Summary
Most of the protocols to analyze metabolic features of cell populations from different tissues rely on in vitro cell culture conditions. Here, we present a flow-cytometry-based protocol for measuring the respiratory chain function in permeabilized mouse microglia ex vivo. We describe microglial cell isolation, followed by analyzing complex I and II using flow cytometry. This optimized protocol requires a low input of permeabilized cells and can be applied to other ex vivo isolated cells or cells derived from cell cultures.
For complete details on the use and execution of this protocol, please refer to Erny et al. (2021).
Subject areas: Cell Biology, Cell isolation, Flow Cytometry/Mass Cytometry, Immunology, Metabolism, Neuroscience
Graphical abstract
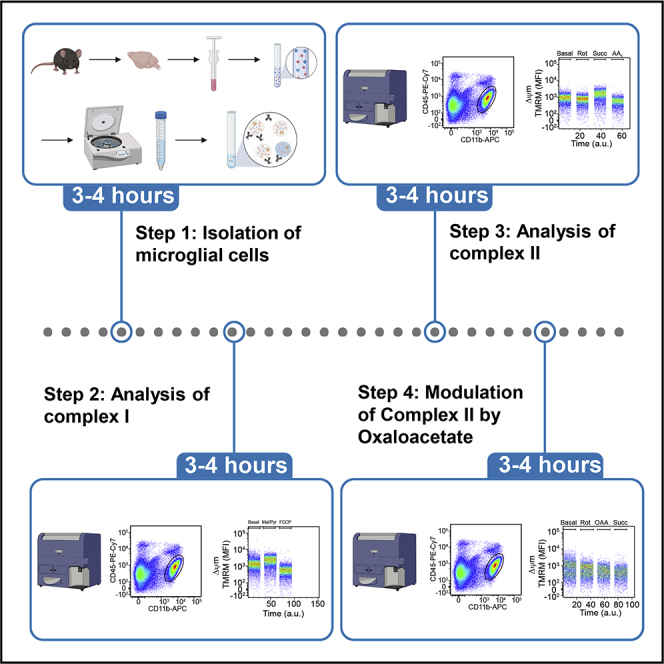
Highlights
-
•
Flow cytometry-based protocol to measure respiratory chain function ex vivo
-
•
Optimized protocol requiring a low input of permeabilized cells
-
•
Analysis of complex I and II in permeabilized mouse microglial cells
-
•
Applicable for all kinds of ex vivo isolated cells or cells derived from cell cultures
Most of the protocols to analyze metabolic features of cell populations from different tissues rely on in vitro cell culture conditions. Here, we present a flow-cytometry-based protocol for measuring the respiratory chain function in permeabilized mouse microglia ex vivo. We describe microglial cell isolation, followed by analyzing complex I and II using flow cytometry. This optimized protocol requires a low input of permeabilized cells and can be applied to other ex vivo isolated cells or cells derived from cell cultures.
Before you begin
For comprehensive functional analysis of mitochondrial function, we describe below a flow cytometry-based protocol which requires a lower cell input compared with other methods such as Seahorse extracellular flux (XF) analysis. In addition, the Seahorse XF approach depends on at least partial culturing conditions over several hours in order to allow e.g., the adherence of the cells to the cell culture wells. Importantly, such culture conditions are known to affect the phenotype of microglia (Gosselin et al., 2014). In the past, most of the data from published studies were generated by the usage of primary microglia cell cultures or microglia-like cell lines (Borst et al., 2018). Therefore, we aimed to establish a comprehensive method for analyzing microglia cells ex vivo within 6–8 h. Furthermore, in this protocol, additional isolation of mitochondria is not needed. After permeabilization of ex vivo isolated microglia cells, the function of specific complexes (C) of the respiratory chain can be investigated. Collectively, this protocol provides robust and detailed profiling of the mitochondrial respiratory chain function, and, after preparative steps, takes only 6–8 h to complete. Notably, this protocol can also be applied for other ex vivo isolated cells or cells derived from cell cultures.
This protocol requires tissue derived from mouse models. Ethical approvals are required prior to starting this procedure. All animal experiments in this study were approved by the Ministry for Nature, Environment and Consumers‘ Protection of the state of Baden-Württemberg and were performed in accordance with the respective national, federal and institutional regulations (G19–02, G19-148 and X16-04A).
Before starting, prepare the necessary buffers and stock solutions for the workflow. Furthermore, label all required conical tubes and FACS tubes.
Prepare Mannitol and Sucrose (MAS) buffer
Timing: 30 min
-
1.
Prepare 1× Mannitol and Sucrose (MAS) buffer (see materials and equipment). Dilute in ddH2O. Adjust the pH to 7.2 with 0.1 M KOH and filter-sterilize the solution. The buffer can be stored for up to 2 months at 4°C.
Prepare MAS-BSA buffer
Timing: 15 min
-
2.
For preparing MAS-BSA add 4 mg BSA per 1 mL of MAS buffer (see materials and equipment).
Prepare MitoTrackerTM Green and TMRM stock solutions
Timing: 15 min
-
3.MitoTrackerTM Green FM:
-
a.Each tube contains 50 μg of lyophilized powder. Before dissolving MitoTrackerTM Green FM (MW: 671.8797 g/mol), bring the powder to room temperature.
-
b.Add 74.4 μL DMSO to a stock concentration of 1 mM. Store at −20°C for up to 12 months.
-
a.
-
4.Tetramethylrhodamine (TMRM):
-
a.Each vial contains 25 mg TMRM. Add 5 mL DMSO to prepare 10 mM TMRM solution.
-
b.Prepare 1 mL 100 μM stock solution by diluting 10 μL TMRM to 990 μL DMSO. Store at −20°C for up to 12 months.
-
a.
Prepare drugs for respiratory chain manipulation
Timing: 1 h
-
5.Drugs for respiratory chain manipulation
-
a.ADP (MW: 501.32) stock solution: 501.32 mg ADP in 10 mL MAS buffer (100 mM). Final concentration 1 mM.
-
b.Pyruvate/malate (MW: 88.06/134.09) stock solution: 440.3 mg pyruvate + 335.2 mg malate in 10 mL MAS buffer (500 mM/250 mM). Final concentration 5 mM/2.5 mM.
-
c.Rotenone (MW: 394.42) stock solution: 5 mg rotenone in 1.267 mL DMSO (10 mM). Working solution (concentration 225 μM): 22.5 μL of the stock solution diluted in 977.5 μL MAS-BSA. Final concentration 1 μM.
-
d.Succinate (MW: 118.09) stock solution: 590 mg succinic acid to 10 mL MAS buffer (500 mM). Final concentration 10 mM.
-
e.Antimycin A (MW: 534.60) stock solution: 25 mg Antimycin A in 1.27 mL DMSO (36 mM). Working stock solution (concentration 2 mM): 16.7 μL of the stock solution diluted in 283.3 μL MAS-BSA). Final concentration 20 μM.
-
f.Carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP; MW: 254.17) stock solution: 10 mg FCCP in 787 μL DMSO (concentration: 50 mM). Working solution (concentration: 100 μM): add 1 μL of the stock solution to 499 μL MAS buffer. Final concentration 1 μM.
-
g.Oxaloacetate (OAA; MW: 132.07) stock solution: 528.28 mg OAA in 10 mL MAS buffer (concentration: 400 mM).
-
a.
Note: Adjust pH of the drugs to 7.2 with KOH. Stock solutions may be stored as aliquots for up to 2 months at –20°C (in the dark since some compounds may be light sensitive).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-mouse CD11b APC conjugated (1:200) | Thermo Fisher Scientific | Cat# 17-0112-83; RRID: AB_469344 |
| Rat anti-mouse CD45 PE-Cy7 conjugated (1:200) | Thermo Fisher Scientific | Cat# 25-0451-82; RRID: AB_2734986 |
| FC receptor blocking antibody CD16/CD32 (1:200) | BD Bioscience | Cat# 564219; RRID: AB_2728082 |
| Chemicals, peptides, and recombinant proteins | ||
| ADP | Sigma-Aldrich | Cat# A5285 |
| Antimycin A | Sigma-Aldrich | Cat# A8674 |
| BSA | Carl Roth | Cat# 2076.3 |
| Dimethyl Sulfoxide (DMSO) | Sigma-Aldrich | Cat# D2438 |
| D-(+)-Glucose solution (45%) | Sigma-Aldrich | Cat# G8769-100ML |
| D-Mannitol | Cayman chemicals | Cat# 21673 |
| EDTA | Sigma-Aldrich | Cat# E6758 |
| EGTA | Sigma-Aldrich | Cat# E6758 |
| FCCP | Sigma-Aldrich | Cat# C2920 |
| Fetal Bovine/Calf Serum (FCS) | Thermo Fisher Scientific | Cat# 10270106 |
| Fixable Viability Dye eFluor™ 780 | Thermo Fisher Scientific | Cat# 65-0865-14 |
| HBSS (Ca2+, Mg2+ free) with phenol red | Thermo Fisher Scientific | Cat# 14170088 |
| HEPES | Thermo Fisher Scientific | Cat# 15630056 |
| Magnesium chloride | Sigma-Aldrich | Cat# M8266 |
| Malate | Sigma-Aldrich | Cat# M0875 |
| MitoTracker™ Green FM | Thermo Fisher Scientific | Cat# M7514 |
| Oxaloacetate | Sigma-Aldrich | Cat# O4126 |
| PBS | Sigma-Aldrich | Cat# D8537 |
| 10× PBS | Thermo Fisher Scientific | Cat# 70013016 |
| Percoll | Sigma-Aldrich | Cat# P1644 |
| Potassium dihydrogen phosphate | VWR Chemicals | Cat# 26936.236 |
| Pyruvate | Sigma-Aldrich | Cat# 107360 |
| Rotenone | Sigma-Aldrich | Cat# R8875 |
| Seahorse XF Plasma Membrane Permeabilizer | Agilent | Cat# 102504-100 |
| Succinate | Sigma-Aldrich | Cat# S3674 |
| Sucrose | Sigma-Aldrich | Cat# S9378 |
| TMRM (tetramethyl-rhodamine methyl ester) | Thermo Fisher Scientific | Cat# T668 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J GF (any age and sex might be used) | University Hospital Bern, Switzerland | N/A |
| Mouse: C57BL/6J SPF (any age and sex might be used) | Janvier labs, France | N/A |
| Mouse: 5×FAD (we used male mice with an age of 4 months. However any age and sex might be used depending on the research question) | Inhouse breeding (CEMT, Freiburg, Germany) | N/A |
| Software and algorithms | ||
| FlowJo software | TreeStar | https://www.flowjo.com |
| Other | ||
| FACS Canto II | Becton Dickinson | N/A |
Materials and equipment
Dissection medium
| Reagent | Final concentration | Amount |
|---|---|---|
| HBSS | - | 48.65 mL |
| HEPES (1 M) solution | 15 mM | 0.75 mL |
| D-(+)-Glucose solution (45%) | 0.54% | 0.60 mL |
| Total | 50 mL |
Dissection medium can be prepared in advance and stored at 4°C for up to 2 days.
37% Percoll solution
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | - | 6.30 mL |
| 10×PBS | 1× | 0.37 mL |
| Percoll | 37% | 3.33 mL |
| Total | 10 mL |
37% Percoll solution can be prepared in advance and stored at 4°C for up to 2 days.
FACS buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | - | 480 mL |
| FCS | 2% | 10 mL |
| EDTA (0.5 M, pH 8.0) | 0.01 M | 10 mL |
| Total | 500 mL |
FACS buffer can be prepared in advance and stored at 4°C after sterile filtration for up to 4 weeks.
FC-Block and live/dead staining solution
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | - | 99.5 μL per sample |
| FC receptor blocking antibody CD16/CD32 | 1:200 | 0.4 μL per sample |
| Fixable Viability Dye eFluor™ 780 | 1:1000 | 0.1 μL per sample |
| Total | 100 μL per sample |
FC-Block and live/dead staining solution is made fresh each time and should be kept at 4°C in the dark.
Surface markers labeling solution
| Reagent | Final concentration | Amount |
|---|---|---|
| FACS buffer | - | 49.5 μL |
| CD11b-APC | 1:200 | 0.25 μL per sample |
| CD45-PE-Cy7 | 1:200 | 0.25 μL per sample |
| Total | 50 μL |
FACS staining solution is made fresh each time and should be kept at 4°C in the dark.
MitoTrackerTM Green FM & TMRM staining solution
| Reagent | Final concentration | Amount |
|---|---|---|
| FACS buffer | - | 4997.4 μL |
| MitoTrackerTM Green FM (1 mM) | 20 nM | 0.1 μL |
| TMRM (10 mM) | 50 nM | 2.5 μL |
| Total | 5,000 μL |
MitoTracker staining solution should be prepared freshly prior to the experiment at 4°C in the dark.
MAS buffer (1×)
| Reagent | Final concentration | Amount |
|---|---|---|
| Mannitol (MW: 182.17 g/mol) | 220 mM | 40.07 g |
| Sucrose (MW: 342.30 g/mol) | 70 mM | 23.96 g |
| KH2PO4 (MW: 136.09 g/mol) | 10 mM | 1.36 g |
| MgCl2 1 M solution | 5 mM | 5 mL |
| HEPES 1 M solution | 2 mM | 2 mL |
| EGTA 0.25 M solution | 1 mM | 4 mL |
| ddH2O | - | 989 mL |
| Total | 1 L |
Adjust the pH to 7.2 with 0.1 M KOH. After filter-sterilization, this buffer may be stored at 4°C for up to 2 months.
MAS-BSA buffer (1×)
| Reagent | Final concentration | Amount |
|---|---|---|
| MAS buffer | - | 1 l |
| BSA | 0.4% | 4 g |
| Total | 1 l |
Adjust the pH to 7.2 with 0.1 M KOH. After filter-sterilization, this buffer may be stored at 4°C for up to 2 months
Permeabilization solution
| Reagent | Final concentration | Amount |
|---|---|---|
| MAS-BSA buffer | - | 5935.8 μL |
| Plasma Membrane Permeabilizer (PMP) | 1 nM | 1.2 μL |
| ADP | 1 mM | 60 μL |
| TMRM (10 mM) | 50 nM | 3.0 μL |
| Total | 6,000 μL |
Permeabilization solution should be prepared freshly prior to the experiment at 4°C in the dark
Drugs for respiratory chain manipulation
| Reagent | Final concentration | Amount |
|---|---|---|
| ADP (MW: 501.32) | 1 mM | 501.32 mg ADP in 10 mL MAS buffer (100 mM). |
| Pyruvate/malate (MW: 88.06/134.09) | 5 mM/2.5 mM | 440.3 mg pyruvate + 335.2 mg malate in 10 mL MAS buffer (500 mM/250 mM). |
| Rotenone (MW: 394.42)∗ | 1 μM | 5 mg rotenone in 1.267 mL DMSO (10 mM). Working solution (concentration 225 μM): 22.5 μL of the stock solution diluted in 977.5 μL MAS-BSA. |
| Succinate (MW: 118.09) | 10 mM | 590 mg succinic acid to 10 mL MAS buffer (500 mM). |
| Antimycin A (MW: 534.60)∗ | 20 μM | 25 mg Antimycin A in 1.27 mL DMSO (36 mM). Working stock solution (concentration 2 mM): 16.7 μL of the stock solution diluted in 283.3 μL MAS-BSA). |
| FCCP (MW: 254.17)∗ | 1 μM | 10 mg FCCP in 787 μL DMSO (concentration: 50 mM). Working solution (concentration: 100 μM): add 1 μL of the stock solution to 499 μL MAS buffer. |
| Oxaloacetate (OAA; MW: 132.07) | 528.28 mg OAA in 10 mL MAS buffer (concentration: 400 mM). |
Adjust pH of the drugs to 7.2 with KOH. Stock solutions may be stored as aliquots for up to 2 months at –20°C (in the dark since some compounds may be light-sensitive).
CRITICAL: Rotenone and antimycin A are respiratory chain inhibitors, and the uncoupler FCCP, can be therefore acutely toxic. Personal protective equipment including gloves, protective clothing, face and eye shields, and respiratory protection should be continuously worn while handling these compounds. Thoroughly clean the bench and other workspace after usage.
Alternatives: Duroquinol as well as tetramethyl-p-phenylenediamine (TMPD) and ascorbate can be used for measuring CIII and CIV function. For further information see also (Salabei et al., 2014).
Step-by-step method details
Percoll gradient and FACS staining of microglial cells
Timing: 3–4 h
Timing: 1–2 hfor dissection and tissue preparation
Timing: 1 hfor Percoll gradient and washing
Timing: 1 hfor FACS labeling
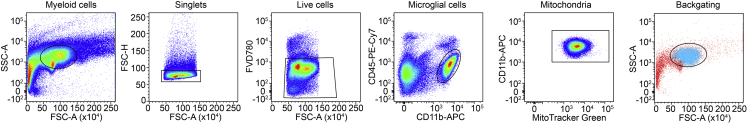
These steps describe how to isolate microglia cells from the CNS. The Percoll-gradient is critical for removal of myelin and separation of the CNS cells including microglia for the following steps. The surface marker staining is necessary to measure the respiratory chain activity specifically in microglial cells. The gating strategy includes single, live, CD11b+CD45lowMitoGreen+ cells (Figure 1).
Note: Here, we provide the flow cytometry-based protocol for respiratory chain analysis for microglial cells. We analyzed individual samples from max. 10 mice in one individual experiment with at least 5,000 microglial cells. Importantly, either CI or CII function should be analyzed in one approach when handling 10 samples. Otherwise, the metabolic function might be impaired when handling more than 10 samples for more than 6–8 h. Alternatively, using less samples would allow the analysis of CI and CII in two smaller approaches from separate replicates. This protocol can be also used for other ex vivo isolated cells or cultured cells.
-
1.
Dissect the required mice and perfuse with 10 mL ice-cold PBS
-
2.
Remove the brain and homogenize the tissue with a Dounce potter in dissection medium at 4°C (total volume 20 mL).
-
3.
Transfer the cell suspension from each sample through a 70 μm cell strainer into a 50 mL Falcon tube respectively and spin at 300 g at 4°C for 5 min (total volume 20 mL).
-
4.
Carefully, remove the supernatant from each sample and resuspend the cell pellets in 10 mL 37% Percoll solution.
-
5.
Transfer the cell suspension to a 15 mL Falcon tube respectively and centrifuge for 30 min at 800 g (without brake) at 4°C.
-
6.
Discard the supernatant and resuspend each cell pellet with 1 mL PBS at 4°C respectively.
-
7.
Transfer the individual cell suspension to FACS tubes and centrifuge at 400 g at 4°C for 5 min.
-
8.
Carefully remove the supernatant.
-
9.
Resuspend each cell pellet in 100 μL FC-Block and live/dead staining solution and incubate for 5 min on ice in the dark.
-
10.
Add 2 mL FACS buffer and centrifuge at 300 g for 5 min at 4°C.
-
11.
Carefully remove the supernatant.
-
12.
Add 50 μL of the surface markers labeling solution per sample (CD11b-APC and CD45-PE-Cy7; diluted in FACS buffer) and incubate for 20 min in dark on ice.
-
13.
Add 2 mL FACS buffer to the samples respectively and centrifuge at 300 g for 5 min at 4°C.
-
14.
Carefully remove the supernatant.
-
15.
Resuspend the cell pellet with 500 μL staining solution containing MitoTrackerTM Green (final concentration 20 nM) and TMRM (final concentration 50 nM) and incubate for 30 min at 37°C in the dark in a water bath.
-
16.
Add 2 mL FACS buffer and centrifuge at 300 g for 5 min at 4°C.
Figure 1.
Gating strategy for flow cytometric analysis of single, live, CD11b+CD45low MitoTracker Green+ microglia
The last dot plot depicts the back gating of CD11b+CD45low cells. Representative dot plots are shown. FSC: forward scatter. SSC: side scatter.
Analysis of complex I
Timing: 3–4 h
The following steps describe how to permeabilize the cells and how to conduct the analysis of complex I activity by flow cytometry. While acquiring the sample for flow cytometry, the temperature should be kept at 37°C.
-
17.
Resuspend each cell pellet in 600 μL MAS buffer + PMP + TMRM solution and incubate for 2 min at 37°C in the dark in order to permeabilize the cells.
-
18.
Gate on single, live (FVD780-), CD11b+CD45low microglia. All microglial cells should contain MitoTrackerTM Green+ mitochondria in the FITC-channel (Figure 1A). (see troubleshooting 1)
-
19.
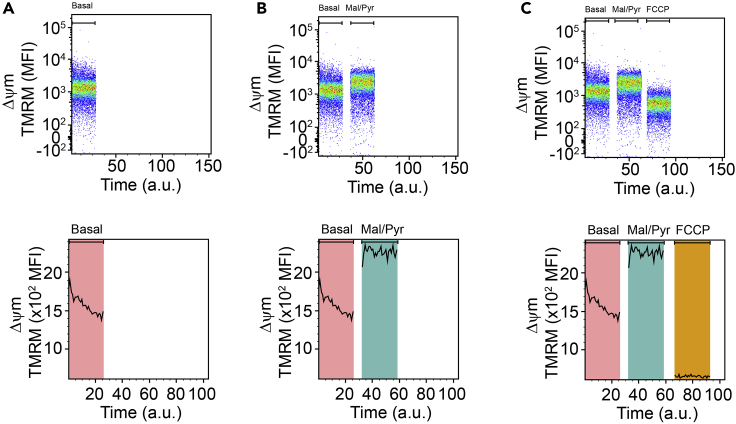
Record the basal mitochondrial membrane potential (Δψm) in the PE-channel for exactly 90 s (Figure 2A).
-
20.
Record for 30 s with ddH2O.
-
21.
Continue with a volume of 550 μL from the previous step and add 5.5 μL Pyruvate/malate working solution (concentration working solution: 500 mM/250 mM → final concentration: 5 mM/2.5 mM), vortex and record the Δψm for 90 s (Figure 2B).
-
22.
Record for 30 s with ddH2O.
-
23.
Continue with a volume of 500 μL from the previous step and add 5 μL FCCP working solution (concentration working solution: 100 μM → final concentration: 1 μM), vortex and record the Δψm for 90 s (Figure 2C).
Optional: Titrate each compound/inhibitor around that range to find the optimal working concentration for the respective setup and/or cells (see troubleshooting 2, 3, 4, and 5). Please see also (Salabei et al., 2014).
Figure 2.
Examination of complex I function
(A) Representative cytometry graphs of the mitochondrial membrane potential Δψm (tetramethylrhodamine, TMRM) which was manipulated by (B) malate & pyruvate (Mal/Pyr) and subsequently by (C) carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP) in CD11b+CD45low microglia.
Analysis of complex II
Timing: 3–4 h
The following steps describe how to permeabilize the cells and how to perform the analysis of complex II activity by flow cytometry. While acquiring the sample for flow cytometry, the temperature should be kept at 37°C.
-
24.
Resuspend each cell pellet in 600 μL MAS buffer + PMP + TMRM solution and incubate for 2 min at 37°C in the dark in order to permeabilize the cells.
-
25.
Gate on single, live (FVD780-), CD11b+CD45low microglia. All microglial cells should contain MitoTracker Green+ mitochondria in the FITC-channel (Figure 1A).
-
26.
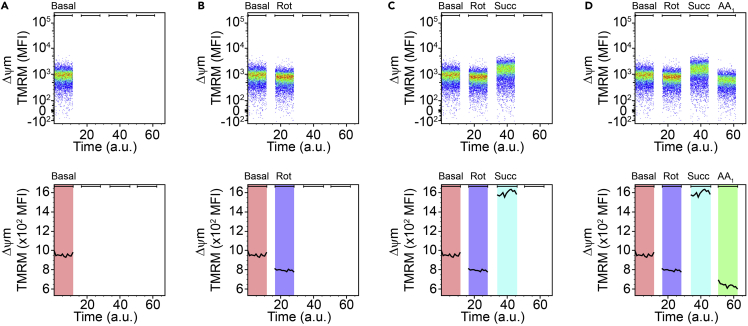
Record the basal mitochondrial Δψm in the PE-channel for exactly 90 s (Figure 3A).
-
27.
Record for 30 s with ddH2O.
-
28.
Continue with a volume of 450 μL from the previous step and add 2 μL rotenone working solution (concentration working solution: 225 μM → final concentration: 1 μM), vortex and record the Δψm for 90 s (Figure 3B).
-
29.
Record for 30 s with ddH2O.
-
30.
Continue with a volume of 400 μL from the previous step and add 8 μL Succinate (concentration stock solution: 500 mM → final concentration: 10 mM), vortex and record the Δψm for 90 s (Figure 3C).
-
31.
Record for 30 s with ddH2O.
-
32.
Continue with a volume of 350 μL from the previous step and add 3.5 μL Antimycin A working solution (working solution concentration: 2 mM → final concentration: 20 μM), vortex and record the Δψm for 90 s (Figure 3D).
Optional: Comparison of microglial cells derived from healthy control and diseased conditions, such as neurodegenerative mouse models (e.g., 5×FAD mice) can be performed (Erny et al., 2021) (MX-O4+ microglia from 5×FAD; Figure S7D).
Figure 3.
Examination of complex II function
(A) Representative cytometry graphs of the mitochondrial membrane potential Δψm which was manipulated by (B) rotenone (Rot) and subsequently by (C) succinate (Succ.) and (D) antimycin A1 (AA1) in CD11b+CD45low microglia.
Modulation of complex II by oxaloacetate
Timing: 3–4 h
Furthermore, additional manipulation e.g., by Oxalacetate (OAA) as inhibitor for CII (Kotlyar and Vinogradov, 1984; Schollmeyer and Klingenberg, 1961) can be performed. The following steps describe how to permeabilize the cells and how to modulate CII activity by OAA. Use increasing concentrations of OAA in order to manipulate CII activity in subsequent samples. While acquiring the sample for flow cytometry, the temperature should be kept at 37°C.
-
33.
Resuspend each cell pellet in 600 μL MAS buffer + PMP + TMRM solution and incubate for 2 min at 37°C in the dark in order to permeabilize the cells.
-
34.
Gate on single, live (FVD780-), CD11b+CD45low microglia. All microglial cells should contain MitoTracker Green+ mitochondria in the FITC-channel (Figure 1A).
-
35.
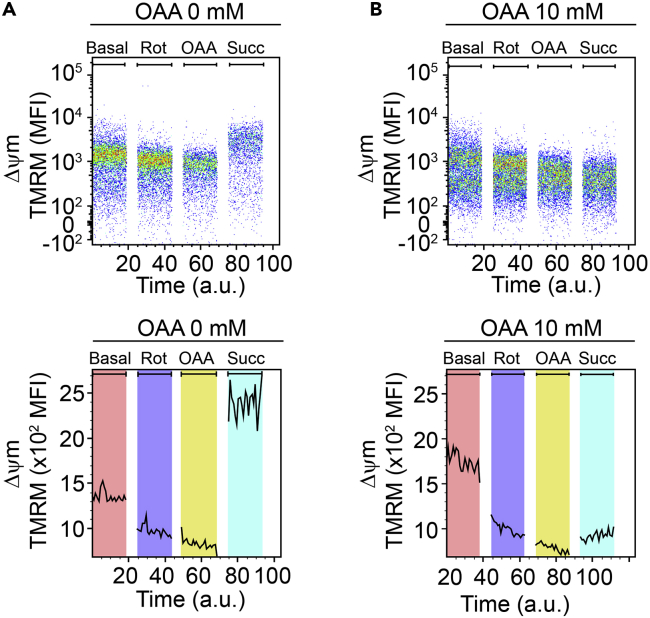
Record the basal mitochondrial membrane potential (Δψm) in the PE-channel for exactly 90 s (Figures 4A and 4B).
-
36.
Record for 30 s with ddH2O.
-
37.
Continue with a volume of 450 μL from the previous step and add 2 μL rotenone working solution (concentration working solution: 225 μM → final concentration: 1 μM), vortex and record the Δψm for 90 s (Figures 4A and 4B).
-
38.
Record for 30 s with ddH2O.
-
39.
Continue with a volume of 500 μL from the previous step and add 12.5 μL OAA (concentration stock solution: 400 mM → final concentration: 10 mM), vortex and record the Δψm for 90 s (Figures 4A and 4B).
-
40.
Record for 30 s with ddH2O.
-
41.
Continue with a volume of 400 μL from the previous step and add 8 μL Succinate working solution (concentration stock solution: 500 mM → final concentration: 10 mM), vortex and record the Δψm for 90 s (Figures 4A and 4B).
Optional: Test various concentrations of OAA in order to inhibit CII function in the cells of interest.
CRITICAL: While acquiring the sample for flow cytometry, the temperature should be kept at 37°C.
Figure 4.
Examination of complex II function
(A) Representative cytometry graphs of the mitochondrial membrane potential Δψm which was manipulated by rotenone (Rot) and subsequently by (B) oxaloacetate (OAA) and succinate (Succ.) in CD11b+CD45low microglia.
Expected outcomes
This protocol is useful for evaluating mitochondrial CI- and CII-mediated respiratory activity. As shown in Figures 2 and 3 and in (Erny et al., 2021), the Δψm can be robustly modulated by the compounds enabling comprehensive analysis of the respiratory chain function. In Figures 3 and 4 we assessed CII function which can be inhibited by increasing concentrations of the endogenous CII inhibitor OAA (Figure 4).
Limitations
One limitation of the method is that the metabolic activity of the cells will be impaired after prolonged procedure. We recommend to process the cells within 6–8 h. Notably, the method does not compensate for potential defects in pathways that feed into tricarboxylic acid cycle (TCA) like glycolysis, amino acid metabolism or fatty acid oxidation. These pathways may be analyzed by additional methods such as glucose uptake (Erny et al., 2021).
Furthermore, this method can be conducted only for cells that can be analyzed by flow-cytometry or other live-cell imaging techniques.
Troubleshooting
Problem 1
Spillover of the MitoTracker Green signal (step 18).
Potential solution
Concentration of MitoTracker Green should be adapted for the cells of interest.
Problem 2
Low Δψm (steps 19–23; 26–32, 35–41).
Potential solution
Metabolic state of cells is not optimal:
Check the cell viability in the live dead staining.
Shorten the time of experimental procedure by processing less samples.
Concentration of PMP was too high and critically damaged the cells. Perform titration gradients on your cells of interest.
Problem 3
No response to the compounds while acquiring the Δψm (steps 19–23; 26–32, 35–41).
Potential solution
Prepare fresh working and/or stock solutions.
The number of cells is too low to detect differences
Problem 4
Shift of the gates while assessing the cells in flow cytometry (steps 19–23; 26–32, 35–41).
Potential solution
Concentration of PMP should be adapted for the cells of interest.
Problem 5
High rate of dead cells (steps 19–23; 26–32, 35–41).
Potential solution
Concentration of PMP should be adapted for the cells of interest (steps 19–23; 26–32, 35–41).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Marco Prinz (marco.prinz@unikklinik-freiburg.de).
Materials availability
This study did not generate new or unique reagents.
Acknowledgments
The graphical abstract and diagrams were created using BioRender.com. D.E. is supported by the DFG (SFB/TRR167) and the Berta-Ottenstein-Programme for advanced Clinician Scientists. T.B. is supported by the DFG (SFB/TRR167). M.P. is supported by the Sobek Foundation, the Ernst-Jung Foundation, the Novo Nordisk Foundation, the DFG (SFB 992, SFB1160, SFB 1479, SFB/TRR167, Gottfried Wilhelm Leibniz-Prize); and the Ministry of Science, Research and Arts, Baden-Württemberg (Sonderlinie “Neuroinflammation”). M.P. was further supported by the DFG under Germany’s Excellence Strategy (CIBSS – EXC-2189 – Project ID390939984).
Author contributions
D.E., N.D., C.M., and O.M. designed, established, and conducted the protocol. D.E. wrote the manuscript. N.D., C.M., O.M., T.B., and M.P. edited the manuscript. D.E. and M.P. conceptualized and supervised the project.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Daniel Erny, Email: daniel.erny@uniklinik-freiburg.de.
Marco Prinz, Email: marco.prinz@uniklinik-freiburg.de.
Data and code availability
The example flow cytometry data were reanalyzed from (Erny et al., 2021). This study did not generate any code.
References
- Borst K., Schwabenland M., Prinz M. Microglia metabolism in health and disease. Neurochem. Int. 2018 doi: 10.1016/j.neuint.2018.11.006. [DOI] [PubMed] [Google Scholar]
- Erny D., Dokalis N., Mezo C., Castoldi A., Mossad O., Staszewski O., Frosch M., Villa M., Fuchs V., Mayer A., et al. Microbiota-derived acetate enables the metabolic fitness of the brain innate immune system during health and disease. Cell Metab. 2021;33:2260–2276.e7. doi: 10.1016/j.cmet.2021.10.010. [DOI] [PubMed] [Google Scholar]
- Gosselin D., Link V.M., Romanoski C.E., Fonseca G.J., Eichenfield D.Z., Spann N.J., Stender J.D., Chun H.B., Garner H., Geissmann F., et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlyar A.B., Vinogradov A.D. Interaction of the membrane-bound succinate dehydrogenase with substrate and competitive inhibitors. Biochim. Biophys. Acta. 1984;784:24–34. doi: 10.1016/0167-4838(84)90168-7. [DOI] [PubMed] [Google Scholar]
- Salabei J.K., Gibb A.A., Hill B.G. Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis. Nat. Protoc. 2014;9:421–438. doi: 10.1038/nprot.2014.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schollmeyer P., Klingenberg M. Oxaloacetate and adenosinetriphosphate levels during inhibition and activation of succinate oxidation. Biochem. Biophys. Res. Commun. 1961;4:43–47. doi: 10.1016/0006-291x(61)90252-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The example flow cytometry data were reanalyzed from (Erny et al., 2021). This study did not generate any code.