Abstract
Chest X-ray becomes one of the most common medical diagnoses due to its noninvasiveness. The number of chest X-ray images has skyrocketed, but reading chest X-rays still have been manually performed by radiologists, which creates huge burnouts and delays. Traditionally, radiomics, as a subfield of radiology that can extract a large number of quantitative features from medical images, demonstrates its potential to facilitate medical imaging diagnosis before the deep learning era. In this paper, we develop an end-to-end framework, ChexRadiNet, that can utilize the radiomics features to improve the abnormality classification performance. Specifically, ChexRadiNet first applies a light-weight but efficient triplet-attention mechanism to classify the chest X-rays and highlight the abnormal regions. Then it uses the generated class activation map to extract radiomic features, which further guides our model to learn more robust image features. After a number of iterations and with the help of radiomic features, our framework can converge to more accurate image regions. We evaluate the ChexRadiNet framework using three public datasets: NIH ChestX-ray, CheXpert, and MIMIC-CXR. We find that ChexRadiNet outperforms the state-of-the-art on both disease detection (0.843 in AUC) and localization (0.679 in T(IoU) = 0.1). We make the code publicly available at https://github. com/bionlplab/lung_disease_detection_amia2021, with the hope that this method can facilitate the development of automatic systems with a higher-level understanding of the radiological world.
1 Introduction
The chest X-ray is one of the most common medical procedures for diagnosis, but the interpretation of chest x-ray images is subject to significant diagnosis variability for important clinical decisions. A radiologist reads about 20,000 images a year, roughly 50-100 per day, and the number is increasing. Each year, the US produces 600 billion images, and 31% of American radiologists have experienced at least one malpractice claim, often missed diagnoses1. The shortage of radiologists and burnout of physicians creates an urgent demand for immediate solutions. Building automatic or semi-automatic approaches to medical imaging diagnosis becomes an unavoidable next step.
The recent development of artificial intelligence, especially deep learning, offers great potential to improve medical imaging diagnosis2. It also sneaks into the radiology reading rooms to build a new paradigm for precision diagnosis3–5. Pioneering work on chest X-rays mainly focused on two problems: disease classification and localization. The recent release of large-scale datasets, such as NIH Chest X-ray4, CheXpert6, and MIMIC-CXR7, have enabled many studies using deep learning for automated chest X-ray diagnosis, such as thorax disease classification3, 8–10 and localization4, 11, 12.
In practice, radiologists use pattern recognition on medical images to make a diagnostic decision13. The knowledge of radiologists can be captured by Radiomics, which has demonstrated the effectiveness of image-based biomarkers for cancer staging and prognostication. Formally, radiomics extracts quantitative data from medical images to represent tumor phenotypes, such as spatial heterogeneity of a tumor and spatial response variations. It plays an important role in precision medicine to support evidence-based clinical decision-making. For example, radiomics can generate the detailed quantification of tumor phenotype14 and acts as a radiographic imaging phenotype which is associated with tumor stage, metabolism, and gene or protein expression profiles15, 16.
While radiomics offer the potential for more precise and accurate clinical predictions, it is surprising that radiomics has not been implemented in the layers of the neural networks, nor to the best of our knowledge in the deep learning workflow for X-ray analysis17, 18. To bridge this gap, in this paper, we propose ChexRadiNet, a new framework that incorporates domain-specific knowledge (radiomics) into deep learning algorithms as soft constraints, and then learns end-to-end to automatically detect thorax diseases and generate bounding boxes on chest X-rays. Compared with previous studies, our proposed model does not need pre-annotated bounding boxes for training and can achieve state-of-the-art performance for thorax disease localization. Therefore, it provides a way to introduce prior information about anticipated explanations, a technique that is widely used in the “Rationale model”19 (Section 2). For ensuring ChexRadiNet is robust and generalizable, three public benchmarking datasets were used for this purpose: NIH Chest X-ray4, CheXpert6, and MIMIC-CXR7. We demonstrate that our model outperforms baseline methods for both thorax disease classification and localization (Section 3).
2 Method
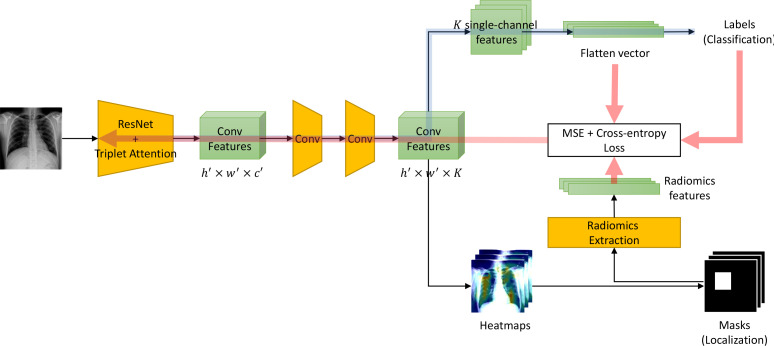
Figure 1 shows our proposed ChexRadiNet, which consists of two branches. The first branch predicts whether the pathology is present or not in the image. The second branch localizes its regions using the radiomic features extracted from the first branch. ChexRadiNet utilizes a multi-task, closed-loop strategy to learn and use radiomic features as soft constraints. Formally, we are learning a two-part latent-variable model of the form Ez∼p(z|x)p(y|x, z), where the latent z is a radiomic-based mask over the image x with the probability p(z|x). p(y|x, z) is a masked version of the classification framework. Therefore, we consider the training process as a weakly-supervised learning. In this section, we first illustrate the architecture of ChexRadiNet and then present the training process.
Figure 1:
Model overview. The model contains three major parts. Blue arrows represents the feedforward multi-label classification part. The below black arrows represents the mask generation and radiomic features extraction part. Red arrows means the radiomic features regularization and backward part.
2.1 Model architecture
2.1.1 Branch I: Multi-label classification
In this branch, we label each image with a 14-dim vector y = [y1, . . . , yk, . . . , yK ], yk ∈ 0, 1, K = 14 for each image. yk indicates the presence with respect to the according pathology in the image while a zero vector represents the status of “Normal” (no pathology is found in the scope of any of 14 disease categories as listed).
We use the residual neural network (ResNet) architecture20, given its dominant performance in ILSVRC competitions and the triplet attention mechanism (see Section 2.1.3). However, our framework can be applied to other CNNs. ResNet-18 and ResNet-50 are used in this paper. After removing the final classification layer and global pooling layer, an input image with shape h × w × c produces a feature tensor with shape h′ × w′ × c′ where h, w, and c are the height, width, and number of channels of the input image, respectively while h′ = h/32, w′ = w/32, c′ = 2048. The output of this network encodes the images into a set of abstracted feature maps. Then through an application of two convolutional layers (each followed by batch normalization and ReLU activation), the number of channels is modified to K, where K is the number of possible disease types. A perchannel probability for each disease class is then derived by a fully-connected layer with a sigmoid activation function; this is denoted p(k|I), where the probability is that whether the image belongs to class k and I denotes the image. Since we intend to build K binary classifiers, we will exemplify just one class k. Note that kth binary classifiers will use the kth-channel features to do prediction. Since all images have their labels, the loss function for class k can be expressed as minimizing the binary-cross entropy as Lk = −yk log p(k|I)−(1−yk ) log(1−p(k|I)), where yk is the ground truth label of the k class. To enable end-to-end training across all classes, we sum up the class-wise losses to define the total loss as LI = ∑kLk .
2.1.2 Branch II: Mask generation
In this branch, we generate bounding boxes (B-Box, or masks) based on the classification result of Branch I to get the most indicative areas using the class activation mappings (CAMs)21. The heatmap produced from the model indicates the approximate spatial location of one particular thoracic disease class each time. Due to the simplicity of intensity distributions in these resulting heatmaps, applying an ad-hoc thresholding-based B-Box generation method for this task is found to be sufficient. Followed by the work of Wang et al.4, the intensities in heatmaps are first normalized to [0, 255] and then thresholded by {60, 180} individually. Finally, B-Boxes are generated to cover the isolated regions in the resulting binary maps.
Radiomic features extraction. With the generated B-Boxes and original images, we extracted radiomic features to regularize the model. Quantitative radiomics can be categorized into the following subgroups:
First-order statistics features describe the distribution of individual pixel values without concerns for spatial relationships. They are histogram-based properties using mean, median, maximum, and minimum values of the pixel intensities on the image, as well as their asymmetry, flatness, uniformity, and entropy.
Shape features describe the shape of the region of interest (ROI) and its geometric properties (e.g., volume, maximum diameter along with different orthogonal directions, maximum surface, tumor compactness, and sphericity).
A Gray Level Co-occurrence Matrix (GLCM) features describe the second-order joint probability function of an image region constrained by the mask. The matrix P (i, j|δ, θ) represents the number of times the combination of levels i and j occurs in two pixels in the image, that are separated by a distance of δ pixels along angle θ.
A Gray Level Size Zone (GLSZM) features quantify gray level zones in an image. A gray level zone is defined as the number of connected pixels that share the same gray level intensity.
A Gray Level Run Length Matrix (GLRLM) features quantify gray level runs, which are defined as the length in number of pixels, of consecutive pixels that have the same gray level value.
A Neighboring Gray Tone Difference Matrix (NGTDM) features quantify the difference between a gray value and the average gray value of its neighbors within distance δ. The sum of absolute differences for gray level i is stored in the matrix.
A Gray Level Dependence Matrix (GLDM) features quantify gray level dependencies in an image. A gray level dependency is defined as the number of connected pixels within distance δ that are dependent on the center pixel.
All above features can be extracted either directly from the images or after applying different filters or transforms (e.g., wavelet transform). In our design, we utilize the Pyradiomics tool to extract radiomic features (https:// pyradiomics.readthedocs.io/).
Finally, we use the pairwise distance between radiomic features and image features as regularization. Therefore, the adjustable loss function is LII = LI + ||IF − RF||p, where IF and RF are the image features and radiomic features, respectively, and l · l denotes the norm and p represents the norm degree, e.g., p = 1 and p = 2 represent the Taxicab norm and Euclidean norm, respectively. In this paper, we set p to 2. Please note that although the original shapes of IF and RF are not equal, we easily adapted one-layer MLP to project them into the same dimension space.
2.1.3 Triplet Attention
To boost the quality of masks, we integrate the triplet-attention mechanism22. Triplet Attention mechanism requires few learnable parameters and could capture important features by taking cross-dimension interaction into account22. In other words, it includes three sub-branches to respectively capture the dependency between spatial dimensions Height (H), Width (W ), and the Channel (C) dimension. For the first branch, in measuring the interactions between dimension H and dimension C, it first performs a Z-pool operation by concatenating the result of average pooling and max pooling across dimension W . This operation can be summarized as where is a 90 degree anti-clockwise rotation along the H axis from the output of the previous convolutional layer is the output of a Z-Pool operation. χ1* then passed through a standard 2D convolutional layer followed by sigmoid activation σ to get attention weights for χ∗1. It would finally rotate back to match the original shape of χ after applying the attention weights. These steps can be represented by the following: where r is the rotation operation to retain the original shape of input. Similarly, y2, y3 are obtained from the last two branches by measuring the interactions between dimensions W and C and between dimensions W and H, respectively. Note that the last branch is similar to the spatial attention in CBAM23, and it requires no rotation. The refined input y is represented by averaging outputs from three branches: y = 1/3 (y1 + y2 + y3).
2.2 Training Strategy of ChexRadiNet
ChexRadiNet adopts an end-to-end multi-task training scheme. Each epoch consists of two tasks. In the first task (Branch I), we use the whole image to fine-tune the ResNet + Triplet Attention network pre-trained on ImageNet. During this process, we feed the generated masks into the radiomics extraction block to get radiomic features. In the second task (Branch II), we use radiomic features as regularization to further fine-tune the whole model. In each epoch, we use the model with the highest AUC on the validation set for testing.
3 Experiments
3.1 Datasets
For the abnormality classification task, we evaluated the ChexRadiNet framework using the NIH Chest X-ray4, CheXpert6, and MIMIC-CXR7 datasets (Table 1). The Chest X-ray dataset contains 112,120 X-ray images collected from 30,805 patients. The disease labels were extracted from radiological reports with Natural Language Processing tools24. There are 15 classes, one for “No findings” and 14 diseases: Atelectasis, Cardiomegaly, Consolidation, Edema, Effusion, Emphysema, Fibrosis, Hernia, Infiltration, Mass, Nodule, Pleural thickening, Pneumonia, and Pneumothorax. The disease labels are expected to have above 90% accuracy. In addition, the Chest X-ray dataset includes 984 bounding boxes for 8 types of chest diseases annotated for 880 images by radiologists.
Table 1:
Descriptions of the datasets.
| Datasets | Patients | Chest X-rays |
|---|---|---|
| NIH Chest X-ray | 30,805 | 112,120 |
| CheXpert | 65,240 | 224,316 |
| MIMIC-CXR | 227,827 | 377,110 |
CheXpert dataset is another large-scale public chest X-ray dataset currently available, which contains 224,316 X-ray scans of 65,240 patients. This dataset was labeled for the presence of 14 observations, including 12 common thoracic pathologies. Each observation can be assigned to either positive (1), negative (0), or uncertain (-1). To simplify the task, we choose to ignore all the uncertain samples. In addition, to compare with previous literature, we follow the same evaluation protocol over 5 observations: Atelectasis, Cardiomegaly, Consolidation, Edema, and Pleural Effusion. MIMIC-CXR is also a large-scale CXR dataset, which contains 377,110 chest X-rays associated with 227,827 imaging studies. Images are provided with 13 labels. Similar to CheXpert, each label can be assigned to either positive (1), negative (0), or uncertain (-1).
3.2 Evaluation metrics and experimental settings
For the abnormality detection task, we randomly split each dataset into training (70%), validation (10%), and test (20%) sets. Note that there is no patient overlap between the sets. We use AUC scores, the area under the ROC curve, to measure the disease identification accuracy. A higher AUC score indicates better performance.
For the abnormality localization task, following the work of Li et al11, we only consider 8 diseases for the evaluation of mask generation because only eight types of diseases are provided with bounding boxes in the NIH Chest X-ray dataset. We use intersection over union (IoU) to evaluate the predicted disease regions against the ground truth bounding boxes.
We use ResNet-50 as the backbone model. We set the batch size as 256 and train the model for 20 epochs. The model is optimized using the stochastic gradient descent (SGD) optimizer with a learning rate of 0.1 and decay the learning rate by 0.1 every 5 epochs of training. We trained our model on AWS with 16 Nvidia K80 GPUs. The model is implemented in PyTorch.
3.3 Results
3.3.1 Disease classification
Table 2 shows the AUC of each class and a mean AUC across the 14 chest diseases. We used ResNet-50 pre-trained on ImageNet as the backbone. Our ChexRadiNet outperforms other models in terms of mean AUC. For every single class, our proposed framework is better than all other models except with DensNet-121 for Fibrosis, Hernia, Mass, Nodule, Pneumonia, and Pneumothorax. Possible reasons can be that Rajpurkar et al’s backbone is much deeper than our ResNet-503, which enables it to capture more discriminative features than our ResNet-50. In addition, “Mass” and “Nodule” parts are small and hard to detect. For “Fibrosis” and “Hernia,” they are not annotated with bounding boxes and diffuse, and thus we cannot apply the weakly-supervised learning with radiomic features.
3.3.2 Disease localization
We compare our disease localization accuracy under varying IoU to other state-of-the-art models, shown in Table 3. Our model predicts well not only for easy tasks but also for hard tasks like localizing “Mass” and “Nodule”, where the disease localization is within a small area. When the IoU is set to 0.1, our model outperforms other models in terms of Atelectasis, Cardiomegaly, Effusion, and Pneumothorax. As the IoU threshold increases, our framework is superior to other models in terms of better accuracy and maintains great performance. For instance, when IoU is set to 0.3, our result for “Cardiomegaly” is 0.73 while the reference model is only 0.46. We get more than 0.15 accuracy improvement for Effusion, Infiltration, Mass, Pneumonia, and Pneumothorax. When IoU is set to 0.5, our result for “Cardiomegaly” is still as high as 0.59 while the reference model drops to barely 0.18.
Table 3:
Disease localization under varying IoU on the NIH Chest X-ray dataset. Please note that since our model doesn’t use any ground truth bounding box information, to fairly evaluate the performance of our model, we only consider the previous methods’ results under the same setting, therefore, for the case T(IoU)=0.1, we have two baselines, but for the rest cases, we only have one baseline.
| T(IoU) | Model | Atelectasis | Cardiomegaly | Effusion | Infiltration | Mass | Nodule | Pneumonia | Pneumothorax | Mean |
|---|---|---|---|---|---|---|---|---|---|---|
| 0.1 | Wang et al., 20174 | 0.69 | 0.94 | 0.66 | 0.71 | 0.40 | 0.14 | 0.63 | 0.38 | 0.569 |
| Li et al., 201811 | 0.63 | 0.89 | 0.78 | 0.91 | 0.70 | 0.29 | 0.31 | 0.44 | 0.619 | |
| ChexRadiNet | 0.72 | 0.96 | 0.81 | 0.88 | 0.67 | 0.33 | 0.59 | 0.47 | 0.679 | |
| 0.2 | Wang et al., 20174 | 0.47 | 0.68 | 0.45 | 0.48 | 0.26 | 0.05 | 0.35 | 0.23 | 0.371 |
| ChexRadiNet | 0.49 | 0.84 | 0.62 | 0.54 | 0.46 | 0.21 | 0.43 | 0.39 | 0.498 | |
| 0.3 | Wang et al., 20174 | 0.24 | 0.46 | 0.30 | 0.28 | 0.15 | 0.04 | 0.17 | 0.13 | 0.221 |
| ChexRadiNet | 0.28 | 0.73 | 0.54 | 0.43 | 0.38 | 0.15 | 0.35 | 0.32 | 0.398 | |
| 0.4 | Wang et al., 20174 | 0.09 | 0.28 | 0.20 | 0.12 | 0.07 | 0.01 | 0.08 | 0.07 | 0.115 |
| ChexRadiNet | 0.17 | 0.65 | 0.42 | 0.32 | 0.29 | 0.09 | 0.21 | 0.19 | 0.293 | |
| 0.5 | Wang et al., 20174 | 0.05 | 0.18 | 0.11 | 0.07 | 0.01 | 0.01 | 0.03 | 0.03 | 0.061 |
| ChexRadiNet | 0.11 | 0.59 | 0.29 | 0.15 | 0.12 | 0.07 | 0.14 | 0.08 | 0.194 | |
| 0.6 | Wang et al., 20174 | 0.02 | 0.08 | 0.05 | 0.02 | 0.00 | 0.01 | 0.02 | 0.03 | 0.029 |
| ChexRadiNet | 0.06 | 0.37 | 0.09 | 0.06 | 0.08 | 0.04 | 0.05 | 0.05 | 0.100 | |
| 0.7 | Wang et al., 20174 | 0.01 | 0.03 | 0.02 | 0.00 | 0.00 | 0.00 | 0.01 | 0.02 | 0.011 |
| ChexRadiNet | 0.02 | 0.21 | 0.04 | 0.02 | 0.07 | 0.01 | 0.03 | 0.04 | 0.055 |
Following Li et al.11, we prefer a higher IoU threshold, i.e., IoU = 0.7, for disease localization because we expect high-accuracy disease localization application in clinical use. To this end, the method we proposed is superior to the baseline by a large margin.
Please note that for some diseases, e.g., Pneumonia and Infiltration, the localization of disease can appear in multiple places while only one bounding box is provided for each image. Thus, it is reasonable that our model doesn’t align well with the ground truth when the threshold is as small as 0.1, especially for Pneumonia and infiltration. Overall, our model outperforms the reference models for all IoU thresholds except for T(IoU)=0.1 (probably because ground truth has missing annotation while ours does not).
4 Discussion
4.1 Ablation study
We conducted an ablation study to demonstrate the performance of radiomics on NIH Chest X-ray (Table 4), CheXpert (Table 5), and MIMIC-CXR (Table 6). We tried ResNet50+Triplet Attention without radiomic features. Table 4 shows that AUC will drop significantly when not using radiomic features. We observe the same trend in the other two datasets. This demonstrates that it is beneficial to include radiomic features.
Table 4:
Comparison of AUC on the NIH Chest X-ray dataset.
| Method | Atelectasis | Cardiomegaly | Consolidation | Edema | Effusion |
|---|---|---|---|---|---|
| w/o radiomics | 0.751 | 0.850 | 0.777 | 0.867 | 0.833 |
| ChexRadiNet | 0.831 | 0.934 | 0.817 | 0.906 | 0.892 |
| Method | Emphysema | Fibrosis | Hernia | Infiltration | Mass |
| w/o radiomics | 0.783 | 0.733 | 0.804 | 0.670 | 0.694 |
| ChexRadiNet | 0.925 | 0.798 | 0.882 | 0.734 | 0.846 |
| Method | Nodule | Pleural Thickening | Pneumonia | Pneumothorax | Mean |
| w/o radiomics | 0.643 | 0.699 | 0.700 | 0.792 | 0.757 |
| ChexRadiNet | 0.748 | 0.867 | 0.737 | 0.889 | 0.842 |
Table 5:
Comparison of AUC on the CheXpert dataset.
| Method | Atelectasis | Cardiomegaly | Consolidation | Edema | Pleural Effusion | Mean |
|---|---|---|---|---|---|---|
| w/o radiomics | 0.781 | 0.813 | 0.893 | 0.918 | 0.921 | 0.865 |
| ChexRadiNet | 0.831 | 0.848 | 0.920 | 0.930 | 0.921 | 0.890 |
Table 6:
Comparison of AUC on the MIMIC-CXR dataset.
| Method | Atelectasis | Cardiomegaly | Consolidation | Edema | Enlarged Card. |
| w/o radiomics | 0.841 | 0.824 | 0.859 | 0.906 | 0.748 |
| ChexRadiNet | 0.851 | 0.831 | 0.866 | 0.900 | 0.767 |
| Method | Fracture | Lung Lesion | Lung Opacity | Pleural Effusion | Pneumonia |
| w/o radiomics | 0.713 | 0.782 | 0.775 | 0.923 | 0.753 |
| ChexRadiNet | 0.735 | 0.814 | 0.810 | 0.933 | 0.831 |
| Method | Pneumothorax | Pleural Other | Support Devices | Mean | |
| w/o radiomics | 0.909 | 0.850 | 0.931 | 0.832 | |
| ChexRadiNet | 0.919 | 0.909 | 0.937 | 0.854 |
|---|
We also report results of ChesxRadiNet using ResNet-18, a relevant small network, as a backbone. Table 7 shows the results with and without using the radiomic features in three datasets. We observe the AUCs drop significantly when not using radiomic features in all cases. This suggests that the generalizability of our proposed method in smaller networks. In addition, the ResNet-18 version still performs better than other models in Table 2 except Rajpurkar et al3. It indicates the superior of our proposed method for using radiomic features.
Table 7:
Comparison of mean AUC on three datasets using ResNet-18 as a backbone.
| NIH Chest X-ray | CheXpert | MIMIC-CXR | |
| w/o radiomics | 0.749 | 0.854 | 0.822 |
| ChesxRadiNet (ResNet-18) | 0.810 | 0.883 | 0.837 |
Table 2:
AUC results on the NIH Chest X-ray dataset.
| Method | Atelectasis | Cardiomegaly | Consolidation | Edema | Effusion |
| Wang et al., 20174 | 0.716 | 0.807 | 0.708 | 0.835 | 0.784 |
| Wang et al., 20185 | 0.732 | 0.844 | 0.701 | 0.829 | 0.793 |
| Yao et al., 20189 | 0.772 | 0.904 | 0.788 | 0.882 | 0.859 |
| Rajpurkar et al., 20173 | 0.821 | 0.905 | 0.794 | 0.893 | 0.883 |
| Kumar et al., 201725 | 0.762 | 0.913 | 0.784 | 0.888 | 0.864 |
| ChexRadiNet | 0.831 | 0.934 | 0.817 | 0.906 | 0.892 |
|---|---|---|---|---|---|
| Method | Emphysema | Fibrosis | Hernia | Infiltration | Mass |
| Wang et al., 20174 | 0.815 | 0.769 | 0.767 | 0.609 | 0.706 |
| Wang et al., 20185 | 0.865 | 0.796 | 0.876 | 0.666 | 0.725 |
| Yao et al., 20189 | 0.829 | 0.767 | 0.914 | 0.695 | 0.792 |
| Rajpurkar et al., 20173 | 0.926 | 0.804 | 0.939 | 0.720 | 0.862 |
| Kumar et al., 201725 | 0.898 | 0.756 | 0.802 | 0.692 | 0.750 |
| ChexRadiNet | 0.925 | 0.798 | 0.882 | 0.734 | 0.846 |
| Method | Nodule | Pleural Thickening | Pneumonia | Pneumothorax | Mean |
| Wang et al., 20174 | 0.671 | 0.708 | 0.633 | 0.806 | 0.738 |
| Wang et al., 20185 | 0.685 | 0.735 | 0.720 | 0.847 | 0.772 |
| Yao et al., 20189 | 0.717 | 0.765 | 0.713 | 0.841 | 0.803 |
| Rajpurkar et al., 20173 | 0.777 | 0.814 | 0.763 | 0.893 | 0.842 |
| Kumar et al., 201725 | 0.666 | 0.774 | 0.715 | 0.859 | 0.795 |
| ChexRadiNet | 0.748 | 0.867 | 0.737 | 0.889 | 0.843 |
4.2 Qualitative analysis
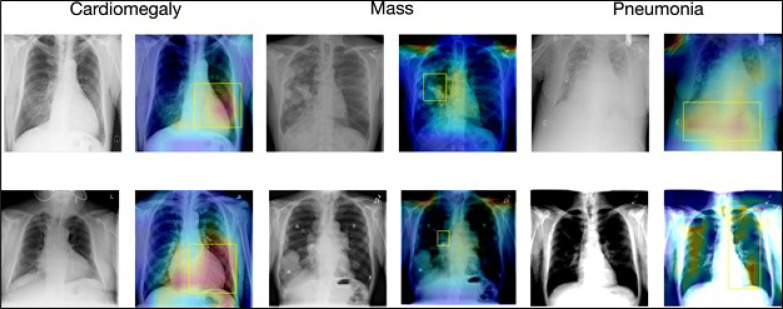
Figure 2 shows the attention map of our model against the ground truth bounding boxes. The visualization provides better explainability of our model. In Figure 2 we visualized our results for Cardiomegaly, Mass, and Pneumonia.
Figure 2:
Visualization of the disease localization on the test images with ChexRadiNet and ground truth bounding boxes. The attention maps are generated from the final output tensor and overlapped on the original radiology images. The left image in each pair is the chest X-ray image and the right one is the generated attention map and the ground truth (in the yellow box).
Cardiomegaly is considered to be present if the cardiothoracic rate is larger than 50% (cardiothoracic Ratio equals “Maximum horizontal cardiac width” over “Maximum horizontal thoracic width”), which means an enlarged heart. The 2nd image in the 1st row as well as the 2nd image in the 2nd row in Figure 2 shows that our model successfully detects cardiomegaly, an enlarged heart, perfectly, and aligns with the yellow bounding box well.
A lung mass is an abnormal spot in the lungs that is more than 3 centimeters. Our results (4th images in the 1st and 2nd rows), although focusing on larger areas, can capture some clues of lung mass.
Note that in the chest X-ray14 dataset, only one bounding box is annotated for one disease image. Though some patients are diagnosed with several diseases, only the most important disease is annotated on the radiology image. This means that ground truth has missing annotations (shown by Pneumonia). Pneumonia inflames the air sacs in one or both lungs. For Pneumonia detection, radiologists will look for white spots in the lungs. For the 6th image in the 2nd row, both lungs are infected and white spots are shown in both lungs. However, the bounding box of the 6th image only annotates the right lung while our model successfully localizes Pneumonia for both lungs.
Overall, our results show that the predicted disease localizations have a great alignment with the ground truth and can even serve as a supplement to the ground truth.
5 Conclusion
We propose a framework that jointly learns radiomic features and predicts 14 thoracic diseases. We evaluated our model on three publicly available corpora. We showed that both our disease identification and localization outperform state-of-the-art models in the quantitative and qualitative analysis.
Our proposed framework has two main limitations. First, chest X-rays are very different from natural images, but we rely on deep learning models (ResNet) that work better on natural images. Second, the robustness of radiomic features relies on the accuracy of bounding boxes, in our work, the bounding boxes are generated by heatmaps. It is not guaranteed that the generated heatmaps are always good and accurate. Our future work will continue to solve these two limitations.
Automatically generating correct bounding boxes can be a milestone to push the agenda for AI-driven medical imaging diagnosis. It can abruptly increase the annotated medical images at a much lower cost so that better CNN models can be trained, therefore better diagnosis models can be obtained. Bounding boxes can increase the interpretability of AI solutions by locating the abnormalities as the visual evidence in medical images, which can build trust between doctors and patients.
Acknowledgment
This work is supported by Amazon Machine Learning Research Award 2020. It also was supported by the National Library of Medicine under Award No. 4R00LM013001.
Figures & Table
References
- 1.Topol Eric J. Deep medicine: how artificial intelligence can make healthcare human again. first edition. New York: Basic Books; 2019. [Google Scholar]
- 2.Ching Travers, Himmelstein Daniel S, Beaulieu-Jones Brett K, Kalinin Alexandr A, Do Brian T, Way Gregory P, Ferrero Enrico, Agapow Paul-Michael, Zietz Michael, Hoffman Michael M, Xie Wei, Rosen Gail L, Lengerich Benjamin J, Israeli Johnny, Lanchantin Jack, Woloszynek Stephen, Carpenter Anne E, Shrikumar Avanti, Xu Jinbo, Cofer Evan M, Lavender Christopher A, Turaga Srinivas C, Alexandari Amr M, Lu Zhiyong, Harris David J, DeCaprio Dave, Qi Yanjun, Kundaje Anshul, Peng Yifan, Wiley Laura K, Segler Marwin H S, Boca Simina M, Swamidass S Joshua, Huang Austin, Gitter Anthony, Greene Casey S. Opportunities and obstacles for deep learning in biology and medicine. Journal of the Royal Society, Interface. April 2018;15(141) doi: 10.1098/rsif.2017.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajpurkar Pranav, Irvin Jeremy, Zhu Kaylie, Yang Brandon, Mehta Hershel, Duan Tony, Ding Daisy, Bagul Aarti, Langlotz Curtis, Shpanskaya Katie, Lungren Matthew P., Andrew Y. Ng. CheXNet: radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv:1711.05225 [cs, stat], December 2017.
- 4.Wang Xiaosong, Peng Yifan, Lu Le, Lu Zhiyong, Bagheri Mohammadhadi, Summers Ronald M. In IEEE Conference on Computer Vision and Pattern Recognition (CVPR) IEEE; 2017. Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases; pp. pages 3462–3471. [Google Scholar]
- 5.Wang Xiaosong, Peng Yifan, Lu Le, Lu Zhiyong, Summers Ronald M. In IEEE Conference on Computer Vision and Pattern Recognition (CVPR) IEEE; June 2018. TieNet: text-image embedding network for common thorax disease classification and reporting in chest x-rays; pp. pages 9049–9058. [Google Scholar]
- 6.Irvin Jeremy, Rajpurkar Pranav, Ko Michael, Yu Yifan, Ciurea-Ilcus Silviana, Chute Chris, Marklund Henrik. CheXpert: a large chest radiograph dataset with uncertainty labels and expert comparison. Proceedings of the AAAI Conference on Artificial Intelligence. 2019;volume 33:pages 590–597. [Google Scholar]
- 7.Johnson Alistair E. W., Pollard Tom J., Greenbaum Nathaniel R., Lungren Matthew P., Deng Chih-ying, Peng Yifan, Lu Zhiyong, Mark Roger G., Berkowitz Seth J., Horng Steven. MIMIC-CXR-JPG, a large publicly available database of labeled chest radiographs. arXiv preprint. January 2019. [DOI] [PMC free article] [PubMed]
- 8.Sowrirajan Hari, Yang Jingbo, Ng Andrew Y, Rajpurkar Pranav. Moco pretraining improves representation and transferability of chest x-ray models. arXiv preprint arXiv. 2020;2010.05352 [Google Scholar]
- 9.Yao Li, Poblenz Eric, Dagunts Dmitry, Covington Ben, Bernard Devon, Lyman Kevin. Learning to diagnose from scratch by exploiting dependencies among labels. arXiv preprint arXiv. 2017;1710.10501 [Google Scholar]
- 10.Gundel Sebastian, Grbic Sasa, Georgescu Bogdan, Liu Siqi, Maier Andreas, Comaniciu Dorin. Progress in pattern recognition, image analysis, computer vision, and applications. Springer International Publishing; 2019. Learning to recognize abnormalities in chest x-rays with location-aware dense networks; pp. pages 757–765. [Google Scholar]
- 11.Li Zhe, Wang Chong, Han Mei, Xue Yuan, Wei Wei, Li Li-Jia, Fei-Fei Li. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2018. Thoracic disease identification and localization with limited supervision; pp. pages 8290–8299. [Google Scholar]
- 12.Hwang Sangheum, Kim Hyo-Eun. Self-transfer learning for fully weakly supervised object localization. arXiv preprint arXiv. 2016;1602.01625 [Google Scholar]
- 13.Bryan R. Nick., editor. Introduction to the science of medical imaging. Cambridge University Press; January 2001. [Google Scholar]
- 14.Nicolasjilwan Manal, Hu Ying, Yan Chunhua, Meerzaman Daoud, Holder Chad A., Gutman David, Jain Rajan, Colen Rivka, Rubin Daniel L., Zinn Pascal O., Hwang Scott N., Raghavan Prashant, Hammoud Dima A., Scarpace Lisa M., Mikkelsen Tom, Chen James, Gevaert Olivier, Buetow Kenneth, Freymann John, Kirby Justin, Flanders Adam E., Wintermark Max. Addition of MR imaging features and genetic biomarkers strengthens glioblastoma survival prediction in TCGA patients. Journal of Neuroradiology. July 2015;42(4):212–221. doi: 10.1016/j.neurad.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganeshan Balaji, Abaleke Sandra, Young Rupert C.D., Chatwin Christopher R., Kenneth A. Miles. Texture analysis of non-small cell lung cancer on unenhanced computed tomography: initial evidence for a relationship with tumour glucose metabolism and stage. Cancer Imaging. 2010;10(1):137–143. doi: 10.1102/1470-7330.2010.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganeshan Balaji, Goh Vicky, Mandeville Henry C., Ng Quan Sing, Hoskin Peter J., Miles Kenneth A. Non–small cell lung cancer: Histopathologic correlates for texture parameters at CT. Radiology. January 2013;266(1):326–336. doi: 10.1148/radiol.12112428. [DOI] [PubMed] [Google Scholar]
- 17.Parekh Vishwa S., Michael A. Jacobs. Deep learning and radiomics in precision medicine. Expert Review of Precision Medicine and Drug Development. March 2019;4(2):59–72. doi: 10.1080/23808993.2019.1585805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Guanxiong, Hsu Tzu-Ming Harry, McDermott Matthew, Boag Willie, Weng Wei-Hung, Szolovits Peter, Ghassemi Marzyeh. In Machine Learning for Healthcare Conference. PMLR; 2019. Clinically accurate chest x-ray report generation; pp. pages 249–269. [Google Scholar]
- 19.Lei Tao, Barzilay Regina, Jaakkola Tommi. Rationalizing neural predictions. In Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing. 2016. pp. pages 107–117.
- 20.He Kaiming, Zhang Xiangyu, Ren Shaoqing, Sun Jian. Deep residual learning for image recognition. In The IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2016. pp. pages 770–778.
- 21.Zhou Bolei, Khosla Aditya, Lapedriza Agata, Oliva Aude, Torralba Antonio. In IEEE Conference on Computer Vision and Pattern Recognition (CVPR) June 2016. Learning deep features for discriminative localization; pp. pages 2921–2929. [Google Scholar]
- 22.Misra Diganta, Nalamada Trikay, Arasanipalai Ajay Uppili, Hou Qibin. Rotate to attend: convolutional triplet attention module. arXiv:2010.03045 [cs] November 2020.
- 23.Woo Sanghyun, Park Jongchan, Lee Joon-Young, Kweon In So. Cbam: Convolutional block attention module. In Proceedings of the European conference on computer vision (ECCV) 2018. pp. pages 3–19.
- 24.Peng Yifan, Wang Xiaosong, Lu Le, Bagheri Mohammadhadi, Summers Ronald, Lu Zhiyong. NegBio: a high-performance tool for negation and uncertainty detection in radiology reports. In AMIA Joint Summits on Translational Science proceedings. 2018;volume 2017:pages 188–196. [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar Pulkit, Grewal Monika, Srivastava Muktabh Mayank. In The International Conference on Computer Vision (ICCV) 2018. Boosted cascaded convnets for multilabel classification of thoracic diseases in chest radiographs; pp. pages 546–552. [Google Scholar]