues were obtained from our institution biobank. We first reviewed CD patients who underwent surgery and we selected patients with evidence of strictures and fibrosis in medical or histological records [n = 44]. Second, we searched for CD patients with no evidence of fibrostenosing disease [pure inflammatory CD, based on endoscopic and imaging evaluations] and who were biopsied during endoscopy in in an inflammatory area or in normal mucosa [normal CD tissues] [n = 29]. Third, non inflammatory bowel disease [IBD] patients who underwent surgery between 2012 and 2017 were included for analysis of ileal and colonic normal tissues extracted at the surgical margin [n = 40]. Whereas for the pilot proteomic study we selected five patients out of the 44 stricturing CD patients, immunochemistry experiments were performed on the 73 CD and the 40 control subjects.
2.2. Histopathological evaluation and classification of tissues
All FFPE tissues were graded for inflammation and fibrosis. Histopathological inflammation evaluation of the tissue specimens was done on haematoxylin and eosin [H&E]-stained sections.18 An expert gastrointestinal pathologist [NB] graded separately chronic and acute inflammation, according to the neutrophil and lymphoplasmacytic infiltrates, respectively, using a classical four grades scale ranging from none [N], mild [I1], moderate [I2], to severe [I3].19–22 Crypt micro-abscesses and cryptitis were considered as hallmarks of the severe acute inflammation grade as acknowledged by others.23–25 Histopathological intestinal fibrosis evaluation of tissue specimens was done after Masson’s trichrome staining [Merck Millipore, GE] performed according to the manufacturer’s recommendations. A four grades scale was used as previously performed,26–28 ranging from none [N; no architectural distortion, no abnormal MT staining], mild [F1; no architectural distortion, mild abnormal MT staining in mucosa/submucosa], moderate [F2; substantial abnormal mucosal/submucosal MT staining with modest distortion of architecture], to severe [F3; transmural fibrosis with abnormal MT staining in all layers, transmural architectural distortion].
2.3. Label-free proteomic study
2.3.1. Patients and tissue selection
For the pilot proteomic experiment we selected five CD patients who had, in the same ileal stricture surgical resection sample, one area with high grade of fibrosis [F2-3], one area with low grade of fibrosis [F1] and, except for two patients, one area with normal aspect [without any sign of fibrosis] [N], allowing a paired comparison of the surface epithelium proteomic profiles according to the fibrosis grade. Thus, 13 samples have been analysed for the proteomic study.
2.3.2. Laser capture microdissection, samples preparation and proteomics by LC-MS/MS
Before the proteomic analysis we collected, by laser capture microdissection, the surface epithelial cells [~20 000] of the different areas selected for each patient’s tissues [five F2-3, five F1, and three N] using a Leica LMD 7000 [Leica Microsystem, GmBh, GE] and treated them for protein extraction according to a standardised procedure29 [see Supplementary data, available at ECCO-JCC online, for details].
The samples were analysed by ultra-performance liquid chromatography/electrospray ionisation tandem mass spectrometry [UPLC-ESI-MS/MS]. This system consists of a 2D nanoAcquity chromatography [Waters] coupled online with a Q Exactive™ Plus Hybrid Quadrupole-Orbitrap™ mass spectrometer [Thermo Fisher Scientific], equipped with a nano-electrospray source operated in positive ion mode [see Supplementary data for details].
2.3.3. Proteins identification and differential analysis
The analysis of the raw spectra was performed using MaxQuant software 1.5.5.1 for proteins identification and label-free quantification [LFQ] [see Supplementary data for search parameters].30 Raw data files have been deposited at the ProteomeXchange Consortium via the PRIDE31 partner repository [http://proteomecentral.proteomexchange.org, identifier PXD022214]. We used Perseus 1.6.0.7 for differential analysis with log2 transformed LFQ values.32,33 Only proteins detected and quantified in at least three out of five replicates for fibrotic tissues, and in at least two out of three replicates for normal tissues, were included in the differential analysis.
First, four comparisons were applied to compare the epithelial proteome in the different areas [N, F1, F2-3]: N versus F1 versus F2-3, N versus F1, N versus F2-3, and F1 versus F2-3. Second, we searched for the ‘absent’ proteins [below the limit of quantitation] in one specific area and ‘present’ in at least three out of five or two out of three samples of the two other tissue types.
2.3.4. Gene ontology, pathways enrichment analyses, functional annotation, and binding partner identifications
Beside differential analysis, the proteins found significantly increased or ‘absent’ [or inversely decreased or ‘present’] in one or several groups in the different comparisons were submitted to gene ontology using different tools: DAVIDTM [bioinformatics version 6.8; https://david.ncifcrf.gov/] for functional enrichment and annotations,34 STRING [consortium 2019; https://string-db.org/] for network and cluster analysis, and the BioGRID database [https://orcs.thebiogrid.org/] for identification of the gene interactors of the proteins of interest [interactors gene list downloaded in November 2019].
2.4. Immunohistochemical confirmation study
2.4.1. IHC characterisation
Due to the limited number of samples presenting the same inflammation and fibrosis grades, we decided to group tissue sections combining all positive inflammatory and/or fibrosis grades together in four groups: normal [N], purely inflammatory [I1-3 referenced as I], fibro-inflammatory [I1-3F1-3 referenced as IF], and purely fibrotic [F1-3 referenced as F].
FFPE tissue sections were treated as previously described35 [see Supplementary data for details]. Three independent observers [M-AM, CM, and CS], scored IHC tissue sections [at least three fields by section], blinded to the degree of fibrosis and inflammation. AGR2 and BiP immuno-stained sections were scored positive if epithelial cells showed specific staining in the cytoplasm. A semi-quantitative IHC score was determined by estimating the global signal intensity detected in the surface and the crypts epithelium. The following staining scores were used: 0 [none], 1 [weak], 2 [medium], 3 [strong], and 4 [very strong]. The final semi-quantitative score attributed for the surface and crypt epithelium for each tissue section was the average value of the three observers in the different fields [n = 3] analysed.35 When for a given segment of intestine of an individual patient, several equal inflammation and fibrosis grades sections of tissue were available, only the averaged IHC score obtained for these redundant tissue sections was considered.
2.5. Functional study
Supplementary Figure 1, available at ECCO-JCC online, details the methodology used for the functional assays.
2.5.1. Cell lines and culture conditions
The human colorectal epithelial cell line HT-29 [ATCC® HTB-38™] and the normal human intestinal fibroblast cell line CCD-18Co [ATCC® CRL-1459™] were grown in high-glucose Dulbecco’s modified Eagle’s medium [DMEM] [Gibco; Thermo Fisher Scientific, USA] supplemented with 10% foetal bovine serum [FBS] [R&D systems, Minneapolis, USA] and 1% penicillin/streptomycin [Sigma, St Louis, USA], and placed at 37°C in a 5% CO2 humidified atmosphere.
2.5.2. Induction of ER stress in HT-29 cells
HT-29 cells were seeded in six-well plates [3 x 106 cells/well] and they were grown during 48 h before treatments. Then, ER stress induction was performed using 2 to 10 µg/mL of Tunicamycin [Tm, solubilised in DMSO] [Sigma] and Tm treatment was applied for 2, 4, 6, 8, 18, and 24 h at the selected dose. DMSO was used as control treatment [at the same final concentration as in the Tm-treated condition]. To assess if ER stress persists after stimulus [Tm] removal, HT-29 supernatant was changed after 18 h of Tm treatment [10 µg/mL] for the stimulated conditions and the relevant control conditions [Supplementary Figure 1A]. Cell extracts and matching supernatants were collected 2, 4, 8, 24, and 32 h after the media change.
2.5.3. Fibroblast to myofibroblast differentiation of the CCD-18Co
CCD-18Co cells were seeded in six-well plates [1 x 106 cells/well] and grown for 48 h before treatments. ER stress was induced on CCD-18Co with Tm [10 µg/mL] and DMSO was used as control condition [at the same final concentration] to monitor the response of CCD-18 to Tm.
The CCD-18Co differentiation-positive control was obtained using a 48 h treatment with 10 ng/mL of TGF-β1 [R&D systems] added in the media of CCD-18Co [after 24 h of serum starvation and addition of 1% FBS at the time of stimulation].
The supernatants of HT-29 cells, pre-conditioned or not by Tm [thus, subject or not to a transient ER stress] and collected 8, 24, and 32 h after the media change, were applied on CCD-18Co, as inducing conditions, for further 48 h of incubation. The HT-29 supernatant was used without any freezing cycle [Supplementary Figure 1B].
The capacity of recombinant AGR2 [rAGR2] to induce fibroblast to myofibroblast differentiation was investigated by application of 40 ng/mL of rAGR2 [Bio-Connect BV, OPCD01192] in the media of CCD-18Co [after 24 h of serum starvation and addition of 1% FBS at the time of stimulation], the control being the same condition without rAGR2 supplementation. The impact of AGR2 blockade, using an anti-AGR2 antibody [rabbit, 1.936 µg/mL, ab76473, Abcam], supplemented to the conditions with rAGR2 or to the HT-29 supernatant pre-conditioned by Tm, was evaluated [Supplementary Figure 1C].
The HT-29 and CCD-18Co cells were harvested and treated for either total protein extracts [stored at -20°C] or for RNA extractions [stored at -80°C]. HT-29 supernatants collected for analysis were stored at -20°C. Methods used for western blot, RNA extraction and RT-qPCR as well as immunofluorescence analyses are detailed in the Supplementary material.
2.6. Statistics
In the proteomic experiment, we performed unpaired analyses to compare N versus F1 versus F2-3 [ANOVA test] and N versus F1 or F2-3 [Welch’s t test], and a paired analysis to compare F1 versus F2-3 [t test] and determine the p-value associated with the difference in abundance between areas [significant if p ≤0.05].
In the immunohistochemical experiments, the statistics of histological data were performed using GraphPad Prism 5. IHC scores of the proteins of interest according to inflammation and fibrosis grades [N, I, IF, F] were compared with ANOVA. When appropriate, Dunn’s post hoc tests were conducted. Correlations between the protein of interest IHC score and inflammation [N, I1, I2, I3] or fibrosis grades [N, F1, F2, F3] were performed using the Spearman correlation analyses.
In the functional experiment, one-way analysis of variance [ANOVA] test was used to assess differences between conditions in real-time quantitative polymerase chain reaction [RT-qPCR], as well as the Turkey’s multiple comparison test when appropriate. All results were considered significant if associated with a p-value ≤0.05.
3. Results
3.1. Patients’ characteristics
The clinical information of the 73 CD studied patients [44 stricturing CD and 29 pure inflammatory CD] are detailed Table 1. The five patients selected for the proteomic study came from the 44 stricturing CD patients also studied in the IHC study. The 40 non-IBD controls included 50% of males, with a median age of 71 years [range: 32–92 years] and underwent surgery for colonic adenocarcinoma [n = 14], diverticular diseases [n = 23], volvulus [n = 2], and colonic perforation [n = 1].
Table 1.
Clinical information of the studied CD patients.
| CD patients [n = 73] | |||
|---|---|---|---|
| Fibrostenosing CD patients [n = 44] | Non fibrostenosing CD patients [n = 29] | ||
| Age [median, range] years | 31 [17–72] | 43 [21–68] | |
| Male gender [n, %] | 22; 50.0 | 12; 41.4 | |
| Disease duration [median, range] years | 8.7 [0.2 - 51.5] | 10.19 [0.07–40.42] | |
| CD treatment [*stopped before surgery] | Infliximab* [n] | 5 | 3 |
| Adalimumab* [n] | 15 | 6 | |
| Vedolizumab [n] | 1 | 0 | |
| Azathioprine [n] | 10 | 5 | |
| Mercaptopurine [n] | 3 | 5 | |
| Methotrexate [n] | 2 | 2 | |
| Corticoids [n] | 7 | 3 | |
| 5ASA [n] | 0 | 5 | |
| None [n] | 11 | 9 | |
| Smoking | Active smokers [n] | 16 | 11 |
| Former smokers [n] | 8 | 6 | |
| Non-smoker [n] | 19 | 11 | |
| Unknown [n] | 1 | 1 | |
The five patients included in the proteomic study came from the 44 fibrostenosing CD patients of the IHC study (60% males; median [range] age: 33 years [22–41]; median [range] disease duration: 8.2 years [4.4–12.3]; 60% active smokers; one patient was treated by infliximab, three patients were treated by adalimumab [in monotherapy for one patient and in a combination therapy with azathioprine for one and with corticoids for the another], and the last patient was treated by corticoids alone.
*In a combination therapy with immunomodulator or corticoids for some patients.
CD, Crohn’s disease; 5-ASA, 5-aminosalicylate; IHC, immunohistochemical.
3.2. Proteomic study
3.2.1. Label-free proteomic analysis
Proteomics identified and quantified 1287 proteins [Supplementary Table 1, available at ECCO-JCC online,] among which 1153 were detected in at least three out of five [for F1/F2-3] or two out of three [for N] patient samples and were subjected to the differential analysis. Supplementary Table 2, available at ECCO-JCC online includes these proteins, their relative distributions, and statistics. We found 203 proteins with significant different abundances in the four comparisons applied. Supplementary Figure 2, available at ECCO-JCC online, shows a Venn diagram with these different comparisons and Supplementary Table 3, available at ECCO-JCC online, shows results of these differences. Supplementary Table 4, available at ECCO-JCC online, details results of the second analysis concerning the research of absent proteins in one specific area and detected in at least three out of five or two out of three samples of the two other tissue types.
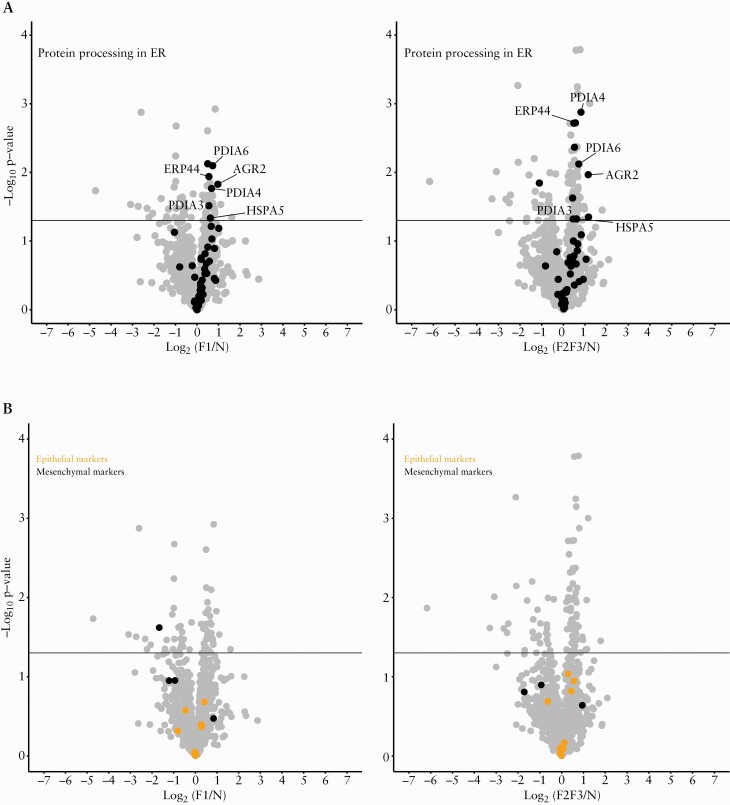
When classifying proteins according to the p-value of the ANOVA test comparing normal ileum [N] with ileum with low-grade [F1] or high-grade [F2-3] of fibrosis, we found five proteins disulphide isomerase [PDIs] among the 25 most differentially expressed proteins according to the fibrosis grade [Table 2], the first being anterior gradient protein 2 homologue [AGR2]. Binding-immunoglobulin protein [BiP], a well-known other endoplasmic reticulum [ER] stress marker, followed the distribution of AGR2 in all performed comparisons. Figure 1A illustrate the volcano plots of proteins involved in ER stress, comparing N versus F1 and N versus F2-3 tissues.
Table 2.
Proteomic results: identification and quantification of the 25 most differentially expressed proteins according to the fibrosis grade, classified according to the p-value of the ANOVA test comparing normal ileum [N] with ileum with low grade [F1] or high grade [F2-F3] of fibrosis with the results of the differential analyses for other comparisons
| Protein IDs | Majority protein IDs | Protein names | Gene names | Peptides | Razor + unique peptides | Unique peptides | MS/MS count | N vs F1 vs F2-3 ANOVA test p-value | N vs F1 Welch’s t test p-value | N vs F2-3 Welch’s t test p-value | F1 vs F2-3 Student’s t test p-value | Gene names for BiP [HSPA5] interactors [n = 123] | Gene names for AGR2 interactors [n = 435] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| O95994 | O95994 | Anterior gradient protein 2 homologue | AGR2 | 10 | 10 | 9 | 470 | 0.0001029 | 0.0148937 | 0.0107909 | 0.231551 | AGR2 | AGR2 |
| Q02818 | Q02818 | Nucleobindin-1 | NUCB1 | 14 | 14 | 14 | 96 | 0.0021707 | 0.0075531 | 0.0001661 | 0.546602 | NUCB1 | |
| P62906 | P62906 | 60S ribosomal protein L10a | RPL10A | 4 | 4 | 4 | 63 | 0.0053065 | 0.0217258 | 0.0007105 | 0.154768 | RPL10A | |
| Q15084 | Q15084 | Protein disulphide-isomerase A6 | PDIA6 | 12 | 12 | 12 | 264 | 0.006122 | 0.0079761 | 0.007557 | 0.8676289 | PDIA6 | PDIA6 |
| Q8N4A0 | Q8N4A0 | Polypeptide N-acetylgalactosaminyltransferase 4 | GALNT4 | 3 | 3 | 3 | 16 | 0.0070341 | 0.0156691 | 0.0045735 | 0.7709886 | ||
| P50914 | P50914 | 60S ribosomal protein L14 | RPL14 | 3 | 3 | 3 | 49 | 0.0070888 | 0.0141703 | 0.0005672 | 0.4814326 | RPL14 | |
| P21399 | P21399 | Cytoplasmic aconitate hydratase | ACO1 | 7 | 7 | 7 | 25 | 0.0087072 | 0.2308691 | 1.0000001 | 1 | ACO1 | |
| Q13510 | Q13510 | Acid ceramidase;acid ceramidase subunit alpha;acid ceramidase subunit beta | ASAH1 | 2 | 2 | 2 | 26 | 0.0094003 | 0.0021152 | 0.0101158 | 0.1999859 | ||
| Q13228 | Q13228 | Selenium-binding protein 1 | SELENBP1 | 14 | 14 | 14 | 94 | 0.0098878 | 0.0451073 | 0.0277982 | 0.4911797 | SELENBP1 | |
| P48735 | P48735 | Isocitrate dehydrogenase [NADP], mitochondrial | IDH2 | 16 | 16 | 15 | 378 | 0.0100318 | 0.0934848 | 0.0898935 | 0.9410172 | IDH2 | |
| P35606 | P35606 | Coatomer subunit beta | COPB2 | 13 | 13 | 13 | 89 | 0.0101221 | 0.0587076 | 0.008697 | 0.1771882 | COPB2 | |
| P63000;P15153;P60763 | P63000;P15153;P60763 | Ras-related C3 botulinum toxin substrate 1;Ras-related C3 botulinum toxin substrate 2;Ras-related C3 botulinum toxin substrate 3 | RAC1;RAC2;RAC3 | 2 | 2 | 2 | 25 | 0.0111521 | 0.1930587 | 0.1433346 | 0.7729319 | ||
| P18085 | P18085 | ADP-ribosylation factor 4 | ARF4 | 5 | 3 | 3 | 55 | 0.0116648 | 0.0998687 | 0.3392676 | 0.0726524 | ARF4 | |
| P30041 | P30041 | Peroxiredoxin-6 | PRDX6 | 7 | 7 | 7 | 30 | 0.0122622 | 0.176258 | 0.1077234 | 0.106493 | PRDX6 | |
| P13667 | P13667 | Protein disulphide-isomerase A4 | PDIA4 | 15 | 15 | 15 | 331 | 0.0128731 | 0.0171605 | 0.0013301 | 0.3133256 | PDIA4 | |
| Q9BS26 | Q9BS26 | Endoplasmic reticulum resident protein 44 | ERP44 | 7 | 7 | 7 | 105 | 0.0136596 | 0.011489 | 0.0018995 | 0.9895542 | ||
| P53992 | P53992 | Protein transport protein Sec24C | SEC24C | 3 | 3 | 3 | 16 | 0.0136843 | 0.1832328 | 0.0019216 | 0.1447004 | ||
| P80404 | P80404 | 4-aminobutyrate aminotransferase, mitochondrial | ABAT | 5 | 5 | 5 | 13 | 0.0143606 | 0.0418549 | 0.4883502 | 0.008795 | ||
| P08758 | P08758 | Annexin A5 | ANXA5 | 12 | 12 | 12 | 120 | 0.0144315 | 0.500855 | 0.0104712 | 0.0513501 | ANXA5 | |
| P11021 | P11021 | 78 kDa glucose-regulated protein | HSPA5 | 24 | 24 | 23 | 661 | 0.0145354 | 0.0462898 | 0.047508 | 0.8154141 | HSPA5 | HSPA5 |
| P04843 | P04843 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1 | RPN1 | 12 | 12 | 12 | 160 | 0.0145686 | 0.0074703 | 0.0042961 | 0.8056194 | RPN1 | RPN1 |
| O75340 | O75340 | Programmed cell death protein 6 | PDCD6 | 3 | 3 | 3 | 30 | 0.0147722 | 0.0024878 | 0.0100022 | 0.6010864 | ||
| Q8NBS9 | Q8NBS9 | Thioredoxin domain-containing protein 5 | TXNDC5 | 10 | 10 | 10 | 85 | 0.0159992 | 0.0651848 | 0.044914 | 0.5333772 | TXNDC5 | |
| O94826 | O94826 | Mitochondrial import receptor subunit TOM70 | TOMM70A | 4 | 4 | 4 | 18 | 0.0181108 | 0.0955309 | 0.9500943 | 0.0418347 | TOMM70A | |
| P09110 | P09110 | 3-ketoacyl-CoA thiolase, peroxisomal | ACAA1 | 8 | 8 | 8 | 34 | 0.0189284 | 0.0229383 | 0.0271681 | 0.1152645 | ACAA1 |
All the proteins identified and quantified by MaxQuant were annotated with the different results of the statistical analysis performed on log2 LFQ intensity used to calculate the p-value associated with the differences in distribution between the compared groups: N vs F1 vs F2-3 [in black], N vs F1 [in blue], N vs F2-3 [in purple], and F1 vs F2-3 [in green]. PDIs, including AGR2, PDIA6, PDIA4, ERP44, and TXNDC5, as well as BiP, are highlighted in bold. Razor peptides are peptides found in several protein sequences sharing at least partial identity of sequence. Unique peptides are peptides that only match with one protein sequence. MS/MS count: number of times that the considered protein entry was associated with an MS/MS identification [or a peptide spectrum match]. The ‘gene name for BiP [HSPA5] interactors’ column details the proteins identified as such using BioGrid according to their gene names [and not their protein name or ID]. The ‘gene name for AGR2 interactors’ column details the proteins identified as such using BioGrid according to their gene names [and not their protein name or ID]. PDI, protein disulphide isomerase.
Figure 1.
ER stress and epithelial-mesenchymal transition proteins. Proteins were represented in volcano plots for the comparison of low grade of fibrosis [F1] with normal tissue [N] and the comparison of high grade of fibrosis [F2-3] with normal tissue [N]. Differential abundance [F1 vs N and F2-3 vs N] of proteins was represented by plotting the log10p-value against the log2 of F1/N and F2-3/N. The horizontal line represents the significance threshold [p-value = 0.05]. [A] The proteins involved in ‘Proteins processing in ER’ were selected according to the KEGG pathway database hsa04141 in which two proteins were manually added [AGR2: anterior gradient protein 2 homologue; ERP44: endoplasmic reticulum resident protein 44]. Among the proteins involved in this pathway and significantly increased in fibrosis, five out seven [N versus F1] and five out of 11 [N versus F2-3] are protein disulphide isomerases [PDIs]: AGR2, ERP44, protein disulphide-isomerase A6 [PDIA6], protein disulphide-isomerase A4 [PDIA4], and protein disulphide-isomerase A3 [PDIA3]. Binding immunoglobulin protein [BiP] was also highlighted. [B] The proteins involved in EMT were selected according to the literature36,37 and are listed in Supplementary Table 7. Epithelial and mesenchymal proteins [highlighted in the figure] were not differentially expressed in epithelium surrounding fibrotic and normal tissues. EMT, epithelial to mesenchymal transition; ER, endoplasmic reticulum.
Search for EMT involved proteins was done for 70 proteins of the EMT process36,37 but only 32 could be identified. Epithelial and mesenchymal proteins highlighted were not differentially expressed in epithelium surrounding fibrotic and normal tissues in the differential analysis [Figure 1B]. Many single apoptosis specific proteins were found differentially expressed in epithelium harbouring fibrotic compared with N tissues [Supplementary Table 2]. Some negative regulator of apoptosis [as ribosomal protein S6 kinase alpha-3 [RPS6KA3] or fatty acid-binding protein 1 [FABP1],38,39 were found decreased in fibrosis areas. Fibrotic areas exhibited higher expression of proteins involved in cell differentiation, a decrease of the normal oxidation-reduction process, and a decrease of some extracellular matrix components in surface epithelium, as well as a decrease of some mitotic checkpoint proteins compared with normal tissues.
3.2.2. Gene ontology, pathways enrichment analyses, functional annotation, and BioGRID interactor identifications based on proteomic results
Supplementary Table 5, available at ECCO-JCC online, contains a compilation of the enrichment’s charts obtained with each list of sets of proteins identified in our experiment [Supplementary Tables 3 and 4] analysed using DAVIDTM. The identified pathways included the metabolism of proteins from production to translation and maturation [ribosomal proteins, translation control, protein maturation, protein transport, protein involved in the spliceosomal complex assembly, mitotic nuclear division, antigen processing and presentation], membrane proteins, but also confirmed the results of our differential analysis with the presence of proteins processing in ER and especially in response to ER stress including many PDIs and several of their complex binding partners. The EMT process was not found to be enriched neither the apoptotic process [Supplementary Table 5].
Supplementary Figure 3 illustrates the STRING network of proteins significantly increased in-high grade fibrosis compared with N and proteins detected only in fibrotic tissues, highlighting ER stress, unfolded protein response [UPR], and structural interconnected proteins.
Based on the BioGRID database, 435 out of 840 [51.79%] known genes coding for AGR2 protein partners and 123 out of 563 [21.85%] BiP interactors could be identified.
3.2.3. Proteins selection strategy for IHC confirmation
As ER stress was found as a predominant pathway in the epithelium of CD stricture, both in the differential analysis and in the pathways enrichment analyses; as the differential analysis highlighted several PDIs, in particular AGR2, among the most discriminating proteins between tissues of different grades of fibrosis; as a pro-fibrotic role of one PDI has already been demonstrated in renal fibrosis40; and as AGR2 has a known action on the MEC in oncology41; we decided to focus the IHC confirmation study on AGR2. BiP was also studied as a well-known ER stress marker.
3.3. Confirmation of AGR2 and BiP distribution by immunohistochemistry
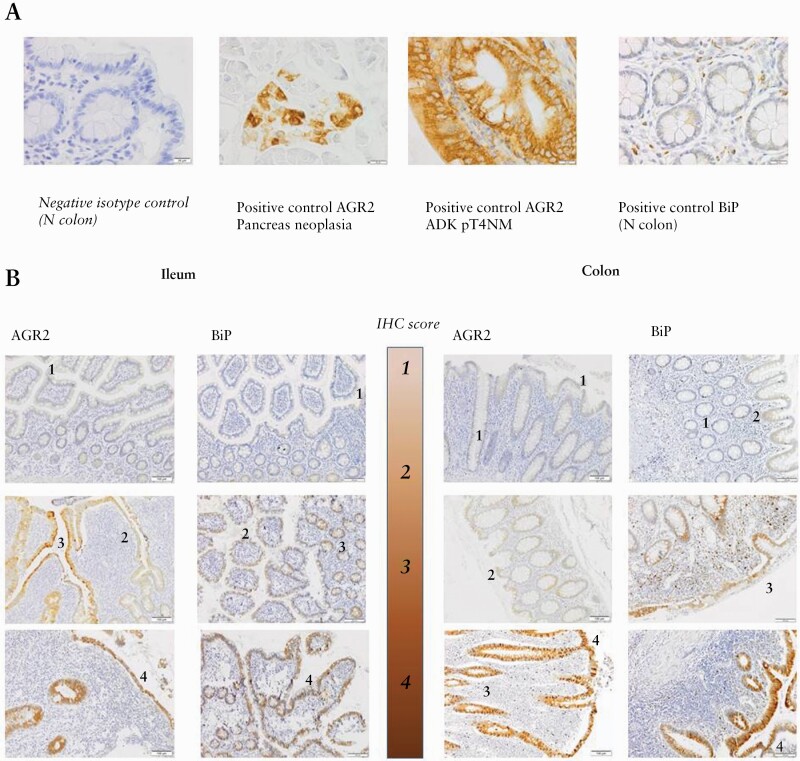
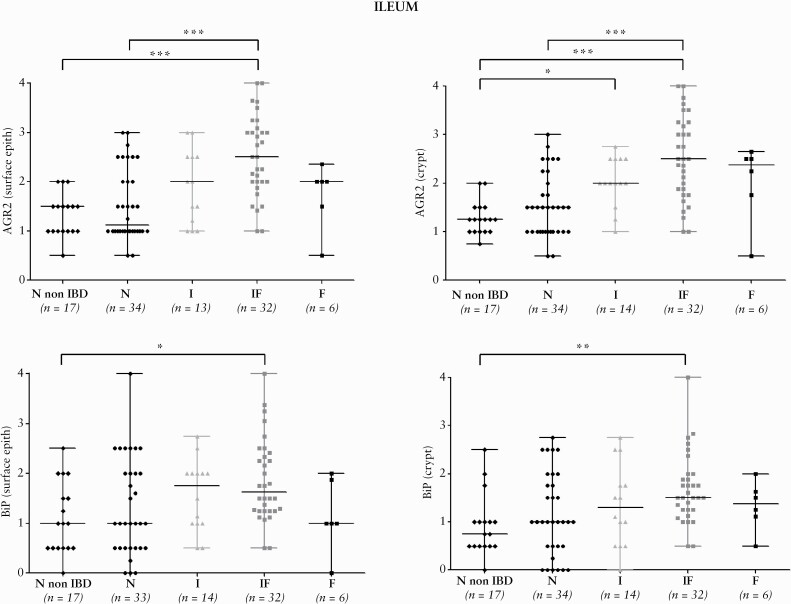
AGR2 and BiP appeared within the cytoplasm of epithelial cells, as previously observed on FFPE tissues,42 and were also detected in some blood vessel endothelial cells as well as some smooth muscle cells. Figure 2 illustrates the isotype antibody [2A] and positive controls performed, as well as the staining scores used [2B]. The results of the IHC semi-quantitative characterisation for AGR2 and BiP in the ileum and the colon epithelium, according to the tissue classification, are summarised in Figure 3 and Supplementary Figure 4, available at ECCO-JCC online, respectively. In the ileum, AGR2 was not significantly increased in I tissues but it was the case in the IF and F tissues, suggesting an association with the fibrotic process. We found weak but significant correlation between AGR2 and fibrosis or acute inflammation grades in the ileum, but not with chronic inflammation grade, for the surface and crypt epithelium [Supplementary Table 6, available at ECCO-JCC online].
Figure 2.
Illustrative pictures of immunohistochemistry scores. A. Negative isotype and positive controls used for AGR2 and BiP IHC protein detection: picture of the negative isotype control is provided for a normal [N] colon. Positive controls were tested on pancreas neoplasia, on colorectal adenocarcinoma [ADK] at pT4NM stage, and on a normal colon extracted at the surgical margin of diverticular disease. B. Illustrative pictures of IHC scores: the 4-grade scale is illustrated for AGR2 and BiP staining performed on ileal and colonic FFPE tissues. IHC,immunohistochemical; FFPE, formalin-fixed paraffin-embedded.
Figure 3.
Distribution of AGR2 and BiP IHC scores for ileal tissues. Distribution of AGR2 and BiP IHC scores for the CD patients groups [N, I, IF, F] and non-IBD controls in the surface and crypt epithelium for ileal tissues. In some samples, due to ulceration associated with the fibrotic area, only the crypt epithelium could be scored. The ANOVA test was significant for AGR2 and BiP in the surface epithelium [p <0.0001 and p <0.05, respectively] and in the crypt epithelium [p < .0001 and p <0.001, respectively]. Horizontal black lines highlight the significant two-by-two comparisons of groups obtained by Dunn’s multiple comparison test. *p <0.05, **p <0.001, ***p <0.0001. IHC,immunohistochemical; CD, Crohn’s disease.
3.4. Functional assays to evaluate the impact of intestinal epithelial ER stress on fibroblast to myofibroblast differentiation
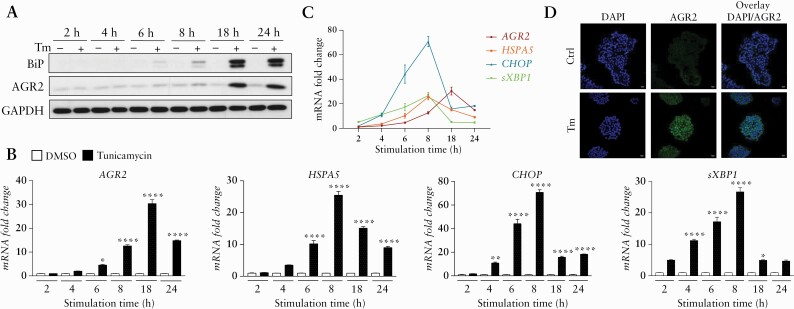
3.4.1. ER stress and secretion of PDIs, including AGR2, induced by tunicamycin in HT-29 cells
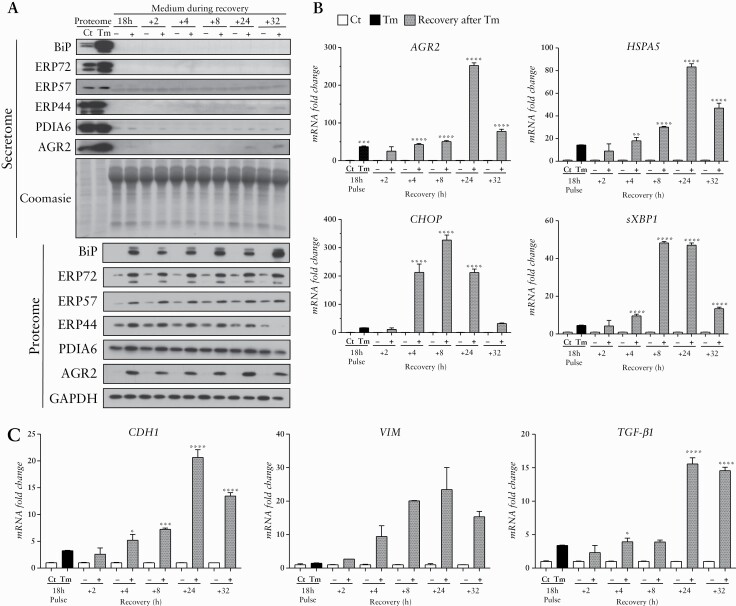
To investigate the role of epithelial ER stress in the development of intestinal fibrosis, we used an in-vitro model. To this end, we treated HT-29 cells with Tm, a chemical ER stress inducer. In response to this Tm treatment, we found an increase of the classical ER stress and UPR-inducible genes and proteins [BiP: binding-immunoglobulin protein; CHOP: C/EBP homologous and sXBP1: the spliced form of X-box-binding protein 1] [Figure 4]. An increase of AGR2 was also highlighted, and the AGR2 maximal induction [at transcript level] was reached after 18 h of Tm treatment [Figure 4] at the dose of 10 µg/mL [data not shown]. To assess if Tm-induced ER stress persists in HT-29 after stimulus removal, we studied ER stress markers and PDI expression during recovery of HT-29 cells in fresh media. A sustained intracellular production of most of the ER stress markers and the PDI tested persisted up to 32 h after the media change, and the maximal intracellular increase of AGR2 was reached 24 h after the transient ER stress [Figure 5A and B]. Several PDI [AGR2, ERP44, and PDIA6] were secreted and found in the supernatant of HT-29, pre-conditioned by Tm, 24 h and 32 h after media change [Figure 5B]. The presence of EMT in these induced ER stress conditions was not clear, since both a mesenchymal [VIM] marker and an epithelial [CDH1] marker were increased [Figure 5C]. Interestingly, the transcript level of TGF-β1 was significantly increased in HT-29 pre-conditioned by Tm, especially 24 and 32 h after the media change [Figure 5C].
Figure 4.
ER stress markers increase after Tm induction on HT-29. [A] Western blot [WB] analysis of AGR2 and BiP in HT-29 according to tunicamycin [Tm] [10 µg/mL] stimulation time course. The GAPDH was used as loading control. [B,C] RT-qPCR expression fold change of AGR2, HSPA5, CHOP, and sXBP1 in HT-29 under Tm treatment time course [black] compared with time control conditions with DMSO [white]. [D] IF analysis of AGR2 in HT-29 after 18 h of Tm treatment at 10 µg/mL and time control DMSO. *p <0.05, **p <0.01, ***p <0.001, ****p <0.0001. ER, endoplasmic reticulum; IF, IL-3F1-3.
Figure 5.
ER stress markers and PDIs persistence in HT-29 during recovery in fresh media after a 18 h Tm treatment. [A] ER stress markers detection in HT-29 during cell recovery after transient ER stress induced by Tm [pulse] or not [DMSO treatment; Ct] and media change: WB analysis of AGR2, BiP, PDIA6, ERP44, ERP57, ERP72 in the proteome [intracellular protein extracts] and in the secretome [extracellular form of proteins]. GAPDH was used as loading control for the proteome and Coomasie blue colouration of the gel was used for normalisation of the secretome loading quantities. [B] RT-qPCR expression fold change of AGR2, HSPA5, CHOP, sXBP1, and [C] EMT markers: E-cadherin [CDH1] and Vimentin [VIM], as well as TGF-β1 in HT-29 cells treated by 18 h of Tm induction [black] or in time control condition with DMSO [Ct] [white], after 2, 4, 8, 24, 32 h of recovery in fresh media [grey]. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. EMT, epithelial to mesenchymal transition; ER, endoplasmic reticulum; WB, western blot; PDI, protein disulphide isomerase; Tm, tunicamycin.
3.4.2. Supernatant of HT-29 cells pre-conditioned by tunicamycin induces the fibroblast to myofibroblast differentiation, and the secretion of AGR2 by epithelial cells can play a role in this differentiation
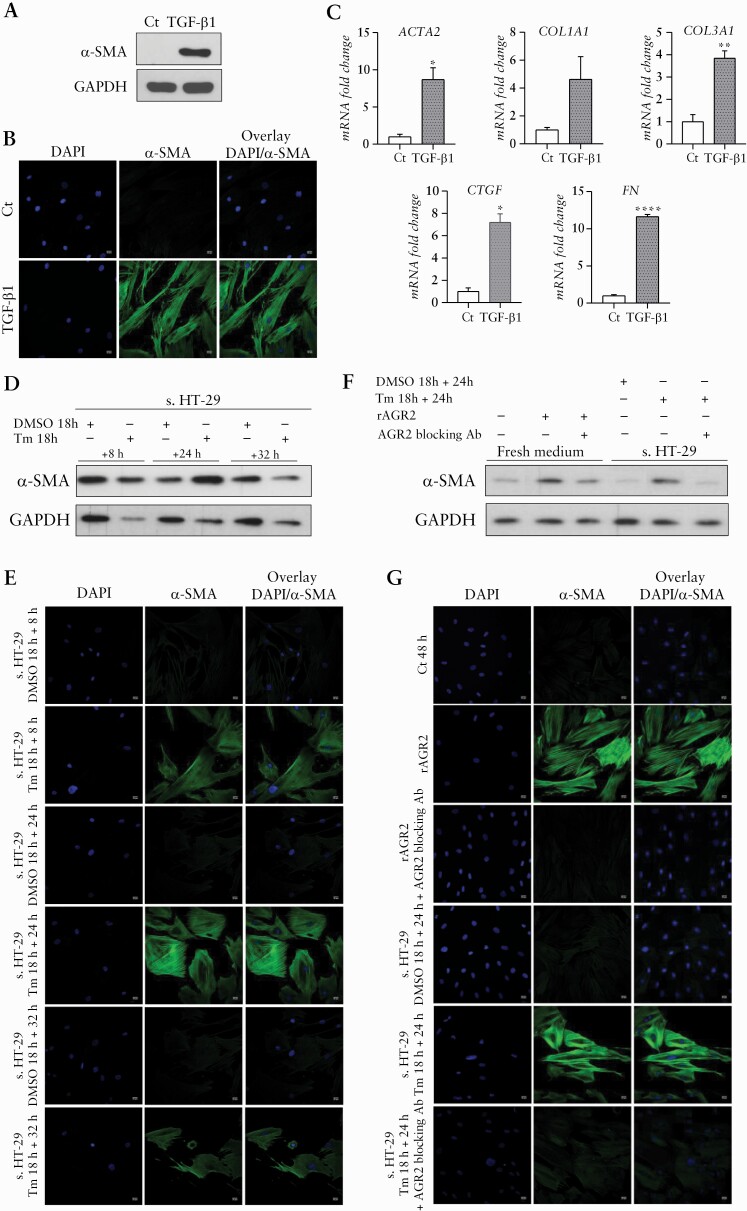
The model of fibroblast to myofibroblast differentiation under TGF-β1 treatment previously described43 was used as positive control and showed an increase of α-SMA marker by western blot [WB], IF together with increased expression of ACTA2, collagen proteins [COL1A1 and COL3A1], fibronectin [FN], and connecting tissue growth factor [CTGF] by RT-qPCR [Figure 6A–C]. Supernatant of HT-29 pre-conditioned by Tm and collected 8, 24 and 32 h after media change induced CCD-18Co differentiation after 48 h of treatment as illustrated by the increased expression of α-SMA obtained by WB and IF [Figure 6D and E] but not by RT-qPCR [data not shown].
Figure 6.
Fibroblast to myofibroblast differentiation obtained with supernatant of HT-29 cells pre-conditioned by tunicamycin and with recombinant AGR2 and reversed by AGR2-blocking antibody. [A,B] WB and IF analysis of α-SMA in CCD-18Co with TGF-β1 treatment as positive control of the myofibroblastic differentiation. [C] RT-qPCR expression fold change of ACTA2, COL1A1, COL3A1, CTGF, and FN in CCD18-Co under TGF-β1 treatment [grey] compared with time control conditions [white]. [D, E] WB and IF analysis of α-SMA in CCD-18Co under supernatant of HT-29 [s. HT-29] either pre-conditioned by Tm or treated with DMSO as control condition [8, 24, and 32 h after media change]. [F,G] WB and IF analysis of α-SMA in CCD-18Co under recombinant AGR2 [rAGR2] and when AGR2 blocking antibody [Ab] is applied in the conditions with rAGR2 or in the supernatant of HT-29 pre-conditioned by Tm. *p <0.05, **p <0.01, ***p <0.001, ****p <0.0001. WB, western blot; IF, IL-3F1-3.
We tested the capacity of recombinant AGR2 [rAGR2] to induce a fibroblast to myofibroblast differentiation and observed an increased expression of α-SMA by WB and IF [Figure 6F and G] but not by RT-qPCR [data not shown].
Blocking AGR2 by using an anti-AGR2 antibody in the supernatant of HT-29 pre-conditioned by Tm or in the condition with rAGR2 showed a mitigation of the increased expression of α-SMA in the CCD-18Co cells [Figure 6F and G].
4. Discussion
Our data show that the secretion of AGR2 by epithelial cells can participate in the development of fibrosis in CD. Our pilot proteomic study showed that the surface epithelium from ileal fibrotic CD strictures exhibits signatures of ER stress, UPR, and likely apoptosis. Importantly, we revealed an increase of several PDIs including AGR2 in fibrotic tissues, leading us to study by IHC links between AGR2, but also BIP and the fibrosis in a larger cohort of CD patients. In the ileum, a significant increase of AGR2 was confirmed by IHC in tissues with a fibrotic component, whereas it was not significant in pure inflammatory samples, highlighting the association of this protein with the fibrostenosing process. We therefore performed functional assays, with a two cell lines model (using HT-29 as intestinal epithelial cells [IEC] and CCD-18Co as intestinal fibroblasts) to understand the capacity of this ER stress in IEC to promote fibrosis, and more specifically, its capacity to induce a myofibroblastic differenciation.43 Our experiments showed that an HT-29 transient ER stress increases intracellular expression of several PDIs including AGR2 and BIP, as well as TGF-β1, and leads to a secretion of AGR2, PDIA6, and ERP44 in the supernatant, with similar kinetics. Finally, a fibroblast to myofibroblast differentiation was obtained with the supernatant of IEC pre-conditioned by ER stress as well as with recombinant AGR2, and its attenuation was obtained after blocking AGR2 with an anti-AGR2 antibody.
The presence of ER stress in fibrotic strictures is consistent with current evidences suggesting a prominent role of these pathways in intestinal fibrosis44 as well as in the fibrotic process of other organs.45,46 IEC, which synthesise high volumes of proteins, are highly susceptible to ER stress, which results in the ER accumulation of unfolded and misfolded proteins under cellular stress conditions. The UPR, and related PDI AGR2, which correctly rearranges the disulphide bonds of the proteins, enables the cell to resolve this ER stress. Although the role of AGR2 is well known in CD inflammation,47 we found a significant increase in fibrotic tissues [whereas it was not significantly increased in pure inflammatory tissues] and its expression was correlated to fibrosis grade in IHC. Although our observations do not allow us to make it a protein purely related to fibrosis, nor to ensure that the observed increase is not partly the reflect of chronic inflammation, this study is the first, to our knowledge, to show a more specific implication of AGR2 in CD fibrosis. Indeed, ER stress is not just related to inflammation in CD patients, but is rather an underlying abnormality in IBD, areas of mucosa without inflammation also presenting an increased mRNA expression of BIP and sXBP1.48
Whereas EMT is well known to contribute to intestinal fibrosis15–17,49 and several studies suggest that ER stress facilitates fibrotic remodelling, notably through induction of EMT transition,45,50,51 our proteomic study, as well as epithelial cells pre-conditioned by Tm in our functional assays, did not clearly highlight EMT. Whereas the absence of clear EMT in the functional assays could be related to the stimulation conditions used and needs further experimentation, there are several possible explanations for its absence in the proteomic study. AGR2 intracellular overexpression [as revealed in our proteomic study] is known to play an important role in maintaining the epithelial phenotype, by preventing the activation of key factors involved in the EMT process52 and, in IEC in particular, by increasing the expression of tight junction proteins and stabilising the cytoskeletal structure [which are reverse mechanisms of EMT].53 Besides a protective role of AGR2 against EMT in specific cell types, it is also possible that, as in mesothelial cells, ER stress leads to EMT or apoptosis in a time- and intensity-dependent way. Whereas mild ER stress might induce a reversible EMT [by a transient CHOP expression], chronic ER stress might induce apoptosis [by a persistent upregulation of CHOP].50 In fibrosis, generally associated with chronic inflammation, there is probably more of a chronic ER stress, which could explain how we found a certain number of apoptosis markers rather than EMT evidence in our proteomic study. Links between ER stress/UPR, AGR2, and EMT are poorly studied in intestinal fibrosis and remain to be further investigated.
As the induction of an ER stress in these IECs did not induce a clear EMT but led to the secretion of several PDIs in the supernatant of IEC, the latter being able to induce a myofibroblastic differentiation, the hypothesis that we support is that epithelial cells under ER stress release a whole set of cytokines into their environment and exercise their pro-fibrotic role through a paracrine action. First of all, epithelial cells are known to release pro-fibrotic cytokines including TGF-β1.54 As we demonstrated that a transient ER stress in epithelial cell increases intracellular TGF-β1 expression and that TGF-β1 is known to be secreted by IEC [in particular by HT-29 cells, and more abundantly when they are the target of pro-inflammatory factors],55 TGF-β1 is probably one of these pro-fibrotic cytokines. The way in which ER stress induces TGF-β1 expression and activation has recently been demonstrated in subepithelial myofibroblasts of fibrostenotic CD patients,44 and we could speculate that a similar mechanism occurs in epithelial cells. The interaction between BiP and latent TGF-β1 increases its activation and XBP1 and ATF-6α [two ER stress sensors] act as transcription factors promoting TGF-β1 gene expression. PDIs can also play a role in this set of pro-fibrotic cytokines. Interestingly, the secretion of ERP57 [PDIA3] in response to ER stress, also found as a predominant pathway in renal fibrosis, was shown as important for ECM accumulation and progression of renal fibrosis.40 If ERP57 was not found to be significantly excreted in the supernatant of IEC preconditioned by ER stress, we highlighted three other PDIs [ERP44, PDIA6, and AGR2] that were significantly increased in fibrotic tissues in our proteomic study. Whereas the role of secreted PDIA6 and ERP44 must be the subject of future experimentations, the role of extracellular AGR2 [eAGR2] has been further studied in oncology. It is implicated in cancer invasion and tumour cell dissemination and is generally associated with worse clinical outcomes.41,56
This secreted AGR2 seems to have a specific pro-fibrotic role, as suggested by the myofibroblastic differenciation obtained using rAGR2 and its attenuation by blocking AGR2 with anti-AGR2 antibody. In oncology, eAGR2 has a whole series of described paracrine actions [often opposed to its intracellular action52,57] and could play a similar role in fibrosis. Fessart et al. demonstrated that eAGR2 plays a pivotal role in tumour microenvironment development [which results from complex interactions between tumour cells and the ECM] by disrupting cellular polarity and epithelial cell-cell adhesion and thus promoting EMT, but also by interacting with the ECM.41 This eAGR2 is also known to interact with core components of the mTORC2 complex [RICTOR], known to be involved in cell proliferation and actin cytoskeleton organisation in fibrosis.57 Finally, secreted AGR2 has been demonstrated to be involved in fibroblasts recruitment and migration [eAGR2 can promote cutaneous wound healing by recruitment of fibroblasts in the wounded area, but can also activate stromal fibroblasts and promote fibroblast-associated cancer invasion in gastric signet-ring cell carcinoma, for example]56,58,59 as well as in the murine fibroblast elongation and proliferation.60 However, to the best of our knowledge, our data are the first to suggest that eAGR2 could be a paracrine inducer of intestinal fibroblast to myofibroblast differentiation.
Our study presents some limitations. The samples used for the proteomic study come from FFPE tissues. This could have an impact on the proteomic coverage, as some proteins might be unavailable for analysis and were therefore not identified in our study. Moreover, only a limited number of samples showed pure fibrotic and pure inflammatory tissues to address a systematic paired comparison of all the possible combinations of inflammation and fibrosis stages with normal tissues. However, this limitation actually reflects the situation met in the vast majority of surgically treated CD strictures harbouring concomitant acute and/or chronic inflammation and fibrosis. Finally, the CCD-18Co from non-IBD patients and HT-29 is a colorectal cancerous cell line. These experiments should be replicated on cells originating form CD patients.
In conclusion, our results suggest, for the first time, that the presence of ER stress in IEC may play a role in intestinal fibrosis through the secretion of AGR2 and its paracrine action on intestinal fibroblasts. Further exploring the role of ER stress and/or AGR2 in intestinal fibrosis may help to discover novel adjuvant therapeutic approaches for CD patients with stricture.
Supplementary Material
Acknowledgements
We thank the different people involved in technical and scientific laboratory assistance provided by D. Morsa, N. Smargiasso, L. Trpziot, N. Rosière [GIGA-proteomics], V. Bertrand, C. Humblet, A. Maquet, T. Di Salvo [GIGA-immunohistology], S. Gofflot, K. El Kandoussi and R. Thonon [biobanking facility], and S. Ormenese [GIGA-imaging and LCM]. A special thanks to S. Azarzar and C. Bachelet, our laboratory technicians, and to G. Paulissen who helped in the setting of IF experiments. A special thanks to S. Speca from the Liric [Lille Inflammation Research International Center] who kindly provided some specific advice on the cell line models and the first CCD-18Co and HT-29 cells batch used in this work.
Contributor Information
Sophie Vieujean, Laboratory of Translational Gastroenterology, University of Liège, Liège, Belgium; Hepato-Gastroenterology and Digestive Oncology, University Hospital CHU of Liège, Liège, Belgium.
Shurong Hu, Laboratory of Translational Gastroenterology, University of Liège, Liège, Belgium; Department of Gastroenterology, Center of Inflammatory Bowel Disease, a Second Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, China.
Emeline Bequet, Laboratory of Translational Gastroenterology, University of Liège, Liège, Belgium; Division of Hepato-Gastroenterology, Department of Paediatrics, University Hospital of Liège, Liège, Belgium.
Catherine Salee, Laboratory of Translational Gastroenterology, University of Liège, Liège, Belgium.
Charlotte Massot, Laboratory of Translational Gastroenterology, University of Liège, Liège, Belgium.
Noëlla Bletard, Pathological Anatomy and Cytology, University Hospital CHU of Liège, Liège, Belgium.
Nicolas Pierre, Laboratory of Translational Gastroenterology, University of Liège, Liège, Belgium.
Florence Quesada Calvo, Laboratory of Translational Gastroenterology, University of Liège, Liège, Belgium.
Dominique Baiwir, GIGA Proteomics Facility, University of Liège, Liège, Belgium.
Gabriel Mazzucchelli, MolSys Research Unit, Laboratory of Mass Spectrometry, University of Liège, Liège, Belgium.
Edwin De Pauw, MolSys Research Unit, Laboratory of Mass Spectrometry, University of Liège, Liège, Belgium.
Carla Coimbra Marques, Abdominal Surgery Department, University Hospital CHU of Liège, Liège, Belgium.
Philippe Delvenne, Pathological Anatomy and Cytology, University Hospital CHU of Liège, Liège, Belgium.
Florian Rieder, Gastroenterology, Hepatology & Nutrition, Cleveland Clinic, Cleveland, OH, USA.
Edouard Louis, Laboratory of Translational Gastroenterology, University of Liège, Liège, Belgium; Hepato-Gastroenterology and Digestive Oncology, University Hospital CHU of Liège, Liège, Belgium.
Marie-Alice Meuwis, Laboratory of Translational Gastroenterology, University of Liège, Liège, Belgium.
The proteomic data that support this study have been deposited into the ProteomeXchange Consortium [http://www.proteomexchange.org/] via the PRIDE partner repository, with the dataset identifier PXD022214, and for the reviewing: user name: reviewer_pxd022214@ebi.ac.uk and password: OB0SiO8g. The Supplementary tables of this article are available at [https://dox.uliege.be/index.php/s/9etZCmwoql5fev1] and can be accessed with the following password: 18112020.
Funding
This work was supported by the Liège University Hospital Center [CHU Liège] [FIRS-2014], the University of Liege, and the Léon Fredericq Foundation. SV was financially supported by the National Fund for Scientific Research [FNRS-FNR] [grant numbers 32729160, 40001034].
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
SV, SH, EL, and M-AM designed the experiment. SV, EB, CM, NB, CCM, PD, EL, and M-AM provided the human samples and the clinical information. FQC and M-AM prepared the samples for the proteomic analysis, MAM performed the differential analysis, DB, GM, and EDP managed the proteomic raw data acquisition and provided advice for the proteomic differential analysis. CM performed sample preparation for immunohistochemistry analysis. CS, CM, NB, and M-AM performed the histological characterisation. SV performed the functional assays. SV, NP, EL, and M-AM analysed the data. SV, EL, and M-AM wrote the paper. All authors revised the manuscript and improved its intellectual content.
Conference presentations: Belgian Week of Gastroenterology, Antwerp 2018; European Crohn’s and Colitis Congress, Vienna 2018; European Crohn’s and Colitis Congress, Vienna 2020; Belgian Week of Gastroenterology, Antwerp 2020.
References
- 1. Thia KT, Sandborn WJ, Harmsen WS, Zinsmeister AR, Loftus EV Jr. Risk factors associated with progression to intestinal complications of Crohn’s disease in a population-based cohort. Gastroenterology 2010;139:1147–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rieder F, Bettenworth D, Imai J, Inagaki Y. Intestinal fibrosis and liver fibrosis: consequences of chronic inflammation or independent pathophysiology? Inflamm Intest Dis 2016;1:41–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rieder F, Fiocchi C, Rogler G. Mechanisms, management, and treatment of fibrosis in patients with inflammatory bowel diseases. Gastroenterology 2017;152:340–50.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bettenworth D, Gustavsson A, Atreja A, et al. A pooled analysis of efficacy, safety, and long-term outcome of endoscopic balloon dilation therapy for patients with stricturing Crohn’s disease. Inflamm Bowel Dis 2017;23:133–42. [DOI] [PubMed] [Google Scholar]
- 5. Rieder F, Zimmermann EM, Remzi FH, Sandborn WJ. Crohn’s disease complicated by strictures: a systematic review. Gut 2013;62:1072–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rieder F, Latella G, Magro F, et al. European Crohn’s and Colitis Organisation topical review on prediction, diagnosis and management of fibrostenosing Crohn’s disease. J Crohns Colitis 2016;10:873–85. [DOI] [PubMed] [Google Scholar]
- 7. Pohlers D, Brenmoehl J, Löffler I, et al. TGF-beta and fibrosis in different organs ‐ molecular pathway imprints. Biochim Biophys Acta 2009;1792:746–56. [DOI] [PubMed] [Google Scholar]
- 8. Massagué J. How cells read TGF-β signals. Nat Rev Mol Cell Biol 2000;1:169–78. [DOI] [PubMed] [Google Scholar]
- 9. Hinz B. Tissue stiffness, latent TGF-beta1 activation, and mechanical signal transduction: implications for the pathogenesis and treatment of fibrosis. Curr Rheumatol Rep 2009;11:120–6. [DOI] [PubMed] [Google Scholar]
- 10. Latella G, Rogler G, Bamias G, et al. Results of the 4th Scientific Workshop of the ECCO. [I]: Pathophysiology of intestinal fibrosis in IBD. J Crohns Colitis 2014;8:1147–65. [DOI] [PubMed] [Google Scholar]
- 11. Lawrance IC, Rogler G, Bamias G, et al. Cellular and molecular mediators of intestinal fibrosis. J Crohns Colitis 2017;11:1491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rieder F, Brenmoehl J, Leeb S, Schölmerich J, Rogler G. Wound healing and fibrosis in intestinal disease. Gut 2007;56:130–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Uehara H, Nakagawa T, Katsuno T, et al. Emergence of fibrocytes showing morphological changes in the inflamed colonic mucosa. Dig Dis Sci 2010;55:253–60. [DOI] [PubMed] [Google Scholar]
- 14. Brittan M, Chance V, Elia G, et al. A regenerative role for bone marrow following experimental colitis: contribution to neovasculogenesis and myofibroblasts. Gastroenterology 2005;128:1984–95. [DOI] [PubMed] [Google Scholar]
- 15. Scharl M, Huber N, Lang S, Fürst A, Jehle E, Rogler G. Hallmarks of epithelial to mesenchymal transition are detectable in Crohn’s disease associated intestinal fibrosis. Clin Transl Med 2015;4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Flier SN, Tanjore H, Kokkotou EG, Sugimoto H, Zeisberg M, Kalluri R. Identification of epithelial to mesenchymal transition as a novel source of fibroblasts in intestinal fibrosis. J Biol Chem 2010;285:20202–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lovisa S, Genovese G, Danese S. Role of epithelial-to-mesenchymal transition in inflammatory bowel disease. J Crohns Colitis 2019;13:659–68. [DOI] [PubMed] [Google Scholar]
- 18.Bancroft J.D., Gamble M. Theory and Practice of Histological Techniques. 6th edn. https://www.elsevier.com/books/theory-and-practice-of-histological-techniques/bancroft/978-0-443-10279-0 Accessed August 14, 2020. [Google Scholar]
- 19. Bataille F, Klebl F, Rümmele P, et al. Morphological characterisation of Crohn’s disease fistulae. Gut 2004;53:1314–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. D’Haens GR, Geboes K, Peeters M, Baert F, Penninckx F, Rutgeerts P. Early lesions of recurrent Crohn’s disease caused by infusion of intestinal contents in excluded ileum. Gastroenterology 1998;114:262–7. [DOI] [PubMed] [Google Scholar]
- 21. Mosli MH, Feagan BG, Sandborn WJ, et al. Histologic evaluation of ulcerative colitis: a systematic review of disease activity indices. Inflamm Bowel Dis 2014;20:564–75. [DOI] [PubMed] [Google Scholar]
- 22. Adler J, Punglia DR, Dillman JR, et al. Computed tomography enterography findings correlate with tissue inflammation, not fibrosis in resected small bowel Crohn’s disease. Inflamm Bowel Dis 2012;18:849–56. [DOI] [PubMed] [Google Scholar]
- 23. Rimola J, Planell N, Rodríguez S, et al. Corrigendum: characterization of inflammation and fibrosis in Crohn’s disease lesions by magnetic resonance imaging. Am J Gastroenterol 2015;110:480. [DOI] [PubMed] [Google Scholar]
- 24. Chen W, Lu C, Hirota C, Iacucci M, Ghosh S, Gui X. Smooth muscle hyperplasia/hypertrophy is the most prominent histological change in Crohn’s fibrostenosing bowel strictures: a semiquantitative analysis by using a novel histological grading scheme. J Crohns Colitis 2017;11:92–104. [DOI] [PubMed] [Google Scholar]
- 25. Chiorean MV, Sandrasegaran K, Saxena R, Maglinte DD, Nakeeb A, Johnson CS. Correlation of CT enteroclysis with surgical pathology in Crohn’s disease. Am J Gastroenterol 2007;102:2541–50. [DOI] [PubMed] [Google Scholar]
- 26. Johnson LA, Luke A, Sauder K, et al. Intestinal fibrosis is reduced by early elimination of inflammation in a mouse model of IBD: impact of a ‘Top-Down’ approach to intestinal fibrosis in mice. Inflamm Bowel Dis 2012;18:460–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Higgins PD, Johnson LA, Sauder K, et al. Transient or persistent norovirus infection does not alter the pathology of Salmonella typhimurium induced intestinal inflammation and fibrosis in mice. Comp Immunol Microbiol Infect Dis 2011;34:247–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dillman JR, Stidham RW, Higgins PD, et al. Ultrasound shear wave elastography helps discriminate low-grade from high-grade bowel wall fibrosis in ex vivo human intestinal specimens. J Ultrasound Med 2014;33:2115–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Longuespée R, Alberts D, Pottier C, et al. A laser microdissection-based workflow for FFPE tissue microproteomics: important considerations for small sample processing. Methods 2016;104:154–62. [DOI] [PubMed] [Google Scholar]
- 30. Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008;26:1367–72. [DOI] [PubMed] [Google Scholar]
- 31. Perez-Riverol Y, Csordas A, Bai J, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 2019;47:D442–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tyanova S, Temu T, Sinitcyn P, et al. The Perseus computational platform for comprehensive analysis of [prote]omics data. Nat Methods 2016;13:731–40. [DOI] [PubMed] [Google Scholar]
- 33. Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 2014;13:2513–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Quesada-Calvo F, Massot C, Bertrand V, et al. OLFM4, KNG1 and Sec24C identified by proteomics and immunohistochemistry as potential markers of early colorectal cancer stages. Clin Proteomics 2017;14:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest 2003;112:1776–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pierre N, Salée C, Massot C, et al. Proteomics highlights common and distinct pathophysiological processes associated with ileal and colonic ulcers in Crohn’s disease. J Crohns Colitis 2020;14:205–15. [DOI] [PubMed] [Google Scholar]
- 38. Anjum R, Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat Rev Mol Cell Biol 2008;9:747–58. [DOI] [PubMed] [Google Scholar]
- 39. Wang G, Gong Y, Anderson J, et al. Antioxidative function of L-FABP in L-FABP stably transfected Chang liver cells. Hepatology 2005;42:871–9. [DOI] [PubMed] [Google Scholar]
- 40. Dihazi H, Dihazi GH, Bibi A, et al. Secretion of ERP57 is important for extracellular matrix accumulation and progression of renal fibrosis, and is an early sign of disease onset. J Cell Sci 2013;126:3649–63. [DOI] [PubMed] [Google Scholar]
- 41. Fessart D, Domblides C, Avril T, et al. Secretion of protein disulfide isomerase AGR2 confers tumorigenic properties. Elife 2016;5:e13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu R, Zhou J, Du XZ, et al. The role of the XBP-1/AGR2 signaling pathway in the regulation of airway Mucin5ac hypersecretion under hypoxia. Exp Cell Res 2019;382:111442. [DOI] [PubMed] [Google Scholar]
- 43. Speca S, Rousseaux C, Dubuquoy C, et al. Novel PPARγ modulator GED-0507-34 Levo ameliorates inflammation-driven intestinal fibrosis. Inflamm Bowel Dis 2016;22:279–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li C, Grider JR, Murthy KS, et al. Endoplasmic reticulum stress in subepithelial myofibroblasts increases the TGF-β1 activity that regulates fibrosis in Crohn’s disease. Inflamm Bowel Dis 2020;26:809–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tanjore H, Lawson WE, Blackwell TS. Endoplasmic reticulum stress as a pro-fibrotic stimulus. Biochim Biophys Acta 2013;1832:940–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Heindryckx F, Binet F, Ponticos M, et al. Endoplasmic reticulum stress enhances fibrosis through IRE1α-mediated degradation of miR-150 and XBP-1 splicing. EMBO Mol Med 2016;8:729–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maurel M, Obacz J, Avril T, et al. Control of anterior GRadient 2 [AGR2] dimerization links endoplasmic reticulum proteostasis to inflammation. EMBO Mol Med 2019;11:e10120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kaser A, Lee AH, Franke A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 2008;134:743–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hausmann M, Rechsteiner T, Caj M, et al. A new heterotopic transplant animal model of intestinal fibrosis. Inflamm Bowel Dis 2013;19:2302–14. [DOI] [PubMed] [Google Scholar]
- 50. Shin HS, Ryu ES, Oh ES, Kang DH. Endoplasmic reticulum stress as a novel target to ameliorate epithelial-to-mesenchymal transition and apoptosis of human peritoneal mesothelial cells. Lab Invest 2015;95:1157–73. [DOI] [PubMed] [Google Scholar]
- 51. McMorrow T, Gaffney MM, Slattery C, Campbell E, Ryan MP. Cyclosporine A induced epithelial-mesenchymal transition in human renal proximal tubular epithelial cells. Nephrol Dial Transplant 2005;20:2215–25. [DOI] [PubMed] [Google Scholar]
- 52. Sommerova L, Ondrouskova E, Vojtesek B, Hrstka R. Suppression of AGR2 in a TGF-β-induced Smad regulatory pathway mediates epithelial-mesenchymal transition. BMC Cancer 2017;17:546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ye X, Sun M. AGR2 ameliorates tumor necrosis factor-α-induced epithelial barrier dysfunction via suppression of NF-κB p65-mediated MLCK/p-MLC pathway activation. Int J Mol Med 2017;39:1206–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Curciarello R, Docena GH, MacDonald TT. The role of cytokines in the fibrotic responses in Crohn’s disease. Front Med 2017;4:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Drygiannakis I, Valatas V, Sfakianaki O, et al. Pro-inflammatory cytokines induce cross-talk between colonic epithelial cells and subepithelial myofibroblasts: implication in intestinal fibrosis. J Crohns Colitis 2013;7:286–300. [DOI] [PubMed] [Google Scholar]
- 56. Tsuji T, Satoyoshi R, Aiba N, et al. Agr2 mediates paracrine effects on stromal fibroblasts that promote invasion by gastric signet-ring carcinoma cells. Cancer Res 2015;75:356–66. [DOI] [PubMed] [Google Scholar]
- 57. Rieder F, de Bruyn JR, Pham BT, et al. Results of the 4th Scientific Workshop of the ECCO. Group II: Markers of intestinal fibrosis in inflammatory bowel disease. J Crohns Colitis 2014;8:1166–78. [DOI] [PubMed] [Google Scholar]
- 58. Zhu Q, Mangukiya HB, Mashausi DS, et al. Anterior gradient 2 is induced in cutaneous wound and promotes wound healing through its adhesion domain. FEBS J 2017;284:2856–69. [DOI] [PubMed] [Google Scholar]
- 59. Wang Z, Hao Y, Lowe AW. The adenocarcinoma-associated antigen, AGR2, promotes tumor growth, cell migration, and cellular transformation. Cancer Res 2008;68:492–7. [DOI] [PubMed] [Google Scholar]
- 60. Mangukiya HB, Negi H, Merugu SB, et al. Paracrine signalling of AGR2 stimulates RhoA function in fibroblasts and modulates cell elongation and migration. Cell Adh Migr 2019;13:332–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.