Abstract
We performed an epidemiological investigation and genome sequencing of severe acute respiratory coronavirus virus 2 (SARS-CoV-2) to define the source and scope of an outbreak in a cluster of hospitalized patients. Lack of appropriate respiratory hygiene led to SARS-CoV-2 transmission to patients and healthcare workers during a single hemodialysis session, highlighting the importance of infection prevention precautions.
Preventing healthcare-associated severe acute respiratory coronavirus virus 2 (SARS-CoV-2) transmission continues to be a critical challenge. Although improved understanding of SARS-CoV-2 biology and transmission has helped define protocols for safe care delivery in inpatient units, these protocols may be more difficult to consistently apply in other care areas such as inpatient hemodialysis units. Hemodialysis represents a unique risk for transmission because it requires 4–6-hour stays close to other patients, usually without solid barriers such as walls, and indirect transmission may occur via contaminated equipment, environmental surfaces, or contact with healthcare workers (HCWs). Moreover, patients receiving hemodialysis have a higher risk of acquiring COVID-19 and higher morbidity and mortality. 1,2
Over a 10-day period in January 2021, our hospital infection prevention department detected a cluster of 8 patients with hospital-onset SARS-CoV-2 infection during routine daily surveillance. We performed epidemiological investigation and SARS-CoV-2 genome sequencing to define the source and scope of the outbreak, to identify exposed HCWs, and to determine remediation strategies.
Materials and Methods
Outbreak investigation
Our 500-bed acute-care hospital in an academic healthcare system in Atlanta, Georgia, performs SARS-COV-2 real-time polymerase chain reaction (RT-PCR) on nasopharyngeal swabs from all patients on admission by protocol. Hospital-onset coronavirus disease 2019 (COVID-19) was defined as a positive SARS-CoV-2 RT-PCR test ≥3 days after admission after testing negative on admission. Upon recognition of an outbreak, infection prevention staff conducted an investigation, including generating a line list to assess for common risk factors and exposures. Electronic medical records (EMRs) were reviewed to obtain clinical and laboratory data, location, and contact with HCWs. Review of HCW schedule logs, HCW interviews, and contact tracing were conducted to determine the outbreak timeline and to identify exposed HCWs.
The study was approved by the Institutional Review Board of Emory University. Residual NP samples were available from 7 individuals; the others had been discarded by the time of investigation. Samples underwent RNA extraction, library construction, Illumina sequencing, and reference-based SARS-CoV-2 genome assembly as previously described. 3,4 SARS-CoV-2 genomes were also sequenced from 10 nonoutbreak patients in the same facility and 58 patients in nearby facilities between December 12, 2020, and January 13, 2021. All locally generated reads (cleaned of human reads) and SARS-CoV-2 genomes are available (NCBI BioProject no. PRJNA634356). All sequences used in this analysis are publicly available in GISAID under the accession numbers provided in Supplementary Table 1 (online). Further details are also available in the Supplementary Material (online).
Results
Between January 4 and 14, 2021, during a period of high community SARS-CoV-2 transmission, 8 patients (cases 1–8) with hospital-onset COVID-19 were identified; all were receiving hemodialysis in our 16-bay hemodialysis unit and 5 were bedded in one 24-bed inpatient renal ward with single patient rooms (Supplementary Fig. 1 online). Each patient had a negative SARS-CoV-2 RT-PCR test on admission within the preceding 60 days. The outbreak investigation identified 53 potentially exposed HCWs who were offered SARS-CoV-2 PCR testing. Of the 29 HCWs who underwent testing, 5 cases (17%) were positive (cases 9–13).
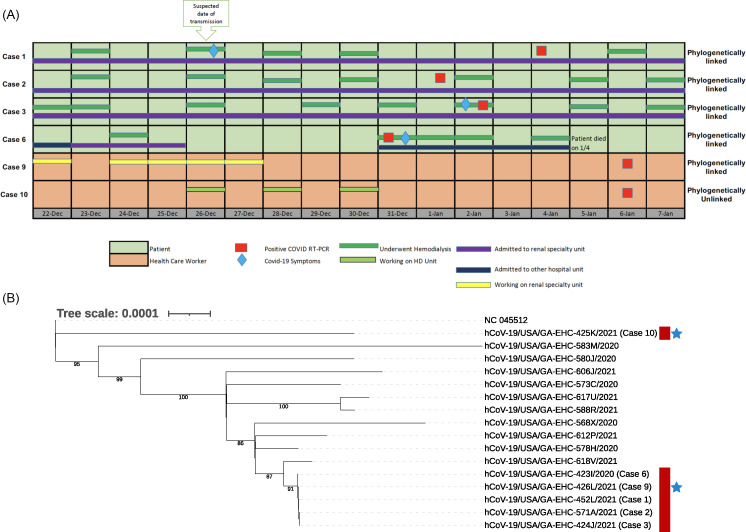
Several plausible transmission paths were identified, all involving a putative index patient, case 1, who was admitted on December 22, 2020, underwent hemodialysis on December 26, 2020, developed fever that evening, and tested positive for SARS-CoV-2 on January 4, 2021 (Fig. 1A; Supplementary Fig. 2 online). In case 1, the infection was deemed community-acquired based on the timing of symptoms. Further investigation revealed that case 1 had a persistent cough during hemodialysis on December 26, 2020, but this patient did not consistently wear a mask over their nose and mouth. Of the 7 other positive patients, 5 (cases 2–5 and 7) were in the hemodialysis unit on December 26, 2020, in bays close to the bay where case 1 was treated (ie, bays 4, 10, 11 and 15) (Supplementary Fig. 1A online). All tested positive for SARS-CoV-2 within 14 days. A sibling of case 1, case 8, was admitted to an inpatient ward in a different building, and the 2 siblings had reportedly visited each other surreptitiously without wearing masks. Case 6 was not in the hemodialysis unit on December 26, 2020, but this patient was bedded in the same inpatient renal ward as case 1 during the preceding 3 days (Fig. 1A; Supplementary Figs. 1B and 2 online).
Fig. 1.
Epidemiological and viral genomic analysis of the 6 outbreak cases with samples available for SARS-CoV-2 genome sequencing. (A) Exposure and onset of symptoms. Four patients with hospital-onset COVID-19 were receiving hemodialysis and bedded in a single inpatient renal ward. A fifth patient (case 6) was bedded in the same inpatient renal ward. Two exposed HCWs were working on that unit (case 9) or in the hemodialysis unit (case 10). (B) Maximum-likelihood phylogenetic tree showing SARS-CoV-2 sequences from 4 patients (cases 1, 2, 3, and 6; red) and 2 HCWs (case 9, 10; blue stars) with samples available as well as 10 patients in the same facility between December 12, 2020, and January 13, 2021. Bootstraps are shown for high confidence branches (>80).
Of the 5 HCWs with positive SARS-CoV-2 tests (cases 9–13), 3 were floor nurses who cared for several of the cases on multiple days in the inpatient renal ward and 2 were nurses on the hemodialysis unit (Fig. 1A; Supplementary Fig. 2 online). Case 10 reported an additional community exposure.
The outbreak investigation also identified lapses in infection prevention including inconsistent (1) eye protection worn by HCWs when interacting with patients; (2) mask use by patients, particularly in the inpatient hemodialysis unit; (3) use of barrier protections (curtains) in the hemodialysis bays; and (4) adherence to occupancy limits in staff break rooms. Recommended remediation strategies were implemented, including wearing consistent eye protection for HCWs, providing masks to patients in their rooms and at hemodialysis and encouraging their use, drawing side curtains during hemodialysis sessions, and identifying alternative break spaces for HCWs. No further cases were identified.
Residual nasopharyngeal samples were available from 7 of the 13 cases, and SARS-CoV-2 genomes were successfully sequenced from 6 of these cases (ie, 4 patients and 2 HCWs). SARS-CoV-2 sequences from all 4 patients (cases 1, 2, 3, and 6) and 1 HCW (case 9) were identical, formed a distinct cluster on a phylogenetic tree (lineage B.1.2), and were at least 4 single-nucleotide polymorphisms (SNPs) removed from the next closest sequence in this study (Fig. 1B, Supplementary Figs. 2 and 3 online). The sixth sequence (case 10) was phylogenetically distinct (lineage A.21). Thus, SARS-CoV-2 sequence analysis supported a common infection source among all 4 patients and 1 of the 2 HCWs for whom sequence data were available.
Discussion
We identified 8 patients undergoing hemodialysis who developed hospital-onset COVID-19, and a detailed epidemiologic investigation identified a probable transmission event in the inpatient hemodialysis unit from 1 patient to 5 other patients on a single day due to suboptimal infection prevention practices. The clinical epidemiology was supported by molecular epidemiology; SARS-CoV-2 sequences from the 3 samples available (cases 1, 2, and 3) were identical to one another and were distinct from contemporary strains in the hospital, local community, and state. Another transmission chain likely involved 2 patients (cases 1 and 6) who were admitted to the same inpatient renal ward and had identical SARS-CoV-2 genome sequences. Although sequence data were not available from case 8, another transmission chain likely involved this patient and their sibling, case 1. Interestingly, SARS-CoV-2 genome sequencing revealed that case 10, an HCW, did not share the same source as the other cases. This individual may instead have acquired SARS-CoV-2 from the community, as has been reported to exceed the risk from patient exposures. 5 Because SARS-CoV-2 sequences were not available from all cases, it is not clear whether case 10 was part of a second transmission cluster.
Unlike in prior published hemodialysis related outbreaks, 1,6–8 we used SARS-CoV-2 genome sequencing to both provide strong support for a transmission cluster linking 5 individuals and to determine that 1 individual was not part of the cluster. This finding underscores the valuable contribution of pathogen genome sequencing to outbreak investigations. 9 Ongoing efforts to expand SARS-CoV-2 genomic surveillance will bolster outbreak investigations as characterization of locally circulating strains provides greater resolution to assess transmission clusters.
Overall, our results highlight how breaks in infection prevention in a high-risk population can result in an outbreak and the importance of pathogen genome sequencing in hospital outbreak investigations. Our findings support the need for continued vigilance and adherence to best practices in hemodialysis centers.
Acknowledgments
We thank Dr Susan Ray, the Emory Clinical Virology Research Laboratory, the Georgia Clinical Research Centers, and the Emory Integrated Genomics Core for support in sample collection and sequencing. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/ice.2021.465.
click here to view supplementary material
Financial support
This study was supported by the Centers for Disease Control and Prevention (contract no. 75D30121C10084 under BAA ERR 20-15-2997). The Emory WHSC COVID-19 Urgent Research Engagement (CURE) Center made generous philanthropic support available from the O. Wayne Rollins Foundation and the William Randolph Hearst Foundation. Further support was provided by the National Center for Advancing Translational Sciences of the National Institutes of Health (grant no. UL1TR002378).
Conflicts of interest
The authors report no relevant conflicts of interest.
References
- 1. Bigelow BF, Tang O, Toci GR, et al. Transmission of SARS-CoV-2 involving residents receiving dialysis in a nursing home—Maryland, April 2020. Morb Mortal Wkly Rep 2020; 69: 1089–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gupta S, Hayek SS, Wang W, et al. Factors associated with death in critically ill patients with coronavirus disease 2019 in the US. JAMA Intern Med 2020; 180: 1436–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Waggoner JJ, Stittleburg V, Pond R, et al. Triplex real-time RT-PCR for severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 2020; 26: 1633–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Babiker A, Bradley HL, Stittleburg VD, et al. Metagenomic sequencing to detect respiratory viruses in persons under investigation for COVID-19. J Clin Microbiol 2020;59:e02142–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jacob JT, Baker JM, Fridkin SK, et al. Risk factors associated with SARS-CoV-2 seropositivity among US healthcare personnel. JAMA Netw Open 2021;4:e211283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schwierzeck V, König JC, Kühn J, et al. First reported nosocomial outbreak of severe acute respiratory syndrome coronavirus 2 in a pediatric dialysis unit. Clin Infect Dis 2021; 72: 265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yau K, Muller MP, Lin M, et al. COVID-19 outbreak in an urban hemodialysis unit. Am J Kidney Dis 2020;76:690–5.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iio R, Kaneko T, Mizuno H, Isaka Y. Clinical characteristics of COVID-19 infection in a dialysis center during a nosocomial outbreak. Clin Exp Nephrol 2021; 25: 652–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Babiker A. mSphere of influence: whole-genome sequencing, a vital tool for the interruption of nosocomial transmission. mSphere 2021;6:e00230–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/ice.2021.465.
click here to view supplementary material