Abstract
Overabundance of information within electronic health records (EHRs) has resulted in a need for automated systems to mitigate the cognitive burden on physicians utilizing today’s EHR systems. We present ProSPER, a Problem-oriented Summary of the Patient Electronic Record that displays a patient summary centered around an auto-generated problem list and disease-specific views for chronic conditions. ProSPER was developed using 1,500 longitudinal patient records from two large multi-specialty medical groups in the United States, and leverages multiple natural language processing (NLP) components targeting various fundamental (e.g. syntactic analysis), clinical (e.g. adverse drug event extraction) and summarizing (e.g. problem list generation) tasks. We report evaluation results for each component and discuss how specific components address existing physician challenges in reviewing EHR data. This work demonstrates the need to leverage holistic information in EHRs to build a comprehensive summarization application, and the potential for NLP-based applications to support physicians and improve clinical care.
Introduction
Electronic health records (EHRs) are one of the primary sources of clinician burnout1–3. Many studies have analyzed physician time spent in EHRs with respect to various interactions such as chart review, documentation, orders, and inbox management4,5, with several identifying time spent on reviewing information within the EHR, i.e. chart review, as a major component of physician interaction with the EHR. A descriptive study on ambulatory medical subspecialists and primary care physicians observed that chart review accounted for the highest proportion of active physician EHR time at 33%4. Another study analyzed audit logs to measure time requirements for EHR use by ophthalmologists and concluded that most of the time spent in the EHR is on reviewing information5.
Several factors contribute to the difficulty in reviewing information in EHRs. Traditionally, the patient medical record was mainly used to document a patient’s medical history and clinical care process to assist physicians in providing informed care. However, the transformation from paper-based to electronic patient records have expanded the scope of documentation to also serve a variety of non-clinical purposes, including administrative, legal, research, and education6. This transformation has resulted in lengthy and complex documentation dominated by content not directly relevant to clinical care, such as text meant for billing purposes, quality improvement measures, avoiding malpractice, and documenting compliance7. Further exacerbating this problem are issues of note bloat, where use of structured data imports, templates and copy-paste have introduced unnecessary, redundant or erroneous data into clinical notes, worsening the problem of information overload and physician stress8–10. A human factors engineering perspective to the information chaos in primary care identified five information hazards: information overload, information underload, information scatter, information conflict, and erroneous information11. These information hazards result in additional workload for physicians, such as expending extra effort to search through charts, asking more questions of the patient to clarify conflicting documentation, or re-working a diagnosis to investigate potentially erroneous information. Together these factors contribute to excess clinician workload and burnout, with adverse outcomes on clinician well-being, patient care, and the health care system12.
Previous research has explored several automated summarization and enhanced visualization methods to enable physicians to efficiently navigate and discern complex data within EHRs13. Summarization efforts include problem identification from a given set of clinical notes14–16, extraction of clinical concepts or relations17,18, and various ways to display such information along with other information in the EHR15,19,20. Another research direction has focused on improved data visualization and interactive displays to facilitate more efficient and accurate information retrieval and an overall more user-friendly interaction with the EHR21–24. Evaluation of such systems in real or simulated clinical settings have received positive feedback from physicians and resulted in improved physician performance15,19,22,25,26. A common theme in many of these systems is the idea of a problem-oriented medical record (POMR)27, and the use of timelines to gain a longitudinal history of the patient. We rely on these principles to build Problem-oriented Summary of the Patient Electronic Record (ProSPER), an application which extends the idea of a POMR by enhancing it with clinically meaningful insights extracted from unstructured clinical notes, such as adverse drug events (ADEs), and displaying them on a timeline with relevant structured elements of the EHR.
During the past several years, we have developed and validated the usability of several components that contribute to the creation of a comprehensive patient record summarization application. The purpose of this paper is to build on our previously published work and present ProSPER, an artificial intelligence-based system that extracts, analyzes, and summarizes patient clinical information from electronic health records, with the goal of reducing the cognitive burden on physicians due to information chaos within the EHR. We present the overall system architecture and pipeline, describe the underlying components needed to build such a system, and discuss how these components address information chaos within the EHR. ProSPER differs from previously published systems in that it presents (1) an auto-generated problem list, (2) disease-specific dashboards, (3) a stacked timeline view incorporating both structured data and insights extracted from unstructured data to allow reasoning across different data elements, and (4) a textual summary of each clinical encounter for a selected disease of interest.
Materials and Methods
Combining the well-established idea of a problem-oriented medical record with the different workflows using EHRs that rely on electronic dashboard summary screens as well as individual progress notes, we envision a workflow for reviewing patient information prior to a clinic visit as follows.
Patient overview: gain an overview of the patient, including the patient’s name, demographics, allergies, and a summary of the active problems, current medications, allergies, vital signs, and laboratory results.
Snapshot of a specific problem: focus on a specific disease by surfacing current medications and recent laboratory results and procedures relevant to the specified disease.
Longitudinal history of a specific problem: investigate a specific disease through a timeline view of relevant medications, laboratory results, procedures, past encounters and clinical events extracted from clinical notes.
Dataset
Effective natural language processing (NLP) techniques are essential to development of the proposed system. To achieve this, we used a set of 1,500 de-identified longitudinal patient records from two large ambulatory multi-specialty medical groups in the United States. Subsets of this corpus were manually annotated by medical students and practicing physicians to generate ground truth for development of smaller NLP components that are used in building the overall system. All annotations were doubly-annotated and adjudicated to ensure the quality and consistency of the ground truth. Details of the ground truth generation process for each task are provided below.
The dataset for problem list generation28 consists of 399 randomly selected patient records annotated by over 17 fourth year medical students. Annotators reviewed each patient record, created a problem list for a comprehensive health assessment, normalized identified problems to the clinical observations recordings and encoding (CORE) problem list subset of Systematized Nomenclature of Medicine-Clinical Terms (SNOMED CT)29 in a one-to-many mapping, and evaluated how closely the SNOMED CT concept represents the problem.
The dataset for relation detection30 consists of 24,537 unique problem-medication pairs, 11,318 unique problem-procedure pairs, and 23,402 unique problem-laboratory test pairs created using structured data from 100 randomly selected patient records. Four fourth year medical students reviewed each pair of entities and indicated if a positive association was present, defined as medications that treat or prevent the problem or its symptoms, and laboratory tests or procedures that can be used to screen, prevent, evaluate, diagnose, or manage the problem.
The dataset for extractive summarization31 consists of 3,453 clinical notes over 762 patients with recent documentation of at least one of five common chronic diseases (hypertension, diabetes mellitus, hyperlipidemia, heart failure, chronic obstructive pulmonary disease) and authored by physicians or nurse practitioners. The corpus was annotated by 12 physicians with emphasis on only annotating the most important insights relevant to physicians’ decision-making.
The ground truth for medication insights, specifically ADE and medication change events, was annotated at the note-level. For ADEs, 602 notes were annotated with medications and their associated adverse events. For medication change events, 38,895 medication mentions were annotated to indicate (1) the presence of a medication change event and (2) the multi-dimensional context (action, temporality, certainty) for identified medication change events.
The dataset for evaluation of semantic find32 is derived from 10 randomly selected patient records. For each patient record, a physician generated a set of relevant search terms based on the patient’s last progress note and problem list. The final dataset consists of 169 search terms, covering a variety of semantic types (e.g. symptoms, drugs), single and multi-word concepts, different parts of speech, and commonly-accepted medical abbreviations.
Pipeline and framework
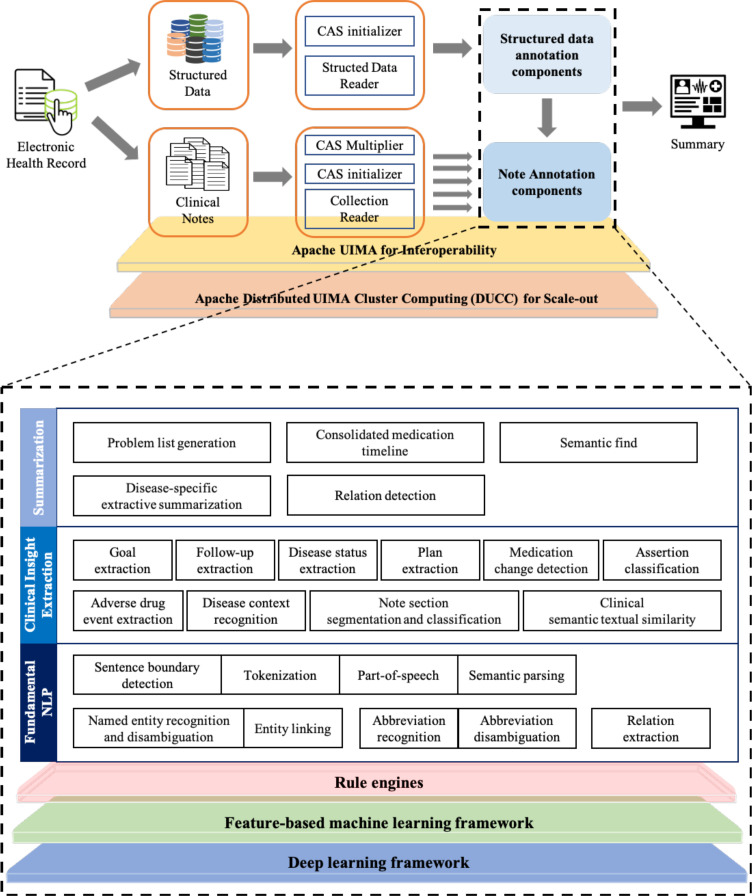
ProSPER was built using the open-source Apache Unstructured Information Management Architecture (UIMA)33 library, a software architecture that provides capabilities to construct pipelines consisting of a sequence of components or analysis engines (AE) that inspect the input data, perform analysis, and store resulting annotations. The system consists of several such components that work together to process an input EHR. These components write their output to a shared data structure called a common analysis structure (CAS). The system achieves high performance by using UIMA Asynchronous Scaleout, a set of capabilities supported for achieving scale-out and Distributed UIMA Cluster Computing, a cluster management system providing tooling, management, and scheduling facilities to automate the scale-out of UIMA applications. Figure 1 shows the high-level flow and underlying NLP components of ProSPER. ProSPER’s NLP components, all tailored towards EHR text, can be broadly divided into three levels, described below.
Figure 1.
A high-level architecture of ProSPER.
Fundamental and clinical NLP components
Despite the success of current open-domain NLP systems for syntactic and semantic analysis, several studies34,35 have shown that substantial effort is needed to adopt existing systems to the clinical domain. Clinical notes are written under considerable time pressure, using a combination of ad-hoc formatting and liberal use of parenthetical expressions, jargon and acronyms to increase the information density35. For example, sentence ends are frequently indicated by layout rather than punctuation, and white space is not always present to indicate token boundaries (eg, “50mg”). To handle such issues, two deep parsing components36 were used: a domain-adapted English Slot Grammar parser for core linguistic analyses (tokenization, segmentation, morpho-lexical analysis, syntactic analysis), and a predicate-argument structure (PAS) builder for simplification and abstraction of the ESG parse. These components incorporate domain-specific rules sensitive to low-level features such as punctuation, capitalization, text-wrap properties, and indentation of clinical documentation.
Rule-based, machine learning (ML)-based and lookup-based methods are three major approaches for extracting useful information from natural language texts. Many components in ProSPER use a combination of hand-crafted rules and ML-based methods for their operation. The rule engine in ProSPER uses a pattern language for describing rules on the output of PAS builder. Details about this rule engine are included in Boguraev et al.37. Two ML frameworks, feature-based learning and deep learning frameworks, were developed to support concept and sentence classification, sequence labeling, sequential sentence labeling, sentence pair scoring, and relation extraction. The feature-based learning framework provides implementations of traditional ML algorithms such as Support Vector Machines38, Conditional Random Fields (CRF)39, and domain-independent and dependent feature extractors. Similarly, the deep learning framework provides implementations of state-of-the-art deep learning architectures, such as Bidirectional Long Short-Term Memory – Conditional Random Fields40 and pre-trained transformer-based models41. These frameworks were tailored to more effectively process clinical notes. They provide the ability to (1) easily integrate feature extractors or architectures, (2) leverage evidence from both structured and unstructured data simultaneously to develop consolidated models, and (3) streamline models or feed predictions from one or more auxiliary tasks to subsequent tasks. ProSPER uses UIMA’s ConceptMapper42, a tool for lookup-based methods, to link text in input documents with entries in a dictionary such as the Unified Medical Language System (UMLS) semantic network43.
Several NLP components that extract useful information from clinical narratives have been developed using these frameworks. Details of these components have been described in our previous studies15,30–32,44–50. These include (1) fundamental NLP components such as named entity recognition (NER) and disambiguation (NED)42,48,49, entity linking51, relation extraction47,48,52 and abbreviation recognition and disambiguation49, and (2) components specific to clinical documentation (i.e. clinical insight extraction components) such as note section segmentation and classification53, clinical semantic textual similarity (STS)50, and assertion classification54.
Clinical insight extraction components can be broadly categorized into (1) concept classification, (2) sentence classification, (3) sequential sentence classification, (4) semantic textual similarity, and (5) relation extraction. Concept classification components, such as assertion classification and medication change extraction, provide one or more categorical labels on a concept based on the surrounding context. Assertion classification uses a deep learning-based model that assigns each disease mention in a note with one of the following labels: present, absent, possible, hypothetical, conditional, associated with someone else; medication change extraction leverages multiple ML models that assign each medication mention in a note with labels corresponding to several dimensions: action (start, stop, increase, decrease), temporality (past, present, future), and certainty (certain, hypothetical, conditional). Sentence classification components assign one or more labels to a sentence in a clinical note; examples include binary classification models such as plan55, follow-up, and goal extraction. Sequential sentence labeling components assign a label to each sentence in a given sequence, which are useful in capturing the inherent dependencies of sentences within a clinical note to assign the correct label for each sentence. For example, the note section classification model identifies the layout of a note with respect to its semantics by assigning one of a set of pre-defined section labels to each sentence (e.g. Review of Systems, Physical Exam). Clinical STS employs a combination of multi-task learning56 and fine-tuned pre-trained language models57,58 to compute the semantic equivalence between a pair of text snippets. Relation extraction components extracts entities from clinical notes and identifies relations between them. An example is ADE extraction which uses a joint deep learning model that extracts medications and related ADEs simultaneously.
Summarization components
Summarization components are high-level analytics built on top of the fundamental NLP and clinical insight extraction components. These include problem list generation, relation detection, disease-specific extractive summarization, consolidated medication timeline, and semantic find.
Problem list generation: To extract an open-ended list of the patient’s active problems from the entire patient record, problem list generation was framed as a multi-label classification and ranking task. The model was based on an alternating decision tree, which used outputs from many upstream analytics (described above) as features, including but not limited to UMLS concepts identified by the NER/NED components and mapped to the CORE problem list subset of SNOMED CT, their assertions and distribution over the longitudinal record, and the degrees of alignment in temporal space between the identified candidate problems and their related medications, laboratory tests, and procedures. This method produced a human interpretable model, which is a desirable property in the clinical domain. To the best of our knowledge, this was the first successful open-ended problem list generation system15,44,45.
Relation detection: An ensemble approach with two supervised ML models was used to associate problems with related medications, laboratory tests, and procedures30. One method used features extracted from distributional semantics and UMLS; the second used features mined from historical, actual patient data. The results of relation detection were used both internally as features to support other components, and as a user interface functionality to sort structured data tables and highlight relevant data elements for a selected problem in the problem list.
Disease-specific extractive summary: Extractive summarization was modeled as a sequential sentence labeling problem, where each sentence in a given note is classified as being relevant (or not) to the management of the specified disease of interest. Outputs from upstream components, such as plan extraction and note section segmentation and classification, were leveraged in a linear-chain CRF model to automatically generate disease-specific summaries from clinical notes. As a post-processing step, boilerplate statements, which are often either not patient-specific or meant for purposes other than clinical care, are downweighted based on frequency analysis on all notes in our corpus. Next, deduplication of semantically equivalent sentences enabling the generation of more salient summaries was achieved by leveraging clinical STS. Details of the extraction summarization system was published in Liang et al.31.
Consolidated medication timeline: Data from structured medication orders, medications extracted from clinical notes and their related insights were aggregated to build a comprehensive medication timeline. In building this timeline, there were two key challenges: (1) some medications are only documented in unstructured data59 and (2) daily dosage, although present in many medication visualization designs21,22, is generally not available as a discrete field and requires understanding free text medication instructions (i.e. Sig). To address this gap, our system leveraged NER and NED components to identify medications and their attributes in clinical notes and Sigs, then normalized these attributes to calculate daily dosage60. To further enhance the timeline, outputs from clinical insight components (e.g. ADE and medication change extraction) were displayed as popovers on the timeline for the relevant medication.
Semantic find: Semantic find is an NLP-based search that leverages UMLS and distributional semantics to perform different kinds of search, such as conceptual search, which leverages UMLS to match clinically equivalent concepts (e.g. lisinopril/Zestril) and ISA relations to match more specific (e.g. cancer/sarcoma) or more general (e.g. gi bleed/hemorrhage) variations of the search term, and associative search, which uses Latent Semantic Analysis61 to measure the probability of an association between the search term and target term, and returns concepts with a score above 0.5 (e.g. asthma/wheezing). Details for semantic find are available in Prager et al.32.
Evaluation metrics
Evaluation of NLP components can be classified into intrinsic and extrinsic methods62. Intrinsic methods evaluate the functionality of a component against a predetermined gold standard, whereas extrinsic methods focus on the component’s contribution to the overall objective of an application and often requires a human-in-the-loop. As highlighted in by Pivovarov et al.13, extrinsic evaluation of NLP systems in a clinical setting is a challenging task due to reasons including (1) vendor EHR systems often do not support interaction with outside applications, and (2) hospitals often request evidence supporting the usefulness of an informatics system before investing resources for implementation. As a result of these constraints, here we report an evaluation of our system based on intrinsic methods, with an extrinsic evaluation of the overall application planned for the future.
As is common in information retrieval, recall (R), precision (P), and F1 score was used to evaluate our components. Recall, also known as sensitivity, is the number of true positives found given the total number of ground truth annotations. Precision, also known as positive predictive value, is the number of true positives given the total number of system predictions. F1 score provides a balanced measure of recall and precision. Specificity, also known as true negative rate, is not useful for our tasks as the number of true negatives are significantly higher than that of true positives, thereby yielding a less meaningful accuracy distinction.
Results
Intrinsic evaluation
Table 1 presents the precision, recall, and F1 score for the summarization components, namely problem list generation, relation detection, disease-specific extractive summarization, consolidated medication timeline (reported separately for ADE and medication change extraction), and semantic find.
Table 1.
Precision, recall and F1 score for problem list generation, relation detection, disease-specific extractive summarization, adverse drug event extraction, medication change extraction, and semantic find.
| Component | Type | Precision | Recall | F1 score |
|---|---|---|---|---|
| Problem list generation | (all problems) | 0.84 | 0.66 | 0.74 |
| Relation detection | Problem – Medication | 0.82 | 0.75 | 0.78 |
| Problem – Procedure | 0.76 | 0.69 | 0.71 | |
| Problem – Laboratory Test | 0.85 | 0.74 | 0.79 | |
| Disease-specific extractive summarization | Hypertension | 0.74 | 0.58 | 0.65 |
| Diabetes mellitus | 0.74 | 0.62 | 0.67 | |
| Hyperlipidemia | 0.67 | 0.47 | 0.55 | |
| Heart failure | 0.74 | 0.50 | 0.60 | |
| COPD | 0.69 | 0.53 | 0.60 | |
| Adverse drug event extraction | Medication – Adverse Event | 0.61 | 0.56 | 0.58 |
| Medication change event extraction | Action | 0.81 | 0.77 | 0.79 |
| Temporality | 0.79 | 0.79 | 0.79 | |
| Certainty | 0.78 | 0.76 | 0.77 | |
| Semantic find | (all search terms) | 0.87 | 0.87 | 0.87 |
ProSPER application
ProSPER consists of (1) a problem-oriented patient summary centered around an auto-generated problem list, and (2) disease-specific views for several chronic conditions. The problem-oriented patient summary allows users to gain a quick overview of the patient, while the disease-specific views allow a deeper dive into a specific problem of interest.
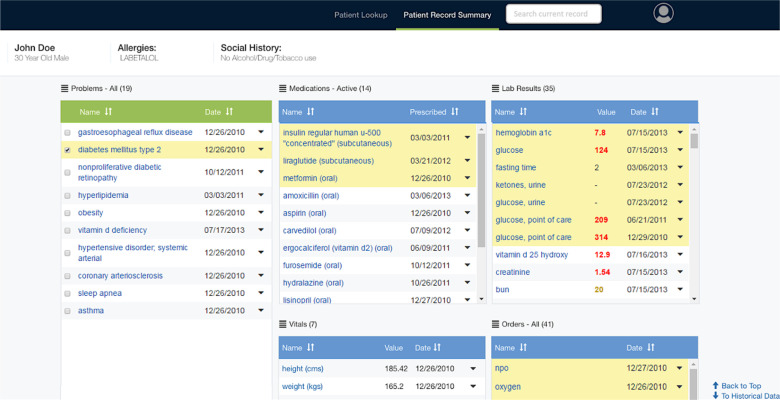
The problem-oriented patient summary presents (1) an auto-generated problem list45, (2) aggregated clinical data by type, (3) relations between problems and other data aggregates30 and (4) a semantic search over the entire EHR32. Figure 2 shows the user interface which presents several panels, one for each data type. When a problem is selected, related medications, laboratory results, and procedures are highlighted and brought to the top for easier review.
Figure 2.
Problem-oriented patient summary with diabetes mellitus selected.
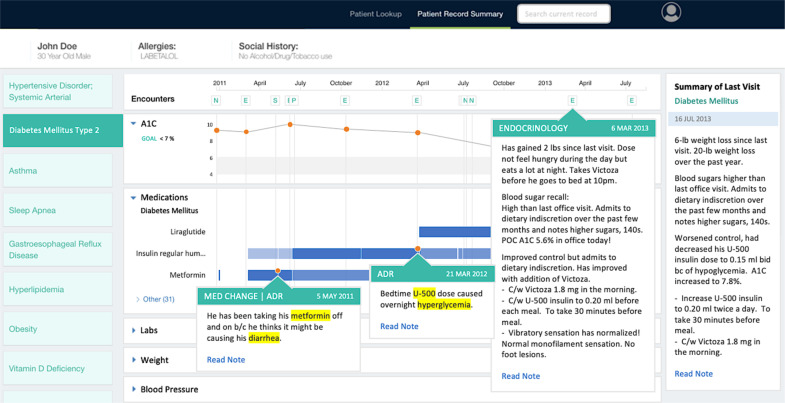
The disease-specific view allows further investigation of a problem by presenting all information relevant to the disease of interest on one screen. In this view, encounters in which the specified disease was discussed are displayed on a timeline, and an extractive summary of each of these encounters is produced containing only the most important information relevant for managing the specified disease31. Relevant medications, test results and their trends are also displayed in timeline form, allowing users to identify potentially informative encounters, and also infer relationships between various elements of the patient record (e.g. medication prescribed after an increase in a particular laboratory result). Figure 3 shows the disease-specific view for diabetes mellitus in a patient.
Figure 3.
Disease-specific view for diabetes mellitus.
Disease-specific extractive summaries for each encounter are displayed by hovering over icons in the encounter timeline, with the summary of the most recent encounter in the right panel for easy viewing. Orange indicators on the medication timeline represent medication insights extracted from clinical notes and are displayed as popovers. Semantic find can be launched using the search box on the top right if the user needs additional information.
Discussion
Challenges in developing an NLP-based application for use in a real-world clinical setting
There is a significant gap between what low-level NLP components can do as compared to the mental work physicians perform during chart review. Previous studies on applying NLP in the clinical domain either focused on advancing the state-of-the-art on a single task or using existing NLP toolkits with more emphasis placed on data visualization. This paper tries to bridge the gap between what low-level NLP components can provide versus what physicians want, by leveraging ML and NLP in all aspects of a chart review scenario.
Several components within our system have been previously published to advance the state-of-the-art47,48,50 on publicly available datasets. However, for many components there are no publicly available datasets, necessitating use of in-house datasets and ground truth. An additional challenge is the need for both structured and unstructured data in creating ground truth for many high-level tasks such as summarization.
Although NLP applications have reported good accuracy, studies have shown that these applications still have low adoption rates in clinical practice63, largely due to uncertainty about minimal performance requirements rather than performance-related issues. The interpretation of a specific component’s accuracy in terms of the overall usefulness of a system as measured by an extrinsic evaluation still warrants more research attention.
How does ProSPER address physician challenges in reviewing EHRs?
To understand how ProSPER can support physicians in their review of EHR data, we discuss how specific components within the system address the five information hazards identified by Beasley et al.11: information overload, information underload, information scatter, information conflict, and erroneous information. Information overload occurs when there is too much data for the user to review, organize, synthesize, and act on. One way ProSPER mitigates this problem is by disease-specific extractive summarization, which leverages components such as note section classification and clinical STS to remove uninformative or redundant text24,64, to produce a succinct summary of a clinical note. Information underload describes the lack of necessary information, such as missing information due to incomplete patient records, or information that is so difficult to find and for all intents and purposes, “missing”. ProSPER targets information underload in several ways, including semantic find, which searches the entire EHR based on a user-provided query, and ADE extraction, which surfaces rare yet important clinical events that have great impact on patient safety. Information scatter describes the problem of having information located in multiple places, thereby requiring additional effort to reconcile this information. The idea of a problem list was conceptualized as a way to address this issue by capturing all of the patient’s important health problems in one centralized location, but in practice it is rarely well-maintained rendering it mostly unusable65. ProSPER supports problem list reconciliation through its problem list generation component, which has been validated by physicians in a small pilot study45. The last two information hazards, information conflict and erroneous information, can be discussed together as conflicts within EHR documentation can be partially attributed to erroneous information. In providing a consolidated medication timeline containing information from both structured and unstructured sources, ProSPER allows users to more easily identify and resolve discrepancies in medication data between structured and unstructured data.
Limitations and future work
We acknowledge several limitations to this work. First, the results presented in this paper are based on an intrinsic evaluation of the NLP components using precision, recall, and F1 score to compare against ground truth created by physicians or medical students. However, these metrics do not fully capture the nuances of what should or should not be included in a problem-oriented patient summary. For instance, a single missed ADE by the system may not have a significant impact when measured using the proposed quantitative metrics, but it may be critically important to patient management and should never be missed. Second, these results are specific to the datasets, which contain only EHRs from two healthcare organizations. Ground truth generation and evaluation on a more diverse dataset is needed to better understand the effectiveness of these approaches. Third, while some of the components were previously evaluated and published using publicly available datasets, others were developed using proprietary datasets not available to the research community. In addition to continuing to adapt and innovate novel methodologies to improve individual NLP components, which in-turn can benefit the overall application, we plan to take the following steps to address the limitations above:
Use extrinsic measures to capture the usefulness of ProSPER for practicing physicians at the point-of-care.
Create ground truth annotations on publicly available datasets such as MIMIC-III66 and organize challenges to invite community-driven efforts.
Conclusion
We present ProSPER, a novel problem-oriented summary of the patient record that leverages 3 levels of NLP informatics, including performing well-studied fundamental NLP tasks such as NER, extracting clinically meaningful events from clinical notes such as ADE, generating an active problem list from the heterogeneous, longitudinal patient record, and creating disease-specific extractive summaries from clinical notes. Components underlying ProSPER target specific information hazards within the EHR that lead to excess physician workload and mental exhaustion. Our work demonstrates the need to leverage holistic information in EHRs to build a comprehensive summarization application, and the potential for NLP-based applications to support physician and improve overall clinical care.
Figures & Table
References
- 1.Gardner RL, Cooper E, Haskell J, et al. Physician stress and burnout: the impact of health information technology. J Am Med Inform Assoc. 2019;26:106–14. doi: 10.1093/jamia/ocy145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reaction Data . Physician Burnout American Fork. UT: 2018. [Google Scholar]
- 3.Physician Burnout Agency for Healthcare Research and Quality. [DOI] [PubMed]
- 4.Overhage JM, McCallie D., Jr Physician Time Spent Using the Electronic Health Record During Outpatient Encounters: A Descriptive Study. Ann Intern Med. 2020;172(3):169–174. doi: 10.7326/M18-3684. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein IH, Hribar MR, Reznick LG, Chiang MF. Analysis of Total Time Requirements of Electronic Health Record Use by Ophthalmologists Using Secondary EHR Data. AMIA Annu Symp Proc. 2018;2018:490–497. [PMC free article] [PubMed] [Google Scholar]
- 6.Ho YX, Gadd CS, Kohorst KL, et al. A qualitative analysis evaluating the purposes and practices of clinical documentation. Appl Clin Inform. 2014;5:153–68. doi: 10.4338/ACI-2013-10-RA-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koopman RJ, Steege LMB, Moore JL, et al. Physician information needs and electronic health records (EHRs): Time to reengineer the clinic note. J Am Board Fam Med. 2015;28:316–23. doi: 10.3122/jabfm.2015.03.140244. [DOI] [PubMed] [Google Scholar]
- 8.Shoolin JS, Ozeran L, Hamann C, et al. Association of medical directors of information systems consensus on inpatient electronic health record documentation. Appl. Clin. Inform. 2013;4:293–303. doi: 10.4338/ACI-2013-02-R-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogel L. Cut-and-paste clinical notes confuse care, say US internists. CMAJ; 2013. p. 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kroth PJ, Morioka-Douglas N, Veres S, et al. Association of Electronic Health Record Design and Use Factors With Clinician Stress and Burnout. JAMA Netw open. 2019;2:e199609. doi: 10.1001/jamanetworkopen.2019.9609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beasley JW, Wetterneck TB, Temte J, et al. Information chaos in primary care: Implications for physician performance and patient safety. J Am Board Fam Med. 2011;24:745–51. doi: 10.3122/jabfm.2011.06.100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel RS, Bachu R, Adikey A, et al. Factors related to physician burnout and its consequences: A review. Behav. Sci. (Basel) 2018;8 doi: 10.3390/bs8110098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pivovarov R, Elhadad N. Automated methods for the summarization of electronic health records. J Am Med Inform Assoc. 2015;22(5):938–947. doi: 10.1093/jamia/ocv032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Vleck TT, Elhadad N. Corpus-Based Problem Selection for EHR Note Summarization. AMIA Annu Symp Proc. 2010;2010:817–21. [PMC free article] [PubMed] [Google Scholar]
- 15.Devarakonda M, Tsou CH. Automated problem list generation from electronic medical records in IBM Watson. Proceedings of the National Conference on Artificial Intelligence; 2015. pp. 3942–7. [Google Scholar]
- 16.Meystre S, Haug PJ. Automation of a problem list using natural language processing. BMC Med Inform Decis Mak. 2005;5:30. doi: 10.1186/1472-6947-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): Architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts A, Gaizauskas R, Hepple M, et al. The CLEF corpus: semantic annotation of clinical text. AMIA Annu Symp Proc. 2007. pp. 625–9. [PMC free article] [PubMed]
- 19.Hirsch JS, Tanenbaum JS, Gorman SL, et al. HARVEST, a longitudinal patient record summarizer. J Am Med Inform Assoc. 2015;22:263–74. doi: 10.1136/amiajnl-2014-002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diomaiuta C, Mercorella M, Ciampi M, et al. A novel system for the automatic extraction of a patient problem summary. Proceedings - IEEE Symposium on Computers and Communications. 2017. pp. 182–6.
- 21.Plaisant C, Mushlin R, Snyder A, et al. LifeLines: using visualization to enhance navigation and analysis of patient records. Proc AMIA Symp. 1998. pp. 76–80. [PMC free article] [PubMed]
- 22.Belden JL, Wegier P, Patel J, et al. Designing a medication timeline for patients and physicians. J Am Med Inform Assoc. 2019;26:95–105. doi: 10.1093/jamia/ocy143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doig AK, Albert RW, Syroid ND, et al. Graphical arterial blood gas visualization tool supports rapid and accurate data interpretation. CIN - Comput Informatics Nurs. 2011;29:204–11. doi: 10.1097/NCN.0b013e3181fc4041. [DOI] [PubMed] [Google Scholar]
- 24.Koopman RJ, Kochendorfer KM, Moore JL, et al. A diabetes dashboard and physician efficiency and accuracy in Accessing data needed for high-quality diabetes care. Ann Fam Med. 2011;9:398–405. doi: 10.1370/afm.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hosseini M, Faiola A, Jones J, Vreeman DJ, Wu H, Dixon BE. Impact of document consolidation on healthcare providers’ perceived workload and information reconciliation tasks: a mixed methods study. J Am Med Inform Assoc. 2019;26(2):134–142. doi: 10.1093/jamia/ocy158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bakken S. Can informatics innovation help mitigate clinician burnout? J Am Med Inform Assoc. 2019;26:93–4. doi: 10.1093/jamia/ocy186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weed LL. Medical records that guide and teach. N Engl J Med. 1968;278:593–600. doi: 10.1056/NEJM196803142781105. [DOI] [PubMed] [Google Scholar]
- 28.Liang JJ, Tsou CH, Devarakonda MV. Ground Truth Creation for Complex Clinical NLP Tasks - an Iterative Vetting Approach and Lessons Learned. AMIA Jt Summits Transl Sci Proc. 2017;2017:203–212. [PMC free article] [PubMed] [Google Scholar]
- 29.Donnelly K. SNOMED-CT: The advanced terminology and coding system for ehealth. Studies in Health Technology and Informatics. 2006. pp. 279–90. [PubMed]
- 30.Dandala B, Devarakonda M, Bornea M, Nielson C. Scoring Disease-Medication Associations using Advanced NLP, Machine Learning, and Multiple Content Sources. BioTxtM@COLING. 2016;2016:125–33. [Google Scholar]
- 31.Liang J, Tsou C-H, Poddar A. A Novel System for Extractive Clinical Note Summarization using EHR Data. Proc 2nd Clinical NLP Workshop. 2019;2019:46–54. [Google Scholar]
- 32.Prager JM, Liang JJ, Devarakonda M V. SemanticFind: Locating What You Want in a Patient Record, Not Just What You Ask For. AMIA Jt Summits Transl Sci proceedings AMIA Jt Summits Transl Sci. 2017;2017:249–58. [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrucci D, Lally A. UIMA: An architectural approach to unstructured information processing in the corporate research environment. Nat Lang Eng. 2004;10:327–48. [Google Scholar]
- 34.Griffis D, Shivade C, Fosler-Lussier E, et al. A quantitative and qualitative evaluation of sentence boundary detection for the clinical domain. AMIA Summits Transl Sci Proc. 2016;2016:88. [PMC free article] [PubMed] [Google Scholar]
- 35.Leaman R, Khare R, Lu Z. Challenges in clinical natural language processing for automated disorder normalization. J Biomed Inform. 2015;57:28–37. doi: 10.1016/j.jbi.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCord MC, Murdock JW, Boguraev BK. Deep parsing in Watson. IBM J Res Dev. 2012;56:1–3. [Google Scholar]
- 37.Boguraev B, Patwardhan S, Kalyanpur A, Chu-Carroll J, Lally A. Parallel and nested decomposition for factoid questions. Natural Language Engineering. 2014;20(4):441–468. [Google Scholar]
- 38.Hearst MA, Dumais ST, Osuna E, et al. Support vector machines. IEEE Intell Syst their Appl. 1998;13:18–28. [Google Scholar]
- 39.Sutton C, McCallum A. An introduction to conditional random fields. Found Tr Mach Learn. 2012;4:267–373. [Google Scholar]
- 40.Huang Z, Xu W, Yu K. Bidirectional LSTM-CRF models for sequence tagging. 2015. arXiv:1508.01991.
- 41.Qiu X, Sun T, Xu Y, Shao Y, Dai N, Huang X. Pre-trained models for natural language processing: A survey. Science China Technological Sciences. 2020;2020:1–26. [Google Scholar]
- 42.Tanenblatt M, Coden A, Sominsky I. The ConceptMapper approach to named entity recognition. Proceedings of the 7th International Conference on Language Resources and Evaluation, LREC 2010; 2010. pp. 546–51. [Google Scholar]
- 43.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Devarakonda M, Zhang D, Tsou CH, et al. Problem-oriented patient record summary: An early report on a Watson application. 2014 IEEE Healthcom. 2014;2014:281–6. [Google Scholar]
- 45.Devarakonda M V, Mehta N, Tsou CH, et al. Automated problem list generation and physicians perspective from a pilot study. Int J Med Inform. 2017;105:121–9. doi: 10.1016/j.ijmedinf.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 46.Mahajan D, Liang JJ, Tsou CH. Toward Understanding Clinical Context of Medication Change Events in Clinical Narratives. 2020. arXiv:2011.08835. [PMC free article] [PubMed]
- 47.Dandala B, Joopudi V, Devarakonda M. Adverse Drug Events Detection in Clinical Notes by Jointly Modeling Entities and Relations Using Neural Networks. Drug Saf. 2019;42:135–46. doi: 10.1007/s40264-018-0764-x. [DOI] [PubMed] [Google Scholar]
- 48.Dandala B, Joopudi V, Devarakonda M, et al. IBM Research System at MADE 2018: Detecting Adverse Drug Events from Electronic Health Records. Proc Mach Learn Res. 2018;90:39–47. [Google Scholar]
- 49.Joopudi V, Dandala B, Devarakonda M. A convolutional route to abbreviation disambiguation in clinical text. J Biomed Inform. 2018;86:71–8. doi: 10.1016/j.jbi.2018.07.025. [DOI] [PubMed] [Google Scholar]
- 50.Mahajan D, Poddar A, Liang JJ, et al. Identification of Semantically Similar Sentences in Clinical Notes: Iterative Intermediate Training Using Multi-Task Learning. JMIR Med Inform. 2020;8(11):e22508. doi: 10.2196/22508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rajani NF, Bornea M, Barker K. Stacking with Auxiliary Features for Entity Linking in the Medical Domain. BioNLP 2017. 2017;2017:39–47. [Google Scholar]
- 52.Wang C, Fan J. Medical relation extraction with manifold models. 52nd Annual Meeting of the Association for Computational Linguistics, ACL 2014 - Proceedings of the Conference; 2014. pp. 828–38. [Google Scholar]
- 53.Rosenthal S, Barker K, Liang JZ. Leveraging medical literature for section prediction in electronic health records. EMNLP-IJCNLP. 2019;2020:4864–73. [Google Scholar]
- 54.Uzuner Ö, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2011;18:552–6. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poddar A, Dandala B, Devarakonda M. Training Models to Extract Treatment Plans from Clinical Notes Using Contents of Sections with Headings. 2019.
- 56.Liu X, He P, Chen W, et al. Multi-task deep neural networks for natural language understanding. ACL 2019 - 57th Annual Meeting of the Association for Computational Linguistics, Proceedings of the Conference.2020. [Google Scholar]
- 57.Devlin J, Chang MW, Lee K, et al. BERT: Pre-training of deep bidirectional transformers for language understanding. NAACL HLT 2019. 2019.
- 58.Lee J, Yoon W, Kim S, et al. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics Published Online First. 2020. [DOI] [PMC free article] [PubMed]
- 59.Turchin A, Shubina M, Breydo E, et al. Comparison of Information Content of Structured and Narrative Text Data Sources on the Example of Medication Intensification. J Am Med Inform Assoc. 2009;16:362–70. doi: 10.1197/jamia.M2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mahajan D, Liang JJ, Tsou C-H. Extracting Daily Dosage from Medication Instructions in EHRs. An Automated Approach and Lessons Learned. 2020. arXiv:200510899.
- 61.Deerwester S, Dumais S, Landauer T, et al. Indexing by latent semantic analysis. J Am Soc Inf Sci. 1990;41:391–407. [Google Scholar]
- 62.Steinberger J, JeŽek K. Evaluation measures for text summarization. Comput Informatics. 2009;28:251–75. [Google Scholar]
- 63.Pons E, Braun LM, Hunink MG, Kors JA. Natural Language Processing in Radiology: A Systematic Review. Radiology. 2016;279(2):329–343. doi: 10.1148/radiol.16142770. [DOI] [PubMed] [Google Scholar]
- 64.Brown PJ, Marquard JL, Amster B, et al. What do physicians read (and ignore) in electronic progress notes? Appl Clin Inform Published Online First. 2014. [DOI] [PMC free article] [PubMed]
- 65.Holmes C. The problem list beyond meaningful use: part I: the problems with problem lists. J AHIMA. 2011;82(2):30–3. [PubMed] [Google Scholar]
- 66.Johnson A, Pollard T, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3:160035. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]