Highlights
-
•
An easy set-up of the co-cultures from 2 different microorganisms (filamentous fungi and bacteria) from different microbial domains resulting into a greater and more diverse metabolic and lignocellulolytic content.
-
•
An over expression of several key enzymatic lignocellulolytic activities is observed during the co-coculture due to elicitation.
-
•
An elicitation of some specific biosynthetic cluster genes is observed due to the activation of those the complexity of the carbon compounds present in the lignocellulose.
-
•
An elicitation of some specific biosynthetic cluster genes is observed only during the co-culture experiment.
-
•
A specific microbial crosstalk and interaction exists at the species level between the 3 Streptomyces and the fungi leading to a specific of lignocellulolytic enzyme and secondary metabolite production.
Keywords: Lignocellulose, Co-culture, Sreptomyces, Comparative genomics, CAZymes
Abstract
Lignocellulose, the most abundant biomass on Earth, is a complex recalcitrant material mainly composed of three fractions: cellulose, hemicelluloses and lignins. In nature, lignocellulose is efficiently degraded for carbon recycling. Lignocellulose degradation involves numerous microorganisms and their secreted enzymes that act in synergy. Even they are efficient, the natural processes for lignocellulose degradation are slow (weeks to months). In this study, the objective was to study the synergism of some microorganisms to achieve efficient and rapid lignocellulose degradation. Wheat bran, an abundant co-product from milling industry, was selected as lignocellulosic biomass. Mono-cultures and co-cultures involving one A.niger strain fungi never sequenced before (DSM 1957) and either one of three different Streptomyces strains were tested in order to investigate the potentiality for efficient lignocellulose degradability. Comparative genomics of the strain Aspergillus niger DSM 1957 revealed that it harboured the maximum of AA, CBM, CE and GH among its closest relative strains. The different co-cultures set-up enriched the metabolic diversity and the lignocellulolytic CAZyme content. Depending on the co-cultures, an over-expression of some enzymatic activities (xylanase, glucosidase, arabinosidase) was observed in the co-cultures compared to the mono-cultures suggesting a specific microbial cross-talk depending on the microbial partner. Moreover, metabolomics for each mono and co-culture was performed and revealed an elicitation of the production of secondary metabolites and the activation of silent biosynthetic cluster genes depending on the microbial co-culture. This opens opportunities for the bioproduction of molecules of interest from wheat bran.
Graphical abstract
1. Introduction
Biorefining is dedicated to produce energy, molecules and materials from renewable feedstocks (Kamm and Kamm, 2007)). The use of lignocellulosic biomass such as agricultural and agro-industrial co-products does not compete with plants dedicated for food. The use of cheap and abundant co-products as substrates for fermentation allows to reduce the costs of the bioprocesses (Sansinenea and Ortiz, 2011). Microbial cultures using lignocellulosic biomass as carbon sources are extensively studied for the production of enzymes and of various molecules of interest (Mäkelä et al., 2014; López-Mondéjar et al., 2019).
"Top-down" approaches, based on the study of natural ecosystems, where the degradation of lignocellulose is significant such as forest soils, compost, have enabled advances in the description and the multiple functions played by multiple microorganisms (Jurado et al., 2014; López-Mondéjar et al., 2016). On the opposite, “bottom-up” approaches consisted in the implementation of co-cultures by a limited number of microorganisms. The use of synthetic microbial co-cultures has also been implemented in order to mimic natural processes and distribute the different functions to specific populations. These microbial co-cultures based on mutualistic relationships, besides the ability to perform different functions, have several advantages such as: the ability to prevent a nutritional deficiency due to the diversity of metabolic pathways present, the ability to exchange metabolites within community (LaPara et al., 2002; Zuroff et al., 2013); stability and robustness within the microbial community (Zuroff et al.; 2013). The proof of concept in co-culture has been demonstrated through various applications such as human health (Steidler, 2004) or pollution control (Chen et al., 2014) and has made it possible to increase the services provided compared to mono-cultures. The use of co-cultures increases the production of enzymes. Several explanations exist: (1) a greater diversity of enzymes is produced allowing a more efficient and more complete degradation of the substrate, this is the enzymatic synergy (Taha et al., 2015), (2) a chemical interaction (interaction molecules, elicitors, secondary metabolites), as well as the sharing of metabolic pathways allows emulation of microbial development, this is the growth synergy (Ren et al., 2015).
In this study, co-cultures were performed between one fungal strain (belonging to the species Aspergillus niger) and either one of three different actinobacteria (belonging to the Streptomyces genus; S. avermitillis ATCC 31,267, S. coelicolor A3(2) and S. griseorubens DSM 40,160). Naturally, the fungi play a role in the carbon cycle through extracellular hydrolytic and oxidative enzymes. This fungus is used in the fermentation industry to produce citric acid, cellulolytic enzymes by fermentation in solid media (Pensupa et al., 2013), or even enzymatic cocktails to pre-treat lignocellulosic biomass (Wang et al., 2019). Enzyme cocktails are easily recovered, since the fungus will secrete the enzymes in the extracellular environment. A.niger enzymes panel includes cellulases, endo/exoglucanases, β-glucosidases, xylanases (Pensupa et al., 2013). Actinobacteria are Gram-positive bacteria characterized by a genome with high G + C ratio and are numerous and widely distributed group of soil microbes, constituting to about 10 – 50% of the soil microflora community and important producers of diversified secondary metabolites which can have several functions such as antifungal and antibacterial activities (Challis, 2014). Streptomyces genus has produced approximately 67% of the natural antibiotics among the actinobacteria and could produce approximately around 7600 bioactive compounds (Olanrewaju and Babalola, 2019). The genus Streptomyces, very widespread in soils, is a major player in the degradation of organic matter and lignocellulose (Lu et al., 2014). This microbial genus has a large enzymatic arsenal encoding carbohydrate esterases, polysaccharide lyases, glycoside hydrolases and enzymes with auxiliary activities (Book et al., 2014; Montella et al., 2017). They act in the catabolism of complex molecules and substances like lignocellulose, xylan, cellulose, and lignin, which are important in soil organic matter catabolism (Malherbe and Cloete, 2002). The ability of Streptomyces to deconstruct lignocellulose has been studied in several ecosystems such as intestinal tracts of insects (Book et al., 2014), grassland (Yeager et al., 2017) and moreover, those bacteria can also act as promotor and increase rice-straw composting by other microorganisms (Feng et al., 2020). Due to their origin (mainly superficial layers of soil), Aspergillus and Streptomyces can present several interactions together. Indeed, the strain Streptomyces leeuwenhoekii C58 has been used to trigger and elicit secondary metabolites production of Aspergillus fumigatus MR2012 (Wakefield et al., 2017).
In the present study, we report on the setup and the co-cultivation of one A.niger strain DSM 1957 and three bacterial strains belonging to the actinomycetes for degrading wheat bran. The genomic sequencing of A. niger DSM 1957 allowed to evaluate the genomic potential of this strain for the production of enzymes such as CAZymes (Carbohydrate Active enZymes) involved in lignocellulose degradation. Genomic comparisons were performed between the four strains in order to highlight their CAZyme and secondary metabolites production potential. The interactions between the strains were first investigated with classical cultivation on Petri dishes. The efficiency of wheat bran degradation was investigated by quantifying the various enzymatic activities produced by the microorganisms and also by analyzing the various metabolites produced during the microbial growth onto wheat bran.
2. Material and methods
2.1. Substrate preparation
Wheat bran (WB) (0.5–2 mm) was provided by the ARD society (https://www.a-r-d.fr/). The WB contained 16% of dry matter (DM) of arabinose, 19% DM of glucose, 26% DM of xylose, 1.1% DM of galactose, 11.6% DM of starch, 15% DM of protein and presents a lignin content of 5% DM”. The% of glucose can be attributed to cellulose and mixed β-glucans present in WB.
2.2. Growth media and inoculation of the microbial partners
In order to characterize the ability of the strains to grow on wheat bran, 250 mL flasks containing 50 mL of M3 media 1X (KH2PO4 1.9 g/L, Na2HPO4 5.1 g/L, MgSO4•7H2O 0.1 g/L, (NH4)2SO4 0.2 g/L) (Vieira and Nahas, 2005) supplemented with dry wheat bran up to 5 g/L were used. After sterilization, 1 mL of a Ca(NO3)2•4H2O solution at 25 g/L and 1 mL of a trace element solution were added. One pellet (2 mm)of actinobacteria (10^8 cells) and 10^6 spores of A. niger DSM 1957 were inoculated into the flasks. Controls were performed using only one carbon source as glucose supplemented up to 40 mM. All flasks were shaken for 144 h at 100 rpm at 30 °C. The experiments were performed in triplicate.
2.3. Defining parameters of the co-cultures
The physico-bio-chemical parameters of the co-cultures were determinated by taking into account several conditions: 1) the biological interactions between the partners (competition, mutualism or neutralism) and, 2) the inoculation parameters of individual partner (simultaneous or shifted inoculation). In order to test a potential antagonism between microorganisms, the interaction between microorganisms was observed on solid medium. Each interaction was carried out on WB + agar medium in duplicate. A first interaction was tested by spreading the Streptomyces all over the Petri dish and inoculating 106 spores of the fungi into the center of the Petri dish (Supplementary Figure 1). Concerning the time of inoculation, different strategies of inoculations were tested (simultaneous or shifted) (Supplementary Figure 2): the parameters selected were based on the observation of two microbial partners at the end of the incubation.
2.4. Enzymatic activities of the mono and co-cultures
To obtain intra- and extracellular enzymes, 1 mL of culture was collected and then centrifuged: 1) the supernatant corresponding to the extracellular enzymes was collected, 2) the pellet obtained was then resuspended in Tris–HCl (pH7, 25 mM) and cells were lysed using FastPrep and lysing matrix B at 6.5 G during 4 times * 40 s (MpBiomedicals, France). All the lignocellulolytic activities were measured in triplicate at the end of the growth curve (144 h) except for the xylanolytic which was measured all along the growth curve. Protein concentrations were determined by the Bradford method using BSA as a standard (Bradford, 1976) in order to normalize the enzymatic activities through time and the different samples. The intracellular activities were expressed as mIU of enzymatic activity /mg of protein. Xylanolytic activity was evaluated as previously described (Kidby and Davidson, 1973) with 0.1 mL of extracted proteins mixed with birchwood xylan (Sigma Aldrich France) at 0.5% w/v in 50 mM phosphate buffer, pH 7.5 at 30 °C for 10 min. Enzymatic activity was expressed in international milliunits (mIU), where 1 IU is defined as the quantity of enzyme required to release 1 µmole of reducing sugar per min.
Total phenol-oxydase/peroxydases activities were evaluated as in (Bach et al., 2013); for this, 0.1 mL of extracted proteins was mixed in a total volume of 2 mL with 8 mM of pyrogallol, 1 mM of EDTA and 0.5 mM of H2O2 at 30 °C during 10 min and measured at 420 nm.
Arabinofuranosidase activity by determining the hydrolysis rate of p-nitrophenyl α-l-arabinofuranoside (0.5 mM) and xylosidase activity was measured by quantifying the rate of hydrolysis of p-nitrophenyl-β-xylopyranoside (0.5 mM) in the same buffer in 1 mL reaction containing 900 µL of buffer (50 mM sodium phosphate buffer, pH 7.5) and 0.1 mL of culture supernatant. β-d-glucosidase activity were measured in the same conditions using as substrate p-nitrophenyl -β-d-glucopyranoside. The extinction coefficient of pNP in the measurement conditions was 15,850 M−1.cm−1. Experiments were measured using the absorbance at 401 nm for 5 min at 30 °C and using recording spectrophotometer (Uvikon 933). All the enzymatic activities will be presented in mIU/mg of protein.
2.5. Genomic annotation: metabolic pathways and CAZYme
The obtained genome sequences were finally annotated by using the NCBI Prokaryotic Genome Annotation Pipeline (https://www.ncbi.nlm.nih.gov/genome/annotation_prok/) in order to get deposited at GenBank. All the Streptomyces genomes studied in that manuscript were also annotated using the Microbial Genome Annotation & Analysis Platform (https://mage.genoscope.cns.fr/) which provided information about: 1) the metabolic profiles present in the genomes using the Metacyc database (Karp et al., 2002), 2) the core and pan-genome shared by the strains using SILIX software (Miele et al., 2011), 3) the presence of syntenic pathways, and 4) the distribution of Cluster of Ortholog groups (COGs). The presence and diversity of the Carbohydrate Active enZyme was demonstrated by using the online resource dbCAN (http://csbl.bmb.uga.edu/dbCAN/annotate.php).The CAZyme secretome of the fungi was analysed by using the SignalP software in order to identify the proteins with a secretion signal to the extracellular medium. The TMHMM server to identify the presence or absence of transmembrane domains in the sequence of these proteins. Only the sequences with no TMHMM topology were obtained and finally submitted to the dbCAN meta server in order to determine the CAZy secretome (Armenteros et al., 2019; Krogh et al., 2001).
2.6. UHPLC/Q-TOF HRMS analysis
Samples extracted from the different cultures on WB and glucose was analyzed by UHPLC/Q-TOF HRMS (Ultra High Performance Liquid Chromatography/ Quadrupole Time-of-Flight High-Resolution Mass Spectrometry) method. The UHPLC–MS analysis was performed on a Waters Acquity UHPLC system coupled with a Waters SYNAPT G2-Si High Resolution Mass Spectrometry equipped with electrospray ionization (ESI) source (Waters Corp., Manchester, UK). Chromatographic separation was carried out on an Acquity UHPLC BEH C18 (50 mm × 2.1 mm, 1.7 µm) column at 40 °C with a flow rate of 0.6 mL/min. The mobile phase A consisted of water containing 0.1% formic acid, while the mobile phase B was acetonitrile. The linear gradient profile started from 10% B, followed by a linear climb to 90% B over 7 min and held for 2 min, and finally returned to10% B for reequilibration for 2 min. Mass detection was conducted in negative ion mode, with the source temperature at 120 °C, capillary voltage and cone voltage were set at 2 KV and 40 V. The desolvation gas was optimized to 900 L/h, the cone gas flow of 50 L/h and the scan range was from 50 to 2500 m/z. Mass was corrected during acquisition using external reference (Lock-Spray) consisting of a 1 ng/µL solution of leucine encephalin at a flow rate of 5 µL/min, in order to make sure the accuracy and reproducibility during the MS analysis. All data collected were acquired using MassLynx™ (V4.1) software in centroid mode.
2.7. Statistical analysis
The values given in this report are the means of triplicates, with error bars corresponding to the standard deviation of this mean. Significance was tested with Student's t-test for two-sided distribution and unpaired samples. All statistical calculations were performed on Microsoft Excel.
2.8. Nucleotide sequence accession numbers
The accession numbers of the whole genome of the strain A.niger DSM 1957 (VYXZ00000000) was deposited at GenBank.
3. Results and discussion
3.1. Defining the parameters of the co-cultures
The growth of the two microbial partners was successfully observed whatever the condition culture used (liquid or solid). For the solid culture type, fungal growth was observed all over the Petri dish after inoculation of the spores in the middle; growth of the bacterial pellet was observed in the same area than the fungal mycelium (Supplementary Figure 3). For the liquid culture type, fungal growth was characterized by the presence of a fungal pellet which could encompass the lignocellulosic substrate and potentially the typical bacterial pellet which confirms that the 2 partners are not antagonistic (Supplementary Figure 4). Concerning the time of inoculation, when the fungi were inoculated prior to the bacteria either 6 h, 24h30 or 30H, no growth of the bacteria was observed. On the opposite, when the bacteria was inoculated either in the same time of before compared to the fungi, both were able to develop and grow in the flasks. However, for the later times of the fungal inoculations (24h30 and 30 h), a lower amount of fungal mycelium was observed. The results were comparable for a fungal inoculation after 6 h or in the same time than the bacterial strains (data not shown). For a practical matter, a simultaneous inoculation was performed.
3.2. Genomic comparison of A.niger DSM 1957
The strain A.niger DSM 1957 was firstly studied by (Steinberg, 1941) in order to describe the sulfur and trace-element nutrition among one strain of the A.niger species. This strain was later described for its ability to express xylanases and cellulases during growth onto different lignocellulosic subtrates (Prasertsan et al., 1997; Prasertsan and Oi, 1992).
The sequencing of the strain A.niger DSM 1957 generated 15,452,965 reads with an ultrasmall percentage of error (0.03%) with a Q30 of 91.4%. The assembly consisted into 138 (> 1000 bp) with a N50 of 544,125 bp. The number of N was low in our contigs with a number of N's per 100 kbp of 1.84.
In order to analyze the strain A.niger DSM 1957, we compared it to relatively closed genome from the strains A.niger An76, A.niger ATCC 1015, A.niger ATCC 13,496, A.niger CBS 513.88, A.niger CBS 101,883 and A.niger ATCC 64,974 N402. The strains have between 10,373 and 13,359 genes encoding proteins, the A.niger strain CBS 101,883 has the most SCP. The length of A.niger DSM 1957 was 35.6 Mbp and its GC content was 47% and was in the range of the compared A.niger strains.
The genome content was annotated according to their CAZyme content. The diversity of structures, compositions and bonds of components forming lignocellulose has led microorganisms, during evolution, to produce large panels of enzymes capable of degrading it (Manavalan et al., 2015). CAZymes are classified according to the CAZy database into five classes (Cantarel et al., 2009): glycoside hydrolases (GH), glycosyltransferases (GT), polysaccharide lyases (PL), carbohydrate esterases (CE) and anxilliary activities (AA). GHs (EC 3.2.1.*) hydrolyze the glycosidic bonds between two carbohydrates, or between a carbohydrate and a non-carbohydrate residue. GTs (EC 2.4.*.*) are involved in the biosynthesis of saccharide chains and have debranching activities. PLs (EC 4.2.2.*) mainly cleave bonds between acids and polysaccharides. The CEs catalyze the hydrolysis of the carbohydrate esters. AAs group together enzymes that act on lignins and polysaccharides (LPMO or Lytic Polysaccharides MonoOxygenases) through redox mechanisms. Another class exists, that of proteins consisting in binding modules (CBM or Carbohydrate Binding Module), which are not enzymes, but increase the efficiency of the latter. The strain A.niger DSM 1957 has the greatest number of hypothetical CAZymes (581 enzymes), it should be noted that A.niger 1957 and A.niger ATCC 64,974 N402 which nevertheless have a smaller number of coding sequences have the same number of CAZymes. The strain with the best CAZymes /Single-coding proteins ratio is A.niger CBS 513.88, its CAZymes representing 5.43% of its total proteins. The strain A.niger 1957 has the most AA, CBM, CE and GH numbers among the compared dataset. The number of CAZymes in A.niger DSM 1957 was the most important (like in A.niger CBS 513.88 and A.niger CBS 101,883) with a total of 581 CAZymes (Table 1).
Table 1.
Genomic characteristics and CAZyme content of the A.niger DSM 1957 strain compared to other Aspergillus strains (GH = Glycoside Hydrolase, CE = Carbohydrate Esterase, CBM = Carbohydrate Binding Module, PL = Polysaccharide lyase, GT = Glycoside Transferase, AA = Auxiliary activity).
| Strain | Length (MBp) | Single-coding proteins | CAZymes number | CAZymes (%) | AA | CBM | CE | GH | GT | PL |
|---|---|---|---|---|---|---|---|---|---|---|
| DSM 1957 | 35.6 | 10,798 | 581 | 5.38 | 106 | 17 | 88 | 262 | 99 | 10 |
| An76 | 34.6 | 10,373 | 551 | 5.31 | 97 | 15 | 86 | 246 | 97 | 10 |
| ATCC 1015 | 34.9 | 10,950 | 565 | 5.16 | 104 | 17 | 81 | 256 | 97 | 10 |
| ATCC 13,496 | 35.7 | 12,194 | 576 | 4.72 | 103 | 16 | 86 | 261 | 101 | 9 |
| CBS 513.88 | 34 | 10,609 | 576 | 5.43 | 104 | 15 | 83 | 249 | 107 | 18 |
| CBS 101,883 | 35.9 | 13,359 | 581 | 4.35 | 106 | 15 | 88 | 262 | 100 | 10 |
| ATCC 64,974 | 35.5 | 11,236 | 581 | 5.17 | 105 | 17 | 87 | 259 | 99 | 14 |
Among the 121 panCAZymes families present in the 7 fungal genomes analysed, a large common core was shared between them; indeed 115 families were present among all the strains. The strain A.niger DSM 1957 had 119 CAZymes families present. Among those families for that genome (with over 15 occurrences), the most important ones were CE10 (59 enzymes), AA7 (42), AA3 (33), GH13 (21), GH28 (21), GH3 (19), GT2 (18) and GH18 (14). Among the GH13 present, those enzymes encoded for several alpha-amylases and glucanotransferases. The GH28 encoded for several rhamnogalacturonases, exo-xylogalacturonan hydrolases and exopolygalacturonases. The GH3 encoded for β-glucosidases and β-xylosidases. The GH18 played a role into chitin degradation. The xylanase activity was carried by the GH10 and GH11. The GT2 were mainly involved in the chitin synthase process. The AA7 encoded for either glucooligosaccharideoxidase, chitooligosaccharide oxidase, cellooligosaccharide dehydrogenase and the AA3 for cellobiose dehydrogenase or glucose 1-oxidase. This result shows that the strain A.niger harbours a wide variety of genes encoding for xylanases, endo-glucanases and cellulases.
The potential CAZyme secretome of the strain A.niger DSM 1957 was studied; 272 among the 581 CAZymes were potentially secreted. Among those secreted CAZymes, 51 AA among the 106 were present, CE 41 among the 88 present, 162 GH among the 262 present which represented more than 46% of each family. The PL was all secreted. On the opposite, only 4 GT and 4 CBM among the 99 and 17 are present in the secretome.
3.3. Genomic comparison of the actinobacterial strains
In comparison to the Aspergillus genome, the actinobacterial strains have a shorter genome. The absolute number of CAZymes was lower compared to the fungal genomes (minimum of 255 and 336 respectively for S. griseorubens and S. avermitilis) so do their relative abundance in CAZymes (between 3.35 and 4.13%). The main difference between the bacterial and fungal CAZyme content was the higher proportions of CBM (Carbohydrate-Binding-Module) in the bacteria (average of 48 per genome) and fungi (16 per genome) (Table 2).
Table 2.
Genomic characteristics and CAZyme content of the actinobacterial strains.
| Length (Mbp) | Single-coding proteins | CAZymes number | CAZymes (%) | AA | CBM | CE | GH | GT | PL | |
|---|---|---|---|---|---|---|---|---|---|---|
| S.avermitillis | 10.5 | 10,003 | 335 | 3,35 | 20 | 53 | 32 | 149 | 69 | 12 |
| S.coelicolor | 9.1 | 8128 | 336 | 4,13 | 15 | 57 | 37 | 156 | 59 | 12 |
| S.griseorubens | 7.7 | 6841 | 255 | 3,73 | 15 | 34 | 34 | 115 | 52 | 5 |
A Venn diagram (Supplementary Figure 5) presents the common CAZymes genes that are present among the three Streptomyces analyzed. A large common core is shared between the 3 genomes with 80 CAZymes families similar. The strain S.avermitillis carried 13 specific CAZymes families which were S.avermitillis (AA6, CBM11, CBM50, CBM61, GH110, GH145, GH27, GH53, GH85, GH88, GH89, PL29, PL4). Among the GH CAZymes only present in S. avermitillis, the GH53, GH110, GH88 were mainly involved in hemicellulose degradation. For the S.coelicolor CAZome, 10 specific CAZymes families were present (CBM12, GH101, GH106, GH117, GH125, GH158, GH50, GH93, PL34, PL6) and involved in the hemicellulose and mannose degradation. The S.griseorubens had only 5 specific CAZymes families (GH136, GH81, GH84, GH97, GT84). The Streptomyces strains were thus well equipped in order to fractionate lignocellulose.
3.4. CAZyme comparison of the co-cultures
A Venn diagram (Fig. 1) describes the richness and diversity of the CAZymes brought by each organism during the mono and co-culture. To do so, CAZyme families with several iterations (such as the AA7 in A.niger which was present 42 times) were only considered once (reducing then the number of CAZymes in the genomes). For the co-culture S. avermitilis and A.niger DSM 1957, the fungal partner brought 60 unique CAZyme family into the genetic pool, whereas the bacteria partner brought 46; 59 were shared in common by the 2 partners. Similar results were obtained for the 2 other co-cultures. Among the CAZyme families only brought by A.niger DSM 1957, AA9 (lytic polysaccharide monooxygenases), AA1 (Laccase-like multicopper oxidase), GH71 (β−1,3-glucanosyltransglycosylase) and GH28 (polygalacturonase) were the most represented. Among the CAZyme families only brought by the actinobacteria, AA10 (laccase-like multicopper oxidase), GH23 (chitinase) and GH42 (β-galactosidase) were the most represented. The Venn diagram represents perfectly that diverse CAZymes were carried by each of the microbial partner and thus that the metabolic CAZymes diversity is drastically increased when a co-culture between Aspergillus and one of the three actinobacteria is performed.
Fig. 1.
Venn diagram representation of the CAZyme family content of each co-culture fungi-bacteria performed.
3.5. Secondary metabolite prediction and production
Among the secondary metabolites predicted by fungal antismash (https://fungismash.secondarymetabolites.org/#!/start), A.niger harboured 66 regions encoding for secondary metabolites encoding majoritary for 19 T1PKS, 14 NRPS-like, 3 T1PKS/NRPS and 2 terpene. A.niger DSM 1957 was able to produce naphthopyrone, pyranonigrin E, clavaric acid and nidulanin. A majority of the secondary metabolites produced by this strain remains unknown and might represent thus a new source of secondary metabolites.
For the actinobacteria, the strains S. coelicolor (Bentley et al., 2002), S. griseorubens and S. avermitillis encoded respectively for 29, 22, 36 regions of secondary metabolite predicted by the Antismash software (https://antismash.secondarymetabolites.org). Among those secondary metabolites, terpenes, NRPS, siderophores, T1PKS and lassopeptides were the most predominant. Among the main secondary metabolites produced, the strain S.avermitillis was able to produce, albaflavenone, avermitilol, carotenoid, citrulassin D, ectoine, filipin, geosmin, informatipeptin, melanin, oligomycin and pentalenolactone. The strain S.coelicolor was able to produce coelichelin, ectoine, melanin, desferrioxamine B, actinorhodin (Bystrykh et al., 1996), albaflavenone, curamycin, undecylprodigiosin, geosmin, hopene and germicidin (Lautru et al., 2005). For the strain S. griseorubens, albaflavenone, alkylresorcinol, ectoin, antimycin and rhizomide were produced (Supplementary Table 1).
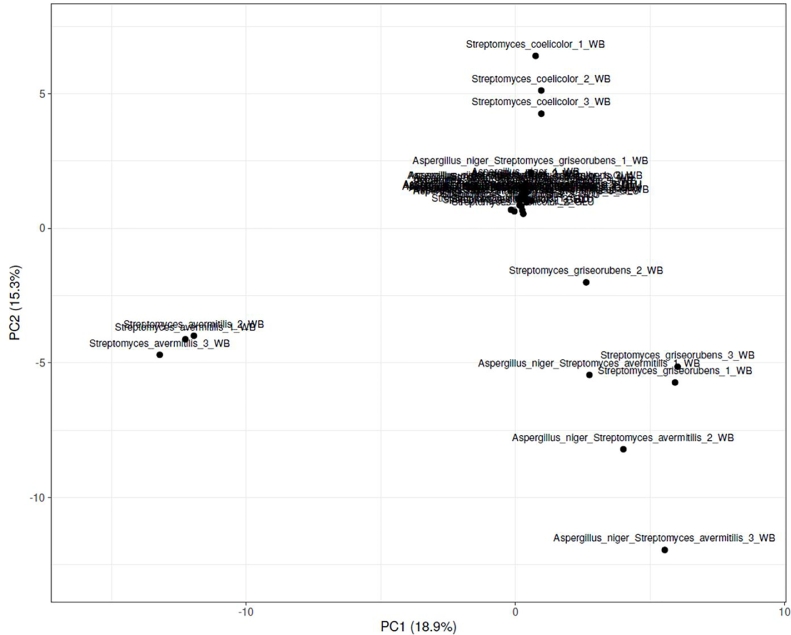
After UHPLC/Q-TOF HRMS analysis, a principal component analysis (Fig. 2) was performed in order to describe the differential secondary metabolite expression by different mono and co-cultures when grown on WB. The results showed that the replicates from each group clustered together showing the reproducibility of the experiment. The groups hardly separate among the principal component axes (PC1 and PC2 axe had percentages of 18.9 and 15.3%). The results of the pcoA strain showed that the actinobacterial strains during their growth on wheat bran separated well from their homolog during their growth on glucose. This result is due do the large number of secondary metabolites produced by those actinobacterial strains and the co-culture A.niger/S.avermitilis on wheat bran compared to the other conditions. Indeed, the strain S.griseorubens was able to produce between 13 and 18 secondary metabolites when grown on wheat bran; however when grown on glucose, a production of 6 to 8 secondary metabolites was observed. For S.coelicolor, between 12 and 16 when grown on WB depending on the replicates; on the opposite, only 8 secondary metabolites were produced. For S.avermitilis, 17 secondary metabolites were produced whatever the number of replicate whereas only 2 or 3 were produced were grown when grown on wheat bran. For the A.niger strain, only one secondary metabolite was detected when growth was perfomed on glucose whereas 5 were detected whatever the replicate when growth was done on glucose (Supplementary Table 1). By comparing those number of secondary metabolites depending on the carbon source, the results showed a lower production when grown on glucose; that result can be due to the carbon catabolite repression which guarantees the sequential utilization of carbon sources when more than one is simultaneously present in the culture media and would thus activate more biosynthetic clusters genes (Romero-Rodríguez et al., 2016). For the strain S.coelicolor, several known compounds were produced and identified by mass spectrometry: coelibactin (m/z = 481, elution time at 3.57), nogalamycin (m/z = 788, elution time at 4.87) and actinorhodin which was clearly detected with a blue pigmentation (m/z = 636, elution time at 3.19). For S.avermitilis and S.griseorubens, none of the predicted secondary metabolites were identified by mass spectrometry suggesting. For A.niger, one compound was produced with a predicted mass identified by mass spectrometry (fumonisin B1, m/z = 722, elution time of 1.1) when grown on wheat bran. The principal components analysis showed that the diversity of secondary metabolites obtained for the different conditions (growth on wheat bran or glucose) is different whatever the mono and co-culture; indeed few secondary metabolites were shared in common during the growth on wheat bran and glucose. Overall, this suggests that some components from WB (carbohydrates, proteins or lignin) could activate some silent biosynthetic cluster genes and will provide a new fingerprint of secondary metabolite production.
Fig. 2.
Principal component analysis of the metabolomics analysis for the different mono and co-cultures when grown on wheat bran (WB) and glucose (GLU).
Regarding the co-cultures, the A.niger/S.griseorubens co-culture, the number of secondary metabolites produced varied between 5 and 7 secondary metabolites (on WB) which were lower compared to the number of obtained for each member alone. By comparing with A. niger/S. griseorubens co-culture on glucose, less secondary metabolites was produced with a different diversity obtained compared to the one on WB. In the A.niger/S.griseorubens co-culture, the number of secondary metabolites produced varied between 5 and 7 secondary metabolites (on WB) which is lower compared to the number of obtained for each member alone. By comparing with the same co-culture on glucose, less secondary metabolites were produced with a different diversity obtained compared to the one on WB. The same trend was observed for the A.niger/S.coelicolor with a lower number of secondary metabolites produced compared to the growth on wheat bran (3 produced) to glucose (2 produced) and none shared together in the different carbon conditions (Supplementary Figure 6).
Contrary, the strain S.avermitilis was able to produce 17 compounds where grown in mono-culture. In the co-culture A.niger/S.avermitilis, 13 compounds in average were produced; 2 were in common with the S.avermitilis alone (at the elution times of 5.3 and 5.91 min) and none of the ones produced by A. niger alone was found in the co-culture. By comparing with A. niger/S. griseorubens co-culture on glucose, less secondary metabolites were produced with a different diversity obtained compared to the one on WB. Eleven new secondary metabolites which were not present in the mono-cultures were detected in the co-culture; among those eleven new secondary metabolites, only 2 were recovered in the coculture with A. niger/S. avermitilis grown on glucose which suggests thus an elicitation and activation of silent biosynthetic cluster genes. None of the molar masses detected in the co-cultures were close to the masses of the secondary metabolites predicted by the Antismash (Blin et al., 2019) and MIBIG algorithms (Kautsar et al., 2020) Previous co-cultures of Aspergillus fungi and Streptomyces showed a suppression of the production of the fungal metabolites (Wakefield et al., 2017). The activation of silent of those silent biosynthetic cluster genes in a second microorganism may be stimulated through microbial crosstalk and may be interpreted as a defense mechanism triggered in response to a chemical signal from the other microorganism (Wakefield et al., 2017)
In our study, it is not possible to confirm which of the microbial partners was able to produce those new secondary metabolites in the co-culture. In order to prove which one the microbial partners is able to produce those new secondary metabolites, 1) either a spiking of the supernatant of one of the microbial partner could be added to the culture of the remaining one, 2) or an elicitation with one of the microbial lysate or the microbial cell components (Abdelmohsen et al., 2015).
Overall, the results of the metabolomic analysis showed a different relationship and crosstalk between the fungal strain and the different actinobacteria. In summary, an inhibition of the secondary metabolites produced by S.griseorubens and S.coelicolor was observed when grown with A.niger whereas when that fungal strain was grown with S. avermitilis, activation and a possible dual elicitation was observed whatever the carbon source (WB or glucose). However, the diversity of the secondary metabolites tend to be different between the growths on the carbon source type revealing the potential role of lignocellulose as elicitor of biosynthetic cluster genes.
3.6. Lignocellulolytic enzymatic activities during mono and co-cultures
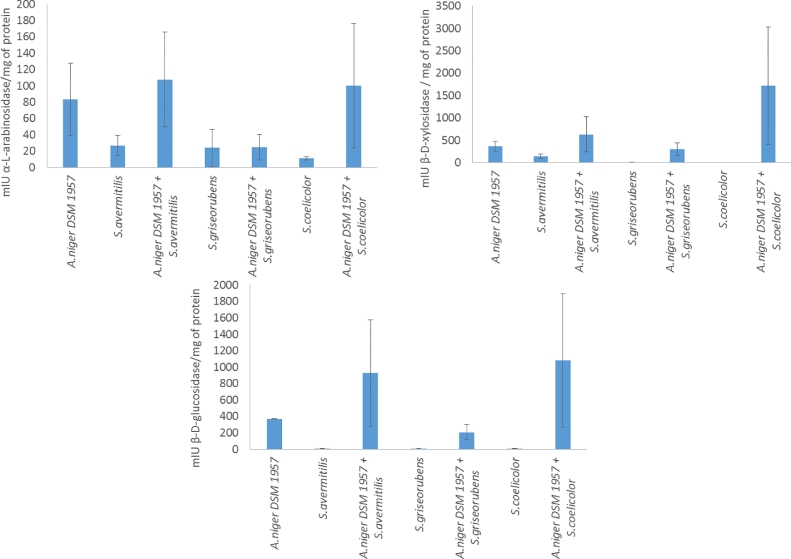
The enzymatic activities were quantified from the mono and co-cultures from wheat bran and not on glucose; indeed, previous preliminary experiments performed at the laboratory showed no lignocellulolytic enzymatic activity produced by the microorganism when grown on glucose on the opposite to wheat bran. Debranching intracellular enzymatic activities were measured for the 3 actinobacterial strains and A.niger in mono and co-cultures (Fig. 3). The results showed that the intracellular β-d-glucosidase and b-d-xylosidase enzymatic activities were the most important with values higher than 1 IU/mg of protein; on the opposite, α-l-arabinosidase enzymatic activities were lower and never reached more than 200 mIU/mg of protein. In all the experiments performed in mono-culture, enzymatic activities were always higher for the fungi compared to the other actinobacteria. The enzymatic activities measured showed different patterns depending on the strain added in the co-culture: 1) more enzymatic activities were detected in the co-cultures with S.avermitilis and S.coelicolor 2) less enzymatic activities were detected in the co-culture with S.griseorubens. Indeed, for the monoculture, intracellular β-d-glucosidase activity was 371.74 ± 3.09 mIU/mg of protein whereas it was lower than 10 mIU/mg of protein for the other actinobacteria; in the co-cultures, β-d-glucosidase activities were 928.7 ± 648.6, 1082.4 ± 812.8 mIU/mg of protein respectively for A.niger/S.avermitilis and A.niger/S.coelicolor. None of the statistical tests were significant. On the opposite, the β-d-glucosidase activity was lower in the A.niger/S.griseorubens co-culture with a value of 205.9 ± 90.9 mIU/mg of protein (p-value < 0.05). The same patterns were also observed for the intracellular xylosidase activity: indeed, the activity quantified for the A.niger was 365.5 ± 112.3 mIU/mg of protein whereas it was much lower for the other actinobacterial strains. A decrease of the b-d-xylosidase activity was observed for the co-culture A.niger/S.griseorubens (303.4 ± 136.9 mIU/mg of protein) and an increase for the others co-culture with respectively 630.3 ± 392.4, 1720.6 ± 1309.2 mIU/mg of protein respectively for A.niger/S.avermitilis and A.niger/S.coelicolor.
Fig. 3.
Intracellular debranching activities of the mono and co-cultures at 144 h.
Regarding the α-l-arabinosidase activity, the enzymatic activities were less important compared to the b-d-xylosidase and β-d-glucosidase. The differences were less important between the activity produced by the fungi (83.25± 44.25 mIU/mg of protein) and the other actinobacteria respectively (26.9 ± 12.1, 23.9 ± 22.6 and 11.2 ± 2.2 S.avermitilis, S.griseorubens and S.coelicolor respectively) compared to the other activities measured. In the A.niger/S.avermitilis and A.niger/S.coelicolor co-cultures, the measured α-l-arabinosidase enzymatic activities were 107.7 ± 58 and 100 ± 76.5 mIU/mg of protein and were superior to the enzymatic activity obtained for the fungi. On the opposite, the value observed for the A.niger/S.griseorubens co-culture was 24.9 ± 15.9 mIU/mg of protein.
In all the co-cultures (exception of the α-l-arabinosidase for A.niger/S.avermitilis) between A.niger with S.avermitilis or S.coelicolor, the enzymatic values obtained were superior to the sum of the enzymatic activity produced by each microbial partner analyzed in its own mono-culture (excepted for the α-l-arabinosidase activity in A.niger/S.avermitilis. Indeed, an overproduction up 1351 mIU/mg of protein was observed in the coculture A.niger/S.coelicolor (1720.6 mIU/mg of protein) for the xylosidase activity compared to the expected activity by the addition of each microbial partner (365.5 ± 4.07 respectively).
For all those debranching activities, extracellular activities were also measured (Supplementary Figure 7). For the strains A.niger and S.avermitilis, elicited enzymatic activities were observed for the arabinosidase and glucosidase. Indeed, the arabinosidase activity was 353 ± 31 mIU/mg of protein in the co-culture whereas they were respectively 7.6 and 34 mIU/mg of protein for S.avermitilis and A.niger. For the glucosidase activity in that same co-culture, the values reached up to 927 ± 198 mIU/mg of protein in the co-culture whereas they were respectively 97 and 60 mIU/mg of protein for S.avermitilis and A.niger.
For A.niger/S.coelicolor coculture, higher enzymatic activities were observed for the xylosidase and glucosidase. For the xylosidase activity in that same co-culture, the values drastically reached up to 1610 ± 101 mIU/mg of protein in the co-culture whereas they were respectively 9 and 57 mIU/mg of protein for and A.niger. A lower elicitation was observed for the glucosidase activity; indeed the activity was 968 ± 400 mIU/mg of protein in the co-culture whereas they were respectively 92 and 60 mIU/mg of protein S.coelicolor for A.niger.
For the A.niger/S.griseorubens coculture, a decrease of the enzymatic activities was observed (except for the arabinofuranosidase) overall for the extracellular activities.
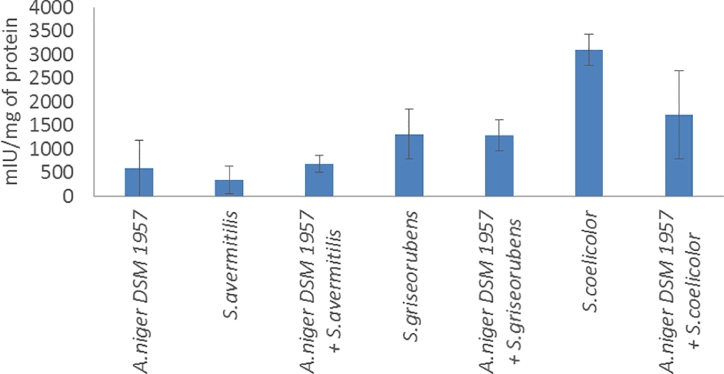
Intracellular peroxidase activities were also measured during mono and co-cultures (Fig. 4). The peroxidase activities mainly responsible of the lignin degradation (Dashtban et al., 2009). Peroxidases catalyze the oxidation of lignin in the presence of hydrogen peroxide as electron acceptor and can involve cytochrome c peroxidase (Dashtban et al., 2009). On opposite to the other enzymatic activities before, all the peroxidase payload was not performed majority by the fungi but also by the bacteria. Indeed, A.niger DSM 1957 showed a peroxidase activity that reached 588 mIU/mg which superior only to the one observed in S.avermitilis (347 mIU/mg) whereas higher activities were present for the 2 remaining bacteria which were S.griseorubens and S.coelicolor (1314 and 3091 mIU/mg of protein respectively). Among all the co-cultures tested, no one showed a superior peroxidase activity compared to each microbial partner separately. This could be due to the low abundance of lignin in wheat bran which would explain this absence of difference. The utilization of more lignified agro-resources would maybe allow differences in term of peroxidase activity. A small decrease was observed for the A.niger/S.avermitilis and A.niger/S.griseorubens co-cultures whereas the decrease was more important for the A.niger/S.coelicolor (1726 mIU/mg of protein the co-culture compared to the sum of each microbial partner individually (3680 mIU/mg of protein).
Fig. 4.
Intracellular peroxidase activity of the mono and co-cultures at 144 h.
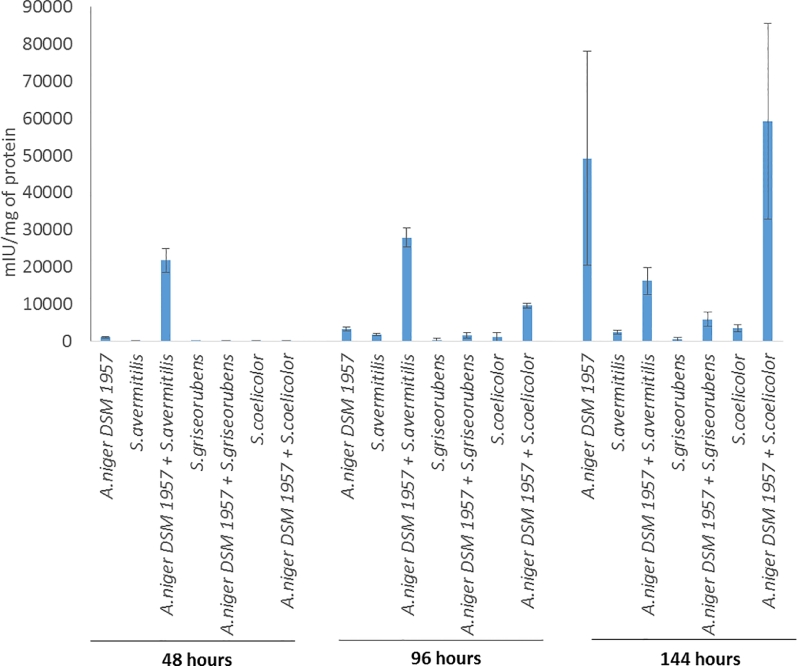
Xylanases catalyze the hydrolysis of xylans and have been widely studied in filamentous fungi such as Aspergillus (Betini et al., 2009; Pal and Khanum, 2010). Aspergillus xylanases have been (Paul et al., 2020) widely used in several industrial processes such as paper pulp biobleaching (Sridevi et al., 2016). Waste management programs make use of xylanases so as to hydrolyze xylan found in industrial and municipal wastes (Motta et al., 2013). Due to the importance of xylanase in the lignocellulolytic, secretion system present in Aspergillus, a dynamic study of the extracellular xylanase activity (at 48, 96 and 144 h) was performed on the compared to the other enzymatic activities (Fig. 5). The results showed that xylanolytic activity load was carried by A.niger and not by the Streptomyces bacteria; indeed, the maximum xylanase activity for one of the Streptomyces member was at 144 h for S.coelicolor (3513 ± 980 mIU/mg of protein). For A.niger, xylanolytic activity increased continuously from 48 h to 144 h reaching up 49,251 ± 28,763 mIU/mg of protein. The same increasing trend was observed for the other actinobacteria through time.
Fig. 5.
Dynamic secreted xylanalytic activity of the mono and co-cultures at 48, 96 and 144 h.
The trend of dynamic xylanalytic activity of the co-culture A.niger/S.griseorubens confirmed the trends obtained from the others enzymatic activities depicted previously. Indeed, the enzymatic activity was 3.5 more time less important for the co-culture A.niger/S.griseorubens compared to the mono-culture of A.niger after 144 h.
On the opposite, for the other actinobacterial strains, an increase factor up to 147% and 229% was found between A.niger and the co-cultures, A.niger/S.avermitilis and A.niger/S.coelicolor respectively. A different dynamic was observed between those two previous co-cultures; 1) indeed, the maximum xylanase activity was observed for A.niger/S.avermitilis was already important at 48 h reaching 21,804 ± 3265 mIU/mg (then slowly increased at 96 h and decreased at 144 h), 2) on the opposite, the xylanalytic activity of A.niger/S.coelicolor was low at 48 and 96 h but reached a peak at 144 h up to 59,226 ± 26,239 mIU/mg. The dynamic lignocellulolytic enzymatic activity was studied for all the co-cultures and showed that through time the xylanase was always over-expressed compared to the other enzymatic activities. Xylanase activity is thus necessary through all the growth and xylan represents consequently the main carbon and energy source of the microbial partners. Those results are correlated to previous studies regarding the dynamic secretion of A.niger An-76 where the expression of xylanase was detected on hydrolysates of lignocellulose polysaccharide at 24 h of inoculation until the end of growth (144 h) (Xing et al., 2013). Debranching activities were detected at the final time point signifying that these enzymes play a crucial role for the hydrolysis of the oligosaccharides produced by the xylanases and other endo-enzymes. Overall, for all the enzymatic activities measured in that manuscript, some standard deviation could be due to the growth type of the microbial partners involved in those co-cultures. Indeed, those microorganisms form pellets of different size with different amount of cells and subsequent protein content. Despite those standard deviations, all the results confirm the different patterns where elicitation is observed between A.niger and S.coelicolor or S. avermitilis and a possible inhibition between A.niger and S.griseorubens. The co-culture growth observation was only conducted in a qualitative manner visually by assessing the presence of the two microbial partners depending on their morphological shape. The main point of our study was to describe the possible elicitation of enzymatic activities and production of secondary metabolites at a final time point when grown on lignocellulose. It is acknowledged that the metabolic profiles into a co-culture will depend on the distribution of the two partners (Karuppiah et al., 2019; Romanens et al., 2020). The morphological patterns of the two microbial partners which form pellets do not allow a spectrophotometric quantification and thus as future experiments, we will develop an approach in follow-up studies about the metabolomic profiles obtained in those co-cultures at different sampling points allowing to generate data for both microorganisms. Here, the enzymatic activities were analyzed as mIU/mg of protein in order to normalize throughout the different samples (through time and the different consortia). The similar enzymatic activities and metabolomics profiles obtained showed that the experiments were reproducible and that the distribution was comparable among the replicates. In our experiment, the precise mechanism behind that microbial interaction is not clear and has not been investigated. The activation of those enzymatic activities can be due to several hypothesis: the presence of signaling molecules or direct contact between the two microbial partners. In the case of signaling molecules, the activation of enzymatic activities in Aspergillus can be due to several factors; indeed the activation can be due to the presence of different types of molecule: 1) previous study revealed indeed that 3 mM Cu2+ supplementation in recombined xylanase A. niger US368 enhanced its activity by 54% (Elgharbi et al., 2015); 2) the presence of secondary microbial metabolites which can over-express xylanase activity by 40% (Andrioli et al., 2012). For the direct contact, previous experiments proved that this was necessary between bacteria/fungi to observe activation of cryptic metabolic pathways (Scherlach and Hertweck, 2009). In order to prove that hypothesis of signaling molecules, further experiments could be performed 1) either by spiking only the secondary metabolites produced by the co-cultures fungi/actinobacteria to another fungal culture the fungi is grown alone, 2) use membrane reactors which would only allow the transfer of the secondary metabolites to each partner without direct contact between them. An accurate description of those molecules will be performed in the future which could be used for the improvement of fractionation by other microorganisms into other biotechnological processes by simple spike. Those activators which would improve lignocellulolytic activities would be then ready-to-use and less expensive compared to other approaches involving genome editing per example (Liu et al., 2017). Overall, the results showed that the consortia tend to have a hemicellulolytic strategy compared to the cellulolytic one. This is related to the chemical structure and diversity of wheat bran which is mainly constituted of arabinoxylans (16% DM of arabinose and 26% DM of xylose) compared to cellulose (19% DM of glucose) which could hypothesis that more energy and carbon source could be available for the consortia from the hemicellulose and notably the arabinoxylans. Moreover, the cellulose degradation pathway requires the expression of more enzymes in A.niger (the main degrader in our consortia) compared to the xylan degradation pathway which could thus represent an energy save in the fungi metabolism (Benoit-Gelber et al., 2017).
The obtained results in that study are promising with: 1) an easy set-up of the co-cultures from 2 different microorganisms from different domains in the “Tree of life” resulting into a greater and more diverse metabolic and lignocellulolytic content, 2) an over expression of several key enzymatic activities, 3) an elicitation of some specific biosynthetic cluster genes observed only in the co-culture experiment, 4) a specific microbial crosstalk and interaction observed at the species level between the 3 Streptomyces and the fungi leading to a specific of lignocellulolytic enzyme and secondary metabolite production. Further experiments will be performed in order to: 1) decipher the regulatory and expression mechanisms at the gene level over-expressed in the co-culture, 2) describe the interaction type (chemical or physical) between the 2 microbial partners, 3) identify the secondary metabolites produced during the co-culture experiments.
4. Author statement
Authors: Julian Detain (JD), Caroline Rémond (CR), Carine Machado Rodrigues (CMR), Dominique Harakat (DH), Ludovic Besaury (LB)
Conception and design of study: LB,
Acquisition of data: JD, CMR, DH, LB
Analysis and/or interpretation of data: JD, CMR, DH, LB
Drafting the manuscript: DH, LB
Revising the manuscript critically for important intellectual content: CR, CMR, DH, LB
Approval of the version of the manuscript to be published (the names of all authors must be listed): Julian Detain (JD), Caroline Rémond (CR), Carine Machado Rodrigues (CMR), Dominique Harakat (DH), Ludovic Besaury (LB)
Declaration of Competing Interest
The authors declare that there are no conflicts of interest.
Acknowledgments
The authors are grateful to the French Region Grand Est, Grand Reims and the European Regional Development Fund for the funding of the Chaire AFERE. This project (FRALICOCMI acronym) was granted by the CEPIA/TRANSFORM department from INRAE.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.crmicr.2022.100108.
Appendix. Supplementary materials
References
- Abdelmohsen U.R., Grkovic T., Balasubramanian S., Kamel M.S., Quinn R.J., Hentschel U. Elicitation of secondary metabolism in actinomycetes. Biotechnol. Adv. 2015;33:798–811. doi: 10.1016/j.biotechadv.2015.06.003. [DOI] [PubMed] [Google Scholar]
- Andrioli W.J., Damásio A.R., Silva T.M., da Silva V.B., Maller A., Nanayakkara N., Silva C.H., Polizeli M.L., Bastos J.K. Endo-xylanase GH11 activation by the fungal metabolite eugenitin. Biotechnol. Lett. 2012;34:1487–1492. doi: 10.1007/s10529-012-0918-3. [DOI] [PubMed] [Google Scholar]
- Armenteros J.J.A., Tsirigos K.D., Sønderby C.K., Petersen T.N., Winther O., Brunak S., von Heijne G., Nielsen H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- Bach C.E., Warnock D.D., Van Horn D.J., Weintraub M.N., Sinsabaugh R.L., Allison S.D., German D.P. Measuring phenol oxidase and peroxidase activities with pyrogallol, l-DOPA, and ABTS: effect of assay conditions and soil type. Soil Biol. Biochem. 2013;67:183–191. [Google Scholar]
- Benoit-Gelber I., Gruntjes T., Vinck A., Van Veluw J., Wösten H.A., Boeren S., Vervoort J., De Vries R. Mixed colonies of Aspergillus niger and Aspergillus oryzae cooperatively degrading wheat bran. Fungal Genetics Biol. 2017;102:31–37. doi: 10.1016/j.fgb.2017.02.006. [DOI] [PubMed] [Google Scholar]
- S.D. Bentley, K.F. Chater, A.-.M. Cerdeño-Tárraga, G.L. Challis, N. Thomson, K.D. James, D.E. Harris, M.A. Quail, H. Kieser, D. Harper, Complete genome sequence of the model actinomycete Streptomyces coelicolor A3 (2), Nature, 417 (2002) 141–147. [DOI] [PubMed]
- Betini J., Michelin M., Peixoto-Nogueira S., Jorge J., Terenzi H., Polizeli M. Xylanases from Aspergillus niger, Aspergillus niveus and Aspergillus ochraceus produced under solid-state fermentation and their application in cellulose pulp bleaching. Bioprocess Biosyst. Eng. 2009;32:819–824. doi: 10.1007/s00449-009-0308-y. [DOI] [PubMed] [Google Scholar]
- Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S.Y., Medema M.H., Weber T. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Book A.J., Lewin G.R., McDonald B.R., Takasuka T.E., Doering D.T., Adams A.S., Blodgett J.A., Clardy J., Raffa K.F., Fox B.G. Cellulolytic Streptomyces strains associated with herbivorous insects share a phylogenetically linked capacity to degrade lignocellulose. Appl. Environ. Microbiol. 2014;80:4692–4701. doi: 10.1128/AEM.01133-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bystrykh L.V., Fernández-Moreno M.A., Herrema J.K., Malpartida F., Hopwood D.A., Dijkhuizen L. Production of actinorhodin-related“ blue pigments” by Streptomyces coelicolor A3 (2) J. Bacteriol. 1996;178:2238–2244. doi: 10.1128/jb.178.8.2238-2244.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel B.L., Coutinho P.M., Rancurel C., Bernard T., Lombard V., Henrissat B. The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis G.L. Exploitation of the Streptomyces coelicolor A3 (2) genome sequence for discovery of new natural products and biosynthetic pathways. J. Ind. Microbiol. Biotechnol. 2014;41:219–232. doi: 10.1007/s10295-013-1383-2. [DOI] [PubMed] [Google Scholar]
- Chen Y., Li C., Zhou Z., Wen J., You X., Mao Y., Lu C., Huo G., Jia X. Enhanced biodegradation of alkane hydrocarbons and crude oil by mixed strains and bacterial community analysis. Appl. Biochem. Biotechnol. 2014;172:3433–3447. doi: 10.1007/s12010-014-0777-6. [DOI] [PubMed] [Google Scholar]
- Dashtban M., Schraft H., Qin W. Fungal bioconversion of lignocellulosic residues; opportunities & perspectives. Int. J. Biol. Sci. 2009;5:578. doi: 10.7150/ijbs.5.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgharbi F., Hlima H.B., Farhat-Khemakhem A., Ayadi-Zouari D., Bejar S., Hmida-Sayari A. Expression of A. niger US368 xylanase in E. coli: purification, characterization and copper activation. Int. J. Biol. Macromol. 2015;74:263–270. doi: 10.1016/j.ijbiomac.2014.12.005. [DOI] [PubMed] [Google Scholar]
- Feng J., Wang B., Zhang D., Chu S., Zhi Y., Hayat K., Wang J., Chen X., Hui N., Zhou P. Streptomyces griseorubens JSD-1 promotes rice straw composting efficiency in industrial-scale fermenter: evaluation of change in physicochemical properties and microbial community. Bioresour. Technol. 2020;321 doi: 10.1016/j.biortech.2020.124465. [DOI] [PubMed] [Google Scholar]
- Jurado M., López M.J., Suárez-Estrella F., Vargas-García M.C., López-González J.A., Moreno J. Exploiting composting biodiversity: study of the persistent and biotechnologically relevant microorganisms from lignocellulose-based composting. Bioresour. Technol. 2014;162:283–293. doi: 10.1016/j.biortech.2014.03.145. [DOI] [PubMed] [Google Scholar]
- Kamm B., Kamm M. Biorefineries–multi product processes. White Biotechnol. 2007:175–204. doi: 10.1007/10_2006_040. [DOI] [PubMed] [Google Scholar]
- Karp P.D., Riley M., Paley S.M., Pellegrini-Toole A. The metacyc database. Nucleic Acids Res. 2002;30:59–61. doi: 10.1093/nar/30.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karuppiah V., Vallikkannu M., Li T., Chen J. Simultaneous and sequential based co-fermentations of Trichoderma asperellum GDFS1009 and Bacillus amyloliquefaciens 1841: a strategy to enhance the gene expression and metabolites to improve the bio-control and plant growth promoting activity. Microb. Cell Fact. 2019;18:1–16. doi: 10.1186/s12934-019-1233-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautsar S.A., Blin K., Shaw S., Navarro-Muñoz J.C., Terlouw B.R., van der Hooft J.J., Van Santen J.A., Tracanna V., Suarez Duran H.G., Pascal Andreu V. MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 2020;48:D454–D458. doi: 10.1093/nar/gkz882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidby D., Davidson D. A convenient ferricyanide estimation of reducing sugars in the nanomole range. Anal. Biochem. 1973;55:321–325. doi: 10.1016/0003-2697(73)90323-0. [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., Von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- LaPara T., Zakharova T., Nakatsu C., Konopka A. Functional and structural adaptations of bacterial communities growing on particulate substrates under stringent nutrient limitation. Microb. Ecol. 2002;44:317–326. doi: 10.1007/s00248-002-1046-8. [DOI] [PubMed] [Google Scholar]
- Lautru S., Deeth R.J., Bailey L.M., Challis G.L. Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat. Chem. Biol. 2005;1:265–269. doi: 10.1038/nchembio731. [DOI] [PubMed] [Google Scholar]
- Liu Q., Gao R., Li J., Lin L., Zhao J., Sun W., Tian C. Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering. Biotechnol. Biofuels. 2017;10:1–14. doi: 10.1186/s13068-016-0693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Mondéjar R., Algora C., Baldrian P. Lignocellulolytic systems of soil bacteria: a vast and diverse toolbox for biotechnological conversion processes. Biotechnol. Adv. 2019;37 doi: 10.1016/j.biotechadv.2019.03.013. [DOI] [PubMed] [Google Scholar]
- López-Mondéjar R., Zühlke D., Becher D., Riedel K., Baldrian P. Cellulose and hemicellulose decomposition by forest soil bacteria proceeds by the action of structurally variable enzymatic systems. Sci. Rep. 2016;6:1–12. doi: 10.1038/srep25279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Zeng G., Fan C., Zhang J., Chen A., Chen M., Jiang M., Yuan Y., Wu H., Lai M. Diversity of two-domain laccase-like multicopper oxidase genes in Streptomyces spp.: identification of genes potentially involved in extracellular activities and lignocellulose degradation during composting of agricultural waste. Appl. Environ. Microbiol. 2014;80:3305–3314. doi: 10.1128/AEM.00223-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkelä M.R., Donofrio N., de Vries R.P. Plant biomass degradation by fungi. Fungal Genetics Biol. 2014;72:2–9. doi: 10.1016/j.fgb.2014.08.010. [DOI] [PubMed] [Google Scholar]
- Malherbe S., Cloete T.E. Lignocellulose biodegradation: fundamentals and applications. Rev. Environ. Sci. Biotechnol. 2002;1:105–114. [Google Scholar]
- Manavalan T., Manavalan A., Heese K. Characterization of lignocellulolytic enzymes from white-rot fungi. Curr. Microbiol. 2015;70:485–498. doi: 10.1007/s00284-014-0743-0. [DOI] [PubMed] [Google Scholar]
- Miele V., Penel S., Duret L. Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformatics. 2011;12:116. doi: 10.1186/1471-2105-12-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montella S., Ventorino V., Lombard V., Henrissat B., Pepe O., Faraco V. Discovery of genes coding for carbohydrate-active enzyme by metagenomic analysis of lignocellulosic biomasses. Sci. Rep. 2017;7:42623. doi: 10.1038/srep42623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- F. Motta, C. Andrade, M. Santana, A review of xylanase production by the fermentation of xylan: classification, characterization and applications, Sustainable Degradation Of Lignocellulosic Biomass-Techniques, Applications and Commercialization, 1 (2013).
- Olanrewaju O.S., Babalola O.O. Streptomyces: implications and interactions in plant growth promotion. Appl. Microbiol. Biotechnol. 2019;103:1179–1188. doi: 10.1007/s00253-018-09577-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal A., Khanum F. Production and extraction optimization of xylanase from Aspergillus niger DFR-5 through solid-state-fermentation. Bioresour. Technol. 2010;101:7563–7569. doi: 10.1016/j.biortech.2010.04.033. [DOI] [PubMed] [Google Scholar]
- Paul M., Nayak D.P., Thatoi H. Optimization of xylanase from Pseudomonas mohnii isolated from Simlipal Biosphere Reserve, Odisha, using response surface methodology. J. Genetic Eng. Biotechnol. 2020;18:1–19. doi: 10.1186/s43141-020-00099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensupa N., Jin M., Kokolski M., Archer D.B., Du C. A solid state fungal fermentation-based strategy for the hydrolysis of wheat straw. Bioresour. Technol. 2013;149:261–267. doi: 10.1016/j.biortech.2013.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasertsan P., Kunghae A., Maneesri J., Oi S. Optimization for xylanase and cellulase production from Aspergillus niger ATTC 6275 in palm oil mill wastes and its application. World J. Microbiol. Biotechnol. 1997;13:555–559. [Google Scholar]
- Prasertsan P., Oi S. Production of cellulolytic enzymes from fungi and use in the saccharification of palm cake and palm fibre. World J. Microbiol. Biotechnol. 1992;8:536–538. doi: 10.1007/BF01201957. [DOI] [PubMed] [Google Scholar]
- Ren D., Madsen J.S., Sørensen S.J., Burmølle M. High prevalence of biofilm synergy among bacterial soil isolates in cocultures indicates bacterial interspecific cooperation. ISME J. 2015;9:81–89. doi: 10.1038/ismej.2014.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanens E., Pedan V., Meile L., Schwenninger S.Miescher. Influence of two anti-fungal Lactobacillus fermentum-Saccharomyces cerevisiae co-cultures on cocoa bean fermentation and final bean quality. PLoS ONE. 2020;15 doi: 10.1371/journal.pone.0239365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Rodríguez A., Rocha D., Ruiz-Villafan B., Tierrafría V., Rodríguez-Sanoja R., Segura-González D., Sánchez S. Transcriptomic analysis of a classical model of carbon catabolite regulation in Streptomyces coelicolor. BMC Microbiol. 2016;16:77. doi: 10.1186/s12866-016-0690-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansinenea E., Ortiz A. Secondary metabolites of soil Bacillus spp. Biotechnol. Lett. 2011;33:1523–1538. doi: 10.1007/s10529-011-0617-5. [DOI] [PubMed] [Google Scholar]
- Scherlach K., Hertweck C. Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 2009;7:1753–1760. doi: 10.1039/b821578b. [DOI] [PubMed] [Google Scholar]
- Sridevi A., Sandhya A., Ramanjaneyulu G., Narasimha G., Devi P.S. Biocatalytic activity of Aspergillus niger xylanase in paper pulp biobleaching. 3 Biotech. 2016;6:165. doi: 10.1007/s13205-016-0480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidler L. Live genetically modified bacteria as drug delivery tools: at the doorstep of a new pharmacology? Expert Opin. Biol. Ther. 2004;4:439–441. doi: 10.1517/14712598.4.4.439. [DOI] [PubMed] [Google Scholar]
- Steinberg R.A. Sulfur and trace-element nutrition of Aspergillus niger. J. Agr. Res. 1941;63:109–127. [Google Scholar]
- Taha M., Shahsavari E., Al-Hothaly K., Mouradov A., Smith A.T., Ball A.S., Adetutu E.M. Enhanced biological straw saccharification through coculturing of lignocellulose-degrading microorganisms. Appl. Biochem. Biotechnol. 2015;175:3709–3728. doi: 10.1007/s12010-015-1539-9. [DOI] [PubMed] [Google Scholar]
- Vieira F.C., Nahas E. Comparison of microbial numbers in soils by using various culture media and temperatures. Microbiol. Res. 2005;160:197–202. doi: 10.1016/j.micres.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Wakefield J., Hassan H.M., Jaspars M., Ebel R., Rateb M.E. Dual induction of new microbial secondary metabolites by fungal bacterial co-cultivation. Front. Microbiol. 2017;8:1284. doi: 10.3389/fmicb.2017.01284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Chen X., Chio C., Yang C., Su E., Jin Y., Cao F., Qin W. Delignification overmatches hemicellulose removal for improving hydrolysis of wheat straw using the enzyme cocktail from Aspergillus niger. Bioresour. Technol. 2019;274:459–467. doi: 10.1016/j.biortech.2018.12.029. [DOI] [PubMed] [Google Scholar]
- Xing S., Li G., Sun X., Ma S., Chen G., Wang L., Gao P. Dynamic changes in xylanases and β-1, 4-endoglucanases secreted by Aspergillus niger An-76 in response to hydrolysates of lignocellulose polysaccharide. Appl. Biochem. Biotechnol. 2013;171:832–846. doi: 10.1007/s12010-013-0402-0. [DOI] [PubMed] [Google Scholar]
- Yeager C.M., Dunbar J., Hesse C.N., Daligault H., Kuske C.R. Polysaccharide degradation capability of Actinomycetales soil isolates from a semiarid grassland of the Colorado plateau. Appl. Environ. Microbiol. 2017;83 doi: 10.1128/AEM.03020-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuroff T.R., Xiques S.B., Curtis W.R. Consortia-mediated bioprocessing of cellulose to ethanol with a symbiotic Clostridium phytofermentans/yeast co-culture. Biotechnol. Biofuels. 2013;6:1–12. doi: 10.1186/1754-6834-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.