Abstract
Analysing electrocardiograms (ECGs) is an inexpensive and non-invasive, yet powerful way to diagnose heart disease. ECG studies using Machine Learning to automatically detect abnormal heartbeats so far depend on large, manually annotated datasets. While collecting vast amounts of unlabeled data can be straightforward, the point-by-point annotation of abnormal heartbeats is tedious and expensive. We explore the use of multiple weak supervision sources to learn diagnostic models of abnormal heartbeats via human designed heuristics, without using ground truth labels on individual data points. Our work is among the first to define weak supervision sources directly on time series data. Results show that with as few as six intuitive time series heuristics, we are able to infer high quality probabilistic label estimates for over 100,000 heartbeats with little human effort, and use the estimated labels to train competitive classifiers evaluated on held out test data.
Introduction
Automatic analysis of electrocardiograms (ECGs) promises substantial improvements in critical care. ECGs offer an inexpensive and non-invasive way to diagnose irregularities in heart functioning. Arrhythmias are abnormal heartbeats which alter both the morphology and frequency of ECG waves, and can be detected in an ECG exam. However, identifying and classifying arrhythmias manually is not only error-prone but also cumbersome. Clinicians may have to analyze each heartbeat in an ECG record, and in critical care settings, carefully analysing each heartbeat is nearly impossible. As a consequence, the medical machine learning (ML) community has worked extensively on computational models to automatically detect and characterize arrhythmias1,2.
Rajpurkar et al.3 demonstrated that modern ML models trained on a large and diverse corpus of patients can exceed the performance of certified cardiologists in detecting abnormal heartbeats. But their Convolutional Neural Network model was trained on a manually annotated dataset of more than 64,000 ECG records from over 29,000 patients. Clearly, research on automated arrhythmia detection has moved the burden of monitoring ECG in critical care to annotating and curating large databases on which ML models can be trained and validated. This currently prevailing process involves laborious manual data labeling that is a major bottleneck of supervised medical ML applications in practice. Popular ML techniques, in particular deep learning, require a large supply of reliably annotated training data, containing records from a diverse cohort of patients. According to Moody et al.4 and our own experience, raw medical data is abundant, but its thorough characterization can be involved and expensive. This reliance on labeled data forces researchers to often use static and older datasets, despite evolving patient populations, systematic improvements in understanding of diseases, and advances in medical equipment.
Recent developments in e.g. web-based tools to visualize and annotate ECG signals have not reduced the annotation time and effort significantly. For example, it took 4 doctors, almost 3 months to annotate 15,000 short ECG records using the LabelECG tool5. In general, gold standard expert annotations can be costly. Conservative estimates place the hourly cost of highly qualified labor for the related task of EEG annotation between $50 and $200 per hour6.
In this work, we explore the use of multiple cheaper albeit perhaps noisier supervision sources to learn an arrhythmia detector, without access to ground truth labels of individual samples. We follow the recently proposed data programming (DP)7 framework in which a factor graph is used to model user-designed heuristics to obtain a probabilistic label for each heartbeat instance. DP has gained attention from the medical imaging and general ML community and has been used for various tasks such as automated detection of seizures from electroencephalography8, intracranial hemorrhage detection with computed tomography, or automated triage of Extremity Radiograph Series9.
Our experiments with ECG data from the MIT-BIH Arrhythmia Database indicate that with as few as 6 heuristics, we are able to train an arrhythmia detection model with only a small amount of human effort. The resulting model is competitively accurate when compared to a model trained on the same data with full supply of pointillistic ground truth annotations. It can also outperform another alternative model trained using active learning, a popular technique used to reduce data labeling efforts when they are expensive. We also show that domain heuristics can be automatically tuned to account for inter-patient variability and further boost reliability of the resulting models.
While many different types of arrhythmias exist, for illustration purposes we focus on identifying heartbeats showing Premature Ventricular Contractions (PVCs). Whereas isolated infrequent PVCs are usually benign, frequent PVCs with exceptionally wide QRS complexes† may be indicative of heart disease and eventually lead to sudden cardiac death10. However, our approach is general and applicable to all classes of abnormal heartbeats.
Related Work
Automated Arrhythmia Detection Automatically detecting abnormal heartbeats is a widely studied problem. Most researchers in the past relied on manually labeled corpora such as the MIT-BIH Arrhythmia Database, the AHA Database for Evaluation of Ventricular Detectors, etc., to train and validate their models1,2. Rajpurkar et al.3 recently demonstrated that a deep Convolutional Neural Network (CNN) can even exceed the performance of experienced cardiologists. However, their model was trained on as many as 64,121 thirty-second ECG records from 29,163 patients, manually-labeled by a group of certified cardiographic technicians. Hence, to fuel advances in automated arrhythmia detection and, more generally, in ML-aided healthcare, there is a clear need to affordably label vast amounts of data.
Some recent studies have attempted to address the annotation bottleneck, albeit at a different context and scale. These studies have used semi-supervised or active learning to incrementally improve the accuracy of models without significant expert intervention. For instance, to overcome inter-patient variability without additional manual labeling of patient specific data, Zhai et al.11 iteratively updated the preliminary predictions of their trained CNN using a semi-supervised approach. Correspondingly, Wang et al.12 used active learning on newly acquired data to choose the most informative unlabeled data points and incorporate them in the training set. Sayantan et al.13 used active learning to improve their model’s classification results with the help of an expert. So far, the work which comes closest to addressing the problem of intelligently labeling vast quantities of ECG data is that of Pasolli et al.14. Starting from a small sub-optimal training set, the authors proposed three active learning strategies to choose additional heartbeat instances to further train an Support Vector Machine (SVM) model. Their work demonstrated that models trained using active learning can achieve impressive performance while using few labeled samples. In this work, we also compare the performance of our weakly supervised method with active learning. As against Pasolli et al.14 who used a manually curated set of ECG features and trained an SVM using margin sampling, we train a Convolutional Neural Network (CNN) which can automatically learn rich feature representations using uncertainty sampling, another popular active learning strategy.
Weak Supervision Of late, some studies have explored the use of multiple noisy heuristics to programmatically label data at scale. The recently proposed Data Programming framework7, where experts express their domain knowledge in terms of intuitive yet perhaps noisy labeling functions (LFs), is a prominent example. Recent studies have used DP for a wide range of clinical applications, ranging from detecting aortic valve malformations using cardiac MRI sequences15, seizures using EEG8 and brain hemorrhage using 3D head CT scans9. With the exception of Khattar et al.16, most prior work on DP has been on image9,15 or natural language11 modalities. Moreover, prior DP research either used weak annotations from lab technicians, students8, or heuristics built on auxiliary modalities (e.g., clinician notes, text reports9, patient videos8), some of which only allow for coarse annotation of the entire time series rather than of the individual segments. On the contrary, we define heuristics directly on time series. This enables seamless labeling of entire time series or their segments using the same framework.
Methodology
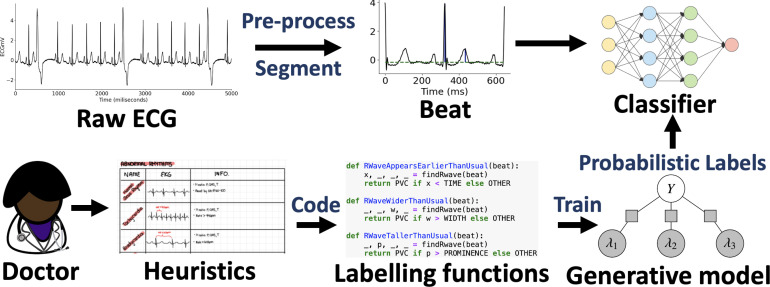
In this section, we will describe how we use domain knowledge to define heuristics to detect PVC in ECG time series. These heuristics will noisily label subsets of data. We will model these noisy labels to obtain an estimate of the unknown true class label for each data point. We then use the estimated labels to train the final classifier, which will be evaluated on held out test data and compared to alternative models trained using ground-truth labels directly. Fig. 1 describes the full workflow we follow to train the end model f.
Figure 1:
Data programming with time series heuristics can affordably train competitive end models for automated ECG adjudication. Instead of labeling each data point by hand (fully supervised setting), experts encode their domain knowledge using noisy labeling functions (LFs). A label model then learns the unobserved empirical accuracy of LFs and uses them to produce probabilistic data label estimates using weighted majority vote.
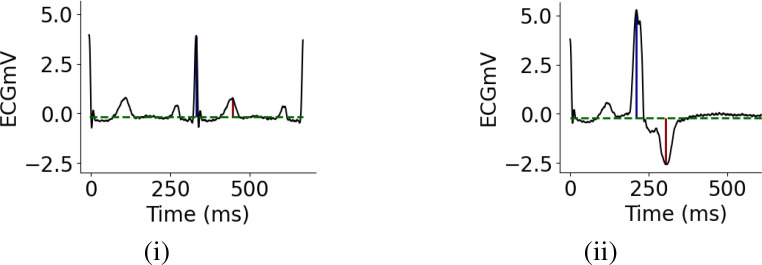
Domain Knowledge to Identify PVC A Premature Ventricular Contraction is a fairly common event when the heartbeat is initiated by an impulse from an ectopic focus which may be located anywhere in the ventricles rather than the sinoatrial node. On the ECG, a PVC beat appears earlier than usual with an abnormally tall and wide QRS-complex, with its ST-T vector directed opposite to the QRS vector, Fig. 2(ii). These generic characteristics allowed one non-domain-expert user to define 6 heuristics in less than 30 minutes. The user was initially unfamiliar with clinical ECG interpretation and referred to an online textbook18 to develop heuristics. Expert clinicians are likely able to define heuristics more rapidly and thoroughly. The heuristics listed below are defined directly on time series. This is in contrast to prior work which uses weak annotations or heuristics defined on an auxiliary modality such as text or images.
Figure 2:
Examples of a normal (i) and PVC (ii) heartbeat. Dotted green horizontal lines represent the ECG baselines detected during pre-processing, blue and red vertical lines mark the QRS-complexes and T-waves.
Heuristics: i. R-wave appears earlier than usual, ii. R-wave is taller than usual, iii. R-wave is wider than usual, iv. QRS-vector is directed opposite to the ST-vector, v. QRS-complex is inverted, vi. Inverted R-wave is taller than usual.
Modeling Labeling Functions over Patient Time Series We will now describe our formal assumptions about the dataset and heuristics, and introduce the modeling procedure. Given an ECG dataset of p patients , where are raw ECG vectors of length T, we can segment each ECG xj into B < T beats such that . Each segment b ∈ {1,..., B} has an unknown class label yb ∈ { — 1,1}, where represents a premature ventricular contraction (PVC). Our goal is to use domain knowledge to model the unknown , without having to annotate the instances individually, to then train an end classifier for automatic detection of PVC. We define m labeling functions (LFs) directly on the time series. These LFs noisily label subsets of beats with corresponding to votes for negative, abstain, or positive. These LFs do not have to be perfect and may conflict on some samples, but must have accuracy better than random19. DP uses this voting behavior to infer true labels by learning the empirical accuracies, propensities and, optionally, dependencies of the LFs via a factor graph. We use a factor graph as introduced in Ratner et al.7 to model the m user defined labeling functions. For simplicity, we assume that the LFs are independent conditioned on the unobserved class label. Let be the concatenated vector of the unobserved class variable for the B beat segments of patient j and be the LF output matrix where is the output of LF k on beat i of patient j. We define a factor for LF accuracy as
We also define a factor of LF propensity as
Then, the label model for a patient j is defined as
(1)
where Zθ is a normalizing constant. We use Snorkel17 to learn θ by minimizing the negative log marginal likelihood given the observed Λj. Finally, as introduced in Ratner et al.7, the end classifier f is trained with a noise aware loss function that uses probabilistic labels .
From Domain Knowledge to an Automated Arrhythmia Detector First, we minimally pre-processed the raw ECG signals by removing baseline wandering using a forward/backward, fourth-order high-pass Butterworth filter20. To segment ECG (xj) into individual beats , we followed a simple segmentation procedure, where we considered the time segment between two alternate QRS-complexes to be a heartbeat.
We had to determine the precise locations of the QRS-complexes and T-waves. As in most prior work, we used the approximate locations of the R-wave available for each ECG record in the database, along with Scipy’s peak finding algorithm21 to find the exact locations of the R and T waves. Further, we used the RANSAC algorithm22 to fit a robust linear regression line to each ECG record, to determine its baseline (horizontal green lines in Fig. 2). The baselines were used to accurately characterize the height and depth of the R and T-waves‡ (blue and red vertical lines in Fig. 2).
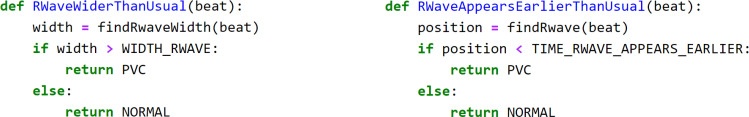
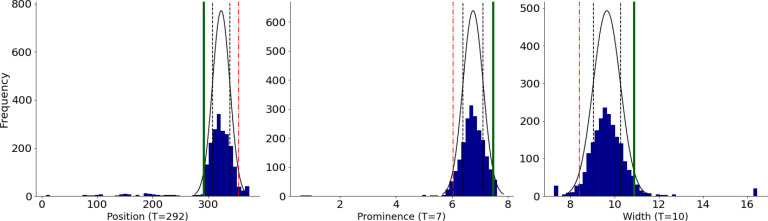
Next, we defined 6 simple LFs based on the domain knowledge to assign probabilistic labels (PVC, OTHER or ABSTAIN) to each beat. Fig. 4 provides example pseudocodes for two of the LFs that were defined. To express the loosely-deined domain knowledge we described previously as LFs, we have to automatically assign thresholds to them. For instance, one heuristic to identify a PVC beat is to check whether its “R-wave appears earlier than usual”. To turn this heuristic into a LF (LFEariy R-wave), one has to determine the “usual” position of the R-wave. For this, we used the Minimum Covariance Determinant algorithm23 to find the covariance of the most-normal subset of the frequency histogram. We then set the threshold to the value 2 standard deviations away from the estimated mean in the direction of interest. For example, for a particular subject (Fig. 3) LFEarly R-wave returns PVC for any beat with the R-wave appearing earlier than TEarly R-wave = 238 ms (vertical green line). To account for inter-patient variability, we automatically compute these subjective thresholds for each heuristic and every patient separately. Note that some of our heuristics did not require estimating any subject-speciic parameters.
Figure 4:
Example Python code for LFWide R-wave and LFEarly R-wave. The findRwaveWidth() and findRwave() sub-routines return the precise width and positions of the R-wave in a beat, while the variables WIDTH_RWAVE and TIME_RWAVE_APPEARS_EARLIER reflect the thresholds TWide R-wave and TEarly R-wave.
Figure 3:
Distribution of the location, height (topological prominence), and width of QRS-complexes of one patient. To turn a loosely defined heuristic such as “R-wave appears earlier than usual” into an LF, we must characterize the “usual” location of the R-wave. To accomplish this, we we fit a robust Gaussian distribution to model the variance of R-wave locations, and assume any beat located two standard deviations earlier (solid green line at TEarly R-wave = 292 ms), than the estimated mean, to be a PVC beat. Since these attributes vary widely from patient-to-patient, we automatically compute these thresholds separately for each patient and heuristic.
The End-Model Classifier With these heuristics, we use the label model in Eq. (1) to obtain probabilistic labels for heartbeats of all training patients in the MIT-BIH Arrhythmia Database. We use these probabilistic labels and the segmented beats to train a noise-aware ResNet classiier, in which we weigh each sample according to the maximum probability that it belongs to either class. Recent studies have shown that ResNet not only performs on par with most of the state-of-the-art time series classiication models24, but also works well for automatic arrhythmia detection1.
Experiments and Results
Data The Massachusetts Institute of Technology - Beth Israel Hospital (MIT-BIH) Arrhythmia Database4 is one of the most commonly used datasets to evaluate automated Arrhythmia detection models. It contains 48 half-hour excerpts of two-lead ECG recordings from 47 subjects. In most records, the first channel is the Modified Limb lead II (MLII), obtained by placing electrodes on the chest. We only used the irst channel to detect PVC events, since the QRS-complex is more prominent in MLII. The second channel is usually lead V1, but may also be V2, V4 or V5 depending on the subject. We refer the interested reader to Moody et al.4 for more details on how the database was curated and originally annotated.
Experimental Setup Our experimental setup follows the evaluation protocol of arrhythmia classification models stipulated by the American Association of Medical Instrumentation (AAMI) as described in25. The AAMI standard, however, does not specify which heartbeats or patients should be used for training classiication models, and which for evaluating them2. Hence, we used the inter-patient heartbeat division protocol proposed by De Chazal et al.26 to partition the MIT-BIH Arrhythmia Database into subsets DS1 and DS2 to make model evaluation realistic. Furthermore, in the MIT-BIH Database, PVCs only account for 8% of the 100, 000 beats, thus to prevent issues stemming from high class imbalance, we randomly oversampled PVC beats in DS1 before using it to train the ResNet classiier. The architecture of our ResNet models is the same as in Fawaz et al.24. To tune the learning rate, batch size, and number of feature maps hyper-parameters, we split the training data into train and validation subsets in the 70/30 proportion. All the models were trained for 25 epochs. In the next subsection, we report the results of the ResNet models which had the best true positive rate (TPR) at low false positive rate (FPR) on the validation data.
We also compare the performance of our weakly supervised model with an active learning (AL) alternative. For AL, we used ResNet to iteratively identify data points for manual labeling using uncertainty sampling27. The model initially had access to a randomly sampled balanced seed set of 100 labeled data points. In each AL iteration, we retrained ResNet using the training data extended with 100 newly labeled data points. We continued this process until the training set consisted of 4,000 points. AL hyper-parameters (the query size and size of the seed set) are similar to Pasolli and Melgani’s setup14. Table 1 reports the performance of the final ResNet model trained on 4000 data points incrementally labeled using AL, averaged over 10 random initializations of the seed set.
Table 1:
Results on held-out test set. Weakly supervised ResNet performs on par with the fully supervised model and outperforms ResNet trained using active learning. FPR50% TPR and FNR50% TNR represent the FPR and FNR at 50% TPR and TNR, respectively. Similarly, TPR1% FPR and TNR1% FNR represent the TPR and TNR at 1% FPR and FNR, respectively. The reported AL results are averaged over 10 independent initializations of the random seed set. All measures are computed with PVC as the positive class.
| Model | TPR | TNR | PPV | FPR | Acc | FPR50% TPR | FNR 50% TNR | TPR 1% FPR | TNR 1% FNR |
|---|---|---|---|---|---|---|---|---|---|
| Fully sup. | 0.884 | 0.970 | 0.664 | 0.030 | 96.25 | 0.005 | 0.028 | 0.793 | 0.266 |
| Pr. labels | 0.645 | 0.960 | 0.523 | 0.039 | 85.84 | 0.019 | 0.140 | 0.165 | 0.252 |
| Active learn. | 0.514 | 0.993 | 0.821 | 0.007 | 94.15 | 0.020 | 0.021 | 0.604 | 0.405 |
| Weak sup. | 0.892 | 0.965 | 0.629 | 0.036 | 97.25 | 0.004 | 0.013 | 0.707 | 0.466 |
Results We trained ResNet models on DS1 as a training set, using either probabilistic labels or the full ground truth, and evaluated them on the held-out set DS2. The results, summarized in Tab. 1, reveal that the end classiier trained using weak supervision is competitive with to the model trained on the full ground truth data. Moreover, our weakly supervised model also outperformed the ResNet trained using 4,000 data points obtained via active learning.
Let us review the key insights stemming from these results. First, the thresholds for the labeling functions that were automatically determined by our proposed auto-thresholding algorithm varied quite drastically across subjects. For instance, the threshold on the position of the R-wave, TEarly R-wave had a mean of 230 ms and a standard deviation of 77.14 ms. This simple personalization of the LF parameters turned out to be the key to good generalization properties of the end-model; it failed to perform well when these parameters were fixed to reasonable global settings. The auto-thresholding algorithm is a practically important contribution of our work, at it allows our methods to scale across diverse cohorts of subjects while mitigating potentially excessive manual effort in tuning LFs to speciic patients. However, unsurprisingly, even with auto-tuning, our LFs and the estimated probabilistic labels (denoted “Pr. labels” in Tab. 1) were not perfect. In fact, we observed high variability in the performance of Pr. labels across different subjects, when compared to ground truth. For example, while they had almost perfect sensitivity for Subject 228 (TPR = 0.994), they performed extremely poorly for Subject 214 (TPR = 0). Overall, Pr. labels had low TPR and high TNR on their own on the training set and held-out test set, which is understandable given the prior class imbalance.
Tab. 1 summarizes performance metrics on the held-out test set (True Positive Rate (TPR), True Negative Rate (TNR), Positive Predictive Value (PPV), False Positive Rate (FPR), and Accuracy (Acc)) measured at the 50-50 class likelihood threshold, chosen for consistency with prior literature on the PVC prediction task. The weakly supervised ResNet (Weak sup.) significantly improves sensitivity to the PVC class compared to just using the LF labels directly (Pr. labels) to directly predict the test data. This illustrates that WS ResNet is able to generalize effectively beyond the hypothesis learned by the noisy weak LFs. Our end model performs on par with the fully supervised ResNet (Fully sup.) trained on the same data but using all the available pointillistic labels in the MIT-BIH Database.
Tab. 1 also compares performance of the four models under consideration at operational settings of pragmatic interest in clinical practice, that is at very low error rates. We report the ability to conidently identify positive cases at FPR of 1%, and the ability to conidently identify negatives at FNR of 1%. We complement these results with the error rates observed at 50% probability of detection of both negative and positive cases. The results show very little operational utility potential from applying the inferred probabilistic labels directly. However, our weakly supervised ResNet model trained on those inferred labels is highly competitive to the equivalently structured ResNet trained on the abundant supply of manually annotated data. Weakly supervised ResNet appears particularly strong at identifying negative cases, while its positive recall performance is close to the ground-truth based equivalent.
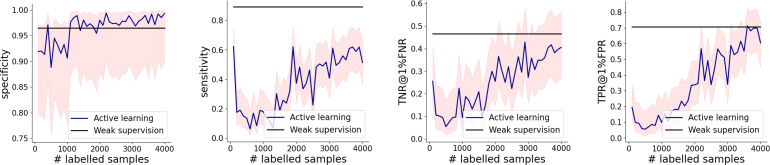
Next, we compare performance of the weakly supervised ResNet versus ResNet trained using active learning (“Active learn.” in Tab. 1), and it looks better on all performance metrics barring TNR, PPV and FPR. Graphs in Fig. 5 show that in the range of up to 4,000 pointillistically labeled training data points, weakly supervised models either outperforms or matches its active learning counterpart, but at a drastically lower requirement of human effort.
Figure 5:
Active learning results. Our weakly supervised model either exceeds or matches the performance of its AL counterpart. The shaded red regions correspond to the 95% Wilson’s score intervals.
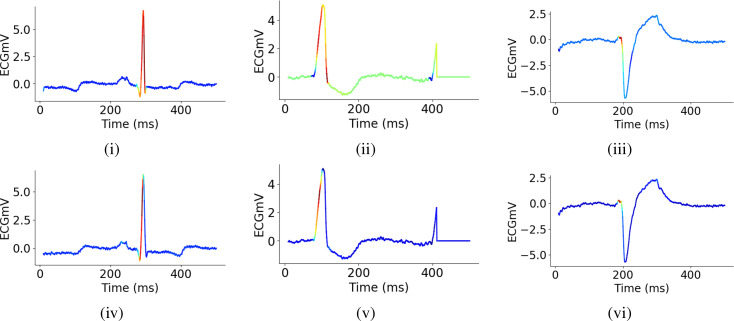
To closely examine what the weakly and fully supervised ResNet models are learning, we plotted Class Activation Maps28 of a normal and PVC beats in Fig. 6. It is evident that to discriminate between PVCs and other beats, our models are primarily paying attention to the QRS complexes in these examples. Moreover, it also appears from these plots that both models not only perform on par, but they also tend to focus on similar signatures of the ECG signals. This observation suggests at least some equivalence between the model trained on ground-truth annotation and the one trained on labels inferred from a small number of simple heuristics. These results reassure us that the more expensive process can be effectively replaced by the proposed framework of weak supervision that uses a few labeling functions based on high-level aspects of domain knowledge derived directly from the time series characteristics.
Figure 6:
Class Activation Maps for weakly [(i) - (iii)] and fully supervised [(iv) - (vi)] ResNet reveal that the models discriminate between PVC and OTHER beats primarily based on the morphology of the QRS-complex. The models appear to have learned to focus on similar regularities. Graphs (i) and (iv) represent an example OTHER beat, while the others show two examples of PVC beats.
Discussion and Conclusion
We demonstrated that weak supervision with domain heuristics defined directly on time series provides a promising avenue for training medical ML models without the need for large, manually annotated datasets. To support this claim, we developed an arrhythmia detection model which performs on par with its fully-supervised counterpart, and does not need point-by-point data annotation. This weakly supervised model has been developed in a fraction of time that would be required to provide a fully labeled training set.
We only needed a handful of heuristics to infer probabilistic labels suficient to yield a reliable end model. These simple heuristics reflected basic clinical intuition that can be gleaned from ECG diagnostics tutorials. We expect that engaging expert clinicians to harvest additional heuristics would allow further improvements. We stipulate that the proposed approach does not only save effort and time, but it also aligns the process of knowledge acquisition from domain experts better with human nature, than its tedious pointillistic data annotation alternative. Further, we show that domain heuristics can be automatically tuned to patient speciic characteristics by deining parameter tuning rules. In our example, auto-tuning of ECG waveform interpretations accounts for inter-patient variability, while keeping manual labor at its minimum.
The ML community has devised several techniques to overcome the limitations of expensive pointillistic labeling such as intelligently choosing the most informative training samples to label27, combining both labeled and unlabeled data29 and harnessing the power of crowds30,31. While semi-supervised learning has been successfully applied to improve arrhythmia detection models without patient-speciic data11, these methods still rely on a signiicant proportion of labeled training data to start with. On the other hand, crowdsourcing has shown promise in generating ground truth for e.g. medical imaging, but prior research30 found several limitations such as the lack of trustworthiness, inability of non-expert workers to annotate ine-grained categories and ethical concerns around patient privacy. Active learning, however, has by far been the most commonly utilized technique in settings where annotating large quantities of data en-masse is prohibitively expensive32.
Multiple avenues of future work include modelling dependencies between LFs to improve both the eficiency and accuracy of label models, and developing a library of time series primitives to streamline development of LFs for such data. We would also like to build interfaces to support interactive discovery of LFs and to rigorously validate resulting end models. Further, we intend to investigate hybrid approaches that will opportunistically combine weak supervision with pointillistic active learning, and conduct user studies with clinicians to better understand the challenges and opportunities for interactive harvesting of domain knowledge. We also aim to enable detection of other types of abnormalities that can be seen in ECG data, and apply our approach to other types of hemodynamic monitoring waveforms.
Time series data is prevalent in healthcare. However the costs of preparing such data for training and validation of new models, as well as for the maintenance of already developed models, prohibit the otherwise realizable beneits from widespread adoption of machine learning in clinical decision support. We believe that approaches similar to the one presented in this paper could help making a decisive push towards proliferating beneicial uses of machine learning in this important ield of its application.
Acknowledgements
This work was partially supported by the Defense Advanced Research Projects Agency award FA8750-17-2-0130, and by the Space Technology Research Institutes grant from National Aeronautics and Space Administration’s Space Technology Research Grants Program.
Footnotes
QRS complexes are generally the most prominent spike seen on a typical ECG. They are a combination of the Q wave, R wave and S wave, which occur in rapid succession, and represent an electrical impulse.
The topological prominence measure returned by Scipy’s peak finding algorithm was imprecise.
References
- [1].Ebrahimi Zahra, Loni Mohammad, Daneshtalab Masoud, Gharehbaghi Arash. A review on deep learning methods for ecg arrhythmia classification. Expert Systems with Applications: X. 2020:page 100033. [Google Scholar]
- [2].Luz Eduardo Joséda S, Schwartz William Robson, Cámara-Chávez Guillermo, Menotti David. Ecg-based heartbeat classification for arrhythmia detection: A survey. Computer methods and programs in biomedicine. 2016;127:144–164. doi: 10.1016/j.cmpb.2015.12.008. [DOI] [PubMed] [Google Scholar]
- [3].Hannun Awni Y, Rajpurkar Pranav, Haghpanahi Masoumeh, Tison Geoffrey H, Bourn Codie, Turakhia Mintu P, Ng Andrew Y. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nature medicine. 2019;25(1):65–69. doi: 10.1038/s41591-018-0268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Moody George B, Mark Roger G. The impact of the mit-bih arrhythmia database. IEEE Engineering in Medicine and Biology Magazine. 2001;20(3):45–50. doi: 10.1109/51.932724. [DOI] [PubMed] [Google Scholar]
- [5].Ding Zijian, Qiu Shan, Guo Yutong, Lin Jianping, Sun Li, Fu Dapeng, Yang Zhen, Li Chengquan, Yu Yang, Meng Long, et al. In Machine Learning and Medical Engineering for Cardiovascular Health and Intravascular Imaging and Computer Assisted Stenting. Springer; 2019. Labelecg: A web-based tool for distributed electrocardiogram annotation; pp. 104–111. [Google Scholar]
- [6].Abend Nicholas S, Wusthoff Courtney J, Goldberg Ethan M, Dlugos Dennis J. Electrographic seizures and status epilepticus in critically ill children and neonates with encephalopathy. The Lancet Neurology. 2013;12(12):1170–1179. doi: 10.1016/S1474-4422(13)70246-1. [DOI] [PubMed] [Google Scholar]
- [7].Ratner Alexander J, De Sa Christopher M, Wu Sen, Selsam Daniel, Re´ Christopher. Data programming: Creating large training sets, quickly. In Advances in neural information processing systems. 2016. pp. 3567–3575. pages . [PMC free article] [PubMed]
- [8].Saab Khaled, Dunnmon Jared, Re´ Christopher, Rubin Daniel, Lee-Messer Christopher. Weak supervision as an efficient approach for automated seizure detection in electroencephalography. npj Digital Medicine. 2020;3(1):1–12. doi: 10.1038/s41746-020-0264-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dunnmon Jared A, Ratner Alexander J, Saab Khaled, Khandwala Nishith, Markert Matthew, Sagreiya Hersh, Goldman Roger, Lee-Messer Christopher, Lungren Matthew P, Rubin Daniel L, et al. Cross-modal data pro-gramming enables rapid medical machine learning. 2020. p. 100019. Patterns, page . [DOI] [PMC free article] [PubMed]
- [10].Moulton Kriegh P, Medcalf Tim, Lazzara Ralph. Premature ventricular complex morphology. a marker for left ventricular structure and function. Circulation. 1990;81(4):1245–1251. doi: 10.1161/01.cir.81.4.1245. [DOI] [PubMed] [Google Scholar]
- [11].Zhai Xiaolong, Zhou Zhanhong, Tin Chung. Semi-supervised learning for ecg classification without patient-specific labeled data. Expert Systems with Applications. 2020;158:113411. [Google Scholar]
- [12].Wang Guijin, Zhang Chenshuang, Liu Yongpan, Yang Huazhong, Fu Dapeng, Wang Haiqing, Zhang Ping. A global and updatable ecg beat classification system based on recurrent neural networks and active learning. Information Sciences. 2019;501:523–542. [Google Scholar]
- [13].Sayantan G, Kien PT, Kadambari KV. Classification of ecg beats using deep belief network and active learning. Medical & Biological Engineering & Computing. 2018;56(10):1887–1898. doi: 10.1007/s11517-018-1815-2. [DOI] [PubMed] [Google Scholar]
- [14].Pasolli Edoardo, Melgani Farid. Active learning methods for electrocardiographic signal classification. IEEE Transactions on Information Technology in Biomedicine. 2010;14(6):1405–1416. doi: 10.1109/TITB.2010.2048922. [DOI] [PubMed] [Google Scholar]
- [15].Fries Jason A, Varma Paroma, Chen Vincent S, Xiao Ke, Tejeda Heliodoro, Saha Priyanka, Dunnmon Jared, Chubb Henry, Maskatia Shiraz, Fiterau Madalina, et al. Weakly supervised classification of aortic valve malformations using unlabeled cardiac mri sequences. Nature communications. 2019;10(1):1–10. doi: 10.1038/s41467-019-11012-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Khattar Saelig, O’Day Hannah, Varma Paroma, Fries Jason, Hicks Jennifer, Delp Scott, Bronte-Stewart Helen, Re Chris. Multi-frame weak supervision to label wearable sensor data. In ICML Time Series Workshop. 2019.
- [17].Ratner Alexander, Bach Stephen H, Ehrenberg Henry, Fries Jason, Wu Sen, Re´ Christopher. In Proceedings of the VLDB Endowment. International Conference on Very Large Data Bases. NIH Public Access; 2017. Snorkel: Rapid training data creation with weak supervision; p. 269. volume 11, page . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].ECG & Echo Learning Clinical ECG Interpretation. 2020. Last accessed: 2021-03-10.
- [19].Boecking Benedikt, Neiswanger Willie, Xing Eric, Dubrawski Artur. Interactive weak supervision: Learning useful heuristics for data labeling. In International Conference on Learning Representations. 2021.
- [20].Lenis Gustavo, Pilia Nicolas, Loewe Axel, Schulze Walther HW, Do¨ssel Olaf. Comparison of baseline wander removal techniques considering the preservation of st changes in the ischemic ecg: a simulation study. Computational and mathematical methods in medicine. 2017;2017 doi: 10.1155/2017/9295029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Virtanen Pauli, Gommers Ralf, Oliphant Travis E, Haberland Matt, Reddy Tyler, Cournapeau David, Burovski Evgeni, Peterson Pearu, Weckesser Warren, Bright Jonathan, van der Walt Ste´fan J, Brett Matthew, Wilson Joshua, Millman K. Jarrod, Mayorov Nikolay, Nelson Andrew R. J, Jones Eric, Kern Robert, Larson Eric, Carey C J, Polat ˙Ilhan, Feng Yu, Moore Eric W, VanderPlas Jake, Laxalde Denis, Perktold Josef, Cimrman Robert, Henriksen Ian, Quintero E. A, Harris Charles R, Archibald Anne M, Ribeiro Antoˆnio H, Pedregosa Fabian, Mulbregt Paul van. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nature Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. , and SciPy 1.0 Contributors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Fischler Martin A, Bolles Robert C. Random sample consensus: a paradigm for model fitting with applications to image analysis and automated cartography. Communications of the ACM. 1981;24(6):381–395. [Google Scholar]
- [23].Rousseeuw Peter J, Driessen Katrien Van. A fast algorithm for the minimum covariance determinant estimator. Technometrics. 1999;41(3):212–223. [Google Scholar]
- [24].Fawaz Hassan Ismail, Forestier Germain, Weber Jonathan, Idoumghar Lhassane, Muller Pierre-Alain. Deep learning for time series classification: a review. Data Mining and Knowledge Discovery. 2019;33(4):917–963. [Google Scholar]
- [25].ANSI AAMI and AAMI EC57. (r) 2008-testing and reporting performance results of cardiac rhythm and st segment measurement algorithms. 2008. American National Standards Institute, Arlington, VA, USA,
- [26].Chazal Philip De, O’Dwyer Maria, Reilly Richard B. Automatic classification of heartbeats using ecg morphology and heartbeat interval features. IEEE transactions on biomedical engineering. 2004;51(7):1196–1206. doi: 10.1109/TBME.2004.827359. [DOI] [PubMed] [Google Scholar]
- [27].Settles Burr. Active learning. Synthesis lectures on AI and ML. 2012;6(1):1–114. [Google Scholar]
- [28].Zhou Bolei, Khosla Aditya, Lapedriza Agata, Oliva Aude, Torralba Antonio. Learning deep features for discriminative localization. In Proceedings of the IEEE conference on computer vision and pattern recognition. 2016. pp. 2921–2929. , pages .
- [29].Van Engelen Jesper E, Hoos Holger H. A survey on semi-supervised learning. Machine Learning. 2020;109(2):373–440. [Google Scholar]
- [30].Rodr´ıguez Antonio Foncubierta, Mu¨ller Henning. Ground truth generation in medical imaging: a crowdsourcing-based iterative approach. In Proceedings of the ACM multimedia 2012 workshop on Crowd-sourcing for multimedia. 2012. pp. 9–14. , pages .
- [31].Ørting Silas Nyboe, Doyle Andrew, Hilten Arno van, Hirth Matthias, Inel Oana, Madan Christopher R, Mavridis Panagiotis, Spiers Helen, Cheplygina Veronika. A survey of crowdsourcing in medical image analysis. Human Computation. 2020;7:1–26. [Google Scholar]
- [32].Wang Donghan, Fiterau Madalina, Dubrawski Artur, Hravnak Marilyn, Clermont Gilles, Pinsky Michael. 797: Interpretable active learning in support of clinical data annotation. Critical Care Medicine. 2014;42(12):A1552. [Google Scholar]