Abstract
Clinical guidelines integrate latest evidence to support clinical decision-making. As new research findings are published at an increasing rate, it would be helpful to detect when such results disagree with current guideline recommendations. In this work, we describe a software system for the automatic identification of disagreement between clinical guidelines and published research. A critical feature of the system is the extraction and cross-lingual normalization of information through natural language processing. The initial version focuses on the detection of cancer treatments in clinical trial reports that are not addressed in oncology guidelines. We evaluate the relevance of trials retrieved by our system retrospectively by comparison with historic guideline updates and also prospectively through manual evaluation by guideline experts. The system improves precision over state-of-the-art literature research strategies while maintaining near-total recall. Detailed error analysis highlights challenges for fine-grained clinical information extraction, in particular when extracting population definitions for tumor-agnostic therapies.
Introduction
With the amount of published medical evidence steadily growing, it becomes increasingly challenging for practitioners to keep up with the latest developments in their field.1 Clinical practice guidelines (CPGs) are designed to summarize the currently available evidence regarding specific clinical questions and provide recommendations based on welldefined and methodologically sound criteria2. Despite advances in the development of living guidelines,3 even the most recent CPGs are ultimately static documents in a constantly evolving landscape of new evidence in the form of primary research articles, e.g., reports of randomized controlled trials (RCTs), or even unpublished results.
Finding and ranking relevant medical evidence is a well-investigated information retrieval problem. Medical literature search engines allow users to apply fine-grained search filters, based on string patterns as well as manually or automatically derived metadata, such as Medical Subject Headings (MeSH). In addition, a ranking of most relevant articles given a user query is desirable, which can be based on additional intrinsic properties of the document. In the case of a clinical trial report, this could be the study design or sample size.
While a growing amount of hand-curated and automatically derived semantic information is available for medical information retrieval, current software systems do not take into account the relationship of new evidence to the currently established CPGs, in particular as their applicability depends on the location of the user. Prospectively, new evidence may disagree with the statements in these CPGs, e.g., when an RCT presents new results that are not (yet) accounted for.
In the remainder of this work, we will refer to such evidence as controversial, which shall broadly incorporate any kind of disagreement with current CPGs. This information is relevant to a variety of audiences, for instance:
Maintainers of CPGs, who wish to identify update signals,4 with the potential to necessitate a CPG update
Readers of CPGs, who need to verify whether a CPG still reflects the latest evidence
Specialists interested in new treatments beyond CPG recommendations, e.g., for specific subgroups of patients, as it is common in precision medicine
In this work, we propose an automatic approach to identify such controversial evidence. The system is based on metadata automatically derived via natural language processing (NLP) from scientific articles and CPGs from clinical guideline repositories.
Due to the country-specific nature of CPGs, they are typically published in their respective national language. Therefore, the underlying NLP problems are inherently multilingual. We address this issue by automatically mapping the extracted information to the Unified Medical Language System (UMLS),5 thereby establishing a novel link between primary and synthesized evidence across languages.
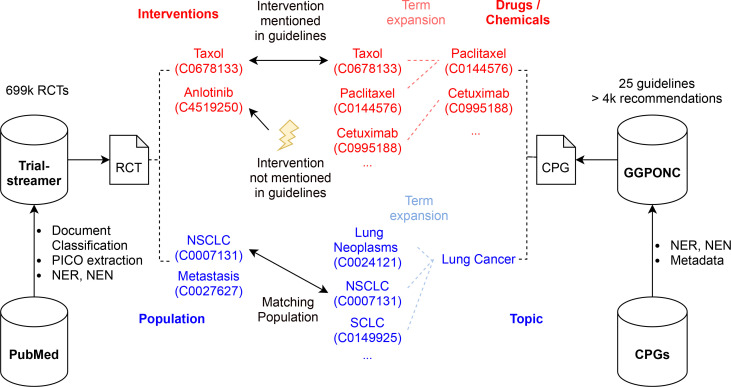
Different types of disagreement can be defined over such information using transparent rules, which enables users to reason about the validity of the identified disagreement. We demonstrate the feasibility of the proposed approach by implementing and evaluating the detection of a highly relevant form of disagreement: RCTs mentioning drug interventions not included in current CPGs, as outlined in Figure 1. To this end, we leverage two recently published resources, the Trialstreamer database of RCTs6 and the German Guideline Program in Oncology NLP Corpus (GGPONC).7
Figure 1:
Overview of our approach to detect RCTs with drug interventions not covered by CPGs. Here, we consider the German CPGs in oncology in GGPONC. CPGs and RCTs from Trialstreamer are matched based on the CPG topic and the extracted population concepts of the RCT. The set of intervention concepts is compared to the drugs / chemicals mentioned in GGPONC to flag RCTs with interventions currently unmentioned in the CPG as controversial. Term expansion is performed using MeSH and SNOMED CT to account for synonymy and hyponymy.
A prototype of the system is publicly available: https://we.analyzegenomes.com/nge.
The remainder of this work is structured as follows: in the following section, we set our approach in the context of related work, followed by a description of the used datasets and components of our system. We proceed by presenting the output of the system for specific clinical indications and the evaluation in an information retrieval setting. Results are interpreted and limitations discussed thereafter, followed by a conclusion and outlook.
Related Work
Information retrieval of the medical research literature has been studied extensively in the last decades.8 A multitude of approaches have been proposed to filter and rank by relevancy collections of RCT reports and other medical publications.
PubMed is probably the most widely used medical search engine and allows fine-grained filtering based on text matches, MeSH terms and other metadata. Recently, PubMed has introduced a new Best Match sorting option, taking into account different metrics and user queries.9 The Trip database allows more advanced search options based on the Population-Intervention-Control-Outcome (PICO) framework and a ranking of articles by relevancy, such as latest and greatest.10 There is a wide selection of systems that enrich PubMed with additional semantic information and employ them for document retrieval, for instance using automatically extracted biomolecular entities and relations.11–13
In practice, literature search strategies for initial CPG creation and updates are based on elaborate Boolean search queries in different literature databases, carefully hand-tailored for the specific clinical questions covered by the CPGs.14 These queries can be continuously applied to identify update signals, i.e., research results that would necessitate an immediate update or amendment to a CPG.4 Reviewing of the results and subsequent data extraction is done manually.
RobotReviewer is a system that automatically extracts structured information from RCT reports and predicts the risk of bias (RoB) of an RCT according the Cochrane RoB tool.15 Recently, Marshall et al. (2020) released Trialstreamer, combining the predictions of RobotReviewer and other components leveraging machine learning (ML) into a living database of RCTs along with their extracted metadata.6 Based on this metadata, the Trialstreamer website allows to filter RCTs by their PICO elements and prioritize large and high-quality RCTs.
To the best of our knowledge, there is currently no system that combines information from published research results and CPGs to expose additional relevancy criteria for information retrieval.
Datasets
Trialstreamer is a publicly available, regularly updated database of RCTs derived from automatically screened PubMed articles and the WHO International Clinical Trials Registry Platform.6 For this work, we focus on PubMed articles, which are included in Trialstreamer according to an automatic classification of articles that describe RCTs in humans. From the corresponding abstracts, structured metadata is extracted using different ML- and rule-based NLP methods.
For our system, we use the extracted sample size, PICO spans and PICO concepts. For the evaluation, we combine the base version of Trialstreamer and all PubMed updates up to December 28th, 2020, resulting in a dataset of around 699k RCTs. In the live version of the system, updates to Trialstreamer are automatically integrated on a weekly basis.
GGPONC is a metadata-enhanced text corpus based on German CPGs in oncology, currently consisting of 25 CPGs with around 1.3M tokens and more than 4k recommendations covering a diverse set of indications.7 It is currently one of the largest publicly available text corpora of CPGs in general and of German medical text in particular. GGPONC has been automatically annotated with entity classes from different UMLS semantic types, for instance, Disorders, Procedures, Chemicals & Drugs, and Anatomy from the German subset of the UMLS. Gold-standard annotations from human experts are available for around half of the corpus.
In addition, GGPONC provides a variety of metadata for individual recommendations, e.g., timestamps, which allow us to simulate past guideline versions by excluding elements introduced after a certain point in time. GGPONC is freely available upon request.† New CPG versions are automatically integrated into the system upon release.
Named Entity Recognition and Normalization
To find drug mentions in GGPONC, we use the same dictionary-based JCORE (i.e., UIMA-based) pipeline as in Borchert et al. (2020).7, 16 In contrast to our earlier work, we configured the pipeline with a larger dictionary of substances derived from the UMLS (version 2020AB) using the JUFIT tool (v1.1).17 The goal of using a larger dictionary is to improve recall (sensitivity), at the expected cost of precision. Therefore, we consider all preferred English terms of the UMLS semantic type Chemicals & Drugs in addition to the German terms, yielding a dictionary of around 1.26M entries compared to only 34.550 from the German UMLS subset. With this extended dictionary, recall for the recognition of Chemicals & Drugs measured on the human-annotated subset of GGPONC increases from 0.600 to 0.788, whereas precision decreases from 0.917 to 0.520. Due to the nature of this dictionary-based approach, all extracted entities are already linked to UMLS CUIs (concept unique identifiers).
The Trialstreamer database already contains automatically derived metadata relating to PICO elements. Entities within PICO spans are linked to UMLS CUIs using a re-implementation of Metamap Lite.18 Recall for PICO concept extraction under relaxed comparison reported by Marshall et al. (2020) is relatively high (0.85 for intervention and 0.78 for population), while precision is rather low (0.57 for intervention and 0.30 for population).6
Identifying Controversial RCTs in Trialstreamer
The current version of our system enables the detection of controversial RCTs in Trialstreamer by linking the extracted PICO concepts to CPGs from GGPONC as outlined in Figure 1. In the following, we describe the rule-based algorithm that detects RCTs concerning drugs unmentioned in current CPGs. Other types of rules can be implemented similarly.
For all RCTs in Trialstreamer, we identify those with intervention CUIs contained in the large UMLS-based dictionary of drugs and chemicals described earlier and retain only such trials where at least one intervention is a drug or chemical, as opposed to other kinds of treatments. For each guideline topic, e.g., lung cancer, we perform a term expansion step to identify synonyms and hyponyms, such as pulmonary neoplasm, lung adenocarcinomas, or bronchogenic carcinoma, which we refer to as topic concepts. This expansion was performed based on the MeSH and SNOMED CT terminologies using PyMedTermino19 and the entity linking module from scispaCy.20
To obtain all potentially relevant RCTs for an existing CPG topic, we filter Trialstreamer by the automatically extracted population CUIs that overlap with the set of topic concepts. For each of these topic-related RCTs, we determine whether at least one of the intervention CUIs is not contained in the complete set of drug CUIs extracted from the corresponding CPG at a particular time point. Such RCTs are flagged as controversial. Term expansion is also performed on the level of interventions, to account for mentions of drug classes, i.e., an RCT mentioning a more general term (e.g., protein kinase inhibitor) should not be considered controversial if a narrower term (e.g., afatinib) is already included in the CPG. This step also accounts for synonymy due to the use of experimental, non-proprietary, and trade names of drugs.
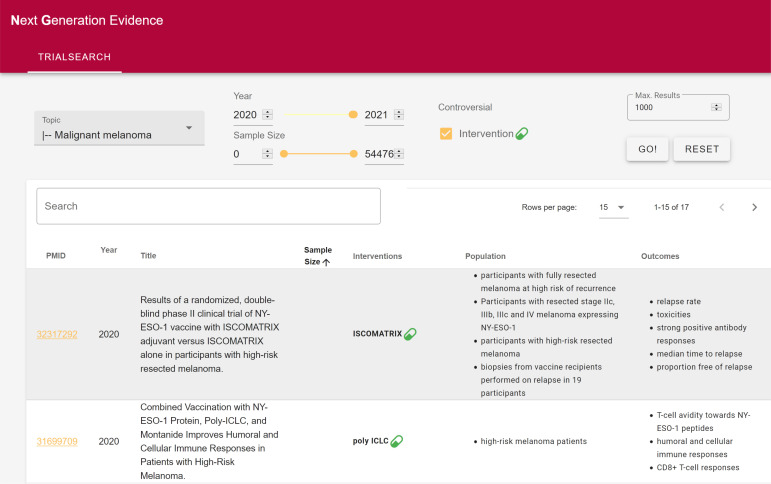
This simple rule over the extracted metadata is completely transparent to the user. Moreover, it is easily extendable and adaptable to different requirements, e.g., when a different trade-off of precision and recall is desired, and can be easily implemented within medical search engines. We provide a prototypical user interface, displayed in Figure 2 that allows users to browse RCTs per CPG topic and to display only controversial results, in addition to filtering based on the publication year, sample size, and free-text matches. It should be noted that even simply filtering by sample size is not available in typical literature search engines and only enabled by the NLP-derived information in Trialstreamer.
Figure 2:
A screenshot of our user interface configured with the parameters used in the prospective evaluation. The user selects a topic (on the top left: Malignant melanoma), applies filters based on metadata such as publication year and sample size and can decide to only view controversial trials (on the top right).
Evaluation
Retrospective Scenario Using the temporal metadata in GGPONC, we simulate CPG versions at a past point in time by manually identifying and removing all drug mentions from the CPGs that were first included after that point in time. Two CPGs and simulated versions are investigated: a 2010 version of the lung cancer (LC) CPG, which has received an update in 2018, as well as a 2017 version of the renal cell carcinoma (RCC) CPG, updated in 2020. For these two topics, we process all RCTs from Trialstreamer and flag them as controversial if at least one of their interventions is a drug not yet included in the historic CPG.
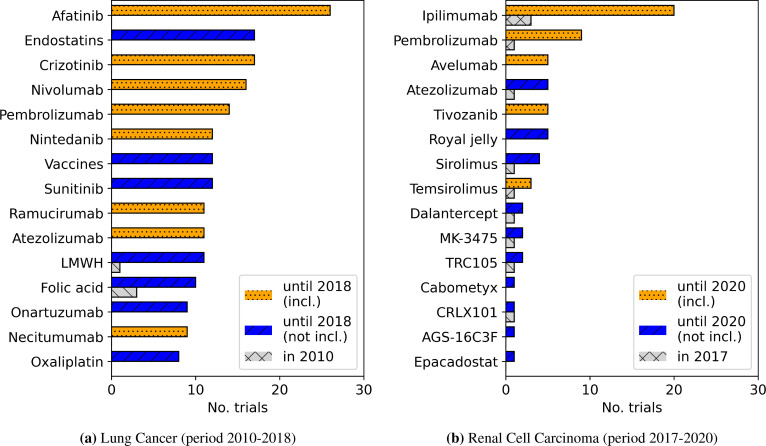
In Figure 3, we show the drugs with highest numbers RCTs identified in this way. At the points in time the CPGs were issued, there are only very few interventions mentioned in RCTs that are going to be included in future CPG updates — the respective CPG can be considered up-to-date. In the following time preceding the next update, evidence relating to the most commonly mentioned drugs has accumulated. However, for both CPGs, there is a noticeable tail of drugs investigated in clinical trials that do not end up in the respective CPG.
Figure 3:
Development of the number of RCTs for the top 15 drug names not mentioned in the previous CPG for two topics and update intervals. We show the initial number of RCTs in the year of the previous CPG update as well as the cumulated number of RCTs until the current update. Drugs marked as incl. have eventually been included in this last update, whereas drugs marked as not incl. are (still) not mentioned in the current CPG version.
Retrospective Evaluation of Retrieval Performance In the considered scenario, where our goal is to find interventions not currently part of CPGs but with the potential to become CPG-relevant in the future, a typical requirement is near-perfect recall, generally at the cost of precision.21
Using the controversial flag as an additional filtering criterion for retrieving RCTs from PubMed, we evaluate retrieval performance in terms of (1) precision with respect to retrieved documents actually describing an RCT with a newly CPG-relevant drug, as well as (2) recall with respect to the proportion of retrieved drugs from the set newly CPG-relevant drugs. The results are given in Table 1. Note that retrospective assessment of recall in terms of documents is actually not possible based on our data, since a particular CPG update cannot be reliably attributed to a single RCT. This would require manual reconstruction of the literature screening process, which might be partially based on existing systematic reviews not included in Trialstreamer.
Table 1:
Retrospective evaluation of retrieval performance under different filters evaluated against past CPG updates. The baseline strategies are filters by Year (Y), by drug interventions only (IDrug), by matching the study population to any sub type of Cancer (PCancer), or to the respective CPG topic (PTopic). We compare these to advanced filters based on the sample size of n from Trialstreamer, and the controversial tag from our method (Controv.). We identify all RCTs in Trialstreamer matched by these criteria over the period between two consecutive updates and report precision (Pr.) in terms of documents and recall (Rec.) in terms of retrieved drugs newly mentioned in the current CPG version. Very high precision can be obtained by our method based on the controversial tag, in particular in combination with a filter based on the sample size (n > 100), while maintaining the same recall as the most specific baseline strategy.
| Lung Cancer (2010-2018) | Renal Cell Carcinoma (2017-2020) | |||||||
|---|---|---|---|---|---|---|---|---|
| Trial Filters | Trials | Interventions | Trials | Interventions | ||||
| Results | Pr. | Results | Rec. | Results | Pr. | Results | Rec. | |
| Baselines | ||||||||
| Y | 271820 | .001 | 34480 | 1.0 | 156244 | .002 | 29914 | 1.0 |
| Y + IDrug | 134188 | .003 | 9058 | 1.0 | 70205 | .004 | 7006 | 1.0 |
| Y + IDrug + PCancer | 11562 | .023 | 2217 | 1.0 | 6731 | .034 | 1802 | 1.0 |
| Y + IDrug + PTopic | 1344 | .089 | 467 | .909 | 140 | .279 | 45 | 1.0 |
| Advanced Filters | ||||||||
| Y + IDrug + PTopic+ n > 100 | 563 | .096 | 244 | .909 | 49 | .306 | 23 | 1.0 |
| Y + IDrug + PTopic+ Controv. | 612 | .194 | 343 | .909 | 113 | .619 | 28 | 1.0 |
| Y + IDrug + PTopic+ n > 100 + Controv. | 235 | .230 | 165 | .909 | 20 | .750 | 13 | 1.0 |
As baselines, we use filters implemented using the extracted PICO information in Trialstreamer, in the spirit of Boolean search queries typically employed when searching PubMed.14 In addition, we consider a filter that is based on the extracted number of participants (n), as a CPG update might be based on a single sufficiently powered RCT.
As expected, filtering by the population attributes per topic results in an increase of precision, a strategy commonly employed when constructing Boolean search queries. Our proposed additional filters based on sample size (here n > 100 as an example) and the controversial flag are able to enhance precision substantially, in particular when combined, while maintaining the same recall as the baseline population filtering strategy PTopic. In the case of the RCC CPG, precision of the combined filter is as high as .750, i.e., three in four retrieved RCTs concern drugs that will be mentioned in the next version of the CPG.
Prospective Evaluation In a real-world deployment, we want our system to detect controversial trials based on the current CPG versions, in contrast to historic versions used in the retrospective evaluation. Therefore, to evaluate the system prospectively, we consider the current versions of the German CPGs on Ovarian Cancer (OC) and Malignant Melanoma (MM), both with a last update in the early 2020, and identify controversial trials with respect to these topics in 2020. As the interventions relevant for the next CPG version are still unknown in the scenario, the output of the system was manually evaluated by guideline experts from the German Cancer Society.
The manual evaluation results are shown in Table 2. 41 RCTs are found by our system for Ovarian Cancer and 21 for Malignant Melanoma in, resulting in a total of 62 articles from 2020 used in this evaluation. 23 of these have been categorized to have direct potential to be relevant for future CPG versions, corresponding to a precision of .355, when applying the same standard as in the retrospective evaluation. When we also consider early-stage RCTs as relevant and also ones reporting negative results, precision increases to .645. In effect, only 35.5% of results are actually clearly irrelevant, because of errors in downstream components of the system. This number is very low compared to usual Boolean searches in PubMed. We will analyze the different error conditions in the next section.
Table 2:
Prospective evaluation for the system configurations based on the current German CPGs on Ovarian Cancer and Malignant Melanoma, both from early 2020. For all identified interventions, we report the expert decision regarding the potential relevance for a future CPG update. We also denote if an RCT does not meet the criteria for CPG inclusion due to an early phase or negative results. If there is more than one publication per intervention, this number is denoted in brackets. Actual false positives are designated in the last column, with the error type distinguished by origin between ontology incompleteness (O) and errors in Trialstreamer: RCT document classification errors (TS-D), intervention extraction errors (TS-I), and population extraction errors (TS-P).
| Topic | Potentially Relevant | Early Stage / Small Study | Negative Results / Study Withdrawn | False Positive (Error Type) | |
|---|---|---|---|---|---|
| Ovarian | Oregovomab (3) | Cabazitaxel | Bio. resp. modifiers | Doxorub. lip. (6) | O |
| Cancer | Durvalumab (2)† Atezolizumab† Cediranib† ProFast Cremophor EL Local anesthetic* Ropivacaine* Apatinib** Lobaplatin** Nedaplatin** |
Fosbretabulin MEDI4736 Metformin Selinexor Taxane |
Guadecitabine Ipilimumab Nivolumab Pimasertib SAR245409 Sorafenib |
Olaparib Denosumab BIDS Epirubicin Fluorouracil Irinotecan Oxaliplatin Prednisone |
O TS-D TS-I TS-P TS-P TS-P TS-P TS-P |
| Malignant Melanoma | Atezolizumab (2) Binimetinib (2) Calcium-EP Encorafenib Bupropion* Varenicline* |
EMD 273063 Selumetinib Peptide vaccines Poly ICLC |
ISCOMATRIX | Dermat. agents Interferon alfa-2b BIDS Sodium DCA Honey prep. Atezolizumab 125 iodine Ranibizumab |
O O TS-I TS-I TS-I TS-P TS-P TS-P |
| # Trials | 22 (35.5 %) | 10 (16.1 %) | 8 (12.9%) | 22 (35.5%) | |
| 40 (64.5%) | 22 (35.5%) | ||||
Notes: †Description of the trial design only. *Relevant for the population, but not the underlying clinical questions used during CPG development. **No or unclear relevance in the EU / Germany.
Error Analysis
Phase I/II Trials, Negative Results, and Study Withdrawal Phase I or phase II trials, e.g., for TRC105 in RCC or cabazitaxel in OC, are not immediately relevant for CPG inclusion. Negative results in such phase I or II trials or study withdrawal occur frequently. These types of results have to be considered as false positives with respect to the goal of identifying CPG update signals, and are counted as such in the retrospective evaluation. However, they can be of great interest to other users of the system. Being able to distinguish the particular type and design of an RCT automatically would be a useful feature in a future version of the system, but would require a more elaborate semantic understanding of study contents through NLP. Negative results for drugs included in current CPGs would constitute a different type of disagreement that we would like to be able to detect.
Misalignment of Topics and Populations When the scope of a CPG is not fully defined by the population alone, some results might be relevant for the topic but not (yet) covered as a clinical question in the CPG. For instance, supportive therapy with royal jelly or the use of local anesthetics might be relevant for the considered population, but covered in a separate CPG on such cross-cutting concerns. Addressing this problem comprehensively would require a complete formal specification of the clinical questions underlying each CPG.
False Negatives For this type of error, drugs would be relevant, e.g., for future CPG versions, but we retrieve no RCTs mentioning them (lower recall). In the retrospective evaluation, recall is < 1 only for LC after applying a population filter, because the drug dabrafenib, newly mentioned in the 2018 guideline, was missed. Dabrafenib could not be found in Trialstreamer after filtering by population, as all RCTs on the drug mention more general population terms, such as solid tumors, which is not captured by our term expansion strategy.
False Positives For this type of error, RCTs are retrieved that mention drug names which are not included in the future CPG version (lower precision). Sources of false positives, by example of the drugs depicted in Figure 3 and Table 2, are:
Ontology incompleteness (O): if no mapping of a concept exists in the UMLS for the language of the CPG (here German), it would not be extracted from the current CPG and therefore not be considered as already mentioned. Incomplete terminologies in the UMLS can also be a source of false negatives, e.g., when novel drugs have not yet been curated. Such a false negative, however, did at least not occur in the retrospective evaluation.
Document classification errors (TS-D): errors introduced due to a misclassification of a publication as an RCT. In one case, a narrative review on denosumab has been misclassified as an RCT.
Intervention extraction errors (TS-I): for example, rapamycin is extracted as an intervention in Trialstreamer and normalized to sirolimus, although it only occurs as part of a larger entity mammalian Target of rapamycin. Given the high reported accuracy of Trialstreamer with respect to the extraction of interventions,6 this error type is only encountered occasionally. This class of errors includes incorrect abbreviation expansion, as in the case of DCA, which was considered as a chemical but supposed to mean decision curve analysis in the RCT.
Population extraction errors (TS-P): in some cases, the study population has been extracted incorrectly in Trialstreamer. These problems can be rather obvious, e.g., for an RCT with breast cancer patients performed at the Center for Familial Breast and Ovarian Cancer. Other instances are more subtle, e.g., when the population malignant melanoma is assigned to an RCT with choroidal melanoma patients in Trialstreamer
Discussion
Limitations Our detailed error analysis highlights limitations of the system as currently implemented. The initial population matching strategy is currently unable to account for drugs like dabrafenib, whose target population is based on molecular characteristics rather than specific tumor entities, i.e., tumor-agnostic therapies.
In general, the performance of our system strongly relies on various downstream components, in particular the ML-based components used to populate Trialstreamer as well as the dictionary-based information extraction from German CPGs. Reliance on currency and completeness of the UMLS is problematic in particular for new concepts and low-resource language communities. While the NER step could be solved using an ML-based approach, matching of entities across languages without access to a multilingual ontology will be challenging. It further needs to be investigated if truly controversial treatments will be detected by current NER solutions, as they might not be adequately represented in terminologies or training data.
An assessment of recall and an error analysis of false negatives in the prospective setting is still missing, as it would require a screening of all articles to account for false negatives in the classification of RCTs by Trialstreamer as well as incompleteness of the UMLS with respect to novel drugs. In a future evaluation, we will apply the system in parallel to an ongoing major update of the German LC CPG, including a full literature screening for relevant RCTs.
The current binary scheme, which flags each RCT as either controversial or not, could be enhanced by softer relevancy criteria. These should account for the inherently probabilistic outputs of downstream NLP components and incorporate relationships to multiple, potentially disagreeing CPGs.22 Using such criteria would enable the implementation of a true ranking of results and the use of ranking-based evaluation criteria, such as P@K or AUC.
Other Types of Evidence While RCTs are the gold standard in evidence-based medicine, they are by far not the only source of medical evidence. For many uses cases, interventions investigated in ongoing RCTs or with unreported results (publication bias) will be relevant and therefore clinical trial registers, such as ClinicalTrials.gov should be considered. This information will be easier to incorporate in the system, as data in these registers is usually available at least in a semi-structured format, and already included in Trialstreamer. Other types of published research, such as case reports, are not included in Trialstreamer and will require custom NLP solutions for information extraction and normalization.
In addition, results which were not published in medical journals and other types of so-called grey literature can be of interest to some users.23 Incorporating these sources of evidence is likely to yield much more controversial information compared to published RCTs. However, these sources will contain a substantial proportion of completely irrelevant information. They are also expected to be more challenging to process via NLP due to large linguistic heterogeneity.
Other Controversial Results There are many types of controversial evidence not yet covered by our methodology. A different type of intervention would be addressed by the UMLS semantic type Procedures, e.g., for radiation therapy or other nonpharmaceutical interventions. Here, we expect cross-lingual matching to be more challenging compared to drug names, which are mostly single-token proper nouns.
Other types of disagreement could occur on the level of outcomes, where new results regarding efficacy and safety of a CPG-recommended drug can influence decision-making. Extraction quality with respect to outcomes is lowest for all PICO elements extracted in Trialstreamer,6 so a detailed investigation and improved NLP solutions are necessary.
Conclusion and Future Work
In this work, we presented a system that allows to detect controversial trials, i.e., ones that disagree with current clinical practice guidelines. The system relies on the NLP-based extraction of metadata from RCTs and CPGs and simple rules to identify disagreement based on this data. While the evaluation results are encouraging in an information retrieval setting, there is ample opportunity for improvement.
With more and more drugs targeting specific genetic alterations, this information should be incorporated into the population matching strategy, in particular for other types of users, e.g., participants of molecular tumor boards. Information extraction of molecular entities has been studied extensively by the BioNLP community,24 albeit rarely in the domain of RCTs. Such an extension would make the system also useful for curators of clinical evidence in precision oncology knowledge bases.25
To enable the implementation of rules for other types of disagreements, future work will focus on the inclusion of information regarding outcomes within the PICO framework as well as different published and non-published types of evidence. To further improve system performance in the investigated multi-lingual setting, we will need to improve clinical NLP methods for languages other than English, which are still restricted by the shortage of publicly available research datasets7 and, in comparison to the English language community, a lack of modern ML-based information extraction solutions.
A prototype of our system is online available for evaluation at: https://we.analyzegenomes.com/nge. We will continuously improve the prototype to allow users to find other types of disagreement, incorporate different kinds of evidence and extend the scope to other CPGs from other medical fields and countries.
Acknowledgements
This work was partially supported by a grant of the German Federal Ministry of Research and Education (01ZZ1802).
Footnotes
https://www.leitlinienprogramm-onkologie.de/projekte/ggponc-english/
Figures & Table
References
- 1.Bastian H, Glasziou P, Chalmers I. Seventy-five trials and eleven systematic reviews a day: how will we ever keep up? PLoS med. 2010;7(9) doi: 10.1371/journal.pmed.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shekelle PG, Woolf S, Grimshaw JM, Sch¨nemann HJ, Eccles MP. Developing clinical practice guidelines: reviewing, reporting, and publishing guidelines; updating guidelines; and the emerging issues of enhancing guide-line implementability and accounting for comorbid conditions in guideline development. Implementation Science. 2012;7(1):62. doi: 10.1186/1748-5908-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akl EA, Meerpohl JJ, Elliott J, et al. Living systematic reviews: 4. living guideline recommendations. Journal of Clinical Epidemiology. 2017;91:47–53. doi: 10.1016/j.jclinepi.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Somerfield MR, Bohlke K, Browman GP, et al. Innovations in American Society of Clinical Oncology practice guideline development. Journal of Clinical Oncology. 2016;34(26):3213–3220. doi: 10.1200/JCO.2016.68.3524. [DOI] [PubMed] [Google Scholar]
- 5.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Research. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall IJ, Nye B, Kuiper J, et al. Trialstreamer: A living, automatically updated database of clinical trial reports. Journal of the American Medical Informatics Association. 2020 09;27(12):1903–1912. doi: 10.1093/jamia/ocaa163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borchert F, Lohr C, Modersohn L, et al. Proceedings of the 11th International Workshop on Health Text Mining and Information Analysis. Association for Computational Linguistics; 2020. GGPONC: A Corpus of German Medical Text with Rich Metadata Based on Clinical Practice Guidelines; pp. p. 38–48. Online. [Google Scholar]
- 8.Hersh W. Information retrieval: a health and biomedical perspective. Springer Science & Business Media; 2008. [Google Scholar]
- 9.Fiorini N, Canese K, Starchenko G, et al. Best match: new relevance search for PubMed. PLoS biology. 2018;16(8) doi: 10.1371/journal.pbio.2005343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trip Database Limited Trip. 2020. Accessed: June 16, 2021. http://www.tripdatabase.com/
- 11.Schapranow MP, Kraus M, Perscheid C, Bock C, Liedke F, Plattner H. 2015 IEEE International Conference on Bioin-formatics and Biomedicine (BIBM) IEEE; 2015. The Medical Knowledge Cockpit: Real-time analysis of big medical data enabling precision medicine; pp. p. 770–775. [Google Scholar]
- 12.Faessler E, Hahn U. Proceedings of ACL 2017, System Demonstrations. Vancouver, Canada: Association for Computational Linguistics; 2017. Semedico: A Comprehensive Semantic Search Engine for the Life Sciences; pp. p. 91–96. [Google Scholar]
- 13.Allot A, Peng Y, Wei CH, Lee K, Phan L, Lu Z. LitVar: a semantic search engine for linking genomic variant data in PubMed and PMC. Nucleic acids research. 2018;46(W1):W530–W536. doi: 10.1093/nar/gky355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karimi S, Pohl S, Scholer F, Cavedon L, Zobel J. Boolean versus ranked querying for biomedical systematic reviews. BMC medical informatics and decision making. 2010;10(1):1–20. doi: 10.1186/1472-6947-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall IJ, Kuiper J, Wallace BC. RobotReviewer: evaluation of a system for automatically assessing bias in clinical trials. Journal of the American Medical Informatics Association. 2016;23(1):193–201. doi: 10.1093/jamia/ocv044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn U, Matthies F, Faessler E, Hellrich J. Proceedings of the 10th International Conference on Language Resources and Evaluation (LREC’16) Portorozˇ, Slovenia: 2016. UIMA-Based JCoRe 2.0 Goes GitHub and Maven Central — State-of-the-Art Software Resource Engineering and Distribution of NLP Pipelines; pp. p. 2502–2509. [Google Scholar]
- 17.Hellrich J, Schulz S, Buechel S, Hahn U. AMIA 2015 — Proceedings of the 2015 Annual Symposium of the American Medical Informatics Association. Vol. 2015. San Francisco, USA: Nov 14-18, 2015. JUFIT : a configurable rule engine for filtering and generating new multilingual UMLS terms; pp. p. 604–610. [PMC free article] [PubMed] [Google Scholar]
- 18.Demner-Fushman D, Rogers WJ, Aronson AR. MetaMap Lite: an evaluation of a new Java implementation of MetaMap. Journal of the American Medical Informatics Association. 2017;24(4):841–844. doi: 10.1093/jamia/ocw177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamy JB, Venot A, Duclos C. PyMedTermino: an open-source generic API for advanced terminology services. MIE. 2015. pp. p. 924–928. [PubMed]
- 20.Neumann M, King D, Beltagy I, Ammar W. ScispaCy: Fast and Robust Models for Biomedical Natural Language Processing. Proceedings of the 18th BioNLP Workshop and Shared Task. 2019. pp. p. 319–327.
- 21.Tsafnat G, Glasziou P, Choong MK, Dunn A, Galgani F, Coiera E. Systematic review automation technologies. Systematic reviews. 2014;3(1):1–15. doi: 10.1186/2046-4053-3-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zadrozny W, Hematialam H, Garbayo L. Towards Semantic Modeling of Contradictions and Disagreements: A Case Study of Medical Guidelines. IWCS 2017 — 12th International Conference on Computational Semantics — Short papers. 2017. pp. p. W17–6943.
- 23.Eysenbach J, Tuische G, Diepgen Thomas L. Evaluation of the usefulness of Internet searches to identify unpublished clinical trials for systematic reviews. Medical Informatics and the Internet in Medicine. 2001;26(3):203–218. doi: 10.1080/14639230110075459. [DOI] [PubMed] [Google Scholar]
- 24.Perera N, Dehmer M, Emmert-Streib F. Named entity recognition and relation detection for biomedical information extraction. Frontiers in Cell and Developmental Biology. 2020;8:673. doi: 10.3389/fcell.2020.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borchert F, Mock A, Tomczak A, et al. Knowledge bases and software support for variant interpretation in precision oncology. Briefings in Bioinformatics. 2021;05 doi: 10.1093/bib/bbab246. bbab134. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]