Abstract
A comprehensive, mapped social determinants of health (SDH) taxonomy in machine readable format was developed. The framework is intended to facilitate the extraction of social risk factors (SRFs) out of electronic health record (EHR) data and categorize them by domain and determinant to facilitate interpretation. Where other SDH frameworks have been focused on data input, this framework is designed from a data extraction point of view using EHR data in conjunction with published literature, public health policy documents, and official crosswalk maps. Frameworks developed by leading public health organizations were reviewed and synthesized to create an SDH framework comprising of 97 distinct SRFs organized under 16 domains. 2,329 medical codes across three standardized medical vocabularies, 10,896 free-text diagnosis descriptors, and 25 health insurance keywords were mapped to individual SRFs in the SDH framework. The framework is available as an open-source resource in Python dictionary or JSON format.
Introduction
Evidence on the impact of social and economic drivers on patient health outcomes has been mounting over the past two decades.1 This research posits that factors such as geography, housing, food, employment, education, and income can substantially shape health and well-being. With the push for value-based, patient-centered medicine in recent years, health care provider institutions have become increasingly focused on addressing these SDHs with the goals to improve patient outcomes and control health care costs.2,3 While much of the work and funding involved with managing social needs occurs outside the health care setting, these institutions recognize that health care providers, especially primary care, can play a pivotal role in identifying at-risk patient populations, screening for social needs, developing care plans that account for social factors, and directing patients to community and government resources.4 Implementation of a carefully considered infrastructure is required to effectively carry out these tasks. An institution’s population health informatics capabilities and its EHR are key elements of this infrastructure, particularly in identifying at-risk populations.5
However, even identifying SDH patients presents a challenge; population health informatics professionals face a three- fold problem. First, there is no standardized definition of what factors are considered social determinants.6 Multiple public health organizations have published frameworks identifying individual SDHs and associated domains including the World Health Organization (WHO)7, National Academy of Medicine (NAM)8, Kaiser Family Foundation (KFF)9, and the United States Department of Health and Human Services’ (DHHS) Healthy People 202010. Further, the International Classification of Diseases, Tenth Revision (ICD-10) includes a section of diagnosis codes (Z55-Z65) which provides a framework of SDH domains and social risk factors (SRFs).11 These frameworks overlap in many respects, but each also uniquely highlights important determinants and domains not addressed by the others. Without a universally agreed upon framework of SDHs and SRFs, an informatics professional must either choose one or attempt to reconcile them.
The second challenge is identifying the multitude of EHR data elements that could potentially record SRFs either directly or indirectly. Failure to carefully consider all the possible ways that SRFs could be documented in the EHR would yield only a partial picture of its prevalence in a population. EHRs in their current form contain a wealth of data elements that can be used to flag patients with social risks. Such SRFs can take the form of standardize medical vocabularies12, unstructured text13, demographic classifications, or administrative information.
The third challenge is the lack of a comprehensive, machine-readable map that connects data elements to a defined determinant or SRF. Creating these linkages is critical so that captured data can be aggregated, no matter how or where in the EHR the data is encoded, at a level that is meaningful from a population health perspective. The Social Interventions Research & Evaluation Network (SIREN) started the development of such a map by conducting a systematic review of standardized medical vocabularies to develop a compendium of medical codes for 20 domains and subdomains. These domains aligned with six widely recognized screening tools used to collect social and economic risk factors.14 SIREN’s compendium aligned key survey questions and patient responses to standardized codes available within the Systematized Nomenclature of Medicine Clinical Terms (SNOMED-CT), ICD-10, the
Logical Observation Identifiers Names and Codes (LOINC®), and the Current Procedural Terminology (CPT®). This pivotal work, and resulting map, facilitates documentation of SDH in clinical settings within the EHR as structured data. While the SIREN provider-focused compendium enables EHR data input, there is no complementary machine-readable tool that enables data extraction. SIREN’s aim was to be thorough in its mapping to ensure providers were able to input all potential patient responses to SDH survey instruments; it does not, however, define target SRFs for data extraction. The SIREN tool also does not address character-limited, free-text data elements or indirect data elements that could be used to identify at-risk patients.
To that end, we conducted literature reviews and utilized EHR data to derive a comprehensive, machine-readable taxonomy of SDH domains and determinants mapped to both standardized and character-limited free-text data elements. The goal was to build upon SIREN’s seminal work to create a mapped SDH taxonomy that can facilitate data extraction and serves as a tool to identify at-risk patients within an EHR. This tool, as well as other materials discussed in this paper, have been made available as an open-source resource for others in the community to use, add to, and refine.
Methods
This study involved two steps. The first step was a review and synthesis of the many SDH frameworks issued by public health organizations with the goal to consolidate them into a broader framework of domains, determinants, and SRFs. The second step focused on identification of data elements and mapping them to this SDH framework in a machine-readable format that can readily be passed into data queries. We prioritized formats that could be applied across a broad range of use cases and analytics approaches.
To aid this twofold endeavor, we introduced a unique aspect to our project: we derived and tested our SDH taxonomy using de-identified data set from three large national ambulatory EHR systems collected and maintained by Veradigm®. Veradigm, a business unit of Allscripts®, is a health information technology, analytics and intervention solutions company that manages the largest source of de-identified ambulatory patient records in the U.S. Its ambulatory EHR dataset contains medical information on patient demographics, prescriptions, problems, laboratory test results, vaccinations and allergies from providers using any of three ambulatory EHRs: Allscripts Professional EHR™ (PRO), Allscripts TouchWorks® EHR (TW), and Practice Fusion EHR (PF). Collectively, these three EHRs represent a nationally diverse range of ambulatory provider organizations, from single-provider or small group practices (PF) to mid-size physician practices (PRO) to large, single or multi-specialty physician practices (TW). By leveraging this vast and growing ambulatory EHR footprint, we were able to develop an SDH taxonomy that is generalizable and relevant to the real-world clinical setting.
Framework Development: Defining Domains and Determinants
We focused our review on commonly cited frameworks promulgated by WHO, NAM, KFF, and DHHS Healthy People 2020 as well as the one outlined in the ICD-10 section Z55 to Z65 code set (“ICD-10’s Z-codes”). The review of ICD-10 was limited to just the ICD-10’s Z-codes as this is the section* of the code set that is generally recognized as containing SDH codes.15
We chose ICD-10 Z-code’s domains and determinants to be the initial outline of our SDH taxonomy. Of the five reviewed, ICD-10’s Z-codes represents the only framework with determinants that are fully articulated as SRFs. For example, while several frameworks mention literacy as a determinant under their education domain, the education domain within ICD-10’s Z-code includes the SRF illiteracy and low-level literacy. Literacy is the SDH that refers to a person’s ability to read or write. Anyone can be evaluated for literacy. Illiteracy, however, refers to a specific subgroup of people whose literacy skills are low or nonexistent. Given the value a society places on literacy skills, illiterate individuals are considered at risk. In this scenario, education is the domain, literary is the SDH, and illiteracy is the target SRF.
The remaining four frameworks by WHO, NAM, KFF, and DHHS Healthy People 2020 were then assessed against this outline. Areas of overlap were noted. We recorded overlaps in both domains and individual SRFs. In some cases, frameworks would reference a domain such as Housing but not include individual SRFs such as Homelessness or Housing Inadequacy. Further, we also evaluated the need for reorganization of SRFs within the outline to improve conceptual groupings under each domain.
Gaps between the four frameworks and ICD-10’s Z-codes were recorded as well. We set a goal to develop an inclusive SDH taxonomy that drew upon unique aspects of the frameworks reviewed rather than a least common denominator approach. We reasoned that such a framework would help future-proof our taxonomy as the definition of SDH evolves over time. Thus, domains and SRFs that were not covered by ICD-10’s Z-codes but were mentioned in the other reviewed frameworks were evaluated for inclusion. This evaluation was conducted by both authors and decisions for adding new domains and SRFs were made on the basis of three factors: (1) the number of frameworks with coverage, (2) the utility of extracted data in health informatics and modeling, and (3) the ability for health institutions to deploy targeted inventions for the SRF.
A full copy of our comparative analysis of SDH frameworks is available online.
Data Elements: Selection and Mapping
Standardized Medical Vocabularies
The SDH taxonomy was populated with mapped ICD-10 codes, ICD-9 codes, and SNOMED codes using a number of methods. First, for areas where our two SDH frameworks overlapped, we used SIREN’s publicly available compendium16 of mapped ICD-10 and SNOMED codes to identify codes to add to the SDH taxonomy. SIREN’s compendium did require modification to accommodate our data extraction-oriented taxonomy. We manually distilled SIREN’s mapped standardized medical vocabularies down to just the codes representing SRFs. For example, SIREN’s Income/Poverty determinant was mapped to SNOMED codes that described a range of income levels, from wealthy to middle-class to destitution. For our taxonomy, however, we only included the codes associated with the SRFs low income, destitution, and poverty. We also eliminated duplicative mappings to ensure that each code was assigned to only one determinant. The ICD-10 code Z60.8 (other problems related to social environment), as an example, was mapped to five different social determinants. Such multiple mappings would make data aggregation difficult and distort comparative results. In these situations, we mapped the code to the most appropriate determinant based on official descriptors and public coding guidelines.
Literature reviews17, value sets18-20, and keyword searches of official code descriptors further supplemented our code search and mapping activities. We also obtained medical codes that were collected as part of our free-text diagnoses descriptors search strategy which is described in greater detail in the next section.
Lastly, we conducted a comprehensive code search using publicly available crosswalk maps: an ICD-10 to ICD-9 map21 and an ICD-10 to SNOMED map22. Our first round of searches identified crosswalk matches for ICD-10’s Z-codes and for all the codes identified using the methods mentioned above. Subsequent rounds used newly identified codes captured from prior searches to find additional crosswalk matches. This was done several times to ensure thoroughness. Code lists obtained from each round of searches were reviewed manually to assess relevancy before inclusion. We also considered alternative mapping if the case could be made that a code had a better fit elsewhere within the taxonomy.
Free-Text Diagnosis Descriptors
While harder to extract, unstructured and character-limited text offers a rich source of SDH data.13,23,24 Documentation of free-text data isn’t necessarily reserved to only the clinical notes section of the EHR. Many EHRs, including PRO and TW, allow free-text capture of diagnoses when a provider is unable to find a code or code descriptor that properly characterizes a patient’s circumstance. In some cases, these free-text diagnosis descriptors may eventually be mapped to discreet medical codes, but most are not.
Table 1 provides an example of how these free-text diagnostic records might appear in a data table within a clinical database for PRO or TW. The descriptor field represents either a code descriptor (when a code is selected by the provider) or a character-limited, free-text diagnosis descriptor (when the provider adds a custom diagnosis).
Table 1.
Example diagnosis records documents in a TW or PRO EHR clinical database
| Patientid | icd9 | icd10 | snomed | descriptor | recorded_dttm |
|---|---|---|---|---|---|
| a | 428078001 | HYSTERECTOMY; TOTAL | 2/12/2013 9:04 | ||
| b | 574.2 | K80.20 | 266474003 | CHOLELITHIASIS | 2/22/2014 8:33 |
| c | 171207006 | DEPRESSION SCREENING (Date is wasdone in comments) | 2/27/2013 10:55 | ||
| d | TETANUS | 6/6/2013 9:10 |
To capture this free-text data, we text mined the descriptor field within our diagnostic tables using string searches of both keywords and word pairings. Ideas for words and word pairings were obtained from multiple sources: (1) definitions found in published literature12,14, (2) common synonyms associated with individual ICD-10 codes25, and (3) official descriptors for mapped ICD26 and SNOMED codes27. Retrieved descriptors were manually reviewed for accuracy. ICD-9, ICD-10, and SNOMED codes that were frequently included in captured records were also collected and evaluated for potential inclusion in the taxonomy. Examples of keywords and word pairing used for text mining have been made available online.
Surrogate Data Elements: Health Insurance Status
Clinical data is not the only source of information on a patient’s SRFs; administrative information can also provide a means to indirectly discern this information as well. It is well documented that Medicaid, for example, can serve as a reliable indicator of a patient’s low-income status.28,29 Thus, we explored the use of health insurance data recorded in the EHR to determine if we could use a patient’s coverage status and/or their health insurance carriers to identify patient SRFs.
Results
SDH Framework
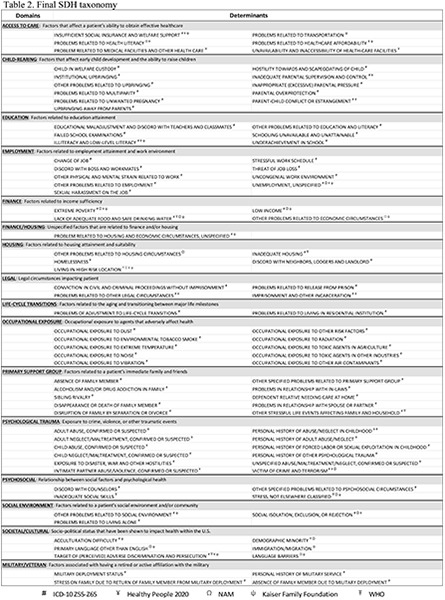
We created a framework that is comprised of 97 distinct SRFs organized under 16 domains (see Table 2). Our comparative evaluation showed that the ICD-10 Z-codes were comprehensive, covering a wide scope of SRFs beyond those addressed by the other frameworks. In line with our broad approach, we included all these ICD-10 topics areas even if they were not addressed in other frameworks.
Table 2.
Final SDH taxonomy
|
# ICD-10 Z55-Z65 ¥ Healthy People 2020 Ω NAM ψ Kaiser Family Foundation Ŧ WHO
Mapped Data Elements
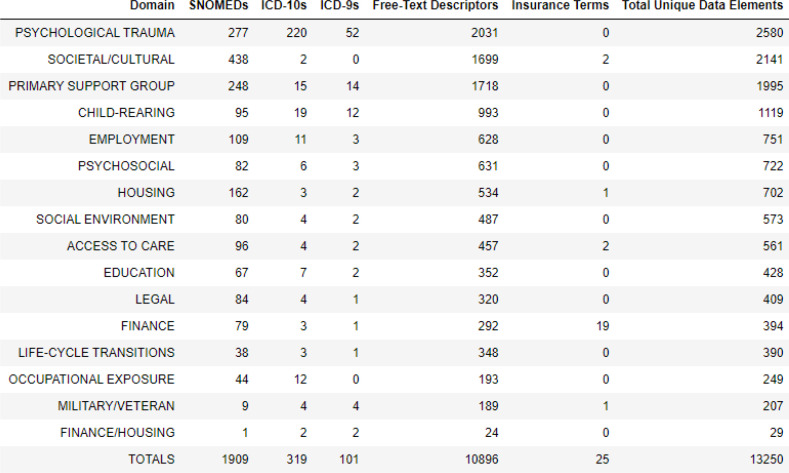
We identified and mapped 2,329 medical codes across three standardized medical vocabularies, 10,896 free-text diagnosis descriptors, and 25 health insurance keywords. Each of these data elements were mapped to a single social determinant. Table 3 provides an aggregate count of mapped data elements by domain. The crosswalk maps were the largest source for standardized medical codes. Approximately 44% (n=852) of SNOMED codes came from the ICD-10 to SNOMED crosswalk map. The ICD-10 to ICD-9 map yielded 37 ICD-9 codes.
Table 3.
Counts of mapped data elements by domain
Through our mining of health insurance names, we found searching with simple keywords did an excellent job in extracting target patient records. Using words like “Medicaid” and “Veteran” was a more efficient extraction approach as compared to collecting thousands of individual health plan names. These words alone were not entirely perfect. We needed to add the local names used by states for their Medicaid programs (e.g. TennCare, CalOptima, Peach State Health…). We also discovered that health institutions often document uninsured status, homelessness, and immigrant status within health insurance data tables.
Machine Readable Formatting
After experimenting with multiple ways of storing our mapped SDH taxonomy, we ultimately organized the information using a nested Python dictionary. The primary keys of the dictionary are the 97 individual SRFs found in the taxonomy. Under each SRF are secondary keys consisting of the five data element categories we examined to which individual data elements (code or text) are linked as mapped dictionary values. Figure 1 provides a visual example of the dictionary’s structure and organization.
Figure 1.
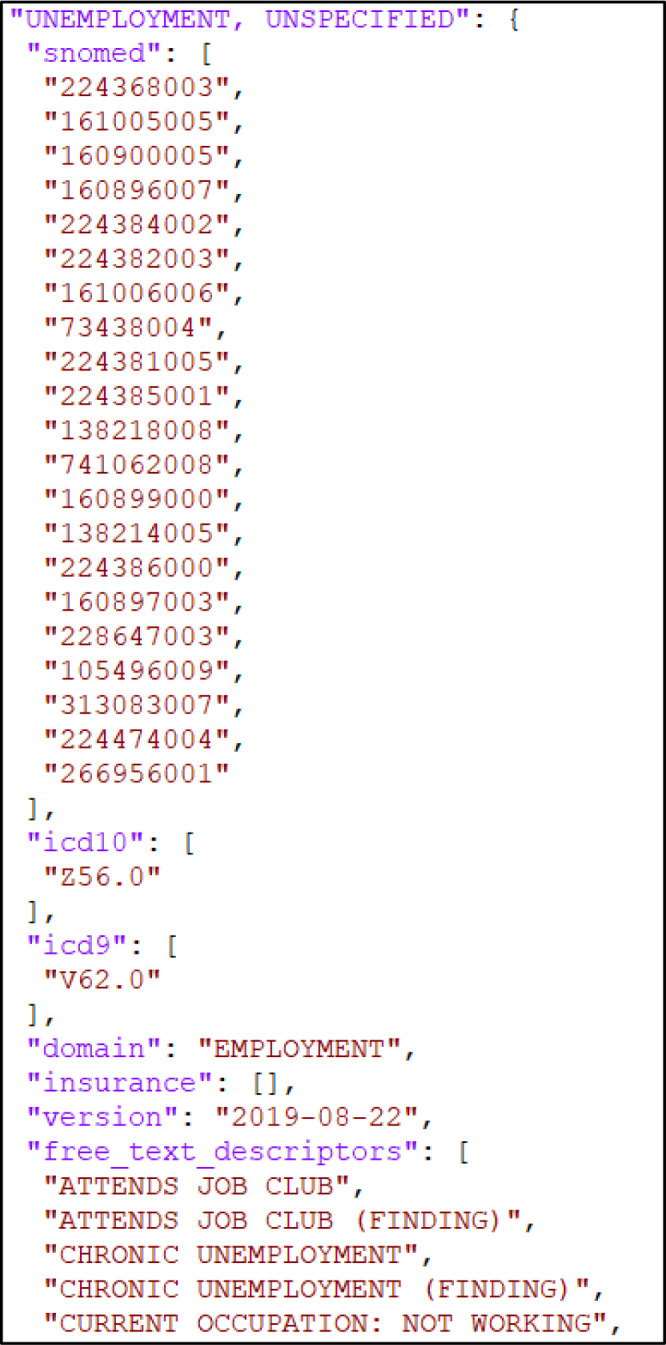
SDH taxonomy layout in JSON format
Discussion
The SDH framework shown in Table 2 is a result of our comparative evaluation of ICD-10’s Z-codes and SDH frameworks promulgated by DHHS Healthy People 2020, WHO, NAM, and KFF. Our analysis showed that ICD-10’s Z-codes codes covered most of the determinants and domains found in the four other SDH frameworks. We did find a few notable domain gaps, specifically access to health care, socio-political status, and exposure to violence. We chose to add the first two domains (and associated SRFs) to our framework because they were extensively covered and recommended by 4 of the 5 reviewed frameworks. We included the last domain (and SRFs) because it was prominently featured within the SIREN compendium under the Safety domain. We did not include all proposed SRFs or SDHs under these three new domains because some lacked clarity on how such risk factors could be captured in the EHR. For example, we chose not to include the SDHs quality of care and provider availability under the new Access to Care domain given the uncertainty on how this information could be measured and documented within a patient’s medical record. Finally, we chose not to include other domains gaps like behavioral/biological factors and gender given that these topics are either typically addressed as medical risk factors (rather than SRFs) or the proposed scope of the domain is so broad that a data extract would have limited usefulness in SDH informatics. The full comparative analysis, made available online, outlines all the domains, SDHs, and SFRs found in the five reviewed frameworks and highlights those that were and were not included in the final taxonomy.
We reorganized and consolidated domains and SRFs to create a more efficient and intuitive framework. Changes we incorporated include: (1) separating out Financing and Housing into two separate domains, (2) consolidating all SRFs: (a) related to violence under a new domain called Psychological Trauma, (b) related to military or veteran issues under a new domain called Military/Veterans, and (c) related to raising children under a new domain called Child Rearing, (3) consolidating ICD-10 “catch-all” codes (e.g. those codes with descriptors starting with Other problems related to or ending in Unspecified) within each domain, and (4) moving 10 SRFs to different or new domains where they thematically aligned with domain definitions. For example, we moved the Acculturation Difficulty and Target of (Perceived) Adverse Discrimination and Persecution SRFs to the newly created Societal/Cultural domain which includes determinants regarding minority status, immigration, and language barriers.
In terms of our data elements mapping activities, not all standardized medical codes were mapped as indicated within the crosswalk map. In some cases, we assigned certain codes to SRFs that were a better fit. We also chose not to use 142 SNOMED codes and 11 ICD-9 codes that came from the crosswalk maps for two general reasons: (1) the code descriptors were so vague that there would be uncertainty about whether it represented an actual SRF; and/or (2) the codes represented concepts that most health care institutions would not consider an at-risk situation. For example, we chose to forego mapping SNOMED codes for inadequate play space and inadequate exercise space to the Inadequate Housing SRF as recommended by the crosswalk maps. The supplemental code search strategies discussed in the Methods section yielded mostly duplicative results; however, each did, to a lesser extent, identify relevant SNOMED codes that would have otherwise been missed. Collecting associated codes as part of the search strategy for free-text diagnosis descriptors, for example, led to the discovery of SNOMED’s Country of Birth and Main Spoken Language code series which were, respectively, mapped to Immigration/Migration and Primary Language Other Than English.
Our search strategy for free-text diagnoses revealed quite a bit of redundancy amongst retrieved descriptors. Many of the descriptors would be considered parallel structures (e.g. loss of job, job loss, lost his job, losing job…), and, thus, the actual count of meaningfully distinct diagnosis descriptors is likely much less than 11,000.
Our choice of machine-readable format for the taxonomy has many advantages. One benefit of a dictionary format is it allows secondary keys to remain empty if there are no data elements to map as values. Further, arranging the taxonomy and its mapped data elements in a modular fashion around a determinant, instead of in multiple lists or tables, improved our ability to curate an unwieldy amount of interconnected information. The dictionary can be saved as a Python file or JSON, making it portable and shareable. Conversion to JSON format also makes the taxonomy code agnostic. The dictionary is also customizable; readers can unzip and extract the entire mapped taxonomy or just the SRFs of interest. To help readers understand what data elements can be found within the taxonomy, we used the Python library python-docx to produce a content catalog that lists all codes (with official descriptors) and all text by domain and SRF. Readers can find a copy of this content catalog at the same site where they can download the taxonomy itself.
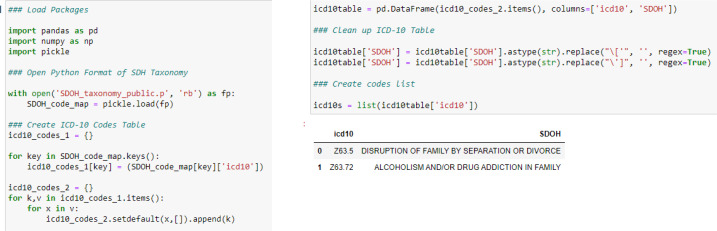
There are a number of ways readers can set up the taxonomy for use in their data platforms. Figure 2 provides a Python example on how we typically access the information, specifically the ICD-10 codes. We first open the *.p file with Python’s pickle library. Then we extract all the ICD-10 codes and their associated SDH/SRF label into a data table. We also create a list of these codes that we feed as inputs for filtering queries run against the database containing our EHR data. For a query involving ICD-10 codes, we’d typically extract unique patient ids, unique provider ids, dates of documentation, and ICD-10 codes for all records that match a code contained within our ICD-10 list. The extract is processed at a data table and then we use a LEFT JOIN to merge our taxonomy ICD-10 table (right table) with our database extract (left table), matching on the ICD-10 field. With this join, the records in our extract of patient ids, provider ids, and dates are each tagged with one of the 97 SDHs/SRFs found within our taxonomy. At this point, one can conduct various aggregations to yield patient counts, provider counts, rankings list, or frequencies.
Figure 2.
Python example for processing the SDH taxonomy’s ICD-10 codes for use
There are a number of weaknesses with our study that should be noted. First, our keyword search strategy was derived from our understanding of how social concepts may be characterized in English within the EHR. Thus, it is limited by “what we know we know”. It is likely diagnosis descriptors were missed because we are not aware of all the possible ways social concepts may be articulated in words and abbreviations. Because the EHR data set is multi-state, there could also be regional differences in use of words and abbreviations. We envision a future role for natural language processing (NLP) to help us gain an understanding of what might have been missed. Second, the taxonomy in its current form solely focuses on identification of documented SRFs. It does not, however, validate the accuracy of this documentation nor does it account for the transient state of many SRFs. Additional work will be required by health informatics professionals to determine if the patients identified by using the taxonomy have a valid and current SRF. Lastly, the taxonomy is only focused on clinical assessment and is binary in nature (i.e. presence or absence of an SRF). It does not currently include a means to capture a richer level of detail of a patient’s social circumstance (e.g. severity, current/past interventions, etc…). These are details that will be needed by those health care providers who are tasked with intervening to improve the patient’s social situation. Currently, such a taxonomy does not exist, but plans should be made to incorporate these data elements once efforts to build one, like the work being done by the Gravity Project, come to fruition.
Available Materials
A copy of the machine-readable SDH taxonomy in a Python dictionary and JSON formats has been made available online as an open-source resource for others in the community to use, add to, and refine. It can be found at https://github.com/Veradigm-Life-Sciences-Research/SDoHTaxonomy. At this site, readers will also find an SDH taxonomy content catalog, a full copy of our comparative analysis of SDH frameworks, Python code examples, and a list of keywords and word pairing that were used for text mining.
Next Steps
Future work will focus on using our EHR data set and the taxonomy to gain an understanding of SDH documentation patterns amongst health care institutions using Allscripts EHRs. We also plan to investigate a way to leverage the free-text diagnosis descriptors we collected to build a corpus to use with our NLP work, especially in regard to conducting analytics and information retrieval on alternative characterizations of social concepts and on the EHR’s unstructured clinical notes. We plan to determine the importance of adding free-text diagnosis descriptors to an SDH data extraction strategy. This is a unique contribution to SDH informatics and we hope to quantify the value it adds. We plan to continue expanding the taxonomy with LOINC codes and zip codes/census tracts. For the latter, geographic data elements could serve as additional surrogates to identify at-risk patients, particularly areas such as food deserts, housing instability, violence, and financial inequities.30 We plan to evolve the taxonomy to incorporate the work of the Gravity Project as it becomes available. Our taxonomy is risk factor and data extraction focused which intentionally complements a subset of the critical work of the Gravity Project. We view it important to adapt it as national standards for documenting SRFs are set by the this consensus group.
Conclusion
Using a large, multi-institutional EHR data set in conjunction with published literature, public health policy documents, and existing crosswalk SDH maps, we assembled a comprehensive, mapped SDH taxonomy designed from a data extraction point of view. This work is unique in that it mapped standardized medical codes, free-text diagnosis descriptors, and surrogate data elements, allowing informatics professionals to search for SRF documentation in multiple areas within the EHR. This multifaceted approach is crucial to characterizing the full scope of the impact of SDHs on an institution’s patient population. The layout of the taxonomy within a machine-readable format enables end-users to efficiently unzip, modify, maintain, and share its mapped content. Our work has the potential to help health care organizations characterize their at-risk populations, a critical first step in addressing social needs that impact their patient’s health and well-being.
Footnotes
Codes for many other social and economic drivers can be found throughout ICD-10, but they are typically contained within sections that, unlike the ICD-10 Z-codes, do not necessarily call attention their status as SRFs and are not organized as such.
References
- 1.Braveman P, Egerter S, Williams DR. The social determinants of health: coming of age. Annual Review of Public Health. 2011;32(1):381–398. doi: 10.1146/annurev-publhealth-031210-101218. [DOI] [PubMed] [Google Scholar]
- 2.Galea S, Tracy M. Estimated deaths attributable to social factors in the United States. Am J Public Health. 2011;101(8):1456–1465. doi: 10.2105/AJPH.2010.300086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spencer A, Freda B. Measuring social determinants of health among Medicaid beneficiaries: early state lessons. Center for Health Care Strategies Brief. Dec 2016. Accessed at www.chcs.org/resource/measuring-social-determinants-health-among-medicaid-beneficiaries-early-state-lessons.
- 4.Center for Health Care Strategies. Addressing health-related social needs among Medicaid beneficiaries: Mapping cross-sector partnership roles. Technical Assistance Tool. Jun 2020. Accessed at www.chcs.org/resource/addressing-health-related-social-needs-among-medicaid-beneficiaries-mapping-cross-sector-partnership-roles.
- 5.Thomas-Henkel C, Schulman M. Screening for social determinants of health in populations with complex needs: implementation considerations. Center for Health Care Strategies Brief, Oct. 2017. Accessed at www.chcs.org/resource/screening-social-determinants-health-populations-complex-needs-implementation-considerations.
- 6.Islam MM. Social determinants of health and related inequalities: confusion and implications. Frontiers in. Public Health. 2019;7(11) doi: 10.3389/fpubh.2019.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solar O, Irwin A. A conceptual framework for action on the social determinants of health: social determinants of health discussion paper 2. Geneva: World Health Organization; 2010. Policy and Practice. [Google Scholar]
- 8.Institute of Medicine of the National Academies Committee on the Recommended Social and Behavioral Domains and Measures for Electronic Health Records. Capturing social and behavioral domains in electronic health records: phase 2. Washington, DC: The National Academies Press; 2014. [PubMed] [Google Scholar]
- 9.Artiga S, Hinton E. Beyond health care: the role of social determinants in promoting health and health equity. Kaiser Family Foundation Issue Brief 2018; May. Accessed at https://www.kff.org/disparities-policy/issue-brief/beyond-health-care-the-role-of-social-determinants-in-promoting-health-and-health-equity. [Google Scholar]
- 10.Office of Disease Prevention and Health Promotion. Accessed at www.healthypeople.gov/2020/topics-objectives/topic/social-determinants-of-health. [Google Scholar]
- 11.Gottlieb L, Tobey R, Cantor J. Integrating social and medical data to improve population health: opportunities and barriers. Health Affairs. 2016;35(11):2116–2123. doi: 10.1377/hlthaff.2016.0723. [DOI] [PubMed] [Google Scholar]
- 12.Monsen KA, Rudenick JM, Kapinos N. Documentation of social determinants in electronic health records with and without standardized terminologies: a comparative study. Proceedings of Singapore Healthcare. 2019;28(1):39–47. [Google Scholar]
- 13.Bettencourt-Silva JH, Mulligan N, Sbodio M. Discovering new social determinants of health concepts from unstructured data: framework and evaluation. Studies in Health Technology and Informatics. 2020;270:173–177. doi: 10.3233/SHTI200145. [DOI] [PubMed] [Google Scholar]
- 14.Arons A, DeSilvey S, Fichtenberg C. Documenting social determinants of health-related clinical activities using standardized medical vocabularies. JAMIA Open. 2019;2(1):81–88. doi: 10.1093/jamiaopen/ooy051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.United Health Care. 2019 Social Determinants of Health ICD-10 Codes. 2019 Network Bulletin: June Accessed at www.chcs.org/resource/screening-social-determinants-health-populations-complex-needs-implementation-considerations.">www.uhcprovider.com/en/resource-library/news/2019-net-bulletin-featured-articles/0619-social-determinants-health.html">www.chcs.org/resource/screening-social-determinants-health-populations-complex-needs-implementation-considerations. [Google Scholar]
- 16.Arons A, DeSilvey S, Fichtenberg C, Gottlieb L. Compendium of Medical Terminology Codes for Social Risk Factors. San Francisco, CA: Social Interventions Research and Evaluation Network; 2018. Accessed at http://sirenetwork.ucsf.edu/tools-resources/mmi/compendium-medical-terminology-codes-social-risk-factors.
- 17.Torres JM, Lawlor J, Colvin JD. ICD social codes: an underutilized resource for tracking social needs. 2017;55(9):810–816. doi: 10.1097/MLR.0000000000000764. Medical Care. [DOI] [PubMed] [Google Scholar]
- 18.NLM. Value Set Authority Center (VSAC) Social Reasons Group. OID: 2.16.840.1.113883.3.666.5.1595. Accessed at https://vsac.nlm.nih.gov/valueset/2.16.840.1.113883.3.666.5.1595/expansion/Latest.
- 19.NLM; VSAC. Social Determinants of Health Problem Observation. OID: 2.16.840.1.113762.1.4.1096.224. Accessed at https://vsac.nlm.nih.gov/valueset/2.16.840.1.113762.1.4.1096.224/expansion/Latest.
- 20.NLM; VSAC. Social Indications. OID: 2.16.840.1.113762.1.4.1029.136 Accessed at https://vsac.nlm.nih.gov/valueset/2.16.840.1.113762.1.4.1029.136/expansion/Latest.
- 21.Centers for Medicare; Medicaid Services (CMS) General Equivalence Mappings (GEMs) 2018. Accessed at www.cms.gov/Medicare/Coding/ICD10/2018-ICD-10-CM-and-GEMs.
- 22.National Library of Medicine (NLM) Unified Medical Language System® (UMLS®): SNOMED CT ICD-10-CM Map. Accessed at www.nlm.nih.gov/research/umls/mapping_projects/snomedct_to_icd10cm.html.
- 23.Bejan C, Angiolillo J, Conway D. Mining 100 million notes to find homelessness and adverse childhood experiences: 2 case studies of rare and severe social determinants of health in electronic health records. JAMIA. 2018;25(1):61–71. doi: 10.1093/jamia/ocx059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bucher BT, Shi J, Pettit RJ. Determination of marital status of patients from structured and unstructured electronic healthcare data. 2020;(2019):267–274. AMIA Annu Symp Proc. [PMC free article] [PubMed] [Google Scholar]
- 25.ICD-10-CM Codes. Accessed at www.icd10data.com/ICD10CM/Codes/Z00-Z99/Z55-Z65.
- 26.Centers for Disease Control and Prevention (CDC) Classification of Diseases, Functioning, and Disability. Accessed at www.cdc.gov/nchs/icd/index.htm.
- 27.NLM. SNOMED CT United States Edition. Accessed at www.nlm.nih.gov/healthit/snomedct/us_edition.html.
- 28.Casey JA, Pollak J, Glymour MM. Measures of SES for Electronic Health Record-based Research. Am J Prev Med. 2018;54(3):430–439. doi: 10.1016/j.amepre.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foraker, Randi E. Neighborhood socioeconomic status, Medicaid coverage and medical management of myocardial infarction: atherosclerosis risk in communities (ARIC) community surveillance. BMC Public Health. 2010;10:632. doi: 10.1186/1471-2458-10-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.CDC. Sources for Data on Social Determinants of Health. Accessed at www.cdc.gov/socialdeterminants/data/index.htm.