Graphical abstract
Keywords: G protein-coupled receptor, Heterobivalent ligand, Accelerated molecular dynamics, Oligomerization
Highlights
-
•
Bivalent ligand;
-
•
modulates conformational preferences of and correlations among microswitches,
-
•
strengthens interaction between G protein and the receptor and also water channel formation.
Abstract
Development of effective bivalent ligands has become the focus of intensive research toward modulation of G protein-coupled receptor (GPCR) oligomers, particularly in the field of GPCR pharmacology. Experimental studies have shown that they increased binding affinity and signaling potency compared to their monovalent counterparts, yet underlying molecular mechanism remains elusive. To address this, we performed accelerated molecular dynamics simulations on bivalent-ligand bound Adenosine 2A receptor (A2AR) dimer in the context of a modeled tetramer, which consists of A2AR and dopamine 2 receptor (D2R) homodimers and their cognate G proteins. Our results demonstrate that bivalent ligand impacted interactions between pharmacophore groups and ligand binding residues, thus modulating allosteric communication network and water channel formed within the receptor. Moreover, it also strengthens contacts between receptor and G protein, by modulating the volume of ligand binding pocket and intracellular domain of the receptor. Importantly, we showed that impact evoked by the bivalent ligand on A2AR dimer was also transmitted to apo D2R, which is part of the neighboring D2R dimer. To the best of our knowledge, this is the first study that provides a mechanistic insight into the impact of a bivalent ligand on dynamics of a GPCR oligomer. Consequently, this will pave the way for development of effective ligands for modulation of GPCR oligomers and hence treatment of crucial diseases such as Parkinson’s disease and cancer.
1. Introduction
G protein-coupled receptors (GPCRs) mediate most of our physiological responses to stimulants by coherent action of various modulators such as orthosteric/allosteric ligands, membrane and receptor partners, collective interaction of which leads to formation of GPCR oligomers. In these supramolecular structures, continuous information flow among the protomers transforms GPCR homo/heteromers into allosteric hubs, thus altering functional behavior of individual receptors. Over the last decades, it has been thought that the supramolecular structure, which is composed of a GPCR homodimer and G protein, constitutes the main functional unit of GPCR signalosomes [1], [2], [3], [4]. Indeed, a recent experimental study has shown that a GPCR heterotetramer is comprised of homodimers of A2AR and D2R along with their cognate G proteins [5].
GPCRs are involved in pathologies of many crucial diseases such as Parkinson‘s disease (PD), Alzheimer’s disease (AD), diabetes, and cancer, thus making them hot targets in drug discovery studies [6], [7], [8], [9]. On the other hand, allosteric interactions present in GPCR oligomers make targeting this class of proteins challenging in the field of GPCR pharmacology [10]. As a notable example, PD can be considered, which is caused by the loss of neurons that produce dopamine in substantia nigra [11]. As an effective therapy, dopaminergic agonists have been considered to increase dopamine level but remained insufficient [12], [13], [14]. It was after the discovery of antagonistic impact of A2AR on D2R, where A2AR decreases affinity and intrinsic efficacy of dopamine at D2R protomer [15], A2AR antagonists have been used as combination therapy along with D2R agonists in treatment of PD [16]. Recently, it has been shown that A2AR-D2R assemble into a tetrameric structure which is composed of a pair of this dimer. Moreover, it has been also demonstrated that simultaneous occupation of both A2AR protomers by an agonist and an antagonist in the tetramer does not induce an allosteric modulation of D2R agonist binding and intrinsic efficacy [5]. Therefore, this suggests that effective modulation of GPCR oligomers requires simultaneous targeting of individual receptors. Also, this shows that simultaneous occupation of one of the dimers in the tetramer impacts the function of the other dimer as well. Hence, bivalent ligands have emerged as useful tools to simultaneously target receptor dimers within the oligomer [17], [18]. A bivalent ligand is composed of two pharmachopores, which are covalently linked by a spacer group and can be categorized into two groups: homo- and heterobivalent ligands which consist of same and different pharmacophore groups, respectively. There has been a couple of experimental studies where the optimum length of the spacer and stability of the bivalent ligand were determined using computational tools, and their biological activities were also tested by in vitro studies [19], [20]. However, none of them provide a mechanistic insight into the impact of these special class of ligands on structure and dynamics of receptors which is crucial for development of effective ligands with precise targeting capabilities.
In this study, we investigated impact of a designed bivalent ligand on dynamics of A2AR dimer in the context of a tetramer, which has been shown to form the signaling unit pertaining to A2AR-D2R oligomer [5] using accelerated molecular dynamics simulations. To do so, we performed simulations on the tetramer model in the presence and absence of the linker (including only pharmacophore groups, but not the spacer and biphenyltriazole moiety) [21], [22], [23]. We investigated how i) conformational preferences of key residues that are involved in receptor activation, ii) allosteric interaction network, iii) membrane interactions of the receptors, iv) water channel formation, and v) coupling with effector G protein are modulated by the bivalent ligand. This study provides a mechanistic insight into the impact of a bivalent ligand on dynamics of the GPCR oligomer, thus assigning additional roles to linkers as modulators of GPCR oligomers.
2. Materials and methods
2.1. Tetramer modeling
The crystal structure of the tetramer is not available but interaction interfaces in homo- and heterodimers were revealed in a recent experimental study [22]. According to that, i) homodimer and heterodimer interfaces were built up of TM6 and TM4/TM5, respectively, and ii) agonist-bound protomers complex with effectors were located at the periphery of the tetramer to accommodate G proteins. Before modeling the dimers, first receptor monomers were modeled. In particular, CGS-21680-bound A2AR mini Gs complex, we have used crystal structures of NECA-bound A2AR mini Gs complex (with RAS-like domain)(PDB ID:55G53) [24] as a template since NECA shares similar chemical groups with CGS-21680. In fact, the crystal structure of CGS-21680-bound A2A (PDB ID:44UHR) [16] is available but it cannot accommodate G protein because of the narrower opening of the intracellular site of the receptor. Therefore, we aligned the two structures and observed that residues that interact with the ligand were aligned well. Consequently, CGS-21680 was transferred from 4UHR to 5G53. For modeling istradefylline-bound A2AR, we used crystal structure of A2AR bound with ZM241385 (PDB ID:44EIY) [25] since it shares similar chemical structure with the target ligand. The Na+ ion which is present in allosteric site of antogonist bound A2AR was kept in our construct from the original crystal structure of 4EIY. We performed induced fit docking, which is implemented in Schrodinger’s Glide docking software [26], [27] to find best possible pose for istradefylline within the binding pocket using the hydrogen bond as a restraint with ASN2536.55 (superscripts denote Ballesteros-Weinstein numbering) [28] residue of the receptor.
As to the D2R dimer, we used crystal structure of D2R bound with risperodine (PDB ID:66CM4) [29] for modeling apo D2R by removing T4 lysozyme which is fused to ICL3 of the receptor. The crystal structure of -opioid receptor complex with mini Gi (PDB ID:6DDF) [30] was used for modeling quinpirole-bound D2R-Gi complex since the crystal structure of D2R-Gi complex was not available at the time of simulations were done. The sequence similarity between -opioid and D2R was 35%. After it’s release, we also compared backbone root mean square deviation (RMSD) between 6DDF and 6VMS (D2R complex with Gi) and found to be 1Å. To find the best possible pose of the ligand we performed induced fit docking by using the hydrogen bond as a restraint with ASP1143.32 residue of the receptor. Herein, it is important to emphasize that D2R has a long ICL3 which is not resolved in crystal structures. We modeled the missing regions as a loop by using the first 60 amino acids of ICL3 to prevent any restraint that may be introduced upon smaller length of loops.
For modeling dimers, we downloaded all the available crystal structures of GPCRs from G protein-coupled receptors database (GPCRdb) (245 structures were present as of April, 2018) to use as templates for generating corresponding homo- and heterodimer interfaces in the tetramer. First, the crystal mates were generated in Maestro, and then, receptor monomers were aligned to corresponding monomers of the templates. In particular, we could get 38 templates with TM4/TM5 and 2 templates with TM6/TM6 dimerization interfaces, the list of which can be found in the Supplementary Table. In fact, various different orientations of the monomers in the dimer can satisfy those interfaces but we made use of the experimental data [22] to further pick up the biologically relevant interfaces. Accordingly, the crystal structures with PDB IDs:5WIU [30] and 5O9H [31] were used to model TM6/TM6 and TM4/TM5 interfaces, respectively. Slight steric clashes in the models were alleviated by using Prime module of Maestro program.
2.2. System preparation
Protein structure preparation was performed using ”Protein Preparation Wizard” implemented in Maestro molecular modeling package [32]. Non-protein entities like BRIL, which is included in the crystal structure of ZM241385-bound A2AR (PDB ID:44EIY), was removed and the missing region was filled by original residues of the receptor using “Cross-Link Protein” module implemented under Bioluminate [33] panel of Schrondinger software. Also, missing regions present either in receptors or in mini G proteins were filled similarly. Missing hydrogen atoms were added using PROPKA [34] to maintain protonation state of residues at pH:7.0. H-atoms were optimized and constrained energy minimization was carried out on the final tetramer model was obtained. The tetramer was aligned in the membrane using Orientations of Proteins in Membrane (OPM) server [35], [36] and then prepared using Membrane Bilayer builder program [37] implemented in CHARMM-GUI server [38]. The membrane was prepared to include 30% cholesterol and 70% sphingomyelin molecules to mimic lipid rafts, thus resulting with a total number of ca 500000 atoms. TIP3P [39] and CHARMM36m force fields [40] were used to model water molecules and protein in the system. Ligand parametrization was done using CGenFF, implemented in CHARMM-GUI server [41]. The output files were examined to check if penalty values associated with ligands exceeded the threshold level but they didn’t exceed.
2.3. Bivalent ligand construction
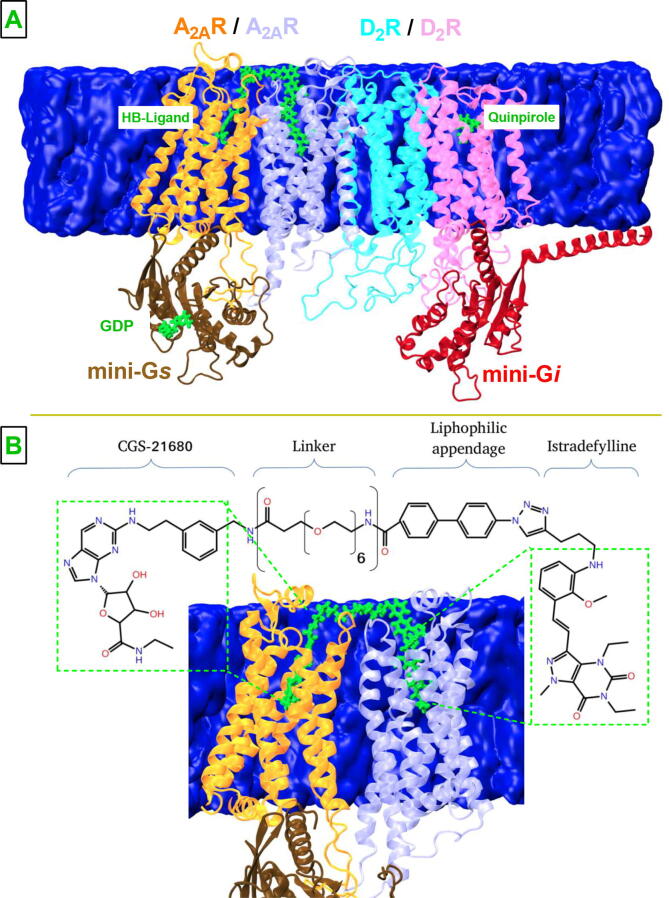
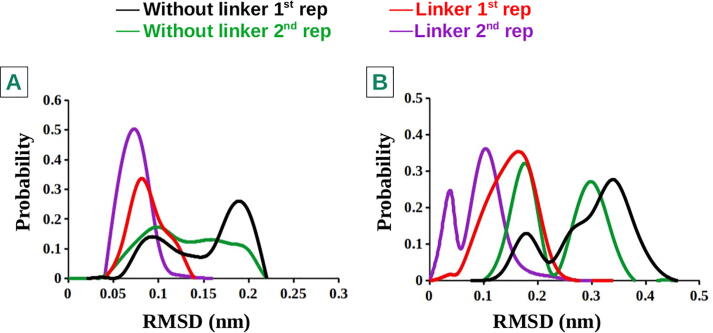
The bivalent ligand was modeled to consist of two pharmacophore groups, namely, istradefylline and CGS-21680 along with the linker, in accordance with the experimental data [5]. The linker, which was shown to bind to a heterodimer [21], was composed of an affinity-generating biphenyltriazole-moiety along with a spacer group. After modeling the tetramer, it was examined to determine possible regions on the pharmacophore groups to which the linker was attached. Consequently, methoxy groups of istradefylline and carboxyl group of CGS-21680 were found to be accessible from the extracellular domain of the receptors and the linker was added to these regions using “build” panel of Schrodinger software. The final chemical composition of the linker was determined by considering possible synthesis opportunities of the bivalent ligand. In addition, to determine the optimum length of the linker we performed MD simulations on the structures having different linker lengths for 250 ns. Analysis of these trajectories showed that linker lengths that were shorter than 48Å were unstable and left the ligand binding pocket during the simulation (See SI Movie). Importantly, as the length was further increased, the RMSD of the linker tended to decrease until the length of 52Å. After that value, we did not observe any further decrease in the RMSD, hence used that linker in our further simulations [21] (See Fig. 1).
Fig. 1.
A2AR/D2R tetramer with bivalent ligand A. Schematic representation of the system used in this study. The membrane is shown in blue. A2AR protomers and mini-Gs are shown in orange, purple and brown, respectively and new cartoon representation whereas bivalent ligand is shown in green and licorice representation. Water molecules are omitted for the sake of simplicity. D2R protomers and mini-Gi are shown in pink, cyan, and red, respectively. B. Representation of the chemical structure of the linker. The pharmacophore groups and the linker are shown in detail together with A2AR dimer.
2.4. Molecular dynamics simulation
In this study, we performed both classical (cMD) and accelerated molecular dynamics (aMD) simulations using Nanoscale Molecular dynamics NAMD [42] package. For aMD, ”dual-boost” was chosen to enhance sampling of the conformational space and performed by getting average potential energy values from cMD simulations. Specifically, the systems studied were heated from 0 K to 310 K within 1 ns and pre-equilibrated using NVT ensemble at 310 K for 50 ns. This was followed by 500 ns cMD simulations which were performed using NPT ensemble at 1 atm and 310 K to get average potential energies, from which the acceleration parameters were calculated. The last snapshots of these simulations were used as starting conformations in aMD simulations and aMD simulations were performed for 1s. Nose–Hoover [43] and Parrinello-Rahman [44] coupling algorithms were used to maintain constant temperature and pressure, respectively throughout simulations. The integration step of 2fs was used in all simulations performed. In the dual-boost aMD, a dihedral potential and a potential boost were added to all the atoms in the systems. The dihedral and potential boost acceleration parameters were calculated using following equations:
| (1) |
| (2) |
| (3) |
| (4) |
where Natoms is the total number of atoms, and and are the average dihedral and potential energies obtained from cMD simulations. is an adjustable acceleration parameter. We used 0.3 for which was shown to perform effectively for GPCR systems [45]. The systems were simulated twice, each of which was simulated starting with a different initial velocity distribution. The reweighted PMF profiles were obtained using cumulant expansion to the 2nd order; however, reweighting caused a large energetic noise because of large system size of the system as shown in SI Fig. 1. Considering that the overall shape of free energy profiles is maintained in aMD simulations, unweighted data should provide an estimate of the free energy differences [46], [47]. Therefore, unweighted free energy profiles are presented in this study (See SI Fig. 1, Fig. 2 for detailed discussion [48], [49]).
Fig. 2.
Root Mean Square Deviation (RMSD) timeline plots. A. Antagonist bound A2AR. B. Agonist bound A2AR.
2.5. Cross-correlation analysis
To calculate the extent of correlation among residues, dynamic cross-correlation maps were obtained. According to that, normalized co-variance values of residues were computed by considering C atoms [50] and using the following equation:
| (5) |
where and represents the coordinates of the and atoms as a function of time indicates the time ensemble average, and . The correlation range of residues lies within −1.0 to 1.0, where positive correlated displacement falls between whereas anti-correlation displacement falls between .
2.6. Network construction
Network analysis was carried out using PSN-Ensemble Software [51] by considering side chain atoms of the residues in A2AR receptor. As an input, two residues were provided as the source and the sink Glu1694.90 and Ile2927.57, which were distant from each other on the protein and between which allosteric communication signal was generated. Herein, cross correlation matrices were used as inputs to calculate and weigh the shortest communication pathways used by the receptor in the presence and absence of the bivalent ligand.
2.7. Binding volume calculation
Ligand binding and G protein binding volumes were calculated using KvFinder plugin of pymol, which is based on geometrical grid-based method combined with space segmentation capabilities [52]. The space defined between molecular surfaces of the two probes is known as the cavity. The probe-in was set to 0.8Å and probe-out was set to 8.0Å with volume of cavity as 200ÅḞor ligand binding pocket Ile161.42, Ala592.57, Thr682.66, His753.23, Val863.34, Phe1835.44, Trp2466.48, Phe2586.60 and His2787.43 were selected and Ser351.61, Gln381.64, Pro1093.57, Ile2005.61, Ala2045.65, Gln2266.27 and Arg2937.58 residues were selected for calculating the volume of G-protein binding pocket.
3. Results
3.1. Optimum linker length is required for stable binding of bivalent ligand to A2AR dimer
As explained in the Methods section tetramer was modeled to consist of dimers of A2AR and D2R along with their corresponding effectors, mini-Gs and mini-Gi, respectively, to be in line with experimental data [5]. In a follow-up study of Navarro et.al [22], homodimer interfaces were shown to consist of TM6 whereas heterodimer interface was shown to consist of TM4/TM5. According to the same study, it was also suggested that protomers which were bound to G protein should be located on the periphery of the quaternary structure to prevent clashes between two G proteins whereas apo D2R and antagonist-bound A2AR were found in the middle. To identify possible attachment points of pharmacophores with the linker, crystal structures of CGS-21680 (PDB ID4UHR) [16] and ZM241385-bound A2AR (PDB ID4EIY) [25] were used. Examination of these structures revealed that methoxy groups of istradefylline and carboxyl group of CGS-21680 were accessible from the extracellular domain of the receptors. Therefore, these two points were linked to each other using an affinity generating biphenyltriazole-moiety and spacer as shown in Fig. 1. Since biphenyltriazole-moiety acts as an affinity-generating group it is anticipated that inclusion of this group in the linker might shorten the time required to observe any possible impact of the linker on the dynamics of the system [60], [61]. To determine the optimum linker length we performed classical molecular dynamics (cMD) simulations with bivalent ligands having different number of spacer units. In fact, we measured the minimum distance (beeline) between the two attachment points of the linker as 45 Å. However, our results showed that bivalent ligand which was shorter than 48Åcould not stably bind to the dimer (See Supplementary Movie) which was in correspondence with a study of Hubner et.al [21]. The reason why the linkers with distance shorter than 48Åcould not stably bind was that they clashed with extracellular loops of the protomers, which were protruded from the receptors. It is also important to emphasize that the linker stabilizes pharmacophores compared to their monovalent counterparts as revealed by smaller RMSD values measured in the presence of the linker (See Fig. 2).
3.2. Extra- and intracellular domains are stabilized and global dynamics is restricted by the linker in A2AR dimer
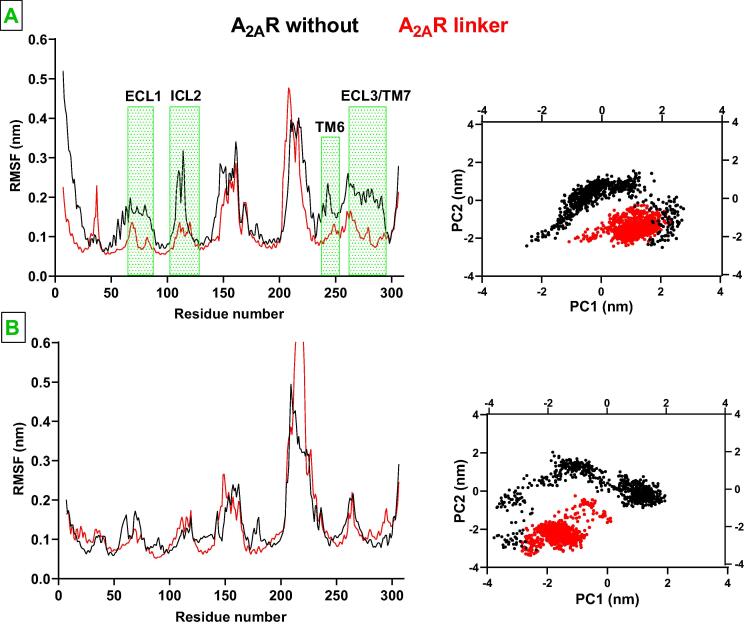
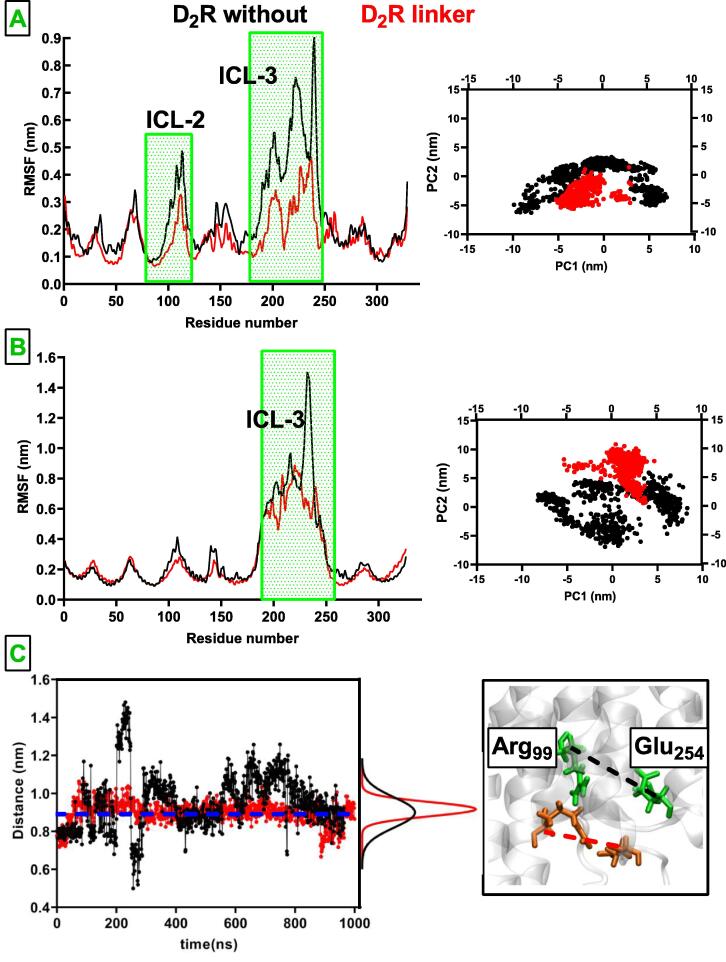
The bivalent ligand was built to composed of an A2AR antagonist (istradefylline) and an agonist (CGS-21680), where affinity-generating biphenyltriazole moiety was connected to istradefylline. Comparative analyses of the trajectories showed that the linker made contacts with extracellular loop (ECL) 1 and 3, thus decreasing fluctuation at these regions in antagonist-bound A2AR as shown in Fig. 3. Specifically, side chains of Thr682.56 and Asp2616.63/Ser2636.65, which are located on ECL1 and ECL3, respectively made hydrogen bonds with nitrogen atoms located on the linker. Moreover, the linker also decreased conformational fluctuation at intracellular loop (ICL) 2, TM1, TM6 and TM7, last two of which were included at homodimer interface. On the other hand the linker did not impact fluctuations in agonist-bound A2AR/mini-Gs complex. Here, it is important to emphasize that the impact of the linker on agonist-bound A2AR/mini-Gs was reflected in the number of contacts made between the ligand and certain residues of ligand binding pocket of receptor. It was increased by 50% in agonist-bound A2AR/mini-Gs while remained the same in antagonist-bound receptor. Specifically, contacts between Trp2466.48 and Ile2747.39, which have been implicated in agonist binding, and CGS-21680 increased in the presence of the linker. Having observed that the linker modulated fluctuation pattern of residues (See Fig. 3B) we also set out to investigate its impact on the global dynamics of A2AR protomers. Results showed that global motion of the receptors was confined in the presence of the ligand as they sampled narrower region on the conformational space, which was revealed by the first and the second eigenvectors of the systems (See Fig. 3).
Fig. 3.
Root mean square fluctuation (RMSF) and 2D principal component analysis (PCA) profiles pertaining to A. antagonist-bound A2AR and B. agonist-bound A2AR/mini-Gs. The regions that showed difference between without and with linker systems are indicated with green rectangles. The RMSF and 2D-(PCA) plots pertaining to 2nd replicate are given in SI Fig. 4.
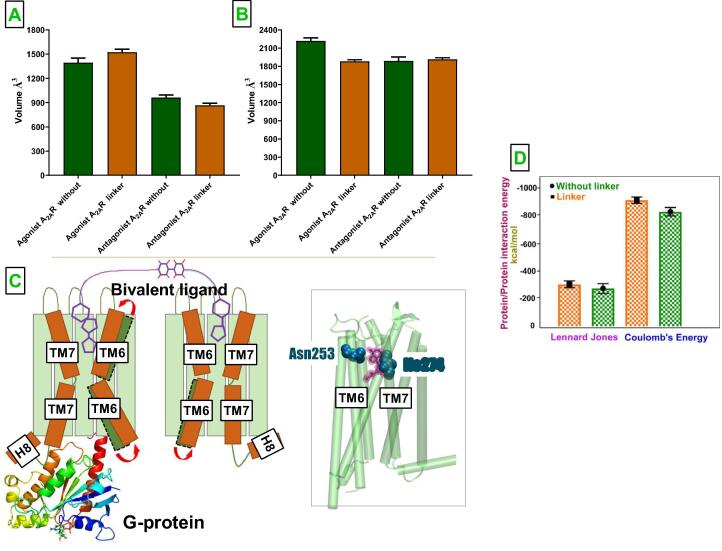
3.3. The linker modulates volume of ligand binding pocket and intracellular domain in A2AR dimer
We set out to investigate possible impact of the linker on structure of ligand binding pocket as the number of contacts made between CGS-21680 and receptor increased in the presence of the linker. Interestingly, we observed that volume of the ligand binding pocket was decreased by in CGS-21680- bound A2AR while this change was not significant for istradefylline-bound A2AR protomer (See Fig. 4A). Moreover, we also measured the volume of the G protein binding site of A2AR protomers to investigate if the change observed in the ligand binding pocket was transmitted to the intracellular domain of the receptors. Indeed, the volume of intracellular domain was increased by in CGS-21680-bound A2AR/mini-Gs (See Fig. 4B). With that, agonist-bound A2AR could achieve such volume range, which was shown to be sampled by active receptors, [53] in the presence of the linker. On the other hand, the volume of the intracellular domain in the absence of the linker was closer to the value which was sampled by the receptor found in an intermediate state between the inactive and active receptor [53]. This change was also reflected in the ionic lock distances as well such that longer distance was observed in agonist-bound A2AR in the presence of the linker, thus having wider intracellular domain. Here, it is important to emphasize that the volume of G protein binding site in istradefylline-bound A2AR decreased despite the absence of a significant change in the volume of ligand binding pocket (See Fig. 4, Fig. 4B). Since istradefylline-bound A2AR was placed between CGS-21680- bound A2AR/mini-Gs and apo D2R in the tetramer (See Methods for detailed information), expansion of the intracellular domain of neighboring A2AR might confine the space available for antagonist-bound A2AR protomer. Here in, we need to elaborate that expansion of intracellular domain of agonist-bound A2AR also impacted interactions between the receptor and mini-Gs: Coulomb energy decreased (became more favorable) by 10% in the presence of the linker, whereas the change in Lennard Jones energy was not significant between without and linker systems.
Fig. 4.
Changes in volumes of A. ligand binding pocket and B. G protein binding domain between without and linker systems C. Schematic representation of conformational changes adapted by A2AR receptors in the presence of the linker. The position of transmembrane (TM) 6 and 7 in the absence and presence of the bivalent ligands is shown in green and orange, respectively. Mini-Gs that is bound to agonist-bound A2AR is shown in new cartoon representation. Right hand side Ile2747.39 interaction with ligand emerge upon adding the linker. D. Protein–protein interaction energies measured between G protein and the receptors are also given.
3.4. Conformational preferences of microswitches and residue correlations are modulated by the linker in A2AR dimer
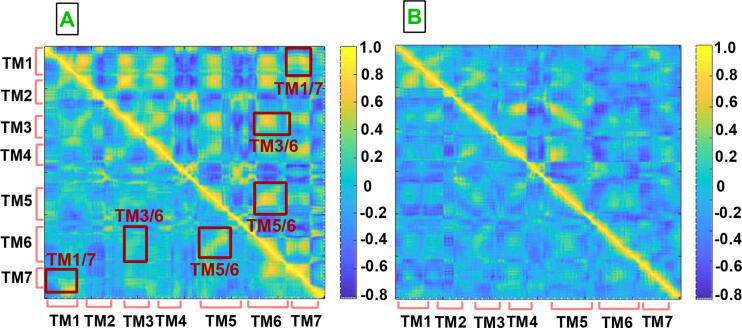
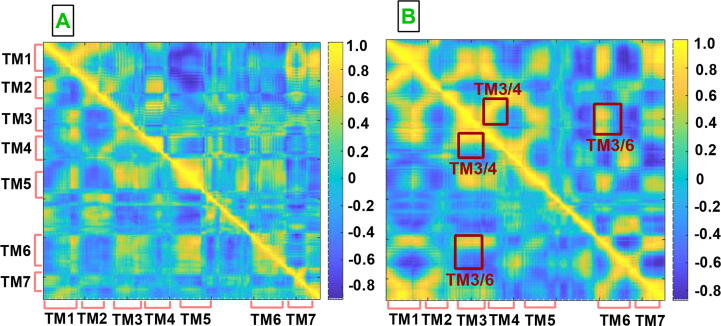
Reciprocal changes observed in the volumes of both ligand binding pocket and G protein binding site of CGS-21680-bound A2AR in the presence of the linker suggested alterations in correlation of interactions at certain TM domains. To corroborate such impact, we calculated dynamics cross correlation matrices for the systems studied (See Fig. 5) and showed that strength of correlated motion between TM1 and TM3, TM1 and TM7, TM5 and TM6 as well as TM6 and TM7 was increased in the presence of the linker. However, the strength of anti-correlated motion was increased at TM3/TM6, all of which were occurred upon receptor activation. That is to say, increment in strength of anti-correlated and correlated motion in TM3/TM6 and TM5/TM6 pairs, respectively, triggered wider opening of the intracellular domain of the receptor, which was also reflected in the volume of G protein binding site at CGS-21680-bound A2AR. This observation was also in line with a recent study where shrinking of the volume of orthosteric ligand binding site and expansion of the intracellular domain of the receptor were shown to be associated with similar changes occurring in TM3, TM6 and TM7 [54].
Fig. 5.
Dynamic cross correlation maps (DCCM) A. agonist-bound A2AR/mini-Gs and B. antagonist-bound A2AR. The upper triangles in A and B correspond to DCCM of the system with linker whereas the lower triangles correspond to those without linker. The DCCM plots are obtained by combining both replicas, and the DCCM plots of each replica is given in SI Fig. 7.
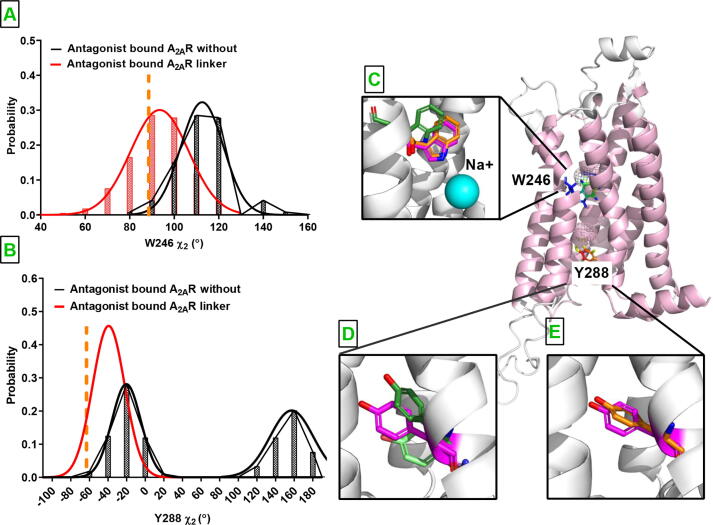
Moreover, these rearrangements also led to stabilization of side chains of certain residues at istradefylline-bound A2AR in the presence of the linker. Importantly, Trp2466.48 adopted 2 angle values which provided close packing of the residue against the sodium ion as seen in the crystal structure of inactive A2AR (PDB ID:4EIY). Also, hydroxyl group of Tyr2887.53, which is involved in G protein binding, is positioned farther from the G protein binding site (See Fig. 6) than in without linker system and crystal structure of inactive A2AR (PDB ID:4UHR) [55].
Fig. 6.
Angle probability distribution of Trp246 and Tyr288. A. 2 of Trp2466.48 is presented for antagonist-bound A2AR in the absence and presence of the linker, in black and red, respectively. The corresponding angle is also measured in crystal structure of antagonist-bound A2AR (PDB ID:44EIY) and indicated with dashed orange line. B. The same as in A but for 2 of Tyr2887.53. Comparison of orientation of C. Trp2466.48 in the absence (green) and presence (orange) of the bivalent ligand. Oreintations of Tyr2887.53 in the D absence of linker (dark and light green) and E. presence of linker (orange). The orientation of Trp6.48 and Tyr7.53 in the crystal structure of antagonist-bound receptor is shown in pink. The residues are shown on the 3D structure of antagonist bound A2AR. Timeline plots of Trp2466.48 and Tyr2887.53 of both replicates are given in the SI Fig. 6.
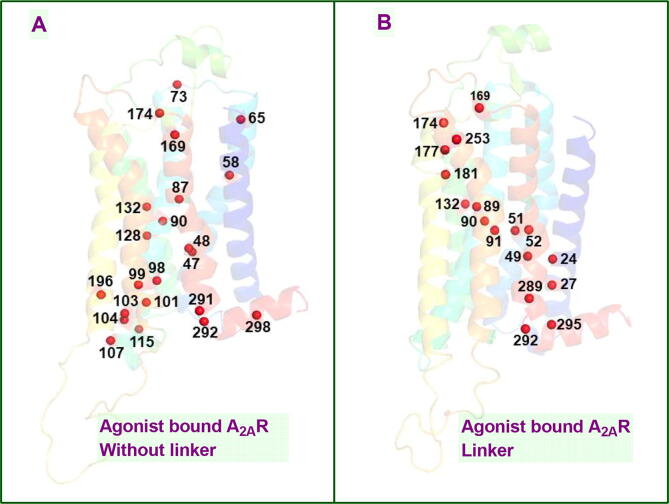
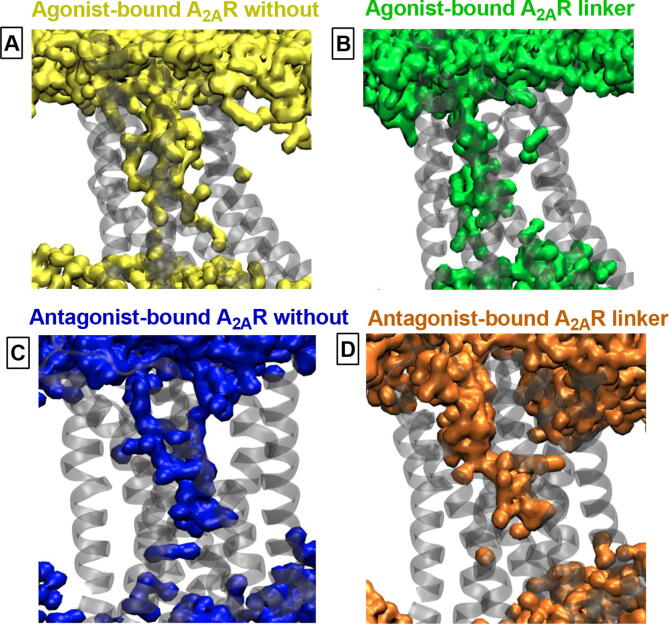
Besides residue correlations, the bivalent ligand also impacted allosteric interaction network in agonist-bound protomer in A2AR dimer. Specifically, TM5 emerged as one of the most significant domain that participated in the interaction network in the presence of the linker. Recalling coupling between orthosteric ligand binding pocket and intracellular domain in agonist-bound A2AR, it can be said that the result was in correspondence with a study where allosteric coupling between extra- and intracellular domain of the receptor was shown to be mediated through TM5 [56]. Moreover, Asn2536.55 residue, which is one of the key residues of the ligand binding pocket that anchors the exocyclic amine of the ligand’s central core, participated to allosteric interaction network in the presence of the linker and interestingly, TM3 didn’t contribute in the presence of the linker (See Fig. 7). We also investigated interactions between A2AR dimer and membrane and showed that Trp1294.50, which was shown to be part of the cholesterol consensus motif, of agonist-bound A2AR made close contacts with a cholesterol molecule in the presence of the linker in one of the replicates [57] (See SI Fig. 9). Therein, the cholesterol moved by almost 5Å throughout the trajectory from its original position and eventually contacted to the receptor, whereas no such interaction was observed for the antagonist-bound A2AR neither in the presence nor the absence of the linker. Lastly, we also compared pattern of water channels formed within the receptors between without and linker systems using the VolMap plugin of VMD. In accordance with a study which showed a correlation between receptor activation and continuous water channel formation [53], we observed a continuous water channel in agonist-bound A2AR whereas it was disrupted in antagonist-bound A2AR (See Fig. 8). Interestingly, continuous water channel could be also maintained in the presence of the linker in agonist-bound receptor; however, water density was decreased in the region between the extracellular domain and orthosteric ligand binding pocket as observed in antagonist-bound A2AR. Analysis of the trajectories showed that the linker precluded water flow from extracellular side to the inside of the receptor. Consequently, water molecules could be clustered only around Na+ ion in antagonist-bound receptor.
Fig. 7.
A2AR Network Pathway.
Fig. 9.
Root mean square fluctuation (RMSF) and 2D principal component analysis (PCA) profiles pertaining to A. apo D2R and B. agonist-bound D2R/mini-Gi. The regions that showed difference between absence and presence of the linker are indicated with green rectangles. C. Time-line ionic lock distance sampled by apo D2R in the presence (red) and absence (black) of the linker. The ionic lock distance measured in the crystal structure of eticlopride-bound D2R (PDB ID:33PBL) is indicated with blue dash line. The RMSF and 2D-(PCA) plots of 2nd replicate is given in SI Fig. 5.
Fig. 8.
Internal Water Channel formation. A. Agonist-bound A2AR, and B. Antagonist-bound A2AR in the absence of the linker, C. Agonist-bound A2AR, and D. Antagonist-bound A2AR in the presence of the linker. Water occupancy maps are computed using the VolMap tool of VMD.
3.5. The linker impacts the dynamics of apo D2R in the neighboring D2R dimer
The tetramer is composed of pairs of A2AR and D2R, where the homodimer interfaces are formed by TM6 and the heterodimer interface is formed by TM4 and TM5. In accordance with experimental data, one of the D2R protomers was modeled as apo and the other protomer was modeled to bound with D2R agonist, quinpirole and mini-Gi (See Methods for more details). The results showed that ICL2, albeit to a lesser extent, and ICL3 were stabilized in apo D2R in the presence of the linker while only a small part of the ICL3 could be stabilized in agonist-bound D2R/mini-Gi(See Fig. 9). Moreover, similar to A2AR dimer, the linker also confined global dynamics of the protomers as revealed by principal component analysis as shown in Fig. 9. Interestingly, the linker also modulated residue correlations at apo D2R despite not being directly bound to the receptor. Specifically, an increase was observed in the strength of correlated motion in residues located on TM3 and TM4, two of which construct the heterodimer interface with antagonist-bound A2AR. Moreover, an increase was also observed in the correlated motion of TM3 and TM6 as well as TM3 and TM4 in the presence of the linker in apo D2R indicating that they tend to get closer to each other as shown in (Fig. 10B).
Fig. 10.
Dynamic cross correlation maps (DCCM) are presented for A. agonist-bound D2R/mini-Gi and B. apo D2R. The upper diagonal in A and B correspond to DCCM of the system with linker whereas the lower diagonal correspond to those in the absence of linker. The DCCM plots are obtained by combining both replicas, and the DCCM plots of all the systems are given in SI Fig. 8.
Herein, it is also important to emphasize that the intracellular domain of apo D2R was modulated by the linker as the receptor could sample a wider range of ionic lock distances which are calculated between Arg1323.50 (C) and Glu3686.30 (C) atoms [58] in the absence of the linker whereas it could only sample shorter distances in the presence of the linker.
4. Discussion
Bivalent ligands have long been thought to act solely as connectors of the two pharmacophore groups. On the other hand, experimental studies have shown that binding affinity and signaling potency are increased by bivalent ligands compared to their monovalent counterparts. Consequently, this suggests that these ligands are capable of modulating dynamics of GPCR oligomers; however, underlying mechanism has remained elusive. In this study, to the best of our knowledge, for the first time, we provided a molecular level understanding to dynamics and functional changes induced by the bivalent ligand. In particular, we demonstrated that the linker made interactions with residues located at the extracellular domains of A2AR protomers. Notably, this has led to stabilization of antagonist-bound A2AR while that impact was not remarkable for agonist-bound A2AR/mini-Gs as revealed by root-mean-square fluctuation patterns. Interestingly; however, this caused an increase in the number of contacts made between ligand binding residues and CGS-21680 in the mini-Gs bound A2AR receptor but not in the antagonist-bound A2AR. In accordance with this, important ligand binding residues, namely M1775.38 ASN2536.55 [59], also emerged on the allosteric pathway that links the ligand binding pocket to the intracellular domain of CGS-21680-bound A2AR in the presence of the linker. Therefore, these results suggest that the coupling between the ligand binding pocket and the intracellular domain of the receptor increased by the bivalent ligand. In accordance with that, volume of the ligand binding pocket was shrunk while the intracellular domain was expanded in agonist CGS-21680-bound A2AR/mini-Gs. In spite of having no remarkable change at the ligand binding site, the volume of the intracellular domain was decreased in istradefylline-bound A2AR. Importantly, modulation of the volume of the G protein binding site impacts contacts formed between intracellular domain and the effector as revealed by an increase in the respective energy term. Moreover, the decrease seen in the intracellular domain of antagonist-bound A2AR might help prevent effector binding to the receptor. Herein, it is also important to emphasize that conformational preferences of some of the microswitches were modulated by the linker. Specifically, conserved Trp2466.48 adopted side chain dihedral angles which provided residue 14 to be tightly packed against the sodium ion in antagonist-bound A2AR. Also, another conserved residue, Tyr2887.53, was oriented far from G protein binding site therein. On the other hand, no such alterations were observed in conformational preferences of these key residues in agonist-bound A2AR mini Gs. We also showed that above mentioned conformational changes led to alterations in residue correlation patterns and allosteric interaction network in the A2AR dimer. Specifically, we observed that conformational rearrangements which occurred during receptor activation were strengthened in the agonist-bound A2AR-mini Gs by the presence of linker. Moreover, TM5 was emerged as the dominant participant of the allosteric interaction network in agonist-bound A2AR-mini Gs in the presence of the linker. A similar trend was observed in A2AR upon complex formation with mini-Gs suggesting that impact of mini-Gs was accentuated by the presence of linker in CGS-21680-bound A2AR. Besides A2AR, the bivalent ligand also impacted apo D2R in the neighboring D2R dimer Notably, apo D2R could sample wider range of ionic lock distances in the absence of the linker, whereas the distance was confined to the value which was sampled by the inactive receptor in the presence of the linker. In a study of Han et.al [23], it was shown that inverse agonist binding to one of the protomers in D2R dimer enhanced D2R signaling from agonist-bound D2R. In addition, it has been also shown in a recent study that simultaneous binding of A2AR agonist/antagonist to A2AR dimer in A2AR/D2R heterotetramer alleviated well-known antagonistic impact of A2AR on D2R signaling. Since apo D2R resembles inactive receptor in the D2R dimer in the presence of the linker it is likely that the bivalent ligand decreases antagonistic impact of A2R on D2R signaling. From that perspective, the bivalent can be considered as a potential therapeutic molecule that can be used for treatment of Parkinson‘s disease, in which the efficacy of D2R agonists is alleviated by antagonistic impact of A2AR [23]. The findings of this study can provide a methodology that can be followed to investigate impact of any bivalent ligand on a GPCR oligomer, thus paving a way for improving design strategies of such ligands and effective modulation of GPCR oligomers.
Author contributions
Samman Mansoor and Ozge Sensoy designed research, Samman Mansoor and Gulru Kayik performed research; Samman Mansoor analyzed data; Samman Mansoor and Ozge Sensoy wrote the paper; Serdar Durdagi and Gulru Kayik edited the paper.
Data and software availability
Classical and accelerated molecular dynamics simulation were performed using NAMD simulation package. Softwares used in the study are owned by their respective developers. The trajectories pertaining to both systems linker and without linker are deposited in OSF repository which can be assessed by the linkhttps://osf.io/tnpgz/?view_only=944246c89c06401d967628b52d71752b
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
“The authors thank TUBITAK (reference number: 216S297) and COST Action CA15135, supported by COST (European Cooperation in Science and Technology) for providing funding. They also thank to TUBITAK ULAKBIM, High Performance and Grid Computing Center and Istanbul Medipol University for providing computational resources.”
Footnotes
Supplementary data associated with this article can be found, in the online version, athttps://doi.org/10.1016/j.csbj.2022.01.016.
Contributor Information
Samman Mansoor, Email: sammanmansoor9@gmail.com.
Gülru Kayık, Email: gulru.kayik@gmail.com.
Serdar Durdagi, Email: serdardurdagi@gmail.com.
Ozge Sensoy, Email: osensoy@medipol.edu.tr.
Supplementary data
The following are the Supplementary data to this article:
References
- 1.Bouvier M. Oligomerization of G-protein-coupled transmitter receptors. Nat Rev Neurosci. 2001;2:274. doi: 10.1038/35067575. [DOI] [PubMed] [Google Scholar]
- 2.Salahpour A., Angers S., Bouvier M. Functional significance of oligomerization of G-protein-coupled receptors. Trends Endocrinol Metab. 2000;11:163–168. doi: 10.1016/s1043-2760(00)00260-5. [DOI] [PubMed] [Google Scholar]
- 3.Jordan B.A., Devi L.A. G-protein-coupled receptor heterodimerization modulates receptor function. Nature. 1999;399:697. doi: 10.1038/21441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ilter M., Mansoor S., Sensoy O. Utilization of biased G protein-coupled receptor signaling towards development of safer and personalized therapeutics. Molecules. 2019;24:2052. doi: 10.3390/molecules24112052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonaventura J., Navarro G., Casadó-Anguera V., Azdad K., Rea W., Moreno E., Brugarolas M., Mallol J., Canela E.I., Lluús C., Cortés A. Allosteric interactions between agonists and antag- onists within the adenosine A2A receptor-dopamine D2 receptor heterotetramer. Proc Natl Acad Sci USA. 2015;27:3609–3618. doi: 10.1073/pnas.1507704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenkins B.G., Zhu A., Poutiainen P., Choi J.-K., Kil K.-E., Zhang Z., Kuruppu D., Aytan N., Dedeoglu A., Brownell A.-L. Functional modulation of G-protein coupled receptors during Parkinson disease-like neurodegeneration. Neuropharmacology. 2016;108:462–473. doi: 10.1016/j.neuropharm.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riddy D.M, Delerive P, Summers R.J, Sexton P.M, Langmead C.J. G. protein- coupled receptors targeting insulin resistance, obesity, and type 2 diabetes mellitus. Pharmacol Rev. 2018;70:39–67. doi: 10.1124/pr.117.014373. [DOI] [PubMed] [Google Scholar]
- 8.Arakaki A.K., Pan W.-A., Trejo J. GPCRs in cancer: protease-activated receptors, endocytic adaptors and signaling. Int J Mol Sci. 2018;19:1886. doi: 10.3390/ijms19071886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mansoor S, Kayik G, Alshahaby E, Guzel M, Durdagi S. O Sensoy. Development of novel therapeutic that can target multiple receptors for treatment of Parkinson’s disease. 2018;67 [Google Scholar]
- 10.Ferré S. The GPCR heterotetramer: challenging classical pharmacology. Molecules. 2015;36:145–152. doi: 10.1016/j.tips.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niethammer M., Tang C.C., Ma Y., Mattis P.J., Ko J.H., Dhawan V., Ei-delberg D. Parkinson’s disease cognitive network correlates with caudate dopamine. Neuroimage. 2013;78:204–209. doi: 10.1016/j.neuroimage.2013.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotzias G.C., Papavasiliou P.S., Gellene R. Modification of Parkinsonism–chronic treatment with L-dopa. N Engl J Med. 1969;280:337–345. doi: 10.1056/NEJM196902132800701. [DOI] [PubMed] [Google Scholar]
- 13.LeWitt P.A. Levodopa therapy for Parkinson’s disease: pharmacokinetics and phar- macodynamics. Mov Disord. 2015;30:64–72. doi: 10.1002/mds.26082. [DOI] [PubMed] [Google Scholar]
- 14.Loonen A.J., Ivanova S.A. New insights into the mechanism of drug-induced dyski- nesia. CNS Spectr. 2013;18:15–20. doi: 10.1017/s1092852912000752. [DOI] [PubMed] [Google Scholar]
- 15.Cieslak M., Komoszynski M., Wojtczak A. Adenosine A2A receptors in Parkinson’s disease treatment. Purinergic Signal. 2008;4:305. doi: 10.1007/s11302-008-9100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lebon G., Edwards P.C., Leslie A.G., Tate C.G. Molecular determinants of CGS21680 binding to the human adenosine A2A receptor. Mol Pharmacol. 2015;87:907–915. doi: 10.1124/mol.114.097360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soriano A., Ventura R., Molero A., Hoen R., Casadó V., Cortés A., Fanelli F., Albericio F., Lluis C., Franco R., Royo M. Adenosine A2A receptor- antagonist/dopamine D2 receptor-agonist bivalent ligands as pharmacological tools to detect A2A–D2 receptor heteromers. J Med Chem. 2009;52:5590–5602. doi: 10.1021/jm900298c. [DOI] [PubMed] [Google Scholar]
- 18.Stanton B.Z., Chory E.J., Crabtree G.R. Chemically induced proximity in biology and medicine. Science. 2018:359. doi: 10.1126/science.aao5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y., Duggineni S., Espitia S., Richman D.D., An J., Huang Z. T.A synthetic bivalent ligand of CXCR4 inhibits HIV infection. Biochem Biophys Res Commun. 2013;435:646–650. doi: 10.1016/j.bbrc.2013.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krall N., Pretto F., Neri D. T.A bivalent small molecule-drug conjugate directed against carbonic anhydrase IX can elicit complete tumour regression in mice. Chem Sci. 2014;24, 5:3640–3644. [Google Scholar]
- 21.Hübner H., Schellhorn T., Gienger M., Schaab C., Kaindl J., Leeb L., Clark T., Möller D., Gmeiner P. Structure-guided development of heterodimer-selective GPCR ligands. Nat Commun. 2016;7:12298. doi: 10.1038/ncomms12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navarro G., Cordomíi A., Casadó-Anguera V., Moreno E., Cai N.-S., Cortés A., Canela E.I., Dessauer C.W., Casadó V., Pardo L., Lluis C., Ferre S. Evidence for functional pre-coupled complexes of receptor heteromers and adenylyl cyclase. Nat Commun. 2018;9:1–12. doi: 10.1038/s41467-018-03522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han Y., Moreira I.S., Urizar E., Weinstein H., Javitch J.A. Allosteric communication between protomers of dopamine class A GPCR dimers modulates activation. Nat Chem Biol. 2009;5:688. doi: 10.1038/nchembio.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carpenter B, Nehmé R, Warne T, Leslie AG, Tate CGT. Structure of the adenosine A2A receptor bound to an engineered G protein. Nature. 2016;536:104–107. doi: 10.1038/nature18966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu W, Chun E, Thompson AA, Chubukov P, Xu F, Katritch V, Han GW, Roth CB, Heitman LH, IJzerman AP, Cherezov VST. Structural basis for allosteric regulation of GPCRs by sodium ions. Publ Am Assoc Adv Sci 2012;337:232–236. [DOI] [PMC free article] [PubMed]
- 26.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, David E. Shaw, PF, Shenkin PST. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem; 2004:47;1739–1749. [DOI] [PubMed]
- 27.Halgren T.A., Murphy R.B., Friesner R.A., Beard H.S., Frye L.L., Pollard W.T., Banks J.L. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrich- ment factors in database screening. J Med Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 28.Ballesteros J.A., Weinstein H. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 29.Wang S., Che T., Levit A., Shoichet B.K., Wacker D., Roth B.L. Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone. Nature. 2018;555:269–273. doi: 10.1038/nature25758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang Y., Kuybeda O., de Waal P.W., Mukherjee S., Van Eps N., Dutka P., Zhou X.E., Bartesaghi A., Erramilli S., Morizumi T. Cryo-EM structure of human rhodopsin bound to an inhibitory G protein. Nature. 2018;558:553–558. doi: 10.1038/s41586-018-0215-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robertson N, Rappas M, Doré AS, Brown J, Bottegoni G, Koglin M, Cansfield J, Jazayeri A, Cooke RM, Marshall FHT. Structure of the complement C5a receptor bound to the extra-helical antagonist NDT9513727. Nature 2018, 553, 111–114. [DOI] [PubMed]
- 32.Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. Molecules. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 33.Zhu K., Day T., Warshaviak D., Murrett C., Friesner R., Pearlman D. Antibody structure determination using a combination of homology modeling, energy-based refinement, and loop prediction. Proteins: Struct, Funct, Bioinf. 2014;82:1646–1655. doi: 10.1002/prot.24551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Søndergaard C.R., Olsson M.H., Rostkowski M., Jensen J.H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. Molecules. 2011;7:2284–2295. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- 35.Lomize A.L., Pogozheva I.D., Mosberg H.I. Anisotropic solvent model of the lipid bilayer. 2. Energetics of insertion of small molecules, peptides, and proteins in mem- branes. J Chem Inf Model. 2011;51:930–946. doi: 10.1021/ci200020k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lomize M.A., Pogozheva I.D., Joo H., Mosberg H.I., Lomize A.L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucl Acids Res. 2011;40:370–376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klauda J.B., Venable R.M., Freites J.A., O’Connor J.W., Tobias D.J., Mondragon-Ramirez C., Vorobyov I., MacKerell A.D., Jr, Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 39.Mark P., Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J Phys Chem. 2001;105:9954–9960. [Google Scholar]
- 40.Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; Sunhwan Jo, V.S.P.; David A. Case, C.L.B.; Alexander D. MacKerell, J.B.K.; Im, W., T. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput 2015, 12, 405–413. [DOI] [PMC free article] [PubMed]
- 41.Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, V.I., P; Jr, M.A., T. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 2010, 31, 671–690. [DOI] [PMC free article] [PubMed]
- 42.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kale L., Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evans D.J., Holian B.L. The nose-hoover thermostat. J Chem Phys. 1985;83:4069–4074. [Google Scholar]
- 44.Parrinello M., Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- 45.Miao Y., Nichols S.E., Gasper P.M., Metzger V.T., McCammon T. Activation and dynamic network of the M2 muscarinic receptor. Proc Natl Acad Sci USA. 2013;110:10982–10987. doi: 10.1073/pnas.1309755110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miao Y., Sinko W., Pierce L., Bucher D., Walker R.C., McCammon J.A. Improved reweighting of accelerated molecular dynamics simulations for free energy calculation. Chem Theory Comput. 2014;10:2677–2689. doi: 10.1021/ct500090q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tikhonova, I.G.; Selvam, B.; Ivetac, A.; Wereszczynski, J.; McCammon, J.A., T. Sim- ulations of biased agonists in the 2 adrenergic receptor with accelerated molecular dynamics. Biochemistry 2013, 52, 5593–5603. [DOI] [PMC free article] [PubMed]
- 48.Miao Y., Nichols S.E., McCammon J.A. Free energy landscape of G-protein cou- pled receptors, explored by accelerated molecular dynamics. Phys Chem Chem Phys. 2014;16:6398–6406. doi: 10.1039/c3cp53962h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang F., Yuan Y., Li H., Shen L., Guo Y., Wen Z., Pu X. Using accelerated molec- ular dynamics simulation to shed light on the mechanism of activation/deactivation upon mutations for CCR5. RSC Adv. 2018;8:37855–37865. doi: 10.1039/c8ra07686c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kasahara K., Fukuda I., Nakamura H. A novel approach of dynamic cross correlation analysis on molecular dynamics simulations and its application to Ets1 dimer-DNA complex. PloS One. 2014;9 doi: 10.1371/journal.pone.0112419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhattacharyya M., Bhat C.R., Vishveshwara S. An automated approach to network features of protein structure ensembles. Protein Sci. 2013;22:1399–1416. doi: 10.1002/pro.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oliveira S.H., Ferraz F.A., Honorato R.V., Xavier-Neto J., Sobreira T.J., de Oliveira P.S. KVFinder: steered identification of protein cavities as a PyMOL plugin. BMC Bioinf. 2014;15:197. doi: 10.1186/1471-2105-15-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan S., Filipek S., Palczewski K., Vogel H. Activation of G-protein-coupled re- ceptors correlates with the formation of a continuous internal water pathway. Nat Commun. 2014;5:1–10. doi: 10.1038/ncomms5733. [DOI] [PubMed] [Google Scholar]
- 54.Miao Y., McCammon J.A. Mechanism of the G-protein mimetic nanobody binding to a muscarinic G-protein-coupled receptor. Proc Natl Acad Sci USA. 2018;115:3036–3041. doi: 10.1073/pnas.1800756115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doré A.S., Robertson N., Errey J.C., Ng I., Hollenstein K., Tehan B., Hurrell E., Bennett K., Congreve M., Magnani F., Tate C.G., Weir M., Marshall F.H. Struc- ture of the adenosine A2A receptor in complex with ZM241385 and the xanthines XAC and caffeine. Structure. 2011;19:1283–1293. doi: 10.1016/j.str.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu F., Wu H., Katritch V., Han G.W., Jacobson K.A., Gao Z.-G., Cherezov V., Stevens R.C. Structure of an agonist-bound human A2A adenosine receptor. Publ Am Assoc Adv Sci. 2011;332:322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McGraw C., Yang L., Levental I., Lyman E., Robinson A.S. Membrane cholesterol depletion reduces downstream signaling activity of the adenosine A2A receptor. Biochim Biophys Acta, Biomembr 1861. 2019:760–767. doi: 10.1016/j.bbamem.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kling R.C., Clark T., Gmeiner P. T.U Comparative MD simulations indicate a dual role for Arg1323. 50 in dopamine-dependent D2R activation. PloS One. 2016;11 doi: 10.1371/journal.pone.0146612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carpenter B., Lebon G. Human adenosine A2A receptor: molecular mechanism of ligand binding and activation. Front Pharmacol. 2017;8:898. doi: 10.3389/fphar.2017.00898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dorfler M., Tschammer N., Hamperl K., Hubner H., Gmeiner P. Novel D3 selective dopaminergics incorporating enyne units as nonaromatic catechol bioisosteres: synthesis, bioactivity, and mutagenesis studies. J Med Chem. 2008;51(21):6829–6838. doi: 10.1021/jm800895v. [DOI] [PubMed] [Google Scholar]
- 61.Tschammer, N., Elsner, J., Goetz, A., Ehrlich, K., Schuster, S., Ruberg, M., and Gmeiner, P. (2011). Highly potent 5-aminotetrahydropyrazolopyridines: Enantioselective dopamine D3 receptor binding, functional selectivity, and analysis of receptor- ligand interactions. J Med Chem 2011 54(7), 2477–2491. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Classical and accelerated molecular dynamics simulation were performed using NAMD simulation package. Softwares used in the study are owned by their respective developers. The trajectories pertaining to both systems linker and without linker are deposited in OSF repository which can be assessed by the linkhttps://osf.io/tnpgz/?view_only=944246c89c06401d967628b52d71752b