Abstract
Findings from randomized controlled trials (RCTs) of behaviour change interventions encode much of our knowledge on intervention efficacy under defined conditions. Predicting outcomes of novel interventions in novel conditions can be challenging, as can predicting differences in outcomes between different interventions or different conditions. To predict outcomes from RCTs, we propose a generic framework of combining the information from two sources - i) the instances (comprised of surrounding text and their numeric values) of relevant attributes, namely the intervention, setting and population characteristics of a study, and ii) abstract representation of the categories of these attributes themselves. We demonstrate that this way of encoding both the information about an attribute and its value when used as an embedding layer within a standard deep sequence modeling setup improves the outcome prediction effectiveness.
1. Introduction
Randomized controlled trials (RCTs) act as a key source of information about intervention outcomes. An RCT in behavioural science usually captures information on the demographic characteristics of each cohort group in the study, the interventions at a broad level defining a general configuration of each cohort, and a configuration for measuring the outcome values, i.e., some measure of the success for each cohort. A predictive model learned from a set of existing literature could potentially find applications in predicting what is likely to happen for new combinations of cohort groups characteristics, interventions, and outcome measurement settings, which could then provide useful insights to facilitate the process of systematic reviews1,2. In addition to help compiling systematic reviews, a predictive model may potentially be useful for policy makers to help prescribe a set of behavioural policies that are likely to be helpful to trigger a behaviour change for societal benefits of a target group of people with a given set of characteristics.
Our Contributions. The objective of the paper is to investigate how effectively can the outcome of a behaviour change RCT be modeled in terms of its characteristics comprising mainly the population settings (the whom), interventions (the what) and outcome measurement criteria (the how). We emphasize that the novelty of the paper is not to develop a new neural end-end architecture for the RCT outcome prediction task, for which we employ an end-to-end neural architecture comprising of bidirectional LSTMs3-5, a model that has been met with considerable success in sequence problems such as those of Natural Language Processing.
The novelty of our work rather lies in enriching the embedded input vectors of an end-to-end neural model with additional useful information, the source of which, in our problem, is the ontology of behaviour change attributes, which in addition to the text features proves effective in improving the down-stream task of modeling the outcome of an RCT. In particular, our predictive model relies on a novel approach of leveraging information from two sources, namely the annotated text and a document-level co-occurrence relationship between the entities in a behavioural science ontology.
2. Related Work
The work related to text mining for RCTs spans domains from Natural Language Processing to medical informatics. Much of this literature begins with information extraction6, which can then be used for summarization7, automating (parts of) systematic reviews8, or prediction. Early work on extraction from RCTs looked at elements from PICO9 and initially classified sentences according to that framework10-12.a Instead, a corpus was annotated with PICO entities to test entity extraction13. The study14 similarly extract entities but use a much larger inventory of entities taken from the Behaviour Change Intervention Ontologyb. This and other work has been undertaken to help in automating systematic reviews of RCTs1,2,8,15, which rely on accurate information extraction.
Not much research has yet been carried out on predictive tasks on RCTs. Related to our use case of behaviour change and smoking cessation, the articles16,17 showed the feasibility of regression approaches to predict the percentage of quitters but this work does not extend to the number of papers and entities that we cover. Probably the closest work to ours is from18. Like our second task of pairwise classification, they look to infer the findings of an RCT based on its intervention, comparator, and outcome entities.
Among existing work that combines text and graph embeddings, joint embeddings of text and relations was employed for link prediction in a knowledge base19. More similar to our embeddings, the authors of20 learn embeddings over a co-occurrence graph of entities and compare them to word embeddings, but they do not explore how those can be combined. A graph-based framework was proposed in21 to incorporate non-local co-occurrences in modeling the semantics between words. While their objective was to improve the effectiveness of word embedding with the help of additional relationships between terms, the objective of our work is to model the relationships between the features in our data with the help of an ontology.
A popular approach towards predictive tasks, such as relation prediction on entities is to use graph convolutional networks22. However, a graph convolution network is suitable for scenarios when an individual instance is modeled as a graph23. In our case, the relations are defined at the level of features and not at the level of each RCT instance. Hence, we leverage the relational information between the features only during the pre-training phase24, so as to generate an enriched set of input vectors to help improve an end-end neural model.
3. Problem Formulation
Ontology Overview. For this study we use the Behaviour Change Intervention Ontology (BCIO)c comprised of hundreds of entities at multiple levels of classification25. Lower-level entities, being more granular, define the features used in our study. Different from25, this paper is not concerned with automated extraction of the values of these entities from the papers, but rather assuming that such values have been extracted, to predict the outcome behaviour values and estimated effects given these values.
An RCT as a set of entity-value pairs. A randomized control trial (RCT) study on behaviour science in our dataset usually contains multiple study arms. Each study arm forms an instance for classification and is associated with an outcome value. We represent each document as a set of entity-value pairs. More specifically, an input document d is a set of 2-tuples of the form (a, xa), where a is one of the entities from the BCIO and xa is the value associated with the entity, i.e., d = {(a, xa) : a ∈ A, xa ≠ ∅}. The cardinality of the set d is the number of different entities for which there exists an annotated value. In addition to the semantic type of an entity, each attribute is also associated with a value-type which is also a part of the ontology, and is one of categorical, numerical, or text.
The value of an entity a (i.e. xa), corresponding to an RCT arm, is annotated by a human expert by highlighting the span of text. In our predictive approach, we consider the string (text span) corresponding to each entity value, generally speaking, as a multi-set (bag) of words. The values of each entity are encoded differently depending on the detected type of their annotated span (text, numeric or categorical). For example, each word of a text value is converted to its embedded representation, whereas a numerical token or a categorical value is appended as an additional dimension to a dense vector input (we will revisit this later in the section on outcome value prediction).
Discretizing RCT outcomes. We assume that in each arm, the relationship between the outcome value and the set of features is given by a function of the form y(d) = ϕ(d), where y(d) ∈ [0, 100] denotes the percentage outcome value. For the sake of readability, from hereon we refer to a study arm of an RCT as a document (denoted by d) in a collection of such arms (denoted as ).
The two types of prediction tasks we address correspond to those of a) predicting a discrete interval or range of outcome values, and b) predicting the relative comparison between two studies. Both these tasks require transforming the real valued y(d)’s into discrete ones. First, we split the range of y(d), i.e., [0, 100] into a number of intervals. We set this number to 7 to achieve a reasonable degree of discriminability26.
We use discrete ranges for the outcome value prediction to simplify the interpretation of the results, e.g., low, moderately low etc. In practice, prediction of a continuous outcome value should be accompanied by a confidence interval, which can be difficult to interpret or even unreliable27. Instead of attempting to predict a single exact value, we fix the intervals and try to figure out a relative notion of the likelihood of a low or a high outcome18. In our experiments, we also report linear regression results in our experiments, i.e. where the outcome value is directly predicted as a continuous variable.
The start and end-points for each interval constituting a partition of [0, 100], is determined from the distribution of the outcome values in the training set (i.e. the values corresponding to input instances that are known to a model), i.e.,
| (1) |
Setting k=7 in Equation 1 partitions the range of y(d)’s into 7 intervals, where the start of the ith interval is specified by ith 100/7 ≈ 14 percentile computed over the distribution of the y(d) values (the percentile points indicating the cut-off points in the cumulative distribution function in Equation 1). Partitioning the range of the outcome values this way seeks to achieve a uniform binning of the values and mitigate effects of any class priors for the classification task. Each is converted to a class label pointing to the index of the interval in which y(d) falls, the intervals being defined as per Equation 1.
Use-case for Point-wise and Pair-wise Models. We now describe how the point-wise and the pair-wise models, trained on the input-output associations of existing RCT studies, could potentially be used in practice by an RCT practitioner. In both the point-wise and the pair-wise case, an RCT practitioner would want to know what is likely to happen for a new combination of whom, what, and how features. These features may be entered into a prediction system in the form of attribute value pairs. In the point-wise case, the user would want to obtain a predicted outcome value percentage range on the target population and interventions specified under certain settings independent of a reference point. Instead of intending to obtain a predicted range, for the pair-wise case, an RCT practitioner would want to know if the new combination of input (test) features is likely to increase the success percentage in comparison to a reference study that already exists in the literature.
4. Outcome Value Prediction
4.1. Model Overview
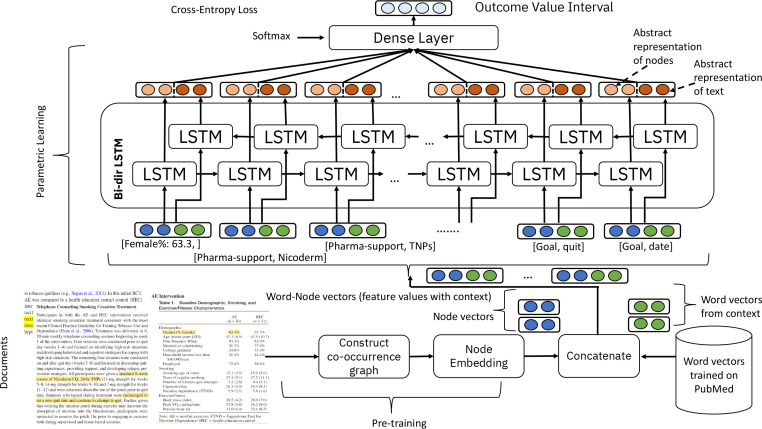
Figure 1 shows a standard deep sequence classification model, comprised of stacked layers of LSTMs the hidden states of which lead to a softmax layer of 7 dimensions (corresponding to one of the 7 classes into which the outcome value is classified as per Equation 1). This so-called bi-LSTM is a standard neural network architecture that has been met with success in domains that feature an input sequence such as time series or sentences in Natural Language Processing28. The novelty lies in defining the scope of the input to this predictive model. More concretely, the input is a set of attribute-value pairs of the form as annotated in document d. The raw input is transformed into a dense vector comprising a pretrained global information about the attribute itself and its text value. Additionally, if the value-type of an attribute is numerical or categorical, its value (a real number or an integer representing the category value) is appended as an additional dimension to the concatenated vector representation comprised of a) the attribute relation and b) the text information. Formally, we denote the transformation from a set of attribute-value pairs to that of dense vectors as
| (2) |
Figure 1:
Neural architectural overview of the proposed outcome range classification model, where the input embedded vectors are from two different modalities, namely the text and the PIQ feature correlations.
where each vector corresponds to two distinct subspaces of sizes kf and kt (and an additional one for the numerical value). The first subspace (of dimension kf ) corresponds to the relations between the entities, whereas the second one (of dimension kt) corresponds to word semantics. The attribute-value set for each arm of a document, d, is transformed to a set of vectors corresponding to these entities, i.e., . The network of Figure 1 is then trained with such sequences of dense vectors xd for each document .
Examples of Input Transformation. To illustrate how the attribute-value pairs annotated in a RCT are transformed into inputs to the network of Figure 1, let us look at the following example annotations from a sample paper on smoking cessation studies from our dataset (the example annotations are also shown as the text highlighted in a document at the bottom-left of Figure 1).
Example 1: For an annotated value (text-span) of ‘(% female) 63.3%’ for the ‘gender’ attribute, after tokenizing the string into ‘female’ and ‘63.3’, we obtain the pretrained vector representation of the word ‘female’. We concatenate the word vector ‘’ with the node vector representation of the attribute ‘’, and append the number ‘63.3’ as an additional dimension.
Example 2: For the sample I attribute with value ‘encouraged to set a new quit date’, either the average is computed over vectors (specifically, pre-trained skipgram on PubMed) for the constituent words ‘’, ‘’ etc., or the context vector (specifically with Bio-BERT) is obtained for the entire piece of text. This context vector is then used substituted into the kt dimensional subspace of Equation 2. The other part (the kf dimensional subspace) is substituted with the node vector for the attribute ‘’. The additional dimension for the numerical value in this example is 0 (since no number exists in the annotated text instance).
4.2. Textual Context Vector Representations
In this study we investigate two different ways of obtaining the vector representation of the textual context around an instance of an attribute occurrence. These two methods correspond to exploring different granularity for embedding text, one at the level of words29,30 and other at the level of sequences of words31,32. Both these approaches are trained on large volumes of unannotated text. While word2vec29 learns a set of linear transformation parameters for each word to predict its context, BERT32 captures term semantics with the help of a transformer architecture33 trained by arbitrarily masking words from text segments.
In our work, we specifically use pretrained word2vec (skipgram) vectors trained on PubMed abstracts. These pretrained vectors are of 200 dimensionsd, i.e. kt = 200 in Equation 2. We used zero vectors used for out-of-vocabulary words (8.9% of our dataset). As the context vector, we used the pre-trained Bio-BERT model34. The vocabulary of the Bio-BERT is initialized from the larger BERT model of32 and then fine-tuned on PubMed abstracts34. The dimensionality of the feature vectors for the Bio-BERT model is 768, i.e., kt = 768 in Equation 2.
Embedding of context text is potentially useful to semantically associate/dissociate instances of different/same feature types (e.g., to discover that while two different interventions can be semantically related, the values for the two instances of the same intervention attribute may in fact be semantically different from one another). Next, we describe how we obtain the vector representations of the entities.
4.3. Learning Node Representations
Motivation. One of the limitations of modeling the outcome value as a (predicted) function value of the set of input feature values (comprising numerical, categorical and text features) is that the predictions are likely to be less effective for a sparse feature space. In the context of our problem, sparsity of the feature values is caused due to a wide range of different population characteristics, or interventions used in the studies, e.g., some studies report the average age of a cohort, while others use median age. Moreover, a predictive model assumes that the features are independent. However, in the context of our domain of behavioural science reports, correlations do exist between the entities. For example, if some interventions are likely to work well on a cohort of young people (with lower values for minimum age), they are also likely to work well on cohorts with lower mean age. As another example, sets of interventions are also correlated with each other, e.g., intuitively speaking, ‘psychological counseling’ often works well with ‘continuous monitoring’.
Embedding nodes as vectors has been reported to improve downstream prediction tasks for the biomedical domain, such as modeling interactions between genes, diseases and drugs35,36. In our case, via embedding nodes as vectors we intend to model the correlations between features of different types. For the purpose of graph construction we group the BCIO entities into broader types for who (P) what (I) and how (Q).
Graph Construction. The first step in our proposed approach is to construct an undirected graph G = (V, E) intending to capture the co-occurrences between different feature instances. Each node in this graph is represented by a tuple v(t, a, xa), a being an attribute of type . Formally,
| (3) |
where and the node set, V, is thus comprised of nodes of unique types with unique values. While constructing a node corresponding to an attribute, only its categorical or numerical value is included as a part of the node. We exclude the string (text span) of the annotation for an attribute because including it would make the graph too fine-grained (e.g., one node for each possible value of an intervention ‘goal setting’). This would lead to sparse edge relations between its nodes, which in turn would not be conducive for modeling the inter-attribute relations. Note that the text information is eventually used in the downstream prediction task because it constitutes a separate subspace of the input vectors (Equation 2). Next, we define the edges in G as follows. Formally, an edge exists between a pair of nodes corresponding to the values of entities of type t and of type , if these values are observed in the same RCT arm (document). To model the likelihood of a correlation between pairs of attribute-values, we set the weight of an edge e, w(e), to reflect the relative number of times such associations between the feature values are observed across a number of different documents in the collection.
Node Embedding. After constructing the graph G = (V, E) from a given collection of documents , the next step in our proposed method is to obtain a dense vector representation for each node of G. Specifically, we applied the random walk based node2vec37 algorithm to learn the vector representation of each node. The choice of visiting a next node in node2vec is controlled by two parameters, namely a) the (inverse) return parameter, p, which if set to a low value makes it more likely for the walk to return to t, and b) the in-out parameter, q, which if set to a high value makes the walk unlikely to visit nodes that are not adjacent to t. A low value of p and a high value of q is thus likely to make the walk more compact. Specifically, for our experiments, we tie the two parameters by setting q = 1 − p.
In our case, after applying node2vec on the weighted graph of Equation 3, attribute-value combinations that are likely to be correlated to each other will be embedded close to each other, because these nodes are likely to be more reachable from each other with a random walk using the edge weights as probabilities.
5. Modeling RCT Comparisons
We now extend the point-wise prediction framework to learn a comparison function between a pair of RCTs. A practical use-case for this pairwise situation arises when an RCT practitioner wants to compare a new combination of population, intervention and outcome settings with reference to an existing study, which we call the reference study involving a target population. The intention of the predictive model in this case is predict if a new combination of interventions is likely to increase the success ratio in comparison to another study, which is different from predicting if the relative comparisons between two arms of the same study yield significant differences18.
For pairwise modeling of RCTs, the input is a pair of RCTs. The attribute-value pairs of each RCT is transformed to a variable length sequence of embedded representations of concatenated node and word vectors identical to the input transformation of Equation 2 (Figure 1). The pairwise prediction model employs a Siamese type architecture38, where we feed in a pair of RCTs as input. The training phase makes use of the annotated attribute-value pairs of existing RCTs reported in the literature. The encoded representation of the LSTM layer for both studies is then concatenated before applying a sigmoid layer. During training, the ground-truth label between a pair of RCTs is 1 if the outcome value of the first element of the pair is less than that of the second, or 0 otherwise. The network is trained with all combinations of pairs of the form (d1, d2) from the training set. A pair (d1, d2) is used in the training set in a unique ordering, i.e., inclusion of (d1, d2) excludes (d2, d1), which means that the total number of pairs used for training is . In the testing set, one of the elements of each pair is a new combination of PIQ features, unseen in the training set, whereas the other is from the training set (i.e. a previously seen reference study).
6. Evaluation
Dataset. For our experiments, we focused on the domain of the smoking cessation behaviour change RCTs.39 reports the compilation of such a dataset (called HBCP) of behaviour change RCTs focused on smoking cessation; however, their dataset is mainly targeted towards addressing information extraction from RCTs. Since we focus on a different task, that of predicting outcomes of RCTs, we construct an extended version of the HBCP dataset for our experiments. Different to39, for our classification task the RCT instances are constructed as described in Equation 2. Each study includes a number of study arms corresponding to a fundamental unit of a study, i.e., a particular population group with certain characteristics and a set of interventions applied on the group. Outcome values are reported separately for each arm and a single RCT can have multiple arms.
Our extended dataset comprises a set of 513 RCTs (PDF documents) on behaviour change for smoking cessation. The annotation schema of our dataset follows the ontology and the guidelines defined in39. A team of in-house domain experts annotated a total of 7451 attributes of different types from the set of 513 PDF documents. Table 1 presents an overview of our dataset.
Table 1:
Dataset Characteristics
| #Docs | 513 |
| #Arms | 1064 |
| #P (whom) attributes | 4808 |
| #I (what) attributes | 5129 |
| #Q (how) attributes | 2554 |
| Mean y(d) (Outcome %) | 16.8 |
| Median y(d) (Outcome %) | 13.9 |
Setup. To assess the effectiveness of the graph-based approach for the point-wise and the pairwise prediction tasks, we compare our proposed approach of joint embedded input representation (text and attribute-value nodes) with two ablation baselinese. As the first ablation baseline, we employ a standard one-hot encoding of each attribute node coupled only with its numeric value (i.e., kf being number of unique attributes, kt = 0 in Equation 2), which is equivalent to standard linear regression and multi-class classification (for the continuous or the interval prediction). Note that this baseline neither uses information from the text around the context of the attribute instances, nor does it use an embedded representation of the attributes themselves. We name this baseline ‘Values-only’.
The second ablation baseline, Text-Only, does not use any information from the attribute-value pair co-occurrences, i.e., we feed in as input vectors to the network of Figure 1 (and its pairwise equivalent) an aggregation (average) of word vectors, each of dimension kt (see Equation 2), from the annotated text spans. For a numeric value, e.g., mean age of a population, we feed in its value as an additional dimension in the input vector along with the word vector representation of its context. We used two different ways of obtaining the feature vectors for the text, namely a) Skipgram, where we used pre-trained skipgram vectors of dimension 200 (i.e. for this baseline kt = 200), and b) Bio-BERT, where we used the pre-trained Bio-BERT modelf to obtain kt = 768 dimensional representation of the context text.
The third ablation baseline, named Text+N2V-1Hot, employs a one-hot encoding of the attribute-values nodes (Equation 3). This baseline treats each graph node as independent ignoring the co-occurrence relations between the edges. The two different text embedding approaches lead to two different settings for the one-hot experiments with different feature dimensions (for the text part).
To test the approaches, we employ 5-fold cross-validation. The intervals to induce the class labels are computed on each training fold instance (Equation 1). For pairwise classification, training proceeds with pairs from the training fold. The test instances are constructed by pairing up each RCT from the test fold with each from the train fold, the objective being to predict if a new study, for which the outcome is not known, is likely to yield a higher or a lower outcome compared with an existing one.
Results Summary. Table 2 summarizes the best results obtained with each method for the point-wise and the pairwise tasks. We observe that a value-only based approach (similar to a simple linear regression or a multi-class classification) produces not too effective results. We observe from the bottom part of Table 2 (‘Text+N2V’) that leveraging information from the co-occurrence likelihoods between the behaviour science attributes in the form of embedded node representations improves significantly (t-test with 95% confidence) the effectiveness of both the point-wise and the pairwise tasks in comparison to the corresponding text-only approaches (e.g. compare the ‘Skipgram Text-Only’ results with ‘Skipgram Text+N2V’ ones). Moreover, the results also improve in comparison to the approach when the node attribute features are treated as independent one-hot vectors (e.g. compare ‘Bio-BERT Text+N2V’ results with ‘Bio-BERT Text+N2V-1Hot’ ones). The pairwise case yields slightly better results when skipgram vectors are combined with the node embeddings.
Table 2:
Summary of the best results with 5-fold cross validation for the point-wise (7-class outcome classification and regression) and the pairwise tasks. The first five are ablation baselines.
| Embedding Method | Point-wise | Pairwise | ||
|---|---|---|---|---|
| Accuracy | RMSE | Accuracy | ||
| None | Values-Only | 0.5532 | 15.05 | 0.6237 |
| Skipgram | Text-Only | 0.6344 | 10.11 | 0.7350 |
| Skipgram | Text+N2V-1Hot | 0.6456 | 7.47 | 0.7282 |
| Bio-BERT | Text-Only | 0.6745 | 7.33 | 0.7479 |
| Bio-BERT | Text+N2V-1Hot | 0.6946 | 7.52 | 0.7429 |
| Skipgram | Text+N2V | 0.6658 | 8.04 | 0.7585 |
| Bio-BERT | Text+N2V | 0.7072 | 7.06 | 0.7553 |
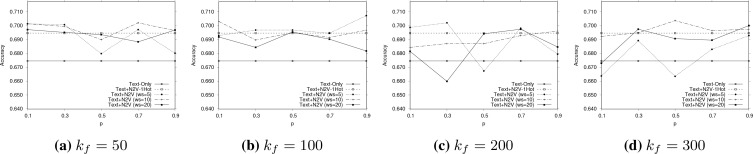
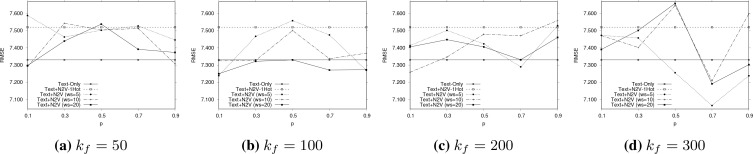
Parameter Sensitivity. In addition to presenting the best results for each method in Table 2, we now investigate the effects of varying the parameters of node2vec for obtaining the embedded vectors that are concatenated as inputs to the architecture of Figure 1, i.e., parameters - the context size (ws), dimension of embedding (kf ) and the return/in-out node2vec parameters (p, q). We explore the parameter space only for the most effective combination method of Table 2, i.e. the ‘Text+N2V’ with the Bio-BERT embedding.
Figures 2 and 3 report parameter sensitivity for the multi-class classification and regression tasks, respectively. From Figure 2, we observe that smaller values of p (and thereby larger values of q = 1 - p) usually result in better outcome value prediction. As per37, small values of p (and large values of q) are likely to yield locally compact walks. In the context of our problem, this means too much exploration on the co-occurrence graph may introduce noise in the form of false long-chain dependencies across entity values of different types.
Figure 2:
Parameter sensitivity effects of Text+N2V (Bio-BERT) for point-wise outcome value classification for different context sizes. It can be observed augmenting pre-trained feature relationship information as a part of the input produces substantially better results in comparison to the Text-Only and the Text+N2V-1Hot approaches (shown as the two constant lines).
Figure 3:
Parameter sensitivity measured in terms of RMSE (lower the better) of Text+N2V (with Bio-BERT) for point-wise outcome value regression for different context sizes. A comparison with Figure 2 shows that regression results are more sensitive to parameter variation effects.
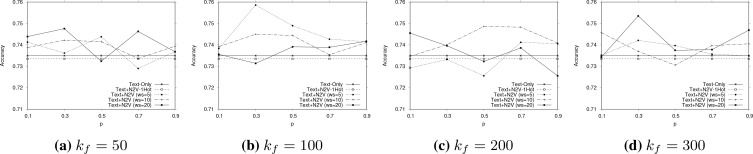
For the pairwise case, we explore the parameter space for the combination of node vectors with skipgram vectors (since this configuration produced better results than the Bio-BERT ones). Figure 4 shows trends that are similar in nature to that of Figure 2, i.e., the optimal results are obtained for smaller values of p.
Figure 4:
Parameter sensitivity effects of Text+N2V (with skipgram PubMed vectors) for pair-wise outcome value comparisons. Similar trends as those in Figure 2 are observed.
6.1. Prediction with Uncertainties
In this section, we investigate the feasibility of a more pragmatic approach where only a small subset of the documents in a collection is annotated with the attribute-value information. This scenario also tests how effectively can a prediction system, trained on a subset of the collection (called the seed set), may subsequently be used to make predictions for newly created research articles on behaviour change (i.e., those for which no manual annotations are available). For each unannotated documents, we employed the unsupervised information extraction method39-41 to automatically extract a set of attribute-value pairs, given its text as an input to the extractor. The prediction system is then trained on a mixture of both manually annotated (hence, clean) and automatically extracted (hence, uncertain) data.
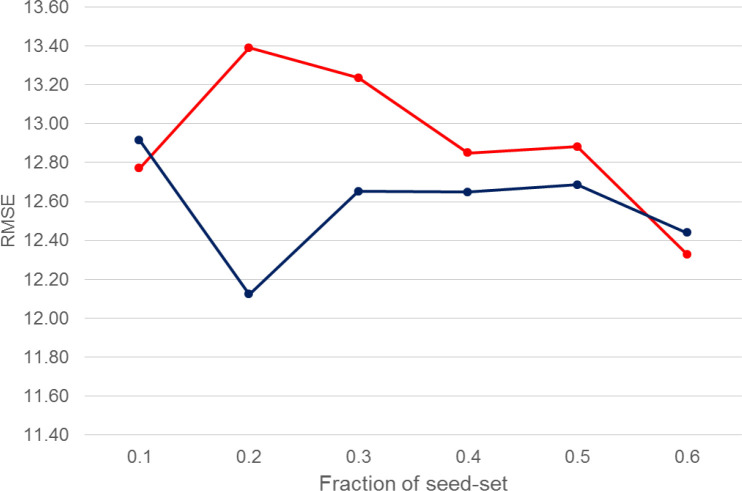
To conduct experiments for predictions with uncertainties, from our static collection of annotated documents, we first use only a fraction of the data as the seed set (signal), and then employ the extractor to automatically infer the attribute values from the remaining set (noise). Figure 5 shows how does the effectiveness of our prediction model (Text + N2V) is affected by the use of automatically extracted values (the RMSE values are averaged over 5-fold CV test splits). The red line plots the RMSE values obtained only with the seed data, whereas the blue line, for each fraction of the seed data, displays the results obtained by augmenting the seed data with extracted information from the remaining fraction of the data.
Figure 5:
Sensitivity of outcome predictions for the regression setting of Bio-BERT (Text + N2V) relative to the proportion of the seed-set. Red: Seed data only, Blue: Seed+Extracted data.
It is seen that too small or too large a seed set (i.e. ≤ 10% or ≥ 60%), the use of additional uncertain data, in the form of automatically extracted attribute values, is not able to outperform the results obtained with clean data only. However, it is seen that using about 20% of clean data, coupled with 80% additional (potentially noisy) data improves the overall outcome value prediction effectiveness. This implies that knowledge gained from new RCTs in the form of extracted attribute-value pairs can potentially be injected into our prediction system for improving its effectiveness.
7. Conclusions
We investigated how effectively can we automatically predict outcomes from RCTs on behaviour change studies. The novelty lies in encoding an RCT instance as a combined representation of the embedded textual context of annotated values coupled with the embedded representation of the relations between attribute-value instances. Our experiments demonstrate that this way of modeling the inputs outperforms the cases which make an oversimplifying assumption that such attribute-value instances are independent. A broader impact of our work is that it shows that the outcome value of a behaviour change study can be predicted within satisfactory levels of accuracy, which implies that AI systems can potentially be used by policy-makers in implementing a set of behaviour change policies (interventions) on a target population.
In future, we would like to investigate outcome prediction for RCTs with automatically extracted attribute values from documents.
Footnotes
The study11 actually uses PIBOSO, which is an extension of PICO.
Implementation of the point-wise and pairwise models, along with the dataset would be made publicly available.
References
- [1].Bashir R, Surian D, Dunn A. Time-to-update of systematic reviews relative to the availability of new evidence. Systematic Reviews. 2018;12:7. doi: 10.1186/s13643-018-0856-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Tsafnat G, Dunn A, Glasziou P, Coiera E. The automation of systematic reviews. BMJ (Clinical research ed) 2013;01:346. doi: 10.1136/bmj.f139. f139. [DOI] [PubMed] [Google Scholar]
- [3].Zhou P, Shi W, Tian J, Qi Z, Li B, Hao H. Attention-Based Bidirectional Long Short-Term Memory Networks for Relation Classification. Proc. of ACL’16. 2016:207–212. [Google Scholar]
- [4].Xu J, Chen D, Qiu X, Huang X. Cached Long Short-Term Memory Neural Networks for Document-Level Sentiment Classification. Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing. 2016:1660–1669. [Google Scholar]
- [5].Zhou X, Wan X, Xiao J. Attention-based LSTM Network for Cross-Lingual Sentiment Classification. Proc. of EMNLP’16. 2016:247–256. [Google Scholar]
- [6].Kiritchenko S, de Bruijn B, Carini S, Martin J, Sim I. ExaCT: Automatic extraction of clinical trial characteristics from journal publications. BMC medical informatics and decision making. 2010 09;10:56. doi: 10.1186/1472-6947-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sarker A, Mollaá D, Paris C. Query-oriented evidence extraction to support evidence-based medicine practice. Journal of Biomedical Informatics. 2016;59:169–184. doi: 10.1016/j.jbi.2015.11.010. [DOI] [PubMed] [Google Scholar]
- [8].Marshall IJ, Wallace BC. Toward systematic review automation: a practical guide to using machine learning tools in research synthesis. Systematic reviews. 2019;8(1):163. doi: 10.1186/s13643-019-1074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Huang X, Lin J, Demner-Fushman D. Evaluation of PICO as a Knowledge Representation for Clinical Questions. AMIA Annu Symp Proc. 2006;02:359–363. [PMC free article] [PubMed] [Google Scholar]
- [10].Kim SN, Martinez D, Cavedon L, Yencken L. Automatic classification of sentences to support Evidence Based Medicine. BMC Bioinformatics. 2011 Mar;12(2):S5. doi: 10.1186/1471-2105-12-S2-S5. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hassanzadeh H, Groza T, Hunter J. Identifying scientific artefacts in biomedical literature: The Evidence Based Medicine use case. Journal of Biomedical Informatics. 2014;49:159–170. doi: 10.1016/j.jbi.2014.02.006. [DOI] [PubMed] [Google Scholar]
- [12].Wallace BC, Kuiper J, Sharma A, Zhu M, Marshall IJ. Extracting PICO Sentences from Clinical Trial Reports Using Supervised Distant Supervision. J Mach Learn Res. 2016 Jan;17(1):4572–4596. [PMC free article] [PubMed] [Google Scholar]
- [13].Nye B, Li JJ, Patel R, Yang Y, Marshall I, Nenkova A. A Corpus with Multi-Level Annotations of Patients, Interventions and Outcomes to Support Language Processing for Medical Literature. Proc. of ACL’18. 2018:197–207. [PMC free article] [PubMed] [Google Scholar]
- [14].Ganguly D, Hou Y, Deleris LA, Bonin F. Information Extraction of Behavior Change Intervention Descriptions. Proceedings of AMIA Joint Summits on Translational Science. 2019:182–191. [PMC free article] [PubMed] [Google Scholar]
- [15].Wallace BC. What Does the Evidence Say? Models to Help Make Sense of the Biomedical Literature. Proc. of IJCAI’19. 2019:6416–6420. doi: 10.24963/ijcai.2019/899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kenford SL, Fiore M, Jorenby DE, Smith SS, Wetter DW, Baker TB. Predicting smoking cessation. Who will quit with and without the nicotine patch. JAMA. 1994;271(8):589–94. doi: 10.1001/jama.271.8.589. [DOI] [PubMed] [Google Scholar]
- [17].Gourlay S, Forbes A, Marriner T, Pethica D, Mcneil J. Prospective study of factors predicting outcome of transdermal nicotine treatment in smoking cessation. BMJ (Clinical research ed) 1994 10;309:842–846. doi: 10.1136/bmj.309.6958.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lehman E, DeYoung J, Barzilay R, Wallace BC. Inferring Which Medical Treatments Work from Reports of Clinical Trials. Proc. of NAACL’19. 2019:3705–3717. [Google Scholar]
- [19].Toutanova K, Chen D, Pantel P, Poon H, Choudhury P, Gamon M. Representing Text for Joint Embedding of Text and Knowledge Bases. Proc. of EMNLP’15. 2015:1499–1509. [Google Scholar]
- [20].Almasian S, Spitz A, Gertz M. Word Embeddings for Entity-Annotated Texts. In: Azzopardi L, Stein B, Fuhr N, Mayr P, Hauff C, Hiemstra D, editors. Advances in Information Retrieval. Cham: Springer International Publishing; 2019. pp. 307–322. [Google Scholar]
- [21].Sen P, Ganguly D, Jones GJF. Word-Node2Vec: Improving Word Embedding with Document-Level Non-Local Word Co-occurrences. NAACL-HLT (1). Association for Computational Linguistics. 2019:1041–1051. [Google Scholar]
- [22].Wu Z, Pan S, Chen F, Long G, Zhang C, Yu PS. A Comprehensive Survey on Graph Neural Networks. IEEE Transactions on Neural Networks and Learning Systems. 2020:1–21. doi: 10.1109/TNNLS.2020.2978386. [DOI] [PubMed] [Google Scholar]
- [23].Wu L, Yang Y, Zhang K, Hong R, Fu Y, Wang M. Joint Item Recommendation and Attribute Inference: An Adaptive Graph Convolutional Network Approach. Proc. of SIGIR’20. 2020:679–688. [Google Scholar]
- [24].Cui P, Wang X, Pei J, Zhu W. A Survey on Network Embedding. IEEE Trans Knowl Data Eng. 2019;31(5):833–852. [Google Scholar]
- [25].Michie S, West R, Finnerty AN, Norris E, Wright AJ, Marques MM. Representation of behaviour change interventions and their evaluation: Development of the Upper Level of the Behaviour Change Intervention Ontology. Wellcome Open Research. 2020;5(123) doi: 10.12688/wellcomeopenres.15902.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dawes J. Do Data Characteristics Change According to the Number of Scale Points Used? An Experiment Using 5-Point, 7-Point and 10-Point Scales. International Journal of Market Research. 2008;50(1):61–104. [Google Scholar]
- [27].Morey RD, Hoekstra R, Rouder JN, Lee MD, Wagenmakers EJ. The fallacy of placing confidence in confidence intervals. Psychonomic Bulletin & Review. 2016;23(1):103–123. doi: 10.3758/s13423-015-0947-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Graves A, Schmidhuber J. Framewise phoneme classification with bidirectional LSTM and other neural network architectures. Neural networks. 2005;18(5-6):602–610. doi: 10.1016/j.neunet.2005.06.042. [DOI] [PubMed] [Google Scholar]
- [29].Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed Representations of Words and Phrases and their Compositionality. Proceedings of NIPS 2013. 2013:3111–3119. [Google Scholar]
- [30].Pennington J, Socher R, Manning CD. GloVe: Global Vectors for Word Representation. Proc. of EMNLP 2014. 2014:1532–1543. [Google Scholar]
- [31].Peters ME, Neumann M, Iyyer M, Gardner M, Clark C, Lee K. Deep contextualized word representations. Proc. of NAACL 2018. 2018:2227–2237. [Google Scholar]
- [32].Devlin J, Chang M, Lee K, Toutanova K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. In: Burstein J, Doran C, Solorio T, editors. Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, NAACL-HLT 2019. Volume 1. Minneapolis, MN, USA: June 2-7, 2019. pp. 4171–4186. (Long and Short Papers) 2019. [Google Scholar]
- [33].Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN. Attention is All You Need. Proceedings of the 31st International Conference on Neural Information Processing Systems. 2017:6000–6010. [Google Scholar]
- [34].Lee J, Yoon W, Kim S, Kim D, Kim S, So CH. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2019;36(4):1234–1240. doi: 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Nelson W, Zitnik M, Wang B, Leskovec J, Goldenberg A, Sharan R. To Embed or Not: Network Embedding as a Paradigm in Computational Biology. Frontiers in Genetics. 2019;10:381. doi: 10.3389/fgene.2019.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kearnes S, McCloskey K, Berndl M, Pande V, Riley P. Molecular graph convolutions: moving beyond fingerprints. Journal of Computer-Aided Molecular Design. 2016;30(8):595–608. doi: 10.1007/s10822-016-9938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Grover A, Leskovec J. Node2Vec: Scalable Feature Learning for Networks. Proc. of ACM SIGKDD 2016. 2016:855–864. doi: 10.1145/2939672.2939754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chechik G, Sharma V, Shalit U, Bengio S. Large Scale Online Learning of Image Similarity Through Ranking. J Mach Learn Res. 2010 Mar;11:1109–1135. [Google Scholar]
- [39].Bonin F, Gleize M, Finnerty A, Moore C, Jochim C, Norris E. HBCP Corpus: A New Resource for the Analysis of Behavioural Change Intervention Reports. Proc. of LREC’20. 2020:1967–1975. [Google Scholar]
- [40].Ganguly D, Hou Y, Deleris LA, Bonin F. Information Extraction of Behavior Change Intervention Descriptions. Proc. of AMIA Symposium. 2019:182–191. [PMC free article] [PubMed] [Google Scholar]
- [41].Ganguly D, Deleris LA, Aonghusa PM, Wright AJ, Finnerty AN, Norris E. Unsupervised Information Extraction from Behaviour Change Literature. Proc. of MIE. vol. 247 of Studies in Health Technology and Informatics. 2018:680–684. [PubMed] [Google Scholar]