Abstract
Chronic diabetes can lead to microvascular complications, including diabetic eye disease, diabetic kidney disease, and diabetic neuropathy. However, the long-term complications often remain undetected at the early stages of diagnosis. Developing a machine learning model to identify the patients at high risk of developing diabetes-related complications can help design better treatment interventions. Building robust machine learning models require large datasets which further requires sharing data among different healthcare systems, hence, involving privacy and confidentiality concerns. The main objective of this study is to design a decentralized privacy-protected federated learning architecture that can deliver comparable performance to centralized learning. We demonstrate the potential of adopting federated learning to address the challenges such as class-imbalance in using real-world clinical data. In all our experiments, federated learning showed comparable performance to the gold-standard of centralized learning, and applying class balancing techniques improved performance across all cohorts.
Introduction
Diseases such as chronic diabetes have evidence of engendering other fatal long-term comorbid complications. For instance, prolonged uncontrolled diabetes can lead to microvascular complications, including diabetic eye disease, diabetic kidney disease, and diabetic neuropathy1-3. Diabetes-related complications resulted in 16 million emergency department visits and 7.8 million hospitalizations, as estimated in 2016, contributing to an increasing burden on the US healthcare system. Diabetes is also the leading cause of end-stage kidney disease, with a crude prevalence of 38% and new cases of vision disability with a crude rate of 11.7%3,4.
However, the long-term complications often remain undetected at the early stages of diagnosis, and most of the treatment plans for addressing the complications are reactive rather than proactive. Developing a state-of-the-art prediction model that can learn from the patient-related factors to identify the patients at high risk of developing diabetes-related complications can help design better treatment interventions. Hence, it can play an essential role in minimizing costs related to hospitalizations, medications, and treatment procedures ensuing from those complications.
Digitizing healthcare data in electronic health records (EHR) evolved into a rich source of patient health history, thus generating more opportunities to innovate data-driven tools and techniques to improve the availability and accuracy of medical services5. Potential utilization of machine learning in predicting health outcomes can involve incorporating large amounts of data generated by independent health systems6,7. However, developing centralized algorithms for centralized data repositories raises privacy, confidentiality, and regulatory concerns such as the HIPAA and HITECH Acts7. For instance, incidences of diabetes-related complications are rare events among the diabetes population. Thus, it is imperative to accumulate data from different healthcare systems to build reliable and robust predictive models using a large representation of the population for such rare complications. To address the persistent concerns related to data-sharing among the healthcare systems, we present a federated learning-based framework that can consolidate predictive models without using central repositories of the actual data itself.
In recent times, the concepts of federated learning have been seamlessly adapted in the healthcare domain to address the privacy concerns on sharing potential patient information among the healthcare systems. A decentralized prediction engine can potentially use data stored from independent health systems without ethical or legal circumstances. This study is the first implementation of federated learning in predicting diabetes-related complications using real-world clinical data to the best of our knowledge. Moreover, predictions of diabetes-related complications using federated learning encounter significant challenges from the inherent class-imbalance characteristics of real-world clinical data. The main objective of this study is to design a decentralized privacy-protected federated learning architecture that can deliver comparable performance to centralized learning. We demonstrate the potential of adopting federated learning to address the challenges in using real-world clinical data.
The key contributions of our study include (i) identifying a diabetes population and three cohorts related to diabetic eye disease, diabetic kidney disease, and diabetic retinopathy from Health Facts EMR data using a structured framework, (ii) implementing a decentralized privacy-controlled federated learning architecture that can utilize the federated datasets from different healthcare systems to predict the complication in each cohort without sharing data among themselves, (iii) performing sampling techniques to address the class-imbalance characteristic of the datasets in the federated learning architecture, and (iv) comparing the performance of federated learning to centralized learning in different sampling conditions.
Background
Federated learning involves iteratively analyzing separate databases and sharing only mathematical parameters (metadata), but not the actual data itself that might reveal potential patient identifiers8. Federated learning mechanisms were initially more popular in image classification and enhancing wireless communication systems9,10. The recent adaptation of federated learning models in the healthcare domain includes predictions on healthcare outcomes such as mortality, hospital stay-time for ICU patients, hospitalization for cardiac events, dyspnea, adverse drug reactions6,11-13. However, most of the implementation of federated learning in predicting healthcare outcomes utilized small datasets and partitioned the data hypothetically (randomly) to mimic the inherent characteristics of real-world data. In this study, we utilize the natural partitions of the Health Facts data using the information of the healthcare systems for each patient data to demonstrate our framework.
In most federated learning applications in the healthcare domain, classification algorithms such as logistic regression, artificial neural network, multi-layer perceptron, support vector machines, random forest were used to build federated predictive models12-16. Existing literature in predicting diabetic retinopathy (eye disease), neuropathy (peripheral nerve disorder), and nephropathy (kidney disease) involve centralized machine learning algorithms using small-size datasets from the US population with limited cases of complications and limited patient information17,18. For our study, we implemented three machine learning models, including logistic regression, 2-layer multiple perceptrons, and 3-layer multi-perceptron in federated learning architecture for binary classification of the incidence of three diabetes-related complications affecting eyes, kidneys, and peripheral nerves, respectively.
Methods
Data Source
We have used Cerner's "Health Facts EMR Data" as our data source in this study. Health Facts is a de-identified electronic health records relational database consolidated from over 90 healthcare systems in the US between 2000 and 2016 in which Cerner has a data use agreement. Health Facts database contains patient-level attributes, such as demographics, encounters, diagnoses, lab results, procedures, prescription orders, and other clinical observations on 69 million unique patients. We identified the diabetes population, defined three cohorts for diabetes-related complications, and extracted patient-level comorbid features for our cohorts using a detailed pipeline on this dataset.
Diabetes Population Selection
We identified the diabetes population in our study using the SUPREME-DM (Surveillance, PREvention, and ManagEment of Diabetes Mellitus algorithm19 based on eight criteria (Table 1). Six criteria were based on lab results, while two criteria were based on International Classification of Disease (ICD-9 and ICD-10) diagnosis codes related to inpatient and outpatient encounters. Patients satisfying at least one criterion or more were selected as the diabetes population. Only patients aged 18 or above at the first encounter were included.
Table 1:
We used the following eight criteria to select the diabetes population from Health Facts. The first six criteria were based on lab results, while the last two were based on ICD-9 and ICD-10 diagnosis codes. The thresholds for the lab tests are chosen based on the SUPREME-DM. Patients satisfying at least one criterion were selected as the diabetes population
| Criterion (at least one) | Value | Notes |
|---|---|---|
| HbA1c | ≥ 2 and ≥ 6.5% | Tests must be within two years apart |
| Fasting plasma glucose | ≥ 2 and ≥ 126 mg/dL | |
| Random plasma glucose | ≥ 2 and ≥ 200 mg/dL | |
| Random plasma plus fasting glucose | 1 at ≥200 mg/dL and 1 at ≥126 mg/dL | |
| HbA1c plus fasting glucose | 1 at ≥6.5% and 1 at ≥126 mg/dL | |
| HbA1c plus random plasma glucose | 1 at ≥6.5% and 1 at ≥200 mg/dL | |
| Inpatient discharge diagnosis | ≥1 inpatient visit with one of the following diagnosis codes primary: 250.x, 357.2, 366.41, 362.01362.07, and E08.x-E13.x | Primary diagnosis code only |
| Outpatient visit diagnosis | ≥2 outpatient visits with one of the following diagnosis codes 250.x, 357.2, 366.41, 362.01362.07, and E08.x-E13.x | Visits must occur on separate days (ambulatory visits only) |
Selection of Cohorts with Diabetes-Related Complications
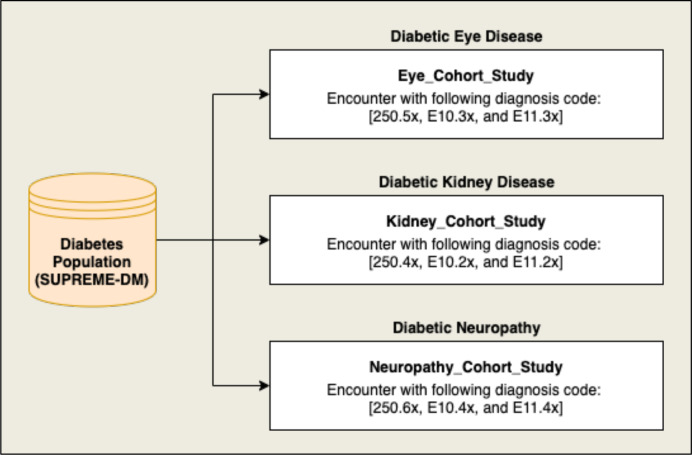
The diabetes-related complications among the selected diabetes population were identified using International Classification of Disease (ICD-9 and ICD-10) diagnosis codes. We selected three cohorts, including diabetic eye disease (EDD), diabetic kidney disease (KDD), and diabetic neuropathy (ND), from the selected diabetes population. Figure 1 shows the flow diagram of the selection of cohorts with ICD codes for each of the diabetes-related complications.
Figure 1:
Flow diagram showing the cohort selection for diabetic eye disease, diabetic kidney disease, and diabetic neuropathy from the diabetes population using the shown diagnosis codes.
Many studies have shown that patients diagnosed with diabetes show higher chances of developing these complications after five years of chronic exposure20-22. Patients diagnosed with any of the above complications who had at least five years of chronic diabetes with total encounters between 25 and 500 were selected as "cases". For the "non-cases", patients with at least seven years of chronic diabetes with no diagnosis of any of the complications and total encounters between 25-500 were selected. The patient-related comorbid features were extracted using the diagnosis table. The diagnosis ICD codes were mapped into the Clinical Classification Software (CCS) tool developed by Healthcare Cost and Utilization Project (HCUP). We used 283 CCS coded features consolidated from the individual ICD-9 and ICD-10 codes by grouping the ICD codes into clinically similar entities, which are used as predictors for the experiments. Table 2 shows the summary statistics for the selected three cohorts of diabetes-related complications.
Table 2:
The summary statistics for each of the three cohorts among the diabetes population are shown below. We excluded the healthcare systems with less than 100 cases from the dataset, which reduced the number of cases in each cohort, as shown in the following table.
| Diabetic Population | Diabetic Eye Disease Cohort | Diabetic Kidney Disease Cohort | Diabetic Neuropathy Cohort | |
| Number of Patients | 102,876 (100) | 10,599 (10.3) | 17,455 (17.1) | 23,682 (23.0) |
| Number of Cases (After Exclusion) | - | 9, 686 (9.4) | 16, 727 (16.3) | 22,477 (21.8) |
| Class-Ratio (Cases: No Cases) | - | 13:100 | 22:100 | 32:100 |
| Number of Independent Healthcare Systems | 70 | 22 | 31 | 31 |
Decentralized Data Architecture
To demonstrate the method of privacy-preserved federated learning using the three selected cohorts, we utilized the hospital system identifiers for each patient in each cohort. The variable "ALT_HEALTH_SYSTEM_ID" was used to extract the hospital identifiers from the "hs_d_hospital" data table for each "PATIENT_ID". The patient IDs were mapped to "PATIENT_SK", which were linked to the CCS table of patient features for each cohort. Table 2 shows the number of hospital systems in each of the cohorts. We distributed the data for each cohort into small datasets for each hospital system to facilitate our federated learning architecture, as discussed below. The healthcare systems with less than 100 cases were discarded from the analysis.
Centralized Predictive Model
A centralized predictive model approach was implemented to represent the characteristics of a centralized data warehouse gathering data from multiple sites. We considered a general binary classification of the diabetes-related complications in this study. Each primary response variable, including diabetic eye disease, diabetic kidney disease, and diabetic neuropathy, is categorized into binary responses, such as cases vs. non-cases. The CCS comorbid features of each patient in each cohort are used to predict the complication as binary responses. For this purpose, we considered logistic regression, 2-layer multiple layer perceptron, and 3-layer multiple layer perceptron models. The machine learning models were trained using 70% of the complete dataset, while 30% were used for testing and validation. The algorithms were implemented using the sci-kit learn module from Python 3.6. The centralized approach to predict each complication from each cohort will act as a benchmark to compare with the federated learning approach.
Federated Predictive Model
The federated learning approach utilized the partitioned data from each cohort to predict the binary cases and non-cases for each complication. We used sci-kit learn, NumPy, pandas for the machine learning tasks, and TensorFlow to create the federated mining pipeline. To demonstrate this approach, separate training and testing datasets were created for each disjointed partitioned dataset, i.e., each healthcare system in each cohort. The labels from training data for each cohort were 1-hot-encoded using the LabelBinarizer() object from sk-learn. The features were then transformed into TensorFlow data objects. We used binary cross-entropy as loss function and stochastic gradient descent as the optimizer to compile three models: logistic regression, a 2-layer multi-perceptron, and a 3-layer multi-perceptron. Since the data in our study are horizontally partitioned, local model parameters will contribute to the global model weighted by the proportion of data points from each participating healthcare system.
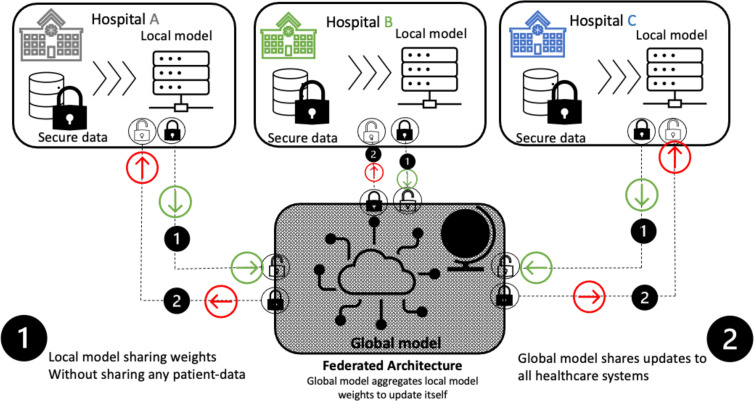
We developed the training module for federated learning using the federated averaging algorithm from McMahan (2017) 8. Algorithm 1 shows the two main functions used to train the federated models. At first, the global model weights are initialized, which serve as the initial weights for all local models. Using these initial weights, the local models for each healthcare system {k = 1, ... , K} are trained to obtain the updated weights. The updated weights are then scaled by a factor nk/N, which is the proportion of data for kth Healthcare system and summed to obtain a new set of weights. These averaged weights (as shown in line 9 from Algorithm 1) are then used to update the global model. The process continues until T rounds of aggregating local model weights to update the global model. Thus, a federated learning mechanism only relies on sharing the weights of the local models without sharing any raw data from the individual healthcare systems. We repeat these steps to predict the three complications in the three cohorts separately. Figure 3 shows the system design of federated learning deployed in our study.
Figure 3:
Federated architecture deployed to predict diabetes-related complications. A global model is initiated at first, and the initial weights are sent to the local models. The local models are trained with the secured data within the healthcare system which generates new set of weights. The updated weights are sent to the global model, where they are aggregated and used to update the global model. In this process, no patient data leave the healthcare system, hence, preserving privacy and confidentiality.


Class-Imbalance Learning
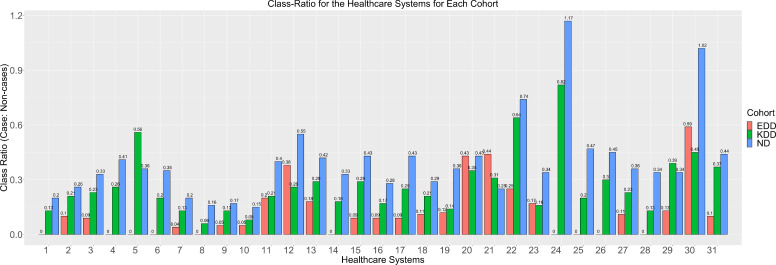
As shown in Figure 2, an imbalance between the cases and non-cases of the three diabetes-related complications among the diabetes population exists. Moreover, the federated datasets for the three cohorts are also subjected to unequal distribution of cases and non-cases. This is consistent with our expectations since the complications are rare for most patients diagnosed with diabetes. Classification algorithms applied in datasets with non-cases as the minority class is more likely to predict new observations in the majority class since they fail to characterize their imbalanced nature. Also, federated averaging would not account for the varying distribution of cases across the different healthcare systems. Thus, it is imperative to consider this inherent characteristic of class distribution in our model training. We applied sampling techniques, such as oversampling and undersampling, to address the class-imbalance attribute of the cohort datasets. Oversampling supplement the minority class, whereas undersampling randomly removes the majority class. For centralized learning, oversampling, undersampling, and no-sampling were performed on the training datasets for each cohort before model compilation. In the federated learning approach, similar sampling strategies were applied on the federated datasets for each cohort before the local model compilations. The module "resample" from sk-learn was used for executing the oversampling and undersampling techniques with "n_samples" parameter set to the class size for both over- and under-sampling.
Figure 2:
A bar plot showing the class-ratio (cases: non-cases) for the three cohorts: EDD (Diabetic Eye Disease), KDD (Diabetic Kidney Disease), and ND (Diabetic Neuropathy) for the 31 Healthcare Systems.
Experimentation and Evaluation Metrics
The three machine learning models were run for each of the cohorts for the three sampling techniques including, no balancing, oversampling, and undersampling. In total, there were 27 experiments for the federated learning architecture and 27 experiments for centralized learning. We combined the testing data from all sites within each cohort to evaluate the performance of the federated model. When the datasets are subjected to class-imbalance, F-1 scores are a more reliable measure compared to accuracy. Performance metrics, such as F-1 score, recall, and precision, were used to compare the performance of the federated learning to centralized learning.
R version 3.4.4 (R Foundation for Statistical Computing, Vienna, Austria) and Python 3.6 were used for data management and all computations were performed on a Mac Book Pro running macOS Catalina version 10.15.2 with 16GB of RAM.
Comparative Analysis
The performance measures across all the classifiers and datasets showed federated learning to exhibit comparable predictive performance with respect to centralized learning. As shown in Table 3, multilayer perceptron consistently performed well compared to logistic regression in predicting diabetic eye disease patients. For logistic regression, applying the undersampling improved the recall by 12% for both federated and centralized learning. The F-1 scores for both multi-layer perceptron models were improved by about 4% for federated learning and 13% for centralized learning. However, precision measures decreased by 20% for both oversampling and undersampling experiments for both the learning mechanisms. The difference between the precision scores between federated and centralized learning varied by 6-7%.
Table 3:
Performance metrics (F-1 score, Precision, and Recall) for centralized learning and federated learning, obtained from three machine learning models: logistic regression, 2-layer multi-perceptron, 3-layer multi-perceptron for the diabetic eye disease cohort.
| Federated Learning | Centralized Learning | ||||||
| Model | Method | F1 | Prec | Recall | F1 | Prec | Recall |
| LR | Oversample | 0.59 | 0.55 | 0.71 | 0.72 | 0.69 | 0.8 |
| LR | Undersample | 0.59 | 0.51 | 0.72 | 0.72 | 0.66 | 0.84 |
| LR | No Balancing | 0.62 | 0.72 | 0.59 | 0.75 | 0.81 | 0.72 |
| MLP (10,1) | Oversample | 0.63 | 0.63 | 0.68 | 0.71 | 0.68 | 0.78 |
| MLP (10,1) | Undersample | 0.59 | 0.54 | 0.71 | 0.7 | 0.61 | 0.85 |
| MLP (10,1) | No Balancing | 0.68 | 0.74 | 0.67 | 0.75 | 0.81 | 0.72 |
| MLP (10,10,1) | Oversample | 0.65 | 0.67 | 0.67 | 0.67 | 0.61 | 0.8 |
| MLP (10,10,1) | Undersample | 0.59 | 0.54 | 0.71 | 0.67 | 0.61 | 0.82 |
| MLP (10,10,1) | No Balancing | 0.69 | 0.73 | 0.68 | 0.71 | 0.66 | 0.82 |
All the classifiers from the two setups performed better in the diabetic kidney disease cohort, as shown in Table 4. Both oversampling and undersampling improved recall for logistic regression and multi-payer perceptron models. For both centralized and federated learning, recall improved by 20%. The maximum difference in recall between federated and centralized learning was within 3% for both the oversampling and undersampling cases. Maximum F-1 scores were obtained by the multi-layer perceptron models and were comparable for both federated and centralized learning. The maximum difference in F-1 scores between federated and centralized learning were about 4% when the balancing techniques were applied. When no balancing technique was implemented, the maximum difference in the performance metrics between the two learning setups is about 10%.
Table 4:
Performance metrics (F-1 score, Precision, and Recall) for centralized learning and federated learning, obtained from three machine learning models: logistic regression, 2-layer multi-perceptron, 3-layer multi-perceptron for the diabetic kidney disease cohort.
| Federated Learning | Centralized Learning | ||||||
| Model | Method | F1 | Prec | Recall | F1 | Prec | Recall |
| LR | Oversample | 0.65 | 0.56 | 0.85 | 0.68 | 0.59 | 0.85 |
| LR | Undersample | 0.62 | 0.53 | 0.83 | 0.67 | 0.56 | 0.86 |
| LR | No Balancing | 0.56 | 0.72 | 0.5 | 0.64 | 0.76 | 0.6 |
| MLP (10,1) | Oversample | 0.67 | 0.58 | 0.85 | 0.66 | 0.56 | 0.84 |
| MLP (10,1) | Undersample | 0.65 | 0.56 | 0.84 | 0.65 | 0.54 | 0.87 |
| MLP (10,1) | No Balancing | 0.61 | 0.69 | 0.6 | 0.65 | 0.74 | 0.61 |
| MLP(10,10,1) | Oversample | 0.64 | 0.54 | 0.84 | 0.63 | 0.53 | 0.84 |
| MLP(10,10,1) | Undersample | 0.64 | 0.53 | 0.85 | 0.67 | 0.57 | 0.85 |
| MLP(10,10,1) | No Balancing | 0.63 | 0.64 | 0.66 | 0.65 | 0.74 | 0.61 |
As shown in Table 5, for logistic regression applying sampling techniques improved the F-1 score by 6-7% for federated learning and about 1% for centralized learning. Recall improved by 34-35% in federated logistic regression, while a 26% increase is observed in centralized logistic regression. The maximum difference in F-1 score and recall between federated and centralized logistic regression is about 2%. In the 2-layer multi-perceptron model, sampling techniques improved F-1 score by 4-5% in federated learning, while an improvement of 6-7% was observed in centralized learning. Recall improved by 24% in federated and 31% in centralized learning setups upon implementing sampling techniques. The maximum difference between F-1 scores is about 3%, while only a 1% difference in the recall. The 3-layer multi-perceptron model is the best performing model in the diabetic neuropathy cohort. Oversampling improved recall by 10% for federated learning vs. centralized learning, while equivalent recall values were obtained for both. F-1 measure improved by 2% and equivalent precision measures were also obtained for federated learning in this scenario. Among the three cohorts, federated learning with sampling performed consistently well in the diabetic neuropathy cohort.
Table 5:
Performance metrics (F-1 score, Precision, and Recall) for centralized learning and federated learning, obtained from three machine learning models: logistic regression, 2-layer multi-perceptron, 3-layer multi-perceptron for the diabetic neuropathy cohort.
| Federated Learning | Centralized Learning | ||||||
| Model | Method | F1 | Prec | Recall | F1 | Prec | Recall |
| LR | Oversample | 0.63 | 0.52 | 0.87 | 0.65 | 0.53 | 0.87 |
| LR | Undersample | 0.64 | 0.54 | 0.86 | 0.65 | 0.53 | 0.87 |
| LR | No Balancing | 0.57 | 0.7 | 0.52 | 0.64 | 0.68 | 0.61 |
| MLP (10,1) | Oversample | 0.65 | 0.54 | 0.86 | 0.64 | 0.53 | 0.85 |
| MLP (10,1) | Undersample | 0.64 | 0.53 | 0.85 | 0.67 | 0.54 | 0.85 |
| MLP (10,1) | No Balancing | 0.6 | 0.65 | 0.61 | 0.58 | 0.69 | 0.54 |
| MLP(10,10,1) | Oversample | 0.64 | 0.54 | 0.84 | 0.62 | 0.55 | 0.74 |
| MLP(10,10,1) | Undersample | 0.64 | 0.53 | 0.85 | 0.64 | 0.54 | 0.84 |
| MLP(10,10,1) | No Balancing | 0.63 | 0.64 | 0.66 | 0.58 | 0.69 | 0.55 |
Discussion
The use of federated learning can bring numerous opportunities to investigate rare clinical events by building decentralized models without exchanging direct raw data. In this paper, we developed a decentralized privacy-protected predictive classifier that can successfully predict diabetes-related complications such as retinopathy, neuropathy, and nephropathy. The federated architecture shares a common global model with the healthcare systems, which utilize their electronic health records to train locally. Updates from the local models are aggregated later to update the global model. In this process, no personal patient identifiers are shared. We used the Health Facts database to demonstrate the process, which is an information-enriched database for patient records. We used the unique identifiers for healthcare systems to partition the data into real federated data sets. We applied class balancing sampling techniques to address the challenge of low class-ratios of cases to non-cases. In all our experiments, federated learning showed comparable performance to the gold-standard of centralized learning.
Moreover, federated learning with 3-layer multi-perceptron model performed consistently better than its centralized counterparts in the diabetic neuropathy cohort. Interestingly, this cohort had fewer class-imbalance issues, hence greater number of cases of neuropathy in the dataset, which accounts for the consistent performance of federated learning in this case. Also, it is evident that applying the class balancing techniques reduced the gap between the measures of federated and centralized learning across all the cohorts. Thus, federated learning models are useful for the healthcare systems where data sharing is a major barrier for building machine-learning based clinical decision support systems.
Conclusion and Future Scope
In conclusion, our results are consistent with the prior and current implementation of federated learning in healthcare domain9,23,24. Currently, our model architecture is limited to multilayer perceptron, whereas logistic regression is also a special case for that. We plan to extend our federated learning architecture to include more classifier algorithms such as support vector machines, decision trees, and random forest for future work. The major limitation of the analysis is the use of a relational database, which leads to under-representation of the challenges in the federated learning process in a real setting. Another key challenge is data inconsistencies, incompleteness, and lack of standardization. As future goals, we plan to use health systems that are already partitioned for better demonstration. Furthermore, we will explore other averaging techniques, optimal methods to address class-imbalance issues, better approaches for hyperparameter tuning of the global model, and extract other important patient-related factors from other data tables to continue our efforts building a more reliable and robust federated predictive model for diabetes-related complications. Our future work will also involve exploring and comparing other privacy-protected learning methods with federated learning.
References
- 1.Young B. A. Diabetes complications severity index and risk of mortality, hospitalization, and healthcare utilization. Am. J. Manag. Care. 2008;14:15–24. [PMC free article] [PubMed] [Google Scholar]
- 2.Deshpande A. D, Harris-Hayes M, Schootman M. Epidemiology of Diabetes and Diabetes-Related Complications. Phys. Ther. 2008;88:1254–1264. doi: 10.2522/ptj.20080020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.National Diabetes Statistics Report 2020. Estimates of diabetes and its burden in the United States. 2020 [Google Scholar]
- 4.Gregg E. W., Hora I, Benoit S. R. Resurgence in Diabetes-Related Complications. JAMA - Journal of the American Medical Association. 2019;321:1867–1868. doi: 10.1001/jama.2019.3471. [DOI] [PubMed] [Google Scholar]
- 5.Xiao C, Choi E, Sun J. Opportunities and challenges in developing deep learning models using electronic health records data: A systematic review. Journal of the American Medical Informatics Association. 2018;vol. 25:1419–1428. doi: 10.1093/jamia/ocy068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brisimi T. S. Federated learning of predictive models from federated Electronic Health Records. Int. J. Med. Inform. 2018;112:59–67. doi: 10.1016/j.ijmedinf.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zerka F. Systematic Review of Privacy-Preserving Distributed Machine Learning From Federated Databases in Health Care. JCO Clin. Cancer Informatics. 2020;4:184–200. doi: 10.1200/CCI.19.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McMahan B., Moore E., Ramage D., Hampson S., Arcas B. A. Artificial Intelligence and Statistics. PMLR; 2017. Communication-Efficient Learning of Deep Networks from Decentralized Data; pp. 1273–1282. [Google Scholar]
- 9.Predd J. B., Kulkarni S. R, Poor H. V. IEEE Signal Processing Magazine. vol. 23. Institute of Electrical and Electronics Engineers, Inc.; 2006. Distributed learning in wireless sensor networks; pp. 56–69. [Google Scholar]
- 10.Ramos-Pollan R. Grid infrastructures for developing mammography CAD systems. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. IEEE Eng. Med. Biol. Soc. Annu. Int. Conf. 2010;2010:3467–3470. doi: 10.1109/IEMBS.2010.5627832. [DOI] [PubMed] [Google Scholar]
- 11.Huang L. Patient clustering improves efficiency of federated machine learning to predict mortality and hospital stay time using distributed electronic medical records. J. Biomed. Inform. 2019;99:103291. doi: 10.1016/j.jbi.2019.103291. [DOI] [PubMed] [Google Scholar]
- 12.Deist T. M. Infrastructure and distributed learning methodology for privacy-preserving multi-centric rapid learning health care: euroCAT. Clin. Transl. Radiat. Oncol. 2017;4:24–31. doi: 10.1016/j.ctro.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choudhury O. Predicting Adverse Drug Reactions on Distributed Health Data using Federated Learning. AMIA Annu. Symp. Proceedings. 2019:313–322. [PMC free article] [PubMed] [Google Scholar]
- 14.Mangold P. A Decentralized Framework for Biostatistics and Privacy Concerns. Stud. Health Technol. Inform. 2020;275:137–141. doi: 10.3233/SHTI200710. [DOI] [PubMed] [Google Scholar]
- 15.Lee G. H, Shin S.-Y. Federated Learning on Clinical Benchmark Data: Performance Assessment. J. Med. Internet Res. 2020;22:e20891. doi: 10.2196/20891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X. Multi-site fMRI analysis using privacy-preserving federated learning and domain adaptation: ABIDE results. Med. Image Anal. 2020;65:101765. doi: 10.1016/j.media.2020.101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dagliati A. Machine Learning Methods to Predict Diabetes Complications. J. Diabetes Sci. Technol. 2018:295–302. doi: 10.1177/1932296817706375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogunyemi O, Kermah D. Machine Learning Approaches for Detecting Diabetic Retinopathy from Clinical and Public Health Records. AMIA Annu. Symp. Proceedings. 2015;2015:983–990. [PMC free article] [PubMed] [Google Scholar]
- 19.Nichols G. A. Construction of a multisite datalink using electronic health records for the identification, surveillance, prevention, and management of diabetes mellitus: The SUPREME-DM project. Prev. Chronic Dis. 2012;9 doi: 10.5888/pcd9.110311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nanayakkara N. Age, age at diagnosis and diabetes duration are all associated with vascular complications in type 2 diabetes. J. Diabetes Complications. 2018;32:279–290. doi: 10.1016/j.jdiacomp.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 21.Zoungas S. Impact of age, age at diagnosis and duration of diabetes on the risk of macrovascular and microvascular complications and death in type 2 diabetes. Diabetologia. 2014;57:2465–2474. doi: 10.1007/s00125-014-3369-7. [DOI] [PubMed] [Google Scholar]
- 22.Ramanathan amnath S. Correlation of duration, hypertension and glycemic control with microvascular complications of diabetes mellitus at a tertiary care hospital. Integr. Mol. Med. 2017;4 [Google Scholar]
- 23.Chang K. Distributed deep learning networks among institutions for medical imaging. J. Am. Med. Informatics Assoc. 2018;25:945–954. doi: 10.1093/jamia/ocy017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McClure P. Distributed Weight Consolidation: A Brain Segmentation Case Study. Adv. Neural Inf. Process. Syst. 2018;2018-Decem:4093–4103. [PMC free article] [PubMed] [Google Scholar]