Summary
This protocol details a staining technique optimized for immunophenotyping of human bone marrow immune populations using mass cytometry. The protocol accounts for the limitations of working with human bone marrow, such as reduced viability, low cell counts, and fragile cell pellets, to successfully acquire single viable cells ready for downstream analysis. This assay can be used to characterize the activation, exhaustion, and cytotoxicity of immune populations and ensure comprehensive immunophenotyping of human bone marrow clinical samples.
Subject areas: Cell Biology, Flow Cytometry/Mass Cytometry, Cell-based Assays, Health Sciences, Immunology
Graphical abstract
Highlights
-
•
Staining protocol for assessment of immune cells in human bone marrow clinical samples
-
•
Validated mass cytometry panel for distinction of major immune populations
-
•
Insights on gating strategy for multiparameter mass cytometry data analysis
This protocol details a staining technique optimized for immunophenotyping of human bone marrow immune populations using mass cytometry. The protocol accounts for the limitations of working with human bone marrow, such as reduced viability, low cell counts, and fragile cell pellets, to successfully acquire single viable cells ready for downstream analysis. This assay can be used to characterize the activation, exhaustion, and cytotoxicity of immune populations and ensure comprehensive immunophenotyping of human bone marrow clinical samples.
Before you begin
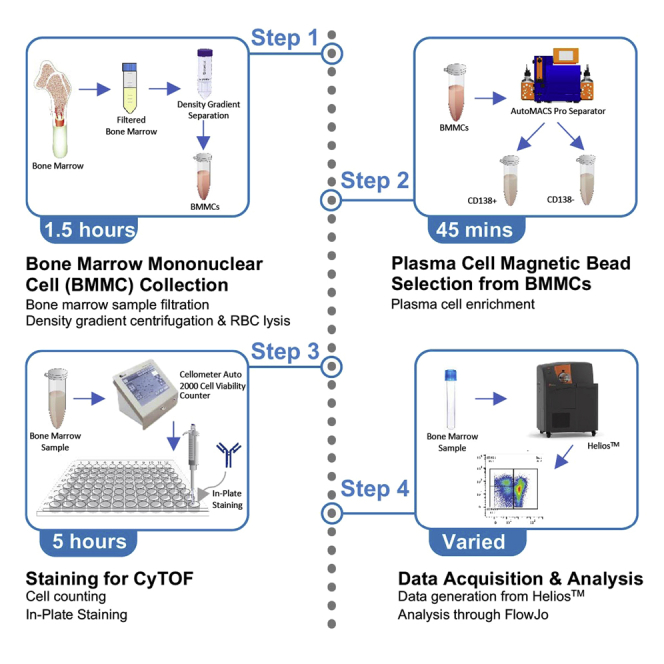
Preparation for bone marrow mononuclear cell (BMMC) collection
Timing: 10 min
Ethics approval was obtained from Dana-Farber/Harvard Cancer Center Institutional Review Board (IRB 14-174) and all patients provided informed consent
-
1.
Bring Ficoll-Paque PLUS, 1× PBS, and 10× RBC Lysis Buffer to room temperature (19°C–22°C)
-
2.
Prepare a working solution of 20 mL 1× RBC Lysis Buffer by diluting 2 mL 10× RBC Lysis Buffer with 18 mL Distilled Ultrapure Water
-
3.
Prepare Freezing Media containing 90% FBS and 10% DMSO by volume and keep at 4°C until use
Note: Each sample requires approximately 1 mL freezing media for the CD138+ cell fraction (expected cell count of 1 × 106 CD138+ cells) and 4 mL of freezing media for the CD138- cell fraction (expected cell count of 4 × 106 CD138+ cells).
Human bone marrow mononuclear cell (BMMC) collection
Timing: 90 min
To isolate bone marrow mononuclear cells (BMMC), BMMCs are first enriched from aspirates by density gradient centrifugation using Ficoll-Paque within SepMate™ PBMC Isolation Tubes. Any remaining erythrocytes are then briefly lysed using 1× RBC Lysis Buffer, and samples are washed with 1× PBS to remove residual lysis buffer and any cellular debris.
-
4.
Pre-wet a 100 μm MACS® SmartStrainer with 1× PBS within a 50 mL conical tube. Discard 1× PBS used to pre-wet MACS® SmartStrainer from your 50 mL conical tube
-
5.
Slowly filter bone marrow sample through the pre-wet MACS® SmartStrainer into 50 mL conical tube, using the end of a syringe plunger to break apart any clots above the filter and to filter out any bone debris
-
6.
Make note of the volume of bone marrow aspirate post-filtering, then dilute bone marrow sample with 1× PBS three times its volume (for example, dilute 10 mL of bone marrow with 30 mL PBS)
-
7.
Mix the 1× PBS and bone marrow by gently inverting the tube 8–10 times
-
8.
Add 15 mL of Ficoll-Paque to a 50 mL SepMate™ tube by carefully pipetting it through the central hole of the plastic insert. The top of the density gradient medium should be just above the insert
Note: Any large bubbles present within the density gradient medium should be removed by spinning at 300 g for 1 min prior to loading your sample.
-
9.
Gently layer the diluted bone marrow onto the Ficoll-Paque by pipetting it down the side of the SepMate™ tube. The bone marrow should overlay on top of the Ficoll-Paque.
Note: If the volume of diluted bone marrow is larger than 30 mL, prepare multiple SepMate™ tubes and evenly distribute the sample
CRITICAL: Do not let the Ficoll-Paque and bone marrow aspirate mix under the plastic insert within the SepMate™ tube, as this will affect the yield, purity and 127I content of the isolated BMMCs.
-
10.
Centrifuge the SepMate™ tube(s) containing the diluted bone marrow aspirate and Ficoll-Paque at 1200 g for 20 min at room temperature, with the brake on
-
11.
Carefully pour the top layer above the plastic insert from the SepMate™ tube(s) (which contains enriched BMMCs) into a new 50 mL conical tube
CRITICAL: Do not hold SepMate™ tube in the inverted position for longer than 2 seconds, as this will cause separated RBCs to pass through the plastic insert into the new 50 mL conical tube.
-
12.
Discard the SepMate™ tube containing RBCs and granulocytes in biohazard waste
-
13.
Centrifuge the new at 300 g for 8 min, with the brake on
-
14.
Pour off the supernatant and resuspend the cell pellet in 20 mL of 1× RBC Lysis Buffer, and gently invert the tube 8–10 times
-
15.
Immediately after centrifuge the 50 mL conical tube containing sample at 300 g for 8 min with the brake on
-
16.
Pour off supernatant and resuspend the pellet in 40 mL PBS. Mix by gently inverting the tube 8–10 times
-
17.
Centrifuge at 300 g for 8 min with the brake on
-
18.
Pour off supernatant and resuspend pellet in 10 mL PBS
-
19.Count cells using 0.4% Trypan Blue (1:1 dilution, 10 μL sample in 10 μL Trypan Blue) with the Invitrogen™ Countess II Automated Cell Counter (or an alternative cell counter)
-
a.Remove 10 μL of resuspended sample and mix well with 10 μL 0.4% Trypan Blue
-
b.Load 10 μL of diluted of the Trypan Blue diluted sample to a Countess Cell Chamber
-
a.
-
20.
The resuspended sample now contains BMMCs. Proceed to plasma cell magnetic bead enrichment using this cell count
Plasma cell magnetic bead selection from BMMCs
Timing: 45 min
Once BMMCs have been isolated from the marrow aspirate, the sample is then enriched for plasma cells, using Miltenyi CD138 Microbeads∗ (Human) and the autoMACS® Pro Separator. This process will produce both CD138+ and CD138- fractions
-
21.
Centrifuge resuspended BMMCs at 300 g for 8 min, with the brake on
-
22.
Aspirate supernatant and resuspend BMMCs in 80 μL of autoMACS Running Buffer and 20 uL CD138 Microbeads per 2.0×107 BMMCs∗∗
Note: ∗Other separation kits are acceptable yet were not validated when developing this protocol. ∗∗Change volume of both beads and running buffer in proportion to BMMC cell count obtained in Step 19 of BMMC Collection.
-
23.
Mix by gently pipetting without introducing bubbles, and incubate at 4°C for 15 min
-
24.
After 15 min, wash the BMMC and magnetic bead mixture by pipetting very gently with 2 mL autoMACS Running Buffer
-
25.
Centrifuge samples at 300 g for 10 min, with the brake on
-
26.
Aspirate the supernatant and resuspend the bead pellet in 500 μL autoMACS Running Buffer
-
27.
Transfer the resuspended sample to a new 15 mL conical tube
-
28.
Label 2 new additional 15 mL conical tubes as “CD138+” and “CD138-”, followed by the sample ID
-
29.
Place the 15 mL tube containing the re-suspended sample in the autoMACS® Pro Separator tube rack in the designated sample slot, and place the empty pre-labeled 15 mL conical tubes in each of the appropriate locations for the positive and negative sample fractions to deposit into
-
30.
Ensure there are adequate volumes of autoMACS Washing Buffer, autoMACS Running Buffer, 70% EtOH, and sufficient space in the waste in reagent reservoirs attached to the autoMACS® Pro Separator
-
31.
Select the protocol named “Possel” on the autoMACS® Pro Separator to enable automated bead selection to begin
-
32.
Once autoMACS® Pro Separator is done with enriching CD138+ and CD138- cells fractions, centrifuge samples at 300 g for 8 min, with the brake on
-
33.
Remove supernatant and resuspend each sample in 1 mL of PBS.
-
34.Count each cell fraction (CD138+/CD138-) using 0.4% Trypan Blue (1:1 dilution, 10 μL sample in 10 μL Trypan Blue) with the Invitrogen™ Countess II Automated Cell Counter (or an alternative cell counter)
-
a.Remove 10 μL of resuspended sample and mix well with 10 μL 0.4% Trypan Blue
-
b.Load 10 μL of diluted of the Trypan Blue diluted sample to a Countess Cell Chamber
-
a.
-
35.
Dilute both CD138+ and CD138- fractions to 10 mL total by adding 9 mL PBS to wash away remaining autoMACS running buffer
-
36.
Centrifuge samples at 300 g for 8 min, with the brake on.
-
37.
Carefully aspirate and discard supernatant.
Note: Given bone marrow research aspirates low volume and the low prevalence of plasma cells within the bone marrow niche, the CD138+ cell pellet is often less than 1 million cells and difficult to visualize after pelleting. Therefore, it is best to aspirate and discard all but the last ∼50 uL of supernatant.
-
38.
Resuspend the CD138+ fraction in 1 mL of Freezing Media (90% FBS + 10% DMSO) and aliquot into a single cryovial
-
39.
Resuspend the CD138- fraction in 4 mL of Freezing Media and aliquot into 4 individual cryovials
-
40.
Place vials in CoolCell FTS30 (or alternative slow-freeze container) and into a -80°C Freezer for 24 h before moving to long-term storage in liquid nitrogen.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Human CD45 (HI30)-89Y—100 Tests (1:100) | Fluidigm | Cat#3089003B |
| Anti-Human CD196/CCR6 (G034E3)-141Pr—50 Tests (1:100) | Fluidigm | Cat#3141003A |
| Anti-Human CD19 (HIB19)-142Nd—100 Tests (1:100) | Fluidigm | Cat#3142001B |
| Anti-Human CD45RA (HI100)-143Nd—100 Tests (1:100) | Fluidigm | Cat#3143006B |
| Anti-Human CD69 (FN50)-144Nd—100 Tests (1:100) | Fluidigm | Cat#3144018B |
| Anti-Human CD4 (RPA-T4)-145Nd—100 Tests (1:100) | Fluidigm | Cat#3145001B |
| Anti-Human IgD (IA6-2)-146Nd—100 Tests (1:100) | Fluidigm | Cat#3146005B |
| Anti-Human CD20 (2H7)-147Sm—100 Tests (1:100) | Fluidigm | Cat#3147001B |
| Anti-Human CD14 (RMO52)-148Nd—100 Tests (1:100) | Fluidigm | Cat#3148010B |
| Anti-Human CD56/NCAM (NCAM16.2)-149Sm—100 Tests (1:100) | Fluidigm | Cat#3149021B |
| Anti-Human CD138/Syndecan-1 (DL-101)-150Nd—25 tests (1:100) | Fluidigm | Cat#3150012C |
| Anti-Human TNFa (mab11)-152Sm—100 Tests (1:100) | Fluidigm | Cat#3152002B |
| Anti-Human TIGIT (MBSA43)-153Eu—100 Tests (1:100) | Fluidigm | Cat#3153019B |
| Anti-Human CD3 (UCHT1)-154Sm—100 Tests (1:100) | Fluidigm | Cat#3154003B |
| Anti-Human CD27 (L128)-155Gd—100 Tests (1:100) | Fluidigm | Cat#3155001B |
| Anti-Human IFNg (B27)-158Gd—100 Tests (1:100) | Fluidigm | Cat#3158017B |
| Anti-Human CD161 (HP-3G10)-159Tb—100 Tests (1:100) | Fluidigm | Cat#3159004B |
| Anti-Human/Mouse MIP1beta (D21-1351)-160Gd—100 Tests (1:100) | Fluidigm | Cat#3160013B |
| Anti-Human/Mouse Tbet (4B10)-161Dy—100 Tests (1:100) | Fluidigm | Cat#3161014B |
| Anti-Human FoxP3 (259D/C7)-162Dy—50 Tests (1:100) | Fluidigm | Cat#3162024A |
| Anti-Human CD183/CXCR3 (G025H7)-163Dy—100 Tests (1:100) | Fluidigm | Cat#3163004B |
| Anti-Human IL-17A (N49-653)-164Dy—100 Tests (1:100) | Fluidigm | Cat#3164002B |
| Anti-Human CD279/PD-1 (EH12.2H7)-165Ho—100 Tests (1:100) | Fluidigm | Cat#3165042B |
| Anti-Human CD197/CCR7 (G043H7)-167Er—50 Tests (1:100) | Fluidigm | Cat#3167009A |
| Anti-Human CD8 (SK1)-168Er—100 Tests (1:100) | Fluidigm | Cat#3168002B |
| Anti-Human CD25 (2A3)-169Tm—100 Tests (1:100) | Fluidigm | Cat#3169003B |
| Anti-Human Granzyme B (GB11)-171Yb—100 Tests (1:100) | Fluidigm | Cat#3171002B |
| Anti-Human CD57 (HCD57)-172Yb—100 Tests (1:200) | Fluidigm | Cat#3172009B |
| Anti-Human HLA-DR (L243)-173Yb—100 Tests (1:200) | Fluidigm | Cat#3173005B |
| Anti-Human CD94 (HP-3D9)-174Yb—100 Tests (1:100) | Fluidigm | Cat#3174015B |
| Anti-Human Perforin (B-D48)-175Lu—100 Tests (1:100) | Fluidigm | Cat#3175004B |
| Anti-Human CD127/IL-7Ra (A019D5)-176Yb—100 Tests (1:100) | Fluidigm | Cat#3176004B |
| Anti-Human CD16 (3G8)-209Bi—100 Tests (1:100) | Fluidigm | Cat#3209002B |
| Anti-Granzyme K antibody [GM-24C3] (ab3771) (1:100) | Abcam | Cat#ab3771 |
| Chemicals, peptides, and recombinant proteins | ||
| Ficoll-Paque Plus | Global Life Sciences | Cat#45001740 |
| 1× Phosphate-Buffered Saline without Calcium & Magnesium | Mediatech | Cat #21040CV |
| 10× RBC Lysis Buffer | BioLegend | Cat #420301 |
| 0.4% Trypan Blue Solution | Life Technologies | Cat #15250061 |
| autoMACS Running Buffer – MACS Separation Buffer | Miltenyi Biotec | Cat #130091221 |
| CD138 MicroBeads, Human | Miltenyi Biotec | Cat #130051301 |
| DMSO | Sigma-Aldrich | Cat #D2650 |
| PBS, pH 7.4 (1×) | Gibco | Cat#10010023 |
| Bovine Serum Albumin (BSA), 30% ± 2% in 0.85% NaCl | Sigma-Aldrich | Cat#A7284 |
| Sodium Azide, 10% (w/v) solution in Ultra-Pure H2O | Teknova | Cat#S0209 |
| RPMI 1640 (1×) Medium | Gibco | Cat#11875093 |
| Fetal Bovine Serum, heat inactivated, qualified (FBS) | Gibco | Cat#10438026 |
| Antibiotic-Antimycotic (100×) | Gibco | Cat#15240096 |
| UltraPureTM DNAse/RNAse-Free Distilled Water | Invitrogen | Cat#10977023 |
| ViaStainTM AOPI Staining Solution | Nexcelom Bioscience | Cat#CS2-0106-5mL |
| Cell-IDTM Intercalator-103Rh—500 μM | Fluidigm | Cat#201103A |
| Cell-IDTM Intercalator-191/193Ir—125 μM | Fluidigm | Cat#201192A |
| Maxpar® Cell Acquisition Solution | Fluidigm | Cat#201237 |
| Maxpar® Cell Staining Buffer | Fluidigm | Cat#201068 |
| EQTM Four Element Calibration Beads | Fluidigm | Cat#201078 |
| eBioscienceTM Permeabilization Buffer (10×) | Invitrogen | Cat#00833356 |
| eBioscienceTM Fixation/Permeabilization Concentrate | Invitrogen | Cat#00512343 |
| eBioscienceTM Fixation/Permeabilization Diluent | Invitrogen | Cat#00522356 |
| Human TruStain FcXTM (FcR Blocking Solution) | BioLegend | Cat#422302 |
| PierceTM 16% Formaldehyde Solution (w/v), Methanol-free | Thermo Scientific | Cat#28906 |
| Critical commercial assays | ||
| Maxpar® X8 Antibody Labeling Kit, 166Er—4 Rxn | Fluidigm | Cat#201166A |
| Biological samples | ||
| Human bone marrow samples (sex, age as required par study) | Any supplier | N/A |
| Software and algorithms | ||
| FlowJo | Becton Dickinson & Company | https://www.flowjo.com/solutions/flowjo/downloads/ |
| CyTOF Software v7.0.8493 | Fluidigm | https://www.fluidigm.com/software |
| Other | ||
| SepMate-50 Tubes | Stemcell Technologies | Cat #85460 |
| 10 mL Serological Pipet | Corning Inc. | Cat #357530 |
| 25 mL Serological Pipet | Corning Inc. | Cat #1367530 |
| 25 mL Serological Pipet | Corning Inc. | Cat #1367530 |
| MACS SmartStrainer 100 μm | Miltenyi Biotec | Cat #130098463 |
| autoMACS Pro | Miltenyi Biotec | Cat #130092545 |
| CoolCell FTS30 | Thermo Fisher ScientificTM | Cat #07210008 |
| Countess II Automated Cell Counter | Thermo Fisher ScientificTM | Cat #AMQAX1000 |
| Countess Cell Counting Chamber Slides | Thermo Fisher ScientificTM | Cat #C10228 |
| Falcon 96-Well Polystyrene Microplate, TC Treated | Corning Inc. | Cat #353296 |
| 15 mL Polypropylene Centrifuge Tubes | Corning Inc. | Cat#430052 |
| 50 mL Polypropylene Centrifuge Tubes | Corning Inc. | Cat#430829 |
| Cellometer® SD100 Cell Counting Chamber Slides | Nexcelom Bioscience | CHT4-SD100-014 |
| 70 μm Cell Strainer, Polypropylene Frame | Biologix | Cat#15-1070 |
| Eppendorf® 1.5 mL Safe-Lock Microcentrifuge Tubes, natural | Eppendorf | Cat#022363204 |
| 96-well V-Bottom Plate, untreated | Corning Inc. | Cat#3896 |
| Polystyrene 96-well Microplate Corner Notch Lid | Corning Inc. | Cat#3930 |
| Falcon® 5 mL Round Bottom Polypropylene Tubes (FACS tubes) | Corning Inc. | Cat#352063 |
| Falcon® 5 mL Round Bottom Polystyrene Tubes with 35 μm Cell Strainer Snap Caps | Corning Inc. | Cat#352235 |
| ThermoFisher ScientificTM SorvallTM LegendTM XTR Refrigerated Centrifuge, 120VAC | Thermo Fisher ScientificTM | Cat#75004521 |
| FisherbrandTM Mini-Centrifuge 100–240V, 50/6-Hz Universal Plug, Grey | Thermo Fisher ScientificTM | Cat#12-006-901 |
| Fisher Scientific Digital Vortex Mixer | Thermo Fisher ScientificTM | Cat#0215370 |
| Thermo ScientificTM PrecisionTM Circulating Water Bath | Thermo Fisher ScientificTM | Cat#TSCIR19 |
| Cellometer® Auto 2000 Cell Viability Counter | Nexcelom Bioscience | Auto 2000 |
| Fluidigm HeliosTM Mass Cytometer | Fluidigm | N/A |
| Nebulizer for HeliosTM Mass Cytometer | Fluidigm | Cat#107144 |
| WB Injector for HeliosTM Mass Cytometer | Fluidigm | Cat#107950 |
CRITICAL: Only the 35 μm cell strainer snap caps included with the Falcon® 5 mL Round Bottom Polystyrene Tubes will be used. Polypropylene tubes are substituted for the polystyrene tubes to minimize cell adhesion to the tubes.
Materials and equipment
Complete RPMI
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI Medium 1640 (1×) | n/a | 500 mL |
| FBS | 10% | 50 mL |
| Antibiotic-Antimycotic (100×) | 1% | 5 mL |
| Total | n/a | 555 mL |
Store at 4°C for up to 2 weeks
CyFACs
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | n/a | 500 mL |
| 30% BSA | 0.5% | 8.3 mL |
| 5% Sodium Azide | 0.02% | 2 mL |
| Total | n/a | 510.3 mL |
Store at 4°C for up to 6 weeks
FoxP3 fixation/permeabilization buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Fixation/Permeabilization Concentrate | 25% | 4 mL |
| Fixation/Permeabilization Diluent | 75% | 16 mL |
| Total | n/a | 20 mL |
Store at 4°C during the day of experiment, prepare fresh every time
CRITICAL: Sodium Azide is carcinogenic and toxic if swallowed, inhaled, or if in contact with skin. Handle in a fume hood using personal protective equipment.
Note: The timing and reagent consumption in this protocol is relative to the handling of 20 samples.
Step-by-step method details
Bone marrow thawing and cell counting
Timing: 1 h
To prepare the cryopreserved bone marrow samples for mass cytometry staining, the samples must first be thawed, counted, and viability assessed.
-
1.
Before staining, fill the appropriate number of 15-mL conical tubes (one per sample) with 9 mL of RPMI +10% FBS +1% Antibiotic-Antimycotic (named Complete RPMI from now on) and place at 37°C
-
2.
Prepare the surface antibody master mix in CyFACs (Table 1)
-
3.
Thaw the samples by placing the frozen vial directly into the 37°C water bath until content is thawed, approximately 45 s, few small ice crystals might remain in the vial.
-
4.
Transfer the totality of thawed cell suspension into the 15 mL conical containing 9 mL of 37°C complete RPMI
-
5.Count cells using AO/PI (1:1 dilution; 20 μL sample in 20 μL AO/PI) with the Cellometer® Auto 2000 Cell Viability Counter
-
a.Gently invert the conical containing the freshly thawed cells to ensure accurate cell counts
-
b.Add 20 μL of the AO/PI-diluted sample to a Cellometer® SD100 Cell Counting Chamber
-
c.Cells should be >90% viable
-
a.
-
6.
Adjust concentration of cell suspension to 3 × 106 cells/sample in complete RPMI
-
7.Centrifuge at 400 g for 10 min
-
a.While the sample is spinning down, prepare the 103Rh Viability Stain (1:500 dilution of 103Rh Cell-ID intercalator in 37°C warm complete RPMI)
-
a.
-
8.
After the sample is spun down, carefully aspirate and discard the supernatant
-
9.
Resuspend the sample in 1 mL of 103Rh Viability Stain
-
10.
Incubate for 15 min at room temperature
-
11.
Once incubation is complete, fill conical up to 5 mL with complete RPMI and spin down (400 g, 10 min)
-
12.
Carefully aspirate and discard supernatant without disturbing the cell pellet
-
13.
Resuspend each cell pellet in 50 μL CyFACs and proceed to Fc-block and surface staining
CRITICAL: If cell viability is below 90%, filter the samples using 70 μM cell pre-wetted strainers to obtain a single cell suspension.
Optional: The surface antibody master mix can be prepared during the viability stain incubation, but we recommend preparing the cocktail prior to the thawing of clinical samples to avoid any critical errors.
Table 1.
Surface antibody master mix
| Marker | Metal | Clone | Volume per sample (μL) |
|---|---|---|---|
| CD45 | 89Y | HI30 | 1.00 |
| CCR6 | 141Pr | G034E3 | 1.00 |
| CD19 | 142Nd | HIB19 | 1.00 |
| CD45RA | 143Nd | HI100 | 1.00 |
| CD69 | 144Nd | FN50 | 1.00 |
| CD4 | 145Nd | RPA-T4 | 1.00 |
| IgD | 146Nd | IA6-2 | 1.00 |
| CD20 | 147Sm | 2H7 | 1.00 |
| CD14 | 148Nd | RM052 | 1.00 |
| CD56 | 149Sm | NCAM16.2 | 1.00 |
| CD138 | 150Nd | DL-101 | 1.00 |
| TNF-α | 152Sm | Mab11 | 1.00 |
| TIGIT | 153Eu | MBSA43 | 1.00 |
| CD3 | 154Sm | UCHT1 | 1.00 |
| CD27 | 155Gd | L128 | 1.00 |
| IFN-γ | 158Gd | B27 | 1.00 |
| CD161 | 159Tb | HP-3G10 | 1.00 |
| CXCR3 | 163Dy | G025H7 | 1.00 |
| IL-17a | 164Dy | N49-653 | 1.00 |
| PD-1 | 165Ho | EH12.2H7 | 1.00 |
| CCR7 | 167Er | G043H7 | 1.00 |
| CD8a | 168Er | SK1 | 1.00 |
| CD25 | 169Tm | 2A3 | 1.00 |
| CD57 | 172Yb | HCD57 | 0.50 |
| HLA-DR | 173Yb | L243 | 0.50 |
| NKG2C | 174Yb | HP-3D9 | 1.00 |
| CD127 | 176Yb | A019D5 | 1.00 |
| CD16 | 209Bi | 3G8 | 1.00 |
| CyFACs | n/a | n/a | 24 μL |
| Total | n/a | n/a | 50 μL |
Fc-block and surface staining
Timing: 1 h
This portion of the protocol stains the sample with pre-selected metal-tagged antibodies to be acquired and used in downstream analysis. Prior to antibody staining, the samples are Fc-blocked to prevent non-specific staining of antibodies.
-
14.Fc-block procedure:
-
a.Add 5 μL undiluted Fc-block to each sample
-
b.Incubate on ice for 10 min
-
i.During the incubation, aliquot the prepared surface antibody cocktail and distribute 50 μL/well according to 96 well plate map
-
i.
-
a.
-
15.Surface staining:
-
a.Once the incubation is complete, distribute the totality of Fc-blocked samples into the appropriate wells (already containing the antibody cocktail) according to the plate map
-
b.Incubate for 30 min at room temperature
-
i.During incubation, prepare the eBioScienceTM FoxP3 Fixation/Permeabilization Buffer as per manufacturer protocol (https://tinyurl.com/foxp3TF) (briefly, mix 1 part fix/perm concentrate (00-5123-43) + 3 parts fix/perm diluent (00-5223-56))
-
ii.During incubation, prepare the eBioScienceTM Perm/Wash solution by diluting 2 mL of 10× Permeabilization Buffer stock into 18 mL of Ultra-Pure H20
-
i.
-
c.Pellet cells by plate centrifugation (400 g, 5 min)
-
d.Carefully aspirate supernatant (∼ 100 μL), wash once with 200 μL CyFACs, spin plate down (400 g, 5 min) and proceed to intracellular staining
-
a.
Optional: The surface antibody master mix can be added individually to each sample well after the Fc-blocked cells have been plated, but this increases risk of technical error and experimental time.
Note: The FoxP3 Fixation/Permeabilization Buffer is prepared in excess and will be needed for the DNA stain at the end of the protocol.
Permeabilization and intracellular staining
Timing: 1.5 h
This portion of the protocol stains the sample with pre-selected metal-tagged antibodies that target intracellular markers. Prior to intracellular staining, the samples are incubated with eBiosciencesTM FoxP3 Fixation/Permeabilization Buffer to preserve cellular morphology while allowing the intracellular antibodies across the plasma membrane to stain intracellularly.
-
16.
Discard the supernatant and resuspend samples thoroughly in 200 μL of FoxP3 Fixation/Permeabilization Buffer
-
17.Incubate for 30 min at room temperature
-
a.During incubation, prepare the intracellular antibody master mix
-
a.
CRITICAL: Intracellular antibody master mix is made in Perm/Wash.
Note: Granzyme K was conjugated to the 166Er metal isotope, prior to this protocol via the Fluidigm Maxpar® X8 Antibody Labeling Kit according to the manufacturer protocol (https://tinyurl.com/maxpAbLabeling).
-
18.
Spin plate down (800 g, 5 min) and wash once with 200 μL of Perm/Wash
-
19.
Carefully aspirate and discard supernatant
-
20.
Resuspend sample in 50 μL of Perm/Wash
-
21.
Add 50 μL of intracellular master mix (Table 2) to each well containing sample
-
22.
Incubate for 30 min at room temperature
-
23.
After incubation is complete, add 100 μL of Perm/Wash and spin plate down (800 g, 5 min)
-
24.
Wash once with 200 μL of Perm/Wash
-
25.
During spin, prepare fresh 4% PFA in Ultra-Pure H2O
-
26.
Add 200 μL of fresh 4% PFA to each sample
-
27.Spin down at 800 g for 10 min and proceed to DNA stain and Pre-Acquisition Wash
-
a.During spin, prepare the 191/193 DNA Intercalator at a 1:5000 dilution of 191/193 Cell-ID Intercalator to FoxP3 Fixation/Permeabilization Buffer
-
a.
CRITICAL: Spin time was increased to 10 min to improve cell recovery post-fixation of cells.
Table 2.
Intracellular antibody master mix
| Marker | Metal | Clone | Volume per sample (μL) |
|---|---|---|---|
| MIP1-β | 160Gd | D21-1351 | 1.00 |
| T-Bet | 161Dy | 4B10 | 1.00 |
| FoxP3 | 162Dy | 259D/C7 | 1.00 |
| Granzyme K | 166Er | 2471A | 0.50 |
| Granzyme B | 171Yb | GB11 | 0.50 |
| Perforin | 175Lu | B-D48 | 0.50 |
| Perm/wash | n/a | n/a | 45.5 μL |
| Total | n/a | n/a | 50 μL |
DNA stain and pre-acquisition wash
Timing: 1.5 h
The final steps of the protocol stain the cell nuclei to allow for single-cell detection and discrimination by the HeliosTM mass cytometer. After completing the DNA stain, the samples undergo a series of washes to prepare for acquisition.
-
28.
Add 200 μL of 191/193 DNA Intercalator to each sample
-
29.
Incubate for 1 h at 4°C
Pause Point: Samples can be incubated in intercalator solution overnight (or 8–16 h) at 4°C and protocol can be resumed the following morning
Optional: If timing of the protocol needs to allow the acquisition within the same day – intercalator staining can be combined with intracellular staining, if extended to 45 min.
-
30.
After the incubation is complete, pellet cells by plate centrifugation (800×g, 5 min)
-
31.
Carefully aspirate and discard supernatant
-
32.
Resuspend the samples in 200 μL of MaxPar Cell Staining Buffer
-
33.
Transfer the sample into a labeled, polypropylene tube with a 35 μm cell pre-wetted strainer cap
-
34.
Add 300 μL of MaxPar Cell Staining Buffer per tube, bringing the total volume to 500 μL
-
35.
Spin the tubes down (800 g, 10 min)
-
36.
Carefully aspirate and discard supernatant
-
37.Resuspend the sample in 500 μL of MaxPar Cell Acquisition Solution (CAS)
-
a.Load 20 μL of sample directly into Cellometer® SD100 Cell Counting Chamber
-
a.
-
38.Spin the tube down (800 g, 5 min)
-
a.Count the cells using the Cellometer® Auto 2000 Cell Viability Counter during spin and record final cell count
-
a.
-
39.
Prepare a 1:5 dilution of MaxPar EQ Calibration beads in CAS (CAS/EQ)
CRITICAL: Vortex the EQ Calibration beads for 1 min before preparing CAS solution.
-
40.
Carefully aspirate and discard supernatant
-
41.
Resuspend samples at a concentration of 1 x 106 cells per 1 mL of CAS/EQ solution.
-
42.
Filter resuspended sample through 35 μm cell pre-wetted strainer cap and proceed to mass cytometer acquisition.
Expected outcomes
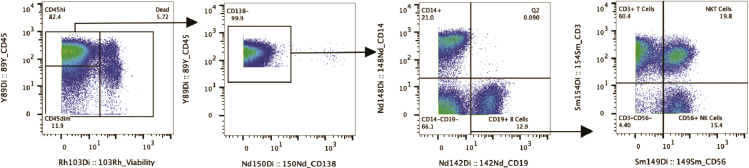
At the end of this protocol, FCS files will be generated for every sample acquired on HeliosTM and normalized using the FCS Processing tab of the Fluidigm CyTOF Software 7.0.8493. Prior to immunophenotyping, the FCS files must be cleaned as described in Figure 1 (Bagwell et al., 2020) (Thrash et al., 2020). Briefly, cell events are gated by event length versus time, beads (to remove EQ calibration beads), and Gaussian parameters (Residual, Width, Offset). Single cells are determined by a bi-axial plot of both DNA Intercalator channels. Specific to bone marrow, we identified live, CD45+ cells, and then confirmed the depletion of CD138 before continuing analysis. As noted in Figure 2, CD45 bone marrow populations are donor-dependent and can vary from a CD45 smear to distinct CD45hi and CD45dim populations. Further analysis focused on the CD45hi lymphocyte population, excluding the non-viable and CD45dim erythroid populations (Cloos et al., 2018).
Figure 1.
Clean up gating strategy used for identifying of viable, CD45+CD138- single cell population using gaussian parameters and 191Ir/193Ir biaxial plots
The single cells were gated against bi-axial plots versus CD45 and CD138 to confirm the population of interest for downstream analysis.
Figure 2.
CD45 expression level, as it is a phosphatase, is expressed in different levels on progenitor/stem cells and is donor dependent
After isolating the CD45hi lymphocyte population, we manually gated our desired lineage populations. This included monocytes (CD45+CD138-CD19-CD14+), B cells (CD45+CD138-CD14-CD19+), T cells (CD45+CD138-CD14-CD19-CD56-CD3+), and NK cells (CD45+CD138-CD14-CD19-CD3-CD56+). The lineage gating strategy is shown in Figure 3. The remaining markers in the panel were used to further characterize the phenotype and function of each cell type.
Figure 3.
Cleaned, CD45hi population manually gated to define lineage populations
Limitations
This protocol was established for staining extracellular proteins and secreted proteins, including cytokines, and other intracellular antigens. It is not applicable for staining of activation-induced phosphorylated antigens.
Troubleshooting
Problem 1
Low cell recovery at the end of staining protocol (step 41)
Potential solution
Do not use microcentrifuge 1.5 mL or 2 mL tubes for this protocol, always use 96 well, V-bottom plate. Do not discard supernatant through plate flicking, but instead aspirate the volume to discard using a multichannel pipette.
Problem 2
High level of 127-iodine contamination in your samples (step 9 and data analysis)
Potential solution
The most popular source of 127-Iodine contamination is Ficoll – Paque, as it contains sodium diatrizoate. It is crucial to carefully remove the immune cells layer without touching the Ficoll - Paque layer or use tubes with a separation membrane. If contamination still occurs, it might be reduced by 3 supplementary washing steps after cell thawing.
Problem 3
Staining seems to “fade” or “streak” over time on certain markers (step 25 and data analysis)
Potential solution
Fixation using freshly prepared PFA is crucial. The fixative agent in the Fix/Perm buffers may degrade over time after repeated exposure to oxygen; freshly prepared PFA assures proper fixation and maintenance of staining signal. We suggest freshly fixing stained cells using 4% PFA. The concentration can be adjusted depending on specific needs of the assay, but not decreased more than 1.6% PFA.
Problem 4
Significantly more than 20 samples to stain and to acquire on mass cytometer (steps 1 and 42)
Potential solution
Up to 80 samples can be stained within the same plate, and the plate can be frozen after the 191-Ir DNA Intercalator incubation in freezing medium containing 90% of FBS and 10% of DMSO (Sumatoh et al., 2017). For long acquisitions, the best solution is to freeze all plates and book the mass cytometer for the number of consecutive days needed. To prepare samples for acquisition, thaw each plate the morning of acquisition, and continue with washes, treating the samples as if they were incubated overnight in a DNA-intercalator.
Problem 5
103Rh Viability Staining seems not to perform when analyzing data (step 9 and data analysis)
Potential solution
Be careful when modifying fixation and permeabilization buffers, as for example 103Rh viability staining is not compatible with methanol fixation. If other fix/perm reagents are being used, staining with Cell-ID™ Cisplatin, Fluidigm #SKU 201064 might replace 103Rh Viability staining. Cisplatin binds covalently to protein and labeling remains strong through subsequent steps of the protocol.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Joanna Baginska, joanna_baginska@dfci.harvard.edu.
Materials availability
There were no new reagents generated.
Acknowledgments
We thank Suzan B. Lazo for technical assistance.
Author contributions
Conceptualization, J.B.; Methodology, M.H., J.D., C.A., J.B., R.S.P., A.N.S., M.S., E.D.L., and D.L.M.; Investigation, M.H. and J.B.; Resources, M.H., R.S.P., A.N.S., D.L.M., M.S.; Writing-Original Draft, M.H., J.D., R.S.P.; Visualization, M.H.; Funding Acquisition I.M.G., F.S.; Supervision, J.B.
Declaration of interests
Hodi FS: Dr. Hodi reports grants and/or personal fees from Bristol-Myers Squibb, Merck, EMD Serono, grants and Novartis, Surface, Compass Therapeutics, Apricity, Sanofi, Pionyr, 7 Hills Pharma, Torque, Bicara, Pieris Pharmaceuticals, Checkpoint Therapeutics, Genentech/Roche, Bioentre, Gossamer, Iovance, Trillium, Catalym, Immunocore, Amgen, Zumutor, outside the submitted work; in addition, Dr. Hodi has a patent (#20100111973) with royalties paid, patents (#7250291) (#9402905) (# 10279021) (#10106611) (#20170248603) (#20160046716) (#20140004112) (#20170022275) (#20170008962) and (#20170343552) pending. Ghobrial IM : Dr Ghobrial has the following potential conflicts of interest to disclose: Honoraria: Celgene, Bristol-Myers Squibb, Takeda, Amgen, Janssen; Consulting or Advisory Role: Bristol-Myers Squibb, Novartis, Amgen, Takeda, Celgene, Cellectar, Sanofi, Janssen, Pfizer, Menarini Silicon Biosystems Oncopeptides, The Binding Site, GlazoSmithKlein, AbbVi Adaptive; Travel, Accommodations, Expenses: Bristol-Myers Squibb, Novartis, Celgene, Takeda, and Janssen Oncology.
Contributor Information
Margaret Hallisey, Email: margaretm_hallisey@dfci.harvard.edu.
Joanna Baginska, Email: joanna_baginska@dfci.harvard.edu.
Data and code availability
FCS files from healthy donors will be available upon request.
References
- Bagwell C.B., Inokuma M., Hunsberger B., Herbert D., Bray C., Hill B., Stelzer G., Li S., Kollipara A., Ornatsky O., Baranov V. Automated data cleanup for mass cytometry. Cytometry A. 2020;97:184–198. doi: 10.1002/cyto.a.23926. [DOI] [PubMed] [Google Scholar]
- Cloos J., Harris J.R., Janssen J., Kelder A., Huang F., Sijm G., Vonk M., Snel A.N., Scheick J.R., Scholten W.J., et al. Comprehensive protocol to sample and process bone marrow for measuring measurable residual disease and leukemic stem cells in acute myeloid leukemia. J. Vis. Exp. 2018 doi: 10.3791/56386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumatoh H.R., Teng K.W., Cheng Y., Newell E.W. Optimization of mass cytometry sample cryopreservation after staining. Cytometry A. 2017;91:48–61. doi: 10.1002/cyto.a.23014. [DOI] [PubMed] [Google Scholar]
- Thrash E.M., Kleinsteuber K., Hathaway E.S., Nazzaro M., Haas E., Hodi F.S., Severgnini M. High-throughput mass cytometry staining for immunophenotyping clinical samples. STAR Protoc. 2020;1:100055. doi: 10.1016/j.xpro.2020.100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
FCS files from healthy donors will be available upon request.