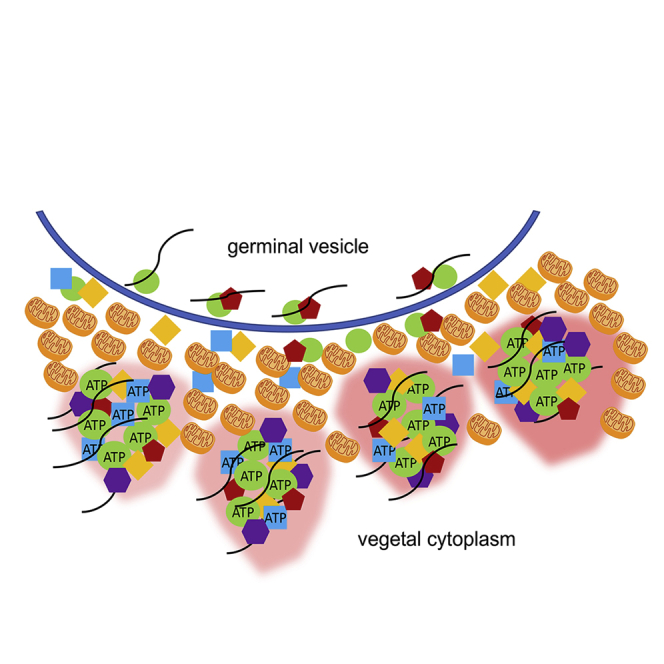
Summary
The Balbiani body (Bb), an organelle comprised of mitochondria, ER, and RNA, is found in the oocytes of most organisms. In Xenopus, the structure is initially positioned immediately adjacent to the nucleus, extends toward the vegetal pole, and eventually disperses, leaving behind a region highly enriched in mitochondria. This area is later transversed by RNP complexes that are being localized to the vegetal cortex. Inhibition of mitochondrial ATP synthesis prevents perinuclear formation of the transport complexes that can be reversed by a nonhydrolyzable ATP analog, indicating the nucleotide is acting as a hydrotrope. The protein composition, sensitivity to hexanediol, and coalescence in the absence of transport provide evidence that the transport RNP complexes are biocondensates. The breakdown of the Bb engenders regions of clustered mitochondria that are used not to meet extraordinary energy demands, but rather to promote a liquid-liquid phase separation.
Subject areas: Molecular biology, Cell biology, Biophysics
Graphical abstract
Highlights
-
•
Mitochondria in remnants of the Bb generate locally elevated levels of ATP
-
•
Mitochondrial activity needed for formation of RNA transport complexes
-
•
Transport complexes are RNP condensates
-
•
ATP functions as a hydrotrope to support the LLPS of RNP transport condensates
Molecular biology; Cell biology; Biophysics
Introduction
The Balbiani body (Bb), an amyloid-like organelle comprised of mitochondria, ER, and RNA, is found in the oocytes of most organisms (Jamieson-Lucy and Mullins, 2019; Kloc et al., 2004; Oh and Houston, 2017). The formation of the Bb is an early event in establishing cell polarization; however, the function of the organelle is not clear. There are phylogenetic differences with regard to whether the organelle participates in germ cell specification (Jamieson-Lucy and Mullins, 2019). In some species (frog, fish, and insect), the Bb contains germ plasm, the maternal determinants that induce germ cell fate; whereas, in other species, including mammals, specification is inductive and independent of this organelle. Another proposed role for the Bb is to select and protect healthy mitochondria that will be passed on to progeny often after a long period of cell dormancy (Bilinski et al., 2017; Jamieson-Lucy and Mullins, 2019); although, there are few details concerning how this process might occur.
In Xenopus, the Bb is initially positioned adjacent to the oocyte nucleus, also known as the germinal vesicle (GV), facing the vegetal pole. Indeed, formation of the Bb is one of the first observable signs of oocyte polarity. In zebrafish oocytes, zygotene telomeres are clustered at one location on the nuclear envelope that coincides with the cytoplasmic location of the centrosome, which in turn marks the site of recruitment of Bucky ball (Buc), the major structural protein that comprises the Bb, initiating its formation (Elkouby et al., 2016). The Xenopus Bb eventually extends toward the vegetal cortex and delivers germ plasm determinants including nanos1 and wnt11 mRNAs, after which it fragments, leaving a wedge-shaped cytoplasmic region rich in mitochondria (Wilk et al., 2005). Shortly after the breakdown of the Bb, a second group of mRNAs begins to move to the vegetal cortex through a process dependent on the cell cytoskeleton (Betley et al., 2004; Gagnon et al., 2013). This latter group of localized mRNAs, which include vg1/gdf1 and vegT, ultimately contribute to axis formation in the embryo.
There is no information addressing whether the polarity established by the Bb has a role in the movement of mRNA through this pathway. We have found that mitochondria-rich remnants of the Bb generate a region of elevated ATP through which vg1 mRNA is being localized to the vegetal cortex. Inhibition of mitochondrial ATP production prevents formation of the vg1 RNP complexes as they enter the cytoplasm. Rescue by a nonhydrolyzable ATP analog indicates that the elevated levels of the nucleotide are not used to fulfill energy demands, but rather may be functioning as a hydrotrope, which are small, amphipathic compounds that increase the solubility of hydrophobic molecules. In this case, ATP appears to maintain protein solubility during formation of the RNP localization complex. We show that the protein composition of the transport complex is quite similar to other macromolecular RNP granules that undergo a liquid-liquid phase separation (LLPS), consistent with this postulated role for ATP.
Results and discussion
Mitochondria of the fragmented Bb generate locally elevated levels of ATP
Oocytes from transgenic frogs that express green fluorescent protein (GFP)-tagged mitochondrial outer membrane protein 25 (Taguchi et al., 2012) were used to examine the relation between the remnants of the dispersed Bb and RNA transported through the late pathway to the vegetal hemisphere (Figure 1). A 453-nucleotide RNA containing the vegetal localization element (VLE) of vg1 mRNA is efficiently transported to the vegetal cortex when injected into stage II/III oocytes and can be used to track movement of RNA through this pathway (Gagnon et al., 2013). The region formerly occupied by the Bb retains a high concentration of mitochondria that coincides with the vg1 RNA transport pathway (Figure 2A). It is especially noteworthy that there is an appreciable perinuclear concentration of mitochondria that overlaps the "cup" structure where the RNA first enters the cytoplasm. It has been proposed that vg1 mRNA transport complexes accumulate at this site prior to engaging dynein motors (Gagnon et al., 2013).
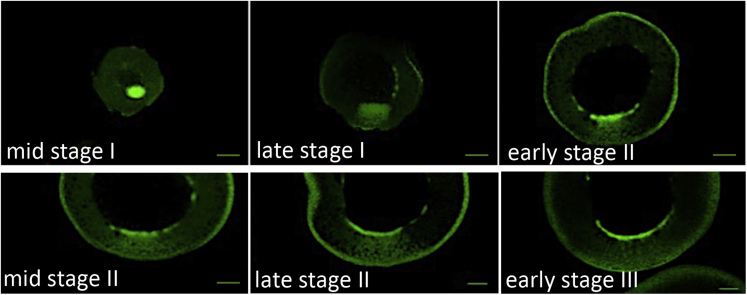
Figure 1.
Expansion and dispersion of the Balbiani body
Oocytes at the designated stage of oogenesis were isolated from a transgenic frog that express GFP-tagged mitochondrial outer membrane protein 25. The Balbiani body forms in early stage I oocytes as a distinct structure on the germinal vesicle surface specifically facing the vegetal pole. During late stage I, the structure extends with the leading-edge delivering germ plasm to the vegetal cortex. During stage II, the Balbiani body fragments, resulting in a region enriched in mitochondria that persists into stage III.
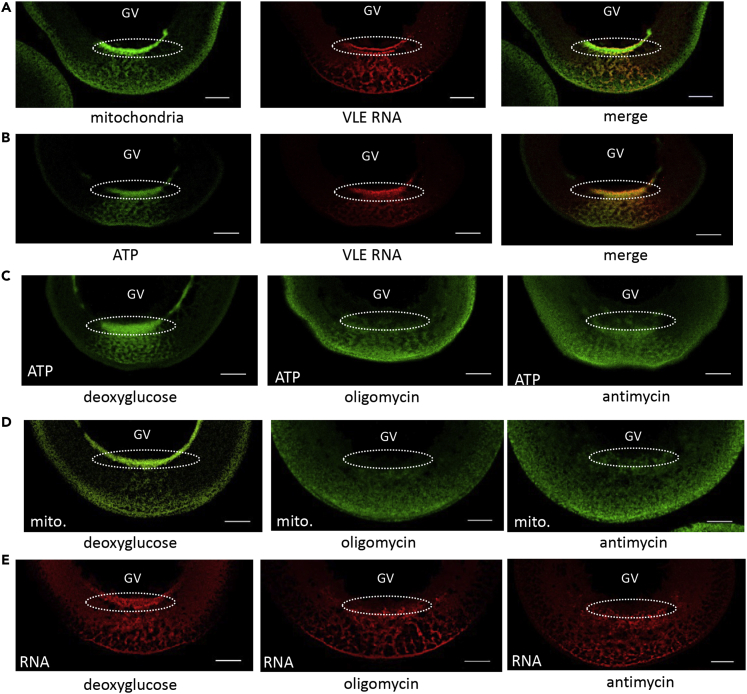
Figure 2.
Remnants of the Balbiani body generate a region of elevated ATP that supports formation of RNA transport granules
GFP-tagged mitochondrial outer membrane protein 25 was used to monitor the position of mitochondria (mito); the FRET-based reporter protein ATeam was used to detect ATP (ATP); Alexa Fluor 546-labeled VLE RNA was used to track RNA localization (RNA)
(A) The area formerly occupied by the Bb is enriched in mitochondria (left); VLE RNA being transported to the vegetal cortex (center) traverses the same area (right). The perinuclear cup structures where the RNA exits the vegetal side of the GV are enclosed in circles.
(B) The clustered mitochondria of the Bb generate a region of elevated ATP concentration that is coincident with VLE RNA.
(C–E). Inhibition of glycolysis (left panels) has no effect on localized ATP production (C), the clustering of mitochondria (D), or RNA transport (E). Inhibition of mitochondrial ATP synthesis with oligomycin (center panels) or antimycin (right panels) eliminates the clustered perinuclear region of ATP (C) and mitochondria (D), and suppresses formation of the RNA granules (E). Scale bars = 50 μm.
There are recent reports of subcellular enrichment of mitochondria in regions where there is a high local demand for ATP that, in one case, supports local protein synthesis during dendritic synaptic plasticity (Rangaraju et al., 2019) or in another case, actin network growth (Kelley et al., 2019). It has been shown that the activity of mitochondria in the Bb and perinuclear cup is 3–4 times higher relative to mitochondria dispersed in the cytoplasm (Wilding et al., 2001). Thus, it appeared possible that these highly active mitochondria might be capable of generating locally elevated amounts of ATP. In order to visualize ATP levels in oocytes, we microinjected mRNA encoding the FRET-based biosensor protein, ATeam (Kotera et al., 2010). The version of the protein (AT1.03) used here has an apparent in vitro dissociation constant of 3 mM; it does not contain a nuclear localization signal and remains cytoplasmic (Imamura et al., 2009). The FRET signal revealed regions of increased ATP within the area containing the remnants of the Bb including the perinuclear cup (Figure 2B).
In order to relate the enriched regions of ATP to mitochondrial activity, we treated oocytes with the inhibitors antimycin A (inhibitor of complex III of the electron transport chain), oligomycin (inhibitor of ATP synthase), or deoxyglucose (inhibitor of glycolysis). Antimycin and oligomycin diminished areas of elevated ATP, whereas deoxyglucose had no detectable influence (Figure 2C). The most pronounced effects of antimycin and oligomycin are on the perinuclear cup structure, although there is some reduction in the number of the RNP transport complexes moving toward the vegetal cortex as well (Figures 2D and 2E). Antimycin and oligomycin trigger the dispersion of the perinuclear cluster of mitochondria into the cytoplasm (Figure 2D), indicating that the integrity of this structure requires active respiration (Wilding et al., 2001).
Assembly of the vg1 transport complexes requires ATP
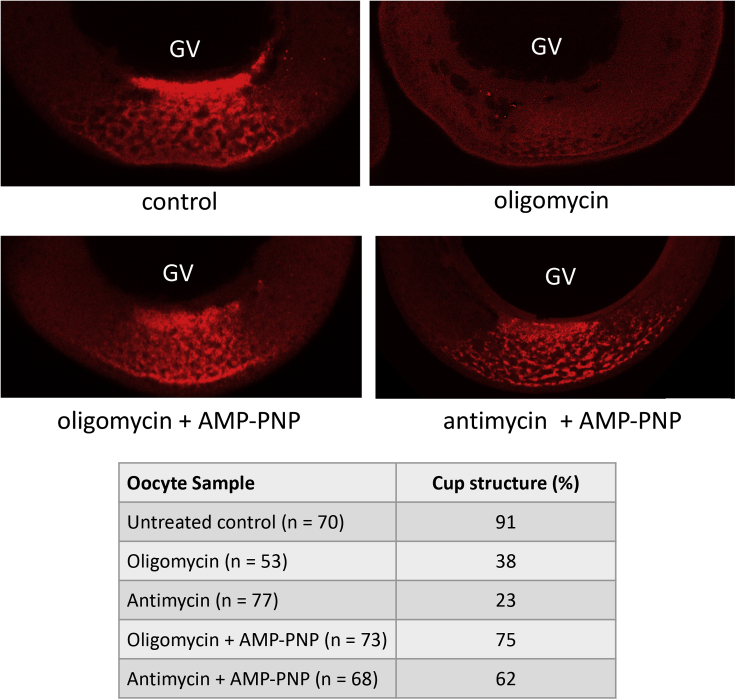
There is evidence that vg1 RNP complexes undergo some type of remodeling event upon entering the cytoplasm that affects protein-RNA and protein-protein interactions (Lewis et al., 2008); however, it is not known whether this process requires ATP. Because a number of RNA helicases mediate changes in RNP structure before and after nuclear export to the cytoplasm (Gilman et al., 2017), it appeared possible that the elevated levels of ATP support RNA helicase activity required for the formation of RNP complexes at the perinuclear cup. To assess the requirement for ATP hydrolysis, we tested the ability of a nonhydrolyzable analog of ATP, adenosine 5'-(β,γ-imido) triphosphate (AMP-PNP), to rescue formation of the cup structure in the presence of oligomycin or antimycin A (Figure 3). AMP-PNP was co-injected with VLE RNA to yield a final in vivo concentration of approximately 4 mM and the oocytes cultured for 12 h. The presence of the nonhydrolyzable analog is sufficient to maintain normal cup formation and stabilize vg1 RNP particles in oocytes incubated with either oligomycin or antimycin A, indicating that the nucleotide provides an activity independent of phosphoryl transfer.
Figure 3.
Rescue of RNA granule formation with a nonhydrolyzable analog of ATP
Oocytes were injected with Alexa Fluor 546-labeled VLE RNA alone (top panels) or coinjection with adenosine 5'-(β,γ-imido) triphosphate (AMP-PNP) to give a calculated intracellular concentration of 4 mM (bottom panels). Oocytes were then cultured for 12 h in the absence (control) or presence of oligomycin (2 ug/mL) or antimycin (1.5 μg/mL).
The cellular concentration of ATP greatly exceeds that of the other three ribonucleotides (Traut, 1994), endowing it with the distinct ability to act as a physiological hydrotrope (Patel et al., 2017); remarkably, ATP is estimated to solubilize over a quarter of the insoluble mammalian proteome (Sridharan et al., 2019). Estimates of ATP concentration in Xenopus oocytes range from approximately 1 to 4 mM (Gabriel and Günzel, 2007; Gribble et al., 1997; Woodland and Pestell, 1972) and there is some evidence that the nucleotide is not entirely uniform throughout the cytoplasm (Gribble et al., 2000). In R. pipiens oocytes, ATP concentration is highest in the GV (∼6 mM) followed by the animal cytoplasm (∼3 mM) and lowest in the vegetal cytoplasm (1.4–1.9 mM) (Miller and Horowitz, 1986). The latter is at the lower end of the effective concentration range (2–8 mM) in which ATP acts to prevent protein aggregation (Patel et al., 2017) or has a stabilizing effect on protein structure (Sridharan et al., 2019). Rescue of cup formation by AMP-PNP indicates that the concentration of ATP generated by mitochondria clustered at the nuclear periphery creates a microenvironment for hydrophobic proteins that is used to maintain their solubility as the RNP complexes enter the cytoplasm and form transport granules. Indeed, the hydrotropic action of ATP has been shown to solubilize protein aggregates in Xenopus oocyte nucleoli (Hayes et al., 2018). The need for elevated amounts of ATP to support helicase activity cannot be fully discounted; however, the sub-millimolar Kd values for ATP binding to DEAD-box helicases makes this situation unlikely (Liu et al., 2014).
vg1 transport complexes behave as liquid-liquid biocondensates
The perinuclear cup structure appears to be a region where localized RNAs such as vg1 are assembling into large structures prior to movement toward the vegetal cortex. The nuclear pore is a selective barrier formed by disordered phenylalanine-glycine (FG) dipeptide motifs that extend into the cytoplasm. It has been proposed that this provides an environment for the integration of newly exported mRNA into P granules in C. elegans that form a stable association with the nuclear membrane (Sheth et al., 2010; Updike et al., 2011). Moreover, the nuclear membrane surface itself can contribute to phase separation and serve as an assembly site for biocondensates (Snead and Gladfelter, 2019; Zhao and Zhang, 2020).
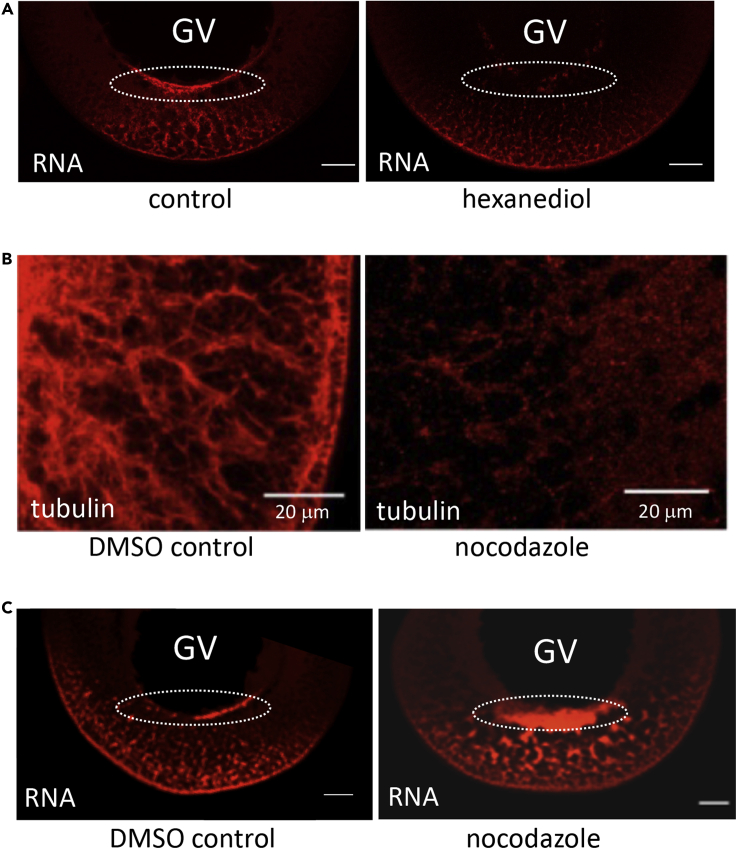
A diagnostic hallmark of liquid-like biocondensates is their dispersion by 1,6-hexanediol. Oocytes injected with VLE RNA were cultured for 16 h and then incubated in 5% 1,6-hexanediol for 1 h. Much of the pattern of VLE RNA is lost upon this treatment, but the effect is especially noticeable at the perinuclear cup structure, which is entirely eliminated (Figure 4A), providing evidence that this is the site of formation of a biocondensate that then moves RNA to the vegetal cortex.
Figure 4.
The VLE RNA transport granules behave as phase-separated biocondensate
(A) Oocytes were taken 16 h after injection with Alexa Fluor 546-labeled VLE RNA and treated with 5% 1,6-hexanediol for 60 min and then fixed.
(B) Depolymerization of microtubules with nocodazole. Oocytes were incubated with DMSO (vehicle control) or nocodazole (5 μg/mL) for 6 h, fixed and bisected prior to immunohistochemical staining using anti-β-tubulin/Alex Fluor 546-conjugated anti-rabbit antibodies.
(C) Following injection with Alexa Fluor 546-labeled VLE RNA, oocytes were placed in culture medium containing DMSO or nocodazole for 48 h and then fixed. Scale bars = 50 μm unless noted otherwise.
One material property of liquid-like condensates compared with hydrogels or amyloid structures is the ability of the former to fuse into larger droplets upon contact (So et al., 2021). The vg1 particles amassed in the perinuclear cup become associated with dynein motors to begin transport to the vegetal cortex (Gagnon et al., 2013). Presumably, this microtubule-dependent movement disrupts their interaction with the GV membrane and, thereby, limits coalescence. We tested the behavior of the vg1 RNP particles in the absence of active transport by disruption of the microtubule cytoskeleton with nocodazole (Figure 4B). Most injected VLE RNA typically clears the perinuclear region after 24 h, but in the presence of nocodazole, a considerable amount of the RNA remains in a greatly expanded cup structure even at 48 h (Figure 4C). In addition to the enlarged size of the cup structure, there is also coalescence of the cytoplasmic VLE transport particles. The irregular shape of the fused vg1 structures at the GV membrane is fully consistent with the expected behavior of a liquid-like condensate (So et al., 2021; Zhao and Zhang, 2020). Indeed, this result is strikingly similar to the ability of nocodazole to induce fusion of RNA P granules on the nuclear membrane of zebrafish germ cells (Strasser et al., 2008). In vitro-induced flow across the nuclear membrane results in a dripping-like release of these P granules whose appearance is remarkably similar to the vg1 structures normally being released from the cup structure (Brangwynne et al., 2009), presumably, in the latter case, as a result of the force generated when these biocondensates engage dynein motors.
The composition of vg1 transport complex is similar to other RNP condensates
Several proteins that bind to the localization element of vg1 mRNA have been identified (Allison et al., 2004; Arthur et al., 2009; Bauermeister et al., 2015; Cote et al., 1999; Czaplinski et al., 2005; Deshler et al., 1998; Havin et al., 1998; Kroll et al., 2002; Loeber et al., 2010; Yoon and Mowry, 2004; Zhao et al., 2001) and most have also been identified in macromolecular RNP granules that undergo LLPS. In order to obtain a more comprehensive inventory of the proteins that comprise the vg1 RNP particle and to compare its composition to other phase-separated RNP granules, we affinity purified complexes that form on biotin-tagged VLE RNA in oocyte extract. Proteins in the purified complexes were identified by tandem mass spectrometry (Figure 5, Table 1). Approximately a third of the proteins in the vg1 complex are solubilized and/or stabilized by ATP (Sridharan et al., 2019), consistent with the proposal that the nucleotide is functioning as a biological hydrotrope to promote LLPS (Table 1).
Figure 5.
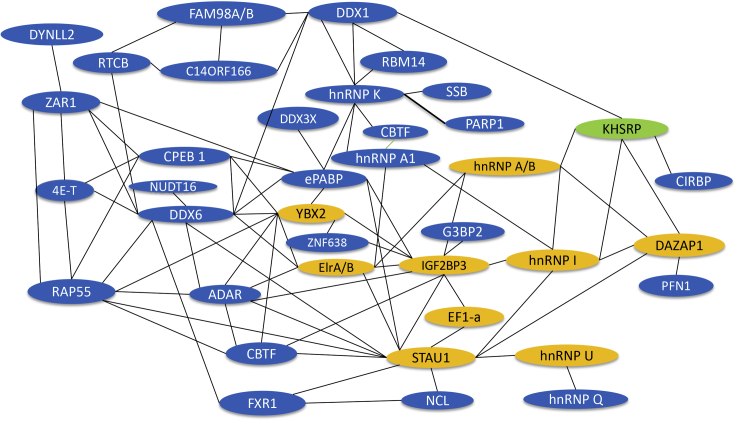
Protein-protein interaction map constructed from proteins identified in the VLE RNP complex affinity purified from oocyte extract
Gold background indicates proteins previously identified as binding directly to VLE RNA; blue background indicates newly identified proteins; VgRBP71 (khsrp), which binds directly to vg1 RNA was not identified in the MS samples.
Table 1.
MS/MS Identification of proteins in the affinity-purified Vg1 RNP complex
| Protein | Normalized LFQ intensity x 108 [au] | Function | |
|---|---|---|---|
| 1 | Nclb | 283.3 | rRNA maturation |
| 2 | Igf2bp3b | 200.5 | mRNA localization |
| 3a | Eef1gb | 250.4 | Translation elongation |
| 4 | Ybx2 (Frgy2) | 146.7 | Translation repression |
| 5 | Hnrnpk | 104.3 | hnRNP |
| 6a | Sart3 | 96.8 | mRNA splicing |
| 7 | Cirbpb | 91.6 | mRNA stabilization |
| 8a | Eef1b2 | 74.6 | Translation elongation |
| 9a | Ssb-b (LaB) | 66.3 | RNA binding |
| 10 | Eef1a1 (EF-1α) | 65.3 | Translation elongation |
| 11 | RtcB | 50.2 | RNA ligase |
| 12 | Pabpn1l/Epabp-2 | 46 | Poly(A) RNA binding |
| 13 | Nudt16 | 45.3 | mRNA decapping |
| 14 | Ddx1 | 41.2 | RNA helicase |
| 15 | Hnrnpab | 38 | hnRNP |
| 16 | Hnrnpdl (40LoVe) | 37.6 | hnRNP |
| 17 | C14orf166 | 35.9 | RNA transporting |
| 18a | Rbm14 | 32.1 | Droplet/Hydrogel forming |
| 19a | Piwil3 | 30.7 | RNA binding |
| 20 | 42Sp50 (EF1-α) | 29 | mRNA localization |
| 21 | Parp1b | 24.5 | Protein modification |
| 22a | Eef1d | 24.1 | Translation elongation |
| 23a | Eif2ab | 23.3 | Translation initiation |
| 24 | G3bp2b | 23.1 | Stress granule assembly |
| 25 | Ptbp1 (Hnrnpl) | 20 | hnRNP |
| 26a | Csnk2a2 | 16.4 | Kinase |
| 27 | Ilf3 (Cbtf)b | 14.9 | mRNA transport |
| 28 | Ddx6 (Xp54)c | 14.4 | RNA helicase |
| 29 | Fam98b | 13.6 | RNA binding |
| 30a | Lsm4 | 13.4 | Droplet/Hydrogel forming |
| 31 | Syncrip (Hnrnpq) | 12.8 | hnRNP |
| 32 | Stau1b | 12.4 | RNA localization |
| 33a | Ptbp3 | 12.1 | RNA binding |
| 34a | Eml4b | 11.4 | Microtubule formation |
| 35a | Csnk2b | 11.1 | Kinase |
| 36a | Csnk2a1 | 9.7 | Kinase |
| 37a | Nemf (Ny-co-1)b | 9.5 | Nuclear export |
| 38a | Abcf1 | 9.3 | Protein transporting |
| 39a | Grsf1 | 8.5 | Translation control |
| 40a | Ilf2b | 8 | Transcription factor |
| 41a | Sub1 | 6.6 | Transcription regulator |
| 42 | Ddx3xb | 6.1 | RNA helicase |
| 43 | Dynll2 | 6 | Microtubule motor |
| 44a | Top1mt | 5.3 | DNA topoisomerase |
| 45 | Hnrnpuc | 5.3 | hnRNP |
| 46 | LSM14B (Rap55b)b | 5.1 | Translation control |
| 47a | Nonob | 4.9 | RNA binding |
| 48a | Map1a | 4.4 | Microtubule association |
| 49 | Tubb4b | 4.2 | Microtubule component |
| 50a | Copa | 4.1 | Protein transport |
| 51a | Sfpqb | 3.9 | RNA binding |
| 52 | Hnrnpd | 3.8 | hnRNP |
| 53a | U2af1b | 3.7 | RNA splicing |
| 54a | Smarca5 (Iswi) | 3.4 | Chromatin remodeling |
| 55 | Map4 | 3.3 | Microtubule association |
| 56 | Eif4enif1 (4E-T) | 3.3 | Nucleoplasmic shuttling |
| 57a | Baz1ab | 3.2 | Chromatin remodeling |
| 58a | POP1-likeb | 3 | Pre-mRNA processing |
| 59 | Elavl2 (ElrB) | 2.6 | mRNA localization |
| 60 | Elavl1 (ElrA) | 2.4 | mRNA localization |
| 61 | Fam98ab | 2.1 | Protein methylation |
| 62 | Dazap1 (Prrp) | 2.1 | RNA anchoring |
| 63a | Srpk1b | 2 | Kinase |
| 64 | Zar1 | 1.9 | Translation repression |
| 65a | Recql | 1.8 | DNA helicase |
| 66 | Cpeb1 | 1.4 | Poly(A) RNA binding |
| 67 | Znf638b | 1.1 | Transcription factor |
| 68 | profilin | 0.8 | Actin binding |
| 69 | Fxr1b | 0.5 | RNA binding |
| 70 | Adarb | 0.2 | RNA editing |
Proteins in bold have been experimentally determined to bind to Vg1 RNA.
These entries have no known interaction with any of the other proteins in the list and were not used to build the protein-protein interaction map in Figure 5.
Proteins shown to be stabilized by ATP. (Sridharan et al., 2019).
Confirmed binding to Vg1 RNA in immunoprecipitation assays. Bertke, M. M., Yang, C., and Huber, P. W. unpublished results.
In addition to the known VLE-binding proteins, most of the newly identified proteins are those commonly found in non-membrane RNP biocondensates (Fritzsche et al., 2013; Han et al., 2012; Hubstenberger et al., 2017; Jain et al., 2016; Kato et al., 2012; Maharana et al., 2018; Wang et al., 2018; Youn et al., 2018). However, a few proteins, e.g., zygote arrest 1 (ZAR1), appear specific to this particle, perhaps reflecting its particular function in the oocyte. While the translation regulatory protein 4E-T and its binding partner in P-bodies, LSM14 (RAP55) (Brandmann et al., 2018), are found in the VLE complex, another major P-body component, the Ccr4-Not complex is not. Indeed, with the exception of Nudt16 decapping enzyme, proteins actively involved in RNA degradation or regulation by miRNA are not present in the VLE complex in accord with its role in transport. It is especially noteworthy that the major structural protein of the Xenopus Bb, Velo1 (Boke et al., 2016), is not found in the VLE particle, nor is Tdrd6a, which drives Bb formation in zebrafish by recruiting the Velo1 ortholog, Bucky ball (Roovers et al., 2018). Neil et al. recently reported using immunoprecipitation to isolate RNP complexes that contain Staufen and hnRNPAB proteins from Xenopus oocytes (Neil et al., 2021). The proteins identified in these complexes, which contain vg1 and vegT RNAs, are strikingly similar to those identified by our strategy that targeted vg1 RNA. Together, these alternative approaches provide a reliable catalog of the proteins in the transport particles.
Text mining for experimental evidence revealed that 42 out of the 70 most abundant proteins contact one or more other proteins in the complex, forming an extensive protein-protein interaction network (Figure 5). Many of the remaining proteins are components of the microtubule cytoskeleton, likely reflecting the transport of vg1 mRNA on these structures. These include Dynll2 (dynein light chain 2), a subunit of the dynein motor that initially moves the vg1 complex from the perinuclear region into the proximal vegetal cytoplasm. Like the vg1 RNP complex, zebrafish P granules also contain Dynll2 where the protein appears on the periphery of these structures (Strasser et al., 2008). Notably, Dynll2 acts as an adaptor between dynein motors and a wide variety of proteins (Barbar, 2008), including various RNP cargos (Navarro et al., 2004; Schnorrer et al., 2000). Xenopus Dynll2 protein is at its maximum developmental level in oocytes (Peshkin et al., 2019) and interacts with ZAR1 (Luck et al., 2020), another protein identified here in the vg1 complex. ZAR1 is an ovary-specific protein that, to our knowledge, has not been detected in other biocondensates. Its presence in the vg1 RNP may account for the concurrent presence of Dynll2 and determine, at least in part, selection of the vg1 biocondensates by dynein motors for initial transport.
After the initial unidirectional movement of vg1 RNP granules using dynein, a subsequent step involving kinesin completes the vg1 transport process (Gagnon et al., 2013). Interestingly, the affinity-purified vg1 complex includes map4, which has been shown to play a role in determining the utilization of the opposing motors, kinesin and dynein (Semenova et al., 2014), suggesting this protein is possibly a key regulator of the vg1 two-step transport mechanism.
The Bb is a feature of nearly all oocytes that have been examined. There is speculation that the organelle may select and store healthy mitochondria that ultimately support the zygote (Bilinski et al., 2017). Indeed, this activity may be universal and operate irrespective of how specification of germ cells is determined. While there is some evidence that the Bb selects healthy mitochondria that will be passed on to progeny via the germ plasm (reviewed in (Bilinski et al., 2017; Jamieson-Lucy and Mullins, 2019), the details of this putative process are not clear. We have shown that the elevated level of ATP generated by the mitochondria of the fragmented Bb is necessary for the formation of RNP transport granules at the surface of the GV facing the vegetal pole. The initial planes of cell cleavage in the Xenopus zygote occur relative to and are determined by the animal-vegetal axis established in the oocyte. In light of our results, it is possible to postulate that, in the absence of the highly active mitochondria that normally constitute the Bb (Wilding et al., 2001), the oocyte would be incapable of proper polarization as a result of defective RNA localization. This failure would likely result either in an egg that cannot be fertilized or a zygote that cannot move through early embryogenesis, thereby providing a selection process that prevents survival of progeny having low quality or aberrant mitochondria. Notably, this mechanism of selection would be independent of how germ cells are specified and account for the universality of Bb structures in oocytes.
Limitations of the study
The experiments presented here show that the late pathway for RNA localization to the vegetal cortex of Xenopus oocytes is functionally tied to the prior expansion and disruption of the Bb. The dispersed clusters of highly active mitochondria produce perinuclear and cytoplasmic regions of elevated ATP. The initial formation of the RNA transport structures does not rely on ATP as an energy source, indicating that the nucleotide functions as a hydrotrope, consistent with our evidence that the vg1 RNP complexes form biocondensates.
An outstanding question not addressed by these experiments is whether mitochondria serve any additional role beyond ATP production that sustains LLPS. The colocalization of the Balbiani fragments and the vg1 condensates raises the possibility that there is a physical association between the two. Loss of the perinuclear cluster of mitochondria in the absence of LLPS due to ATP depletion also suggests an interaction between the two organelles. Furthermore, mitochondria and vg1 particles are transported on microtubules using the same motors (Boldogh and Pon, 2006; MacAskill and Kittler, 2010), consistent with a mechanism in which one structure is associated with the other during transport. Bb is also enriched in ER whose potential role in the localization process was not examined.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Polyclonal anti-β-tubulin | Novus | NB600-936; RRID:AB_10000656 |
| Rabbit polyclonal anti-GFP | Invitrogen | PA1-980A; RRID:AB_325960 |
| Chemicals, peptides, and recombinant proteins | ||
| Liberase TH collagenase | Sigma Aldrich | 5401135001 |
| Antimycin A | Sigma Aldrich | A8674 |
| RNasin | Promega | N2511 |
| T7 RNA polymerase | Promega | P2075 |
| SP6 RNA polymerase | Promega | P1085 |
| Ribo m7G Cap Analog | Promega | P1711 |
| Oligomycin A | Sigma Aldrich | 75351 |
| Nocodazole | Sigma Aldrich | M1404 |
| Leibovitz L-15 medium | Sigma Aldrich | L1518 |
| Insulin | Sigma Aldrich | I0516 |
| Penicillin G | Sigma Aldrich | P3032 |
| Streptomycin | Sigma Aldrich | S9137 |
| L-glutamine | Fisher Biotech | BP379 |
| Nystatin | Sigma Aldrich | N1638 |
| Gentamycin | Dot Scientific | DSG38000 |
| Biotin-GMP | Tri-Link BioTechnologies | N-6003 |
| Streptavidin-agarose beads | Sigma Aldrich | S1638 |
| Alexa Fluor 546-14-UTP | Thermo Fisher | C11404 |
| Alexa Fluor 488-5-UTP | Thermo Fisher | C11403 |
| adenosine 5'-(β,γ-imido) triphosphate | Sigma Aldrich | A2647 |
| 1,6-hexanediol | Sigma Aldrich | 240117 |
| Vitellogenin | This paper | N/A |
| S-Trap columns | Protifi | CO2-Mini-80 |
| Oasis HLB Vac Cartridge | Waters | 186000383 |
| Critical commercial assays | ||
| Primescript Master Mix | Promega | A5001 |
| Experimental models: Organisms/strains | ||
| Xenopus laevis: eGFP-OMP25 | Marine Biological Lab | NXR_0059 |
| Xenopus laevis: Albino | Nasco | LM00531(B) |
| Xenopus laevis: Wild type | Nasco | LM00531 |
| Recombinant DNA | ||
| Plasmid: pBSVg1-B | This paper | N/A |
| ATeam 1.03-nD/nA/pcDNA3 | Kotera et al., 2010 | Addgene #51958; RRID:Addgene_51958 |
| Deposited data | ||
| MS/MS data | MassIVE | ftp://massive.ucsd.edu/MSV000087737 |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Paul W. Huber (phuber@nd.edu).
Materials availability
This study did not generate new unique reagents. Plasmids generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Cell culture
Oocytes used in this study were isolated from wild-type albino (Nasco) or OMP-25 transgenic (Marine Biological Laboratory, Woods Hole, MA) frogs. Oocytes were cultured in 50% Leibovitz’|'s L-15 medium, 15 mM HEPES pH 7.6, 1 mg/mL L-glutamine, 5 mg/mL vitellogenin, 1 μg/mL insulin, 100 U/mL penicillin G, 100 μg/mL streptomycin, 100 μg/mL gentamicin, 50 U/mL nystatin at 18°C. Media reagents were purchased from Sigma Aldrich.
Method details
Plasmids and RNA synthesis
RNA was synthesized by run-off transcription in a 50 μL reaction containing rNTP (0.5 mM), m7G(5′)ppp(5′)G cap analog (2 mM), DTT (10 mM), linearized plasmid (1 μg/10μL), RNasin (1 U/μL), T7 or SP6 RNA polymerase (1 U/μL, Promega) and transcription buffer (Promega). An additional 0.5 U/μL of RNA polymerase was added after 2 h for a total reaction time of 5 h at 37°C. Following RNA synthesis, the DNA template was degraded by addition of RQ1 RNase-free DNase (0.5 U/μL, Promega) and the mixture was incubated for 25 min at 37°C. The reaction was stopped by adding 1 μL of 0.5 M EDTA. The volume of the RNA solution was brought up to 200 μL by adding DEPC-treated H2O and extracted once with an equal volume of phenol (pH 4.5) and once with equal volume of chloroform:isoamyl alcohol (24:1) at room temperature, followed by ethanol precipitation (2.5 volume of ice-cold ethanol, 0.1 volume of 3 M sodium acetate) for 1 h at −80°C. The precipitate was washed once with 70% ice-cold ethanol and the pellet dissolved in 100 μL renaturation buffer (50 mM HEPES pH 7.4, 0.3 M KCl, 10 mM MgCl2), incubated for 10 min at 55°C followed by slowly cooling to room temperature to allow RNA renaturation. Spin columns (1 mL bed volume) prepared with G-50 Sephadex were used to remove unincorporated nucleotides.
The plasmid pBSVg1-B was used to synthesize a 453 nt transcript that carries the complete VLE sequence (nt 1440–1816) (Zhao et al., 2001). The plasmid ATeam1.03-nD/nA/pcDNA3 (linearized with NotI) was used to prepare mRNA for microinjection and expression of the ATP indicator protein. Synthesis reactions were modified for the following RNAs: (1) RNA carrying biotin at the 5′-terminus contained 3 mM biotin-GMP (Tri-Link BioTechnologies) in place of m7G(5′)ppp(5′)G; (2) fluorophore-labeled RNA contained 0.05 mM Alexa Fluor 546-14-UTP or 488-14-UTP and 0.45 mM UTP. RNA was quantified by measuring absorbance at 260 nm using a NanoDrop 2000C.
Oocyte preparation, microinjection, and culturing
Xenopus oocytes were obtained by surgically removing part of the ovary tissue and treated with 0.1 mg/mL Liberase TH collagenase (Sigma) in 25 mL OR– buffer (5 mM HEPES pH 7.8, 82.5 mM NaCl, 2.5 mM KCl, 1 mM Na2HPO4) with gentle rotation at room temperature until oocytes were visibly separated from ovary matrix. Dispersed oocytes were washed 3 times with OR2– buffer and 3 times with OR2+ buffer (OR2– buffer supplemented with 1 mM MgCl2 and 1 mM CaCl2), 10 min per wash. The oocytes were manually separated according to developmental stage (Dumont, 1972) and kept in OR2+ buffer from 3 h to overnight to allow oocytes to recover from collagenase treatment.
A Narishige micromanipulator was used for microinjection (volumes ranged from 2 to 12 nL) of stage II/III oocytes in OR2+ buffer. After injection, oocytes were placed in oocyte culture medium (OCM) [50% Leibovitz L-15 medium, 15 mM HEPES pH 7.6, 1 mg/mL L-glutamine, 5 mg/mL vitellogenin, 1 μg/mL insulin, 100 U/mL penicillin G, 100 μg/mL streptomycin, 100 μg/mL gentamicin, 50 U/mL nystatin]. OCM was made fresh and sterilized by filtration (Whatman 0.2 μm). Oocytes were cultured using 24-well plates with up to 50 oocytes per well (500 μL OCM). Culture medium was changed daily. Damaged oocytes were removed and the healthy oocytes were placed in a new well each day. The plate was placed in a hydrated container and kept at 18°C.
For RNA localization experiments, 150 pg VLE RNA (4 nL) Alexa Fluor 546-14-UTP labeled VLE RNA was microinjected into early stage III oocytes and the oocytes were kept in 18°C for 4 to 36 h. For expression of ATeam protein, 250 pg mRNA (2 nL) was microinjected and oocytes cultured in OCM for 5 h before subsequent injection of Alexa Fluor labeled VLE RNA.
Microscopy
Oocytes were fixed for 1 h in MEMFA (0.1 M MOPS, pH 7.4, 2 mM EGTA, 1 mM MgSO4, 3.7% formaldehyde), washed once with OR2+ buffer and dehydrated by successive 5 min washes in 25% methanol-75% OR2+, 50% methanol-50% OR2+, 75% methanol-25% OR2+, and stored in 100% methanol at −20°C. Before imaging, oocytes were cleared by replacing methanol with clearing medium (2:1 benzyl benzoate:benzyl alcohol).
Imaging was done using a Nikon A1R confocal microscope. For detection of Alexa Fluor 546 labeled RNA, oocyte samples were excited at 561 nm laser light, and emission from 570 to 620 nm was collected. For imaging of the FRET based ATP indicator (ATP bound form), cyan fluorescent protein (CFP) was excited with 457 nm laser light, and emission from 500 to 550 nm was measured.
Inhibitor studies
For ATP inhibition experiments, oocytes were injected with VLE RNA and placed in culture medium containing either oligomycin (2 μg/mL), antimycin (1.5 μg/mL), or 2-deoxyglucose (10 mM) plus pyruvate (5 mM). In rescue experiments, adenosine 5'-(β,γ-imido) triphosphate (100 mM neutralized stock) was co-injected with VLE RNA. Oocytes were fixed 12 to 16 h after injection. For inhibition of microtubule polymerization, oocytes were injected with VLE RNA and cultured in medium containing 2 μg/mL nocodazole for 48 h.
To confirm the depolymerization activity of nocodazole on microtubules, control or treated oocytes were fixed in MEMFA for 1 h and washed twice in PBST (137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, and 2 mM KH2PO4, pH 7.4, 0.1% Tween 20) for 10 min. Oocytes were bisected and incubated in PBST containing 2 mg/mL BSA and 10 ug/mL β-tubulin antibody (Novus) overnight at 4°C. The oocytes were washed 3 times in PBST, 3 h for each wash at 4°C and then incubated with Alex Fluor 546-conjugated anti-rabbit IgG (Invitrogen, 1:300 dilution in PBST with 2 mg/mL BSA) overnight at 4°C. The oocytes were washed 3 times in PBST again for 3 h for each wash at 4°C. The samples were dehydrated with methanol dehydration prior to imaging.
Affinity purification of the VLE RNP complex
Oocyte extract was made by homogenizing 500 oocytes (late stage II/stage II) in 1 mL TGKED buffer (50 mM Tris-HCl pH 7.5, 25% glycerol, 50 mM KCl, 0.1 mM EDTA, 0.5 mM DTT) supplemented with 1 mM PMSF and 1X protease inhibitor (Roche cOmplete) with a pestle in a microcentrifuge tube. The homogenate was cleared by centrifuging at 13,200 rpm for 3 min at 4°C. Extract was then pre-cleared with 400 μL agarose 4B beads for 1 h at 4°C. Streptavidin-agarose beads (100 μL per sample, Sigma) were pre-blocked with 50 μg acetylated BSA, 30 μg yeast tRNA in 200 μL S binding buffer (50 mM HEPES pH 7.4, 150 mM NaCl, 10 mM MgCl2, 0.05% NP-40) for 1 h at 4°C with gentle rotation followed by washing in S binding buffer (5 times for 10 min each). Pre-blocked streptavidin beads were incubated with 6–10 μg of biotin-labeled RNA in 500 μL S binding buffer with 2 μL RNasin for 2 h at 4°C. The RNA-bound streptavidin-agarose beads were added to the pre-cleared oocyte extract and incubated overnight at 4°C with gentle inversion to allow protein binding. The RNP-bound streptavidin-agarose beads were washed 5 times with buffer (20 mM Tris-HCl pH 7.5, 0.25 mM EDTA, 6.25 mM MgCl2, 10% glycerol, 0.05% NP40), 10 min for each wash, and then incubated for 1 h at 4°C in 30 μL elution buffer (50 mM NH4HCO3, 8 mM biotin) with gentle pipetting up and down every 5 min to release biotin-RNP. All the steps involving collection of beads by centrifugation at 200 rpm for 1 min.
Mass spectrometry
Protein Preparation and Digestion. Reagents and chemicals were obtained from Sigma Aldrich (St. Louis, MO) unless otherwise specified. Protein bound NA-beads were extracted, reduced, alkylated and digested with trypsin. Briefly, washed beads were resuspended in 100 mM TEAB pH 8.5, 20 mM DTT, containing 6% SDS and heated at 95°C for 5 min and then incubated at 60°C for 15 min. Samples were alkylated with 2.5-fold molar excess of iodoacetamide for 20 min in the dark and the reaction quenched by addition of phosphoric acid to 1.2% final (v/v). A 7-fold volume excess of 100 mM TEAB in 90% methanol was added to flocculate and each sample (including beads) was loaded onto an S-Trap midi column (Protifi, Farmingdale, NY). The sample was washed twice with the same buffer and rehydrated with 2 μg sequencing grade trypsin (Promega, Madison WI) in 100 mM TEAB and digested overnight at 37°C. Peptides were desalted using an Oasis HLB 1 mL SPE cartridge (Waters, Beverly, MA) according to the manufacturer’|'s instructions.
LC-MS/MS
LC-MS/MS was performed on a Q-Exactive HF (Thermo, San Jose, CA) running a TOP18 DDA.
Quantification and statistical analysis
MS RAW files were searched and quantified using MaxLFQ and Andromeda within MaxQuant (v 1.6.2.3) against the current Xenopus FASTA sequences (Xenbase) concatenated with common contaminants. False Discovery Rate was set to 0.01. Protein abundances were normalized against common abundant proteins co-associated with control RNA beads and data reduction was performed with a Benjamini-Hochberg corrected P-value to determine significantly enriched/reduced proteins.
Acknowledgments
We thank Dr. Michelle Joyce, Mr. William Phillips, and Mr. William Archer for their assistance with the experiments; Drs. Daniel Weeks and Kevin Vaughn for expert advice; Dr. Jessica Brown for comments on the manuscript. The research was supported by the University of Notre Dame and the National Institutes of Health (5R01 HD084399).
Author contributions
Conceptualization, C.Y. and P.W.H.; Methodology, C.Y., M.M.C., and P.W.H.; Investigation, C.Y., G.M.D., and M.M.C.; Writing – Original Draft, C.Y. and P.W.H.; Writing – Review & Editing, G.M.D., M.M.C., and P.W.H.; Funding Acquisition, P.W.H.; Supervision, P.W.H.
Declaration of interests
The authors declare no competing interests.
Published: March 18, 2022
Data and code availability
-
•
Processed and raw MS/MS data reported in this paper have been deposited at MassIVE: ftp://massive.ucsd.edu/MSV000087737 and are publicly available as of the date of publication with accession number MSV000087737.
-
•
No original code was created as a part of this work.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Allison R., Czaplinski K., Git A., Adegbenro E., Stennard F., Houliston E., Standart N. Two distinct Staufen isoforms in Xenopus are vegetally localized during oogenesis. RNA. 2004;10:1751–1763. doi: 10.1261/rna.7450204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur P.K., Claussen M., Koch S., Tarbashevich K., Jahn O., Pieler T. Participation of Xenopus Elr-type proteins in vegetal mRNA localization during oogenesis. J. Biol. Chem. 2009;284:19982–19992. doi: 10.1074/jbc.M109.009928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbar E. Dynein light chain LC8 is a dimerization hub essential in diverse protein networks. Biochemistry. 2008;47:503–508. doi: 10.1021/bi701995m. [DOI] [PubMed] [Google Scholar]
- Bauermeister D., Claussen M., Pieler T. A novel role for Celf1 in vegetal RNA localization during Xenopus oogenesis. Dev. Biol. 2015;405:214–224. doi: 10.1016/j.ydbio.2015.07.005. [DOI] [PubMed] [Google Scholar]
- Betley J.N., Heinrich B., Vernos I., Sardet C., Prodon F., Deshler J.O. Kinesin II mediates Vg1 mRNA transport in Xenopus oocytes. Curr. Biol. 2004;14:219–224. doi: 10.1016/j.cub.2004.01.028. [DOI] [PubMed] [Google Scholar]
- Bilinski S.M., Kloc M., Tworzydlo W. Selection of mitochondria in female germline cells: is Balbiani body implicated in this process? J. Assist. Reprod. Genet. 2017;34:1405–1412. doi: 10.1007/s10815-017-1006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boke E., Ruer M., Wühr M., Coughlin M., Lemaitre R., Gygi S.P., Alberti S., Drechsel D., Hyman A.A., Mitchison T.J. Amyloid-like self-assembly of a cellular compartment. Cell. 2016;166:637–650. doi: 10.1016/j.cell.2016.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh I.R., Pon L.A. Interactions of mitochondria with the actin cytoskeleton. Biochim. Biophys. Acta (Bba) - Mol. Cell Res. 2006;1763:450–462. doi: 10.1016/j.bbamcr.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Brandmann T., Fakim H., Padamsi Z., Youn J.-Y., Gingras A.-C., Fabian M.R., Jinek M. Molecular architecture of LSM14 interactions involved in the assembly of mRNA silencing complexes. EMBO J. 2018;37:e97869. doi: 10.15252/embj.201797869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Julicher F., Hyman A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Cote C.A., Gautreau D., Denegre J.M., Kress T.L., Terry N.A., Mowry K.L. A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol. Cell. 1999;4:431–437. doi: 10.1016/s1097-2765(00)80345-7. [DOI] [PubMed] [Google Scholar]
- Czaplinski K., Kocher T., Schelder M., Segref A., Wilm M., Mattaj I.W. Identification of 40LoVe, a Xenopus hnRNP D family protein involved in localizing a TGF-β-related mRNA during oogenesis. Dev. Cell. 2005;8:505–515. doi: 10.1016/j.devcel.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Deshler J.O., Highett M.I., Abramson T., Schnapp B.J. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr. Biol. 1998;8:489–496. doi: 10.1016/s0960-9822(98)70200-3. [DOI] [PubMed] [Google Scholar]
- Dumont J.N. Oogenesis in Xenopus laevis (Daudin) I. Stages of oocyte development in laboratory maintained animals. J. Morphol. 1972;136:153–180. doi: 10.1002/jmor.1051360203. [DOI] [PubMed] [Google Scholar]
- Elkouby Y.M., Jamieson-Lucy A., Mullins M.C. Oocyte polarization is coupled to the chromosomal bouquet, a conserved polarized nuclear configuration in meiosis. Plos Biol. 2016;14:e1002335. doi: 10.1371/journal.pbio.1002335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsche R., Karra D., Bennett K.L., Ang F.Y., Heraud-Farlow J.E., Tolino M., Doyle M., Bauer K.E., Thomas S., Planyavsky M., et al. Interactome of two diverse RNA granules links mRNA localization to translational repression in neurons. Cell Rep. 2013;5:1749–1762. doi: 10.1016/j.celrep.2013.11.023. [DOI] [PubMed] [Google Scholar]
- Gabriel T.E., Günzel D. Quantification of Mg2+ extrusion and cytosolic Mg2+-buffering in Xenopus oocytes. Arch. Biochem. Biophys. 2007;458:3–15. doi: 10.1016/j.abb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Gagnon J.A., Kreiling J.A., Powrie E.A., Wood T.R., Mowry K.L. Directional transport is mediated by a Dynein-dependent step in an RNA localization pathway. PLoS Biol. 2013;11:e1001551. doi: 10.1371/journal.pbio.1001551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman B., Tijerina P., Russell R. Distinct RNA-unwinding mechanisms of DEAD-box and DEAH-box RNA helicase proteins in remodeling structured RNAs and RNPs. Biochem. Soc. Trans. 2017;45:1313–1321. doi: 10.1042/BST20170095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble F.M., Ashfield R., Ammala C., Ashcroft F.M. Properties of cloned ATP-sensitive K+ currents expressed in Xenopus oocytes. J. Physiol. 1997;498:87–98. doi: 10.1113/jphysiol.1997.sp021843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble F.M., Loussouarn G., Tucker S.J., Zhao C., Nichols C.G., Ashcroft F.M. A novel method for measurement of submembrane ATP concentration. J. Biol. Chem. 2000;275:30046–30049. doi: 10.1074/jbc.M001010200. [DOI] [PubMed] [Google Scholar]
- Han T.W., Kato M., Xie S., Wu L.C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Havin L., Git A., Elisha Z., Oberman F., Yaniv K., Schwartz S.P., Standart N., Yisraeli J.K. RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev. 1998;12:1593–1598. doi: 10.1101/gad.12.11.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes M.H., Peuchen E.H., Dovichi N.J., Weeks D.L. Dual roles for ATP in the regulation of phase separated protein aggregates in Xenopus oocyte nucleoli. Elife. 2018;7:e35224. doi: 10.7554/eLife.35224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubstenberger A., Courel M., Benard M., Souquere S., Ernoult-Lange M., Chouaib R., Yi Z., Morlot J.B., Munier A., Fradet M., et al. P-body purification reveals the condensation of repressed mRNA regulons. Mol. Cell. 2017;68:144–157. doi: 10.1016/j.molcel.2017.09.003. [DOI] [PubMed] [Google Scholar]
- Imamura H., Nhat K.P., Togawa H., Saito K., Iino R., Kato-Yamada Y., Nagai T., Noji H. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc. Natl. Acad. Sci. U S A. 2009;106:15651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Wheeler J.R., Walters R.W., Agrawal A., Barsic A., Parker R. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson-Lucy A., Mullins M.C. The vertebrate Balbiani body, germ plasm, and oocyte polarity. Curr. Top Dev. Biol. 2019;135:1–34. doi: 10.1016/bs.ctdb.2019.04.003. [DOI] [PubMed] [Google Scholar]
- Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J., et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L.C., Chi Q., Caceres R., Hastie E., Schindler A.J., Jiang Y., Matus D.Q., Plastino J., Sherwood D.R. Adaptive F-actin polymerization and localized ATP production drive basement membrane invasion in the absence of MMPs. Dev. Cell. 2019;48:313–328. doi: 10.1016/j.devcel.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M., Bilinski S., Etkin L.D. In: Current Topics in Developmental Biology. Schatten G.P., editor. Academic Press; 2004. The Balbiani body and germ cell determinants: 150 Years later; pp. 1–36. [DOI] [PubMed] [Google Scholar]
- Kotera I., Iwasaki T., Imamura H., Noji H., Nagai T. Reversible dimerization of Aequorea victoria fluorescent proteins increases the dynamic range of FRET-based indicators. ACS Chem. Biol. 2010;5:215–222. doi: 10.1021/cb900263z. [DOI] [PubMed] [Google Scholar]
- Kroll T.T., Zhao W.-m., Jiang C., Huber P.W. A homolog of FBP2/KSRP binds to localized mRNAs in Xenopus oocytes. Development. 2002;129:5609–5619. doi: 10.1242/dev.00160. [DOI] [PubMed] [Google Scholar]
- Lewis R.A., Gagnon J.A., Mowry K.L. PTB/hnRNP I is required for RNP remodeling during RNA localization in Xenopus oocytes. Mol. Cell. Biol. 2008;28:678–686. doi: 10.1128/MCB.00999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Putnam A.A., Jankowsky E. DEAD-box helicases form nucleotide-dependent, long-lived complexes with RNA. Biochemistry. 2014;53:423–433. doi: 10.1021/bi401540q. [DOI] [PubMed] [Google Scholar]
- Loeber J., Claussen M., Jahn O., Pieler T. Interaction of 42Sp50 with the vegetal RNA localization machinery in Xenopus laevis oocytes. FEBS J. 2010;277:4722–4731. doi: 10.1111/j.1742-4658.2010.07878.x. [DOI] [PubMed] [Google Scholar]
- Luck K., Kim D.K., Lambourne L., Spirohn K., Begg B.E., Bian W., Brignall R., Cafarelli T., Campos-Laborie F.J., Charloteaux B., et al. A reference map of the human binary protein interactome. Nature. 2020;580:402–408. doi: 10.1038/s41586-020-2188-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAskill A.F., Kittler J.T. Control of mitochondrial transport and localization in neurons. Trends Cell Biol. 2010;20:102–112. doi: 10.1016/j.tcb.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Maharana S., Wang J., Papadopoulos D.K., Richter D., Pozniakovsky A., Poser I., Bickle M., Rizk S., Guillen-Boixet J., Franzmann T.M., et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 2018;360:918–921. doi: 10.1126/science.aar7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D.S., Horowitz S.B. Intracellular compartmentalization of adenosine triphosphate. J. Biol. Chem. 1986;261:13911–13915. [PubMed] [Google Scholar]
- Navarro C., Puthalakath H., Adams J.M., Strasser A., Lehmann R. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat. Cell Biol. 2004;6:427–435. doi: 10.1038/ncb1122. [DOI] [PubMed] [Google Scholar]
- Neil C.R., Jeschonek S.P., Cabral S.E., O'Connell L.C., Powrie E.A., Otis J.P., Wood T.R., Mowry K.L. L-bodies are RNA-protein condensates driving RNA localization in Xenopus oocytes. Mol. Biol. Cell. 2021;32:ar37. doi: 10.1091/mbc.E21-03-0146-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D., Houston D.W. RNA localization in the vertebrate oocyte: establishment of oocyte polarity and localized mRNA assemblages. Results Probl. Cell Differ. 2017;63:189–208. doi: 10.1007/978-3-319-60855-6_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A., Malinovska L., Saha S., Wang J., Alberti S., Krishnan Y., Hyman A.A. ATP as a biological hydrotrope. Science. 2017;356:753–756. doi: 10.1126/science.aaf6846. [DOI] [PubMed] [Google Scholar]
- Peshkin L., Lukyanov A., Kalocsay M., Gage R.M., Wang D., Pells T.J., Karimi K., Vize P.D., Wühr M., Kirschner M.W. The protein repertoire in early vertebrate embryogenesis. bioRxiv. 2019:571174. [Google Scholar]
- Rangaraju V., Lauterbach M., Schuman E.M. Spatially stable mitochondrial compartments fuel local translation during plasticity. Cell. 2019;176:73–84. doi: 10.1016/j.cell.2018.12.013. [DOI] [PubMed] [Google Scholar]
- Roovers E.F., Kaaij L.J.T., Redl S., Bronkhorst A.W., Wiebrands K., de Jesus Domingues A.M., Huang H.Y., Han C.T., Riemer S., Dosch R., et al. Tdrd6a regulates the aggregation of buc into functional subcellular compartments that drive germ cell specification. Dev. Cell. 2018;46:285–301 e9. doi: 10.1016/j.devcel.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorrer F., Bohmann K., Nüsslein-Volhard C. The molecular motor dynein is involved in targeting Swallow and bicoid RNA to the anterior pole of Drosophila oocytes. Nat. Cell Biol. 2000;2:185–190. doi: 10.1038/35008601. [DOI] [PubMed] [Google Scholar]
- Semenova I., Ikeda K., Resaul K., Kraikivski P., Aguiar M., Gygi S., Zaliapin I., Cowan A., Rodionov V. Regulation of microtubule-based transport by MAP4. Mol. Biol. Cell. 2014;25:3119–3132. doi: 10.1091/mbc.E14-01-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Pitt J., Dennis S., Priess J.R. Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development. 2010;137:1305–1314. doi: 10.1242/dev.044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snead W.T., Gladfelter A.S. The control centers of biomolecular phase separation: how membrane surfaces, PTMs, and active processes regulate condensation. Mol. Cell. 2019;76:295–305. doi: 10.1016/j.molcel.2019.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So C., Cheng S., Schuh M. Phase separation during germline development. Trends Cell Biol. 2021;31:254–268. doi: 10.1016/j.tcb.2020.12.004. [DOI] [PubMed] [Google Scholar]
- Sridharan S., Kurzawa N., Werner T., Gunthner I., Helm D., Huber W., Bantscheff M., Savitski M.M. Proteome-wide solubility and thermal stability profiling reveals distinct regulatory roles for ATP. Nat. Commun. 2019;10:13. doi: 10.1038/s41467-019-09107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser M.J., Mackenzie N.C., Dumstrei K., Nakkrasae L.-I., Stebler J., Raz E. Control over the morphology and segregation of Zebrafish germ cell granules during embryonic development. BMC Dev. Biol. 2008;8:58. doi: 10.1186/1471-213X-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A., Tak ii M., Motoishi M., Or ii H., Moch ii M., Watanabe K. Analysis of localization and reorganization of germ plasm in Xenopus transgenic line with fluorescence-labeled mitochondria. Dev. Growth Differ. 2012;54:767–776. doi: 10.1111/dgd.12005. [DOI] [PubMed] [Google Scholar]
- Traut T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- Updike D.L., Hachey S.J., Kreher J., Strome S. P granules extend the nuclear pore complex environment in the C. elegans germ line. J. Cell Biol. 2011;192:939–948. doi: 10.1083/jcb.201010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Choi J.M., Holehouse A.S., Lee H.O., Zhang X., Jahnel M., Maharana S., Lemaitre R., Pozniakovsky A., Drechsel D., et al. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell. 2018;174:688–699 e16. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding M., Carotenuto R., Infante V., Dale B., Marino M., Di Matteo L., Campanella C. Confocal microscopy analysis of the activity of mitochondria contained within the ‘mitochondrial cloud’ during oogenesis in Xenopus laevis. Zygote. 2001;9:347–352. doi: 10.1017/s096719940100140x. [DOI] [PubMed] [Google Scholar]
- Wilk K., Bilinski S., Dougherty M.T., Kloc M. Delivery of germinal granules and localized RNAs via the messenger transport organizer pathway to the vegetal cortex of Xenopus oocytes occurs through directional expansion of the mitochondrial cloud. Int. J. Dev. Biol. 2005;49:17–21. doi: 10.1387/ijdb.041906kw. [DOI] [PubMed] [Google Scholar]
- Woodland H.R., Pestell R.Q. Determination of the nucleoside triphosphate contents of eggs and oocytes of Xenopus laevis. Biochem. J. 1972;127:597–605. doi: 10.1042/bj1270597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y.J., Mowry K.L. Xenopus Staufen is a component of a ribonucleoprotein complex containing vg1 RNA and kinesin. Development. 2004;131:3035–3045. doi: 10.1242/dev.01170. [DOI] [PubMed] [Google Scholar]
- Youn J.-Y., Dunham W.H., Hong S.J., Knight J.D.R., Bashkurov M., Chen G.I., Bagci H., Rathod B., MacLeod G., Eng S.W.M., et al. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mole. Cell. 2018;69:517–532. doi: 10.1016/j.molcel.2017.12.020. [DOI] [PubMed] [Google Scholar]
- Zhao W.M., Jiang C., Kroll T.T., Huber P.W. A proline-rich protein binds to the localization element of Xenopus Vg1 mRNA and to ligands involved in actin polymerization. EMBO J. 2001;20:2315–2325. doi: 10.1093/emboj/20.9.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.G., Zhang H. Phase separation in membrane biology: the interplay between membrane-bound organelles and membraneless condensates. Dev. Cell. 2020;55:30–44. doi: 10.1016/j.devcel.2020.06.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
-
•
Processed and raw MS/MS data reported in this paper have been deposited at MassIVE: ftp://massive.ucsd.edu/MSV000087737 and are publicly available as of the date of publication with accession number MSV000087737.
-
•
No original code was created as a part of this work.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.