Abstract
SARS-CoV-2 is an RNA virus that was identified for the first time in December 2019 in Wuhan, China. The World Health Organization (WHO) labeled the novel coronavirus (COVID-19) outbreak a worldwide pandemic on March 11, 2020, due to its widespread infectivity pattern. Because of the catastrophic COVID-19 outbreak, the development of safe and efficient vaccinations has become a key priority in every health sector throughout the globe. On the 13th of January 2021, the vaccination campaign against SARS-CoV-2 was launched in India and started the administration of two types of vaccines known as Covaxin and Covishield. Covishield is an adenovirus vector-based vaccine, and Covaxin was developed by a traditional method of vaccine formulation, which is composed of adjuvanted inactivated viral particles. Each vaccine’s utility or efficiency is determined by its formulation, adjuvants, and mode of action. The efficacy of the vaccination depends on numeral properties like generation antibodies, memory cells, and cell-mediated immunity. According to the third-phase experiment, Covishield showed effectiveness of nearly 90%, whereas Covaxin has an effectiveness of about 80%. Both vaccination formulations in India have so far demonstrated satisfactory efficacy against numerous mutant variants of SARS-CoV-2. The efficacy of Covishield may be diminished if the structure of spike (S) protein changes dramatically in the future. In this situation, Covaxin might be still effective for such variants owing to its ability to produce multiple antibodies against various epitopes. This study reviews the comparative immunogenic and therapeutic efficacy of Covaxin and Covishield and also discussed the probable vaccination challenges in upcoming days.
Keywords: COVID-19, Immune response, Vaccine, Covaxin, Covishield
Introduction
Coronavirus is an RNA virus belongs to the big family of Coronaviridae (order: Nidovirales), responsible for severe sickness in both humans and animals [1]. There are seven types of coronavirus that have been discovered and are responsible for producing illnesses in people worldwide, while the most prevalent human coronaviruses among them are 229E, NL63, OC43, and HKU1 [2]. Three zoonotic coronaviruses (CoV) have been identified in humans: severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome CoV (MERS-CoV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [3]. The WHO termed the SARS-CoV-2 clinical syndrome COVID-19 on February 11, 2020 (short form of coronavirus disease-19) [2]. This virus affects the lower respiratory tracts of humans, resulting in inflammatory pneumonia [3]. These viruses can easily be transmitted by symptomatic, asymptomatic, and presymptomatic peoples, mainly through respiratory droplets when they come into close contact with each other [4]. Given the severity of the condition, COVID-19 was declared a pandemic by WHO on March 11, 2020. [5]. In India, in the month of September 2020, the first wave of the SARS-CoV-2 infection rate reached its maximum spike and gradually declined over time. In March 2021, again, new cases of infection started, and as of 17 December 2021, a total number of 271,963,258 confirmed cases, a total of 5,331,019 deaths were reported and 8,337,664,456 vaccine doses have been administered as recorded by WHO [6]. Both the innate and adaptive immune responses to the SARS-CoV-2 virus have evolved rapidly. In innate immunity responses, the cytokine, interleukin-6 (IL-6), and complement system work against the virus, and in adaptive immunity, the monoclonal antibody is focused against the S proteins of the virus along with massive CD4+/CD8+ T cell, natural killer (NK) cell, memory B cell, and follicular T cell responses have been pragmatic within exposed persons [7]. SARS-CoV-2, having a crown-like appearance and a diameter of 60–140 nm can form a binding interface with the N-terminal end of the ACE2 receptor with higher affinity. After binding, an enzyme, Furin, found in the host cell, helps these viruses enter the respiratory tract after 4 to 5 days. [8]. ACE2 plays a physiological role in blood pressure regulation; metabolize angiotensin II (vasoconstrictor) to form angiotensin 1–7 (vasodilator). ACE2 is extensively disseminated in the cells of the mucosal surfaces of the nose and lungs, which makes the progression of infection in the respiratory tract easier. In addition to the respiratory tract, ACE2 is found on cells throughout the body, such as the endothelium, the cardiovascular system, the gastrointestinal system, and the kidneys. These organs can be infected by viruses, making them prone to infection. [9]. There is currently no specific COVID-19 responsive drug commercially available for the treatment of COVID-19-associated diseases. Certain medicines, like antiviral/retroviral drugs and some kinds of steroids, are being used to reduce swelling and inflammation of the lungs and other parts of the body. In India, treatment with remdesivir and favipiravir has been found to speed up recovery in some patients; while lopinavir/ritonavir has not been shown to be effective in treating severe patients. The early viral negative conversion was seen with triple therapy. Ribavirin, lopinavir, and interferon-1β are used in triple therapy [10]. Arbidol inhibits the “S” protein/ACE2 interaction in various patients with SARS-CoV-2 infection, resulting in improved therapeutic efficacy [11]. But after a certain period of time, contradictory results of various popular antiretroviral drugs were observed, which dramatically limits their wide application in the management of COVID-19. In the first phase of the initiative, India began providing free COVID-19 vaccinations and aimed to vaccinate its 30 million healthcare and frontline workers on January 16, 2021. In this phase, Covishield (Oxford-AstraZeneca vaccine, the name designated by India) and Covaxin (Bharat Biotech, India, an indigenous vaccine) were approved under restricted emergency use [12]. Both vaccines function by stimulating the immune system in the same way against the S protein of SARS-CoV-2, and both need two doses to achieve effective seroprotection. Nonreplicating adenovirus is used in Covishield to express SARS-CoV-2 S protein towards the antigen-presenting cells of the host, while Covaxin uses SARS-CoV-2, which has been inactivated and taken from an asymptomatic patient and mixed with alum-adjuvant for better antibody response. In both types of vaccination, stimulation of immunogen produces significant cell-mediated and humoral immune responses [13]. The Drug Controller General of India (DCGI) recently approved Sputnik V, a third coronavirus vaccine, for use against the second wave of SARS-CoV-2 illnesses, alongside two previous vaccines. It is originally produced by the Gamaleya Research Institute (GRI) of Russia, acts similar to Covishield, and gives around 92% protection. Two types of nonreplicating (E1 gene removed) human adenoviruses, Ad26 (serotype 26) and Ad5 (serotype 5), were employed to produce the gene that encodes the entire S protein of SARS-CoV-2 (serotype 5) [14]. In the general public, as well as in the scientific community, there are many doubts, misunderstandings, and misinterpretations about which vaccine is the better of the two. There is also lots of information about the composition, therapeutic efficacy, and adverse effects of the two vaccine formulations on the web. Some review reports based on the types of vaccines available against SARS-CoV-2 are also accessible. However, no review article has included a rigorous comparison of the immunologic and therapeutic efficacy of Covaxin and Covishield. So, in this review, an attempt has been made to provide detailed information about the efficacy of those two vaccine formulations in a comparative manner.
Epidemiology of COVID-19
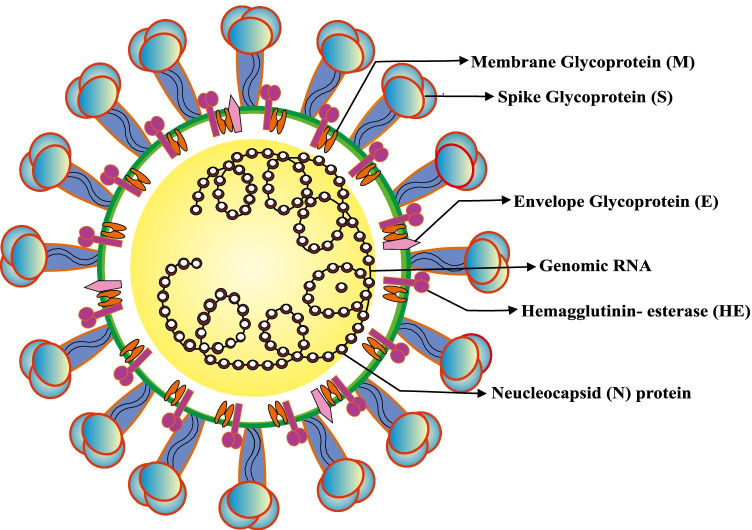
Coronavirus (CoV) is named after the Latin word corona, which means “crown”. This virus has several crown-shaped spike patterns on its surface [15]. It rapidly infects the human respiratory tract, causing mild to severe respiratory distress syndrome. CoV was discovered for the first time in the 1960s. The last significant coronavirus epidemics were SARS (severe acute respiratory syndrome) driven by SARS-CoV in 2002–2003 and MERS (Middle East respiratory syndrome) caused by MERS-CoV in 2012. [16]. Following a pneumonia outbreak in Wuhan, China, in December 2019 and considering its infectivity pattern, the WHO renamed it the novel coronavirus-19 (nCOVID-19) [3]. The size of SARS-CoV-2 virus particles is 29.9 kb, with spikes. The diameter of the spike is 60–140 nm, and the length is 9–12 nm, which are found on and are made up of four nanostructural proteins (NSPs) [17]. Spike (S) glycoprotein, tiny envelope (E) glycoprotein, membrane (M) glycoprotein, and nucleocapsid (N) glycoprotein are the four types of glycoproteins found in the viral body, and they are the main structural proteins of this virus [18] (Fig. 1). Among all known RNA viruses, coronavirus has the largest genome (26.4–31.7 kb) [19]. The “S” protein aids in the virus’ binding to the host cell, allowing it to enter the cell [20]. The M protein, which has three transmembrane domains, helps the virus bend and become spherical, as well as maintain the virus’ basic structure [21]. The viral assembly and release are aided by the E protein [22]. It is an RNA virus with a positive strand. When they bind to cellular receptors and enter the host cell, the viral essential protein is produced due to RNA translation, which is required for viral replication. The new viral particle will be released from the host cell and infect additional host cells [23]. The human ACE2 receptor is activated by the spike of this virus, which binds to it via its receptor and receptor-binding domain (RBD). Using biochemical and pseudovirus entry, the binding of receptor and protease activation of the SARS-CoV-2 S protein was studied [24].
Fig. 1.
The core structural proteins of SARS-CoV-2. Spike (S), envelope (E), membrane (M), and nucleocapsid (N) are the four major structural proteins found in the virus (N). The lipid bilayer envelope contains the S, E, and M proteins, while the N protein encases the virus RNA genome
Natural immunity against COVID-19
During infection, the structural protein S of SARS-CoV-2 interacts with the host cell’s target receptor ACE2. S1 and S2 are two subunits of glycoprotein S. S1 contains a receptor-binding domain, while S2 mediates viral-host cell fusion [25]. T and B cells activate and mediate adaptive immunity, which collectively become effective against the coronavirus [26]. Cytotoxic T cells and NK cells are both required for immune responses to viruses. In comparison to a healthy person, a COVID-19 patient has low lymphocyte and high neutrophil count, with the number of CD8 + and NK cells, in particular, being reduced [27] (Fig. 2). The pathogen is recognized by macrophages, dendritic cells, and monocytes after it enters the body, owing to pattern recognition receptors (PRRs), which recognize the microbe-associated molecular pattern (MAMPs) [25]. After infection, the incubation period has been estimated in symptomatic patients approximately 5 days of symptom onset. Approximately 97.5% of patients then develop symptoms such as fever, dry cough (less common), difficulty breathing, headache, dizziness, diarrhea, nausea, loss of taste (dysgeusia), muscle joint discomfort, and loss of smell (anosmia) within 11.5 days [4]. The viral loads in mouth swabs and sputum specimens reached their maximum level at 5 to 6 days following the onset of symptoms, ranging from 104 to 107 copies per milliliter of fluid [28]. Then, an infected person, as well as an asymptomatic patient, can develop acute respiratory distress syndrome (ARDS), which occurs within 7 days after the inception of symptoms and a decline in oxygen levels [29].
Fig. 2.
Summarization of various immune responses against COVID-19. PRRs identify PAMPs and DAMPs in the innate immune response, causes macrophage activation and the release of inflammatory cytokines. T and B cells are also activated, and differentiation is aided. T cells come in a variety of subtypes that release cytokines. IL-1β activates neutrophils, causing them to produce IFN-γ, TNF-α, perforin, granzymes, as well as activates DCs. Infected epithelial cells may give virus antigens to naïve T cells (TH0), which then release IL-12 and differentiate into TH1 (CD4 + T cells) and TH2 (CD8 + T cells). Apoptosis occurs when NK cells become cytotoxic to virus-infected epithelial cells. DCs and macrophages use the MHC-TCR interaction to present viral antigen to CD4 + T cells. Memory T cells are developed, and they may confer protection against reinfection with the same viral strain for a period of time that has yet to be determined. The activated B cell also acts as an APC, presenting the antigen to the TH2 cell via MHC-II and TCR interaction. The TH2 cell then generates IL-4, IL-5, IL-6, IL-10, and TGF-β, while the B cell differentiates into plasma cells and memory B cells, which produce anti-SARS-CoV-2 specific IgM, IgA, and IgG antibodies
Innate immune response
Epithelial cells and hyperactive macrophages are activated by the virus’ innate immune response. When these cells are infected, they interact with damage-associated molecular patterns (DAMPs) and pathogen-associated molecular patterns (PAMPs) via PRRs like Toll-like receptor (TLR), C-type lectin-like receptors (CLRs), NOD-like receptors (NLR), RIG-1-like receptors (RLR), and free molecule receptors in the cytoplasm, for instance, Cgas, IFI 16, STING, DAI, etc., on the immune cell, which activate the inflammasome and release pro-inflammatory and inflammatory mediators [30]. In the immune system, myeloid-derived cells such as macrophages and monocytes play a crucial role. TLRs are found in macrophages that identify the nucleic acid pattern (ssRNA) of invading viruses. TLR4, TLR7, TLR8, and TLR9 are among these TLRs, but no specific TLRs involved in SARS-COV-2 pathogenesis have yet been identified [31]. TLRs attach to viruses, which can activate the NF-ĸB and MAPK signaling pathways, to clean the virus and produce antiviral responses. TLRs also begin the processes of phagocytosis, cytokine release, and several other mediators in order to augment the local anti-inflammatory processes as part of immune control [32] (Fig. 2).
Pattern recognition receptors (PRRs)
PAMPs are microbial structures that can be detected by PRRs in the innate immune system, such as TLRs and RIG-I like receptors, NLR, and others, triggering downstream signaling cascades that finally prompt an efficient antimicrobial response [33]. The type I and III interferon and several other chemokines can be produced more efficiently when the signaling pathways are targeted by the ligand-binding activity, which is notably targeted the various transcription factor (AP-1, IRF3, and NF-κB). These chemokines stimulate the recruitment of more innate immunity responsive cells (polymorphonuclear leukocytes, monocytes, NK cells, and DCs), which in turn stimulate the production of chemokines (MIG, IP-10, and MCP-1) to further promote the recruitment of lymphocytes to the site of infection, which is provided by DCs. [34, 35]. These cytokines act on nearby cells in a paracrine manner through the IFN-α/β receptor (IFNAR) and cause the production of interferon-stimulated genes (ISGs). IFNAR mediates the activation of the Jak/STAT pathway, which leads to the attachment of STAT1 homodimers and STAT1/2 heterodimers to the promoter region of ISGs, which is triggered by interferon-stimulated gene expression. [34].
PAMPs and TLRs
TLRs recognize PAMPs in the cell membrane, endosomes, lysosomes, endocytosomes, and other locations [33]. TLRs activate a variety of adaptor proteins (TRAM, TIRAP, MyD88, and TRIP), which cause a variety of biological responses, all of which share the Toll/interleukin-1 receptor (TIR) structure [36]. Except for TLR3, all TLRs act as adaptor proteins for MyD88 (myeloid differentiation factor 88), which activates the NF-κB and MAPKs pathways and results in the generation of pro-inflammatory molecules [37]. TRIF binds to TLR3 and TLR4 and supports an alternate pathway for the generation of inflammatory cytokines and IFN-1 genes by activating IRF3, NF-κB, and MAPKs [36]. With the help of TLR-7/8, the virus activates monocytes or macrophages, stimulating IFN-I responses [38]. In activated inflammatory macrophages, CCL2, CCL3, CCL20, CXCL1, CXCL3, CXCL10, IL1, IL8, and TNF levels are all increased [39] (Fig. 2).
RIG-1-like receptors (RLRs)
RLRs recognize the nucleic acid of RNA viruses and can be utilized to program the adaptive immune response via a cross talk with the innate immune system’s parallel arms [40]. RIG-1 recognizes triphosphine RNA at the 5′ end of viral nucleocapsid proteins, as well as double-stranded RNA containing basic acid [41]. Ultimately, it stimulates some downstream intracellular pathways to produce cytokines, chemokines, and chemotactic factors for effective antiviral action, in addition to MDA5 and LGP2 [40].
NOD-like receptors (NLR)
NOD-like receptors (NLRs) are cytoplasmic receptors that aid in the innate immune response by detecting PAMPs and DAMPs [42]. In response to PAMPs identification, these receptors can trigger the expression of pro-inflammatory cytokines; they are also required for reproduction and embryo regeneration, regulation of apoptosis, and participation in acquired immune system reactions. NLRs are mostly expressed in macrophages, DCs, and lymphocytes, but they have also been recognized in nonimmune cells like epithelial cells [43].
Innate immune response through specialized cells
DCs, one of the professional antigen presenting cells (APCs), play a critical role in adaptive and innate immune responses [44]. DCs are classified into two types: plasmacytoid DC (pDCs) and conventional DC [45]. These cells include the generation of immune-increasing cytokines like IL-12 and IFN-I and also the mobilization of innate lymphocytes. Despite the fact that DCs have phagocytic properties, limited particle absorption potency and their phagocytic ability are not known to contribute to microbial minimization or death; their phagocytic ability is effective for antigen presentation and processing on MHC I and II [46]. pDCs boost the production of IFN-I to remove viruses from circulation through the macrophages and also to preserve the integrity of pDCs for the priming of adaptive immunological responses [47]. NK cells, also known as cytotoxic cells, provide virus immunity by recognizing pathogens using natural cytotoxic receptors (NCRs) such as NKp46 and NKp44 [48]. NK cells are yet another important component of innate immunity. It was quickly discovered that the overall number of NK cells in COVID-19 patients is significantly reduced [49]. NKG2A expression on NK cells was dramatically enhanced in individuals infected with SARS-CoV-2 [50]. It binds to a nonclassical HLA class I molecule with negligible polymorphism (HLA-E). NKG2A transduces inhibitory signaling through two inhibitory immune-receptor tyrosine-based inhibition motifs after being ligated by peptide-loaded HLA-E, reducing NK cytokine production and cytotoxicity [51]. The killing of bacteria by neutrophils via phagocytosis and degranulation promotes the first-line defense of innate immunity [52]. To improve the severity of COVID-19, this cell forms neutrophil extracellular traps (NETs). CXCR2 is higher in neutrophils, and the type I IFN response is delayed [53]. The complement system is also engaged in innate immune response, which is a mechanism that does not require the use of antibodies (lectin pathway). The mannose-binding lectin-associated serine protease 2 (MASP-2) is involved in this mechanism, which causes the complement system to be questioned [54]. MASP-2 activation by SARS-CoV-2 causes the complement system to become hyperactive, causing damage to the infected tissue [55] (Fig. 2).
Adaptive immune response
Cell-mediated immunity
Severe lymphopenia was observed in non-survivor COVID-19 patients. Despite having normal or high total B lymphocyte counts, up to 80% of SARS patients in the hospital had severe leukopenia and a decrease in CD4 and CD8 T cells [50, 56]. Internalized by phagocytic cells like macrophages and DCs, the virus is digested and peptide fragments known as antigens (epitopes) are represented by MHC II on the surface of this cell by specific CD4 + /helper T cells [57]. The CD4 + cells then secrete IL-2 and IFN-γ, which activate many macrophages and increase B-cell proliferation, allowing antigen-specific antibodies to be produced [58](Fig. 2). Then, the antibody forms a coat around the bacteria, and they become easier to phagocytosis. In addition to phagocytosis, the innate immunity, cells recognize the pathogen by PRRs and trigger inflammation signaling and begin cytokine production, such as TNF, IL-1 and IL-12 [59]. When a virus infects a cell, certain cells promote the release of type I IFN-α and IFN-β, which induces antiviral resistance and activates NK cells [60]. The immune system generates memory cells to provide defense against reinfection. The memory CD8 + and CD4 + T cells and B cells remained in a resting condition during reinfection but became active and quickly eliminated the source of antigen. Vaccine strategy is largely based on immunological memory [61]. When normal cells become infected by a virus, the number of MHC-I decreases, then, the infected respiratory epithelial represent the viral epitopic part through MHC-I to the CD8 + cytotoxic T cell. When a viral epitope interacts with a CD8 + cytotoxic T cell, the T cell produces perforin and granzyme, which kills the virally infected cell [62] (Fig. 2).
Humoral immunity
The humoral immune response is a neutralizing mechanism by which it prevents viral infection. The host humoral response to SARS-CoV-2 (IgA, IgM, and IgG) was studied using an ELISA-based test based on the recombinant viral nucleocapsid protein. The post-symptomatic onset (PSO) of IgM is observed within 5 days, IgA between 3 and 6 days and IgG of 14 days [63, 64]. IgA and IgG antibodies against S protein subunit 1 of SARS-CoV-2 were measured in the serum of infected people [65]. After roughly 28 days, IgM and IgA levels drop, whereas IgG amounts reach their peak after around 49 days [66]. Along with neutralization, these antibodies also can perform antiviral protection through other mechanisms, including antibody-dependent cell cytotoxicity (ADCC), antibody-dependent phagocytosis (ADCP), etc. The ADCC is a result of FcγRIIIa cross-linking in NK cells, and ADCP is mediated by professional phagocytes (monocytes, macrophages, neutrophils, and eosinophils), which are attached to the antibody-covered virus via special Fc receptor, and IgM and IgG also activate the complement system [67]. Long-term memory T cell and B cell work against SARS-CoV-2 the same as adaptive immune responses. During subsequent infection, T cells aid in the destruction of infected cells. Without the presence of any antigens, case investigations in recovered SARS patients revealed that both CD4 + and CD8 + memory T cells were effective in inducing immunological responses from 3 months to 6 years [68]. After the acute phase of infection, the antibody level declines because most of the plasmoblasts are induced during the first week after a short-lived infection. Smaller number of long-lived plasma cells maintain the serological memory. The long-lived plasma cell is found in the bone marrow and secretes antibodies when there is no antigen present to recognize. The pool of long-lived memory B cells generates this response [68]. During the first week after infection with SARS-CoV-2, T cells become activated, and virus-specific memory is generated in the body. CD4 + and CD8 + T cells reach their peak level in 2 weeks but remain low for 100 days or more. SARS-CoV-2 memory CD4 + T cells were detected in up to 100% of COVID-19 patients, while CD8 + T cells were observed in roughly 70% [69] (Fig. 2).
Infection mechanism of SARS-CoV-2
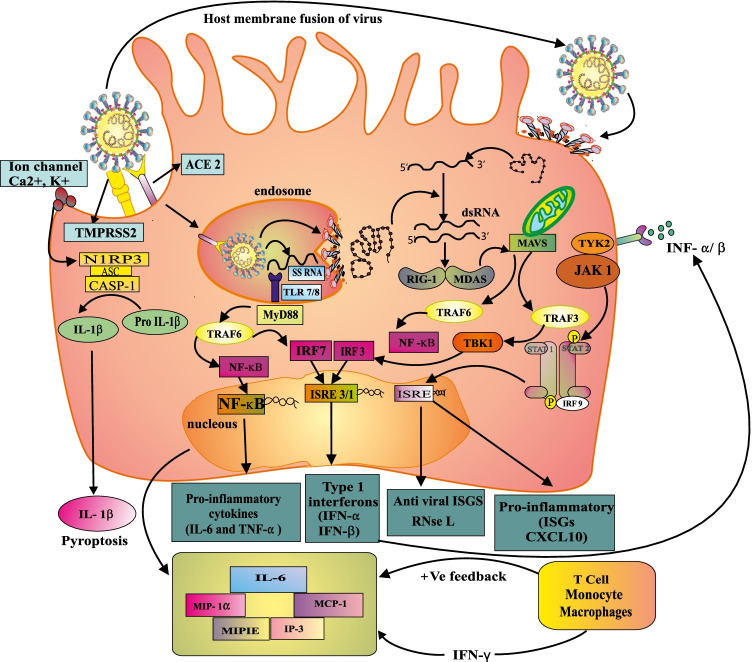
Despite the fact that SARS-CoV-2 is assumed to primarily infect respiratory epithelial cells, a recent study revealed that it could also affect T lymphocytes, spleen, and lymph nodes. [70]. The infection is mediated through the ACE2 receptor in the same way as SARS and the receptor-binding motif (RBM), which is a part of the RBD of SARS-CoV-2 that establishes contact with that receptor [71]. ACE2 is largely expressed on type II alveolar epithelial cells, but it is also identified on the surface of epithelial cells from the nasal and oral mucosa, in addition to the nasopharynx, implying that SARS-CoV-2 is primarily concentrated on the lungs. This ACE2 is highly expressed in enterocytes of the small intestine, notably in the ileum, as well as heart, kidney (proximal tube), and bladder urothelial cells [72, 73]. The entry of the first virus may be strongly dependent on TMPRSS2 expression because even small quantities of ACE2 can support SARS-CoV-2 entry if TMPRSS2 is present [74]. RNA is injected within the host cell by viruses, which is translated into viral replicase polyproteins, which then cleave into NSPs of individuals and form RNA-dependent RNA polymerase [75]. Enzyme replicase requires a full-length positive-strand RNA genome as a template in order to create full-length negative-strand RNA copies of the viral genome. During transcription, the RNA polymerase enzyme transforms subgenomic mRNA to viral protein (S, E, N, and M). The viral protein and genome RNA are formed into virions in the Golgi and endoplasmic reticulum, which then migrate into ERGIC (ER-Golgi-intermediated compartment) and exit the cell via vesicles [76, 77] (Fig. 3).
Fig. 3.
Infection mechanism of COVID-19. When alveolar epithelium cells are invaded by SARS-CoV-2, the cells express the surface receptors ACE 2 and TMPRSS2, and the virus is recognized by innate immune receptors such as the endosomal RNA sensors TLR7/8, RIG-I, and MDA5. This TLR7/8 can detect viral RNA species created during viral replication, such as viral genomic RNA and dsRNA. While RIG-I and MDA5 are responsible for sensing cytoplasmic viral RNAs such as 5′-3’ RNA or dsRNA. This causes NF-κB and IRF3/7 to become activated, resulting in the generation of proinflammatory cytokines and type I interferons, respectively. Type I interferons are important in restricting viral multiplication, and their effect is exacerbated by the production of ISGs like RNAse L. IL-6, IP-10, MIP-1α, MIPIE and MCP1 are the major pro-inflammatory cytokines and chemokines. These proteins draw monocytes, macrophages, and T cells to the infection site, causing further inflammation and triggering a pro-inflammatory feedback loop. In the case of a faulty immune response (left side), this can lead to increase inflammatory immune cell trafficking in the lungs, generating an overproduction of pro-inflammatory cytokines, and eventually causing lung damage. The cytokine storm that results spreads to other organs, causing multi-organ damage
PAMPs and DAMPs
Alveolar epithelial cells and alveolar macrophages identify and release PAMPs (viral RNA), and DAMPs (ASC oligomers, DNA, and ATP) via a number of PRRs [78]. This detection process affects the activation of intracellular signaling pathways, which is able to produce IFN-I, GM-CSF, TNF, IL-1, IL-6, IL-8, and also many other pro-inflammatory cytokines, which systematically employ activated leukocytes within the lungs and amplify microvascular permeability [79]. The RLRs include RIG-I and melanoma differentiation-associated gene 5 (MDA-5), TLR 3, 7, and 8 that trigger IFN pathways and the production signaling of cytokines. When PRRS recognizes a viral particle, it acts downstream via the TANK-binding kinase-1 (TBK1) and inhibitor-κB kinases (IKKs), which activate IRF-3, IRF-7, and the nuclear factor of the kappa light chain enhancer in activated B cells. This entire cascade promotes the nuclear translocation process. The activation of these proteins promotes the type I IFNs and IFN-stimulated genes; many of them have antiviral properties; on the other hand, the pro-inflammatory mediators, such as chemokines, cytokines, and antimicrobial peptides, they all are important for the trigger immune response (innate and adaptive) in the host. Moreover, the synthesis of IL-18, IL-1, and inflammasomes is triggered by the missing in melanoma 2 (AIM2)-like receptors and NLRs, resulting in pyroptosis [80].
Downregulation of antiviral cytokines
The antiviral is based on IFN-I (particularly IFN-β and IFN-α), and type III IFNs serve as a basic component of innate immune response and facilitate the development of adaptive immune response against viral infection [81]. This infection promotes low levels of antiviral cytokines IFN-α/β. This virus suppresses IFN synthesis by covering viral RNA from cellular components in the host cell [82]. SARS-CoV nsp-1 (nanostructural protein 1) suppresses the immune response along with the generation of IFNs (IFN-I). nsp demonstrates an elevated expression of IFN-I [83]. Activation of IFN-I also minimize the expression of viral proteins such as nsp1, nsp7, and nsp15, as well as ORFs such as ORF3B, ORF6, and ORF9b [84]. IFR3, which promotes IFN production, is activated by IFN gene signaling, which is disrupted by the virus’ ability to prevent IFN production [85]. Transmembrane inhibitor 1(TM1), which binds to RIG-1, TANK-binding kinase-1, TNF receptor-associated factor-3 (TRAF-3) and inhibits M protein production (TBK-1). After binding these molecules to M protein, it prevents them from binding to any other downstream effectors to prevent IFN-1 induction [86]. SARS-CoV-2 produces the abnormal level of the following cytokines and chemokines such as GM-CSF, M-CSF, G-CSF, MCP-1, MIP 1-α, IFN-γ, TNF-α, IP-10, hepatocyte growth factor (HGF), IL-1, IL-2, IL-4, IL-6, IL-7, IL-10, IL-12, IL-13, IL-17, and vascular endothelial growth factor (VEGF) [87]. The abnormal production of these cytokines and chemokines pulls immune cells from the bloodstream, particularly monocytes and T lymphocytes. But they are not able to pull neutrophils into the infected site. An elevated level of neutrophil–lymphocyte ratio and lymphopenia were observed approximately in 80% of COVID-19 patients, and this observation could be the scientific explanation of pulmonary recruitment of immune cells from the bloodstream [78]. The dysregulation of innate immunity is caused by the elevated production of TNFα, IL-6, and a low amount of type I and III IFNs in people who are affected by COVID-19 [87].
Cytokine storm
COVID-19-associated lung pathogenesis is common during manifestation. The infection and overproduction of pro-inflammatory cytokines, which are referred to as ‘cytokine storm’, are mainly responsible for this [88]. In COVID-19, the cytokine storm is characterized by elevated levels of expression of IFN-γ, IL-1, IL-6, IL-8, IL-12, TGF-β, and also the upregulation of CXCL9, CXCL10, etc. In addition, plasma levels of IL-2, IL-7, IL-10, GM-CSF, MIP1-α, TNF-α, and MCP-1 were also raised in patients suffering from SARS-CoV-2 infection [89]. The cytokine storm follows the angiotensin 2 (AngII) pathways, and IL-6 has been found to be the central mediator in this event [90]. GM-CSF activates inflammatory monocytes (CD14+ CD16+), causing them to generate significant amounts of IL-6, TNF-α, and other pro-inflammatory cytokines [91]. Poor IFN induction could be a significant cytokine amplifier, Fc and Toll-like receptors, etc. may lead to an imbalanced inflammatory response [92]. The IL-6-sIL-6R complex is generated when IL-6 bind to the sIL-6R through gp130, and in the case of nonimmune cells, it can activate the signaling cascade and act as the activator of STAT3 [90]. The acquired immune system and innate immune system both have pleiotropic effects when cis signaling (classical signaling) is activated, which can play an important role in cytokine release syndrome [93]. The cytokine storm impacts negatively on the body, which is responsible for the ARDS, several organ malfunctions, and death [94]. A cytokine storm causes an uncontrolled influx of immune cells into the lungs, particularly monocytes and neutrophils. IL-1, IL-6, TNF-α, CCL2, CCL7, and CCL12 are among the inflammatory cytokines and chemokines secreted by these cells, which contribute to the severity of the condition. IL-1 has previously been shown to trigger pyroptosis, which is defined as non-programmed cell death in response to pathogenic infection [95]. Due to the increased release of harmful chemicals, such as proteases and reactive oxygen species (ROS), uncontrolled cell infiltrations cause serious lung injury, including pneumocyte desquamation, alveolar injury, hyaline membrane development, and pulmonary edema, all of which are early indications of ARDS [96].
Disturbance in complement activation
COVID-19 people have excessive coagulation, which leads to more thrombosis and a poor clinical outcome [97]. Complement activation is a key factor in the host intermediary of SARS-CoV-2-mediated illness because it controls a systemic pro-inflammatory response to it [98]. The levels of complement components like soluble C5b-9 and C5a in plasma of the severely ill patient increased notably [99]. The virus’ N protein and MASP-2 interact to initiate the lectin pathway, and this interaction also causes abnormal complement activation [98]. The complement system uses pattern recognition molecules from the canonical classical and lectin pathways (CP and LP) to detect invading infections along with environmental or self-derived antigens. A sequence of proteolytic activities results in pathway-specific canonical C3 convertase cleaving C3 into the anaphylatoxin (AT) C3a and the opsonin (C3b). The foundation for the C5 convertase that cleaves C5 into the AT C5a and C5b is formed by the C3 convertase. Apart from CP and LP activation, any nucleophilic attack on the thioester in C3 could activate it directly, resulting in the activation of the so-called alternative route (AP) and the synthesis of substantial amounts of C3a and C5a [100]. When complement hyperactivation was revealed in SARS-CoV-2 infected patients, anti-C5a monoclonal antibodies were injected into them, which led to a suppressive effect [101].
Dysregulation of immune cells
SARS-CoV-2 engages monocytes/macrophages through TLR7 and TLR8, which can produce type I IFN responses along with cytokine production. These collective immune responses gave inhibitory outcomes towards viral infection [38]. The activities of monocytes and macrophages are dysregulated by SARS-CoV-2 causing lung damage and respiratory disorders, including ARDS [102]. During the early stages of infection, the number of CD14 + monocytes and CD14 + IL-1 + monocytes are increased in the blood circulation of infected patients [103]. Macrophages play an important role in the induction of cytokine storms. In COVID-19 patients, IL-1 and IL-6 from circulating monocytes and macrophages cause macrophage activation syndrome (MAS), which leads to severe respiratory failure [104]. The number of CD4 + and CD 8 + T lymphocytes also decreased. In the patient’s body, the delayed IFN responses can lead to enhanced pro-inflammatory cytokine responses, altering the dynamics of antigen presentation and cytokine generation in innate immune cells and perhaps contributing to T cell response dysregulation [105]. SARS-CoV-2 similarly avoids the host immune system via reducing the number of NK cells. From the 16th day after the onset of the disease, the number of NK cells begins to decline. The deficiency of this cell is responsible for the number of viral infections and viruses protect themselves by enhancing their own survival mechanism [106]. As similar to the NK cells, T and B cells also showed gene signatory modification, which is linked with the high IFN-based immune responses and also to the activation of the apoptotic signaling cascade in the infected patient. According to the prior research, the responsible genes for B cell activation were found lower in the severely infected group as compared to the moderate group, these may be associated with the delayed humoral response contributed to the disease severity [107]. Infection of DCs with SARS-CoV-2 led to the inhibition of MHC-I expression on DCs, delaying the development of IFN-α [108]. This virus causes DCs to produce too many chemokines, which causes DCs to migrate to lymph nodes, while increased TRAIL expression causes lymphocytes to apoptosis when they come into contact with DCs. Due to the DC-associated death of T cells, the virus-specific adaptive immune response is declined. SARS infection slowed DC activation and migration to draining lymph nodes, resulting in a poor adaptive T cell response [109].
Overview of COVID-19 vaccines
The alarming SARS-CoV-2 outbreak necessitated a rapid response and vaccine development in an unprecedented time frame, and vaccine development was initiated [110]. The S protein of the SARS-CoV-2 virus has been used for the production of several COVID-19 vaccines. Over 214 numbers of unique vaccines were developed against SARS-CoV-2 by various pharmaceutical companies and academic institutes as of December 10, 2020, using different vaccine platforms such as recombinant viral-vectored vaccines, live attenuated viruses, protein subunit vaccines, inactivated vaccines, nucleic acid-based vaccinations, and virus-like particles [111]. The novel vaccination must pass three steps before being approved: There are three stages in the research process: exploratory, preclinical, and clinical (phase 1, phase 2, and phase 3) [112]. Moderna’s mRNA-1273, Ad5-nCoV (CanSino BIO), ChAdOx1 (University of Oxford), AstraZeneca’s AZD1222 and Pfizer, and BioNTech’s BNT162 are currently in phase 3 clinical trials [113].
Inactivated vaccine
In an inactivated vaccination, the virulence properties of live viruses are reduced by heat, UV rays and chemical treatment (formaldehyde or propiolactone). The pathogen in this vaccine remains in a dormant state. Although this deadly virus cannot cause disease, it can trigger an immune response in the event of a future infection. The inactivation form of the organism creates a secure vaccine, especially for immunocompromised individuals. Moreover, these COVID-19 vaccines showed a lower immune response as compared to the live vaccines, and they also may need many booster doses. These inactivated vaccines take a large amount of time to make, since the virus must first be cultivated in the laboratory before being inactivated [114, 115]. In addition, in the host body, the vaccine is unable to initiate a replicating process. It is less dangerous than a live attenuated vaccination (LAVs). Adjuvant in vaccine formulations is required to improve vaccination efficacy [116]. CoronaVac (Sinovac), Sinopharm, BBIBP-CorV, and Covaxin are the inactivated vaccines [117].
Live attenuated vaccine
LAV is the most traditional technique. A live attenuated vaccine is created by a live virus until it weakens as it transits through an animal or human cell, rendering the genome incapable of producing disease. This virus may multiply and replicate like a normal infection, resulting in a powerful immunological response from T and B cells. However, there is a slight risk that the mutation will revert to virulence and cause diseases [118]. Medicago Inc. and Codagenix are live attenuated vaccines. It is an influenza virus vaccine that is developed to utter the RBD domain in the SARS-CoV-2 S protein. This vaccine has the ability to induce TLRs (TLR 3, TLR 7/8, and TLR 9), which interact with B cells to generate an immune response [119].
Protein subunit vaccine
Long-lasting immune responses require recombinant antigenic proteins mediated sub-unit vaccines. SARS-CoV RBD (sRBD) holds antigenic epitopes, which elicit the neutralization process of antibodies as well as T cell responses [117]. RBD vaccinations were more immunogenic than full-length S protein vaccines. In the area beyond the RBD of the full-length S protein, super-antigens are present there. As a result, the RBD-based sub-unit vaccine may be the most suitable and secure way to make the COVID vaccine [120]. Sub-unit proteins are a considerably safer option, although their efficiency is limited. Adjuvanted systems have already been shown to efficiently improve the immunogenicity potency of these sub-unit vaccines by protecting and improving antigen targeting to professional antigen-presenting cells [121]. Recently, the Chinese Academy of Science developed a subunit vaccine known as Novavax.
Nucleic acid vaccine
The development of COVID-19 vaccines using nucleic acid is a cost-effective strategy. The nucleic acid-coded antigen is significantly used in vaccine development research. After the failure of the initial clinical study, plasmid DNA emerged as a potential platform despite the volatility of mRNA. A new set of clinical trials to make vaccinations against COVID-19 has been sparked by advancements in vaccine delivery. Several research organizations are now working on nucleic acid-based vaccinations [122]. The mRNA-based vaccination is a new, noninfectious, non-integrating platform that has a lower risk of insertional mutagenesis. Because of its less expensive and high safety issue in animal trials, it is the most promising option [123]. The immunogenic potency of the mRNA can be reduced, and changes can improve the vaccine’s stability. The mRNA-1273 is a synthetic viral mRNA vaccine that covers the entire SARS-CoV-2 S protein and simulates natural infection. Synthesized viral mRNA is recognized by the host body, which then translates the viral protein [124]. sRBD and the entire spike are encoded by the BNT162b1/BNT162b2 vaccines, which are codon-optimized mRNA vaccines [125]. In the case of antigen-encoded DNA vaccine, it is potent enough to stimulate the adaptive immune response, and it is the most revolutionary technique for immunization. Similar to the living virus, the transfected cells convey the transgene and allow a gradual supply of transgene-specific proteins. DNA vaccines that target the S, M, and N proteins elicit both humoral and cellular immune responses. Although the DNA-based vaccine platform is temperature stable and can be produced quickly, its efficacy and immunogenicity in humans have yet to be established. The injection of genome integration, vector mutations, and DNA vaccines within the host gene are still challenges [117]. INOVIO Pharmaceuticals has developed INO-4800, a DNA vaccination candidate. It was developed with the intention of improving the S protein sequence of the SARS-CoV-2 virus [126].
Viral vector vaccine
Recombinant DNA technology is used to create viral vector vaccines. Also introduced into the virus is DNA that encodes an antigen from the pathogen [118]. These types of vaccines are able to transfer the genes to the targeted cells in a very precise way. It was also observed that these kinds of vaccines produce a high level of immune response along with the massive gene transduction process. Due to the long half-life and peak level of antigenic protein expression, viral vector vaccines are more effective compared to other types of vaccines. They activate cytotoxic T cells, which kill virus-affected cells. Replicating (vesicular stomatitis virus and measles virus) and nonreplicating vectors (adenoviruses and poxviruses) are the two types of viral vectors. Interestingly, this adenovirus vector is used as the key element in some kinds of vaccine formulations. Due to its unique characteristics (genetically and physically stable), it does not combine with the host DNA. With DCs, entire dividing and nondividing cells can be easily infected by the Ads vector. But adenovirus vectors need high concentrations or doses to produce a satisfactory immune response. Also introduced into the virus is DNA that encodes an antigen from the pathogen. The viral vectors then enter the host cell and express the antigen. Within the host cell, some viruses can replicate and some fail to do so. In the biotechnology field, the S protein gene of SARS-CoV-2 was encoded into a replicating virus, such as measles or adenovirus. It is a safe virus and is able to induce a strong immune response through T and B cells (such as hepatitis B and HPV) [118, 121]. SARS-CoV-2 is also encoded by nonreplicating measles or adenoviruses with dormant genes. This vaccination is also safe, but long-term immunity requires a booster dose [118]. In this case, chimpanzee adenovirus (ChAdOx1) is used as a vector, which has been engineered to express the S protein of SARS-CoV-2 [13]. The University of Oxford and AstraZeneca are working together to develop AZD1222. This adenovirus has been genetically altered to prevent it from replicating in humans [127].
Immunological efficacy of COVID-19 vaccines currently used in India
Covaxin
With the collaboration of the Indian Council of Medical Research (Indian government-funded biomedical research agency), Bharat Biotech developed the Covaxin, India’s first indigenous COVID-19 vaccine [128]. Covaxin gained DCGI permission for human clinical trials in June 2020, and the first phase of the trials was completed by July [129]. Covaxin phase I clinical trials were conducted on 365 healthy volunteers across 12 institutes in India, including the All India Institute of Medical Sciences (AIIMS) in New Delhi, AIIMS Patna, and the Post Graduate Institute (PGI) of Medical Sciences in Rohtak [117]. It is an inactivated vaccine, and it cannot cause serious sickness in the host body. It makes use of a fully infective SARS-CoV-2 viral particle, which is composed of RNA, encased in a protein capsid that has been modified to hinder the viral reproduction process. It is comprised mostly of an entire inactivated SARS-CoV-2 antigen (strain NIV-2020–770), 250-g aluminum hydroxide gel as an adjuvant, 15 g TLR 7/8 agonist (imidazoquinolinone), 2.5 mg TM 2-phenoxyethanol, and phosphate buffer saline to a concentration of 0.5 ml. The vaccine does not require subzero storage or reconstitution, and it is available in multidose vials that are stable at temperatures ranging from 2 to 6 °C [130]. An adjuvant is used to boost up the specific immune response against the particular antigen, which is present in vaccine formulation [131]. In the case of Covaxin, an aluminum hydroxide-based adjuvant is formed to generate a crystalline aluminum oxyhydroxide by the addition of alkali to the solution of aluminum salt [132].
Covishield
Covishield (ChAdOx1-nCOV or AZD1222) is a recombinant, replication-deficient chimpanzee adenovirus vector encoding the SARS-CoV-2 S glycoprotein vaccine currently being developed by the Serum Institute of India (SII), Pune, based on the AstraZeneca-Oxford model [133, 134]. The AZD1222 COVID-19 vaccine was licensed by the UK on December 30, 2020, and India on January 2, 2021 [135]. This contains nonreplicating SARS-CoV-2 and adenovirus strains (causative of the common cold) that have been genetically engineered and weakened. According to the interim research, AZD1222 is 70.4% effective against COVID-19 prevention with no significant side [13]. A number of 5 × 1010 ChAdOx1-S (recombinant) virus particles are contained in one dose (0.5 ml). This vaccine also comprises the recipients’ magnesium chloride hexahydrate, L-histidine, L-histidine hydrochloride monohydrate, disodium edetate dihydrate, sodium chloride, ethanol, sucrose, polysorbate 80, and water for injection, in addition to ChAdOx1-S (recombinant) [130]. Both vaccines are stored and transferred at 2–8 °C [136, 137]. The immunization regimen of Covaxin and Covishield consists of two doses; in the case of Covaxin, 28 days apart; intramuscular injections are advised, and Covishield (0.5 ml in each dose) should be given 4–6 weeks apart [137].
Immunological response of both vaccines
Immune response of Covaxin
Individuals cannot be infected by inactivated vaccines because they cannot multiply, and the adjuvants are used to boost immunogenicity [138]. One of the most important findings from the phase I/II studies was the evidence of improved humoral and cell-mediated immune responses in Covaxin recipients [139]. Though CD4 + and CD8 + T cell responses were only detected in a subgroup of patients in the phase I study, the cell-mediated immune response was considerably improved in the phase II trial. Covaxin improved T cell memory, as seen by a rise in CD4 + , CD45RO + , and CD27 + T cells, supporting the antigen recall memory response. The adjuvant molecule Covaxin has a Toll-like receptor (TLR 7/8) agonist [140]. TLR7 and TLR8 agonists enhance the Th1 immune response but suppress the TH2 response, which is good for COVID-19 vaccines. Additionally, employing TLR 7/8 agonists as adjuvants may boost CD8 + T cell responses. Antiviral cytokines like TLR 7/8 agonists boost Th1 responses while suppressing Th2 responses, which is good for COVID-19 vaccines. Additionally, employing TLR 7/8 agonists as adjuvants may boost CD8 T cell responses. On days 7 and 14, antiviral cytokines such as IL-2, IL-4, IL-6, IL-10, IL-17, TNF-α, and IFN were also released, possibly contributing to increased APC activation such as DCs and macrophages. TLR recognition in the innate cell population has also been linked to the generation of early-IFN I, which aids viral clearance and the generation of pro-inflammatory cytokines [141] (Fig. 4).
Fig. 4.
Immune response of Covaxin and Covisheild. Covaxin: the virus is used in its inactive form, but its S protein remained intact. Aluminium-based adjuvant was also used here. Some of the inactivated viruses are captured by APCs inside the host. The coronavirus is processed apart by the antigen-presenting cell and displays some of the fragments on its surface. The fragment could be detected by helper T cell and become activated, which can help recruit other immune cells to respond to the vaccine if the fragment fits into one of its surface proteins. Covisheild: the genetic instruction of the SARS-CoV-2 S protein has infiltrated the adenovirus. In the body, the virus is engulfed in a bubble by the cell, which pulls it within. Once inside, the adenovirus breaks free from the bubble and proceeds to the nucleus and pushes into it. The coronavirus S protein gene may be read by the cell and copied into mRNA, which begins constructing S proteins after it leaves the nucleus. Some of the cell’s S proteins make spikes that migrate to the cell’s surface and stick out their tips. Some of the proteins are also broken down into pieces by the vaccinated cells, which are present on their surface. The immune system can then recognize these protruding spikes and S protein fragments
APCs (DCs, macrophages, neutrophils) pick up the S protein via phagocytosis, and they have the ability to trigger the proliferation and secretion of cytokines by naïve T cells [142]. APCs can break the protein into fragments and present the peptides on the surface through MHC I and MHC II with CD8 + and CD4 + T cells in lymph nodes [143]. Then, interleukin-12 and interleukin-4 are released by the interaction, which differentiates the naïve T cell to T helper cell-1 and T helper cell-2, respectively. T helper cell-1 acts in cell-mediated immunity and releases cytokines, IFN-γ, interleukin-2, and TNF-β, which make these cells particularly effective at preventing intracellular viral infections. T helper-2 cells can identify by secretion of IL-4, IL-5, IL-10, and IL-13 [144]. Helper T cells are important in a variety of immunologic processes, including cytotoxic T cell and macrophage activation, B cell maturation into plasma cells and memory B cells and produce B cell antibodies [145]. Cytotoxic CD8 T cells also secrete the cytokines IFN-γ, TNF-α, and TNF-β, which help the host defend itself in a variety of ways. Some B cells and T cells become memory cells, and they are ready to be reactive when they come into contact with some antigens [146]. Memory B cell plays a role in long-term immunity [147]. When SARS-CoV-2 enters the body, antibodies stop the viruses from binding to cells or tag them for destruction, and cytotoxic T cells recognize and destroy infected cells. Antibodies also stick to them and target S proteins and prevent the virus to enter cells [108].
Immune response of Covishield
The adenovirus vector ChAdOx1 nCoV-19 adenoviral vector-based vaccine (AZD1222) carries the entire structural surface glycoprotein (S protein) of the SARS-CoV-2. The S protein encoded by ChAdOx1 nCoV-19 has a codon-optimized coding sequence [148]. Human adenovirus, often known as the common cold virus, is the most prevalent nonreplicating viral vector used in the COVID-19 vaccine, i.e., Covishield [149]. ChAdOx1 nCoV-19 generates a widespread and robust T cell response in the host. There was a considerable increase in B cell activation and proliferation, as well as anti-IgA and anti-IgG antibodies, to the SARS-CoV-2 virus after immunization. S proteins were easily recognized in vaccinated people’s serum [150]. CD4 + T cells mainly produced Th1 cytokines (IFN-γ, IL-2, and TNF-α/β) instead of Th2 cytokines (IL-5 and IL-13). Importantly, it is shown, using a number of approaches, that immunization with ChAdOx1 nCoV-19 induces mostly a Th1 response [148]. ChAdOx1 nCoV-19 was found to be safe, tolerable, and immunogenic, with paracetamol being found to reduce reactogenicity and tolerability. A single dose of this vaccine can develop both humoral and cellular responses, and a booster dose generates neutralizing antibody titers [150].
In the vaccine, the adenovirus of chimpanzees is used. Some genes (E1 and E3) of this adenovirus are removed by a biotechnological method that makes adenovirus replication incompetent [151]. The gene of RNA of SARS-CoV-2, which synthesizes S protein through the process of reverse transcription from double-stranded DNA, S protein. Then, the DNA gene is inserted into the adenovirus, and the adenovirus is converted into the vector-based DNA vaccine and, after entering into the cell, activates the immune response. Then, adenovirus enters into the host cell and forms an endocytotic vesicle. Extrachromosomal DNA is converted into SARS CoV’s S protein mRNA, and it comes out of the nucleus. Next, the mRNA of S protein is translated by using the cellular translation machinery into the S protein [152]. Then, the S protein is processed, and some fragment of S protein (epitope) is displayed by MHC-I on the surface of this host cell. Next, the cytotoxic T cell (CD8 + lymphocyte) interacts with the MHC-I receptor and becomes active. Some S protein particles are picked up by B lymphocytes. APC engulfs the S protein particle and the viral DNA transcribed and translated by the cell machinery and produces S proteins. Some of these S proteins break into small pieces and are represented by the MHC-II on the surface of this cell to the CD4 + T cell. Through these interactions, some chemokines and interleukins are released and activate the growth and proliferation of CD8+ T cells and B cells. When the cytotoxic T cells become active, some of them destroy the vaccine-infected host cell by releasing granzyme and perforin, and some convert them to memory T cells. Then, the B cells are converted into plasma cells and memory B cells. The plasma cell produces antibodies against the S protein. This vaccine is based on highly immunogenic technology and stimulates a strong antibody and cell-mediated immune response, which provides long-term protection [153].
Efficacy of Covaxin and Covishield against mutant variants
WHO has renamed some variants “Variants of Concerns (VOCs)” or “Variants of Interests (VOIs)” because of their potency to notably change the virus’ properties. Recently, WHO has designated the dominantly spreading virus variants by Greek alphabets, i.e., Alpha (α) for B.1.1.7 (UK variant), Beta (β) for B.1.351 (South Africa), Gamma (γ) for P.1 (Brazil), Delta (γ) for B.1.617.2 (India), etc. [154]. SARS-CoV-2 Brazil variant, known as B.1.1.28 variant originated in Brazil, COVID-19 South Africa variant, known as the B.1.351 variant outbreak in South Africa, and UK variant, known as B.1.1.7 variant, are around in Delhi and Punjab, also a British variant [155]. A number of cases in the second wave have been gradually increasing in India. SARS-CoV-2 L452R, T478K, E484Q, D614G, and P681R variant cause for a large number of coronavirus cases in Maharashtra. The B.1.36 variant of SARS-CoV-2 cases was found in much higher numbers in Bangalore [156]. The known SARS-CoV-2 mutation in India is the SARS-CoV-2 double mutant strain, a combination of more than 2 COVID-19 variants, found in Punjab, Delhi, and Maharashtra and is classified as a B.1.617 variant, which contains L452R and E484Q variants [157]. SARS-CoV-2 triple mutant strains are also found recently in Maharashtra and Delhi during the second wave, which are more complex based on their severe infectivity pattern [158]. The B.1.617.2 variant has been given the name Delta. Delta, a highly contagious (and probably more severe) SARS-CoV-2 virus strain initially found in India in December 2020, is currently a major source of concern. It subsequently spread quickly throughout that country, as well as throughout Great Britain. The first Delta case in the USA was discovered just a few months ago (in March). Initially, the Delta variant was thought to be a variant of interest. However, due to its rapid spread over the world, the WHO classified this strain as a VOC in May 2021 [159]. Along with the mutations identified in the Beta and Gamma variations, the Delta Plus variant carries extra three mutations, K417N, V70F, and W258L on the coronavirus S protein. It was found for the first time in South Africa and Brazil, respectively. According to a previous analysis, Beta was associated with higher hospitalization and fatalities during South Africa’s first wave of illnesses, whereas Gamma was thought to be highly transmissible at the time. When comparing infectivity and transmission qualities amongst people who were previously infected earlier in the pandemic or who had weak or incomplete vaccination immunity, Delta Plus may have a minor advantage [160]. On November 25, WHO received the first report of the novel SARS-CoV-2 variant, Omicron, from South Africa. According to WHO, the first confirmed B.1.1.529 infection was discovered in a sample obtained on November 9 of this year. The new SARS-CoV-2 variation B.1.1.529, which was discovered in South Africa, was given the name ‘Omicron’ by WHO on November 26. Omicron has been categorized under ‘Variant of Concern,’ according to the WHO [161]. A summary of the information on the variants of SARS-CoV-2 has been provided in Table 1.
Table 1.
Characteristics of different SARS-CoV-2 variants and efficacy of COVID-19 vaccines
| Mutant strains of SARS-CoV-2 | Pango lineage | First identified | GISAID clade | Site of mutation | Key mutation | Additional amino acid changes observed | Alteration of vaccine effectiveness | Antibody response | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Alpha variant | B.1.1.7 | UK, September 2020 | GRY | Spike protein |
N501Y mutation: enhanced the viral binding potency with ACE2 receptor. H69del/V70del mutation: responsible for S gene targeting failure. P681H mutation: effectively increase the viral cell entry |
+ S:484 K + S:452R |
The efficacy of the AstraZeneca vaccine’s 70% and of the Pfizer vaccine is roughly 90% | High | [162, 163] |
| Beta variant | B.1.351 | South Africa, May 2020 | GH/501Y.V2 | Spike protein | There are 23 mutations with 17 amino acid changes, but the notable mutations in this variant are K417N, E484K, and N501Y on the spike protein. These mutations are able to increase their binding efficacy to the ACE2 receptor | + S:L18F | The Pfizer vaccine was 75% effective against any infection caused by the Beta variant after two doses. Novavax clinical trials in the UK revealed 89% efficacy, compared to merely 60% in South Africa, where the Beta strain was prevalent. Similarly, trials of the Johnson & Johnson vaccine in South Africa found lower levels of protection against moderate-to-severe COVID-19 than in the USA | Reduced | [163, 164] |
| Gamma variant | P.1 | Brazil, November 2020 | GR/501Y.V3 | Spike protein | 17 numbers of novel amino acid changes were observed, where 10 numbers of mutation in its S protein. The three main alarming mutations are N501Y, E484K, and K417T | + S:681H | After vaccination with Moderna or Pfizer, the variant has been demonstrated to be relatively resistant to neutralisation by convalescent plasma and vaccinee sera. The severity of the loss, however, was minor (3.8 to 4.8-fold) | High | [165] |
| Delta variant | B.1.617.2 | India, October 2020 | G/478 K.V1 | Spike protein | L452R and P681R mutations were observed in spike protein. Some extra mutations were observed such as Y145H, A222V, etc., in the Delta Plus variant | + S:417 N + S:484 K |
mRNA-1273 Vaccine effectiveness against infection with the Delta variant declined from 94.1% (90.5 to 96.3%) 14–60 days after vaccination to 80.0% (70.2 to 86.6%) 151–180 days after vaccination |
High | [166, 167] |
| Epsilon variant | B.1.429B.1.427 | Cedars-Sinai Medical Center, California July 2020 | CAL.20C | Spike protein | I4205V in ORF1a; D1183Y in ORF1b; L452R; W152C and L452R; these mutations were identified in spike protein | + S:13I | Neutralizing antibody titers against the B.1.427/B.1.429 variations are reduced (3 to sixfold) as compared to wild-type pseudoviruses, according to an analysis of neutralising antibody responses following spontaneous infection or mRNA vaccination | High | [168, 169] |
| Lambda variant | C.37 | Peru, December2020 | GR/452Q.V1 | Spike protein | G75V,T76I, L452Q, F490S, D614G, and T859N amino acid mutations were observed in S protein | – | Lambda variant of interest confers increased infectivity and immune escape from neutralizing antibodies elicited by CoronaVac | High | [170, 171] |
| Mu variant | B.1.621 | Colombia, January 2021 | GH | Spike protein | T95I, Y144S, Y145N, R346K, E484K, or the escape mutation, N501Y, D614G, P681H, and D950N were observed | – | Mu variant shows a pronounced resistance to antibodies elicited by natural SARS-CoV-2 infection and by the BNT162b2 mRNA vaccine | High | [172, 173] |
| Omicron variant | B.1.1.529 | Multiple countries, November 2021 | GRA | Spike protein | A67V, Δ69-70, T95I, G142D, Δ143-145, Δ211, L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F; these mutations were observed in spike protein. Many other mutations were located in RBD 319 to 541 | + R346K | Three doses of the Pfizer BioNTech Vaccine potently minimize the Omicron variant. After two significant doses of this vaccine, reduction of titer neutralization was noted | High | [174, 175] |
Comparative therapeutic advantages
In the Covaxin (BBV152), the virus strain (NIV-2020–770) with the Asp614Gly mutation, was isolated and sequenced from the patient who is infected by COVID-19, is used. The vaccine is injected intramuscularly into the deltoid muscle with 0.5 ml/dose volume. Two doses are needed in the gap of 28 days [133]. In this vaccine, the entire virion β-propiolactone-inactivated SARS-COV 2 vaccine is absorbed into the alum. By the 3 phases’ trail, it is observed that the efficacy is 81% [140]. This vaccine is also found to be effectively neutralized against the B1.1.7 variant of SARS-CoV-2 [176]. Covishield (AZD1222), the ChAdOxl nCoV-19 vaccine, is made with the help of an adenovirus vector; it carries a full length of this virus, which is inserted the coding sequence of the S protein of SARS-CoV-2. It is also administrated in two doses and 0.5 ml, which contain 5 × (10)10 viral particles. It is injected intramuscularly into the deltoid muscle, and the second dose is generally given 12 weeks apart after the first dose [148]. The general efficacy of this vaccine is 70% in Brazil and in the UK. In South Africa, the same vaccine showed only 22% efficacy for the new variant 501Y.V2 (B.1.351), which can resist both innate and adaptive immunity [177]. Covaxin is applied only to those people above 18, and Covishield is applied to people over 12 years old. The first dose of Covaxin was administrated among 96 health workers, and Covishield was administrated in 456 individuals in India. Covishield had a larger responder rate and a higher median increase in anti-spike antibodies than Covaxin (86.8% vs 43.8%), as well as a higher median increase in anti-spike antibodies (61.5 vs 6 AU/ml). Age, sex, and BMI had no effect on the results. With hypertension, the response rate was lower (65.5 vs 82.3%), and Covishield recipients experienced more side effects than Covaxin recipients (46.7 vs 31.2%). After the first dosage of Covishield, seropositivity rates to anti-spike antibody are higher in Covishield recipients than in Covaxin recipients [133]. Covishield vaccine has completed three trials, Covaxin has completed phase I and phase II studies, and phase-III is enrolled. The two doses of Covishield improve protection with a gap of 12 weeks, and Covaxin is recommended to be taken as a second dose after 4 weeks of the first dose [178]. Mild symptoms occur as a side effect of Covishield vaccine administration. Headache, nausea, myalgia, arthralgia, injection site tenderness, injection site pain, injection site warmth, injection site pruritus, weariness, malaise, feverishness, and chills are all very prevalent (around 10% of participants). Injection site edema, erythema, and fever are common (1–10% of people). Bharat Biotech also highlighted the hazards and negative effects of Covaxin. These include injection site pain, swelling, redness, itching, stiffness in the upper arm, weakness in the injection arm, body ache, headache, fever, malaise, weakness, rashes, nausea, vomiting, and body ache, headache, fever, malaise, weakness, rashes, nausea, and vomiting [130, 138]. If someone has a severe allergic problem with any ingredient in a vaccine, then he/she should not get the Covishield vaccine. And if anybody shows an allergic reaction after taking the first dose of Covishield, then he/she should not take the second dose of this vaccine. In the case of Covaxin, the same conditions apply, including acute infection and fever, pregnant women, and if someone is on blood thinner [179]. In Covaxin, the entire virion is propiolactone inactivated, imidazoquinoline molecule (Algel-IMDG) is used as TLR 7/8 agonist, but in the case of the Covishield vaccine, the adjuvant is not part of this vaccine formulation [139]. Here, the authors, Singh et al., conducted a study to increase the humoral response in the kinetics of antibody production against SARS-CoV-2 S protein as a result of the first and second doses of both vaccinations, Covishield and Covaxin, until 6 months after the trial was completed. SARS-CoV-2 naïve and SARS-CoV-2 recovered people are both involved in this study. The binding antibody against S protein kinetics after the first dose (day 21 or more till the second dose of vaccine) of two vaccines were reported in this experiment from the continuing coronavirus vaccine-induced antibody titer (COVAT) investigation. According to a survey, there is no significant difference in response rates based on age, sex, BMI, blood group, or other comorbidities, including any of the previously mentioned variables. Females had a greater median antibody titer than males. In the SARS-CoV-2 naïve individual, the median anti-spike antibody titer is higher in the SARS-CoV-2 vaccine-induced naïve individual. Post-vaccination side effects and the median elevation in antibody against SARS-CoV-2 S protein are higher in Covishield recipients than in Covaxin recipients. In the case of Covaxin, there is an increase in IgG antibodies (peaked by 28 days), and in the case of Covishield, there is an increase in IgG anti-spike antibodies after the second dose. The first dosage of Covishield generates binding antibody immunoreactivity by raising antibodies against the S protein in most individuals 21 days after the first dose, whereas Covaxin shows lower efficacy after the first dose. The structure of this virus’ spike glycoprotein and nucleocapsid protein is critical for the development of a vaccine [133]. In India, two vaccines are used, Covaxin and Covishield. These two main types of vaccines are produced by two different mechanisms; Covaxin, which is an inactivated virus vaccine, and Covishield, which is a nonreplicating virus vector vaccine. They induced immune responses and protective mechanisms in various ways [130, 139]. In the body, Covishield can produce 98.1% antibodies, and Covaxin can produce 80% antibodies. A total of 5.5% of people has become infected with the virus after taking Covishield, and 2.2% of people has become infected after taking Covaxin [133]. Multiple doses are required to induce adaptive immunity with these vaccines, which is one of their major drawbacks. Vaccines given with adjuvants cause a significant local reaction, which increases the antigenicity [180]. These vaccines necessitate booster shots as well as a large amount of virus to maintain the immunogenic particle’s integrity [181]. The therapeutic effectiveness of Covaxin and Covishield has been given in the comparative form in Table 2.
Table 2.
Summary of the comparative therapeutic advantages of Covaxin and Covishield
| Properties | Covaxin | Covishield |
|---|---|---|
| Developed by | Bharat Biotech, Hyderabad, and ICMR | Based on the AstraZeneca-Oxford model, Serum Institute of India |
| Types | Inactivated virus vaccine | Nonreplicating viral vector vaccine |
| Composition | It consists primarily of the entire inactivated SARS-CoV-2 antigen (strain NIV-2020–770) and a 250-g aluminium hydroxide gel adjuvant [140] | Covishield (AZD1222), the ChAdOxl nCoV-19 vaccine is formulized by the adenovirus vector, and it carries an entire length of virus, which is inserted the coding sequence of the spike protein of SARS-CoV-2 [148] |
| Efficacy rate | Covaxin has been found 78–80% effective | Covishield can be up to 90% effective |
| Antibody response | After the second dose of Covaxin, the level of IgG anti-spike antibody increased. Covaxin has lower efficacies after the first dose than Covishield. Covaxin can produce 80% antibodies in the body [133] | Single dose of Covishield, the number of IgG antibodies increased (peaking after 28 days). Covishield produces binding antibody immunoreactivity by increasing antibodies against the spike protein in most people 21 days after the first dose. Covishield can produce 98.1% antibodies [133] |
| Side effects | Localized side effects of Covaxin include pain and swelling at the injection site, whereas widespread side effects include upper arm stiffness, fever, fatigue, weakness, headache, nausea, vomiting, body discomfort, and rashes [182] | Localized effects include pain and tenderness, warmth, redness, swelling, or brushing at the injection site, and generalized effects include headache, fatigue, myalgia (muscle pain), malaise (generalized weakness), pyrexia (fever), chills, arthralgia (joint pain), nausea, and in rare cases, neurological complications [171] |
| Post-vaccination side effect | Lower than Covishield [133] | Higher in the case of Covishield [133] |
| Limitation | A person having acute illness and fever, pregnant women, and people who are on blood thinners should not take this vaccine [183] | If a person has a severe allergic reaction to any vaccine ingredient, he or she should not receive Covishield vaccination, and if a person has an allergic reaction after receiving the first dose of Covishield, he or she should not receive the second dose of this vaccine |
| Level the titer of antibodies | 51 AU/ml | 115 AU/ml |
| Doses | 2-dose gap intervals 28 days or 4 weeks [133] | 2 doses gap in 12 weeks [148] |
| Infected after vaccination | Approx. 2.2% [130] | Approx. 5.5% [130] |
Upcoming vaccines in India
Sputnik V
Sputnik V, a nonreplicating viral vector vaccine developed by the Gamaleya Research Institute in collaboration with the Russian Federation’s Ministry of Health, is now being studied in a phase 3 study in the Russian Federation. On August 11, 2020, Russia granted a license to the anti-SARS-CoV-2 vaccine Sputnik V, which was developed by the Gamaleya Institute in Moscow [179]. It is an adenoviral vector-delivered antigen same as Covishield. It also induces both cellular and humoral immunity after a single immunization, and after double immunization, it gives a double and long-lasting immune response [184]. This is a rAd type 26 and rAd type 5 combination vector vaccination. They both have the full-length SARS-CoV-2 glycoprotein S gene. They are administrated intramuscular and separately with a gap of 21 days. The first and second phase of the trial of this vaccine is done in August 2020 [185].
Biological E vaccine
Biotechnological E vaccine (Corbevax) is a new vaccination in India that is similar to the Novavax vaccine. It will be available in the country by the end of the year. It is a form of protein subunit vaccination that requires two separate doses to be effective. The Bio-E vaccine is a made-in-India vaccine that is projected to have 90% effectiveness against COVID-19 variations and will likely be a game changer in the fight against the pandemic, according to the manufacturer. Novavax will be manufactured in India by the Serum Institute of India, which also manufactures Covishield [186].
ZyCoV-D vaccine
ZyCov-D vaccine is developed by Ahmedabad-based Zydus-Cadila. Zydus Cadila, focused on discovering and developing NCEs, Novel Biologicals, Biosimilars, and Vaccines. Zocovirus-D is a DNA vaccine that is designed to function against the key viral membrane protein that is responsible for the novel coronavirus’ entrance into human cells. A plasmid DNA molecule, a small circular, and extrachromosomal bacterial DNA molecule that is employed in genetic engineering serve as the foundation for this technology. Additionally, the vaccine will be the first COVID-19 vaccine in India to have been tested in an adolescent population in the 12–18 years age bracket, which will be a significant benefit for the country. In addition, ZyCoV-D is a three-dose, intradermal vaccine [187].
Nasal spray vaccine
COVID-19 nasal vaccine BBV154 is developed by Bharat Biotech based in Hyderabad, India. Syringes are not used to administer other vaccines. In the area where they are immediately taking from the nose, COVID-19 nasal immunizations are more effective than syringe-based vaccines. Vaccination with BBV154 is administered intravenously and results in a broad immune response that includes neutralizing IgG, mucosal IgA, and T cell responses. Immune responses at the injection site (in the mucosa of the nose) are crucial for preventing both COVID-19 infection and transmission [188].
Future perspectives
Covishield is a type of vaccine that provokes antibodies against the S protein of the coronavirus. The immunological stimulation is better as it consists of a single epitope. As there is no significant mutation-related alteration in the S protein epitopic structure, Covishield is still widely effective against all mutant variants of COVID-19, including the Delta strain in India. On the other hand, Covaxin can develop antibodies against several epitopes, which are mostly similar to natural COVID-19 infections. But the overall effectiveness of this vaccine is lower than the Covishield. However, if the epitopic structure of spike protein is altered significantly in the future owing to spontaneous mutation, the Covishield may not function correctly. Therefore, immunization with one dosage of Covaxin and another dose of Covishield, or vice versa, may provide superior effectiveness. If a vaccine is developed based on the virus’ conserved sequence, it will be more effective than others. Even if many new variants or mutant strains are discovered, the efficacy rate of those vaccines will not be reduced. But it needs more experimental trials and should meet the entire desired standard for a successful vaccine formulation.
Conclusion
Since the emergence of SARS-CoV in 2002 and its propagation throughout 32 nations and areas, the globe has seen the outbreak of MERS-CoV and now the COVID-19. CoVs pose a constant threat to human health since they appear on a regular and unpredictable basis, spread quickly, and cause deadly infectious diseases. The first case of SARS-CoV-2 in India was reported in Kerala on 27 January 2020. There is currently no specific COVID-19 responsive drug commercially available for the treatment of SARS-CoV-2 infection. Some medicines and therapies like antiviral/retroviral drugs and certain kinds of steroids are being used to reduce swelling and inflammation of the lungs and other parts of the body. There are many mutant strains that have been detected in India, but still, the Covaxin and Covishield work against them. To date, in India, the effectiveness of Covishield is much higher than Covaxin. In Covishield, only the S protein work as a single epitope, so the immunogenicity and antigenicity are more. Because Covaxin contains the entire SARS-CoV-2 virus in an inactivated form, the body generates antibodies against various epitopes. Several studies reported that there is reduced efficacy of both vaccines against the Delta variant (B.1.617.2). Following Delta, the Omicron variant of SARS-CoV-2 (classified as B.1.1.529 by WHO) has recently emerged as another global menace, and investigations are being conducted to search out the efficacy of existing COVID-19 vaccines against it. Finally, the present review covers detailed insights into the immunological and therapeutic efficacy of two widely administered vaccines in India, which will enrich the knowledge in the field of concern.
Acknowledgements
The University of Gour Banga is greatly acknowledged for providing the opportunity to work in the Department of Physiology.
Abbreviations
- ARDS
Acute respiratory distress syndrome
- CAR
Coxsackievirus and adenovirus receptor
- CLRs
C-type lectin-like receptors
- CXCL
Chemokine (C-X-C motif) ligand
- DAMPs
Damage-associated molecular patterns
- DCGI
Drugs Controller General of India
- GM-CSF
Granulocyte-macrophage colony-stimulating factor
- LAV
Live attenuated vaccine
- MAMPs
Microbe-associated molecular pattern
- MASP-2
Mannan-binding lectin-associated serin protease-2
- MCP-1
Monocyte chemoattractant protein-1
- MERS
Middle East respiratory syndrome
- MIP 1α
Macrophage inflammatory protein 1-α
- MyD88
Myeloid differentiation factor 88
- NIV
National Institute of Virology
- NLR
NOD-like receptor
- NOD
Nucleotide-binding oligomerization domain
- PAMPs
Pathogen-associated molecular patterns
- PD1
Programmed cell death protein 1
- PRRs
Pattern recognition receptors
- RBD
Receptor-binding domain
- RLR
RIG-1-like receptor
- SARS
Severe acute respiratory syndrome
- SII
Serum Institute of India
- TCRs
Toll-like receptors
- TGFβ
Transforming growth factor β
- TIR
Toll/interleukin-1 receptor
- TLR
Toll-like receptor
- TNF-α
Tumor necrosis factor α
Author contribution
SD and SS: acquisition of information, drafted the article, designed the figures, and final approval of the version to be submitted. SKD and SSK: the conception and design of the study, revising it critically for important intellectual content, designed the figures, and final approval of the version to be submitted. JB and BG: interpretation of data, and revising it critically for important intellectual content. All the authors gave final approval of the version to be submitted.
Data availability
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Corman VM, Muth D, Niemeyer D, Drosten C. Hosts and sources of endemic human coronaviruses. Adv Virus Res. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hafeez A, Ahmad S, Siddqui SA, Ahmad M, Mishra S. A review of COVID-19 (coronavirus disease-2019) diagnosis, treatments and prevention. EJMO. 2020;4(2):116–25. doi: 10.14744/ejmo.2020.90853. [DOI] [Google Scholar]
- 3.Sohrabi C, Alsafi Z, O'neill N, Khan M, Kerwan A, Al-Jabir A, Iosifidis C, Agha R. World Health Organization declares global emergency: a review of the novel coronavirus (COVID-19) Int J Surg. 2019;2020(76):71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324(8):782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 5.Cucinotta D, Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91(1):157. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. WHO coronavirus (COVID-19) Dashboard. https://covid19.who.int/ (Accessed 19th December, 2020).
- 7.Jordan SC. Innate and adaptive immune responses to SARS-CoV-2 in humans: relevance to acquired immunity and vaccine responses. Clin Exp Immunol. 2021;204(3):310–320. doi: 10.1111/cei.13582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chowdhury MA, Hossain N, Kashem MA, Shahid MA, Alam A. Immune response in COVID-19: a review. J Infect Public Health. 2020;13(11):1619–1629. doi: 10.1016/j.jiph.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Subbarao K, Mahanty S. Respiratory virus infections: understanding COVID-19. Immunity. 2020;52(6):905–909. doi: 10.1016/j.immuni.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen PL, Lee NY, Cia CT, Ko WC, Hsueh PR. A review of treatment of coronavirus disease 2019 (COVID-19): therapeutic repurposing and unmet clinical needs. Front Pharmacol. 2020;1110.3389/fphar.2020.584956 [DOI] [PMC free article] [PubMed]
- 11.Sanders JM, Monogue ML, Jodlowski TZ, Cutrell JB. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;323(18):1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- 12.Kumar VM, Pandi-Perumal SR, Trakht I, Thyagarajan SP. Strategy for COVID-19 vaccination in India: the country with the second highest population and number of cases. NPJ Vaccines. 2021;6(1):1–7. doi: 10.1038/s41541-021-00327-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiagarajan K. COVID-19: India is at centre of global vaccine manufacturing, but opacity threatens public trust. BMJ. 2021;372:n196. doi: 10.1136/bmj.n196. [DOI] [PubMed] [Google Scholar]
- 14.Logunov DY, Dolzhikova IV, Zubkova OV, Tukhvatullin AI, Shcheblyakov DV, Dzharullaeva AS, Grousova DM, Erokhova AS, Kovyrshina AV, Botikov AG, Izhaeva FM. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. The Lancet. 2020;396(10255):887–897. doi: 10.1016/S0140-6736(20)31866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss SR, Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev. 2005;69(4):635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Binns C, Low WY, Kyung LM. The COVID-19 pandemic: public health and epidemiology. Asia Pac J Public Health. 2020;32(4):140–144. doi: 10.1177/1010539520929223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Astuti I. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes MetabSyndr: Clin Res Rev. 2020;14(4):407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mousavizadeh L, Ghasemi S. Genotype and phenotype of COVID-19: their roles in pathogenesis. J Microbiol Immunol Infect. 2021;54(2):159–163. doi: 10.1016/j.jmii.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol. 2022;23(1):3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, Connelly S, Droese B, Klaus JP, Makino S, Sawicki SG, Siddell SG. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011;174(1):11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeDiego ML, Álvarez E, Almazán F, Rejas MT, Lamirande E, Roberts A, Shieh WJ, Zaki SR, Subbarao K, Enjuanes L. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J Virol. 2007;81(4):1701–1713. doi: 10.1128/JVI.01467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mei J, Zhao D, Strotmann A. Intellectual structure of coronavirus research: a perspective from an author cocitation analysis. Front Res Metr. 2020;5:13. doi: 10.3389/frma.2020.595370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dos Santos WG. Impact of virus genetic variability and host immunity for the success of COVID-19 vaccines. Biomed Pharmacother. 2021;12:111272. doi: 10.1016/j.biopha.2021.111272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephens DS, McElrath MJ. COVID-19 and the path to immunity. JAMA. 2020;324(13):1279–1281. doi: 10.1001/jama.2020.16656. [DOI] [PubMed] [Google Scholar]
- 27.Yaqinuddin A, Kashir J. Innate immunity in COVID-19 patients mediated by NKG2A receptors, and potential treatment using Monalizumab, Cholroquine, and antiviral agents. Med Hypotheses. 2020;140:109777. doi: 10.1016/j.mehy.2020.109777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan Y, Zhang D, Yang P, Poon LL, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu C, Chen X, Cai Y, Zhou X, Xu S, Huang H, Zhang L, Zhou X, Du C, Zhang Y, Song J. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan. China JAMA Int Med. 2020;180(7):934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amarante-Mendes GP, Adjemian S, Branco LM, Zanetti LC, Weinlich R, Bortoluci KR. Pattern recognition receptors and the host cell death molecular machinery. Front Immunol. 2018;9:2379. doi: 10.3389/fimmu.2018.02379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manik M, Singh RK. Role of toll‐like receptors in modulation of cytokine storm signaling in SARS‐CoV‐2‐induced COVID‐19. J Med Virol. 2021;21:1–9. doi: 10.1002/jmv.27405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livia O, Caraglia M, Facchini G, Margherita V, De Placido S, Buonerba C. Toll-like receptors and COVID-19: a two-faced story with an exciting ending. Future Sci OA. 2020;6(8):FSO605. doi: 10.2144/fsoa-2020-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 34.Taefehshokr N, Taefehshokr S, Hemmat N, Heit B. COVID-19: perspectives on innate immune evasion. Frontiers in immunology. 2020;11. [DOI] [PMC free article] [PubMed]
- 35.Manjili RH, Zarei M, Habibi M, Manjili MH. COVID-19 as an acute inflammatory disease. J Immunol. 2020;205(1):12–19. doi: 10.4049/jimmunol.2000413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 37.Banerjee J, Dey S, Ali KM, Giri B, Dash SK. Advancements in adjuvanticity of bioactive inorganic and organic compounds. Endocr Metab Immune Disord. 2021;21(10):1721–1743. doi: 10.2174/1871530321666210225104819. [DOI] [PubMed] [Google Scholar]
- 38.de Marcken M, Dhaliwal K, Danielsen AC, Gautron AS, Dominguez-Villar M. TLR7 and TLR8 activate distinct pathways in monocytes during RNA virus infection. Sci Signal. 2019; 29;12(605). 10.1126/scisignal.aaw1347 [DOI] [PubMed]
- 39.Chua RL, Lukassen S, Trump S, Hennig BP, Wendisch D, Pott F, Debnath O, Thürmann L, Kurth F, Völker MT, Kazmierski J. COVID-19 severity correlates with airway epithelium–immune cell interactions identified by single-cell analysis. Nat Biotechnol. 2020;38(8):970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 40.Kell AM, Gale M., Jr RIG-I in RNA virus recognition. Virology. 2015;479:110–121. doi: 10.1016/j.virol.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goubau D, Schlee M, Deddouche S, Pruijssers AJ, Zillinger T, Goldeck M, Schuberth C, Van der Veen AG, Fujimura T, Rehwinkel J, Iskarpatyoti JA. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature. 2014;514(7522):372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim YK, Shin JS, Nahm MH. NOD-like receptors in infection, immunity, and diseases. Yonsei Med J. 2016;57(1):5–14. doi: 10.3349/ymj.2016.57.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kozlov EM, Ivanova E, Grechko AV, Wu WK, Starodubova AV, Orekhov AN. Involvement of oxidative stress and the innate immune system in SARS-CoV-2 infection. Diseases. 2021;9(1):17. doi: 10.3390/diseases9010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Charles TP, Shellito JE. Human immunodeficiency virus infection and host defense in the lungs. Semin Respir Crit Care Med. 2016;37(02):147–156. doi: 10.1055/s-0036-1572553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villadangos JA, Schnorrer P. Intrinsic and cooperative antigen-presenting functions of dendritic-cell subsets in vivo. Nat Rev Immunol. 2007;7(7):543–555. doi: 10.1038/nri2103. [DOI] [PubMed] [Google Scholar]
- 46.Steinman RM, Hemmi H. Dendritic cells: translating innate to adaptive immunity. From innate immunity to immunological memory. 2006:17–58. 10.1007/3-540-32636-7_2 [DOI] [PubMed]
- 47.Cervantes-Barragán L, Kalinke U, Züst R, König M, Reizis B, López-Macías C, Thiel V, Ludewig B. Type I IFN-mediated protection of macrophages and dendritic cells secures control of murine coronavirus infection. J Immunol. 2009;182(2):1099–1106. doi: 10.4049/jimmunol.182.2.1099. [DOI] [PubMed] [Google Scholar]
- 48.Glasner A, Zurunic A, Meningher T, LenacRovis T, Tsukerman P, Bar-On Y, Yamin R, Meyers AF, Mandeboim M, Jonjic S, Mandelboim O. Elucidating the mechanisms of influenza virus recognition by Ncr1. PLoS ONE. 2012;7(5):e36837. doi: 10.1371/journal.pone.0036837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D, Xu Y, Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020;17(5):533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guihot A, Litvinova E, Autran B, Debré P, Vieillard V. Cell-mediated immune responses to COVID-19 infection. Front Immunol. 2020;11:1662. doi: 10.3389/fimmu.2020.01662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Braud VM, Allan DS, O'Callaghan CA, Söderström K, D'Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, Lanier LL. HLA-E binds to natural killer cell receptors CD94/NKG2A. B and C Nature. 1998;391(6669):795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 52.Rosales C. Neutrophil: a cell with many roles in inflammation or several cell types. Front Physiol. 2018;9:113. doi: 10.3389/fphys.2018.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zuo Y, Zuo M, Yalavarthi S, Gockman K, Madison JA, Shi H, Woodard W, Lezak SP, Lugogo NL, Knight JS, Kanthi Y. Neutrophil extracellular traps and thrombosis in COVID-19. J Thromb Thrombolysis. 2021;51(2):446–453. doi: 10.1007/s11239-020-02324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ali YM, Ferrari M, Lynch NJ, Yaseen S, Dudler T, Gragerov S, Demopulos G, Heeney JL, Schwaeble WJ. Lectin pathway mediates complement activation by SARS-CoV-2 proteins. Front Immunol. 2021;12:2645. doi: 10.3389/fimmu.2021.714511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jarlhelt I, Nielsen SK, Jahn CX, Hansen CB, Pérez-Alós L, Rosbjerg A, Bayarri-Olmos R, Skjoedt MO, Garred P. SARS-CoV-2 antibodies mediate complement and cellular driven inflammation. Front. Immunol. 2021;12. 10.3389/fimmu.2021.767981 [DOI] [PMC free article] [PubMed]
- 56.Chen G, Wu DI, Guo W, Cao Y, Huang D, Wang H, Wang T, Zhang X, Chen H, Yu H, Zhang X. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Investig. 2020;1 130(5):2620–9. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jansen JM, Gerlach T, Elbahesh H, Rimmelzwaan GF, Saletti G. Influenza virus-specific CD4+ and CD8+ T cell-mediated immunity induced by infection and vaccination. J Clin Virol. 2019;119:44–52. doi: 10.1016/j.jcv.2019.08.009. [DOI] [PubMed] [Google Scholar]
- 58.Jenkins MK, Khoruts A, Ingulli E, Mueller DL, McSorley SJ, Reinhardt RL, Itano A, Pape KA. In vivo activation of antigen-specific CD4 T cells. Annu Rev Immunol. 2001;19(1):23–45. doi: 10.1146/annurev.immunol.19.1.23. [DOI] [PubMed] [Google Scholar]
- 59.Newton K, Dixit VM. Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol. 2012;4(3):a006049. doi: 10.1101/cshperspect.a006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14(1):36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, Grifoni A, Ramirez SI, Haupt S, Frazier A, Nakao C. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021; 371(6529). 10.1126/science.abf4063 [DOI] [PMC free article] [PubMed]
- 62.Azkur AK, Akdis M, Azkur D, Sokolowska M, van de Veen W, Brüggen MC, O’Mahony L, Gao Y, Nadeau K, Akdis CA. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy. 2020;75(7):1564–1581. doi: 10.1111/all.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo L, Ren L, Yang S, Xiao M, Chang D, Yang F, Dela Cruz CS, Wang Y, Wu C, Xiao Y, Zhang L. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin Infect Dis. 2020;71(15):778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu HQ, Sun BQ, Fang ZF, Zhao JC, Liu XY, Li YM, Sun XZ, Liang HF, Zhong B, Huang ZF, Zheng PY. Distinct features of SARS-CoV-2-specific IgA response in COVID-19 patients. Eur Respir J. 2020; 56(2). 10.1183/13993003.01526-2020 [DOI] [PMC free article] [PubMed]
- 65.Fourati S, Hue S, Pawlotsky JM, Mekontso-Dessap A, de Prost N. SARS-CoV-2 viral loads and serum IgA/IgG immune responses in critically ill COVID-19 patients. Intensive Care Med. 2020;46(9):1781–1783. doi: 10.1007/s00134-020-06157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stephens DS, McElrath MJ. COVID-19 and the path to immunity. Jama. 2020;6 324(13):1279–8. doi: 10.1001/jama.2020.16656. [DOI] [PubMed] [Google Scholar]
- 67.Lu LL, Suscovich TJ, Fortune SM, Alter G. Beyond binding: antibody effector functions in infectious diseases. Nat Rev Immunol. 2018;18(1):46–61. doi: 10.1038/nri.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shah VK, Firmal P, Alam A, Ganguly D, Chattopadhyay S. Overview of immune response during SARS-CoV-2 infection: lessons from the past. Front Immunol. 2020;7(11):1949. doi: 10.3389/fimmu.2020.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol. 2020;20(6):355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020;12:1–8. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang H, Li HB, Lyu JR, Lei XM, Li W, Wu G, Lyu J, Dai ZM. Specific ACE2 expression in small intestinal enterocytes may cause gastrointestinal symptoms and injury after 2019-nCoV infection. Int J Infect Dis. 2020;96:19–24. doi: 10.1016/j.ijid.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harrison AG, Lin T, Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;41(12):1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Snijder EJ, Van Der Meer Y, Zevenhoven-Dobbe J, Onderwater JJ, Van Der Meulen J, Koerten HK, Mommaas AM. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J Virol. 2006;80(12):5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu HY, Brian DA. Subgenomic messenger RNA amplification in coronaviruses. Proc Natl Acad Sci U S A. 2010;107(27):12257–12262. doi: 10.1073/pnas.1000378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LF. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martin TR, Wurfel MM, Zanoni I, Ulevitch R. Targeting innate immunity by blocking CD14: novel approach to control inflammation and organ dysfunction in COVID-19 illness. EBio Med. 2020;57:102836. doi: 10.1016/j.ebiom.2020.102836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Amor S, Fernández Blanco L, Baker D. Innate immunity during SARS-CoV-2: evasion strategies and activation trigger hypoxia and vascular damage. Clin Exp Immunol. 2020;202(2):193–209. doi: 10.1111/cei.13523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Min YQ, Huang M, Sun X, Deng F, Wang H, Ning YJ. Immune evasion of SARS-CoV-2 from interferon antiviral system. Comput Struct Biotechnol J. 2021;19:4217–4225. doi: 10.1016/j.csbj.2021.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoshikawa T, Hill TE, Yoshikawa N, Popov VL, Galindo CL, Garner HR, Peters CJ, Tseng CT. Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection. PLoS ONE. 2010;5(1):e8729. doi: 10.1371/journal.pone.0008729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shemesh M, Aktepe TE, Deerain JM, McAuley JL, Audsley MD, David CT, Purcell DF, Urin V, Hartmann R, Moseley GW, Mackenzie JM. SARS-CoV-2 suppresses IFNβ production mediated by NSP1, 5, 6, 15, ORF6 and ORF7b but does not suppress the effects of added interferon. PLoSPathog. 2021;17(8):e1009800. doi: 10.1371/journal.ppat.1009800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lei X, Dong X, Ma R, Wang W, Xiao X, Tian Z, Wang C, Wang Y, Li L, Ren L, Guo F. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat Commun. 2020;11(1):1–2. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jefferies CA. Regulating IRFs in IFN driven disease. Front Immunol. 2019;10:325. doi: 10.3389/fimmu.2019.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Siu KL, Chan CP, Kok KH, Woo PC, Jin DY. Suppression of innate antiviral response by severe acute respiratory syndrome coronavirus M protein is mediated through the first transmembrane domain. Cell Mol Immunol. 2014;11(2):141–149. doi: 10.1038/cmi.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Costela-Ruiz VJ, Illescas-Montes R, Puerta-Puerta JM, Ruiz C, Melguizo-Rodríguez L. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lucas C, Wong P, Klein J, Castro TB, Silva J, Sundaram M, Ellingson MK, Mao T, Oh JE, Israelow B, Takahashi T. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584(7821):463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hirano T, Murakami M. COVID-19: a new virus, but a familiar receptor and cytokine release syndrome. Immunity. 2020;19 52(5):731–3. doi: 10.1016/j.immuni.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou Y, Fu B, Zheng X, Wang D, Zhao C, Sun R, Tian Z, Xu X, Wei H. Aberrant pathogenic GM-CSF+ T cells and inflammatory CD14+ CD16+ monocytes in severe pulmonary syndrome patients of a new coronavirus. BioRxiv.2020 Jan 1. 10.1093/nsr/nwaa041
- 92.Yeap WH, Wong KL, Shimasaki N, Teo EC, Quek JK, Yong HX, Diong CP, Bertoletti A, Linn YC, Wong SC. CD16 is indispensable for antibody-dependent cellular cytotoxicity by human monocytes. Sci Rep. 2016;6(1):1–22. doi: 10.1038/srep34310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moore JB, June CH. Cytokine release syndrome in severe COVID-19. Science. 2020;368(6490):473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 94.Li G, Fan Y, Lai Y, Han T, Li Z, Zhou P, Pan P, Wang W, Hu D, Liu X, Zhang Q. Coronavirus infections and immune responses. J Med Virol. 2020;92(4):424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rayamajhi M, Zhang Y, Miao EA. Detection of pyroptosis by measuring released lactate dehydrogenase activity. Methods Mol Biol. 2013;1040:85–90. doi: 10.1007/978-1-62703-523-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J ThrombHaemost. 2020;18(4):844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Java A, Apicelli AJ, Liszewski MK, Coler-Reilly A, Atkinson JP, Kim AH, Kulkarni HS. The complement system in COVID-19: friend and foe?. JCI insight. 2020; 6: 5(15). 10.1172/jci.insight.140711 [DOI] [PMC free article] [PubMed]
- 99.Cugno M, Meroni PL, Gualtierotti R, Griffini S, Grovetti E, Torri A, Panigada M, Aliberti S, Blasi F, Tedesco F, Peyvandi F. Complement activation in patients with COVID-19: a novel therapeutic target. J Allergy Clin Immunol. 2020;146(1):215. doi: 10.1016/j.jaci.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jodele S, Köhl J. Tackling COVID-19 infection through complement-targeted immunotherapy. Br J Pharmacol. 2021;178(14):2832–2848. doi: 10.1111/bph.15187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mastellos DC, da Silva BG, Fonseca BA, Fonseca NP, Auxiliadora-Martins M, Mastaglio S, Ruggeri A, Sironi M, Radermacher P, Chrysanthopoulou A, Skendros P. Complement C3 vs C5 inhibition in severe COVID-19: early clinical findings reveal differential biological efficacy. Clin Immunol. 2020;220:108598. doi: 10.1016/j.clim.2020.108598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liao M, Liu Y, Yuan J, et al. The landscape of lung bronchoalveolar immune cells in COVID-19 revealed by single-cell RNA sequencing. Nat Med. 2020;26:842–844. doi: 10.1101/2020.02.23.20026690. [DOI] [PubMed] [Google Scholar]
- 103.Wen W, Su W, Tang H, et al. Immune cell profiling of COVID-19 patients in the recovery stageby single-cell sequencing. Cell Discov. 2020;6(1):31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Giamarellos-Bourboulis EJ, Netea MG, Rovina N, et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27(6):992–1000. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Diao B, Wang C, Tan Y. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19). Front Immunol. 2020; 11(827). 10.3389/fimmu.2020.00827 [DOI] [PMC free article] [PubMed]
- 106.van Erp EA, van Kampen MR, van Kasteren PB, et al. Viral infection of human natural killer cells. Viruses. 2019;11(3):243. doi: 10.3390/v11030243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yao C, Bora SA, Parimon T, Zaman T, Friedman OA, Palatinus JA, Surapaneni NS, Matusov YP, Chiang GC, Kassar AG, Patel N. Cell-type-specific immune dysregulation in severely ill COVID-19 patients. Cell Rep. 2021;34(1):108590. doi: 10.1101/2020.07.23.20161182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shah VK, Firmal P, Alam A, Ganguly D, Chattopadhyay S. Overview of immune response during SARS-CoV-2 infection: lessons from the past. Front Immunol. 2020;11:1949. doi: 10.3389/fimmu.2020.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lau YL, Peiris J, Law HKW, et al. Role of dendritic cells in SARS coronavirus infection. Hong Kong Med J. 2012;18:28–30. [PubMed] [Google Scholar]
- 110.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 111.World Health Organization. DRAFT landscape of COVID-19 candidate vaccines. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines Accessed 4 March 2020.
- 112.Sharma O, Sultan AA, Ding H, Triggle CR. A review of the progress and challenges of developing a vaccine for COVID-19. Front Immunol. 2020;11:2413. doi: 10.3389/fimmu.2020.585354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dash P, Mohapatra S, Ghosh S, Nayak B. A scoping insight on potential prophylactics, vaccines and therapeutic weaponry for the ongoing novel coronavirus (COVID-19) pandemic-a comprehensive review. Front Pharmacol. 2021;11:2471. doi: 10.3389/fphar.2020.590154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jeyanathan M, Afkhami S, Smaill F, Miller MS, Lichty BD, Xing Z. Immunological considerations for COVID-19 vaccine strategies. Nat Rev Immunol. 2020;20(10):615–632. doi: 10.1038/s41577-020-00434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xia S, Duan K, Zhang Y, Zhao D, Zhang H, Xie Z, Li X, Peng C, Zhang Y, Zhang W, Yang Y. Effect of an inactivated vaccine against SARS-CoV-2 on safety and immunogenicity outcomes: interim analysis of 2 randomized clinical trials. Jama. 2020;8 324(10):951–60. doi: 10.1001/jama.2020.15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ciabattini A, Nardini C, Santoro F, Garagnani P, Franceschi C, Medaglini D. Vaccination in the elderly: the challenge of immune changes with aging. Semin Immunol. 2018;40:83–94. doi: 10.1016/j.smim.2018.10.010. [DOI] [PubMed] [Google Scholar]
- 117.Belete TM. Review on up-to-date status of candidate vaccines for COVID-19 disease. Infect Drug Resist. 2021;14:151. doi: 10.2147/IDR.S288877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Khuroo MS, Khuroo M, Khuroo MS, Sofi AA, Khuroo NS. COVID-19 vaccines: a race against time in the middle of death and devastation. J Clin Exp Hepatol. 2020;10:610–21. doi: 10.1016/j.jceh.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kaur SP, Gupta V. COVID-19 vaccine: a comprehensive status report. Virus Res. 2020;288:198114. doi: 10.1016/j.virusres.2020.198114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dai L, Zheng T, Xu K, Han Y, Xu L, Huang E, An Y, Cheng Y, Li S, Liu M, Yang M. A universal design of betacoronavirus vaccines against COVID-19, MERS, and SARS. Cell. 2020;182(3):722–733. doi: 10.1016/j.cell.2020.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Perrie Y, Mohammed AR, Kirby DJ, McNeil SE, Bramwell VW. Vaccine adjuvant systems: enhancing the efficacy of sub-unit protein antigens. Int J Pharm. 2008;364:272–280. doi: 10.1016/j.ijpharm.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 122.Piyush R, Rajarshi K, Chatterjee A, Khan R, Ray S. Nucleic acid-based therapy for coronavirus disease 2019. Heliyon. 2020;19:e05007. doi: 10.1016/j.heliyon.2020.e05007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough MP, Chappell JD, Denison MR, Stevens LJ, Pruijssers AJ. An mRNA vaccine against SARS-CoV-2—preliminary report. N Engl J Med. 2020;14. 10.1056/NEJMoa2022483 [DOI] [PMC free article] [PubMed]
- 124.Wang F, Kream RM, Stefano GB. An evidence based perspective on mRNA-SARS-CoV-2 vaccine development. Med Sci Monit. 2020;26:e924700–1. doi: 10.12659/MSM.924700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Walsh EE, Frenck R, Falsey AR, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Mulligan MJ, Bailey R, Swanson KA. RNA-based COVID-19 vaccine BNT162b2 selected for a pivotal efficacy study. Medrxiv. 2020 10.1056/NEJMoa2027906
- 126.ClinicalTrials.gov. Identifier: NCT04336410. 2020. Available from: https://clinicaltrials.gov/ct2/show/NCT04336410. Accessed May2, 2020
- 127.Saini P. COVID-19 pandemic: potential phase III vaccines in development. T Appl Biol Chem J. 2020;1(1):21-33. https://doi.org/10.52679/tabcj.2020.0004
- 128.Thiagarajan K. What do we know about India’s Covaxin vaccine?. BMJ: British Medical Journal (Online). 2021 Apr 20;373. 10.1136/bmj.n997 [DOI] [PubMed]
- 129.Zhang N, Li C, Hu Y, Li K, Liang J, Wang L, Du L, Jiang S. Current development of COVID-19 diagnostics, vaccines and therapeutics. Microbes Infect. 2020;22(6–7):231–235. doi: 10.1016/j.micinf.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gharate JS, Daitkar SA, Aher KA. Approved COVID 19 vaccines: a review. World J Pharm Res. 2021;10(10):523–542. [Google Scholar]
- 131.Vogel FR. Improving vaccine performance with adjuvants. Clin Infect Dis. 2000; 30(Supplement_3):S266–70. 10.1086/313883 [DOI] [PubMed]
- 132.Liang Z, Zhu H, Wang X, Jing B, Li Z, Xia X, Sun H, Yang Y, Zhang W, Shi L, Zeng H. Adjuvants for coronavirus vaccines Front Immunol. 2020;11:2896. doi: 10.3389/fimmu.2020.589833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Singh AK, Phatak SR, Singh R, Bhattacharjee K, Singh NK, Gupta A, Sharma A. Antibody response after first and second-dose of ChAdOx1-nCOV (CovishieldTM®) and BBV-152 (CovaxinTM®) among health care workers in India: the final results of cross-sectional coronavirus vaccine-induced antibody titre (COVAT) study. Vaccine. 2021;39(44):6492–6509. doi: 10.1016/j.vaccine.2021.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Malabadi RB, Meti NT, Chalannavar RK. Applications of nanotechnology in vaccine development for coronavirus (SARS-CoV-2) disease (Covid-19) Int J Res Sci Inno. 2021;8(2):191–198. [Google Scholar]
- 135.Voysey M, Clemens SA, Madhi SA, Weckx LY, Folegatti PM, et al. Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet. 2021;397(10277):881–91. 10.1016/S0140-6736(21)00432-3. [DOI] [PMC free article] [PubMed]
- 136.Thiagarajan K. What do we know about India’s Covaxin vaccine. bmj. 2021;373:n997. doi: 10.1136/bmj.n997. [DOI] [PubMed] [Google Scholar]
- 137.Misra SK, Pathak K, Pathak D, Yadav R. Current updates on COVID-19 vaccines. Asian J Pharm Clin Res. 2021;7:17–23. doi: 10.22159/ajpcr.2021v14i5.41061. [DOI] [Google Scholar]
- 138.World Health Organization. Interim recommendations for use of the Bharat Biotech BBV152 COVAXIN® vaccine against COVID-19: interim guidance, 3 November 2021.
- 139.Ella R, Reddy S, Jogdand H, Sarangi V, Ganneru B, Prasad S, Das D, Raju D, Praturi U, Sapkal G, Yadav P, Reddy P, Verma S, Singh C, Redkar SV, Gillurkar CS, Kushwaha JS, Mohapatra S, Bhate A, Rai S, Panda S, Abraham P, Gupta N, Ella K, Bhargava B, Vadrevu KM. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBV152: interim results from a double-blind, randomised, multicentre, phase 2 trial, and 3-month follow-up of a double-blind, randomised phase 1 trial. Lancet Infect Dis. 2021;21(7):950–961. doi: 10.1016/S1473-3099(21)00070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ella R, Vadrevu KM, Jogdand H, Prasad S, Reddy S, Sarangi V, Ganneru B, Sapkal G, Yadav P, Abraham P, Panda S, Gupta N, Reddy P, Verma S, Kumar Rai S, Singh C, Redkar SV, Gillurkar CS, Kushwaha JS, Mohapatra S, Rao V, Guleria R, Ella K, Bhargava B. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBV152: a doubleblind, randomised, phase 1 trial. Lancet Infect Dis. 2021;21(5):637–646. doi: 10.1016/S1473-3099(20)30942-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ganneru B, Jogdand H, Daram VK, Das D, Molugu NR, Prasad SD, Kannappa SV, Ella KM, Ravikrishnan R, Awasthi A, Jose J. Th1 skewed immune response of whole virion inactivated SARS CoV 2 vaccine and its safety evaluation. Iscience. 2021;24(4):102298. doi: 10.1016/j.isci.2021.102298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.García LF. Immune response, inflammation, and the clinical spectrum of COVID-19. Front Immunol. 2020;11:1441. doi: 10.3389/fimmu.2020.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Steinman RM, Hawiger D, Nussenzweig MC. Tolerogenic dendritic cells. Annu rev immunol. 2003;21:685711. doi: 10.1146/annurev.immunol.21.120601.141040. [DOI] [PubMed] [Google Scholar]
- 144.Kaiko GE, Horvat JC, Beagley KW, Hansbro PM. Immunological decision-making: how does the immune system decide to mount a helper T-cell response? Immunology. 2008;123(3):326–338. doi: 10.1111/j.1365-2567.2007.02719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Abbas AK, Burstein HJ, Bogen SA. Determinants of helper T cell-dependent antibody production. Semin Immunol. 1993;5(6):441–447. doi: 10.1006/smim.1993.1050. [DOI] [PubMed] [Google Scholar]
- 146.Pennock ND, White JT, Cross EW, Cheney EE, Tamburini BA, Kedl RM. T cell responses: naive to memory and everything in between. Adv Physiol Educ. 2013;37(4):273–283. doi: 10.1152/advan.00066.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dörner T, Radbruch A. Antibodies and B cell memory in viral immunity. Immunity. 2007;21 27(3):384–92. doi: 10.1016/j.immuni.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 148.Folegatti PM, Ewer KJ, Aley PK, Angus B, Becker S, Belij-Rammerstorfer S, Bellamy D, Bibi S, Bittaye M, Clutterbuck EA, Dold C. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Flanagan KL, Best E, Crawford NW, Giles M, Koirala A, Macartney K, Russell F, Teh BW, Wen SC. Progress and pitfalls in the quest for effective SARS-CoV-2 (COVID-19) vaccines. Front Immunol. 2020;11:2410. doi: 10.3389/fimmu.2020.579250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Goyal K, Goel H, Baranwal P, Tewary A, Dixit A, Pandey AK, Benjamin M, Tanwar P, Dey A, Khan F, Pandey P. Immunological mechanisms of vaccine-induced protection against SARS-CoV-2 in humans. Immuno. 2021;1(4):442–456. doi: 10.3390/immuno1040032. [DOI] [Google Scholar]
- 151.Kovesdi I, Hedley SJ. Adenoviral producer cells. Vaccine. 2010;2:1681–1703. doi: 10.3390/v2081681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Doerfler W. Adenoviral vector DNA-and SARS-CoV-2 mRNA-based COVID-19 vaccines: possible integration into the human genome-are adenoviral genes expressed in vector-based vaccines? Virus Res. 2021;302:198466. doi: 10.1016/j.virusres.2021.198466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dong Y, Dai T, Wei Y, Zhang L, Zheng M, Zhou F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct Target Ther. 2020;5(1):1–4. doi: 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Khateeb J, Li Y, Zhang H. Emerging SARS-CoV-2 variants of concern and potential intervention approaches. Crit Care. 2021;25(1):1–8. doi: 10.1186/s13054-021-03662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Garcia-Beltran WF, Lam EC, Denis KS, Nitido AD, Garcia ZH, Hauser BM, Feldman J, Pavlovic MN, Gregory DJ, Poznansky MC, Sigal A. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184(9):2372–2383. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cherian S, Potdar V, Jadhav S, Yadav P, Gupta N, Das M, Rakshit P, Singh S, Abraham P, Panda S, Team NI. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms. 2021;9(7):1542. doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Joshi SR, Pandit R, Parikh P, Gulia A. Containing COVID-19 second surge in India. Indian J Med Sci. 2021;73:1. doi: 10.25259/IJMS_157_2021. [DOI] [Google Scholar]
- 158.Sahoo JP, Mishra AP, Samal KC. Triple mutant Bengal strain (B. 1.618) of coronavirus and the worst COVID outbreak in India. Biol Res Tod. 2021;3(4):261–5. [Google Scholar]
- 159.Aleem A, AB AS, Slenker AK. Emerging variants of SARS-CoV-2 and novel therapeutics against coronavirus (COVID-19). [PubMed]
- 160.Kannan SR, Spratt AN, Cohen AR, Naqvi SH, Chand HS, Quinn TP, Lorson CL, Byrareddy SN, Singh K. Evolutionary analysis of the Delta and Delta Plus variants of the SARS-CoV-2 viruses. J Autoimmun. 2021;124:102715. doi: 10.1016/j.jaut.2021.102715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mahase E. COVID-19: do vaccines work against omicron—and other questions answered. BMJ 2021; 375 10.1136/bmj.n3062 [DOI] [PubMed]
- 162.Zuckerman NS, Fleishon S, Bucris E, Bar-Ilan D, Linial M, Bar-Or I, Indenbaum V, Weil M, Lustig Y, Mendelson E, Mandelboim M. A unique SARS-CoV-2 spike protein P681H variant detected in Israel. Vaccines. 2021;9(6):616. doi: 10.3390/vaccines9060616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Duong D. Alpha, Beta, Delta, Gamma: what’s important to know about SARS-CoV-2 variants of concern? Can Med Assoc J. 2021;193(27):E1059–E1060. doi: 10.1503/cmaj.1095949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Madhi SA, Baillie V, Cutland CL, Voysey M, Koen AL, Fairlie L, Padayachee SD, Dheda K, Barnabas SL, Bhorat QE, Briner C. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B. 1.351 variant. N Engl J Med. 2021;384(20):1885–98. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Faria NR, Claro IM, Candido D, Franco LM, Andrade PS, Coletti TM, Silva CA, Sales FC, Manuli ER, Aguiar RS, Gaburo N. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virological. 2021;12(372):815–821. doi: 10.1101/2021.02.26.21252554. [DOI] [Google Scholar]
- 166.American society of microbiology. How-Dangerous-is-the-Delta-Variant-B-1–617–2. https://asm.org/Articles/2021/July. (Accessed in 15th December, 2021).
- 167.Bruxvoort K, Sy LS, Qian L, Ackerson BK, Luo Y, Lee GS, Tian Y, Florea A, Aragones M, Tubert JE, Takhar HS. Effectiveness of mRNA-1273 against Delta, Mu, and other emerging variants. medRxiv. 2021;375:e068848. doi: 10.1136/bmj-2021-068848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wadman M. California coronavirus strain may be more infectious—and lethal. Science. 2021 (Accessed in 15th December, 2021).
- 169.Zhang W, Davis BD, Chen SS, Martinez JM, Plummer JT, Vail E. Emergence of a novel SARS-CoV-2 variant in Southern California. JAMA Netw Open. 2021;325(13):1324–1326. doi: 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.World Health Organization. Tracking SARS-CoV-2 variants. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ WHO. (Accessed in 15th December, 2021).
- 171.Kimura I, Kosugi Y, Wu J, Yamasoba D, Butlertanaka EP, Tanaka YL, Liu Y, Shirakawa K, Kazuma Y, Nomura R, Horisawa Y. SARS-CoV-2 Lambda variant exhibits higher infectivity and immune resistance. BioRxiv.2021 Jan 1. 10.1101/2021.07.28.454085
- 172.Outbreak. Info. Lineage. Mutation Tracker, Mu Variant Report. https://outbreak.info/situation-reports/Mu (Accessed in 15th December, 2021).
- 173.Fratev F. The R346K mutation in the Mu variant of SARS-CoV-2 alter the interactions with monoclonal antibodies from class 2: a free energy of perturbation study. BioRxiv. 2021 10.1101/2021.10.12.463781 [DOI] [PubMed]
- 174.UK Health Security Agency. SARS-CoV WH. Variants of concern and variants of interest, updated 31 May 2021. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1042367/technical_briefing-31-10-december-2021.pdf. (Accessed in 19th December, 2021).
- 175.Pfize. Pfizer and BioNTech provide update on Omicron variant. https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-provide-update-omicron-variant (Accessed in 19th December, 2021).
- 176.Sapkal GN, Yadav PD, Ella R, Deshpande GR, Sahay RR, Gupta N, Vadrevu KM, Abraham P, Panda S, Bhargava B. Inactivated COVID-19 vaccine BBV152/COVAXIN effectively neutralizes recently emerged B. 1.1. 7 variant of SARS-CoV-2. J. Trav Med. 2021;28:51. doi: 10.1093/jtm/taab051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Karim SS. Vaccines and SARS-CoV-2 variants: the urgent need for a correlate of protection. Lancet. 2021;397:1263–1264. doi: 10.1016/S0140-6736(21)00468-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bhasker B. COVID 19 vaccination: latest guidelines on blood donor deferral in India. Transfus Clin Biol. 2021;28(3):299. doi: 10.1016/j.tracli.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kashte S, Gulbake A, El-Amin SF, III, Gupta A. COVID-19 vaccines: rapid development, implications, challenges and future prospects. Hum cell. 2021;7:1–23. doi: 10.1007/s13577-021-00512-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Coffman RL, Sher A, Seder RA. Vaccine adjuvants: putting innate immunity to work. Immunity. 2010;33(4):492–503. doi: 10.1016/j.immuni.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kaur SP, Gupta V. COVID-19 vaccine: a comprehensive status report. Virus Res. 2020;13:198114. doi: 10.1016/j.virusres.2020.198114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Jayadevan R, Shenoy RS, Anithadevi TS. Survey of symptoms following COVID-19 vaccination in India.Medrxiv.2021 .10.1101/2021.02.08.21251366
- 183.COVAXIN Factsheet-V4. Restricted use in emergency situation of COVID-19. https://www.bharatbiotech.com/images/covaxin/covaxin-factsheet.pdf (Accessed 19th December, 2021).
- 184.Jones I, Roy P. Sputnik V COVID-19 vaccine candidate appears safe and effective. Lancet. 2021;397:642–643. doi: 10.1016/S0140-6736(21)00191-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Logunov DY, Dolzhikova IV, Shcheblyakov DV, Tukhvatulin AI, Zubkova OV, Dzharullaeva AS, Kovyrshina AV, Lubenets NL, Grousova DM, Erokhova AS, Botikov AG. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021;397(10275):671–681. doi: 10.1016/S0140-6736(21)00234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Pagliusi S, Jarrett S, Hayman B, Kreysa U, Prasad SD, Reers M, Thai PH, Wu K, Zhang YT, Baek YO, Kumar A. Emerging manufacturers engagements in the COVID− 19 vaccine research, development and supply. Vaccine. 2020;38(34):5418–5423. doi: 10.1016/j.vaccine.2020.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sharun K, Dhama K. India’s role in COVID-19 vaccine diplomacy. J. Travel Med. 2021 10.1093/jtm/taab064 [DOI] [PMC free article] [PubMed]
- 188.Chavda VP, Vora LK, Pandya AK, Patravale VB. Intranasal vaccines for SARS-CoV-2: from challenges to potential in COVID-19 management. Drug Discov Today. 2021;26(11):2619–2636. doi: 10.1016/j.drudis.2021.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.