Graphical abstract
Keywords: Microplastics, Plastisphere, Microorganisms, Biodegradation
Abstract
The pollution of plastic waste has become an increasingly serious environmental crisis. Recently, plastic has been detected in various kinds of environments, even in human tissues, which is an increasing threat to the ecosystems and humans. In the ocean, the plastic waste is eventually fragmentized into microplastics (MPs) under the disruption of physical and chemical processes. MPs are colonized by microbial communities such as fungi, diatoms, and bacteria, which form biofilms on the surface of the plastic called “plastisphere”. In this review, we summarize the studies related to microorganisms in the plastisphere in recent years and describe the microbial species in the plastisphere, mainly including bacteria, fungi, and autotrophs. Secondly, we explore the interactions between MPs and the plastisphere. The depth of MPs in the ocean and the nutrients in the surrounding seawater can have a great impact on the community structure of microorganisms in the plastisphere. Finally, we discuss the types of MP-degrading bacteria in the ocean, and use the “seed bank” theory to speculate on the potential sources of MP-degrading microorganisms. Challenges and future research prospects are also discussed.
1. Introduction
Plastics have been widely used in industry and daily life due to their excellent durability, plasticity, corrosion resistance, and low cost [1]. However, the corrosion resistance of plastics also makes them difficult to degrade. In the past few years, global plastic waste has reached about 6.3 billion tons [2], [3]. Without intervention at the current rate of plastic waste emissions, the ever-increasing plastic pollution could double by 2030 [4]. Carbon-based polymers such as polyethylene (PE), polypropylene (PP), polystyrene (PS), polyvinyl chloride (PVC), and polyethylene terephthalate (PET) are the most common plastics found in the environment, accounting for approximately 80% of global plastic waste [2], [5], [6], [7]. In addition to these compounds, plastic products made of polyurethane (PU) and polyamides (PA) are also widely distributed [8]. Depending on the degree of degradation, plastics can be divided into biodegradable and non-biodegradable plastics. Biodegradable plastics are mainly made from biological sources, such as starch or cellulose, which can be either bio-based or fossil-based [9]. A class of biodegradable plastics, such as polylactic acid (PLA), polylcaprolactone (PCL), polyhydroxybutyrate (PHB), and Polyurethanes (PU), have been developed [10]. At present, landfill is the main treatment of plastic waste, which inevitably results in secondary pollution to the environment [11], [12], [13]. 70% to 80% of the plastic debris is transferred through rivers to the ocean [14], and distributed along the coastline, at the surface and seafloor, and even in remote areas such as open oceans far from land [15], [16]. Currently, about 51 trillion plastic particles weighing 236,000 tons were estimated in the ocean [17]. The longer half-life and hydrophobic surface of plastic promote the colonization of microbes and transportation of harmful algal species and persistent organic pollutants [18].
Microplastics (MPs) are defined by Frias and Nash [19] as “Microplastics are any synthetic solid particle or polymeric matrix, with regular or irregular shape and with size ranging from 1 μm to 5 mm, of either primary or secondary manufacturing origin, which are insoluble in water”. Plastic particles that are produced to microscopic dimensions are called primary MPs [20]. An estimated 1.5 million tons of primary MPs are released into the ocean each year globally, accounting for approximately 15–31% of all MPs in the ocean [21]. Smaller MP particles are more harmful in general, and more difficult to remove than larger plastic fragments of the same weight [22], [23]. The MPs formed from fiber fragments released from synthetic fibers during the washing process, the degradation of agricultural mulch, and the weathering and decomposition of plastic waste in the marine environment become secondary MPs, which are much more abundant in the ocean than primary MPs [20], [24]. Once plastic waste reaches the marine environment, buoyancy will determine the distribution of the plastic. Some plastics such as PE and PP that are less dense than seawater will float. They will be transported over long distances by wind and surface currents [25]. In contrast, MPs with greater density, such as PVC, are more likely to sink in seawater [26]. Due to microbial degradation and various physicochemical effects, plastics floating on the sea may lose their surface hydrophobicity and increase in density over weeks to months, eventually sinking to the sea floor [27]. These plastics may last hundreds to thousands of years because of their stability and durability.
The most frequently collected MPs in the surface water sample are in the form of fragments, fibers, films, foams, and pellets [28]. In the Yellow Sea of China, researchers found that the predominant type of MPs in the area was PE, while the most frequently shape appeared to be fiber [29]. In addition to the variety of shapes, the surface of MPs particles is rough and porous due to various physical effects such as wave friction. The larger surface area to volume ratio of MPs makes it easier for the attachment of hydrophobic organic substances in seawater [30], [31]. Previous studies found that the adsorption capacity of MPs for organic fouling in the ocean is about two orders of magnitude and six orders of magnitude higher than that of sediments and surrounding seawater, respectively [32], [33]. The organic compounds of the plastic coupled with the various substances adsorbed make MPs a unique substrate for microbial attachment in the ocean. Assuming one plastic particle with a diameter of 1 mm per cubic meter of seawater, these MPs could provide a surface area of 4.2 million square kilometers, which would provide a large space for the adsorption of harmful substances and microorganisms [34]. This amount, however, is considerably lower than the current estimation of the total amount of MPs in the ocean, which is around 12.7 million tons [35]. In addition to providing ample organic matter, the rough surface of the MPs also provides a stable habitat that helps microorganisms to resist environmental pressure [36]. Thus, plastics might be an ideal substrate for microorganisms in the environment [37], [38], [39].
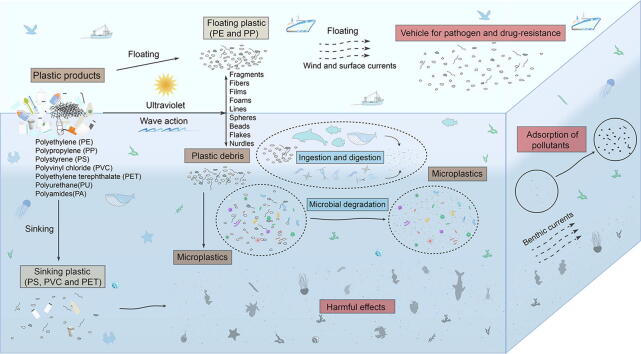
The large amount of MPs in the ocean has an impact on the ecology of the ocean and the food chain. In the ocean, MPs could affect the photosynthesis and growth of phytoplankton [40], slow down the swimming of zooplankton and thus reduce their reproductive efficiency [41], and may also impact the ocean carbon stock [42]. In addition, floating MPs could become a transport vehicle for microorganisms and even some pathogenic microorganisms, accelerating the spread of infectious diseases [43]. Drifting MPs, when dispersed by currents and waves, move the microbiota they carry to a new habitat and are able to spread even across oceans [43]. In the Arctic Ocean, which has been less affected by anthropogenic activities, researchers have also found large amounts of MPs in the sea surface and sediments [44], [45]. MPs are likely to have been enriched in the food chain and food web. They have been found in fish, shellfish, Antarctic krill and coral in the ocean and are contaminating the food web to a much greater degree than we know [46], [47], [48], [49]. Most common MPs type is mainly blue-colored [50]. These colored MPs are particularly harmful in aquatic environments since they can be mistaken for food and ingested directly by marine animals [50], [51]. MPs that enter biological cells may have a greater impact on living organisms, capable of causing weight loss, local inflammation and interfering with energy redistribution (Fig. 1) [52], [53].
Fig. 1.
The possible fate of MPs and plastisphere in the ocean ecosystem.
In addition to the hazards of the MPs own compounds, the organic pollutants adsorbed on their surface can also cause a lot of ecological damage, such as phenanthrene, diazinon, and nonylphenol [54]. MPs tend to adsorb and accumulate pollutants from surrounding water, such as dichloro-diphenyl-trichloroethane (DDTs), polychlorinated biphenyls (PCBs), polybrominated diphenyl ethers (PBDEs), and alkylphenols and bisphenol A (BPA) [55], [56], [57]. The adsorption capacity of MPs is related to their own properties, and the adsorption capacity of MPs of different materials varies for the same volume and surface area, with PE reported to absorb more organic pollutants than other types of MPs [58], [59]. Antibiotics, which are heavily abused in mariculture, are highly susceptible to adsorption by MPs, which will accelerate the emergence of drug-resistant bacteria and the transfer of drug-resistant genes [60]. As a result, MPs create a potential stress on the environment, both biotic and abiotic, in the ocean [61], [62].
Microorganisms can also colonize the surface of MPs, forming biofilms called plastisphere [18]. In this review, we provide a holistic view of current knowledge about the composition and distribution of the plastisphere, with an emphasize on its role in plastic biodegradation. As the treatment and recycling of plastic waste is of great interest, future challenges and perspectives are also highlighted.
2. Plastisphere biodiversity
Plastisphere, first termed by Zettler et al., describes a novel microbial community attached to plastic and distinct from the surroundings [18]. There are two main strategies to study plastisphere on marine MPs, namely environmental sampling and laboratory incubation. Reports of environmental sampling are limited due to the difficulty and expense for sampling, especially for benthic ocean. By using laboratory incubation, several studies have been performed in which plastic fragments are incubated in collected seawater or sediment under artificially created laboratory conditions, which exclude the disturbance of variable natural environments [63]. Due to the more stable environment, this kind of study is usually suitable to observe the plastic degradation ability of marine microorganisms and the function of degradation enzymes. Early studies of the plastisphere relied primarily on microscopy or scanning electron microscopy (SEM) to morphologically identify different organisms [4]. High-throughput sequencing has been used to investigate plastisphere only recently [18]. In most studies of microorganisms on the surface of marine MPs, the small ribosomal subunit 16S gene (16S rRNA) and the eukaryotic 18S rRNA gene have been used for MetabarCoding with second-generation sequencing technologies, mainly MiSeq Illumina sequencing as well as 454 pyrosequencing (Roche) [64]. The internal transcribed spacer (ITS) technique is also used for the classification of fungi in plastisphere [65], [66]. For prokaryotes, the most commonly used genetic barcodes in metabarcoding studies are located at the high-variable 16S rRNA V4-V5 locus of bacteria, and the V3 locus has also been used in some early studies (Table 1). Due to the shorter barcode sequence length that limits barcode resolution, the second-generation sequencing technology can usually identify to the genus level. Long-read sequencing technologies, such as third-generation sequencing, should be used to enable the sequencing of longer regions of the same barcode, leading to more accurate species classification [67]. 18 s rRNA is the typical target for the identification of microbial eukaryotes. However, the 18S rRNA gene has limited taxonomic resolution for some fungal groups, so fungal-specific primers, such as those targeting the Internal Transcribed Space regions (ITS) of the rRNA operon, are used for the detection of fungi to ensure the accuracy (Table 2). Studies focused on eukaryotes in the marine plastisphere are relatively few. However, as a natural part of the plastisphere, future studies are needed for the detection of eukaryotic taxa.
Table 1.
Bacteria in the marine plastisphere.
| Species | Type of plastic | Studied area | Incubation time | Sample type | Method | Reference |
|---|---|---|---|---|---|---|
| Bryozoa, Cyanobacteria, Alphaproteobacteria, and Bacteroidetes | – | North Pacific subtropical Gyre | – | sea surface | Metagenomic sequencing | Bryant et al., 2016 [73] |
| Flavobacteriaceae, Cryomorphaceae, Saprospiraceae | PET | North Sea | 1 month | sea surface | V4 16S rRNA sequencing | Oberbeckmann et al., 2016 [110] |
| Alpha- and gammaproteobacteria | PE | Belgian part of the North Sea | 1–44 months | seafloor | V3–V4 16S rRNA | De Tender et al., 2017 [66] |
| Rhodobacterales, Rhizobiales, Streptomycetales and Cyanobacteria | – | North Atlantic subtropical gyre | – | seafloor | V4 16S rRNA sequencing | Debroas et al., 2017 [167] |
| Alphaproteobacteria, Flavobacteria, Cyanobacteria, and Actinobacteria | PE | Mediterranean Sea | 7–45 days | sea surface | V3-V5 16S rRNA sequencing | Dussud et al., 2018 [91] |
| Alphaproteobacteria, Cyanobacteria, Flavobacteria andGammaproteobacteria | – | Mediterranean Sea | – | sea surface | V3-V5 16S rRNA sequencing | Dussud et al., 2018 [144] |
| Proteobacteria, Nitrospira, Planctomycetacia, Caldilineae and Acidimicrobiia | PE, PP, PS, PET, PLA | North Sea, Germany | 15 months | – | V3-V4 16S rRNA sequencing | Kirstein et al., 2018 [168] |
| Pirellula, Planctomyces, Pseudomonas, Synechococcus, and Ilumatobacter and Blastopirellula | PE, PET | Arabian Sea | 30 days | sea surface | V4 16S rRNA sequencing | Muthukrishnan et al., 2018 [72] |
| Erythrobacteraceae, Rhodobacteraceae and Cyanobacteria | PE, PP, PS | East China Sea | – | sea surface | V3-V4 16S rRNA sequencing | Jiang et al., 2018 [169] |
| Actinobacteria, Cyanobacteria and Proteobacteria | – | East China Sea | – | deepwater | V5–V6 16S rRNA sequencing | Wu et al., 2018 [170] |
| Alcanivorax, Marinobacter and Arenibacter genera | – | Mediterranean Sea | – | sea surface and sediment | V3-V4 16S rRNA | Delacuvellerie et al., 2019 [171] |
| Alphaproteobacteria, Gammaproteobacteria and Bacteroidia | PE | Offshore of Yantai, China | 30 days, 75 days, and 135 days | 2 m, 6 m, and 12 m | V4 16S rRNA sequencing | Chen et a., 2020 [87] |
| Proteobacteria, Bacteroidetes and Cyanobacteria | Herzliya marina | 1 months | sea surface | full 16S rRNA sequencing | Davidov et a., 2020 [65] | |
| Bacteroidetes, Proteobacteria, Firmicutes and Cyanobacteria | – | Hikine Island, Japan | – | 7 m | V4–V5 16S rRNA sequencing | Harvey et al., 2020 [172] |
| Flavobacteriia, Saprospirae, and Cytophagia | PETE, HDPE, PVC, LDPE, PP, PS | Coast of Bocas del Toro | 6 weeks | sea surface | V4–V5 16S rRNA sequencing | Dudek et al., 2020 [173] |
| Bacteriodes and Proteobacteria | PE, PUF, PVC, PLA | York River estuary | 7 days, 16 days | – | V4–V5 16S rRNA sequencing | Seeley et al., 2020 [63] |
| Flavobacteriales, Rhodobacterales, Cytophagales, Rickettsiales, Alteromonadales, Chitinophagales, and Oceanospirillales | PE, PP, PE | Mediterranean Sea | – | sea surface | V4–V5 16S rRNA sequencing | Vaksmaa et al., 2021 [174] |
| Methylologellaceae, Colwelliaceae, Pseudomonadaceae, Haliangiaceae, Micrococcaceae, Halieceaea | PE | North Atlantic | 719 days | 3300 m | V4 16S rRNA sequencing | Agostini et al., 2021 [175] |
| Idiomarina, Marinobacter, Exiguobacterium, Halomonas and Ochrobactrum | PET, PE | Huiquan Bay (Qingdao, China) | several weeks to months | sea surface | V4-V5 16S rRNA sequencing | Gao et al., 2021 [176] |
| Bdellovibrio and Pseudomonas | PS | Artificial seawater | 60 days | – | V4 16S rRNA sequencing | Ye et al., 2021 [177] |
| Rhodobacteriaceae and Flavobacteriaceae | PVC | North-Western Mediterranean Sea, Atlantic Ocean and Indian Ocean | 28 days, 75 days | seafloor | V4-V5 16S rRNA sequencing | Catao et al., 2021 [178] |
| Alteromonadaceae, Thalassospiraceae and Vibrionaceae | PET | Porthcawl beach | 6 weeks | laboratory incubations | V4-V5 16S rRNA sequencing | Wright et al., 2021 [179] |
| Actinobacteria, Gammaproteobacteria, Alphaproteobacteria, Opitutae and Sphingobacteriaf | PE, PP | Baltic, Sargasso and Mediterranean seas | – | sea surface | V3-V4 16S rRNA sequencing | Scales et al., 2021 [180] |
| Proteobacteria, Bacteroidetes, Acidobacteria, Cyanobacteria and Actinobacteria | PP, PET, PE | South China Sea | – | sea surface | V4-V5 16S rRNA sequencing | Chen et al., 2021 [181] |
Table 2.
Fungi and autotrophs in the marine plastisphere.
| Species | Type of plastic | Studied area | Incubation time | Sample type | Method | Reference |
|---|---|---|---|---|---|---|
| Fungus | ||||||
| Chytridiomycota, Cryptomycota and Ascomycota | PE,PS | Baltic Sea | 15 days | sea surface | V4 18S rRNA sequencing | Kettner et al., 2017 [34] |
| Paramoeba permaquidensis, Paramoeba aestuarina, Pleurobrachia pileus, Sugiura chengshanense, Sagartia elegans, and Rhizostoma pulmo | PE | Belgian part of the North Sea | 1–44 months | seafloor | ITS2 | De Tender et al., 2017 [66] |
| Chytridiomycetes | PE, PP, PS, PET, PLA | North Sea, Germany | 15 months | V4 18S rRNA sequencing | Kirstein et al., 2018 [168] | |
| Chytridium, Rhizidiomyces and Pythium | PE, PS | Baltic Sea | 15 days | sea surface | V4 18S rRNA sequencing | Kettner et al., 2019 [69] |
| Aspergillus, Cladosporium and Wallemia | PE, PA, PU, PP, PS | western South Atlantic and Antarctic Peninsula | – | sea surface | V9, V4 18S rRNA and ITS2 | Lacerda et al., 2020 [84] |
| Pleosporales | PE | Herzliya marina | 1 months | sea surface | ITS | Davidov et a., 2020 [65] |
| Autotroph | ||||||
| Coscinodiscophytina, Bacillariophytina | PET | North Sea | 1 month | sea surface | V9 18S rRNA sequencing | Sonja et al., 2016 [110] |
| Cryptophyceae, Haptophyta and Chloroplastida | PE, PS | Baltic Sea | 15 days | sea surface | V4 18S rRNA sequencing | Kettner et al., 2019 [69] |
| Archaeplastida | – | East China Sea | – | deepwater | V4 18S rRNA sequencing | Wu et al., 2018 [170] |
| Navicula, Achnanathes, Amphora, Nitzschia, Rhaphoneis, Cylindrotheca, Aneumastus and Ochrophyta | PE | Herzliya marina | 1 months | sea surface | 18S rRNA and tufA | Davidov et a., 2020 [65] |
| diatoms, dinoflagellates, red, green, and brown algae | PETE, HDPE, PVC, LDPE, PP, PS | Coast of Bocas del Toro | 6 weeks | sea surface | V4 18S rRNA sequencing | Dudek et al., 2020 [173] |
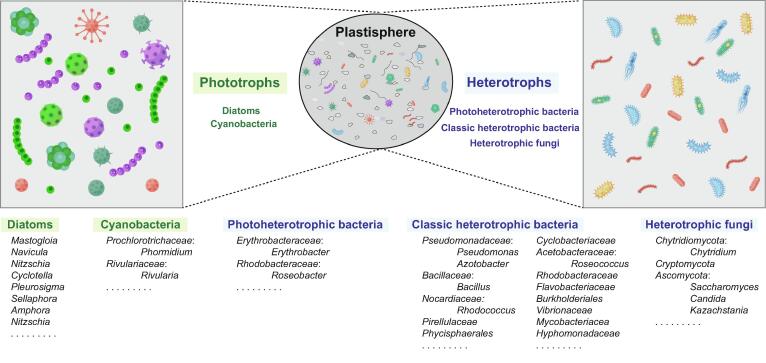
Microscopic and molecular sequence data indicate that plastisphere is composed of primary producers, heterotrophs, symbionts, and predators. Interactions between phytoplankton and bacteria play a key role in mediating the ecological cycles of the Earth and the structure of food webs in the oceans. This association between autotrophic organisms and other microorganisms is also present in the plastisphere [68]. Studies have shown that diatoms are almost ubiquitous on plastic debris and sometimes even dominate the plastic surface [69], [70], [71]. These autotrophic organisms can provide a source of organic matter to the plastisphere and regulate the microbial community [68]. Diatoms have been widely found in the plastisphere, including Mastogloia, Navicula, Nitzschia, Cyclotella, Pleurosigma, Sellaphora, Amphora, and Nitzschia [18], [72]. In addition to them, cyanobacteria (such as Phormidium and Rivularia) were also identified from the plastisphere [18], [73]. It was found that cyanobacteria colonizing the plastic surface used a completely different light-harvesting mechanism compared to those in the surrounding seawater, with an increased abundance of phycobilisome antenna encoding genes in the former and a higher expression of genes encoding Chlorophyll a/b-binding light-harvesting protein in the latter [73]. This suggests that cyanobacterial photosynthesis in seawater takes place mainly in the Chlorophyll-binding complexes, whereas cyanobacteria on plastic surfaces are photosynthesized through the phycobilisome complexes. In terms of the number of amino acids required to bind the chromophore, Chlorophyll a/b proteins require less than that of phycobilisome, and therefore less nitrogen is required to synthesize Chl a/b proteins [74]. The high expression of phycobilisome gene seems to be very detrimental to the survival of cyanobacteria on plastic surfaces that are already inadequate of nitrogen sources. The advantage of phycobilisome proteins is that they are easily broken down in the absence of nitrogen, and can quickly reorganize once a suitable nitrogen source is available [75]. In addition to acting as a light-harvesting complex, phycobilisome serves as a reservoir for nitrogen, increasing the viability of cyanobacteria and providing significant advantages in MP surface nitrogen-limited environments. Besides, high expression of nitrogen fixing enzymes (nifH, nifD and nifK) was also found in microbial communities on the surface of MPs [73]. With the help of producers like cyanobacteria, the nutrient limitation in the MPs community is not as severe as we think.
In addition to photoautotrophs, photoheterotrophic bacteria, such as the Erythrobacter and Roseobacter, are present in the plastisphere (Fig. 2) [76]. In addition to heterotrophic energy acquisition by oxidation of organic substrates, some of these heterotrophic bacteria possess aerobic anoxygenic phototrophic (AAP) apparatus for light harvesting and carbon cycling [76]. Except for photoheterotrophic bacteria, classic heterotrophic bacteria have also been identified on plastic debris. Previous culturing studies with plastics as a sole carbon source exhibited accumulation of members in the Gammaproteobacteria (such as Pseudomonas and Azotobacter), Firmicutes (such as Bacillus), and Actinobacteria (such as Rhodococcus) [4]. An in situ study have demonstrated higher abundance of Pirellulaceae, Phycisphaerales, Cyclobacteriaceae and Roseococcus on the MPs than on the natural substrates [77]. Rhodobacteraceae, Flavobacteriaceae, and Burkholderiales also exhibited higher abundance on the plastic [78], [79]. Notably, Vibrionaceae and Mycobacteriacea were also identified in the plastisphere [18], [80], [81].
Fig. 2.
The microbial community of plastisphere.
Bacterial diversity is the most concerned target of plastisphere studies. Each species of bacteria plays a very important role in the plastisphere, for example, Hyphomonadaceae is able to adhere tightly to the MPs surface through its production of polysaccharide scaffolds, while also producing carotenoids and bacteriochlorophyll a, which provide nutrition and support to neighbors in the plastisphere [82]. Compared to bacteria, relatively little research has been done targeting fungi in the plastisphere. Recently, an increasing number of studies focused particularly on the fungal diversity of MPs have been reported [34], [83]. A metabarcoding study of plastic samples from the western South Atlantic and Antarctic Peninsula was performed by Lacerda et al. [84]. They found that Ascomycota and Basidiomycota were dominant in both samples, but with a highly varied phylogenetic assemblage. Furthermore, Marie et al. reported that members of Chytridiomycota (such as Chytridium), Cryptomycota (lineage LKM11) and Ascomycota (such as Saccharomyces, Candida and Kazachstania) dominating were dominant on the surface of PE and PS [34]. Fungi account for about 2.8–3% of eukaryotes in the plastisphere, and may have significant ecological impacts [81].
Previous studies suggested possible symbiontic or predatory relationships in the plastisphere [4]. The possible ectosymbiontic relationship between Ephelota and sulfide-oxidizing Gammaproteobacteria has been reported [18], [69]. The positively associations between Amoebophyra and Suessiaceae on PE, Micromonas and Eudoraea (Flavobacteriaceae) on PS, and Micromonas and Litoreibacter (Rhodobacteraceae) on PE were also observed. Although co-occurrence of organisms can only imply but not confirm the interaction. These results give hints to the inter communications in the plastisphere.
3. The formation of plastisphere
3.1. Assembly of the community
Similar with other biofilms, the plastisphere generally involves microbial attachment, secretion of extracellular polymeric substances, and microbial proliferation [18]. Normally microorganisms in the plastisphere are detected in the surrounding seawater or in the sediments of the seafloor [66]. The physical properties of plastics, such as hydrophobicity, particle shape, roughness, crystallinity and surface charge, play a role in the selection of bacterial communities early in microbial colonization, and these pioneer bacteria can also have an impact on the selection of subsequent communities within the plastic circle [85]. First, pioneer bacteria occupy the plastic surface by reversible attachment, forming the first layer of the initial biofilm. With the colonization of these microorganisms, the surface hydrophobicity of the MPs decreases over time [86], [87]. They can also affect the vertical transport of MPs in the ocean and provide a new ecological niche for other microorganisms [39], [88], [89]. In addition, the pioneer bacteria can secret exopolysaccharides that increase the adhesion of the plastic surface, allowing further colonization of more microorganisms [90]. Subsequently, secondary microorganisms can promote irreversible attachment by forming pili, adhesion proteins, and other active mechanisms such as the production of extracellular polymeric substrates, which provide more attachable sites on the plastic surface [91]. Further recruitment or loss of species on the biofilm occurs with increased adhesion to the plastic surface, eventually resulting in the formation of a mature biofilm due to competition or synergistic effects of different microorganisms [92].
Microorganisms colonize the surface of MPs but in a sequential order. To colonize the surface of MPs, microorganisms must first overcome the hydrophobic surface of MPs [93]. Microorganisms can easily colonize on the surface of MPs within a few minutes, but it takes a long to form a stable biofilm [16]. The early MP settling microorganisms are members of Gammaproteobacteria and Alphaproteobacteria, which are called pioneer colonizers [16]. It is reported that Gammaproteobacteria dominate on the early biofilms on most plastic polymer types [66], [91], [94]. It was also found that Alteromonas, Thalassobius, Neptuniibacter, Poseobacter, and Phodobacteriaceae appeared in the early stages of biofilm formation [80]. Diatoms and cyanobacteria were also identified at the early stage of the biofilm [95], [96]. Over time, there is a tendency for the number of Bacteroidetes (especially Flavobacteriaceae) to increase due to their wide distribution, adaptability, and ability to utilize the released organic substrates by pioneer colonizers [16], [80]. These bacteria are called secondary colonizers (Fig. 3). The primary biofilm usually takes about one week or less to be observed, while secondary biofilm takes several months to form [66], [97]. Chen et al. showed that the main colonizers of biofilms in the early phase (30 days) of PE surface were Flavobacteriaceae (Bacteroidia), Rhodobacteraceae (Alphaproteobacteria), and Microtrichaceae (Acidimicrobiia) [87]. In the mid-term (75 days), the dominant populations gradually shifted from Flavobacteriaceae and Erythrobacteriaceae to Bacillaceae (Bacilli) and Moraxellaceae (Gammaproteobacteria). At the later stage of biofilm formation (135 days), the dominant family of PE colonizers shifted again to the Flavobacteriaceae (Bacteroidia), while a significant increase in the number of these bacteria occurred in the Rhodobacteraceae (Alphaproteobacteria), Microtrichaceae (Acidimicrobiia), and Pirellulaceae (Planctomycetes). There is still no agreement in different experiments regarding the formation time of different stages of biofilm, and the rate of biofilm formation is likely to be related to the total nutrients and total microorganisms in the marine environment where the plastic is located.
Fig. 3.
The scheme of the formation of plastisphere.
3.2. Vertical distribution of plastisphere
The vertical distribution of different MPs varies in the ocean, with PE and PP mostly observed in the upper waters, while PA, PVC and PET dominate in the sediments [25]. This is related to the physical properties of MP particles such as density, surface area, and volume. The microorganisms in the plastisphere are dynamic according to the state of the substrate. When a biofilm forms, the physical and chemical properties of the MPs, including hydrophobicity, density, specific surface area, roughness, surface micro-morphology, and surface charge, would be affected [93], [98], [99]. These changes will in turn affect the vertical migration, weathering, and sorption of MPs to other materials, which will eventually affect the biofilm community. Smaller particles are more likely to be distributed deeper in the ocean, which are less efficiently captured and therefore relatively less studied [100]. Small particles of plastics should be more concerned as they have generally been immersed in the ocean for a longer time, covering with more stable biofilms. The vertical distribution of MPs depends on their hydrophobicity, which decreases when the biofilm is colonized and allows them to sink to deeper water [78]. The slower benthic currents in the ocean transport MPs deposited on the seafloor and spread them to larger areas of the seafloor [101].
Microbial communities on the surface of MPs are altered due to the low temperature and lack of sunlight in deep water, and biodegradation by bacteria and other microorganisms dominates the decomposition of MPs [85]. Studies have shown that Bacteroidetes and Proteobacteria are constantly present on the surface of plastics even in deep water [91], [102]. In addition to the plastic floating on the surface and sinking on the sea floor, a small percentage of plastic debris is washed ashore by the waves and deposited on land along the coast [103]. Settling of plastic polymers in seawater can increase access to nutrients for microorganisms on their surfaces, but also lead to more environmental stresses. Some MPs may adhere to macroalgae and thus be enriched in reefs or transferred to animal organisms along the food chain [29], [104]. In China, a large number of Ulva prolifera carrying MPs land off the coast of Qingdao City annually with the ocean currents [105]. When MPs are transferred to land by these ways, the microorganisms adsorbed to MPS are transferred along with them.
3.3. Environmental impact on plastisphere
In the natural environment, the composition of plastisphere is related to temperature, seasonal and geographical factors as well as the type of plastic [106]. The results of the large-scale sampling in the Pacific Ocean showed that Bryozoa, Cyanobacteria, Alphaproteobacteria, and Bacteroidetes were found on particles with different size [73]. Environmental factors such as salinity, pressure, oxygen, and current velocity vary in different regions of the sea, which leads to differences in plastisphere communities between different marine areas [107], [108]. However, Wu et al. found similar operational taxonomic units (OTUs) from MPs at different sampling sites, and speculated that MPs provide a strong stress tolerance, attenuating the influence of geographic variation [109]. However, their study was confined to different locations in the Haihe Estuary with limited distribution of latitude and longitude. The differences between plastispheres would extend when the distance spans a larger area. In contrast to this result, Sonja et al. reported that biofilms formed on plastics from different marine sites had significantly different community structures [110]. It is still controversial whether the microbial communities in the plastisphere are approximately the same in different seas.
The community composition of biofilms can be largely influenced by the nutrients in the surrounding seawater. It was found that low nutrient levels trigger the attachment of many bacterial species to the substrate surface, while at high nutrient levels, the ability of many microorganisms to attach is reduced, which is not conducive to specialized biofilm formation [111]. High level of nutrients in the seawater results to relative low level of nutrients provided by plastics, thus reducing the substrate specificity of the plastisphere. More available nutrients also facilitate faster formation of primary and secondary biofilms, which leads to a convergence of microbial species in the plastisphere. Communities of the plastisphere differ between geography, time, and environment, but there is a controversy about whether biofilms differ between substrates. A previous study reported no significant differences between PET-colonizing biofilms and glass biofilms [110] Similar study showed that inert surfaces such as glass slides, ceramic tiles, reef sediments, and coral skeletons have little effect of the composition of marine plastisphere [112]. However, a recent study reported that the introduction of polyhydroxyalkanoate (PHA) resulted to an accumulation of sulfate-reducing microorganisms (SRM) families [113]. Another research also supports this finding with PS showing a distinct composition to those of PE and PP. Furthermore, the size of MP has no influence on the plastisphere regardless of substrate type, site and date of collection [114]. It has also been found that the bacterial adhesion on PE and PVC surfaces is much higher than PP and PET, and the dominant factor is the hardness of the plastic surface [39]. In the early stage of pellet colonization, different substrates produce a strong sorting effect on the customized biofilms, and even the same initial aquatic microbial community can produce different bacterial communities when grown on different substrates.
4. Biodegradation of plastics
4.1. Potential MP degrading microorganisms in the plastispheres
The recycling of plastics is a primary subject of concern. Currently, mechanical (typically leading to regranulate) and chemical (typically leading to monomer building blocks) recycling are common approaches for plastic recycling [115], [116]. However, the mechanical recycling is usually a “downcycling” process which converts the waste materials into products with lower quality and value. Plastics are also used as secondary fuel. But the energy recovery from plastic waste produces toxic and noxious dioxins [117]. Converting the plastics into smaller molecules through chemical reactions is an implement approach for plastic recycling [118]. But the harsh reaction conditions and large energy requirement impede the large-scale application of chemical recycling. Furthermore, a large amount of carbon dioxide is emitted during these processes and many toxic compounds may be released [102].
Biorecycling is a fast-growing and promising approach to meet the demand for plastics recycling in the coming years. Similar to the biodegradation of other polymers, microbial-mediated MPs degradation is mainly the breakdown of plastic macromolecules into smaller environmentally harmless metabolites, such as H2O, CO2, and CH4, through various enzymes secreted in biofilms [119]. The use of microorganisms to degrade MPs enhances biodegradability without harming the environment, and the ability of microorganisms to adapt to almost any environment also ensures the degradation of MPs [120], [121]. With the recent increase in plastic waste, more attentions are paid on efficient degrading microorganisms or highly effective degrading enzymes to alleviate the plastic pollution. The small size and huge amount of plastic particles present great challenges to remove MPs from the ocean. Even high-precision capture efforts for scientific research are unable to capture all of the small particles of MPs or those deposited deep into the ocean [122].
Some studies have shown that capturing shoreline or floating macroalgae can reduce MPs enrichment on macroalgae, but this is a drop in the bucket compared to MP pollution in the ocean as a whole [29]. Therefore, biodegradation, especially microbial degradation, has become a very important method to solve marine plastic pollution. The microorganisms in the plastisphere provide great convenience for the isolation and enrichment of plastic degradation bacteria, but existence of degradation bacteria varies on the surface of different types of plastic particles. The hydrophobicity of MPs allows microorganisms in the plastisphere to degrade plastics more effectively than marine planktonic strains [123]. Ogonowski et al. found a high similarity of microbial species in the plastispheres on PE and PP, and they hypothesized that similar degrading microorganisms existed among different types of plastics [78]. Some studies have found that MPs biofilm communities have the potential to degrade hydrocarbons or break down and fragmentize plastic polymers incompletely [7], [78]. Along with degrading MPs, microbial communities associated with plastic debris may also degrade the adsorbed organic pollutants [124], [125]. However, not all microorganisms in the plastisphere are efficient at degrading MPs; many only adhere temporarily to the surface of plastic particles, while others must rely on autotrophs in the plastisphere to survive. It has been found that these plastic-related microbial communities are largely dependent on carbon and other nutrients accumulated by filter-feeding bryozoans, other marine eukaryotes, and autotrophic activities, rather than using plastic as the only carbon source [73].
The studies of MP biodegradation have focused on the laboratory culture of microorganisms isolated from the natural environment [119]. These microorganisms are usually derived from terrestrial dumps, wastewater, mangrove sediment and sludge, but rarely studied directly obtained the microorganisms from seawater [20], [126]. Currently, several bacteria (e.g. Bacillus, Phododoccus, Enterobacter asburiae, and Vibrio natrigens) and fungi (e.g. Aspergillus, Penicillium, Pestalotiopsis, and Zalerion) have been showed with biodegradable potential [119], [127]. However, the microorganisms that can be cultured under laboratory conditions are probably not even 1% [128]. Further researches about the identification of plastic degrading microorganisms are still needed.
4.2. Evaluation of the degradation effect of MPs
The detection of MP degradation can currently be assessed from different perspectives, such as physical or chemical. Changes of the surface appearance state, weight loss, or mechanical properties of plastics are relatively simple and intuitive physical changes to assess biodegradation of MPs [99]. Weight loss is the most frequently used index to expound the degradation rate with different treatments [129], [130]. The assessment of plastic degradation by gravity may not be sensitive enough if the slow rate of biodegradation does not lead to a massive weight loss of the plastic [8]. Changes in the physical properties of plastics, such as tensile strength, thermal stability, and surface hydrophilicity, are used as a basis for evaluating their degradation [131], [132], [133]. Plastic surfaces are usually smooth and flat, and degradation results in sunken, cracked, or rough surfaces. Thus optical, atomic force, and SEM can be used to assess surface biological degradation caused by microbial activity or biofilm formation. Besides, surface hydrolysis, chromatographic techniques (gas chromatography, liquid chromatography, gel permeation chromatography) measurements combined with spectroscopic techniques (mass spectrometry, nuclear magnetic resonance spectroscopy, Fourier transform infrared spectroscopy) can also assess the degradation of plastics [20].
4.3. Biodegradable plastics and non-biodegradable plastics
According to the production process and the difficulty of degradation, plastics can be divided into biodegradable plastics and non-biodegradable plastics. The most important feature of biodegradable plastics is that they can be broken down into CO2 and H2O by microbial actions in industrial or municipal composting facilities. Biodegradable plastics can be divided into bio- and petroleum-based plastics, depending on the synthesis methods and materials [134]. Biodegradable plastics have been made for decades to alleviate plastic waste overload and are considered a potential solution for plastic waste. Polylactic acid (PLA), PHA and cellulose are all biodegradable plastics. The production of biodegradable plastics does not require oil, and is more efficient in terms of degradation. Since biodegradable plastic products have been utilized for a relatively short time, few of this type of MPs are found in the ocean. Most of the studies based on such plastic involve artificially putting them into the ocean for a period of time, and then removing them to observe the changes in the surface biofilms [98], [135]. The main reason for the lack of research on degradable plastics in the ocean is likely that a large proportion of these degradable plastics are harmlessly degraded through composting and other means, with only a small proportion reaching the ocean. However, as the production and use of degradable plastic products increases, more studies are still needed to investigate their hazards and fate in the ocean [131].
Non-biodegradable plastics are designed to be durable, and resistant to abiotic and biotic degradation. However, once these plastics are released into the environment, their durability becomes a serious issue. Most of the MPs found in marine, such as PS, PP, PE, PVC and PU, are non-biodegradable plastics. These plastics contain only C–C chains or ether bonds connecting building blocks, which are usually difficult to be decomposed by microorganisms, but they are not completely undegradable [136]. The solid nature of non-biodegradable results in extremely low bioavailability, with only a small fraction of the polymer being exposed to potentially degrading organisms [8]. The large molecular structure of plastic materials also poses a great challenge for microbial degradation, which requires breaking down the plastic into small molecular compounds that suitable for cellular uptake and further metabolism before more complete decomposition. The hydrophobicity and the high molecular weight determine the insolubility of MPs in seawater, and also provide challenges for rapid microbial degradation [8], [137]. Hydrophobic polymer surfaces have also been shown to hinder the effective adsorption and catalytic properties of polymer degradation enzymes [138], [139].
The crystallinity of polymers has a significant impact on their biodegradability. Most petroleum-based plastics contain both crystalline and amorphous regions, with the amorphous region being vulnerable to microbial attack [137], [140]. Hydrolysable plastics, such as PUR, PA, and PET, which have ester or amide bonds on their surfaces, can be attacked by extracellular hydrolytic enzymes secreted by various microorganisms, resulting in harmless degradation [141], [142]. The process of extracellular microbial invasion becomes more complex for non-hydrolysable synthetic polymers, such as PE, PP, PS, and PVC. However, their bioavailability can be increased by the action of abiotic factors, including sunlight exposure, thermal, and chemical treatments, which can produce various functional groups (for example, carbonyl group and nitro group) on the carbon chains of MPs, as well as reduce hydrophobicity [143]. Most of the production of non-degradable plastics is associated with petroleum, therefore degrading microorganisms are often associated with petroleum degradation as well. By modifying common plastic products (such as PE, PP, and PVC) with starch in the form of mixtures and processing them into compounds such as starch-based plastics, cellulose-based plastics, and protein-based plastics, the biodegradability of the original plastics can be enhanced. The abundance, diversity and activity of bacteria vary greatly between non-biodegradable and biodegradable plastics due to the vast differences in degradation performance [144], [145].
4.4. Microbial degradation of plastics
To be degraded by microorganisms, the plastics need to be first fragmented by UV light, hydrolysis, and abrasion. The growth of mycelium plays an important role in this process. The macromolecules are then hydrolyzed and/or oxidatively cleaved by various enzymes secreted by the microorganisms, releasing low molar mass molecules, such as zwitterions and monomers. Further degradation requires inner-cellular oxidization by microorganisms, so the molecular weight of the polymer must be low enough to cross the cell membrane. These molecules are eventually assimilated and used by more microorganisms, which eventually turns complex compounds into CO2 and H2O [146]. Fungi have the ability to adhere to and degrade microplastics, and the mycelium they produce disrupts the physical structure of the plastic, which is more conducive to the biodegradation of microplastics [127]. Most studies of plastic-degrading fungi have been conducted under terrestrial conditions instead of in the ocean [147], [148]. A previous study found that the filamentous fungi Fusarium oxysporum and F. solani could degrade PET, but many of these strains came from landfill soil [149]. MPs degradation is the result of the joint metabolism of multiple microorganisms in the plastisphere [150]. Studies with single degrading microorganisms are often difficult to produce significant degradation, diverse microorganisms should be combined when such studies are conducted.
Through the analysis of rRNA genes, the researchers identified hydrocarbon-degrading members among the marine plastisphere including Phormidium, Muricauda, Hyphomonadaceae, and Rhodobacteraceae, which are associated with oil degradation [73], [151]. It is also thought that microorganisms such as Rhodobacteraceae strains, which can also degrade lignin, may be involved in plastic degradation [33], [152]. Zettler, et al. identified Phormidium sp. And Pseudoalteromonas sp. In the plastisphere and speculated that these documented hydrocarbon-degrading bacteria may play a role in the degradation of MPs [18]. Studies in marine fungi have found that obligately marine, lignicolous fungi, Corollospora lacera, C. maritima, and Lulworthia sp. are able to use hydrocarbons as their sole source of carbon for growth and these fungi may potentially have the ability to efficiently degrade marine MPs [153].
The essence of MP degradation by either fungi or bacteria is the function of various enzymes. These enzymes are collectively known as plastic degrading enzymes which can be divided into extracellular and intracellular enzymes [154], [155]. Extracellular enzymes are mainly involved in the pre-degradation phase of MPs, where they enable the depolymerization of long carbon chains to form oligomers, dimers, or monomers, which can be more easily absorbed. Peroxidases, lipases, esterases, amidases, oxidases, and laccases, have been found to be involved in the extracellular degradation of plastics [90], [156], [157]. Some of the extracellular enzymes secreted by the pioneer bacteria could degrade the hydrophobic groups, thus reducing the hydrophobicity of the plastic surface, and allowing more microorganisms to colonize. Eventually, the plastic is degraded by intracellular degradation enzymes into harmless substances that are returned to the biogeochemical cycle, such as CO2, H2O and N2 [158]. The process may involve enzymes including esterases, lipases, cutinases, peroxidases and laccase [8]. Fungi are able to degrade plastics using intracellular enzymes mediated by cytochrome P450 family epoxidases (phase I enzyme) and transferases (phase II enzyme) [127]. Few studies have been performed in situ on degrading enzymes in the marine plastisphere, and most plastic degrading enzymes have been found under laboratory conditions using strains from enriched cultures. Like the biodegrading microorganisms, it is difficult for individual enzyme to degrade plastic well.
4.5. The sources of plastic degrading microorganisms
Human interference with Earth’s biomes has been accelerating since the mid-20th century, and led to major changes in the structure and function of microbial communities, evidenced by significant changes in the microbiota or specific genes (for example, antibiotic resistance genes) in the environment [159], [160]. Plastics began to be used in daily life within the last 100 years, with large scale production starting after 1950 [161]. In just 100 years, bacteria have evolved the ability to degrade plastics that did not exist in nature before.
Microbial communities in different environments are dynamic, and can quickly respond and adapt to environmental changes caused by anthropogenic pressures or climate change [36]. This tremendous amount of species, as well as the strong adaptability, also provides the foundation for the emergence of MP-degrading microorganisms. Several commonly reported bacteria in plastic-associated biofilms have possible ability to metabolize and assimilate petroleum-derived carbon, or are functionally related to these processes, therefore able to degrade plastics [137]. The percentage of functional microorganisms that degrade plastics is less than 0.1% of marine microbial diversity [91]. Members of microbial communities that can live on plastics or metabolize oil are rare in seawater, but there are some dormant functional microbes. These opportunistic microbes do not appear to survive well in the marine environment, but can grow rapidly and become plastic-dependent “core species” after exposure to plastics. The “seed bank” theory seems to explain these early colonizing microorganisms. By entering a dormant state, individual microorganism can tolerate unfavorable conditions, and allow for the continuation of the population. However, dormancy comes at a cost, as individuals not only miss the opportunity to reproduce, but also expend a certain amount of energy to maintain dormant requirements [162]. The accumulation of these dormant microorganisms in the ocean forms a “seed bank”. Of course, the microorganisms can quickly switch between dormant and active states, and when they encounter a suitable plastic substrate, they can quickly rejuvenate and start to colonize [163], [164]. In addition to the “seed bank” theory, some plastics that are structurally similar to natural compounds are also available to some microorganisms. For example, the enzymes secreted by microorganisms that degrade plant surface cutin in the rumen of cattle can degrade PET [165]. These microorganisms secrete enzymes attack chemical bonds in plastics that are similar to those of natural substrates. Usually, biodegradable plastics are synthesized based on the principle of structural analogues of enzymatic substrates, therefore degradable by microorganisms.
The ultimate source of MP degradation genes is genetic mutations, which is the ultimate source of genetic diversity. The mutation rate of microorganisms in the ocean has been gradually accelerated by human activities [166]. The plastic degrading bacteria derived by mutation may get into a dormant state as they temporarily cannot find a suitable substrate, and become part of the “seed bank”. However, there is little data to support the origin of plastic degrading microorganisms, and further understanding is still needed.
5. Summary and outlook
Plastics have been increasingly detected in the environment and even humans, which make them an unneglectable risk. In nature, plastics are fragmented through exposure to UV light, mechanical disruption by waves and winds, or grinding on rocks and sediments. The resulting MPs have a larger surface area, which facilitate the colonization of massive microorganisms known as plastisphere. In order to deal with the increasing amount of marine MPs, the harmless degradation of microplastics by marine microorganisms is essential. In this review, we summarize current findings about the biodiversity and formation of plastisphere, with an emphasize on the biodegrading potence of plastisphere microorganisms. These advances promote better understand the effect of plastics on the marine ecosystem and pave the way to fundamentally solve the issue of plastic pollution.
Conventional recycling approaches are usually not very effective in degrading plastics and have more or less side effects. Biodegradation of plastics is a promising and environmental-friendly approach for recycling. Researches on plastisphere help to explore natural plastic-degrading enzymes and microorganisms. Traditional approaches relied on culture-based methods in laboratory conditions, while over 99% of the total microorganisms are unculturable. The past decades have witnessed the tremendous progress of sequencing techniques and computational tools. The developments of high-throughput metagenome facilitate the exploration of genetic information of non-cultured microorganisms. An amount of plastic degrading enzymes have been identified and modified for effective degradation of plastics. However, limited database of known plastic-degrading enzymes causes poor discovery of novel families of enzymes. New screening platforms to efficiently and effectively identify novel plastic-degrading enzymes and consequent functional verifications are needed to address the challenge. Another challenge is the lack of genetic engineering tools for the microorganisms that naturally degrade plastics. With the development of molecular biology techniques, this problem will be solved one day. For now, heterogenous expression of valuable enzymes in conventional hosts, such as Escherichia coli, is a potential solution of the issue.
6. Methods
Bibliographic research of peer-reviewed international articles was conducted on NCBI Pubmed and Web of Science with the following keywords: “microplastics” and “marine”, “plastisphere” and “marine”, “microplastics” and “plastisphere”, “plastics” and “plastisphere”, “microplastics” and “microbial community”, “microplastics” and “degradation”. These selected articles reported on biodiversity, formation, and biodegradation of microplastics in the marine environment. A first manual selection of articles of interest was based on the available information in the title and abstract. A second round selected the final articles based on a thorough content check. Then, the references of the selected articles were also analyzed. Studies on biodiversity, formation, and role in degradation of marine plastisphere were selected up to 20 October 2021. There were no lower time limits.
CRediT authorship contribution statement
Yuhui Du: Conceptualization, Data curation, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. Xinbei Liu: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. Xusheng Dong: Formal analysis, Investigation, Software, Visualization. Zhiqiu Yin: Conceptualization, Funding acquisition, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded by Shandong Provincial Natural Science Foundation (Grant No. ZR2021QC208) and the Scientific Research Foundation of Shandong Agricultural University (Grant No. 010/72100).
References
- 1.Xu S., Ma J., Ji R., Pan K., Miao A.-J. Microplastics in aquatic environments: occurrence, accumulation, and biological effects. Sci Total Environ. 2020;703:134699. doi: 10.1016/j.scitotenv.2019.134699. [DOI] [PubMed] [Google Scholar]
- 2.Huang D., Xu Y., Lei F., Yu X., Ouyang Z., Chen Y., et al. Degradation of polyethylene plastic in soil and effects on microbial community composition. J Hazard Mater. 2021;416:126173. doi: 10.1016/j.jhazmat.2021.126173. [DOI] [PubMed] [Google Scholar]
- 3.Zhang L., Xie Y., Zhong S., Liu J., Qin Y., Gao P. Microplastics in freshwater and wild fishes from Lijiang River in Guangxi, Southwest China. Sci Total Environ. 2021;755:142428. doi: 10.1016/j.scitotenv.2020.142428. [DOI] [PubMed] [Google Scholar]
- 4.Amaral-Zettler L.A., Zettler E.R., Mincer T.J. Ecology of the plastisphere. Nat Rev Microbiol. 2020;18(3):139–151. doi: 10.1038/s41579-019-0308-0. [DOI] [PubMed] [Google Scholar]
- 5.Rochman C.M., Kross S.M., Armstrong J.B., Bogan M.T., Darling E.S., Green S.J., et al. Scientific evidence supports a ban on microbeads. Environ Sci Technol. 2015;49(18):10759–10761. doi: 10.1021/acs.est.5b03909. [DOI] [PubMed] [Google Scholar]
- 6.Wright S.L., Thompson R.C., Galloway T.S. The physical impacts of microplastics on marine organisms: a review. Environ Pollut. 2013;178:483–492. doi: 10.1016/j.envpol.2013.02.031. [DOI] [PubMed] [Google Scholar]
- 7.Urbanek A.K., Rymowicz W., Mirończuk A.M. Degradation of plastics and plastic-degrading bacteria in cold marine habitats. Appl Microbiol Biotechnol. 2018;102(18):7669–7678. doi: 10.1007/s00253-018-9195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krueger M.C., Harms H., Schlosser D. Prospects for microbiological solutions to environmental pollution with plastics. Appl Microbiol Biotechnol. 2015;99(21):8857–8874. doi: 10.1007/s00253-015-6879-4. [DOI] [PubMed] [Google Scholar]
- 9.Hernández-García E, Vargas M, González-Martínez C, Chiralt A. Biodegradable antimicrobial films for food packaging: effect of antimicrobials on degradation. Foods (Basel, Switzerland) 2021;10. 10.3390/foods10061256. [DOI] [PMC free article] [PubMed]
- 10.Zhu J., Wang C. Biodegradable plastics: green hope or greenwashing? Mar Pollut Bull. 2020;161:111774. doi: 10.1016/j.marpolbul.2020.111774. [DOI] [PubMed] [Google Scholar]
- 11.Gross M. Our planet wrapped in plastic. Curr Biol. 2017;27(16):R785–R788. doi: 10.1016/j.cub.2017.08.007. [DOI] [Google Scholar]
- 12.Corradini F., Meza P., Eguiluz R., Casado F., Huerta-Lwanga E., Geissen V. Evidence of microplastic accumulation in agricultural soils from sewage sludge disposal. Sci Total Environ. 2019;671:411–420. doi: 10.1016/j.scitotenv.2019.03.368. [DOI] [PubMed] [Google Scholar]
- 13.Schwabl P., Köppel S., Königshofer P., Bucsics T., Trauner M., Reiberger T., et al. Detection of various microplastics in human stool: a prospective case series. Ann Intern Med. 2019;171(7):453–457. doi: 10.7326/M19-0618. [DOI] [PubMed] [Google Scholar]
- 14.Horton A.A., Walton A., Spurgeon D.J., Lahive E., Svendsen C. Microplastics in freshwater and terrestrial environments: evaluating the current understanding to identify the knowledge gaps and future research priorities. Sci Total Environ. 2017;586:127–141. doi: 10.1016/j.scitotenv.2017.01.190. [DOI] [PubMed] [Google Scholar]
- 15.Cau A., Alvito A., Moccia D., Canese S., Pusceddu A., Rita C., et al. Submarine canyons along the upper Sardinian slope (Central Western Mediterranean) as repositories for derelict fishing gears. Mar Pollut Bull. 2017;123(1-2):357–364. doi: 10.1016/j.marpolbul.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 16.Quero G.M., Luna G.M. Surfing and dining on the “plastisphere”: microbial life on plastic marine debris. Adv Oceanogr Limnol. 2017;8:199–207. doi: 10.4081/aiol.2017.7211. [DOI] [Google Scholar]
- 17.van Sebille E., Wilcox C., Lebreton L., Maximenko N., Hardesty B.D., van Franeker J.A., et al. A global inventory of small floating plastic debris. Environ Res Lett. 2015;10(12):124006. doi: 10.1088/1748-9326/10/12/124006. [DOI] [Google Scholar]
- 18.Zettler E.R., Mincer T.J., Amaral-Zettler L.A. Life in the “plastisphere”: microbial communities on plastic marine debris. Environ Sci Technol. 2013;47(13):7137–7146. doi: 10.1021/es401288x. [DOI] [PubMed] [Google Scholar]
- 19.Frias J.P.G.L., Nash R. Microplastics: finding a consensus on the definition. Mar Pollut Bull. 2019;138:145–147. doi: 10.1016/j.marpolbul.2018.11.022. [DOI] [PubMed] [Google Scholar]
- 20.Park S.Y., Kim C.G. Biodegradation of micro-polyethylene particles by bacterial colonization of a mixed microbial consortium isolated from a landfill site. Chemosphere. 2019;222:527–533. doi: 10.1016/j.chemosphere.2019.01.159. [DOI] [PubMed] [Google Scholar]
- 21.Boucher J, Friot D. Primary Microplastics in the Oceans: A Global Evaluation of Sources. Primary Microplastics in the Oceans: A Global Evaluation of Sources; 2017.
- 22.Wang W., Gao H., Jin S., Li R., Na G. The ecotoxicological effects of microplastics on aquatic food web, from primary producer to human: a review. Ecotoxicol Environ Saf. 2019;173:110–117. doi: 10.1016/j.ecoenv.2019.01.113. [DOI] [PubMed] [Google Scholar]
- 23.Wang J., Liu X., Liu G., Zhang Z., Wu H., Cui B., et al. Size effect of polystyrene microplastics on sorption of phenanthrene and nitrobenzene. Ecotoxicol Environ Saf. 2019;173:331–338. doi: 10.1016/j.ecoenv.2019.02.037. [DOI] [PubMed] [Google Scholar]
- 24.Andrady A.L. The plastic in microplastics: a review. Mar Pollut Bull. 2017;119(1):12–22. doi: 10.1016/j.marpolbul.2017.01.082. [DOI] [PubMed] [Google Scholar]
- 25.Andrady A.L. Microplastics in the marine environment. Mar Pollut Bull. 2011;62(8):1596–1605. doi: 10.1016/j.marpolbul.2011.05.030. [DOI] [PubMed] [Google Scholar]
- 26.Cook S., Chan H.-L., Abolfathi S., Bending G.D., Schäfer H., Pearson J.M. Longitudinal dispersion of microplastics in aquatic flows using fluorometric techniques. Water Res. 2020;170:115337. doi: 10.1016/j.watres.2019.115337. [DOI] [PubMed] [Google Scholar]
- 27.Woodall L.C., Sanchez-Vidal A., Canals M., Paterson G.L.J., Coppock R., Sleight V., et al. The deep sea is a major sink for microplastic debris. R Soc Open Sci. 2014;1(4):140317. doi: 10.1098/rsos.140317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koelmans A.A., Mohamed Nor N.H., Hermsen E., Kooi M., Mintenig S.M., De France J. Microplastics in freshwaters and drinking water: critical review and assessment of data quality. Water Res. 2019;155:410–422. doi: 10.1016/j.watres.2019.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng Z., Zhang T., Shi H., Gao K., Huang W., Xu J., et al. Microplastics in bloom-forming macroalgae: distribution, characteristics and impacts. J Hazard Mater. 2020;397:122752. doi: 10.1016/j.jhazmat.2020.122752. [DOI] [PubMed] [Google Scholar]
- 30.Engler R.E. The complex interaction between marine debris and toxic chemicals in the ocean. Environ Sci Technol. 2012;46(22):12302–12315. doi: 10.1021/es3027105. [DOI] [PubMed] [Google Scholar]
- 31.Li H., Wang F., Li J., Deng S., Zhang S. Adsorption of three pesticides on polyethylene microplastics in aqueous solutions: kinetics, isotherms, thermodynamics, and molecular dynamics simulation. Chemosphere. 2021;264:128556. doi: 10.1016/j.chemosphere.2020.128556. [DOI] [PubMed] [Google Scholar]
- 32.Teuten E.L., Rowland S.J., Galloway T.S., Thompson R.C. Potential for plastics to transport hydrophobic contaminants. Environ Sci Technol. 2007;41(22):7759–7764. doi: 10.1021/es071737s. [DOI] [PubMed] [Google Scholar]
- 33.Fu L., Li J., Wang G., Luan Y., Dai W. Adsorption behavior of organic pollutants on microplastics. Ecotoxicol Environ Saf. 2021;217:112207. doi: 10.1016/j.ecoenv.2021.112207. [DOI] [PubMed] [Google Scholar]
- 34.Kettner M.T., Rojas-Jimenez K., Oberbeckmann S., Labrenz M., Grossart H.-P. Microplastics alter composition of fungal communities in aquatic ecosystems. Environ Microbiol. 2017;19(11):4447–4459. doi: 10.1111/1462-2920.13891. [DOI] [PubMed] [Google Scholar]
- 35.Oberbeckmann S., Labrenz M. Marine microbial assemblages on microplastics: diversity, adaptation, and role in degradation. Ann Rev Mar Sci. 2020;12(1):209–232. doi: 10.1146/annurev-marine-010419-010633. [DOI] [PubMed] [Google Scholar]
- 36.Bowley J., Baker-Austin C., Porter A., Hartnell R., Lewis C. Oceanic Hitchhikers - assessing pathogen risks from marine microplastic. Trends Microbiol. 2021;29(2):107–116. doi: 10.1016/j.tim.2020.06.011. [DOI] [PubMed] [Google Scholar]
- 37.Li H.-X., Orihuela B., Zhu M., Rittschof D. Recyclable plastics as substrata for settlement and growth of bryozoans Bugula neritina and barnacles Amphibalanus amphitrite. Environ Pollut. 2016;218:973–980. doi: 10.1016/j.envpol.2016.08.047. [DOI] [PubMed] [Google Scholar]
- 38.Foulon V., Le Roux F., Lambert C., Huvet A., Soudant P., Paul-Pont I. Colonization of polystyrene microparticles by vibrio crassostreae: light and electron microscopic investigation. Environ Sci Technol. 2016;50(20):10988–10996. doi: 10.1021/acs.est.6b02720. [DOI] [PubMed] [Google Scholar]
- 39.Cai L., Wu D., Xia J., Shi H., Kim H. Influence of physicochemical surface properties on the adhesion of bacteria onto four types of plastics. Sci Total Environ. 2019;671:1101–1107. doi: 10.1016/j.scitotenv.2019.03.434. [DOI] [Google Scholar]
- 40.Sjollema S.B., Redondo-Hasselerharm P., Leslie H.A., Kraak M.H.S., Vethaak A.D. Do plastic particles affect microalgal photosynthesis and growth? Aquat Toxicol. 2016;170:259–261. doi: 10.1016/j.aquatox.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 41.Lim X. Microplastics are everywhere - but are they harmful? Nature. 2021;593(7857):22–25. doi: 10.1038/d41586-021-01143-3. [DOI] [PubMed] [Google Scholar]
- 42.Wu Y., Tam W.-S., Chau H.-F., Kaur S., Thor W., Aik W.S., et al. Solid-phase fluorescent BODIPY-peptide synthesis via in situ dipyrrin construction. Chem Sci. 2020;11(41):11266–11273. doi: 10.1039/d0sc04849f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keswani A., Oliver D.M., Gutierrez T., Quilliam R.S. Microbial hitchhikers on marine plastic debris: human exposure risks at bathing waters and beach environments. Mar Environ Res. 2016;118:10–19. doi: 10.1016/j.marenvres.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 44.Ramasamy E.V., Sruthy S., Harit A.K., Mohan M., Binish M.B. Microplastic pollution in the surface sediment of Kongsfjorden, Svalbard. Arctic. Mar Pollut Bull. 2021;173:112986. doi: 10.1016/j.marpolbul.2021.112986. [DOI] [PubMed] [Google Scholar]
- 45.Collard F., Husum K., Eppe G., Malherbe C., Hallanger I.G., Divine D.V., et al. Anthropogenic particles in sediment from an Arctic fjord. Sci Total Environ. 2021;772:145575. doi: 10.1016/j.scitotenv.2021.145575. [DOI] [PubMed] [Google Scholar]
- 46.Hipfner J.M., Galbraith M., Tucker S., Studholme K.R., Domalik A.D., Pearson S.F., et al. Two forage fishes as potential conduits for the vertical transfer of microfibres in Northeastern Pacific Ocean food webs. Environ Pollut. 2018;239:215–222. doi: 10.1016/j.envpol.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 47.Savinelli B., Vega Fernández T., Galasso N.M., D'Anna G., Pipitone C., Prada F., et al. Microplastics impair the feeding performance of a Mediterranean habitat-forming coral. Mar Environ Res. 2020;155:104887. doi: 10.1016/j.marenvres.2020.104887. [DOI] [PubMed] [Google Scholar]
- 48.Gonçalves C., Martins M., Sobral P., Costa P.M., Costa M.H. An assessment of the ability to ingest and excrete microplastics by filter-feeders: a case study with the Mediterranean mussel. Environ Pollut. 2019;245:600–606. doi: 10.1016/j.envpol.2018.11.038. [DOI] [PubMed] [Google Scholar]
- 49.Dawson A., Huston W., Kawaguchi S.o., King C., Cropp R., Wild S., et al. Uptake and depuration kinetics influence microplastic bioaccumulation and toxicity in Antarctic krill (Euphausia superba) Environ Sci Technol. 2018;52(5):3195–3201. doi: 10.1021/acs.est.7b05759. [DOI] [PubMed] [Google Scholar]
- 50.Mattan-Moorgawa S., Chockalingum J., Appadoo C. A first assessment of marine meso-litter and microplastics on beaches: where does Mauritius stand? Mar Pollut Bull. 2021;173:112941. doi: 10.1016/j.marpolbul.2021.112941. [DOI] [PubMed] [Google Scholar]
- 51.Naidoo T., Sershen, Thompson R.C., Rajkaran A. Quantification and characterisation of microplastics ingested by selected juvenile fish species associated with mangroves in KwaZulu-Natal, South Africa. Environ Pollut. 2020;257:113635. doi: 10.1016/j.envpol.2019.113635. [DOI] [PubMed] [Google Scholar]
- 52.Li S., Wang P., Zhang C., Zhou X., Yin Z., Hu T., et al. Influence of polystyrene microplastics on the growth, photosynthetic efficiency and aggregation of freshwater microalgae Chlamydomonas reinhardtii. Sci Total Environ. 2020;714:136767. doi: 10.1016/j.scitotenv.2020.136767. [DOI] [PubMed] [Google Scholar]
- 53.Sussarellu R., Suquet M., Thomas Y., Lambert C., Fabioux C., Pernet M.E.J., et al. Oyster reproduction is affected by exposure to polystyrene microplastics. Proc Natl Acad Sci USA. 2016;113(9):2430–2435. doi: 10.1073/pnas.1519019113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seidensticker S., Grathwohl P., Lamprecht J., Zarfl C. A combined experimental and modeling study to evaluate pH-dependent sorption of polar and non-polar compounds to polyethylene and polystyrene microplastics. Environ Sci Eur. 2018;30:30. doi: 10.1186/s12302-018-0155-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khalid N., Aqeel M., Noman A., Hashem M., Mostafa Y.S., Alhaithloul H.A.S., et al. Linking effects of microplastics to ecological impacts in marine environments. Chemosphere. 2021;264:128541. doi: 10.1016/j.chemosphere.2020.128541. [DOI] [PubMed] [Google Scholar]
- 56.Li J., Zhang K., Zhang H. Adsorption of antibiotics on microplastics. Environ Pollut. 2018;237:460–467. doi: 10.1016/j.envpol.2018.02.050. [DOI] [PubMed] [Google Scholar]
- 57.Hodson M.E., Duffus-Hodson C.A., Clark A., Prendergast-Miller M.T., Thorpe K.L. Plastic bag derived-microplastics as a vector for metal exposure in terrestrial invertebrates. Environ Sci Technol. 2017;51(8):4714–4721. doi: 10.1021/acs.est.7b00635. [DOI] [PubMed] [Google Scholar]
- 58.Wang W., Wang J. Comparative evaluation of sorption kinetics and isotherms of pyrene onto microplastics. Chemosphere. 2018;193:567–573. doi: 10.1016/j.chemosphere.2017.11.078. [DOI] [PubMed] [Google Scholar]
- 59.Wang W., Wang J. Different partition of polycyclic aromatic hydrocarbon on environmental particulates in freshwater: microplastics in comparison to natural sediment. Ecotoxicol Environ Saf. 2018;147:648–655. doi: 10.1016/j.ecoenv.2017.09.029. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y., Lu J., Wu J., Wang J., Luo Y. Potential risks of microplastics combined with superbugs: enrichment of antibiotic resistant bacteria on the surface of microplastics in mariculture system. Ecotoxicol Environ Saf. 2020;187:109852. doi: 10.1016/j.ecoenv.2019.109852. [DOI] [PubMed] [Google Scholar]
- 61.Haward M. Plastic pollution of the world’s seas and oceans as a contemporary challenge in ocean governance. Nat Commun. 2018;9:667. doi: 10.1038/s41467-018-03104-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Galloway T.S., Cole M., Lewis C. Interactions of microplastic debris throughout the marine ecosystem. Nat Ecol Evol. 2017;1:116. doi: 10.1038/s41559-017-0116. [DOI] [PubMed] [Google Scholar]
- 63.Seeley M.E., Song B., Passie R., Hale R.C. Microplastics affect sedimentary microbial communities and nitrogen cycling. Nat Commun. 2020;11:2372. doi: 10.1038/s41467-020-16235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Santos A., van Aerle R., Barrientos L., Martinez-Urtaza J. Computational methods for 16S metabarcoding studies using Nanopore sequencing data. Comput Struct Biotechnol J. 2020;18:296–305. doi: 10.1016/j.csbj.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Davidov K., Iankelevich-Kounio E., Yakovenko I., Koucherov Y., Rubin-Blum M., Oren M. Identification of plastic-associated species in the Mediterranean Sea using DNA metabarcoding with Nanopore MinION. Sci Rep. 2020;10:17533. doi: 10.1038/s41598-020-74180-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.De Tender C., Devriese L.I., Haegeman A., Maes S., Vangeyte J., Cattrijsse A., et al. Temporal dynamics of bacterial and fungal colonization on plastic debris in the north sea. Environ Sci Technol. 2017;51(13):7350–7360. doi: 10.1021/acs.est.7b00697. [DOI] [PubMed] [Google Scholar]
- 67.Krehenwinkel H, Pomerantz A, Henderson JB, Kennedy SR, Lim JY, Swamy V, et al. Nanopore sequencing of long ribosomal DNA amplicons enables portable and simple biodiversity assessments with high phylogenetic resolution across broad taxonomic scale. Gigascience 2019;8. 10.1093/gigascience/giz006. [DOI] [PMC free article] [PubMed]
- 68.Mayali X. Editorial: metabolic interactions between bacteria and phytoplankton. Front Microbiol. 2018;9:727. doi: 10.3389/fmicb.2018.00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kettner M.T., Oberbeckmann S., Labrenz M., Grossart H.-P. The eukaryotic life on microplastics in brackish ecosystems. Front Microbiol. 2019;10:538. doi: 10.3389/fmicb.2019.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Michels J., Stippkugel A., Lenz M., Wirtz K., Engel A. Rapid aggregation of biofilm-covered microplastics with marine biogenic particles. Proc R Soc B. 2018;285(1885):20181203. doi: 10.1098/rspb.2018.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng J., Jacquin J., Conan P., Pujo-Pay M., Barbe V., George M., et al. Relative influence of plastic debris size and shape, chemical composition and phytoplankton-bacteria interactions in driving seawater plastisphere abundance, diversity and activity. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.610231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Muthukrishnan T., Al Khaburi M., Abed R.M.M. Fouling microbial communities on plastics compared with wood and steel: are they substrate- or location-specific? Microb Ecol. 2019;78(2):361–374. doi: 10.1007/s00248-018-1303-0. [DOI] [PubMed] [Google Scholar]
- 73.Bryant J.A., Clemente T.M., Viviani D.A., Fong A.A., Thomas K.A., Kemp P., et al. Diversity and activity of communities inhabiting plastic debris in the north pacific gyre. mSystems. 2016;1(3) doi: 10.1128/mSystems.00024-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stadnichuk IN, Tropin I V. [Antenna replacement in the evolutionary origin of chloroplasts]. Mikrobiologiia 2014;83:385–402. [PubMed]
- 75.Levy I., Gantt E. Development of photosynthetic activity in porphyridium purpureum (Rhodophyta) following nitrogen starvation. J Phycol. 1990;26:62–68. doi: 10.1111/j.0022-3646.1990.00062.x. [DOI] [Google Scholar]
- 76.Luo H., Moran M.A. Evolutionary ecology of the marine Roseobacter clade. Microbiol Mol Biol Rev. 2014;78(4):573–587. doi: 10.1128/MMBR.00020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miao L., Wang P., Hou J., Yao Y., Liu Z., Liu S., et al. Distinct community structure and microbial functions of biofilms colonizing microplastics. Sci Total Environ. 2019;650:2395–2402. doi: 10.1016/j.scitotenv.2018.09.378. [DOI] [PubMed] [Google Scholar]
- 78.Ogonowski M., Motiei A., Ininbergs K., Hell E., Gerdes Z., Udekwu K.I., et al. Evidence for selective bacterial community structuring on microplastics. Environ Microbiol. 2018;20(8):2796–2808. doi: 10.1111/1462-2920.14120. [DOI] [PubMed] [Google Scholar]
- 79.De Tender C., Schlundt C., Devriese L.I., Mincer T.J., Zettler E.R., Amaral-Zettler L.A. A review of microscopy and comparative molecular-based methods to characterize “Plastisphere” communities. Anal Methods. 2017;9(14):2132–2143. [Google Scholar]
- 80.Zhang S.-J., Zeng Y.-H., Zhu J.-M., Cai Z.-H., Zhou J. The structure and assembly mechanisms of plastisphere microbial community in natural marine environment. J Hazard Mater. 2022;421:126780. doi: 10.1016/j.jhazmat.2021.126780. [DOI] [PubMed] [Google Scholar]
- 81.Rogers K.L., Carreres‐Calabuig J.A., Gorokhova E., Posth N.R. Micro-by-micro interactions: how microorganisms influence the fate of marine microplastics. Limnol Oceanogr Lett. 2020;5(1):18–36. doi: 10.1002/lol2.10136. [DOI] [Google Scholar]
- 82.Oberbeckmann S., Kreikemeyer B., Labrenz M. Environmental factors support the formation of specific bacterial assemblages on microplastics. Front Microbiol. 2017;8:2709. doi: 10.3389/fmicb.2017.02709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xue N., Fang Q., Pan X., Zhang D. Distinct fungal plastisphere across different river functional zones: a watershed scale study. Sci Total Environ. 2021;752:141879. doi: 10.1016/j.scitotenv.2020.141879. [DOI] [PubMed] [Google Scholar]
- 84.Lacerda A.L.d.F., Proietti M.C., Secchi E.R., Taylor J.D. Diverse groups of fungi are associated with plastics in the surface waters of the Western South Atlantic and the Antarctic Peninsula. Mol Ecol. 2020;29(10):1903–1918. doi: 10.1111/mec.15444. [DOI] [PubMed] [Google Scholar]
- 85.Rummel C.D., Jahnke A., Gorokhova E., Kühnel D., Schmitt-Jansen M. Impacts of biofilm formation on the fate and potential effects of microplastic in the aquatic environment. Environ Sci Technol Lett. 2017;4(7):258–267. doi: 10.1021/acs.estlett.7b00164. [DOI] [Google Scholar]
- 86.Lobelle D., Cunliffe M. Early microbial biofilm formation on marine plastic debris. Mar Pollut Bull. 2011;62(1):197–200. doi: 10.1016/j.marpolbul.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 87.Tu C., Chen T., Zhou Q., Liu Y., Wei J., Waniek J.J., et al. Biofilm formation and its influences on the properties of microplastics as affected by exposure time and depth in the seawater. Sci Total Environ. 2020;734:139237. doi: 10.1016/j.scitotenv.2020.139237. [DOI] [PubMed] [Google Scholar]
- 88.Fazey F.M.C., Ryan P.G. Biofouling on buoyant marine plastics: an experimental study into the effect of size on surface longevity. Environ Pollut. 2016;210:354–360. doi: 10.1016/j.envpol.2016.01.026. [DOI] [PubMed] [Google Scholar]
- 89.Kooi M., Nes E.H.V., Scheffer M., Koelmans A.A. Ups and downs in the ocean: effects of biofouling on vertical transport of microplastics. Environ Sci Technol. 2017;51(14):7963–7971. doi: 10.1021/acs.est.6b04702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.A. G.K., K. A., M. H., K. S., G. D. Review on plastic wastes in marine environment - Biodegradation and biotechnological solutions. Mar Pollut Bull. 2020;150:110733. doi: 10.1016/j.marpolbul.2019.110733. [DOI] [PubMed] [Google Scholar]
- 91.Dussud C., Hudec C., George M., Fabre P., Higgs P., Bruzaud S., et al. Colonization of non-biodegradable and biodegradable plastics by marine microorganisms. Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.01571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lorite G.S., Rodrigues C.M., de Souza A.A., Kranz C., Mizaikoff B., Cotta M.A. The role of conditioning film formation and surface chemical changes on Xylella fastidiosa adhesion and biofilm evolution. J Colloid Interface Sci. 2011;359(1):289–295. doi: 10.1016/j.jcis.2011.03.066. [DOI] [PubMed] [Google Scholar]
- 93.Gong M., Yang G., Zhuang L., Zeng E.Y. Microbial biofilm formation and community structure on low-density polyethylene microparticles in lake water microcosms. Environ Pollut. 2019;252:94–102. doi: 10.1016/j.envpol.2019.05.090. [DOI] [PubMed] [Google Scholar]
- 94.Harrison J.P., Schratzberger M., Sapp M., Osborn A.M. Rapid bacterial colonization of low-density polyethylene microplastics in coastal sediment microcosms. BMC Microbiol. 2014;14:232. doi: 10.1186/s12866-014-0232-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Amaral-Zettler L.A., Zettler E.R., Slikas B., Boyd G.D., Melvin D.W., Morrall C.E., et al. The biogeography of the Plastisphere: implications for policy. Front Ecol Environ. 2015;13(10):541–546. doi: 10.1890/150017. [DOI] [Google Scholar]
- 96.Dang H., Lovell C.R. Microbial surface colonization and biofilm development in marine environments. Microbiol Mol Biol Rev. 2016;80(1):91–138. doi: 10.1128/MMBR.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wright R.J., Erni-Cassola G., Zadjelovic V., Latva M., Christie-Oleza J.A. Marine plastic debris: a new surface for microbial colonization. Environ Sci Technol. 2020;54(19):11657–11672. doi: 10.1021/acs.est.0c02305. [DOI] [PubMed] [Google Scholar]
- 98.Richard H., Carpenter E.J., Komada T., Palmer P.T., Rochman C.M. Biofilm facilitates metal accumulation onto microplastics in estuarine waters. Sci Total Environ. 2019;683:600–608. doi: 10.1016/j.scitotenv.2019.04.331. [DOI] [PubMed] [Google Scholar]
- 99.Jacquin J., Cheng J., Odobel C., Pandin C., Conan P., Pujo-Pay M., et al. Microbial ecotoxicology of marine plastic debris: a review on colonization and biodegradation by the “plastisphere”. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reisser J., Slat B., Noble K., du Plessis K., Epp M., Proietti M., et al. The vertical distribution of buoyant plastics at sea: an observational study in the North Atlantic Gyre. Biogeosciences. 2015;12(4):1249–1256. doi: 10.5194/bg-12-1249-2015. [DOI] [Google Scholar]
- 101.Kane I.A., Clare M.A., Miramontes E., Wogelius R., Rothwell J.J., Garreau P., et al. Seafloor microplastic hotspots controlled by deep-sea circulation. Science. 2020;368(6495):1140–1145. doi: 10.1126/science.aba5899. [DOI] [PubMed] [Google Scholar]
- 102.Emadian S.M., Onay T.T., Demirel B. Biodegradation of bioplastics in natural environments. Waste Manag. 2017;59:526–536. doi: 10.1016/j.wasman.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 103.Tekman M.B., Krumpen T., Bergmann M. Marine litter on deep Arctic seafloor continues to increase and spreads to the North at the HAUSGARTEN observatory. Deep Res Part. 2016;I(12):88–99. doi: 10.1016/j.dsr.2016.12.011. [DOI] [Google Scholar]
- 104.Gao G., Clare A.S., Chatzidimitriou E., Rose C., Caldwell G. Effects of ocean warming and acidification, combined with nutrient enrichment, on chemical composition and functional properties of Ulva rigida. Food Chem. 2018;258:71–78. doi: 10.1016/j.foodchem.2018.03.040. [DOI] [PubMed] [Google Scholar]
- 105.Song W., Li Y., Fang S., Wang Z., Xiao J., Li R., et al. Temporal and spatial distributions of green algae micro-propagules in the coastal waters of the Subei Shoal, China. Estuar Coast Shelf Sci. 2015;163:29–35. doi: 10.1016/j.ecss.2014.08.006. [DOI] [Google Scholar]
- 106.Oberbeckmann S., Loeder M.G.J., Gerdts G., Osborn A.M. Spatial and seasonal variation in diversity and structure of microbial biofilms on marine plastics in Northern European waters. FEMS Microbiol Ecol. 2014;90:478–492. doi: 10.1111/1574-6941.12409. [DOI] [PubMed] [Google Scholar]
- 107.Wang J., Peng C., Li H., Zhang P., Liu X. The impact of microplastic-microbe interactions on animal health and biogeochemical cycles: a mini-review. Sci Total Environ. 2021;773:145697. doi: 10.1016/j.scitotenv.2021.145697. [DOI] [PubMed] [Google Scholar]
- 108.Harrison JP, Hoellein TJ, Sapp M, Tagg AS, Ojeda JJ. Microplastic-associated biofilms: a comparison of freshwater and marine environments. Freshw Microplast 2018.
- 109.Wu N., Zhang Y., Zhao Z., He J., Li W., Li J., et al. Colonization characteristics of bacterial communities on microplastics compared with ambient environments (water and sediment) in Haihe Estuary. Sci Total Environ. 2020;708:134876. doi: 10.1016/j.scitotenv.2019.134876. [DOI] [PubMed] [Google Scholar]
- 110.Oberbeckmann S., Osborn A.M., Duhaime M.B., Carter D.A. Microbes on a bottle: substrate, season and geography influence community composition of microbes colonizing marine plastic debris. PLoS One. 2016;11(8):e0159289. doi: 10.1371/journal.pone.0159289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stanley N.R., Lazazzera B.A. Environmental signals and regulatory pathways that influence biofilm formation. Mol Microbiol. 2004;52:917–924. doi: 10.1111/j.1365-2958.2004.04036.x. [DOI] [PubMed] [Google Scholar]
- 112.Witt V., Wild C., Uthicke S. Effect of substrate type on bacterial community composition in biofilms from the Great Barrier Reef. FEMS Microbiol Lett. 2011;323(2):188–195. doi: 10.1111/j.1574-6968.2011.02374.x. [DOI] [PubMed] [Google Scholar]
- 113.Pinnell L.J., Turner J.W. Shotgun metagenomics reveals the benthic microbial community response to plastic and bioplastic in a coastal marine environment. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.01252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Frère L., Maignien L., Chalopin M., Huvet A., Rinnert E., Morrison H., et al. Microplastic bacterial communities in the Bay of Brest: Influence of polymer type and size. Environ Pollut. 2018;242:614–625. doi: 10.1016/j.envpol.2018.07.023. [DOI] [PubMed] [Google Scholar]
- 115.Ignatyev I.A., Thielemans W., Vander Beke B. Recycling of polymers: a review. ChemSusChem. 2014;7(6):1579–1593. doi: 10.1002/cssc.201300898. [DOI] [PubMed] [Google Scholar]
- 116.Ragaert K., Delva L., Van Geem K. Mechanical and chemical recycling of solid plastic waste. Waste Manag. 2017;69:24–58. doi: 10.1016/j.wasman.2017.07.044. [DOI] [PubMed] [Google Scholar]
- 117.Brems A., Baeyens J., Dewil R. Recycling and recovery of post-cet alonsumer plastic solid waste in a European context. Therm Sci. 2012;16:669–685. doi: 10.2298/TSCI120111121B. [DOI] [Google Scholar]
- 118.Angyal A., Miskolczi N., Bartha L. Petrochemical feedstock by thermal cracking of plastic waste. J Anal Appl Pyrolysis. 2007;79(1-2):409–414. doi: 10.1016/j.jaap.2006.12.031. [DOI] [Google Scholar]
- 119.Yuan J., Ma J., Sun Y., Zhou T., Zhao Y., Yu F. Microbial degradation and other environmental aspects of microplastics/plastics. Sci Total Environ. 2020;715:136968. doi: 10.1016/j.scitotenv.2020.136968. [DOI] [PubMed] [Google Scholar]
- 120.Brooks A.N., Turkarslan S., Beer K.D., Yin Lo F., Baliga N.S. Adaptation of cells to new environments. Wiley Interdiscip Rev Syst Biol Med. 2011;3(5):544–561. doi: 10.1002/wsbm.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Qi X., Ren Y., Wang X. New advances in the biodegradation of Poly(lactic) acid. Int Biodeterior Biodegrad. 2017;117:215–223. doi: 10.1016/j.ibiod.2017.01.010. [DOI] [Google Scholar]
- 122.Turner A., Ostle C., Wootton M. Occurrence and chemical characteristics of microplastic paint flakes in the North Atlantic Ocean. Sci Total Environ. 2022;806:150375. doi: 10.1016/j.scitotenv.2021.150375. [DOI] [PubMed] [Google Scholar]
- 123.Pérez-Álvarez L., Ruiz-Rubio L., Azua I., Benito V., Bilbao A., Vilas-Vilela J.L. Development of multiactive antibacterial multilayers of hyaluronic acid and chitosan onto poly(ethylene terephthalate) Eur Polym J. 2019;112:31–37. doi: 10.1016/j.eurpolymj.2018.12.038. [DOI] [Google Scholar]
- 124.Osborn A.M., Stojkovic S. Marine microbes in the plastic age. Microbiol Aust. 2014;35(4):207. [Google Scholar]
- 125.Edwards S.J., Kjellerup B.V. Applications of biofilms in bioremediation and biotransformation of persistent organic pollutants, pharmaceuticals/personal care products, and heavy metals. Appl Microbiol Biotechnol. 2013;97(23):9909–9921. doi: 10.1007/s00253-013-5216-z. [DOI] [PubMed] [Google Scholar]
- 126.Auta H.S., Emenike C.U., Jayanthi B., Fauziah S.H. Growth kinetics and biodeterioration of polypropylene microplastics by Bacillus sp. and Rhodococcus sp. isolated from mangrove sediment. Mar Pollut Bull. 2018;127:15–21. doi: 10.1016/j.marpolbul.2017.11.036. [DOI] [PubMed] [Google Scholar]
- 127.Mishra S., Swain S., Sahoo M., Mishra S., Das A.P. Microbial colonization and degradation of microplastics in aquatic ecosystem: a review. Geomicrobiol J. 2021:1–11. doi: 10.1080/01490451.2021.1983670. [DOI] [Google Scholar]
- 128.Hugenholtz P., Hooper S.D., Kyrpides N.C. Kyrpides NC. Focus: synergistetes. Environ Microbiol. 2009;11(6):1327–1329. doi: 10.1111/j.1462-2920.2009.01949.x. [DOI] [PubMed] [Google Scholar]
- 129.Luo Y., Zhang Y., Xu Y., Guo X., Zhu L. Distribution characteristics and mechanism of microplastics mediated by soil physicochemical properties. Sci Total Environ. 2020;726:138389. doi: 10.1016/j.scitotenv.2020.138389. [DOI] [PubMed] [Google Scholar]
- 130.Kong X., Jin D., Jin S., Wang Z., Yin H., Xu M., et al. Responses of bacterial community to dibutyl phthalate pollution in a soil-vegetable ecosystem. J Hazard Mater. 2018;353:142–150. doi: 10.1016/j.jhazmat.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 131.Tosin M., Weber M., Siotto M., Lott C., Degli I.F. Laboratory test methods to determine the degradation of plastics in marine environmental conditions. Front Microbiol. 2012;3:225. doi: 10.3389/fmicb.2012.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jeyakumar D., Chirsteen J., Doble M. Synergistic effects of pretreatment and blending on fungi mediated biodegradation of polypropylenes. Bioresour Technol. 2013;148:78–85. doi: 10.1016/j.biortech.2013.08.074. [DOI] [PubMed] [Google Scholar]
- 133.Yang Z., Peng H., Wang W., Liu T. Crystallization behavior of poly(ε-caprolactone)/layered double hydroxide nanocomposites. J Appl Polym Sci. 2010;116:2658–2667. doi: 10.1002/app. [DOI] [Google Scholar]
- 134.Müller C.A., Perz V., Provasnek C., Quartinello F., Guebitz G.M., Berg G., et al. Discovery of polyesterases from moss-associated microorganisms. Appl Environ Microbiol. 2017;83(4) doi: 10.1128/AEM.02641-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Anderson G., Shenkar N. Potential effects of biodegradable single-use items in the sea: polylactic acid (PLA) and solitary ascidians. Environ Pollut. 2021;268:115364. doi: 10.1016/j.envpol.2020.115364. [DOI] [PubMed] [Google Scholar]
- 136.Wierckx N., Narancic T., Eberlein C., Wei R., Drzyzga O., Magnin A., et al. In: Consequences of Microbial Interactions with Hydrocarbons, Oils, and Lipids: Biodegradation and Bioremediation. Steffan R.J., editor. Springer International Publishing; Cham: 2019. Plastic Biodegradation: Challenges and Opportunities. Consequences of Microbial Interactions with Hydrocarbons; pp. 333–361. [DOI] [Google Scholar]
- 137.Restrepo-Flórez J.M., Bassi A., Thompson M.R. Microbial degradation and deterioration of polyethylene - A review. Int Biodeterior Biodegrad. 2014;88:83–90. doi: 10.1016/j.ibiod.2013.12.014. [DOI] [Google Scholar]
- 138.Ribitsch D., Herrero Acero E., Przylucka A., Zitzenbacher S., Marold A., Gamerith C., et al. Enhanced cutinase-catalyzed hydrolysis of polyethylene terephthalate by covalent fusion to hydrophobins. Appl Environ Microbiol. 2015;81(11):3586–3592. doi: 10.1128/AEM.04111-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sammond D.W., Yarbrough J.M., Mansfield E., Bomble Y.J., Hobdey S.E., Decker S.R., et al. Predicting enzyme adsorption to lignin films by calculating enzyme surface hydrophobicity. J Biol Chem. 2014;289(30):20960–20969. doi: 10.1074/jbc.M114.573642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Webb H.K., Arnott J., Crawford R.J., Ivanova E.P. Plastic degradation and its environmental implications with special reference to poly(ethylene terephthalate) Polymers (Basel) 2013;5:1–18. doi: 10.3390/polym5010001. [DOI] [Google Scholar]
- 141.Roth C., Wei R., Oeser T., Then J., Föllner C., Zimmermann W., et al. Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca. Appl Microbiol Biotechnol. 2014;98(18):7815–7823. doi: 10.1007/s00253-014-5672-0. [DOI] [PubMed] [Google Scholar]
- 142.Negoro S., Shibata N., Tanaka Y., Yasuhira K., Shibata H., Hashimoto H., et al. Three-dimensional structure of nylon hydrolase and mechanism of nylon-6 hydrolysis. J Biol Chem. 2012;287(7):5079–5090. doi: 10.1074/jbc.M111.321992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Krueger M.C., Seiwert B., Prager A., Zhang S., Abel B., Harms H., et al. Degradation of polystyrene and selected analogues by biological Fenton chemistry approaches: opportunities and limitations. Chemosphere. 2017;173:520–528. doi: 10.1016/j.chemosphere.2017.01.089. [DOI] [PubMed] [Google Scholar]
- 144.Dussud C., Meistertzheim A.L., Conan P., Pujo-Pay M., George M., Fabre P., et al. Evidence of niche partitioning among bacteria living on plastics, organic particles and surrounding seawaters. Environ Pollut. 2018;236:807–816. doi: 10.1016/j.envpol.2017.12.027. [DOI] [PubMed] [Google Scholar]
- 145.Eich A., Mildenberger T., Laforsch C., Weber M., Carter D.A. Biofilm and diatom succession on polyethylene (PE) and biodegradable plastic bags in two marine habitats: early signs of degradation in the pelagic and benthic zone? PLoS One. 2015;10(9):e0137201. doi: 10.1371/journal.pone.0137201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Magnin A., Pollet E., Phalip V., Avérous L. Evaluation of biological degradation of polyurethanes. Biotechnol Adv. 2020;39:107457. doi: 10.1016/j.biotechadv.2019.107457. [DOI] [PubMed] [Google Scholar]
- 147.Magnin A., Hoornaert L., Pollet E., Laurichesse S., Phalip V., Avérous L. Isolation and characterization of different promising fungi for biological waste management of polyurethanes. Microb Biotechnol. 2019;12(3):544–555. doi: 10.1111/1751-7915.13346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gajendiran A., Krishnamoorthy S., Abraham J. Microbial degradation of low-density polyethylene (LDPE) by Aspergillus clavatus strain JASK1 isolated from landfill soil. 3 Biotech. 2016;6(1) doi: 10.1007/s13205-016-0394-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Barros J., Seena S. Plastisphere in freshwaters: An emerging concern. Environ Pollut. 2021;290:118123. doi: 10.1016/j.envpol.2021.118123. [DOI] [PubMed] [Google Scholar]
- 150.Wayman C., Niemann H. The fate of plastic in the ocean environment - a minireview. Environ Sci Process Impacts. 2021;23(2):198–212. doi: 10.1039/d0em00446d. [DOI] [PubMed] [Google Scholar]
- 151.Lamendella R., Strutt S., Borglin S., Chakraborty R., Tas N., Mason O.U., et al. Assessment of the deepwater horizon oil spill impact on gulf coast microbial communities. Front Microbiol. 2014;5 doi: 10.3389/fmicb.2014.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sivan A. New perspectives in plastic biodegradation. Curr Opin Biotechnol. 2011;22(3):422–426. doi: 10.1016/j.copbio.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 153.Raghukumar, Seshagiri. Fungi in Coastal and Oceanic Marine Ecosystems || The Macroalgal Ecosystem 2017;10.1007/978-3-319-54304–8:111–34.
- 154.Amobonye A., Bhagwat P., Singh S., Pillai S. Plastic biodegradation: frontline microbes and their enzymes. Sci Total Environ. 2021;759:143536. doi: 10.1016/j.scitotenv.2020.143536. [DOI] [PubMed] [Google Scholar]
- 155.Zhu B., Wang D., Wei N. Enzyme discovery and engineering for sustainable plastic recycling. Trends Biotechnol. 2022;40(1):22–37. doi: 10.1016/j.tibtech.2021.02.008. [DOI] [PubMed] [Google Scholar]
- 156.Gan Z., Zhang H. PMBD: a comprehensive plastics microbial biodegradation database. Database (Oxford) 2019;2019 doi: 10.1093/database/baz119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gómez-Méndez L.D., Moreno-Bayona D.A., Poutou-Piñales R.A., Salcedo-Reyes J.C., Pedroza-Rodríguez A.M., Vargas A., et al. Biodeterioration of plasma pretreated LDPE sheets by Pleurotus ostreatus. PLoS One. 2018;13(9):e0203786. doi: 10.1371/journal.pone.0203786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ho B.T., Roberts T.K., Lucas S. An overview on biodegradation of polystyrene and modified polystyrene: the microbial approach. Crit Rev Biotechnol. 2018;38(2):308–320. doi: 10.1080/07388551.2017.1355293. [DOI] [PubMed] [Google Scholar]
- 159.Gillings M.R., Paulsen I.T. Microbiology of the Anthropocene. Anthropocene. 2014;5:1–8. doi: 10.1016/j.ancene.2014.06.004. [DOI] [Google Scholar]
- 160.Zhu Y.-G., Gillings M., Simonet P., Stekel D., Banwart S., Penuelas J. Human dissemination of genes and microorganisms in Earth’s Critical Zone. Glob Chang Biol. 2018;24(4):1488–1499. doi: 10.1111/gcb.14003. [DOI] [PubMed] [Google Scholar]
- 161.Geyer R., Jambeck J.R., Law K.L. Production, use, and fate of all plastics ever made. Sci Adv. 2017;3 doi: 10.1126/sciadv.1700782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Locey K.J., Fisk M.C., Lennon J.T. Microscale insight into microbial seed banks. Front Microbiol. 2016;7:2040. doi: 10.3389/fmicb.2016.02040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ishiguro K., Washio J., Sasaki K., Takahashi N. Real-time monitoring of the metabolic activity of periodontopathic bacteria. J Microbiol Methods. 2015;115:22–26. doi: 10.1016/j.mimet.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 164.Shoemaker W.R., Lennon J.T. Evolution with a seed bank: the population genetic consequences of microbial dormancy. Evol Appl. 2018;11(1):60–75. doi: 10.1111/eva.12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Quartinello F., Kremser K., Schoen H., Tesei D., Ploszczanski L., Nagler M., et al. Together is better: the rumen microbial community as biological toolbox for degradation of synthetic polyesters. Front Bioeng Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.684459. [DOI] [Google Scholar]
- 166.He Q., Silliman B.R. Climate change, human impacts, and coastal ecosystems in the Anthropocene. Curr Biol. 2019;29:R1021–R1035. doi: 10.1016/j.cub.2019.08.042. [DOI] [PubMed] [Google Scholar]
- 167.Debroas D., Mone A., Ter Halle A. Plastics in the North Atlantic garbage patch: a boat-microbe for hitchhikers and plastic degraders. Sci Total Environ. 2017;599–600:1222–1232. doi: 10.1016/j.scitotenv.2017.05.059. [DOI] [PubMed] [Google Scholar]
- 168.Kirstein I.V., Wichels A., Krohne G., Gerdts G. Mature biofilm communities on synthetic polymers in seawater - Specific or general? Mar Environ Res. 2018;142:147–154. doi: 10.1016/j.marenvres.2018.09.028. [DOI] [PubMed] [Google Scholar]
- 169.Jiang P., Zhao S., Zhu L., Li D. Microplastic-associated bacterial assemblages in the intertidal zone of the Yangtze Estuary. Sci Total Environ. 2018;624:48–54. doi: 10.1016/j.scitotenv.2017.12.105. [DOI] [PubMed] [Google Scholar]
- 170.Wu W., Lu H.-P., Sastri A., Yeh Y.-C., Gong G.-C., Chou W.-C., et al. Contrasting the relative importance of species sorting and dispersal limitation in shaping marine bacterial versus protist communities. ISME J. 2018;12:485–494. doi: 10.1038/ismej.2017.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Delacuvellerie A., Cyriaque V., Gobert S., Benali S., Wattiez R. The plastisphere in marine ecosystem hosts potential specific microbial degraders including Alcanivorax borkumensis as a key player for the low-density polyethylene degradation. J Hazard Mater. 2019;380 doi: 10.1016/j.jhazmat.2019.120899. [DOI] [PubMed] [Google Scholar]
- 172.Harvey B.P., Kerfahi D., Jung Y., Shin J.-H., Adams J.M., Hall-Spencer J.M. Ocean acidification alters bacterial communities on marine plastic debris. Mar Pollut Bull. 2020;161 doi: 10.1016/j.marpolbul.2020.111749. [DOI] [PubMed] [Google Scholar]
- 173.Dudek K.L., Cruz B.N., Polidoro B., Neuer S. Microbial colonization of microplastics in the Caribbean Sea. Limnol Oceanogr Lett. 2020;5:5–17. doi: 10.1002/lol2.10141. [DOI] [Google Scholar]
- 174.Vaksmaa A., Knittel K., Abdala Asbun A., Goudriaan M., Ellrott A., Witte H.J., et al. Microbial communities on plastic polymers in the Mediterranean sea. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.673553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Agostini L., Moreira J.C.F., Bendia A.G., Kmit M.C.P., Waters L.G., Santana M.F.M., et al. Deep-sea plastisphere: long-term colonization by plastic-associated bacterial and archaeal communities in the Southwest Atlantic Ocean. Sci Total Environ. 2021;793 doi: 10.1016/j.scitotenv.2021.148335. [DOI] [PubMed] [Google Scholar]
- 176.Gao R., Sun C. A marine bacterial community capable of degrading poly(ethylene terephthalate) and polyethylene. J Hazard Mater. 2021;416 doi: 10.1016/j.jhazmat.2021.125928. [DOI] [PubMed] [Google Scholar]
- 177.Ye G., Zhang X., Yan C., Lin Y., Huang Q. Polystyrene microplastics induce microbial dysbiosis and dysfunction in surrounding seawater. Environ Int. 2021;156 doi: 10.1016/j.envint.2021.106724. [DOI] [PubMed] [Google Scholar]
- 178.Catão C.P.E., Pollet T., Garnier C., Barry-Martinet R., Rehel K., Linossier I., et al. Temperate and tropical coastal waters share relatively similar microbial biofilm communities while free-living or particle-attached communities are distinct. Mol Ecol. 2021;30:2891–2904. doi: 10.1111/mec.15929. [DOI] [PubMed] [Google Scholar]
- 179.Wright R.J., Bosch R., Langille M.G.I., Gibson M.I., Christie-Oleza J.A. A multi-OMIC characterisation of biodegradation and microbial community succession within the PET plastisphere. Microbiome. 2021;9:141. doi: 10.1186/s40168-021-01054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Scales B.S., Cable R.N., Duhaime M.B., Gerdts G., Fischer F., Fischer D., et al. Cross-hemisphere study reveals geographically ubiquitous, plastic-specific bacteria emerging from the rare and unexplored biosphere. MSphere. 2021;6 doi: 10.1128/mSphere.00851-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chen L., Li J., Tang Y., Wang S., Lu X., Cheng Z., et al. Typhoon-induced turbulence redistributed microplastics in coastal areas and reformed plastisphere community. Water Res. 2021;204 doi: 10.1016/j.watres.2021.117580. [DOI] [PubMed] [Google Scholar]