Abstract
All cells in multicellular organisms are housed in the extracellular matrix (ECM), an acellular edifice built up by more than a thousand proteins and glycans. Cells engage in a reciprocal relationship with the ECM; they build, inhabit, maintain, and remodel the ECM, while, in turn, the ECM regulates their behavior. The homeostatic balance of cell-ECM interactions can be lost, due to ageing, irritants or diseases, which results in aberrant cell behavior. The ECM can suppress or promote disease progression, depending on the information relayed to cells. Instructions come in the form of biochemical (e.g., composition), biophysical (e.g., stiffness), and topographical (e.g., structure) cues. While advances have been made in many areas, we only have a very limited grasp of ECM topography. A detailed atlas deciphering the spatiotemporal arrangement of all ECM proteins is lacking. We feel that such an extracellular matrix architecture (matritecture) atlas should be a priority goal for ECM research. In this commentary, we will discuss the need to resolve the spatiotemporal matritecture to identify potential disease triggers and therapeutic targets and present strategies to address this goal. Such a detailed matritecture atlas will not only identify disease-specific ECM structures but may also guide future strategies to restructure disease-related ECM patterns reverting to a normal pattern.
Keywords: Extracellular matrix, Matrictecture, Mapping
Abbreviations: ECM, ExtraCellular Matrix; 3D, Three-Dimensional; IMS, Imaging Mass Spectrometry; AI, Artificial Intelligence
Introduction
The genes coding for ECM structural proteins are well conserved throughout the evolutionary tree [1], likely because acellular organ scaffolds organizing different cell types and giving shape to tissues has proven to be an efficient solution to support multicellular organisms. These genes are translated into ECM proteins that are secreted to the extracellular space, where they are woven into basement membranes, an adhesive sheet based on interconnected networks of laminins and collagen IV, linked by accompanying proteins, and an interstitial matrix providing stiffness and recoil where fibronectin, elastin, and fibrillar collagens assemble into a 3D architecture.
Pioneering work using mass spectrometry techniques has determined the detailed composition of the ECM from many mouse and human tissues and indexed the results in a “matrisome” database (http://matrisomeproject.mit.edu) that has been enormously useful to understand ECM physiology [2], [3], [4]. The under- or over-abundance of a protein may be a guide to its importance in homeostasis or disease, however, mass spectrometry requires the destruction of the tissue and thereby its topographical information. Moreover, protein abundance does not necessarily have to be altered for a protein to play a pivotal role, e.g., cryptic domains in ECM proteins [5] regulate growth factor signaling by sinking and in turn liberating them. Thus, their 3D positioning, even in a restricted space, can have a direct impact on spatiotemporal growth factor activity. Protein structure can be altered, either by post-transcriptional or post-translational modifications, and they have a wide range of turnover rates, from practically lifetime (e.g., elastin) to several days-weeks [6], [7], [8], [9], [10], [11], [12]. It is conceivable that when we talk about “ECM structure” we are really describing a dynamic state, where cells constantly replace and remodel, aiming to maintain homeostasis under the influence of ageing, chronic pro-inflammatory stimuli [13], disease, external irritants, and pathogenic agents. This dynamism sets forth several challenges: how to determine the 3D location of ECM structural components; how to detect, in that space, the presence of post-transcriptional and post-translational modifications; how to link these structures to cellular responses; and finally, how to assign these features to external impaction. Here, we will propose avenues to explore and map the matritecture throughout lifetime.
Tools to map the ECM
Pioneering work on kidney, using stochastic optical reconstruction microscopy, demonstrates the feasibility to precisely locate and thereby map molecules inside the ECM [14]. This study emphasizes that gene expression origin does not ultimately fit the protein location. Protein-protein interactions can be estimated by such detailed mapping, highlighting the potential significance of a precise matritecture atlas. This atlas will open novel perspectives and reveal unknown interactions and functions.
Remarkably, structural analysis resolved the assembly of mucin and von Willebrand factor polymers combining cryo-electron microscopy and crystal structures [15]. Thus, enhancing super-resolution imaging with structural analysis is a powerful tool to dissect ECM 3D organization.
Imaging Mass Spectrometry (IMS) [16], [17], [18] is another emerging tool to map the spatial distribution of ECM components with important advantages: fixed tissue can be used allowing detection of soluble proteins and growth factors and imaging of more than 2000 proteins potentially including post-translational modifications can be processed. Nevertheless, IMS resolution is relatively low (≈100 µm) and produces 2D maps.
The development of Artificial Intelligence (AI) - powered tools that predict protein folding by leveraging genomics data [19] herald a new era that will predictably tackle complex, multi-molecular assemblies such as the ECM. Both imaging and biochemical data currently being compiled could well be part of the input necessary for AI tools.
Decellularization, the removal of cells from a tissue, results in an ECM organ scaffold capable of microscopic and biochemical exploration. The initial efforts to decellularize tissues, more than a quarter century ago [20], aimed to produce organ scaffolds for regenerative medicine and tissue engineering. The use of decellularized ECM organ scaffolds in regenerative medicine has rapidly evolved; in the last 3 years, 20% of all biomedical and tissue engineering publications report the use of decellularized ECM scaffolds [21]. Broadly, these ECM organ scaffolds are repopulated with cells to engineer transplantable constructs. Groundbreaking work in decellularization-based regenerative medicine successfully implanted different decellularized ECM organ scaffolds repopulated with specialized cells [22], [23]. Although the implanted organ scaffolds show limited functionality, these studies emphasize the advantage of using repopulated decellularized organ scaffolds and gives the rational to further optimize repopulation strategies enabling the transplantation of these implants into humans in the future. A trailblazing report describes a first-in-human bioimplant based on such decellularized and repopulated ECM scaffold [24]. Of note, the authors used the natural vasculature to perfuse reagents, leaving an ECM that retains the shape of the original organ and thus the topography of the ECM. Other decellularization techniques, notably agitation, severely disrupt tissue architecture. Thus, perfusion-based decellularization techniques are superior to map the matritecture. This technology opens the way to determine the spatiotemporal organization of ECM proteins in critical locations, like the invasive front of solid tumors, the organ parenchyma destroyed by progressive fibrosis, and tissue undergoing repair after wounding and therapy.
A profound limitation of decellularization as a mapping tool is the removal of soluble ECM components (e.g. ECM modulating enzymes, growth factors, etc.). However, using a “naked” scaffold as a biomaterial presents opportunities to functionalize the ECM by adding those soluble components and exploring adhesion patterns and binding sites. It also offers an opportunity to isolate cell-ECM structure relationships during disease progression [25]. Notably, decellularized ECM scaffolds provide beneficial features for imaging, such as penetration depth, antibody accessibility, and light scattering. Moreover, a combination of confocal and 2-photon microscopy can map several ECM components (as many as different antibodies and fluorophores can fit in a microscope setup) with sub-micron resolution [26], [27], [28]. There is an urgent need to map the matritecture across organs and diseases as this represents a major gap in our knowledge of the ECM and its regulation of cell behavior. Although other techniques provide a representation of ECM structure, studying decellularized organ scaffolds has provided the first 3D, high-resolution imaging of the ECM (Fig. 1).
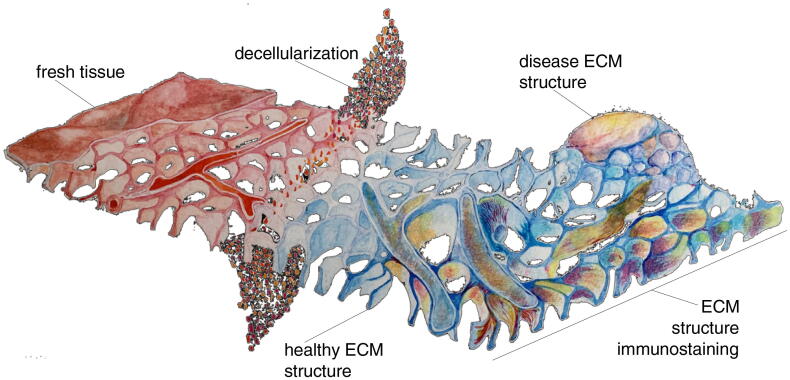
Fig. 1.
ECM mapping schematic. In fresh tissue, the ECM is covered by cells, hampering both the tagging of ECM proteins with antibodies and the visualisation of ECM structure. Removing cells (decellularization) lifts this obstacle, opening the way for precise imaging, down to sub-micron scale, of ECM structure. The development of method to decellularise and map disease-associated structures opens a window into disease progression.
These studies highlight that we should shed light on the matritecture by using an innovative combination of different methods, which will synergistically enable us to succeed and open new opportunities for the ECM field (Fig. 2).
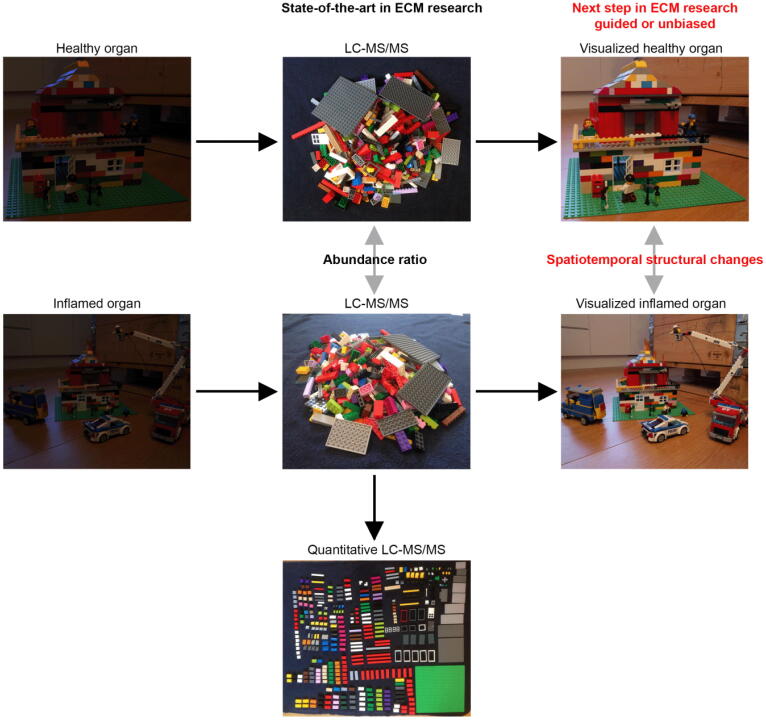
Fig. 2.
Scheme depicts the current state-of-the-art approach in ECM research, in which mass spectrometry techniques are used to determine protein abundance changes in two different situations within a tissue piece or an entire organ. After isolation of an organ the compositional and the spatial information of ECM protein components are not visible (left hand side). LC-MS/MS technology enables researcher to determine ECM protein abundance between two different conditions and/or quantitative protein amount. We believe that the next step in ECM research will be the spatiotemporal mapping of the ECM protein arrangement in decellularized healthy organs guided by LC-MS/MS experiments or unbiased for example during development or in comparison to respective inflamed situations such as cancer.
A call to action
Decoding the 3D organization of ECM components across developmental stages, ageing, and disease is a monumental, but necessary task that will have a direct impact on future therapy. Modifying therapeutic strategies to leverage ECM structure [29], [30], [31] could well represent a way to design “silver bullet” agents able to recognize and bind to tissue- or disease-specific ECM, thus focusing their effect in space and time. On the other hand, directly targeting ECM structures to normalize their properties is a promising therapeutic avenue [32], [33], [34].
Our group has successfully mapped ECM proteins and their respective structural arrangement in murine primary and metastatic tumors [26]. We have further started to compile a validated antibody library to detect a vast number of matrisome and matrisome-associated proteins [27]. Furthermore, our studies demonstrate how to map structural ECM components in almost every tissue during different developmental and disease stages. Another pioneering work has remarkably explored the mouse kidney ECM by an approach similar to our ISDoT study. Here, the authors performed immunofluorescence staining of decellularized tissue (iStainDoT) guided by mass-spectrometry analysis [35]. This study supports our vision of matritecture mapping as the next step in ECM research.
We could start by calling on the ECM research community to team up in generating an antibody database to enable precise mapping of the ECM architecture [36]. Such a catalogue could be integrated into databases such as RRID (https://www.rrids.org) or MatrixDB (http://matrixdb.univ-lyon1.fr/). Here, the society should take care of full transparency and validation of deposited antibodies, which could be supported by antibody-specific commentary sections. We believe that this mapping effort can decisively influence the future of therapy development, revealing, just as a land map does, the crucial structures that foster disease, and where to target them to collapse the support and normalize organ function.
A profound obstacle to map human tissues is the scarcity of samples coming from representative cohorts. Collaboration between ECM scientists, pathologists, and surgeons could result in biobanks containing surgical and autopsy samples (e.g. residual tissue from surgical excision) preserved without altering ECM structure (e.g. snap frozen). In the case of decellularization-based mapping, samples containing vessels and their territory are indispensable. Mapping the ECM involves more than decoding the matritecture during disease progression, generating disease-specific and -progressive maps; ultimately, it is necessary to understand the impact of the matritecture on cell behavior. The main limitation of decellularization-based methods is the absence of cells. Although we need to do the next step in ECM research by drawing these matritecture maps, we should simultaneously draft multimodal strategies to understand the impact of matritecture alterations on cellular programs. Here, IMS could guide decellularization and can further be complemented by transcriptomics assays such as Slide-seqV2 [37], [38] to trace back the gene expression information on the single cell level. In addition, Slide-seqV2 can be combined with scRNA-seq to increase sensitivity. Such innovative combinations of cutting-edge methods will synergistically yield the instructive role of the matritecture on cell behavior with spatiotemporal high-resolution at the single cell level. Finally, AI tools will enable us to integrate these imaging, genomics, and proteomics data to draw in silico models of disease relevant ECM organization guiding future strategies to normalize altered matritecture structures.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We want to thank the entire matrix biology community for providing custom made ECM antibodies and input for our research. Additionally, we thank Joshua Nikodemus for providing his Lego bricks and skills. This commentary was funded by the European Research Council (ERC-2015-CoG-682881-MATRICAN; A.E.M.-G., J.T.E.), German Cancer Aid (R.R.), and the Danish Cancer Society (R204-A12454; R.R.).
References
- 1.Sebe-Pedros A., Degnan B.M., Ruiz-Trillo I. The origin of Metazoa: a unicellular perspective. Nat. Rev. Genet. 2017;18(8):498–512. doi: 10.1038/nrg.2017.21. [DOI] [PubMed] [Google Scholar]
- 2.Naba A., Clauser K.R., Hoersch S., Liu H., Carr S.A., Hynes R.O. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol. Cell. Proteomics. 2012;11(4):M111–014647. doi: 10.1074/mcp.M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naba A., Hoersch S., Hynes R.O. Towards definition of an ECM parts list: an advance on GO categories. Matrix Biol. 2012;31(7–8):371–372. doi: 10.1016/j.matbio.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.R.O. Hynes, A. Naba, Overview of the matrisome--an inventory of extracellular matrix constituents and functions, Cold Spring Harb. Perspect. Biol. 4(1) (2012) a004903. [DOI] [PMC free article] [PubMed]
- 5.Zhu J., Clark R.A.F. Fibronectin at select sites binds multiple growth factors and enhances their activity: expansion of the collaborative ECM-GF paradigm. J, Invest. Dermatol. 2014;134(4):895–901. doi: 10.1038/jid.2013.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trier J.S., Allan C.H., Abrahamson D.R., Hagen S.J. Epithelial basement membrane of mouse jejunum. Evidence for laminin turnover along the entire crypt-villus axis. J. Clin. Invest. 1990;86(1):87–95. doi: 10.1172/JCI114720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verzijl N., DeGroot J., Thorpe S.R., Bank R.A., Shaw J.N., Lyons T.J., Bijlsma J.W.J., Lafeber F.P.J.G., Baynes J.W., TeKoppele J.M. Effect of collagen turnover on the accumulation of advanced glycation end products. J. Biol. Chem. 2000;275(50):39027–39031. doi: 10.1074/jbc.M006700200. [DOI] [PubMed] [Google Scholar]
- 8.Verzijl N., DeGroot J., Bank R.A., Bayliss M.T., Bijlsma J.W., Lafeber F.P., Maroudas A., TeKoppele J.M. Age-related accumulation of the advanced glycation endproduct pentosidine in human articular cartilage aggrecan: the use of pentosidine levels as a quantitative measure of protein turnover. Matrix Biol. 2001;20(7):409–417. doi: 10.1016/s0945-053x(01)00158-5. [DOI] [PubMed] [Google Scholar]
- 9.Halfter W., Dong S., Dong A., Eller A.W., Nischt R. Origin and turnover of ECM proteins from the inner limiting membrane and vitreous body. Eye (Lond) 2008;22(10):1207–1213. doi: 10.1038/eye.2008.19. [DOI] [PubMed] [Google Scholar]
- 10.Pesch M., König S., Aumailley M. Targeted disruption of the Lama3 gene in adult mice is sufficient to induce skin inflammation and fibrosis. J, Invest. Dermatol. 2017;137(2):332–340. doi: 10.1016/j.jid.2016.07.040. [DOI] [PubMed] [Google Scholar]
- 11.Keeley D.P., Hastie E., Jayadev R., Kelley L.C., Chi Q., Payne S.G., Jeger J.L., Hoffman B.D., Sherwood D.R. Comprehensive endogenous tagging of basement membrane components reveals dynamic movement within the matrix scaffolding. Dev. Cell. 2020;54(1):60–74.e7. doi: 10.1016/j.devcel.2020.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Y. Ariosa-Morejon, A. Santos, R. Fischer, S. Davis, P. Charles, R. Thakker, A.K. Wann, T.L. Vincent, Age-dependent changes in protein incorporation into collagen-rich tissues of mice by in vivo pulsed SILAC labelling, Elife 10 (2021). [DOI] [PMC free article] [PubMed]
- 13.Furman D., Campisi J., Verdin E., Carrera-Bastos P., Targ S., Franceschi C., Ferrucci L., Gilroy D.W., Fasano A., Miller G.W., Miller A.H., Mantovani A., Weyand C.M., Barzilai N., Goronzy J.J., Rando T.A., Effros R.B., Lucia A., Kleinstreuer N., Slavich G.M. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 2019;25(12):1822–1832. doi: 10.1038/s41591-019-0675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suleiman H., Zhang L., Roth R., Heuser J.E., Miner J.H., Shaw A.S., Dani A. Nanoscale protein architecture of the kidney glomerular basement membrane. Elife. 2013;2:e01149. doi: 10.7554/eLife.01149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Javitt G., Khmelnitsky L., Albert L., Bigman L.S., Elad N., Morgenstern D., Ilani T., Levy Y., Diskin R., Fass D. Assembly mechanism of mucin and von willebrand factor polymers. Cell. 2020;183(3):717–729.e16. doi: 10.1016/j.cell.2020.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Angel P.M., Schwamborn K., Comte‐Walters S., Clift C.L., Ball L.E., Mehta A.S., Drake R.R. Extracellular matrix imaging of breast tissue pathologies by MALDI-imaging mass spectrometry. Proteomics Clin. Appl. 2019;13(1):1700152. doi: 10.1002/prca.201700152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piehowski P.D., Zhu Y., Bramer L.M., Stratton K.G., Zhao R., Orton D.J., Moore R.J., Yuan J., Mitchell H.D., Gao Y., Webb-Robertson B.M., Dey S.K., Kelly R.T., Burnum-Johnson K.E. Automated mass spectrometry imaging of over 2000 proteins from tissue sections at 100-mum spatial resolution. Nat. Commun. 2020;11(1):8. doi: 10.1038/s41467-019-13858-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchberger A.R., DeLaney K., Johnson J., Li L. Mass spectrometry imaging: a review of emerging advancements and future insights. Anal. Chem. 2018;90(1):240–265. doi: 10.1021/acs.analchem.7b04733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Senior A.W., Evans R., Jumper J., Kirkpatrick J., Sifre L., Green T., Qin C., Žídek A., Nelson A.W.R., Bridgland A., Penedones H., Petersen S., Simonyan K., Crossan S., Kohli P., Jones D.T., Silver D., Kavukcuoglu K., Hassabis D. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577(7792):706–710. doi: 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- 20.Battle T., Arnal J.-F., Michel J.-B. Hyperproliferation of aortic smooth muscle cells and fibroblasts from young SHR rats is not shared by endothelial cells. Clin. Exp. Pharmacol. Physiol. 1994;21(12):981–989. doi: 10.1111/j.1440-1681.1994.tb02660.x. [DOI] [PubMed] [Google Scholar]
- 21.Behmer Hansen R.A., Wang X., Kaw G., Pierre V., Senyo S.E. Accounting for material changes in decellularized tissue with underutilized methodologies. Biomed Res. Int. 2021;2021:6696295. doi: 10.1155/2021/6696295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitano K., Schwartz D.M., Zhou H., Gilpin S.E., Wojtkiewicz G.R., Ren X., Sommer C.A., Capilla A.V., Mathisen D.J., Goldstein A.M., Mostoslavsky G., Ott H.C. Bioengineering of functional human induced pluripotent stem cell-derived intestinal grafts. Nat. Commun. 2017;8(1):765. doi: 10.1038/s41467-017-00779-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song J.J., Guyette J.P., Gilpin S.E., Gonzalez G., Vacanti J.P., Ott H.C. Regeneration and experimental orthotopic transplantation of a bioengineered kidney. Nat. Med. 2013;19(5):646–651. doi: 10.1038/nm.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prat-Vidal C., Rodriguez-Gomez L., Aylagas M., Nieto-Nicolau N., Gastelurrutia P., Agusti E., Galvez-Monton C., Jorba I., Teis A., Monguio-Tortajada M., Roura S., Vives J., Torrents-Zapata S., Coca M.I., Reales L., Camara-Rosell M.L., Cediel G., Coll R., Farre R., Navajas D., Vilarrodona A., Garcia-Lopez J., Munoz-Guijosa C., Querol S., Bayes-Genis A. First-in-human PeriCord cardiac bioimplant: Scalability and GMP manufacturing of an allogeneic engineered tissue graft. EBioMedicine. 2020;54 doi: 10.1016/j.ebiom.2020.102729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rafaeva M., Horton E.R., Jensen A.R.D., Madsen C.D., Reuten R., Willacy O., Brøchner C.B., Jensen T.H., Zornhagen K.W., Crespo M., Grønseth D.S., Nielsen S.R., Idorn M., Straten P.T., Rohrberg K., Spanggaard I., Højgaard M., Lassen U., Erler J.T., Mayorca‐Guiliani A.E. Modeling metastatic colonization in a decellularized organ scaffold‐based perfusion bioreactor. Adv Healthcare Mater. 2022;11(1):e2100684. doi: 10.1002/adhm.202100684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayorca-Guiliani A.E., Madsen C.D., Cox T.R., Horton E.R., Venning F.A., Erler J.T. ISDoT: in situ decellularization of tissues for high-resolution imaging and proteomic analysis of native extracellular matrix. Nat. Med. 2017;23(7):890–898. doi: 10.1038/nm.4352. [DOI] [PubMed] [Google Scholar]
- 27.Mayorca-Guiliani A.E., Willacy O., Madsen C.D., Rafaeva M., Elisabeth Heumüller S., Bock F., Sengle G., Koch M., Imhof T., Zaucke F., Wagener R., Sasaki T., Erler J.T., Reuten R. Decellularization and antibody staining of mouse tissues to map native extracellular matrix structures in 3D. Nat. Protoc. 2019;14(12):3395–3425. doi: 10.1038/s41596-019-0225-8. [DOI] [PubMed] [Google Scholar]
- 28.Mayorca-Guiliani A.E., Rafaeva M., Willacy O., Madsen C.D., Reuten R., Erler J.T. Decellularization of the murine cardiopulmonary complex. J Vis Exp. 2021;171 doi: 10.3791/61854. [DOI] [PubMed] [Google Scholar]
- 29.Mansurov A., Ishihara J., Hosseinchi P., Potin L., Marchell T.M., Ishihara A., Williford J.-M., Alpar A.T., Raczy M.M., Gray L.T., Swartz M.A., Hubbell J.A. Collagen-binding IL-12 enhances tumour inflammation and drives the complete remission of established immunologically cold mouse tumours. Nat. Biomed. Eng. 2020;4(5):531–543. doi: 10.1038/s41551-020-0549-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martino M.M., Briquez P.S., Güç E., Tortelli F., Kilarski W.W., Metzger S., Rice J.J., Kuhn G.A., Müller R., Swartz M.A., Hubbell J.A. Growth factors engineered for super-affinity to the extracellular matrix enhance tissue healing. Science. 2014;343(6173):885–888. doi: 10.1126/science.1247663. [DOI] [PubMed] [Google Scholar]
- 31.Momin N., Mehta N.K., Bennett N.R., Ma L., Palmeri J.R., Chinn M.M., Lutz E.A., Kang B., Irvine D.J., Spranger S., Wittrup K.D. Anchoring of intratumorally administered cytokines to collagen safely potentiates systemic cancer immunotherapy. Sci. Transl. Med. 2019;11(498) doi: 10.1126/scitranslmed.aaw2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.K.K. McKee, S.C. Crosson, S. Meinen, J.R. Reinhard, M.A. Ruegg, P.D. Yurchenco, Chimeric protein repair of laminin polymerization ameliorates muscular dystrophy phenotype, J. Clin. Invest. 127(3) (2017) 1075-1089. [DOI] [PMC free article] [PubMed]
- 33.Reinhard J.R., Lin S., McKee K.K., Meinen S., Crosson S.C., Sury M., Hobbs S., Maier G., Yurchenco P.D., Rüegg M.A. Linker proteins restore basement membrane and correct LAMA2-related muscular dystrophy in mice. Sci. Transl. Med. 2017;9(396) doi: 10.1126/scitranslmed.aal4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reuten R., Zendehroud S., Nicolau M., Fleischhauer L., Laitala A., Kiderlen S., Nikodemus D., Wullkopf L., Nielsen S.R., McNeilly S., Prein C., Rafaeva M., Schoof E.M., Furtwängler B., Porse B.T., Kim H., Won K.J., Sudhop S., Zornhagen K.W., Suhr F., Maniati E., Pearce O.M.T., Koch M., Oddershede L.B., Van Agtmael T., Madsen C.D., Mayorca-Guiliani A.E., Bloch W., Netz R.R., Clausen-Schaumann H., Erler J.T. Basement membrane stiffness determines metastases formation. Nat. Mater. 2021;20(6):892–903. doi: 10.1038/s41563-020-00894-0. [DOI] [PubMed] [Google Scholar]
- 35.Lipp S.N., Jacobson K.R., Hains D.S., Schwarderer A.L., Calve S. 3D mapping reveals a complex and transient interstitial matrix during murine kidney development. J. Am. Soc. Nephrol. 2021 doi: 10.1681/ASN.2020081204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rickelt S., Hynes R.O. Antibodies and methods for immunohistochemistry of extracellular matrix proteins. Matrix Biol. 2018;71-72:10–27. doi: 10.1016/j.matbio.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 37.Rodriques S.G., Stickels R.R., Goeva A., Martin C.A., Murray E., Vanderburg C.R., Welch J., Chen L.M., Chen F., Macosko E.Z. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363(6434):1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stickels R.R., Murray E., Kumar P., Li J., Marshall J.L., Di Bella D.J., Arlotta P., Macosko E.Z., Chen F. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat. Biotechnol. 2021;39(3):313–319. doi: 10.1038/s41587-020-0739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]