Abstract
Mass spectrometry-based metaproteomics has emerged as a prominent technique for interrogating the functions of specific organisms in microbial communities, in addition to total community function. Identifying proteins by mass spectrometry requires matching mass spectra of fragmented peptide ions to a database of protein sequences corresponding to the proteins in the sample. This sequence database determines which protein sequences can be identified from the measurement, and as such the taxonomic and functional information that can be inferred from a metaproteomics measurement. Thus, the construction of the protein sequence database directly impacts the outcome of any metaproteomics study. Several factors, such as source of sequence information and database curation, need to be considered during database construction to maximize accurate protein identifications traceable to the species of origin. In this review, we provide an overview of existing strategies for database construction and the relevant studies that have sought to test and validate these strategies. Based on this review of the literature and our experience we provide a decision tree and best practices for choosing and implementing database construction strategies.
Keywords: Metaproteome, Metagenomics, Microbiome, Microbial community, Multi-omics, Microbiota, Microbial ecology
1. Introduction
Metaproteomics is an umbrella term encompassing approaches for the large-scale identification and quantification of proteins from microbial communities [1]. Metaproteomics provides insights into the expressed genes and thus actual phenotypes on the molecular level whereas the more popular DNA sequencing approaches can only determine functional potential by profiling gene content [2], [3], [4]. As part of the molecular phenotype of the cell, proteins provide more direct insight into what is happening physiologically in the cells within microbial communities [5]. For example, a study of symbiotic marine worms discovered abundant enzymes for the use of carbon monoxide as an energy source in the symbionts, revealing the first animal known to be able to use this poisonous gas [6]. Another study identified an increase in the abundances of iron sequestration enzymes in the microbiota of human preterm infants with necrotizing enterocolitis (NEC), suggesting an association between iron homeostasis and NEC [7]. In mouse gut communities, Patnode et al. found distinct expression of polysaccharide utilization loci (PUL) in the presence of different food grade fibers and showed that these PULs were necessary for the competitive fitness of specific Bacteroides species in the presence of these fibers [8]. Finally, Li et al. confirmed the assimilation of methanol by microbes in the plant rhizosphere by first detecting abundant methanol dehydrogenases and associated oxidation pathways, then using 13C-labeled methanol to confirm the incorporation of labeled carbon into proteins of organisms that expressed these proteins [9].
The leading technique for identifying and quantifying proteins in biological samples is called shotgun proteomics [10]. For shotgun proteomics proteins are first isolated from samples and then (sometimes) separated by gel electrophoresis. Isolated proteins are digested into peptides using trypsin, and these peptides are separated by liquid chromatography based on physicochemical properties before analysis in a mass spectrometer. Both intact peptide masses (MS1) and, after fragmentation, the masses of their fragments (MS2) are measured in the mass spectrometer. This technique is called tandem mass spectrometry. To identify proteins, a bioinformatics method called database search matches peptide tandem mass spectra to theoretical spectra derived from an in silico digested protein database (Fig. 1) [11], [12], [13], [14]. Tens of thousands of peptides can be analyzed using this method [15], [16], [17]. These peptides can subsequently be used to infer the presence of thousands of proteins in the sample.
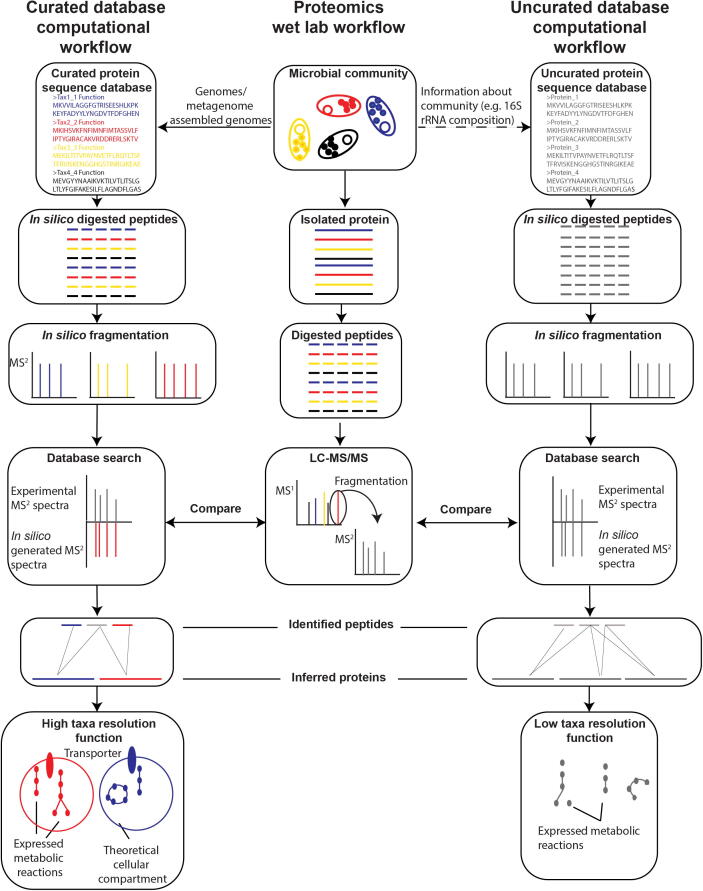
Fig. 1.
Overview of how protein sequence database construction impacts protein identification. The figure shows an overview of the wetlab experimental portion of acquiring metaproteomic mass spectrometry data and the computational steps done on the database that mirror the wet lab steps. In the database search step the experimental MS2 spectra are then compared with the in silico generated spectra from the database. We differentiate in the figure between two distinct database quality levels (curated and uncurated) and their ultimate impact on the output and interpretation of metaproteomic experiments.
While the shotgun proteomics approaches described above were originally developed to analyze proteomes of individual organisms, they have been adapted in a very similar form for metaproteomics [1], [5], [18]. However, metaproteomics comes with unique challenges not encountered when working with single organisms including (1) the difficulty of obtaining protein sequences from the organisms in the often highly diverse microbial communities and (2) the fact that the presence of homologous sequences from related organisms can make protein inference much more difficult [19], [20]. Some researchers avoid the protein inference problem by using a peptide-centric approach, which skips the protein inference step and infers taxonomy and function directly from detected peptides by matching the peptide sequences to peptide sequences generated from public protein sequence reference databases [21], [22]. The peptide-centric approach has the advantage that it avoids the protein inference step, however, it comes at the cost of being unable to know which specific proteins are present in a sample and thus functions and presence of taxonomic groups have to be aggregated at a relatively unspecific level.
In this review, we focus on a protein-centric approach, which seeks to identify and quantify specific proteins, which are ultimately the biological unit investigated with metaproteomics. The protein-centric approach, when coupled with a well curated protein sequence database, is more sensitive and selective for taxonomic and functional annotation than the peptide-centric approach as key information for the taxonomic and functional classification of a protein can be accessed [23], [24]. This information includes the genome of origin of the protein sequence [24], [25], [26], which provides information on the taxon of origin and the neighborhood of the expressed gene, which, in prokaryotes, can often be very informative for deriving protein function [27], [28]. Our intended audiences for this review are (1) proteomics experts who have not previously worked with microbial communities and as such may be unfamiliar with some of the additional challenges involved in database construction for metaproteomics, (2) microbiologists who are interested in metaproteomics and are looking for concise guidance on specific elements of the metaproteomic process, and (3) metaproteomics experts seeking an overview of which database construction strategies have been developed and validated so far and where further need for development and validation still exists. Here we focus on construction of the protein sequence database, a key element of any metaproteomic study. This review is divided into two sections. In the first section we describe how protein database source and construction can impact peptide identification, protein inference, and taxonomic assignment. In the second section we provide a decision making framework for constructing protein databases for metaproteomics. For other topics, we refer the reader to articles on the overall metaproteomic workflow [18], [29], and on methodological considerations for specific components of the metaproteomic workflow [30], [31], [32], [33], [34], [35], [36].
Box 1: Definitions of key terms used in the manuscript.
Tandem mass spectrometry: A mass spectrometry technique where (peptide) ions are isolated by their mass to charge ratio and then fragmented in the mass spectrometer, both the initial mass of the ion and the masses of the resulting fragment ions are recorded.
MS1 spectrum: Mass spectrum of intact (peptide) ions i.e. not fragmented.
MS2 or MS/MS spectrum: Mass spectrum of (peptide) fragment ions generated by tandem mass spectrometry.
Database search algorithm: An algorithm that identifies peptides from tandem mass spectrometry data by matching experimental MS2 spectra to MS2 spectra generated in silico from a protein database. The algorithm also scores and ranks the best matches for each MS2 spectrum.
Protein database: Database of protein sequences in FASTA file format used by a database search algorithm.
Peptide-spectrum match (PSM): Match between an MS2 spectrum and a peptide sequence found by a database search algorithm.
False discovery rate (FDR): Proportion of PSMs, peptides or proteins passing selection criteria (e.g. search algorithm score threshold) that are incorrect.
Target-decoy search strategy: A database search strategy to estimate false discovery rates in which the mass spectrometry data is searched against a database made up of correct protein sequences (targets) and incorrect protein sequences (decoys). Commonly decoy sequences are generated by reversing the sequences in the target database.
Metagenome-assembled genome (MAG): Genome fragments (contigs) extracted from metagenomic assemblies and combined into what is thought to be a close representation of an actual individual genome that matches a specific strain/species in the sequenced sample. Sometimes the product of the initial contig grouping is called a “bin” and only after various quality checks for completeness and contamination is the bin then considered a “MAG” if quality thresholds are met [37].
Genome-resolved metagenomics: A metagenomic processing strategy in which the goal is to extract metagenome-assembled genomes. At the center of this strategy are binning methods, which group sequenced genome fragments into MAGs based on characteristics such as tetranucleotide frequency and sequencing coverage.
Matched metagenome database: Protein database that is derived from metagenomic sequencing of samples that match the ones used for metaproteomics.
Reference protein database: Protein database derived from public repositories such as NCBI or UniProt.
Unmatched metagenome: Protein database derived from previous metagenomics and isolate sequencing efforts of a specific system. Sometimes these metagenomes are published as gene catalogs. Examples include the mouse gene catalog [38] or the IGC [39].
Protein unique peptide: A peptide identified by a database search algorithm that is unique to a single protein sequence in the database.
Average nucleotide identity (ANI): A measure of genome similarity that is commonly used to classify genomes into species [40], [41].
Protein group: A group of similar, but not identical, protein sequences that share identified peptides and which cannot be distinguished due to the lack of protein unique peptides.
Lowest common ancestor (LCA) : A taxonomy assignment strategy that matches a sequence (raw read, protein, or contig) to a reference database and assigns the taxonomy of the lowest unambiguous taxonomic rank of similar sequences in the reference database.
2. Protein database source and construction affects peptide identification, protein inference, and taxonomic assignment in metaproteomics
Assigning peptide sequences to tandem mass spectra and inferring proteins depends on the sequence database provided to the database search algorithm. In this section, we provide an in depth review of the interconnection of the sequence database with peptide identification and protein inference and how the source of sequences in the database influences peptide identification and protein inference. Furthermore we review the current literature on studies that have sought to evaluate the impact of different database sources and construction strategies on the quality and information content of metaproteomic data.
2.1. Peptide identification by database search algorithms
To understand the importance of protein sequence databases for shotgun proteomics it is critical to understand how database search algorithms work. For a detailed explanation we would like to refer readers to an excellent article by Marcotte [42]. Briefly, database search relies on a target protein sequence database to provide a search space of theoretical MS2 spectra for peptides that might be in the sample. The algorithm tries to match the experimental peptide MS2 spectra to these theoretical spectra. If a spectrum is successfully matched to a peptide, this is referred to as a peptide-spectrum match (PSM). Database search algorithms score and rank PSMs based on the similarity of the match between theoretical and experimental MS2 spectra. Since the implementation of the first 1994 algorithm - SEQUEST - many additional algorithms have been developed with accompanying improvements in the scoring scheme and search speed [11], [43], [44], [45], [46]. Metaproteomic studies tend to have more mass spectra and a larger search space than single organism studies, making certain search algorithms unable to handle the data due to time or memory limitations. There are, however, many database search platforms able to process metaproteomic data. These platforms include MetaproteomeAnalyzer [47], MetaProIQ [48], and Sipros Ensemble [49], which have been built specifically for metaproteomic data, and more general proteomics pipelines, such as the open source Crux toolkit [50] and commercial pipelines such as Thermo Fisher’s Proteome Discoverer and Bioinformatics Solutions’ PEAKS.
After database search, PSMs are filtered to only retain quality PSMs. Commonly PSMs are filtered based on database search algorithm scores and peptide properties such as length and missed cleavages to meet a specified false discovery rate (FDR). To calculate the FDR, a decoy database made up of reversed or randomized sequences from the target database is included in the database search [51]. The FDR is calculated using a target-decoy competition, where the top PSM (target or decoy) for a MS2 spectrum is retained and the FDR is the proportion of total PSMs that are decoy hits at the used score threshold [51]. Crude FDR filtering using individual database search algorithm scores can lead to biased removal of PSMs with specific properties. This issue has been addressed by the development of machine learning algorithms, such as Percolator [52] and Sipros Ensemble [49], which consider a large diversity of scores and peptide properties for FDR-based PSM filtering. Advice on FDR thresholds is out of scope for this article, but thresholds typically range between one and five percent. PSMs from the target database whose score passes the FDR threshold are considered identified and used for protein inference. A problem with the target-decoy competition for FDR calculation that the literature has just started to address is that it assumes that there is only one peptide per MS2 spectrum; however, sometimes the mass spectrometer co-isolates multiple peptides with similar mass to charge ratios. The higher complexity of metaproteomics studies increases the probability of these co-isolation events. Though some solutions have been suggested [53], [54], [55], further discussion is beyond the scope of this review.
The size and comprehensiveness of the protein database impacts the number of PSMs that can be identified. The comprehensiveness of the database (i.e. how many of the proteins in the sample are represented as sequences in the database) constrains the maximum number of peptides that can be detected in a sample as peptides not present in the database cannot be identified by database search. Increased database size, especially with regards to sequences not expected to be present in the sample, increases the number of high-scoring random hits to both the target and decoy portion of the database. These high-scoring random hits are false positives that lead to a tighter score threshold needed to attain the desired false discovery rate [56], [57]. The tighter score threshold needed for large databases with unnecessary sequences leads to the filtering out of true PSMs that would be retained with score thresholds needed for a smaller, better fitting database. Small non-comprehensive databases experience a similar problem, when not handled carefully, as similar sequences (very similar precursor mass and some shared y- and b-ions) can receive the best score when the real sequence is absent [58]. Thus, a large database, especially with many sequences not relevant to the sample, or a small non-comprehensive database limit the potential for peptide identifications.
A solution for issues with very large databases that has been proposed is a two-step or multi-step search approach. In this approach, searches made with higher (>5%) or no FDR thresholding are used to generate a protein database restricted to just sequences that had a match in the initial search against the very large database. Two-step approaches have been shown to increase peptide identifications when databases are very large [59]; however, the validity of this kind of approach is debated because it takes advantage of prior information to improve the FDR [32], [60], [61]. Some solutions to this problem have been suggested but not fully validated [58], [62].
Automated peptide de novo sequencing from MS2 spectra represents an alternative to database search algorithms for obtaining peptide sequences from mass spectrometry data [63], [64], [65]. While no longer constrained by the database search space for peptide identification [66], peptide de novo sequencing approaches typically generate fewer peptides than database search [67], and still depend on a protein sequence database for protein inference [68] and subsequent biological interpretation [66]. Thus, de novo peptide sequencing approaches do not overcome the need for high quality protein sequence databases in metaproteomics.
2.2. Protein inference after database search
Peptides that pass the FDR threshold can be used to infer proteins by mapping identified peptide sequences back to proteins in the protein database. A challenge with this approach, known as the protein inference problem, is that some peptides are shared between protein sequences making it difficult to determine which protein was the actual source of the peptide and should thus be identified as present in the sample [20]. Metaproteomics exacerbates this problem on two levels. First, metaproteomic samples often contain many relatively closely related strains/species which have a partially shared set of homologous proteins. Depending on the sequence similarity between these homologs, a set of peptides that can be derived from these proteins will be identical between multiple strains/species, making them “non-unique” to a protein from a specific strain/species. These “non-unique” peptides can therefore only be used to determine the presence of a protein, but not its source species/strain. Second, sequence databases often contain very similar or identical sequences. This sequence redundancy can either be caused by having multiple identifiers for identical or highly similar sequences caused by bringing together data from multiple metagenomic assemblies or public databases, or the presence of strains/species with very similar sequences in a sequenced sample. Ultimately, both presence of proteins that yield identical peptides and sequence database redundancy lead to the same outcome of protein inference, namely the ambiguous matching of peptides to multiple sequences in the database. While no perfect approach exists to address the protein inference challenge in metaproteomics and even for single organisms proteomics, there are several approaches to limit the impact of protein inference challenges on metaproteomic data interpretation.
To address the protein inference challenge, several metrics can be employed to improve confidence in protein identification. The most critical is filtering of inferred proteins to attain a specific FDR cutoff. FDRs on the protein level are estimated with the same target-decoy approach described above for PSM identification. Different protein inference methods and algorithms use a diversity of parameters to filter inferred proteins. These parameters can include the number of peptides matching to a protein [15], [69], [70], uniqueness of peptides matching to a protein (unique peptides) [26], [71], and the quality scores or FDR of peptides matching to a protein [72], [73]. In addition to FDR filtering, parsimony methods are frequently used to group proteins that share peptides, but have no independent evidence in the form of unique peptides, into protein groups of shared evidence [74], [75], [76]. Thus the detection of peptides unique to a protein sequence critically impacts the interpretation of identified proteins in metaproteomics [24]. Proteins with unique peptides have the advantage of being unambiguously identified and can be directly linked back to the taxon of origin if the protein sequence came from a taxonomically classified genome, whereas protein groups can potentially not be associated with a specific taxon only with a wider group of taxa. Protein groups, however, can still provide a clear identification of a particular protein function. To deal with unnecessary sequence redundancy in databases and to increase the number of identified unique peptides metaproteomics researchers frequently use a sequence clustering algorithm, such as CD-HIT [77] or UCLUST [78], to group highly similar or identical protein sequences, adding only one representative protein sequence with a single identifier to the protein sequence database [18]. This approach has been applied in many studies [7], [26], [30], [70], [79] and is discussed in detail in section 3.3.
2.3. Sources of sequence information to construct protein databases for metaproteomic studies
In metaproteomics, creating a protein database that is both comprehensive and not larger than it needs to be is particularly challenging. In a proteomics study with only one organism, the associated protein sequence database comes from predicted or known protein sequences derived from the organism's genome [56]. Ideally, this protein database comes from publicly available and reviewed reference proteomes, such as those found in NCBI RefSeq [80] or UniProt [81]. In metaproteomics, sequences from multiple organisms need to be acquired to create a comprehensive representation of the protein sequences likely to be in the sample. With the exception of artificially created, fully defined, communities [8], [82] or symbioses with a limited number of highly specific symbionts [6], [83], [84], [85], [86], it is often not possible to create this database by just combining the relevant RefSeq/UniProt reference proteomes or by using previous sequencing initiatives because the composition of environmental microbial communities cannot be known in advance without some form of prior sequencing. Even if the composition of a community is known (e.g. from amplicon sequencing), genomes for the organisms in the community are often not available in public databases [87], [88], [174]. Adding complication, in many cases, the microbial composition of a system can be different from sample to sample, as is the case for intestinal microbiome samples [89], [90]. All this makes assembling the set of sequences needed for a metaproteomic study a task that requires careful consideration to obtain and combine the best possible set of sequences.
In the following we will describe the different types of sources of sequence information that have been used in the past to create metaproteomic protein databases.
-
1.
Matched metagenomes: sequences collected from metagenomes assembled from a set of samples that match the metaproteomic samples [26], [70], [79]. The main advantage of this sequence source is that it provides a set of protein sequences derived from the genomes of the organisms present in the samples interrogated by metaproteomics. With extensive processing, these matched metagenomes can be made genome-resolved by extracting metagenome-assembled genomes (MAGs) using a variety of binning methods to assign genome fragments (contigs) from a metagenomic assembly to individual genomes (see section 3.2) [120], [121], [122]. Protein sequences predicted from MAGs have the advantage that more information is available for analyses after identification, such as genomic neighborhood and taxonomic classification by the genome of origin [7], [25], [26]. Genome-resolved metagenomics, however, has the limitation that it has historically required extensive metagenomics expertise often unavailable to mass spectrometry groups. It is also currently infeasible to bin all the sequences in a metagenome into MAGs, which leads to loss of information if only binned sequences are used [6]. To save time, the protein database can also be assembled from genes predicted directly from raw sequence information [88], [94] or unbinned metagenomic assemblies [70], [79], [88]. This comes at the cost of lost taxonomic resolution as discussed in section 3.4.
-
2.
Unmatched metagenomes: sequences collected from metagenomic data from the same system (e.g., human or mouse gut) but different samples potentially from different studies and laboratories [48], [169], sometimes called gene catalogs [38], [39], [91]. Use of sequences from unmatched metagenomes is most common in human microbiome studies where there have already been massive sequencing efforts [48], [169]. The use of results from these sequencing initiatives for metaproteomics results in databases of millions of sequences that can make it difficult to achieve a high number of identifications at a low false discovery rate without a multi-step search strategy [48], [59], [170]. This approach also limits the taxonomic resolution to higher levels (e.g. phylum or genus level) when MAGs are not included [169], [170], [171]. In instances where the community is known to be the same despite different samples, such as microbial symbioses with highly specific symbionts, this approach can be equivalent to a matched metagenome [6], [83], [84], [85], [86].
-
3.
Unrestricted reference databases: this approach uses all of the sequences from one of the major sequence repositories (e.g. NCBI RefSeq). Large unrestricted databases have the advantage of covering a large sequence space; however, they suffer from not being specific to the sample, leading to the possibility of false hits [57], [102] and low identification numbers [88], [92], [93], [94] due to the tight PSM identification scores needed to achieve a desired FDR with a large database (see section 2.1). Also, public databases are currently very incomplete with regards to genome coverage for microbial communities [87], [88], [174]. As evidenced by the fact that 80%-90% of the MAGs in metagenome projects belong to unnamed species absent from public repositories [172], [173].
-
4.
Restricted reference databases: this approach uses prior knowledge of the community’s composition to acquire taxonomically relevant reference proteomes. For artificially defined communities (e.g., germ-free mice inoculated with a set of bacterial isolates), this approach is equivalent or better than matched metagenomes in terms of taxonomic resolution and completeness because the exact community composition is known in advance and reference proteomes of the specific members can be used [8], [82]. When the exact community composition is not known, an alternative approach is to use results from a phylogenetic marker gene analysis of the sample, such as 16S rRNA gene sequencing, to identify reference genomes that correspond most closely to the phylogeny of the marker genes [88], [106]. This approach relies on the relevant reference genomes being present in a public repository, and depends on strains from the same species, let alone genus, having similar gene content. Many strains from the same species, however, do not have the same gene content as evidenced by studies on the massive pangenomes of some microbial species such as Legionella pneumophila [175] and Escherichia coli [176], which have variable gene content between strains.
We outlined the advantages and disadvantages of these different sources of sequence information in Table 1. As mentioned in the above text, there are some specific cases where previous sequence information or reference databases can provide equivalent or better sequence information than a matched metagenome. The matched metagenome, however, is often a critical component, along with some specific reference genomes for creating a database that is comprehensive without adding too many extraneous sequences.
Table 1.
Characteristics, advantages and disadvantages of sequence sources for metaproteomic databases.
| Matched metagenome | Unmatched metagenome | Unrestricted reference database | Restricted database amplicon sequencing | Restricted database defined community | |
|---|---|---|---|---|---|
| Monetary cost | Sample type dependent $100-$2,000/sample or pooled samples | Free | Free | $50-$100/sample | Free |
| Time cost (labor & computation) | Genome-resolved month-year, otherwise weeks | Days | Days | Weeks | Days |
| Presence of sequences representing proteins not actually in the sample | Low, sequences are derived from sample | Medium, sequences are derived from system but not specific sample | High, sequences represent all of sequenced life | Medium, sequences are derived from same taxa as the sample, but not the same genomes | Low, exact composition is known and reference database is used |
| Likelihood of sequences missing | Low to medium, Dependent on depth of sequencing and inclusion of unbinned sequences. | Medium to high, dependent on similarity between previously sequenced samples and samples measured by metaproteomics. | Medium to high, even if relatives of community members are present in public repositories, even closely related strains differ significantly in gene content. | Medium to high, even if representative genomes for identified taxa are available, closely related strains differ significantly in gene content. | None to low |
| Potential sources for redundant (highly similar or identical) sequences | Artificial: bringing together sequences from sequential gene prediction and multiple assemblies. Biological: similar genes in different strains from the same species or genus. | Artificial: bringing together sequences from sequential gene prediction and multiple assemblies. Biological: similar genes in different strains from the same species or genus. | Artificial: bringing together sequences from multiple sources. Biological: similar genes in different strains from the same species or genus. |
Artificial: bringing together sequences from multiple sources. Biological: similar genes in different strains from the same species or genus. | Biological: similar genes in different strains from the same species or genus. |
| Taxonomic resolution | If genome-resolved subspecies to species, otherwise genus to phylum based on LCA to reference databases | If genome-resolved subspecies to species, otherwise genus to phylum based on LCA to reference databases | Genus to phylum based on LCA of all matches in the reference databases | Genus to phylum based on LCA to reference databases | Subspecies to species |
| Likelihood of misidentifying taxa | Low | Medium, dependent on relevance of metagenome to sample | High, many sequences missing from database and many sequences in the database are not in the sample | Medium, dependent on relevance of selected reference genomes to actual genomes in sample | Low |
2.4. Studies on the effects of protein database construction on metaproteomic studies
Comparisons of the impact of different database construction strategies on peptide identification, protein inference, taxonomic assignment, and functional assignments in metaproteomic studies are critical for making informed decisions for protein database construction. Several studies have evaluated the effects of protein database construction on peptide identification, particularly with regards to source of sequence information. These studies focused on the number of peptides identified and generally found that protein databases from matched or unmatched metagenomes yielded more peptide identifications than protein databases from reference proteomes [48], [88], [92], [93], [94]. A 2016 article written by Tanca et al. presents a thorough evaluation of the effects of different database construction approaches on peptide identification [88]. In that paper, Tanca et al. compared databases derived from matched metagenomes to databases derived from UniProt [81]. These UniProt databases were made up of either all bacteria sequences in UniProt or were restricted to taxa identified by 16S rRNA gene sequencing at the family, genus, or species level. Tanca et al. found that the matched metagenomes yielded more peptides from human stool samples than any database constructed from UniProt. They also compared matched and unmatched metagenomes to a UniProt derived protein database for mouse and human fecal samples, and found that the matched metagenomes yielded more peptide identifications than unmatched metagenomes. In this evaluation, Tanca et al. found that the the mouse microbiome was underrepresented in UniProt and as a result the UniProt database had even fewer peptide identifications for the mice as compared to humans, indicating that reference proteomes may not be a good source of sequence information for environmental samples in general.
Furthermore, Tanca et al. evaluated the effects on peptide identification of databases that combined protein sequences from multiple sources. They found that combining metagenomes from multiple human or mouse subjects yielded more peptide identifications, as long as the matched-metagenome was included. Furthermore, they found that combining all the databases, including the UniProt one, did not decrease the number of identifications. This result indicates that missing sequences have a greater impact on the peptides identified than increased database size. This is in line with a 2013 study, in which Tanca et al. found that a protein database made up of genomes sequenced from isolates in a mock community yielded more peptides than a database derived from metagenomic sequencing of the mock community [92]. In another study on arctic ocean samples, May et al. also found that combining the results from reference database and metagenome derived protein databases yielded more peptides than metagenome derived databases alone; however, these databases were not searched together making this result inadequate for determining impact of database size on peptide identification [94]. The results from May et al., however, do provide some insight on the impact of database size on peptide identification. The authors found that thousands of peptides identified in the smaller metagenome-derived database were missed when searching against the larger NCBI environmental database despite those peptides being present in the NCBI database, which shows that increased database size leads to peptides not being identified. A contrasting result was obtained by Zhang et al., who found that an unmatched metagenomic database made up of millions of sequences from the extensive metagenomic sequencing efforts previously done in humans yielded more peptide identifications than a matched metagenome [39], [48]. This result could suggest that with enough sequencing efforts, as has been done in humans, unmatched metagenomes could be equivalent to a matched metagenome in terms of number of peptides identified; however, Zhang et al. conducted this comparison using a two-step search approach, which blunts the effects of large protein database size using techniques that have not been fully validated (see subsection 2.1). Together these studies show the importance of matched metagenomes for creating complete protein databases when sequences for proteins in the sample are not present in public databases. These studies also show that matched-metagenomes alone do not necessarily provide complete databases.
One feature that has a major impact on the completeness of protein databases from matched metagenomes is sequencing depth. In their 2016 article, Tanca et al. showed that increasing sequencing depth had a positive linear relationship with the number of peptides identified. Sequencing depth in this evaluation was limited to eighteen megabase pairs (Mbps) per sample. Later in the study, when comparing matched to unmatched metagenomes in mice and humans, they sequenced metagenomes that were six gigabase pairs (Gbps) per sample. These sequencing depths are not high enough to produce a complete evaluation of the effects of sequencing depth on peptide identification based on simple calculations (expected number of organisms X average length of an organism’s genome X desired sequence coverage). For comparison, assuming an average genome length of 3.8 Mbps [95] and a desired coverage of 20-fold [96], 7.6 Gbps are needed to obtain good coverage of a 100 species community. As such, further evaluations of sufficient depth are needed to find how much sequencing is really needed to generate a protein database using matched metagenomes that cover all detectable proteins in metaproteomic samples.
How protein sequences are predicted from a metagenome and whether they are predicted from raw reads or assembled contigs also impacts the completeness of a metagenome derived protein database, and as a result, how many peptides can be identified by database search. Proteins can be predicted from raw reads or contigs using brute-force six-frame translations or dedicated gene prediction softwares [88]. Six-frame translations extract all possible open reading frames (ORFs) above a certain length cutoff from a contiguous DNA sequence even if the ORFs overlap. Gene prediction softwares use models of prokaryotic genes to predict non-overlapping ORFs likely corresponding to true genes [97]. In their 2016 article, Tanca et al. found that six-frame translation yielded fewer peptides than gene predictions on both assembled contigs and raw reads, and gene predictions from raw reads yielded slightly more peptides than gene predictions from contigs [88]. In contrast, significantly more peptides were assigned a functional annotation when genes were predicted from contigs instead of raw reads. May et al. identified substantially more peptides when predicting genes on raw reads versus contigs, but more peptides were identified in total when combining the two approaches [94]. The effect of raw reads or contigs on taxonomic assignment was inconclusive and neither of these approaches looked at the effects of these database construction approaches on protein inference nor the effect of protein inference on functional annotation. Our assumption would be that protein inference and gene predictions on contigs would yield better functional annotations since length or completeness of a predicted gene has been shown to yield more sensitive and accurate annotations [23], [98], [99], [100]. Additionally, none of the evaluations mentioned in this paragraph processed their assembled metagenomes into MAGs, thus they were not able to evaluate whether MAGs improved taxonomic assignment or functional annotations. Benefits such as improved taxonomic classification, assignment of complete pathways to individual organisms, and the ability to analyze genes in their genomic context have led to MAGs being a critical component of the many studies that have investigated function with metaproteomics at the species and genome level [6], [7], [9], [25], [26], [90], [101]. Therefore, the use of genome-resolved metagenomes in metaproteomics databases deserves more careful future evaluations.
The studies described in this section provide insight into the effect of database size and completeness on the number of peptides identified, but offer only limited information on the effects of protein database construction on protein inference, taxonomic resolution and accuracy, functional assignment and interpretation, and whether low FDR peptides are actually being identified accurately. More evaluations are needed, especially in light of an article by Timmins-Schiffman et al., which showed that protein databases generated from assembled metagenomes versus reference databases yielded very different taxonomic compositions and functional results [102]. These different taxonomic compositions were observable even at the phylum level, and the 10 functions that changed the most varied depending on the database used. Timmins-Schiffman et al. suggested that metagenome derived databases were likely the safer option based on these results, but they did not further evaluate if the metagenome was indeed the most accurate database in this study. In their 2013 study, Tanca et al. showed that assembled (but not genome-resolved) matched metagenomes had lower mismatches at the species level than protein databases made up of all bacteria, fungi, and viruses in UniProt or NCBI, when evaluating taxonomy of a mock community of known composition [92]. Since this study was not genome-resolved, taxonomy was assigned separately to individual peptides using the UniPept [21] or MEGAN [103] softwares, which use LCA methods to identify a consensus taxonomy based on matching the sequences to taxonomically classified sequences in UniProt or NCBI. This leads to somewhat circular logic as the taxonomy is being evaluated using the databases to which peptides are compared to in the analysis. Further evaluations are needed to investigate the accuracy of species level assignments independent of these reference databases. This sort of evaluation would need to be done in the context of MAGs as discussed in the previous paragraph. Beyond taxonomic accuracy, the impact of different protein database construction strategies on FDR estimation of peptides and proteins still needs to be studied. Since FDR is just an estimation of peptide or protein identification accuracy, the actual accuracy needs to be evaluated empirically. Kumar et al. provide some insight into how to do this by including a set of sequences known to not be in the sample in the target database (an entrapment database) [62]. They then evaluated the number of identifications after database search that were from the entrapment database to estimate FDR calculation accuracy in their evaluation of multi-step search methods. An alternative approach could be to use spiked-in peptides or proteins in various quantities to create a population of peptides known to be in the sample. Spiked-in peptides are a form of ground truth typically used in the evaluation of proteomic quantification methods [104]. By spiking peptides into some samples, but not into others, spiked-in peptides could be used as a way to evaluate whether peptides known to be in the sample or absent from the sample are being detected by database search. Finally, studies are also needed to evaluate the effect of database construction on peptide and protein identification beyond just source of sequence information. For example, evaluations on the effect of sequencing depth and removal of sequence redundancy on peptide identification and particularly protein inference are still needed. Despite these limitations, the information above can be used to inform the construction of a protein database based on the current standards of the field, which we explore in section 3.
3. Considerations and best practice suggestions for constructing a metaproteomics protein database
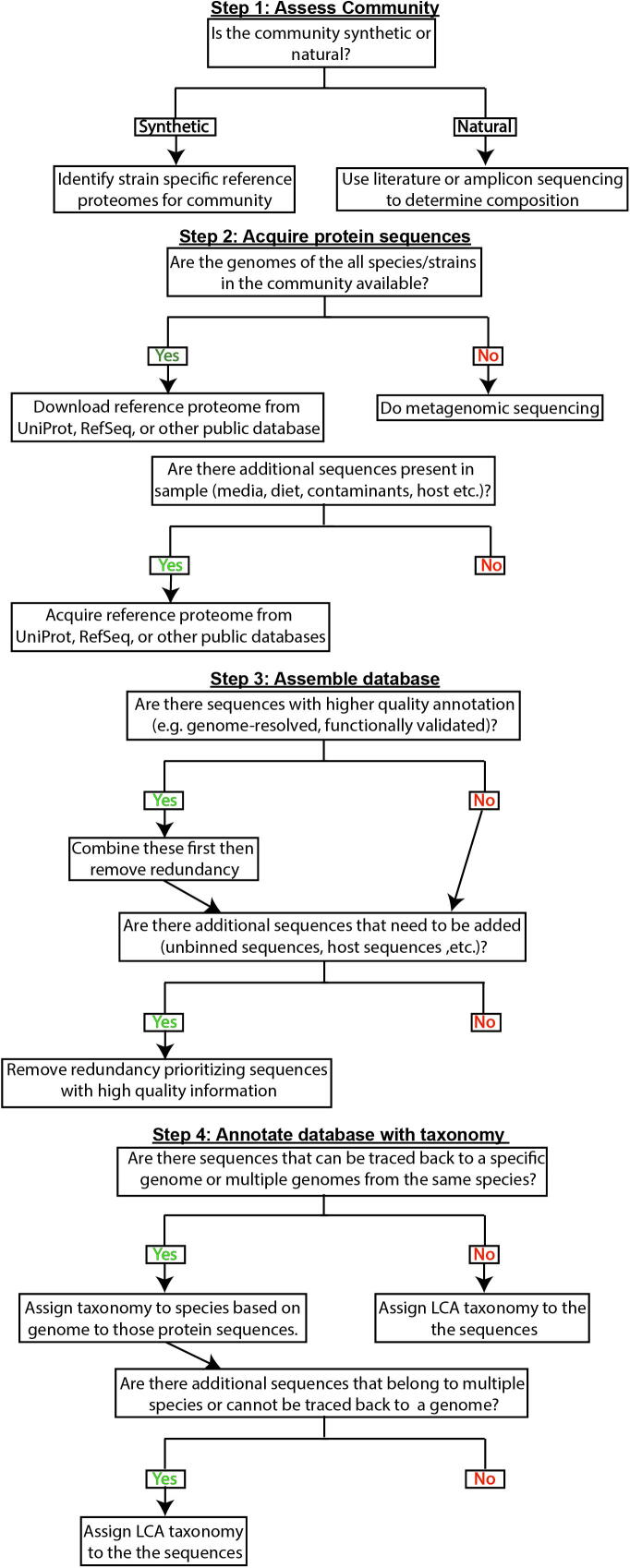
The information provided in section 2 can guide decision making for constructing a metaproteomic protein database. A well-constructed database has three main elements: (1) comprehensive sequence coverage while minimizing irrelevant sequences (covered in subsections 3.1 and 3.2), (2) a link to the genome of origin for each protein sequence when possible (covered in 3.2 and 3.4), and (3) curation of redundant sequences to facilitate unambiguous protein inference and annotation (covered in 3.3 and 3.4). In Fig. 2 we provide a decision tree that divides database construction into these four main steps: (1) community assessment, (2) sequence acquisition, (3) database construction, and (4) annotation. We discuss these steps in detail in the following subsections. In addition to the steps represented in the decision tree, we provide an additional section that discusses functional annotation of the protein database (section 3.5).
Fig. 2.
Decision tree reflecting the steps to take when constructing a protein sequence database for metaproteomics. We define a synthetic community as one that is designed by the researcher (e.g. defined communities, mock communities, gnotobiotic systems). We define a natural community as a community taken from the environment (e.g. soil, fecal, ocean).
3.1. Assessing the community prior to protein database construction
The first step of any metaproteomics study should be to determine the composition of the studied microbial community to create a protein database. In most cases this can be done using prior literature, amplicon sequencing [105], [108], [109] or metagenomic sequencing [7], [107], [108]. In specific cases, the exact composition of a microbial community in a sample is known as in the case of constructed, fully defined communities [8], [82] or well-characterized highly specific symbioses [6], [83], [84], [85], [86]. Often, however, the exact community composition is unknown, as discussed in section 2.3. In these cases, sequencing the samples can provide insight into the steps that need to be taken. Though not typically done, it can be beneficial to conduct a preliminary amplicon sequencing analysis, prior to shotgun metagenomic sequencing. Amplicon sequencing results can be analyzed using robust analysis platforms such as mothur [177] or QIIME2 [178]. In contrast to metaproteomics, which allows the analysis of proteins from all domains of life in a single analysis, determining microbial community composition with amplicon sequencing may require separate analysis of multiple different marker genes to obtain a comprehensive overview of community composition (e.g. 16S rRNA gene for Archaea and Bacteria, and 18S rRNA gene and Internal Transcribed Spacer (ITS) for various eukaryotes) [109]. These preliminary analyses can provide insight into the availability of genomes from community members in public repositories, if and how much metagenomic sequencing is needed, and metagenomic processing steps needed to cover the community (see section 3.2).
3.2. Acquiring sequences for protein database construction
Once the community in the samples in question has been assessed, the next step is to gather protein sequences that cover all sequences of proteins potentially present in the samples. If the microbial community composition is known in advance and genomes of specific strains are publically available, these microbial protein sequences can be acquired by going to the source and downloading them. In the case of a defined/synthetic community with reference genomes, this involves going to RefSeq or UniProt and downloading the relevant FASTA files for the reference proteomes of the strains in question. If genomes are not available in reference repositories, this involves acquiring the sequences from sources found through the data accessibility statement of previous manuscripts [6] or by sequencing the isolates making up the defined community [92]. Please note that we suggest only collecting reference protein sequences from public repositories if you can be certain that they correspond to the strains in your samples. We do not recommend the use of genomes from relatives based on phylogenetic marker gene analysis as gene content of even closely related strains can differ widely (see section 2.3).
In addition to microbial sequences, additional sequences of proteins that may be in the sample need to be gathered, for example, host sequences for host-associated microbiomes, culture media components, dietary components if working with gut microbiomes, and common laboratory contaminants [6], [26], [79]. The cRAP database provides many of the common contaminants found in proteomics studies [179]. In the case of studies on host-associated microbiomes, such as from humans, mice, or Arabidopsis, complete protein sequence sets (reference proteomes) can be acquired from UniProt’s reference proteomes [81].
In most cases protein sequences for the studied microbial community are not available from repositories and then metagenomic and sometimes metatranscriptomic sequencing is the most straightforward way to obtain sequences for metaproteomics. The first decisions to make when starting a metagenomic study are the sequencing technology to use and the sequencing depth to aim for. Currently most of the tools for analyzing shotgun metagenomic data are built to use paired-end reads from Illumina sequencers and how much sequencing is needed can be calculated based on the assessment of microbial community composition suggested in section 3.1 (See section 2.4 for details on how this could be calculated). After DNA extraction, library preparation and sequencing of the samples can be done at a core sequencing facility or commercial service provider. Once raw sequence data are provided by the sequencing facility, publicly available tools that are relatively easy to install and have good documentation can be used to acquire a comprehensive set of predicted protein sequences, many of which come from MAGs. These tools can be used in individual steps as detailed below or as easily installable bundled workflows such as Anvi’o [110], MetaWRAP [111], and ATLAS [112].
-
1.
Decontamination: sequencing reads are quality checked and trimmed for adapters and low quality regions if necessary. This can be done automatically using, for example, Trim Galore [113]. Additionally, undesired contaminating sequences, such as host derived sequences and the Illumina control spike-in PhiX, can also be removed using an aligner, such as BBMap in the BBtools suite [114], BWA [115], or Bowtie [116].
-
2.
Assembly: metagenome-specific assemblers extend short read sequences into contigs using iterative de Bruijn graph assembly. Generally, metaSPAdes [117] generates the most accurate assemblies , while MEGAHIT [118] generates reasonably good assemblies but has the advantage that it is an order of magnitude more efficient in terms of time and memory usage [119]. MEGAHIT also has the ability to assemble reads from multiple samples in tandem to form a consensus assembly (co-assembly) between all the samples in a study.
-
3.
Contig binning: To obtain genome-resolved metagenomes, contigs are grouped into bins using so-called binning approaches. Binning approaches use contig-intrinsic information such as read coverage in several samples (differential coverage) and tetranucleotide frequencies. These binning approaches are implemented in automated software tools such as the frequently used CONCOCT [122], MetaBat2 [121], and MaxBin [120]. Performance of the different binning approaches is sample specific and can be empirically evaluated with tools like DASTool [123] and MetaWRAP [111].
-
4.
Evaluation of bin quality to determine which bins can be considered metagenome-assembled genomes: the most common approach for evaluating if a bin potentially corresponds to a partial or complete genome is the assessment of single copy gene content of bins [124], [125]. Essentially a list of genes that have been empirically shown to be present as a single copy in genomes of specific phylogenetic groups is used to taxonomically classify bins at a higher taxonomic level and to evaluate the percent completion and contamination (redundancy) of each bin. The tools Anvi’o, BUSCO, and CheckM produce this evaluation automatically [110], [124], [125]. Other metrics, such as number of tRNAs and the presence of rRNA genes can also be used to evaluate MAG quality, as well as general assembly quality metrics, such as N50 and circularity [37], [107]. There are additional steps that can be taken to improve bin quality, such as manual curation and re-assembly [107], facilitated by tools such as Anvio and MetaWRAP [110], [111]. For a good review on generating high quality MAGs see [107]. The specific completeness and contamination levels for when a bin can be considered a MAG vary in the literature. Generally, following the recommendations set forth by the Genomic Standards Consortium (GSC) is recommended for any MAGs that will be submitted to a public repository, such as the NCBI databases [37], [126]. These recommendations include a > 50% completion and < 10% contamination cutoff for MAGs to be considered medium quality genomes [37]. It's still not clear, however, if these cutoffs are ideal for the construction of protein databases for metaproteomics, as there can still be useful information about gene neighborhoods and a protein’s organism of origin for organisms whose genome could not be assembled into a medium quality MAG. Further evaluations are needed to investigate the impacts of different MAG quality thresholds on protein database construction from MAGs, as discussed in section 2.4. It is possible that there will not be one hard set of rules, as these cutoffs may end up being system or study specific.
-
5.
Organizing MAGs into species and subspecies groups: once a set of acceptable MAGs has been selected, they can be grouped into species and subspecies groups by average nucleotide identity (ANI), with tools such as dRep [7], [127]. The genomic delimiter of bacterial species has been shown to be 95% ANI [40], [41]. Higher ANI thresholds, such as 98% have been used to delineate subspecies groupings [7]. dRep outputs the highest quality MAG, by single copy gene metrics, for an ANI group as a representative genome. dRep also outputs a table containing the information about which MAGs were grouped by ANI.
-
6.
Gene annotation: after a set of MAGs has been selected, prokaryotic gene calling algorithms can be used to predict genes on binned and unbinned contigs. Many metagenomic studies set a contig length cutoff of 1000 bp to reduce the number of predicted genes that are fragmented, but it can be beneficial to use a lower cutoff for a metaproteomic protein database in order to not lose potentially identifiable peptide sequences. Prodigal [97] and MetaGeneMark [128] are common gene annotators used in metagenomics and both softwares output translated amino acid sequences for the predicted genes. Many bioinformatics tools for processing and analyzing metagenomic data or providing functional annotation to genes use Prodigal for gene prediction (more details in section 3.5) [125], [127], [129], [130], [131], [132].
The above steps favor the detection of bacterial and archeal genes and MAGs. If assessment of the community, as described in section 3.1, indicates the substantial presence of one or more eukaryotic organisms in the community that has no or low quality public data, then additional steps need to be taken to acquire those protein sequences. Eukaryotic contigs will often be present in the unbinned fraction of a metagenomic assembly, and gene calls from a prokaryotic gene caller, like Prodigal, can still be used to identify eukaryotic proteins as was done with green algae by Kleiner et al. [24]. These gene predictions are, however, often highly fragmented and incomplete due to the presence of introns in the genes of many eukaryotes, nevertheless they can potentially be classified as eukaryotic using an LCA approach as described later in section 3.4. To acquire better gene annotations for eukaryotes there are two options: (1) use de novo assembled transcripts from RNA-seq to identify eukaryotic transcripts and predict their complete encoded protein sequences as was done for crustacean [133] and gutless worm hosts [134], or (2) retrieve eukaryotic contigs from the metagenome and apply eukaryotic specific gene prediction algorithms. A workflow for retrieving eukaryotic contigs using machine learning, and applying binning methods to assemble eukaryotic MAGs, was proposed by West et al. and benchmarked using a variety of data sets [135]. West et al. further used this approach to identify proteins of the yeast Candida in preterm infants using metaproteomics [101]. The BUSCO tool can be used to classify bins as eukaryotic, bacterial, or archaeal, and provides gene predictions and completion versus contamination metrics for those bins [124]. None of the approaches described here to obtain eukaryotic protein sequences are ideal either in terms of quality in the case of Prodigal gene predictions, or labor in the case of the West et al. approach; however, they represent the current state of the field for acquisition of eukaryotic protein sequences for metaproteomics.
In summary, assessment of community composition in advance provides a powerful framework with which to select the sources of sequence information needed for a metaproteomics study, whether it be reference proteomes for known communities or metagenomic/metatranscriptomic sequences for understudied communities.
3.3. Assembling and curating the protein database
After acquiring the protein sequences the next step is to assemble the database. For this, all sequences from different sources (in FASTA format) acquired in the sequence acquisition step need to be combined into a single database. A simple linux utility like “cat” can be used for simply combining all fasta files. Sometimes simply combining fasta files is not the best mode of action, for example, when the same sequence may be present multiple times in the various sequence sources used. In these cases sequences can be combined in a stepwise fashion considering their annotation quality (see below). The next step after combining sequences is to remove redundant sequences, i.e. highly similar or identical sequences that have different identifiers (accession numbers). This can be done by clustering sequences based on amino acid identity (percentage of amino acids that match between sequences) with algorithms, such as UCLUST [78] or CD-HIT [77], which in addition to a FASTA file with representative sequences also provide an output file that specifies which sequences were clustered together.
For sequence clustering the choice of the identity threshold at which sequences are clustered is critical. Various studies have used sequence identity thresholds for clustering that range from 90 to 100 percent amino acid identity [7], [18], [26], [70], [79], [86]. Lower identity thresholds will result in smaller databases and allow for a greater number of possible unique peptides at the price of losing peptide sequences that are distinct between similar protein sequences, as well as the ability to differentiate proteins from very closely related organisms. Higher identity thresholds allow for better species and subspecies level resolution by retaining more similar sequences, at the cost of increased database size and identification of proteins without unique peptides. The optimal clustering threshold needs to be determined specifically for each study and sample type, for example, by searching a subset of the data against databases with different clustering levels and then evaluating quality metrics such as number of protein unique peptides identified, number of unambiguously identified proteins, and percentage of proteins traceable to a specific species or subspecies group.
If sequences from multiple sources are used and some sources have more useful information associated with them, such as being derived from taxonomically classified MAGs or reference proteomes, it can be beneficial to cluster sequences in a way that will preferentially retain the better annotated sequences as the representative sequence of a cluster. This was, for example, done in a study by Kleiner et al. when combining protein sequences from MAGs and unbinned contigs [6]. Kleiner et al. preferentially retained the well annotated sequences from the MAGs and only added sequences from unbinned contigs if no similar sequence from a MAG was available. For this, the authors used CD-HIT-2D, an extension of CD-HIT, which allows users to compare databases and output sequences that are unique to one of the databases. Specifically after combining and clustering the well annotated sequences with CD-HIT, the authors compared the sequences from unbinned contigs against this initial “higher-quality” database using CD-HIT-2D and then added sequences absent from the “higher-quality” database to the final database.This approach will maximize unambiguous identification of protein sequences, while favoring better annotated sequences.
3.4. Annotating the protein database with taxonomic information
Once the protein database has been assembled, the database needs to be annotated with functional and taxonomic information. Taxonomy can be assigned to the protein sequences that make up a metaproteomic protein database based on their genome of origin or through a consensus taxonomy acquired from similarity searches against reference databases (i.e. LCA). For proteins that originate from a genomic unit (i.e., acquired from strain specific reference proteomes or MAGs), the most straightforward course of action is to assign the taxonomy of the genome. Proteins acquired from a strain-specific reference proteome can simply be assigned the species of that reference proteome; however, proteins acquired from MAGs require the MAG to be taxonomically classified. For MAGs, if the species has already been discovered and has a representative in a reference database, the MAG’s species can be assigned by matching with an ANI of 95% or greater to its representatives in a reference database. If the species of the MAG does not have a representative genome in a reference database, then the lowest possible taxonomy can be predicted using the consensus taxonomy from similarity searches of all the genes in the genome, as done by BAT [136], or by using phylogenomic methods to place the genome in a tree of life. GTDB-Tk does the ANI and phylogenetic comparison automatically for bacterial and archaeal MAGs based on the Genome Taxonomy Database [129]. The Genome Taxonomy Database is built from genomes of sufficient quality in NCBI’s genbank along with some additional MAGs [129], [137], [138]. While MAGs that do not have a representative in a reference database cannot be assigned a species name, proteins from these MAGs can still be traced to an unnamed species using the species and subspecies groupings described in section 3.2. This unnamed species can be assigned a unique identifier for the study, which can then be used in the submission of the MAG to NCBI. Since proteins are often gathered from multiple sources and clustered to remove redundancy, as discussed in section 3.3, the taxonomic origin of all the sequences that make up a cluster should be considered when assigning taxonomy based on the genome of origin. Based on our experience, we suggest doing this as follows. If all protein sequences in a cluster come from genomes of the same subspecies or species (see section 3.2), then that species or subspecies can be assigned to the representative sequence of the cluster. If there are protein sequences in a cluster that come from genomes that are not the same species, then the representative protein could be labeled as multi-species, while retaining information about all the possible origin species. If none of the sequences that make up a cluster can be linked to a genome then the taxonomy can be determined using LCA approaches as described in the next paragraph. For proteins that can be traced to a specific species, a predefined species code can be added to the identifier of the protein to facilitate interpretation of the data once the metaproteomic data has been processed, as described here [24].
For proteins that do not retain their genome of origin, for example, unbinned contigs in a metagenome, unbinned gene catalogs built from previous metagenomic studies [38], [39], or from a general download of one of the major reference databases like UniProt, taxonomy can only be acquired by doing similarity searches against reference databases using LCA algorithms. If proteins come from an unbinned contig with multiple genes, then the consensus taxonomy of all the genes in the contig can be used. This is done automatically by the algorithm CAT [136]. If proteins are independent singletons, they can be assigned a taxonomy using a standard metagenomic LCA method, such as those provided by MEGAN [103], Kaiju [139], Centrifuge [140], or Kraken2 [141]. For LCA approaches, if a sequence similar (>95% identity) to the protein in question is not present in any of the genomes present in a public reference database, then it is impossible to assign a species specific taxonomy.
3.5. Annotating the protein database with functional information
To assign putative functions to protein sequences, they are compared to sequences or profiles/models of sequence groups in public reference databases of protein function, for example eggNOG [142], KEGG [143], UniProt [81], InterPro [144], MEROPS [145], MetaCyc [146], and CAZy [147], among others. The KEGG and MetaCyc databases are mostly focused on enzymes though they do provide information about other cellular processes, such as transporters. Other databases such as eggNOG, UniProt, and InterPro are more comprehensive, including information for many cellular processes. In addition to these more general databases there are specialized databases such as MEROPS and CAZy that focus on peptidases and carbohydrate active enzymes, respectively. The quality of the functional annotations in these databases, and their link to the metaproteomic protein database, plays a major role in determining functional output of any metaproteomics study. An example of high quality annotations would be the reviewed fraction of the UniProt database (Swiss-Prot) as compared to the computationally generated unreviewed fraction [81]. Functional information from these databases comes in the form of functional descriptions found in the header of protein sequences in FASTA files or in tables provided by these databases, and in the form of functional classification systems. These functional classification systems can be general, such as Gene Ontology terms (GO) [148]; based on protein families, like eggNOG [142], COG [149], or KEGG ORTHOLOGY (KO) [143]; or more focused on specific metabolism, like the enzyme commission (EC) numbers [150], Transporter Classification (TC) [151], or Carbohydrate-Active enZYmes (CAZy) [147].
Several tools provide automated functional annotation of protein sequences with the above classifications. Evaluating these tools is outside of the scope of this review, but several studies have been conducted that provide some insight [152], [153], [154]. There are full-service genome annotation tools, such as RAST [155], Prokka [130], DRAM [131], and MetaErg [132], that work on contigs and genomes predicting genes and their functions in tandem. These tools also predict other non-protein coding genetic features, such as tRNAs and rRNAs. RAST and Prokka are older softwares, originally developed for single prokaryotic genomes, and are limited to functional descriptions and EC numbers, while DRAM, and MetaErg were more recently developed for unbinned metagenome and MAG annotation and provide a wide array of functional classifications. Web-based tools such as RAST have the advantage of providing easy access to visualizations of the gene neighborhood of all the genes predicted on a contig, providing insight into the potential function of unknown proteins and easy comparison with other genomes.
If the protein database has already been compiled, but more functional annotations are needed, other annotation tools can be used to annotate protein sequences directly, such as eggNOG-mapper [152], InterProScan [156], dbCAN2 [157] and GhostKOALA [158]. These tools can be run on a curated protein database or on only the protein sequences that have been identified, saving computational time. InterProScan and eggNOG-mapper provide a wide array of functional annotation information, while dbCAN2 and GhostKOALA are more specialized, focusing on CAZy enzymes and KO terms, respectively. In the end, the choice of annotation tool depends on the desired functional outcomes of any given metaproteomic study.
4. Perspectives and concluding remarks
Construction of the protein sequence database plays a critical role in the outcome of any metaproteomics study. In this review, we have provided a comprehensive overview of the effects of protein databases on peptide identification and protein inference, as well as their subsequent taxonomic and functional interpretation. Existing evidence indicates that peptides and proteins are best identified, and taxonomically classified, when the database is complete and has minimal extraneous sequences, which is usually best accomplished through sequencing sample-matched metagenomes or comprehensive prior sequencing of a specific system. For continued improvement of protein sequence databases, future evaluations should focus on how different metagenomic processing methods used for protein database construction affect peptide identification numbers, peptide identification accuracy, and taxonomic accuracy (e.g. discussed in 2.4). For example, one option in database generation that has not been evaluated is if and how combining metagenomes from multiple samples impacts peptide identification numbers and accuracy i.e. will having sample specific databases or databases that combine all metagenomes for the whole experiment be better. Improving the accuracy of peptide and protein identifications in the context of multi-step search strategies, such as two-step searches [59], is also needed because their validity has recently been put into question [62], and they are needed for the identification of peptides and proteins using the massive databases from previous sequencing initiatives [48].
As large genome-resolved gene catalogs become available for more biological systems, such as mice [159], it will become all the more critical to evaluate the utility of these databases for metaproteomic studies and to develop better database reduction strategies. We still do not fully understand if distinct construction strategies produce databases that perform differently on biological systems of variable complexity since the most thorough evaluations described in this paper were done only on mice and human gut samples [88]. Community efforts such as the Metaproteomics Initiative [160], which, for example, recently carried out an interlaboratory comparison of metaproteomic workflows [19], may represent an excellent mechanism to evaluate the impact of database construction approaches on metaproteomics.
Emerging technologies in the realms of DNA sequencing, peptide mass spectrometry and novel protein measurement approaches will likely impact protein database construction for metaproteomics. With regards to DNA sequencing technologies, long-read sequencing technologies, such as Oxford Nanopore [161] and PacBio [162], as well as sequencing technologies that connect DNA reads based on their cell of origin, such as Hi-C sequencing [163], provide avenues for obtaining higher quality MAGs with better taxonomic resolution. With regards to protein identification, new technologies, such as ion mobility spectrometry TOF mass spectrometers [164], data-independent acquisition (DIA) [165], or actual sequencing of proteins independent of mass spectrometry using nanoPores [166] provide new avenues to improve metaproteomic depth, quantification and protein inference. These technologies are likely to change how protein databases affect the outcome of a metaproteomic study. In the case of the new mass spectrometry technologies, however, recent publications indicate that identification of proteins using these technologies will follow similar principles with regards to spectral matching and FDR calculations as database search [167], [168], indicating that many of the principles described in this review will still apply. In the case of protein sequencing technologies, protein databases will likely no longer be needed for the identification of proteins; however, protein databases will still be important for taxonomic and functional classification. Therefore, at least for the foreseeable future, protein database construction remains critical for investigating molecular phenotypes of microbial communities.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We would like to thank Heather Maughan and four anonymous reviewers for thoughtful comments and feedback, and the members of the Kleinerlab for feedback on the figures and table.
Sources of funding
This work was funded by the National Institutes of Health grant 2T32DK007737-26, and the National Institute Of General Medical Sciences of the National Institutes of Health under Award Number R35GM138362. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Contributor Information
J. Alfredo Blakeley-Ruiz, Email: jablakel@ncsu.edu.
Manuel Kleiner, Email: mkleine@ncsu.edu.
References
- 1.Wilmes P., Bond P.L. The application of two-dimensional polyacrylamide gel electrophoresis and downstream analyses to a mixed community of prokaryotic microorganisms. Environ Microbiol. 2004;6(9):911–920. doi: 10.1111/j.1462-2920.2004.00687.x. [DOI] [PubMed] [Google Scholar]
- 2.Blakeley-Ruiz J.A., McClintock C.S., Lydic R., Baghdoyan H.A., Choo J.J., Hettich R.L. Combining integrated systems-biology approaches with intervention-based experimental design provides a higher-resolution path forward for microbiome research. Behav Brain Sci. 2019;42 doi: 10.1017/S0140525X18002911. [DOI] [Google Scholar]
- 3.Heintz-Buschart A., Wilmes P. Human gut microbiome: function matters. Trends Microbiol. 2018;26(7):563–574. doi: 10.1016/j.tim.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Moya A., Ferrer M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Spec Issue Microb Endur. 2016;24(5):402–413. doi: 10.1016/j.tim.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Kleiner M. Metaproteomics: much more than measuring gene expression in microbial communities. MSystems. 2019;4:e00115–e119. doi: 10.1128/mSystems.00115-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kleiner M., Wentrup C., Lott C., Teeling H., Wetzel S., Young J., et al. Metaproteomics of a gutless marine worm and its symbiotic microbial community reveal unusual pathways for carbon and energy use. Proc Natl Acad Sci. 2012;109(19):E1173–E1182. doi: 10.1073/pnas.1121198109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown C.T., Xiong W., Olm M.R., Thomas B.C., Baker R., Firek B., et al. Hospitalized premature infants are colonized by related bacterial strains with distinct proteomic profiles. MBio. 2018;9(2) doi: 10.1128/mBio.00441-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patnode M.L., Beller Z.W., Han N.D., Cheng J., Peters S.L., Terrapon N., et al. Interspecies competition Impacts Targeted Manipulation of Human Gut Bacteria by Fiber-Derived Glycans. Cell. 2019;179(1):59–73.e13. doi: 10.1016/j.cell.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z., Yao Q., Guo X., Crits-Christoph A., Mayes M.A., Hervey W.J., et al. Genome-resolved proteomic stable isotope probing of soil microbial communities using 13CO2 and 13C-methanol. Front Microbiol. 2019;10:2706. doi: 10.3389/fmicb.2019.02706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y., Fonslow B.R., Shan B., Baek M.-C., Yates J.R. Protein analysis by shotgun/bottom-up proteomics. Chem Rev. 2013;113(4):2343–2394. doi: 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eng J.K., McCormack A.L., Yates J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 12.Link A.J., Eng J., Schieltz D.M., Carmack E., Mize G.J., Morris D.R., et al. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17(7):676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 13.Yates J.R., Eng J.K., McCormack A.L., Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67(8):1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 14.Eng JK, Searle BC, Clauser KR, Tabb DL. A face in the crowd: recognizing peptides through database search. Mol Cell Proteomics MCP 2011;10:R111.009522-R111.009522. https://doi.org/10.1074/mcp.R111.009522. [DOI] [PMC free article] [PubMed]
- 15.Verberkmoes N.C., Russell A.L., Shah M., Godzik A., Rosenquist M., Halfvarson J., et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;3(2):179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 16.Washburn M.P., Wolters D., Yates J.R. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X.u., Chen W., Ning Z., Mayne J., Mack D., Stintzi A., et al. Deep metaproteomics approach for the study of human microbiomes. Anal Chem. 2017;89(17):9407–9415. doi: 10.1021/acs.analchem.7b02224. [DOI] [PubMed] [Google Scholar]
- 18.Xiong W., Abraham P.E., Li Z., Pan C., Hettich R.L. Microbial metaproteomics for characterizing the range of metabolic functions and activities of human gut microbiota. Proteomics. 2015;15:3424–3438. doi: 10.1002/pmic.201400571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Den Bossche T., Kunath B.J., Schallert K., Schäpe S.S., Abraham P.E., Armengaud J., et al. Critical Assessment of MetaProteome Investigation (CAMPI): a multi-laboratory comparison of established workflows. Nat Commun. 2021;12(1) doi: 10.1038/s41467-021-27542-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nesvizhskii A.I., Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Mol Cell Proteomics. 2005;4(10):1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Gurdeep Singh R., Tanca A., Palomba A., Van der Jeugt F., Verschaffelt P., Uzzau S., et al. Unipept 4.0: functional analysis of metaproteome data. J Proteome Res. 2019;18(2):606–615. doi: 10.1021/acs.jproteome.8b00716. [DOI] [PubMed] [Google Scholar]
- 22.Mesuere B., Devreese B., Debyser G., Aerts M., Vandamme P., Dawyndt P. Unipept: tryptic peptide-based biodiversity analysis of metaproteome samples. J Proteome Res. 2012;11(12):5773–5780. doi: 10.1021/pr300576s. [DOI] [PubMed] [Google Scholar]
- 23.Barrett K., Lange L. Peptide-based functional annotation of carbohydrate-active enzymes by conserved unique peptide patterns (CUPP) Biotechnol Biofuels. 2019;12:102. doi: 10.1186/s13068-019-1436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleiner M., Thorson E., Sharp C.E., Dong X., Liu D., Li C., et al. Assessing species biomass contributions in microbial communities via metaproteomics. Nat Commun. 2017;8(1) doi: 10.1038/s41467-017-01544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brooks B., Mueller R., Young J., Morowitz M., Hettich R., Banfield J. Strain-resolved microbial community proteomics reveals simultaneous aerobic and anaerobic function during gastrointestinal tract colonization of a preterm infant. Front Microbiol. 2015;6:654. doi: 10.3389/fmicb.2015.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong W., Brown C.T., Morowitz M.J., Banfield J.F., Hettich R.L. Genome-resolved metaproteomic characterization of preterm infant gut microbiota development reveals species-specific metabolic shifts and variabilities during early life. Microbiome. 2017;5:72. doi: 10.1186/s40168-017-0290-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Overbeek R., Fonstein M., D'Souza M., Pusch G.D., Maltsev N. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci U S A. 1999;96(6):2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dandekar T., Snel B., Huynen M., Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 29.Kunath BJ, Minniti G, Skaugen M, Hagen LH, Vaaje-Kolstad G, Eijsink VGH, et al. Metaproteomics: Sample Preparation and Methodological Considerations. In: Capelo-Martínez J-L, editor. Emerg. Sample Treat. Proteomics, Cham: Springer International Publishing; 2019, p. 187–215. https://doi.org/10.1007/978-3-030-12298-0_8. [DOI] [PubMed]
- 30.Hinzke T., Kouris A., Hughes R.-A., Strous M., Kleiner M. More Is Not always better: evaluation of 1D and 2D-LC-MS/MS methods for metaproteomics. Front Microbiol. 2019;10:238. doi: 10.3389/fmicb.2019.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian C., Hettich R.L. Optimized extraction method to remove humic acid interferences from soil samples prior to microbial proteome measurements. J Proteome Res. 2017;16(7):2537–2546. doi: 10.1021/acs.jproteome.7b00103. [DOI] [PubMed] [Google Scholar]
- 32.Schiebenhoefer H., Van Den Bossche T., Fuchs S., Renard B.Y., Muth T., Martens L. Challenges and promise at the interface of metaproteomics and genomics: an overview of recent progress in metaproteogenomic data analysis. Expert Rev Proteomics. 2019;16(5):375–390. doi: 10.1080/14789450.2019.1609944. [DOI] [PubMed] [Google Scholar]
- 33.Xiong W., Giannone R.J., Morowitz M.J., Banfield J.F., Hettich R.L. Development of an enhanced metaproteomic approach for deepening the microbiome characterization of the human infant gut. J Proteome Res. 2015;14(1):133–141. doi: 10.1021/pr500936p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mordant A., Kleiner M. Evaluation of sample preservation and storage methods for metaproteomics analysis of intestinal microbiomes. Microbiol Spectr. 2021;9(3) doi: 10.1128/Spectrum.01877-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen M., Wippler J., Kleiner M. Evaluation of RNAlater as a field-compatible preservation method for metaproteomic analyses of bacterium-animal symbioses. Microbiol Spectr. 2021;9(2) doi: 10.1128/Spectrum.01429-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X., Li L., Mayne J., Ning Z., Stintzi A., Figeys D. Assessing the impact of protein extraction methods for human gut metaproteomics. J Proteomics. 2018;180:120–127. doi: 10.1016/j.jprot.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 37.Bowers R.M., Kyrpides N.C., Stepanauskas R., Harmon-Smith M., Doud D., Reddy T.B.K., et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol. 2017;35(8):725–731. doi: 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao L., Feng Q., Liang S., Sonne S.B., Xia Z., Qiu X., et al. A catalog of the mouse gut metagenome. Nat Biotechnol. 2015;33(10):1103–1108. doi: 10.1038/nbt.3353. [DOI] [PubMed] [Google Scholar]
- 39.Li J., Jia H., Cai X., Zhong H., Feng Q., Sunagawa S., et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32(8):834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 40.Konstantinidis K.T., Tiedje J.M. Towards a genome-based taxonomy for prokaryotes. J Bacteriol. 2005;187(18):6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olm MR, Crits-Christoph A, Diamond S, Lavy A, Matheus Carnevali PB, Banfield JF. Consistent Metagenome-Derived Metrics Verify and Delineate Bacterial Species Boundaries. MSystems 2020;5. https://doi.org/10.1128/mSystems.00731-19. [DOI] [PMC free article] [PubMed]
- 42.Marcotte E.M. How do shotgun proteomics algorithms identify proteins? Nat Biotechnol. 2007;25(7):755–757. doi: 10.1038/nbt0707-755. [DOI] [PubMed] [Google Scholar]
- 43.Tabb D.L., Fernando C.G., Chambers M.C. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J Proteome Res. 2007;6:654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 45.Bjornson R.D., Carriero N.J., Colangelo C., Shifman M., Cheung K.-H., Miller P.L., et al. X!!Tandem, an improved method for running X!tandem in parallel on collections of commodity computers. J Proteome Res. 2008;7(1):293–299. doi: 10.1021/pr0701198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Howbert J.J., Noble W.S. Computing exact p-values for a cross-correlation shotgun proteomics score function. Mol Cell Proteomics MCP. 2014;13(9):2467–2479. doi: 10.1074/mcp.O113.036327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muth T., Behne A., Heyer R., Kohrs F., Benndorf D., Hoffmann M., et al. The MetaProteomeAnalyzer: a powerful open-source software suite for metaproteomics data analysis and interpretation. J Proteome Res. 2015;14(3):1557–1565. doi: 10.1021/pr501246w. [DOI] [PubMed] [Google Scholar]
- 48.Zhang X.u., Ning Z., Mayne J., Moore J.I., Li J., Butcher J., et al. MetaPro-IQ: a universal metaproteomic approach to studying human and mouse gut microbiota. Microbiome. 2016;4(1) doi: 10.1186/s40168-016-0176-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo X, Li Z, Yao Q, Mueller RS, Eng JK, Tabb DL, et al. Sipros Ensemble improves database searching and filtering for complex metaproteomics. Bioinforma Oxf Engl 2018;34:795–802. https://doi.org/10.1093/bioinformatics/btx601. [DOI] [PMC free article] [PubMed]
- 50.McIlwain S., Tamura K., Kertesz-Farkas A., Grant C.E., Diament B., Frewen B., et al. Crux: rapid open source protein tandem mass spectrometry analysis. J Proteome Res. 2014;13(10):4488–4491. doi: 10.1021/pr500741y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elias J.E., Gygi S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 52.Käll L., Canterbury J.D., Weston J., Noble W.S., MacCoss M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4(11):923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 53.McCain J.S.P., Bertrand E.M. Prediction and consequences of cofragmentation in metaproteomics. J Proteome Res. 2019;18(10):3555–3566. doi: 10.1021/acs.jproteome.9b00144. [DOI] [PubMed] [Google Scholar]
- 54.Keich U., Kertesz-Farkas A., Noble W.S. Improved false discovery rate estimation procedure for shotgun proteomics. J Proteome Res. 2015;14(8):3148–3161. doi: 10.1021/acs.jproteome.5b00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dorfer V., Maltsev S., Winkler S., Mechtler K. CharmeRT: boosting peptide identifications by chimeric spectra identification and retention time prediction. J Proteome Res. 2018;17(8):2581–2589. doi: 10.1021/acs.jproteome.7b00836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar D, Yadav AK, Dash D. Choosing an Optimal Database for Protein Identification from Tandem Mass Spectrometry Data. In: Keerthikumar S, Mathivanan S, editors. Proteome Bioinforma., New York, NY: Springer New York; 2017, p. 17–29. https://doi.org/10.1007/978-1-4939-6740-7_3. [DOI] [PubMed]
- 57.Knudsen G.M., Chalkley R.J., Aloy P. The effect of using an inappropriate protein database for proteomic data analysis. PLoS ONE. 2011;6(6):e20873. doi: 10.1371/journal.pone.0020873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin A., Plubell D.L., Keich U., Noble W.S. Accurately assigning peptides to spectra when only a subset of peptides are relevant. J Proteome Res. 2021;20(8):4153–4164. doi: 10.1021/acs.jproteome.1c00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jagtap P., Goslinga J., Kooren J.A., McGowan T., Wroblewski M.S., Seymour S.L., et al. A two-step database search method improves sensitivity in peptide sequence matches for metaproteomics and proteogenomics studies. Proteomics. 2013;13:1352–1357. doi: 10.1002/pmic.201200352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bern M., Kil Y.J. Comment on “unbiased statistical analysis for multi-stage proteomic search strategies”. J Proteome Res. 2011;10(4):2123–2127. doi: 10.1021/pr101143m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Everett L.J., Bierl C., Master S.R. Unbiased statistical analysis for multi-stage proteomic search strategies. J Proteome Res. 2010;9(2):700–707. doi: 10.1021/pr900256v. [DOI] [PubMed] [Google Scholar]
- 62.Kumar P., Johnson J.E., Easterly C., Mehta S., Sajulga R., Nunn B., et al. A sectioning and database enrichment approach for improved peptide spectrum matching in large, genome-guided protein sequence databases. J Proteome Res. 2020;19(7):2772–2785. doi: 10.1021/acs.jproteome.0c00260. [DOI] [PubMed] [Google Scholar]
- 63.O'Bryon I., Jenson S.C., Merkley E.D. Flying blind, or just flying under the radar? the underappreciated power of de novo methods of mass spectrometric peptide identification. Protein Sci. 2020;29(9):1864–1878. doi: 10.1002/pro.v29.910.1002/pro.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tran N.H., Zhang X., Xin L., Shan B., Li M. De novo peptide sequencing by deep learning. Proc Natl Acad Sci. 2017;114(31):8247–8252. doi: 10.1073/pnas.1705691114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tran N.H., Qiao R., Xin L., Chen X., Liu C., Zhang X., et al. Deep learning enables de novo peptide sequencing from data-independent-acquisition mass spectrometry. Nat Methods. 2019;16(1):63–66. doi: 10.1038/s41592-018-0260-3. [DOI] [PubMed] [Google Scholar]
- 66.Kleikamp H.B.C., Pronk M., Tugui C., Guedes da Silva L., Abbas B., Lin Y.M., et al. Database-independent de novo metaproteomics of complex microbial communities. Cell Syst. 2021;12(5):375–383.e5. doi: 10.1016/j.cels.2021.04.003. [DOI] [PubMed] [Google Scholar]
- 67.Muth T., Renard B.Y. Evaluating de novo sequencing in proteomics: already an accurate alternative to database-driven peptide identification? Brief Bioinform. 2018;19:954–970. doi: 10.1093/bib/bbx033. [DOI] [PubMed] [Google Scholar]
- 68.Zhang J, Xin L, Shan B, Chen W, Xie M, Yuen D, et al. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol Cell Proteomics MCP 2012;11:M111.010587-M111.010587. https://doi.org/10.1074/mcp.M111.010587. [DOI] [PMC free article] [PubMed]
- 69.Carr S., Aebersold R., Baldwin M., Burlingame A.l., Clauser K., Nesvizhskii A. The need for guidelines in publication of peptide and protein identification data: working group on publication guidelines for peptide and protein identification data. Mol Cell Proteomics. 2004;3(6):531–533. doi: 10.1074/mcp.T400006-MCP200. [DOI] [PubMed] [Google Scholar]
- 70.Erickson A.R., Cantarel B.L., Lamendella R., Darzi Y., Mongodin E.F., Pan C., et al. Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn’s disease. PLoS ONE. 2012;7(11):e49138. doi: 10.1371/journal.pone.0049138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao Y., Lin Y.-H. Whole-cell protein identification using the concept of unique peptides. Genomics Proteom. Bioinform. 2010;8(1):33–41. doi: 10.1016/S1672-0229(10)60004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Savitski M.M., Wilhelm M., Hahne H., Kuster B., Bantscheff M. A scalable approach for protein false discovery rate estimation in large proteomic data sets. Mol Cell Proteomics MCP. 2015;14(9):2394–2404. doi: 10.1074/mcp.M114.046995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Serang O., MacCoss M.J., Noble W.S. Efficient marginalization to compute protein posterior probabilities from shotgun mass spectrometry data. J Proteome Res. 2010;9(10):5346–5357. doi: 10.1021/pr100594k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langella O., Valot B., Balliau T., Blein-Nicolas M., Bonhomme L., Zivy M. X!TandemPipeline: a tool to manage sequence redundancy for protein inference and phosphosite identification. J Proteome Res. 2017;16(2):494–503. doi: 10.1021/acs.jproteome.6b00632. [DOI] [PubMed] [Google Scholar]
- 75.Ma Z.-Q., Dasari S., Chambers M.C., Litton M.D., Sobecki S.M., Zimmerman L.J., et al. IDPicker 2.0: Improved protein assembly with high discrimination peptide identification filtering. J Proteome Res. 2009;8(8):3872–3881. doi: 10.1021/pr900360j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tabb D.L., McDonald W.H., Yates J.R., 3rd. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 78.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 79.Blakeley-Ruiz J.A., Erickson A.R., Cantarel B.L., Xiong W., Adams R., Jansson J.K., et al. Metaproteomics reveals persistent and phylum-redundant metabolic functional stability in adult human gut microbiomes of Crohn’s remission patients despite temporal variations in microbial taxa, genomes, and proteomes. Microbiome. 2019;7(1) doi: 10.1186/s40168-019-0631-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2014;42:D7–D17. doi: 10.1093/nar/gkt1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.The UniProt Consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 2021;49:D480–9. https://doi.org/10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed]
- 82.Müller D.B., Schubert O.T., Röst H., Aebersold R., Vorholt J.A. Systems-level proteomics of two ubiquitous leaf commensals reveals complementary adaptive traits for phyllosphere colonization. Mol Cell Proteomics MCP. 2016;15(10):3256–3269. doi: 10.1074/mcp.M116.058164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ponnudurai R., Kleiner M., Sayavedra L., Petersen J.M., Moche M., Otto A., et al. Metabolic and physiological interdependencies in the Bathymodiolus azoricus symbiosis. ISME J. 2017;11(2):463–477. doi: 10.1038/ismej.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gruber-Vodicka H.R., Leisch N., Kleiner M., Hinzke T., Liebeke M., McFall-Ngai M., et al. Two intracellular and cell type-specific bacterial symbionts in the placozoan Trichoplax H2. Nat Microbiol. 2019;4(9):1465–1474. doi: 10.1038/s41564-019-0475-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Assié A., Leisch N., Meier D.V., Gruber-Vodicka H., Tegetmeyer H.E., Meyerdierks A., et al. Horizontal acquisition of a patchwork Calvin cycle by symbiotic and free-living Campylobacterota (formerly Epsilonproteobacteria) ISME J. 2020;14(1):104–122. doi: 10.1038/s41396-019-0508-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hinzke T., Kleiner M., Breusing C., Felbeck H., Häsler R., Sievert S.M., et al. Host-microbe interactions in the chemosynthetic riftia pachyptila symbiosis. MBio. 2019;10(6) doi: 10.1128/mBio.02243-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lloyd K.G., Steen A.D., Ladau J., Yin J., Crosby L., Neufeld J.D. Phylogenetically novel uncultured microbial cells dominate earth microbiomes. MSystems. 2018;3:e00055–e118. doi: 10.1128/mSystems.00055-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tanca A., Palomba A., Fraumene C., Pagnozzi D., Manghina V., Deligios M., et al. The impact of sequence database choice on metaproteomic results in gut microbiota studies. Microbiome. 2016;4(1) doi: 10.1186/s40168-016-0196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.David L.A., Maurice C.F., Carmody R.N., Gootenberg D.B., Button J.E., Wolfe B.E., et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Young J.C., Pan C., Adams R.M., Brooks B., Banfield J.F., Morowitz M.J., et al. Metaproteomics reveals functional shifts in microbial and human proteins during a preterm infant gut colonization case. Proteomics. 2015;15:3463–3473. doi: 10.1002/pmic.201400563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chibani C.M., Mahnert A., Borrel G., Almeida A., Werner A., Brugère J.-F., et al. A catalogue of 1,167 genomes from the human gut archaeome. Nat Microbiol. 2022;7(1):48–61. doi: 10.1038/s41564-021-01020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tanca A., Palomba A., Deligios M., Cubeddu T., Fraumene C., Biosa G., et al. Evaluating the impact of different sequence databases on metaproteome analysis: insights from a lab-assembled microbial Mixture. PLoS ONE. 2013;8(12):e82981. doi: 10.1371/journal.pone.0082981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Heyer R., Schallert K., Zoun R., Becher B., Saake G., Benndorf D. Challenges and perspectives of metaproteomic data analysis. Bioinforma Solut Big Data Anal Life Sci Present Ger Netw Bioinforma Infrastruct. 2017;261:24–36. doi: 10.1016/j.jbiotec.2017.06.1201. [DOI] [PubMed] [Google Scholar]
- 94.May D.H., Timmins-Schiffman E., Mikan M.P., Harvey H.R., Borenstein E., Nunn B.L., et al. An alignment-free “metapeptide” strategy for metaproteomic characterization of microbiome samples using shotgun metagenomic sequencing. J Proteome Res. 2016;15(8):2697–2705. doi: 10.1021/acs.jproteome.6b00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.diCenzo G.C., Finan T.M. The divided bacterial genome: structure, function, and evolution. Microbiol Mol Biol Rev MMBR. 2017;81(3) doi: 10.1128/MMBR.00019-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Luo C., Tsementzi D., Kyrpides N.C., Konstantinidis K.T. Individual genome assembly from complex community short-read metagenomic datasets. ISME J. 2012;6(4):898–901. doi: 10.1038/ismej.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hyatt D., LoCascio P.F., Hauser L.J., Uberbacher E.C. Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics. 2012;28:2223–2230. doi: 10.1093/bioinformatics/bts429. [DOI] [PubMed] [Google Scholar]
- 98.Klassen J.L., Currie C.R. Gene fragmentation in bacterial draft genomes: extent, consequences and mitigation. BMC Genomics. 2012;13(1):14. doi: 10.1186/1471-2164-13-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Treiber M.L., Taft D.H., Korf I., Mills D.A., Lemay D.G. Pre- and post-sequencing recommendations for functional annotation of human fecal metagenomes. BMC Bioinf. 2020;21:74. doi: 10.1186/s12859-020-3416-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Prakash T., Taylor T.D. Functional assignment of metagenomic data: challenges and applications. Brief Bioinform. 2012;13(6):711–727. doi: 10.1093/bib/bbs033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.West P.T., Peters S.L., Olm M.R., Yu F.B., Gause H., Lou Y.C., et al. Genetic and behavioral adaptation of Candida parapsilosis to the microbiome of hospitalized infants revealed by in situ genomics, transcriptomics, and proteomics. Microbiome. 2021;9(1) doi: 10.1186/s40168-021-01085-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Timmins-Schiffman E., May D.H., Mikan M., Riffle M., Frazar C., Harvey H.R., et al. Critical decisions in metaproteomics: achieving high confidence protein annotations in a sea of unknowns. ISME J. 2017;11(2):309–314. doi: 10.1038/ismej.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huson D.H., Auch A.F., Qi J.i., Schuster S.C. MEGAN analysis of metagenomic data. Genome Res. 2007;17(3):377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Argentini A., Staes A.n., Grüning B., Mehta S., Easterly C., Griffin T.J., et al. Update on the moFF algorithm for label-free quantitative proteomics. J Proteome Res. 2019;18(2):728–731. doi: 10.1021/acs.jproteome.8b00708. [DOI] [PubMed] [Google Scholar]
- 105.Hugerth L.W., Andersson A.F. Analysing microbial community composition through amplicon sequencing: from sampling to hypothesis testing. Front Microbiol. 2017;8:1561. doi: 10.3389/fmicb.2017.01561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Morris BEL, Herbst F-A, Bastida F, Seifert J, von Bergen M, Richnow H-H, et al. Microbial interactions during residual oil and n-fatty acid metabolism by a methanogenic consortium. Environ Microbiol Rep 2012;4:297–306. https://doi.org/10.1111/j.1758-2229.2012.00333.x. [DOI] [PubMed]
- 107.Chen L.-X., Anantharaman K., Shaiber A., Eren A.M., Banfield J.F. Accurate and complete genomes from metagenomes. Genome Res. 2020;30(3):315–333. doi: 10.1101/gr.258640.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pérez-Cobas A.E., Gomez-Valero L., Buchrieser C. Metagenomic approaches in microbial ecology: an update on whole-genome and marker gene sequencing analyses. Microb. Genomics. 2020;6 doi: 10.1099/mgen.0.000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Uyaguari-Diaz M.I., Chan M., Chaban B.L., Croxen M.A., Finke J.F., Hill J.E., et al. A comprehensive method for amplicon-based and metagenomic characterization of viruses, bacteria, and eukaryotes in freshwater samples. Microbiome. 2016;4(1) doi: 10.1186/s40168-016-0166-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Eren A.M., Kiefl E., Shaiber A., Veseli I., Miller S.E., Schechter M.S., et al. Community-led, integrated, reproducible multi-omics with anvi’o. Nat Microbiol. 2021;6(1):3–6. doi: 10.1038/s41564-020-00834-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Uritskiy G.V., DiRuggiero J., Taylor J. MetaWRAP—a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome. 2018;6:158. doi: 10.1186/s40168-018-0541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kieser S., Brown J., Zdobnov E.M., Trajkovski M., McCue L.A. ATLAS: a Snakemake workflow for assembly, annotation, and genomic binning of metagenome sequence data. BMC Bioinf. 2020;21:257. doi: 10.1186/s12859-020-03585-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Krueger F. The Babraham Institute; 2021. Trim Galore. [Google Scholar]
- 114.Bushnell B. BBMap: a fast, accurate, splice-aware aligner. Lawrence Berkeley National Lab.(LBNL), Berkeley, CA (United States); 2014.
- 115.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma Oxf Engl. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nurk S., Meleshko D., Korobeynikov A., Pevzner P.A. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27(5):824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li D., Liu C.-M., Luo R., Sadakane K., Lam T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 119.van der Walt A.J., van Goethem M.W., Ramond J.-B., Makhalanyane T.P., Reva O., Cowan D.A. Assembling metagenomes, one community at a time. BMC Genomics. 2017;18:521. doi: 10.1186/s12864-017-3918-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wu Y.-W., Tang Y.-H., Tringe S.G., Simmons B.A., Singer S.W. MaxBin: an automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome. 2014;2:26. doi: 10.1186/2049-2618-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 2019;7:e7359–e7359. https://doi.org/10.7717/peerj.7359. [DOI] [PMC free article] [PubMed]
- 122.Alneberg J., Bjarnason B.S., de Bruijn I., Schirmer M., Quick J., Ijaz U.Z., et al. Binning metagenomic contigs by coverage and composition. Nat Methods. 2014;11(11):1144–1146. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- 123.Sieber C.M.K., Probst A.J., Sharrar A., Thomas B.C., Hess M., Tringe S.G., et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol. 2018;3(7):836–843. doi: 10.1038/s41564-018-0171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol 2021. https://doi.org/10.1093/molbev/msab199. [DOI] [PMC free article] [PubMed]
- 125.Parks D.H., Imelfort M., Skennerton C.T., Hugenholtz P., Tyson G.W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25(7):1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Field D., Garrity G., Gray T., Morrison N., Selengut J., Sterk P., et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26(5):541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Olm M.R., Brown C.T., Brooks B., Banfield J.F. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017;11(12):2864–2868. doi: 10.1038/ismej.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res 2010;38:e132. https://doi.org/10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed]
- 129.Chaumeil P.-A., Mussig A.J., Hugenholtz P., Parks D.H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2020;36:1925–1927. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinforma Oxf Engl. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 131.Shaffer M, Borton MA, McGivern BB, Zayed AA, La Rosa SL, Solden LM, et al. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res 2020;48:8883–900. https://doi.org/10.1093/nar/gkaa621. [DOI] [PMC free article] [PubMed]
- 132.Dong X., Strous M. An integrated pipeline for annotation and visualization of metagenomic contigs. Front Genet. 2019;10:999. doi: 10.3389/fgene.2019.00999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gouveia D., Pible O., Culotta K., Jouffret V., Geffard O., Chaumot A., et al. Combining proteogenomics and metaproteomics for deep taxonomic and functional characterization of microbiomes from a non-sequenced host. Npj Biofilms Microbiomes. 2020;6(1) doi: 10.1038/s41522-020-0133-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wippler J., Kleiner M., Lott C., Gruhl A., Abraham P.E., Giannone R.J., et al. Transcriptomic and proteomic insights into innate immunity and adaptations to a symbiotic lifestyle in the gutless marine worm Olavius algarvensis. BMC Genomics. 2016;17(1) doi: 10.1186/s12864-016-3293-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.West P.T., Probst A.J., Grigoriev I.V., Thomas B.C., Banfield J.F. Genome-reconstruction for eukaryotes from complex natural microbial communities. Genome Res. 2018;28(4):569–580. doi: 10.1101/gr.228429.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.von Meijenfeldt F.A.B., Arkhipova K., Cambuy D.D., Coutinho F.H., Dutilh B.E. Robust taxonomic classification of uncharted microbial sequences and bins with CAT and BAT. Genome Biol. 2019;20:217. doi: 10.1186/s13059-019-1817-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Parks D.H., Chuvochina M., Waite D.W., Rinke C., Skarshewski A., Chaumeil P.-A., et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36(10):996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 138.Rinke C., Chuvochina M., Mussig A.J., Chaumeil P.-A., Davín A.A., Waite D.W., et al. A standardized archaeal taxonomy for the Genome Taxonomy Database. Nat Microbiol. 2021;6(7):946–959. doi: 10.1038/s41564-021-00918-8. [DOI] [PubMed] [Google Scholar]
- 139.Menzel P., Ng K.L., Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun. 2016;7:11257. doi: 10.1038/ncomms11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim D., Song L.i., Breitwieser F.P., Salzberg S.L. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 2016;26(12):1721–1729. doi: 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 2019;47:D309–14. https://doi.org/10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed]
- 143.Kanehisa M., Furumichi M., Sato Y., Ishiguro-Watanabe M., Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Blum M, Chang H-Y, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res 2021;49:D344–54. https://doi.org/10.1093/nar/gkaa977. [DOI] [PMC free article] [PubMed]
- 145.Rawlings N.D., Barrett A.J., Thomas P.D., Huang X., Bateman A., Finn R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018;46:D624–D632. doi: 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Caspi R, Billington R, Keseler IM, Kothari A, Krummenacker M, Midford PE, et al. The MetaCyc database of metabolic pathways and enzymes - a 2019 update. Nucleic Acids Res 2020;48:D445–53. https://doi.org/10.1093/nar/gkz862. [DOI] [PMC free article] [PubMed]
- 147.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(D1):D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.The Gene Ontology Consortium The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., et al. The COG database: an updated version includes eukaryotes. BMC Bioinf. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Saier M.H., Reddy V.S., Tamang D.G., Västermark Å. The transporter classification database. Nucleic Acids Res. 2014;42(D1):D251–D258. doi: 10.1093/nar/gkt1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, et al. Fast Genome-Wide Functional Annotation through Orthology Assignment by eggNOG-Mapper. Mol Biol Evol 2017;34:2115–22. https://doi.org/10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed]
- 153.Jiang Y., Oron T.R., Clark W.T., Bankapur A.R., D’Andrea D., Lepore R., et al. An expanded evaluation of protein function prediction methods shows an improvement in accuracy. Genome Biol. 2016;17(1) doi: 10.1186/s13059-016-1037-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sajulga R, Easterly C, Riffle M, Mesuere B, Muth T, Mehta S, et al. Survey of metaproteomics software tools for functional microbiome analysis. PLOS ONE 2020;15:e0241503. https://doi.org/10.1371/journal.pone.0241503. [DOI] [PMC free article] [PubMed]
- 155.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9(1) doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jones P., Binns D., Chang H.-Y., Fraser M., Li W., McAnulla C., et al. InterProScan 5: genome-scale protein function classification. Bioinforma Oxf Engl. 2014;30(9):1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 2018;46:W95–101. https://doi.org/10.1093/nar/gky418. [DOI] [PMC free article] [PubMed]
- 158.Kanehisa M., Sato Y., Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. Comput Resour Mol Biol. 2016;428(4):726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 159.Zhu J., Ren H., Zhong H., Li X., Zou Y., Han M.o., et al. An expanded gene catalog of mouse gut metagenomes. MSphere. 2021;6(1) doi: 10.1128/mSphere.01119-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Van Den Bossche T., Arntzen M.Ø., Becher D., Benndorf D., Eijsink V.G.H., Henry C., et al. The Metaproteomics Initiative: a coordinated approach for propelling the functional characterization of microbiomes. Microbiome. 2021;9(1) doi: 10.1186/s40168-021-01176-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ciuffreda L., Rodríguez-Pérez H., Flores C. Nanopore sequencing and its application to the study of microbial communities. Comput Struct Biotechnol J. 2021;19:1497–1511. doi: 10.1016/j.csbj.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Xie H., Yang C., Sun Y., Igarashi Y., Jin T., Luo F. PacBio long reads improve metagenomic assemblies, gene catalogs, and genome binning. Front Genet. 2020;11:1077. doi: 10.3389/fgene.2020.516269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bickhart DM, Kolmogorov M, Tseng E, Portik DM, Korobeynikov A, Tolstoganov I, et al. Generating lineage-resolved, complete metagenome-assembled genomes from complex microbial communities. Nat Biotechnol 2022. https://doi.org/10.1038/s41587-021-01130-z. [DOI] [PubMed]
- 164.Aballo TJ, Roberts DS, Melby JA, Buck KM, Brown KA, Ge Y. Ultrafast and Reproducible Proteomics from Small Amounts of Heart Tissue Enabled by Azo and timsTOF Pro. J Proteome Res 2021;20:4203–11. https://doi.org/10.1021/acs.jproteome.1c00446. [DOI] [PMC free article] [PubMed]
- 165.Aakko J., Pietilä S., Suomi T., Mahmoudian M., Toivonen R., Kouvonen P., et al. Data-independent acquisition mass spectrometry in metaproteomics of gut microbiota—implementation and computational analysis. J Proteome Res. 2020;19(1):432–436. doi: 10.1021/acs.jproteome.9b00606. [DOI] [PubMed] [Google Scholar]
- 166.Brinkerhoff H., Kang A.S.W., Liu J., Aksimentiev A., Dekker C. Multiple rereads of single proteins at single–amino acid resolution using nanopores. Science. 2021;374(6574):1509–1513. doi: 10.1126/science:abl4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Prianichnikov N., Koch H., Koch S., Lubeck M., Heilig R., Brehmer S., et al. MaxQuant software for ion mobility enhanced shotgun proteomics. Mol Cell Proteomics MCP. 2020;19(6):1058–1069. doi: 10.1074/mcp.TIR119.001720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sinitcyn P., Hamzeiy H., Salinas Soto F., Itzhak D., McCarthy F., Wichmann C., et al. MaxDIA enables library-based and library-free data-independent acquisition proteomics. Nat Biotechnol. 2021;39(12):1563–1573. doi: 10.1038/s41587-021-00968-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang X., Deeke S.A., Ning Z., Starr A.E., Butcher J., Li J., et al. Metaproteomics reveals associations between microbiome and intestinal extracellular vesicle proteins in pediatric inflammatory bowel disease. Nature Communications. 2018;9(1) doi: 10.1038/s41467-018-05357-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kolmeder C.A., de Been M., Nikkilä J., Ritamo I., Mättö J., Valmu L., et al. Comparative Metaproteomics and Diversity Analysis of Human Intestinal Microbiota Testifies for Its Temporal Stability and Expression of Core Functions. PLOS ONE. 2012;7(1) doi: 10.1371/journal.pone.0029913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kolmeder C.A., Salojärvi J., Ritari J., de Been M., Raes J., Falony G., et al. Faecal Metaproteomic Analysis Reveals a Personalized and Stable Functional Microbiome and Limited Effects of a Probiotic Intervention in Adults. PLOS ONE. 2016;11(4) doi: 10.1371/journal.pone.0153294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lesker T.R., Durairaj A.C., Gálvez E.J.C., Lagkouvardos I., Baines J.F., Clavel T., et al. An Integrated Metagenome Catalog Reveals New Insights into the Murine Gut Microbiome. Cell Reports. 2020;30(9) doi: 10.1016/j.celrep.2020.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Royo-Llonch M., Sánchez P., Ruiz-González C., Salazar G., Pedrós-Alió C., Sebastián M., et al. Compendium of 530 metagenome-assembled bacterial and archaeal genomes from the polar Arctic Ocean. Nature Microbiology. 2021;6(12) doi: 10.1038/s41564-021-00979-9. [DOI] [PubMed] [Google Scholar]
- 174.Choi J., Yang F., Stepanauskas R., Cardenas E., Garoutte A., Williams R., et al. Strategies to improve reference databases for soil microbiomes. The ISME Journal. 2017;11(4) doi: 10.1038/ismej.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.D’Auria G., Jiménez-Hernández N., Peris-Bondia Francesc, Moya A., Latorre A. Legionella pneumophila pangenome reveals strain-specific virulence factors. BMC Genomics. 2010;11(1) doi: 10.1186/1471-2164-11-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Horesh G., Blackwell G.A., Tonkin-Hill G., Corander J., Heinz E., Thomson N.R. A comprehensive and high-quality collection of Escherichia coli genomes and their genes. Microbial Genomics. 2021;7(2) doi: 10.1099/mgen.0.000499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and environmental microbiology. 2009;75(23) doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bolyen E., Rideout J.R, Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature biotechnology. 2019;37(8) doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.https://www.thegpm.org/crap/ v2012.01.01.