Abstract
Objective
To investigate the abundance and the prevalence of Dysosmobacter welbionis J115T, a novel butyrate-producing bacterium isolated from the human gut both in the general population and in subjects with metabolic syndrome. To study the impact of this bacterium on host metabolism using diet-induced obese and diabetic mice.
Design
We analysed the presence and abundance of the bacterium in 11 984 subjects using four human cohorts (ie, Human Microbiome Project, American Gut Project, Flemish Gut Flora Project and Microbes4U). Then, we tested the effects of daily oral gavages with live D. welbionis J115T on metabolism and several hallmarks of obesity, diabetes, inflammation and lipid metabolism in obese/diabetic mice.
Results
This newly identified bacterium was detected in 62.7%–69.8% of the healthy population. Strikingly, in obese humans with a metabolic syndrome, the abundance of Dysosmobacter genus correlates negatively with body mass index, fasting glucose and glycated haemoglobin. In mice, supplementation with live D. welbionis J115T, but not with the pasteurised bacteria, partially counteracted diet-induced obesity development, fat mass gain, insulin resistance and white adipose tissue hypertrophy and inflammation. In addition, live D. welbionis J115T administration protected the mice from brown adipose tissue inflammation in association with increased mitochondria number and non-shivering thermogenesis. These effects occurred with minor impact on the mouse intestinal microbiota composition.
Conclusions
These results suggest that D. welbionis J115T directly and beneficially influences host metabolism and is a strong candidate for the development of next-generation beneficial bacteria targeting obesity and associated metabolic diseases.
Keywords: obesity, probiotics, intestinal microbiology
Significance of this study.
What is already known on this subject?
Numerous bacteria from the human gut remain to be identified.
The vast majority of bacteria detected by sequencing methods have never been cultured.
Several bacteria have been shown to be positively or negatively associated with metabolic health but few of them have been tested in preclinical studies and then validated in human studies.
What are the new findings?
Dysosmobacter welbionis is a newly isolated commensal bacterium found in 70% of the general population.
D. welbionis is inversely correlated with body mass index (BMI), glycaemia and glycated haemoglobin in overweight and obese subject with a metabolic syndrome.
Mice treated with live D. welbionis J115T are partially protected against diet-induced obesity and fat mass gain, have improved glucose tolerance and lower insulin resistance.
Live D. welbionis J115T reduces white adipose tissue hypertrophy and inflammation together with increased number of mitochondria in brown-adipose tissue and non-shivering thermogenesis.
Pasteurisation of D. welbionis J115T abolishes its beneficial effects.
How might it impact on clinical practice in the foreseeable future?
These findings suggest that D. welbionis J115T is a next-generation beneficial bacterium that should be investigated in other clinical paradigm than obesity and diabetes. This bacterium should be tested in humans for its positive effects on host metabolism.
Introduction
Variations of the intestinal microbiota composition and functionality have an impact on host physiology by influencing metabolism, immunity, ageing and behaviour.1 2 The intestinal microbiota is mainly studied by culture-independent methods based on faecal DNA sequencing such as 16S rRNA gene sequencing or metagenomic sequencing. These observational studies provide initial evidence of an association between gut microbiota composition and/or functionality and host pathophysiology.3–5 Yet, moving from association to causation for specific taxa requires growing the bacteria of interest in pure culture. Despite large-scale efforts to cultivate previously uncultured prokaryotic members of the human intestinal microbiota,6–10 recent estimates indicate that it comprises at least 4664 species, of which only 1332 have a cultivated representative.7 Moreover, the effect of specific species on the development of obesity and associated metabolic disorders has been demonstrated only for a handful of species. For example, it has been shown that the colonisation of germ-free mice with a strain of Enterobacter cloacae aggravates obesity and insulin resistance on high-fat diet (HFD)11 and that supplementation of conventionally raised mice with Bilophila wadsworthia aggravates glucose intolerance and hepatic steatosis on HFD without effect on the body weight.12 Conversely, a beneficial impact of other bacteria has been observed: administration of Anaerobutyricum soehngenii (formerly designated as Eubacterium hallii strain L2-7) to genetically obese/diabetic mice increased energy expenditure and decreased adiposity.13 In humans, A. soehngenii administration is associated with improved peripheral insulin sensitivity.14 Colonisation of germ-free mice with the microbiota of an obese human with low levels of Christensenellaceae amended with Christensella minuta reduces adiposity in recipient mice.15 In 2013, we showed that Akkermansia muciniphila counteracts diet-induced obesity and related disorders, such as glucose intolerance, insulin resistance and hepatic steatosis in rodents.16 Later, we demonstrated that bacterial viability was not required to maintain its health-promoting properties, as pasteurised A. muciniphila was as effective as the live bacteria.17 Noteworthy, we demonstrated that A. muciniphila was also beneficial when administered to overweight and obese humans.18 The aforementioned examples are showing that correlations between the abundance of specific bacteria in human cohorts19 with clinical variables can then be successfully translated to a beneficial or harmful effect in animal models. Nevertheless, one needs to move from correlation to causality. Indeed, we found that Subdoligranulum spp was strongly correlated with A. muciniphila and metabolic health, or increased on dietary fibres intervention or metformin administration in numerous human cohorts.20 However, treating mice with Subdoligranulum variabile did not improve diet-induced metabolic disorders,20 thereby reinforcing the urgent need to directly challenge the real physiological relevance of correlative observations.
We recently isolated and described a novel Ruminococcaceae species: Dysosmobacter welbionis.21 This bacterium is closely related to a not yet cultured species that has been consistently associated to leanness: Oscillospira guillermondii.22 In the present study, we aimed to decipher whether D. welbionis has an impact on host’s metabolic health. First, we measured the relative abundance and prevalence of D. welbionis in the faecal microbiota of the general population using several cohorts (ie, human microbiome project (HMP), American Gut Project (AGP), Flemish Gut Flora Project (FGFP) and Microbes4U).18 23–26 We discovered that D. welbionis abundance correlates negatively with body mass index (BMI) and fasting glycaemia. Then, we showed that supplementation of D. welbionis strain J115T to HFD-fed mice reduced body weight and fat mass gain. This was associated with improved glucose homeostasis and increased non-shivering thermogenesis, coupled to a higher number of mitochondria in the brown adipose tissue (BAT).
Results
Prevalence and abundance of D. welbionis in the human microbiota
We recently isolated a new species from human stools that belongs to the Ruminococcaceae family that we named D. welbionis.21 This is the first species isolated from this new genus.21 Beyond the taxonomic interest of the description of a novel genus and species, it emerges that D. welbionis is closely related to several taxa associated with lower body weight. Indeed, D. welbionis has 92% of 16S rRNA gene sequence similarity with the uncultured species O. guillermondii that has repeatedly been associated with leanness in human studies based on faecal microbiota sequencing.22 In addition, the Anaerotruncus genus, also belonging to the Ruminococcaceae family, is also negatively correlated with body weight in pigs and humans.27 28 Considering the taxonomic proximity of D. welbionis with these taxa, we sought to decipher the association between Dysosmobacter abundance in the intestinal microbiota and host health.
We first investigated whether D. welbionis was present in the general population. To this aim, we used several independent cohorts such as the HMP,25 the AGP23 24 and the FGFP.26 Based on the 16S rRNA gene sequence of D. welbionis J115T, this sequence variant had a prevalence of 62.7% and a relative abundance range of 0%–3.9% in the HMP cohort (n=161). For the AGP cohort (n=9511) the prevalence was 69.8% with a relative abundance range of 0%–9.2%. Finally, the sequence variant was detected in 65.9% of individuals of the cross-sectional FGFP (n=2259) with a relative abundance ranging between 0% and 0.9%.
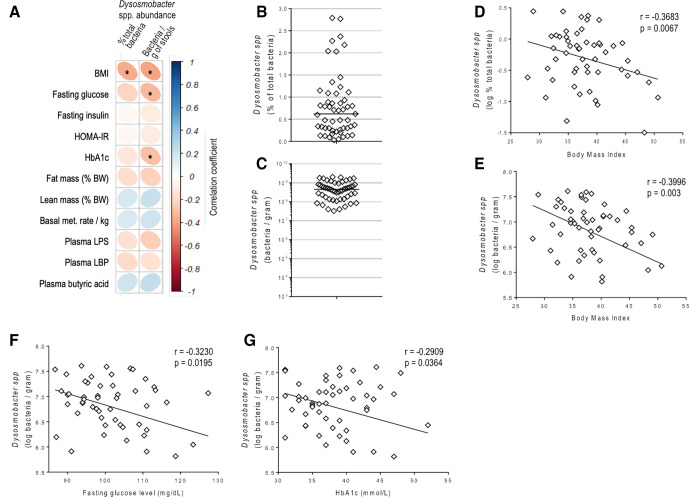
We further explored whether this bacterium was correlated with BMI in obese subjects or any specific metabolic markers using a previously recruited cohort of 53 non-diabetic, insulin-resistant and overweight or obese individuals: the Microbes4U cohort.18 Using Dysosmobacter genus-specific primers, we quantified it in the faecal DNA of the Microbes4U cohort. Dysosmobacter spp was detected in the stools of all individuals and ranged between 0.00035% and 2.7% of the total microbiota with an average of 0.75 %, corresponding to a range of 1.7×105 to 2.0×109 with an average of 5.9×108 bacteria per gram of stools (figure 1).
Figure 1.
Dysosmobacter spp correlates negatively with BMI in humans. (A) Pearson’s correlation matrix between Dysosmobacter spp abundance in the faecal microbiota and clinical variables in the Microbes4U cohort. *P<0.05. (B) Dysosmobacter spp relative abundance in the faecal microbiota of a cohort of overweight and obese humans. (C) Dysosmobacter spp concentration in stool samples from a cohort of overweight and obese humans. (D) Pearson’s correlation between Dysosmobacter spp relative abundance and BMI. (E) Pearson’s correlation between Dysosmobacter spp absolute concentration and BMI. (F) Pearson’s correlation between Dysosmobacter spp concentration and fasting blood glucose. (G) Pearson’s correlation between Dysosmobacter spp. and glycated haemoglobin. Results are represented as dot-plots with median for figure parts B, C. BMI, body mass index; HbA1c, glycated haemoglobin.
Interestingly, in the Microbes4U cohort, the relative and absolute abundance of Dysosmobacter spp correlated negatively with BMI, while absolute abundance of Dysosmobacter spp correlated negatively with fasting glucose and glycated haemoglobin (HbA1c) (figure 1A, D–G, online supplemental table S1). This suggests that Dysosmobacter genus may protect against obesity and obesity-associated glucose metabolism alteration.
gutjnl-2020-323778supp001.pdf (31.6KB, pdf)
D. welbionis J115T prevents diet-induced obesity in mice
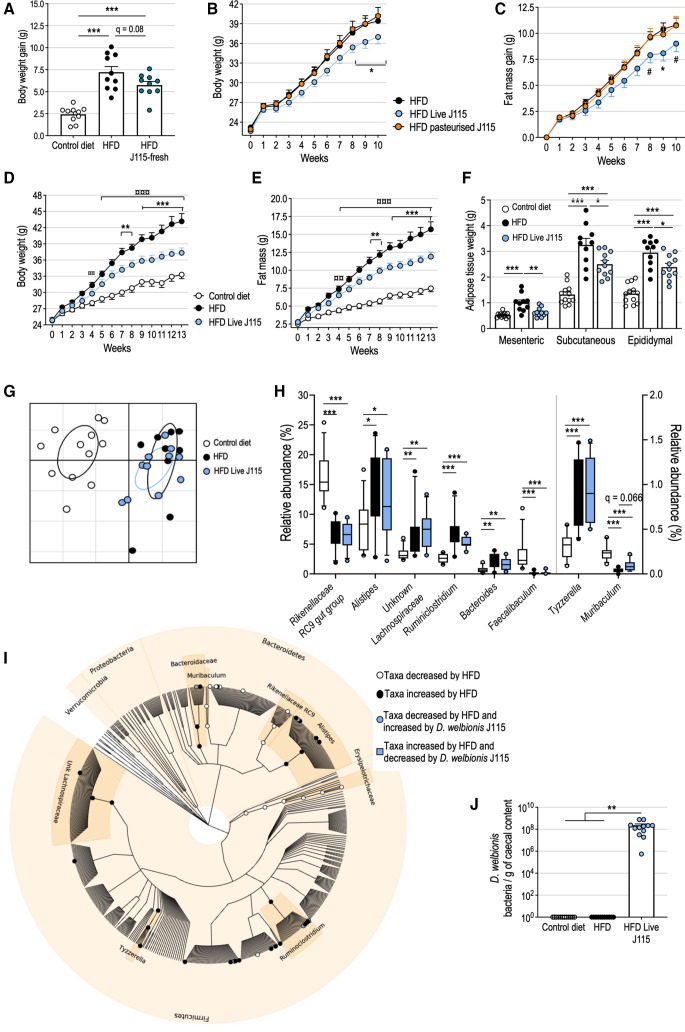
In a first experiment, we found that gavaging high-fat diet (HFD)-fed mice with daily and freshly prepared live D. welbionis J115T (1.0×109 cultivable, live bacteria per day and per mouse) for 6 weeks (HFD J115-fresh group) decreased the HFD-induced weight gain by 29% (ie, 1.5 grams) (figure 2A). We then investigated whether the viability of D. welbionis J115T impacted its anti-obesity effects. Killing D. welbionis J115T using pasteurisation (30 min at 70°C) completely abolished its beneficial effects on both body weight and fat mass gain (figure 2B, C). Then, we tested whether treating mice with live D. welbionis J115T for a longer period of time (ie, 13 weeks) could maintain its beneficial effects. We found that live D. welbionis J115T remains active when frozen in trehalose (1.0×109 cultivable, live bacteria per day and per mouse) and profoundly decreased HFD-induced weight gain and fat mass gain than HFD-fed mice after 13 weeks of treatment (figure 2D, E). This corresponded to a significant reduction in the weight of mesenteric, subcutaneous (inguinal) and epididymal fat deposits (figure 2F). No impact on the weight of several muscles of the mice was observed (online supplemental figure S1). It is worth noting that treating mice with a dose lower than 1.0×109 live bacteria (ie, 108) had no beneficial effects (online supplemental figure S2).
Figure 2.
Live Dysosmobacter welbionis J115T prevents diet-induced obesity in mice without major alterations of the faecal microbiota composition. (a) Body weight gain of mice fed a HFD and treated during 6 weeks by daily oral gavage with 1.0×109 colony forming units (cfus) of freshly prepared D. welbionis J115T (HFD J115-fresh) and mice fed a control diet or a high-fat diet (HFD) and treated by daily oral gavage with vehicle. (B, C) Body weight and fat mass gain of mice treated during 10 weeks by daily oral gavage with live D. welbionis J115T frozen in trehalose (1.0×109 cultivable, live bacteria per day and per mouse) and fed a HFD (HFD Live J115) or pasteurised D. welbionis J115T (HFD pasteurised J115) (1.0×109 heat-killed bacteria per day and per mouse) and mice fed a HFD and treated by daily oral gavage with vehicle. (D, E) Body weight and fat mass of mice treated during 13 weeks by daily oral gavage live D. welbionis J115T frozen in trehalose (1.0×109 cultivable, live bacteria per day and per mouse) and fed a HFD (HFD Live J115) and mice fed a control diet or a HFD and treated by daily oral gavage with vehicle. (F) Mesenteric, subcutaneous (inguinal) and epididymal fat pads weight at the end of the 13-week period. (G) Principal coordinates analysis of the microbiota composition of experiment 2. Mice microbiota were clustered and the centre of gravity computed for each group. (H) Relative abundance of the bacterial genera significantly altered by HFD or live D. welbionis J115T treatments. (I) Cladogram representing mice microbiota with white clade markers highlighting bacterial groups significantly more abundant in control mice than in HFD mice, black clade markers markers highlighting bacterial groups significantly more abundant in HFD mice than in control mice and light blue clade markers highlighting bacterial groups significantly increased (circle) or decreased (square) by live D. welbionis J115T administration in HFD-fed mice as assessed by figure part H. (J) Dysosmobacter spp concentration estimated by quantitative PCR in the caecal content of the mice. Number of mice per group: 10–12. Data were analysed using one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test for figure parts A, F and J and two-way repeated measures ANOVA for figure parts B–E. Data were analysed using Kruskal–Wallis test followed by Dunn’s pairwise multiple comparison procedure for H–I. *q<0.05; **q<0.01; ***q<0.001. Results are represented as dot plots and bar plots with mean±SEM for figure parts A, F and J, and as boxes and whiskers (first quartile, median and third quartile) for figure part H. In figure parts B–E *q < 0.05; **q<0.01; ***q<0.001 for HFD versus HFD Live J115 comparisons and ¤¤¤q<0.001 for control versus HFD comparisons. In figure part C, #p=0.06. HFD, high-fat diet.
gutjnl-2020-323778supp002.pdf (7.5MB, pdf)
D. welbionis J115T supplementation has minor impact on mouse intestinal microbiota composition
To investigate if the protection against HFD-induced weight gain is a direct effect of D. welbionis J115T or results from the remodelling of the intestinal microbiota ecosystem, we analysed the mouse caecal microbiota composition by 16S rRNA gene amplicon sequencing. Principal coordinates analysis based on generalised UniFrac dissimilarity matrix indicated that HFD profoundly affected the overall composition of the ecosystem. The microbiota of HFD mice and HFD mice that received live D. welbionis were very similar (figure 2G). Richness, Simpson’s and Shannon’s alpha diversity indices were similar between the three groups (online supplemental figure S3). We found that 31 operational taxonomic units (OTUs) had a significantly different relative abundance between two or more mouse groups (figure 2H, I).
Bacteroidaceae family as well as Alistipes, Ruminoclostridium, Tyzerella and unidentified Lachnospiraceae genera were increased by the HFD while Erysipelotrichaceae family along with Rikenellaceae RC9 gut group and Muribaculum genus were decreased by the HFD in comparison with the control group (figure 2I). The relative abundance of 2 OTUs was increased by the HFD and restored to control diet mice levels by D. welbionis J115T treatment: OTUs 11 and 16, belonging to Ruminoclostridium and Alistipes genera, respectively. OTUs 54 and 71, associated to Bacteroides and Ruminoclostridium genera, were respectively increased and decreased by live D. welbionis J115T treatment but were not affected by the diet. Quantitative PCR (qPCR) indicated that D. welbionis was not present in untreated mice and reached 2.3×108 bacteria per gram of caecal content (corresponding to 0.76% of the microbiota) in mice force fed with live D. welbionis J115T (figure 2J). D. welbionis J115T did not durably colonise the mouse gut as it was not detected in the faeces of any mouse after a washout period of 3 days (online supplemental figure S3).
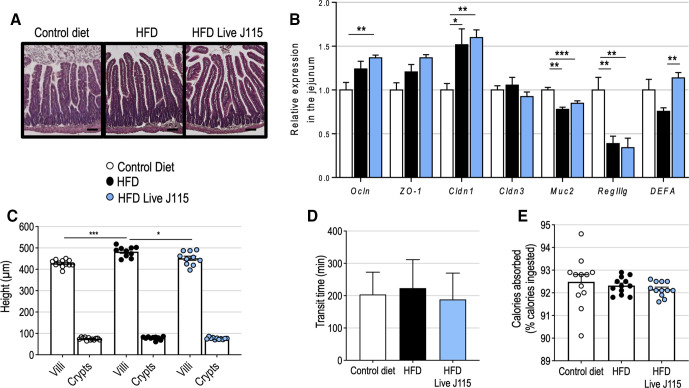
D. welbionis J115T slightly affects gut morphology without impacting the absorptive function
As Dysosmobacter spp is not detected in the gut microbiota of specific pathogen free mice, we sought to confirm that live D. welbionis J115T administration was well tolerated by the mice. No sign of inflammation was histologically observed in the jejunum of the mice treated with live D. welbionis J115T (figure 3A). This was supported by the measurement of the relative expression of pro-inflammatory markers in different segments of the gut (online supplemental figure S3) and the absence of deleterious effects on gut barrier markers (eg, antimicrobial peptides, mucus production and tight-junction proteins) (figure 3B). Live D. welbionis J115T administration slightly decreased the height of the villi in the jejunum (542.7 vs 482.3 µm, q=0.024), but the depth of the crypts was not affected (figure 3C). A decrease of the villi’s height could potentially result in a reduction of the absorption surface and affect energy absorption, as would an acceleration of the transit time. However, measurement and calculation of both transit time and percentage of dietary calories absorbed from the diet showed that none of these parameters were affected by live D. welbionis J115T administration (figure 3D–E). Our results show that live D. welbionis J115T was well tolerated by the mice and did not alter gut physiology.
Figure 3.
Live Dysosmobacter welbionis J115T moderately alters gut physiology. (A) Representative H&E-stained pictures of the jejunum. Scale bar=100 µm. (B) Relative expression of genes related to gut barrier function in the jejunum. (C) Mean crypts and villi’s height in the jejunum. (D) Transit time. (E), Percentage of calories absorbed from the food. Number of mice per group: 10–12. Data were analysed using one-way analysis of variance followed by Tukey’s post hoc test for figure part B. *q<0.05; ***q<0.001. Results are represented as bar plots with mean±SEM for figure parts B–D and dot plots and bar plots with mean±SEM for figure parts C–E. HFD, high-fat diet.
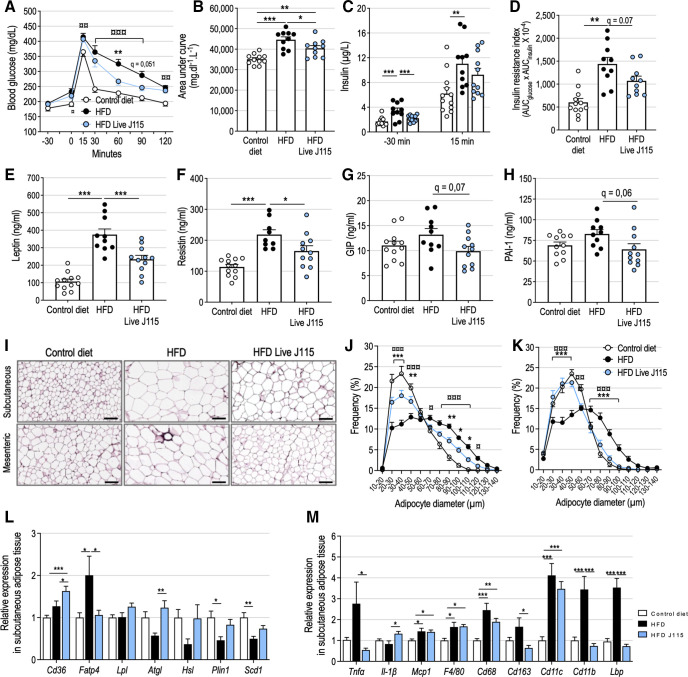
D. welbionis J115T reduces adipose tissue expansion on HFD and improves glucose control
The lower adiposity observed with live D. welbionis J115T administration was associated with better glucose tolerance during an oral glucose tolerance test (OGTT), with faster return to baseline glucose levels after the initial blood glucose peak 15 min after glucose administration (figure 4A, B). This was associated to significantly lower fasting insulin levels and lower insulin resistance index (figure 4C, D). Moreover, mice supplemented with live D. welbionis J115T had significantly lower plasma levels of leptin and resistin than untreated HFD-fed mice (figure 4E, F). A similar trend was observed for plasma glucose-dependent insulinotropic polypeptide (GIP) and plasminogen activator inhibitor-1 (PAI-1) levels (figure 4G, H). Conversely, plasma lipids were not modified by the treatment (online supplemental figure S1).
Figure 4.
Live Dysosmobacter welbionis J115T reduces adipose tissue expansion and inflammation on high-fat diet (HFD) and improves altered metabolic profile. (A) Plasma glucose profile and (B) mean area under the curve measured during an oral glucose tolerance test (OGTT). (C) Plasma insulin measured 30 min before and 15 min after glucose administration during the OGTT. (D) Insulin resistance index. (E) Leptin, (F) resistin, (G) glucose-dependent insulinotropic polypeptide (GIP) and (H) plasminogen activator inhibitor-1 (PAI-1) plasma levels after a 6 hours fasting period. (I) Representative H&E-stained pictures of subcutaneous and mesenteric adipose tissues (SAT and MAT, respectively). Scale bar=100 µm. (J) Adipocytes diameter (µm) distribution in the SAT. (K) Adipocytes diameter (µm) distribution in the MAT. (L) Relative expression of genes related to lipid metabolism in the SAT. (M) Relative expression of genes related to inflammation and immune system in the SAT. Number of mice per group: 9–12. Results are represented as dot plots and bar plots with mean±SEM for figure parts B–H and as bar plots with mean±SEM for figure parts L and M. Data were analysed using one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test for figure parts B–H, L and M and two-way repeated measures ANOVA for figure parts A, J and K. *q<0.05; **q<0.01; ***q<0.001 for HFD versus HFD Live J115 comparisons and ¤¤q<0.01 control versus HFD, ¤¤¤q<0.001 for control versus HFD comparisons. HFD, high-fat diet.
D. welbionis J115T supplementation is associated with reduced adipocyte diameter and lower adipose tissue inflammation
Obesity is associated with adipocyte hypertrophy, impaired lipolysis and pro-inflammatory phenotype, which contribute to adverse adipokine secretion, insulin resistance and ectopic fat deposition. Thus, we measured adipocyte diameter in the subcutaneous adipose tissue (SAT) and mesenteric adipose tissue (MAT) by histology and image analysis (figure 4I). D. welbionis J115T treatment significantly increased the proportion of small adipocytes (diameter ≤50 µm) and decreased the proportion of large adipocytes (diameter ≥70 µm) in SAT (figure 4J). This effect was even more pronounced in MAT (figure 4K).
D. welbionis J115T increases the expression of lipolysis markers in association with altered inflammation markers expression in white adipose tissue
Smaller adipocyte size in live D. welbionis J115T treated mice was associated with an overall lipid metabolism less prone to fat storage. Indeed, the fatty acids transport protein 4 (Fatp4), involved in fatty acids uptake from the bloodstream, was two times less expressed in the SAT of treated mice (figure 4L). Conversely, genes involved in lipolysis and fatty acid mobilisation, such as the adipose triglycerides lipase (Atgl) and the hormone sensitive lipase (Hsl), were more expressed in the SAT of treated mice than in HFD mice. Moreover, the relative expression of the proinflammatory cytokine Tnfα and several macrophages infiltration markers such as Cd68, Cd163, Cd11b and Lbp was increased by the HFD and decreased by live D. welbionis J115T supplementation, whereas some other markers were not affected (figure 4M).
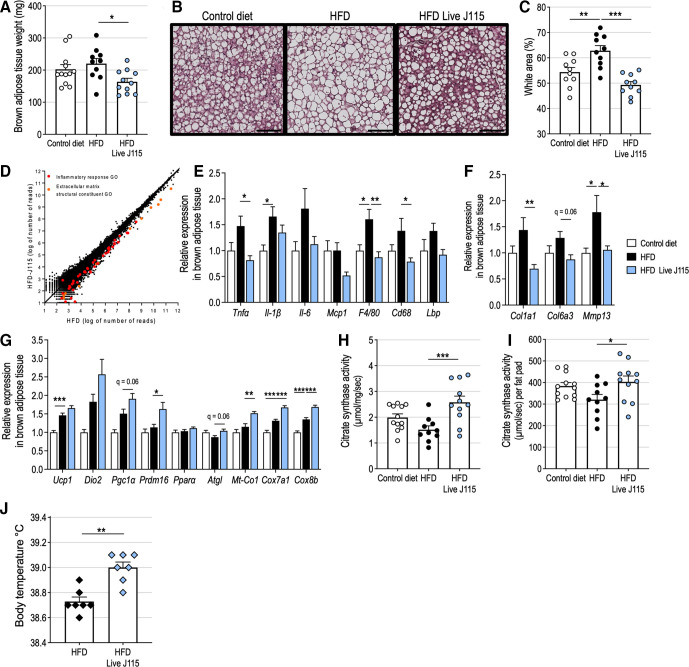
D. welbionis J115T counteracts HFD-induced alterations and increases mitochondria number in BAT
The weight of the interscapular BAT of HFD-fed mice supplemented with live D. welbionis J115T was significantly lower than that one of the HFD-fed mice and even slightly lower than that of control diet fed mice (figure 5A). Histology analysis indicated that BAT expansion induced by the HFD was associated with increased lipid droplet size and a unilocular, white-like appearance of the adipocytes (figure 5B). The surface occupied by the lipids droplets in the slices was significantly increased by the HFD (figure 5C). Supplementation of HFD-fed mice with live D. welbionis J115T restored the brown adipocyte morphology and reduced the lipid content. Then, we performed a RNA-seq analysis on a pool of RNA extracted from the BAT of HFD-fed mice treated or not with live D. welbionis J115T (figure 5D). Among the differences between the two pools, it appeared in particular that transcripts belonging to the inflammatory response process and extracellular matrix gene ontologies (GO:0006955 and GO:0030020) were more expressed in the BAT of HFD mice than in that of HFD live D. welbionis J115T treated mice (figure 5D, online supplemental figure S4, online supplemental table S2). Quantitative PCR on several markers of inflammation and macrophage infiltration performed on all mice confirmed that HFD increased inflammation in comparison with control diet-fed mice and that inflammation of the BAT on HFD was completely prevented by live D. welbionis J115T supplementation (figure 5E). In addition, the expression of profibrotic factors such as collagens and matrix metalloproteinases genes was upregulated by the HFD and normalised by live D. welbionis J115T (figure 5F), suggesting a protection against HFD-induced remodelling and/or fibrosis of the BAT.
Figure 5.
Live Dysosmobacter welbionis J115T reduces high fat diet (HFD)-induced brown adipose tissue (BAT) dysfunction and increases mitochondria number. (A) Interscapular BAT weight of mice treated by daily oral gavage with live D. welbionis J115T frozen in trehalose and fed an HFD (HFD Live J115) or mice fed a control diet or a HFD and treated by daily oral gavage with an equivalent volume of vehicle. (B) Representative H&E-stained pictures of BAT. Scale bar=100 µm. (C) Percentage of white area on the slices, corresponding to lipid droplets, in the BAT. (D) Scatter dot plot between RNA-seq expression data of a pool of RNA from the BAT of HFD mice and a pool of RNA from the BAT of HFD J115 mice. (E) Relative expression of genes related to inflammation and immune system in the BAT. (F) Relative expression of genes related to extracellular matrix and fibrosis in the BAT. (G) Relative expression of genes related to mitochondria number and function in the BAT. (H) citrate synthase activity per mg of BAT. (I) Citrate synthase activity per brown fat pad. (J) Body temperature of mice treated by daily oral gavage with live D. welbionis J115T and fed a high fat-diet (HFD Live J115) or mice fed a high-fat diet (HFD) and gavaged daily with vehicle for 3 weeks (experiment 4, see methods). Number of mice per group: 10–12 in figure parts A, C and E–I). Number of mice per group: 7 in figure part J. Results are represented as dot plots and bar plots with mean±SEM for figure part A, C and H–J and as bar plots with mean±SEM for figure parts E–G. Data were analysed using one-way analysis of variance followed by Tukey’s post hoc test for figure parts A, C and E–I and Mann-Whitney test for figure part J. *q<0.05; **p or q<0.01; ***q<0.001.HFD, high-fat diet.
gutjnl-2020-323778supp003.pdf (619.2KB, pdf)
Inflammation and brown-to-white conversion are associated with mitochondrial dysfunction in the BAT and impaired thermogenesis. The expression of BAT-specific uncoupling protein Ucp1, specific BAT markers (Prdm16 and Dio2) and mitochondrial biogenesis marker Pgc1α (figure 5G) was increased by D. welbionis J115T supplementation, although statistical significance was reached only for Prdm16. To confirm the possible BAT activation and higher number of mitochondria, we measured the expression of several genes of the mitochondrial respiratory chain (Mt-Co1, Cox7a1 and Cox8b). Their expression was significantly increased by the HFD and was further raised by live D. welbionis J115T (figure 5G).
To further investigate whether the genetic signature observed when investigating mRNA could be linked with specific changes at the metabolic level, we measured citrate synthase activity. This activity (expressed per mg of tissue), a proxy of mitochondria content, was 70% higher in the BAT of HFD live J115 mice than in that of HFD mice (figure 5H). Despite the reduction of the BAT size, the total citrate synthase activity of the HFD live J115 mice fat pads remained 25.7% higher than the activity in fat pads of mice kept on HFD alone (figure 5I), suggesting that live D. welbionis J115T supplementation increases mitochondrial content and activity in the BAT. As the BAT produces heat by non-shivering thermogenesis, we sought to determine if live D. welbionis J115T supplementation increases body temperature. In a separate set of experiments (experiment 4, detailed in Material and methods) during which mice were subjected to HFD or HFD and live D. welbionis J115T supplementation for 3 weeks (online supplemental figure S5), we observed that mice supplemented with live D. welbionis J115T had a rectal temperature on average 0.25°C higher than HFD mice (figure 5J).
Material and methods
See online supplemental material and methods.
gutjnl-2020-323778supp004.pdf (134.9KB, pdf)
Discussion
In this study, we first discovered that the newly isolated bacterium D. welbionis J115T is highly prevalent in the general population and that this bacterium is inversely associated with BMI, fasting plasma glucose and HbA1c in obese humans suffering from metabolic syndrome. We next demonstrated that supplementation with live D. welbionis J115T partially protects mice against HFD-induced metabolic disorders and low-grade inflammation notably through a mechanism involving the recovery of BAT metabolism and activity.
Although numerous bacteria have been shown to be positively or negatively associated with obesity and cardiometabolic disorders, the vast majority have never been cultured,7 making it difficult to demonstrate the proof of concept of their causal involvement in adverse or beneficial phenotypes. In this study, we moved from the simple correlation to the demonstration that the newly identified bacteria D. welbionis J115T is a promising candidate. It is important to note that the bacterium has been grown in a well-defined medium suitable for use of the bacterium in humans. This makes it possible to circumvent the difficulties previously faced when investigating A. muciniphila, for which all the initial data in mice were obtained using a medium containing animal-derived compounds not suitable for administration to humans.17
Interestingly, although D. welbionis had initially been isolated from the human gut of one healthy subject,21 the presence of this bacterium is widely spread into the general population. Indeed, by using several cohorts such as the HMP, the AGP and the FGFP (ie, 11 984 subjects in total), we discovered that at least two-thirds of the general population harbour this newly discovered bacterium in their intestine.
As this bacterium abundance correlates negatively with metabolic parameters and BMI in obese humans suffering from metabolic syndrome, we tested whether the bacterium could affect host metabolism on HFD feeding. To do so, we treated mice with fresh daily prepared live D. welbionis J115T (1.0×109 cultivable, live bacteria per day and per mouse) or using bacteria that were alive but that had been frozen in trehalose (1.0×109 cultivable, live bacteria per day and per mouse) or pasteurised D. welbionis J115T (ie, heat-killed bacteria). We found that both treatment of mice with the live bacteria, either obtained from a fresh culture or frozen in a cryoprotectant, reduced the body weight and the fat mass gain of HFD-treated mice. However, killing the bacteria by using pasteurisation completely abolished its beneficial effects. We found that treating the mice for a longer period of time (ie, 13 weeks) was associated with a stronger effect and further reduced fat mass gain, improved glucose tolerance and reduced insulin resistance. Nevertheless, when treated with a lower dose of D. welbionis J115T (ie <1.0×109 cultivable and viable cells), the mice did not respond to the beneficial effects of the bacteria (online supplemental figure S2).
The viability of probiotics and beneficial bacteria has often been considered as a prerequisite for their health-promoting properties. While it is known to be true for certain species, for example, A. soehngenii whose beneficial effect is associated with its capacity to replicate in the gut,14 it has also been shown that heat-killed A. muciniphila and several Lactobacillus and Bifidobacterium species are as potent as their live form.17 29 In the present study, we found that pasteurised D. welbionis J115T was inefficient. We also found that the live bacterium was unable to permanently colonise the gastrointestinal tract of the mice. Indeed, after 72 hours of wash-out, the faecal content of D. welbionis J115T was below qPCR detection limit (online supplemental figure S3). Whether the beneficial effects of D. welbionis J115T are mediated by constitutive components or metabolites present both in the fresh and frozen preparation but not by metabolites produced in the gut warrants further investigation. Altogether, this study provides a proof of concept that the correlation observed in obese humans is fully supported by mouse experiments showing beneficial effects of D. welbionis J115T.
To further explore the mechanisms underlying how live D. welbionis J115T improves metabolic parameters, we sequenced the gut microbiota of mice and confirmed that HFD changes the gut microbiota composition.30 31 However, we observed only subtle shifts in the gut microbiota composition between treated and untreated groups, thereby suggesting a direct effect of the bacteria on host metabolism rather than through the modulation of the whole gut ecosystem. This is a finding that we previously observed for A. muciniphila.16 18 Hence, we investigated whether the lower body weight and fat mass gain observed on live D. welbionis J115T treatment could be the consequence of acute inflammation or decreased energy absorption. We found that the chronic administration of live D. welbionis J115T did not affect intestinal inflammation, transit time and energy absorption therefore suggesting that bacterium administration is well tolerated and that the metabolic improvements did not result from decreased energy absorption. We found that live D. welbionis J115T completely normalised visceral fat cell morphology, as well as several markers of inflammation known to contribute to the development of insulin resistance and glucose intolerance. These data are also supported by lower circulating levels of resistin and, to a lower extent, of PAI-1. Both were shown to be involved in the onset of insulin resistance.32 33
Besides the white adipose depots, the brown adipose tissue (BAT) is of utmost importance in the control of energy and glucose metabolism. It has been largely described that HFD feeding induces alteration of BAT, with a brown-to-white conversion of the morphology and functionality of the adipocytes, a process referred to as whitening of the brown adipose tissue that eventually leads to lower BAT thermogenesis and energy expenditure.34 We found that live D. welbionis J115T completely prevented high-fat diet-induced brown adipose tissue whitening, characterised by a lower weight and lower abundance of white adipocytes. In addition, RNAseq and qPCR analyses showed that the administration of live D. welbionis J115T reduced macrophages infiltration, expression levels of several markers of inflammation and profibrotic markers, the latter being associated with a dysfunctional adipose tissue and impaired energy production through substrate oxidation.35 Transcriptomic data suggesting an increased number of mitochondria were substantiated by a higher activity of the citrate synthase, an exclusive marker of the mitochondrial matrix.36 This increase in the number of mitochondria is triggered by D. welbionis and leads to higher body temperature, associated to a better energy metabolism and a higher energy expenditure in live D. welbionis J115T treated mice as compared with HFD-treated mice. Although our experiments strongly suggest important effects of live D. welbionis J115T on energy metabolism, it would be interesting to further investigate whether live D. welbionis J115T could induce even stronger effects on metabolism when exposed to thermoneutrality.37
The exact mechanisms of action, however, remain to be deciphered. Among the potential candidate-mediators, we can mention butyrate, which is produced by D. welbionis J115T from myo-inositol and known to increase BAT thermogenesis,38 but also other specific bioactive lipids controlling glucose metabolism and inflammation.39 40 However, we did not find any increase in the levels of butyrate in the plasma or in the caecal content (online supplemental figure 6).
Altogether, our data strongly support that the human gut commensal D. welbionis J115T is a bacterium contributing to the regulation of host’s metabolism in response to an obesogenic diet. Using different complementary in vivo approaches, we demonstrate that D. welbionis J115T regulates host’s energy metabolism likely through the modulation of fat mass development and BAT metabolism. As for other next-generation beneficial bacteria, the final mechanisms explaining how the bacterium is dialoguing with the host deserve additional exploration.41 42 In the present study, although we found that the bacterium is a butyrate producer, we cannot rule out that other specific secreted factors might be involved. Finally, we found that the adminstration of D. welbionis J115T at the dose of 1.0×109 cultivable, viable cells to genetically and severely obese ob/ob mice for 6 weeks did not significantly reduce body weight and fat mass gain (online supplemental figure S7). Altogether, our data strongly support the beneficial effects of live D. welbionis J115T on diet-induced obesity and diabetes. However, whether higher doses of live D. welbionis J115T and/or longer duration of treatment are required to observe a beneficial effect in a specific model such as ob/ob (also known as a hyperphagic model) warrants further investigation. Moreover, we may not exclude that the administration of live D. welbionis J115T induces beneficial effects on insulin resistance independently from changes in adiposity in these mice. Indeed, it has been shown that host exposure time to microbiota is a critical factor to contribute to change insulin sensitivity.43 Therefore, exposure time is an important concern in bacteria targeted strategies aimed at attenuating diabetes versus overweight and obesity.
In conclusion, D. welbionis J115T is a newly isolated human bacterium, which is highly prevalent in the general population and inversely associated with BMI, fasting plasma glucose and HbA1c in overweight or obese subjects suffering from a metabolic syndrome. The daily administration of live D. welbionis J115T to mice for at least 6 weeks and up to 13 weeks blunted the high-fat diet induced metabolic disorders, by mechanisms independent of specific changes in the gut microbiota, but rather acting on both white and brown adipose tissue metabolism.
gutjnl-2020-323778supp005.pdf (32KB, pdf)
Acknowledgments
We are grateful to Alexandre Barrois, Anthony Puel, Rose-Marie Goebbels, Henri Danthinne and Isabelle Blave for excellent technical help. We would like to thank Hubert Plovier for fruitful technical discussions. We would like to thank the Institut de Recherche Expérimentale et Clinique (IREC) imagery platform (2IP) from the IREC for the use of the Leica SCN400 slide scanner.
Footnotes
Twitter: @matthias_vanhul, @Laure_Bindels, @MicrObesity
Contributors: PDC, TLR and EMdH conceived the project. PDC supervised all the preclinical and clinical part. PDC, AE, CDr, CDe, MdB, JPT, AL, DM and MPH contributed to the clinical study Microbes4U. SV, GF and JR contributed to the analysis of the bacterium in the cohort FGFP. RP analysed the human microbiome project and American Gut Project cohort. AP and GGM performed short chain fatty acids measurements. TLR, EMdH, MVH and PDC performed experiments and interpreted all the results. TLR and EMdH generated figures and tables. NMD, LBB, MR and JR contributed to scientific discussions. PDC, TLR, EMdH and MVH wrote the manuscript. All authors discussed the results and approved the manuscript.
Funding: This work was supported by the Fonds de la Recherche Scientifique – FNRS-FNRS via the FRFS-WELBIO under grant WELBIO-CR-2017C-02, WELBIO-CR-2019C-02R and Projet de Recherche PDR convention: FNRS T.0030.21. PDC is a recipient of the Funds Baillet Latour (Grant for Medical Research 2015). This work was supported by the Fonds Wetenschappelijk Onderzoek – Vlaanderen (FWO) and the Fonds de la Recherche Scientifique – FNRS under EOS Project No. (EOS programme no. 30770923). Funding support for the development of NIH Human Microbiome Project – Core Microbiome Sampling Protocol A (HMP-A) was provided by the NIH Roadmap for Medical Research. Clinical data from this study were jointly produced by the Baylor College of Medicine and the Washington University School of Medicine. Sequencing data were produced by the Baylor College of Medicine Human Genome Sequencing Centre, The Broad Institute, the Genome Centre at Washington University and the J. Craig Ventor Institute. These data were submitted by the EMMES Corporation, which serves as the clinical data collection site for the HMP.
Competing interests: PDC is cofounder of A-Mansia biotech. TLR and PDC are inventors on patent applications dealing with the use bacteria in the treatment of obesity and related disorders.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information. All datasets and raw data generated and/or analysed during the current study are available from the corresponding author on reasonable request. The 16S rRNA gene sequencing raw sequences of the mouse study can be accessed in Sequence Read Archive database with accession code PRJNA606762. The raw 16S data of the FGFP cohort are available at European Genome-Phenome Archive (https://ega-archive.org/) under accession no. EGAS00001004420, and for the Microbes4U cohort under accession no. EGAS00001003585. For the HMP, healthy human subjects cohort was downloaded from the human microbiome project data portal (https://portal.hmpdacc.org/).
Ethics statements
Patient consent for publication
Not required.
Ethics approval
The mouse experiments were approved by and performed following the guidelines of the local ethics committee for animal care of the Health Sector of the Université catholique de Louvain under the specific agreement number 2017/UCL/MD/005. Animal housing conditions were as specified by the Belgian Law of 29 May 2013 regarding the protection of laboratory animals (Agreement number LA 1230314). FGFP procedures were approved by the Medical Ethics Committee UZ Brussels-VUB (BUN 143201215505) and the Commissie Medische Ethiek, UZ/KU Leuven (S58125). The cohort Microbes4U was registered at https://clinicaltrials.gov as trial number NCT02637115. Written informed consent was obtained from each participant, and the study protocol was approved by the Commission d’Ethique Biomédicale Hospitalo-facultaire of the Université catholique de Louvain.
References
- 1. Cani PD, Van Hul M, Lefort C, et al. Microbial regulation of organismal energy homeostasis. Nat Metab 2019;1:34–46. 10.1038/s42255-018-0017-4 [DOI] [PubMed] [Google Scholar]
- 2. Marx W, Lane M, Hockey M, et al. Diet and depression: exploring the biological mechanisms of action. Mol Psychiatry 2021;26:134–50. 10.1038/s41380-020-00925-x [DOI] [PubMed] [Google Scholar]
- 3. Cani PD. Gut microbiota — at the intersection of everything? Nat Rev Gastroenterol Hepatol 2017;14:321–2. 10.1038/nrgastro.2017.54 [DOI] [PubMed] [Google Scholar]
- 4. Aron-Wisnewsky J, Vigliotti C, Witjes J, et al. Gut microbiota and human NAFLD: disentangling microbial signatures from metabolic disorders. Nat Rev Gastroenterol Hepatol 2020;17:279–97. 10.1038/s41575-020-0269-9 [DOI] [PubMed] [Google Scholar]
- 5. Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013;500:541–6. 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- 6. Browne HP, Forster SC, Anonye BO, et al. Culturing of 'unculturable' human microbiota reveals novel taxa and extensive sporulation. Nature 2016;533:543–6. 10.1038/nature17645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Almeida A, Mitchell AL, Boland M, et al. A new genomic blueprint of the human gut microbiota. Nature 2019;568:499–504. 10.1038/s41586-019-0965-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Forster SC, Kumar N, Anonye BO, et al. A human gut bacterial genome and culture collection for improved metagenomic analyses. Nat Biotechnol 2019;37:186–92. 10.1038/s41587-018-0009-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zou Y, Xue W, Luo G, et al. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat Biotechnol 2019;37:179–85. 10.1038/s41587-018-0008-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lagier J-C, Khelaifia S, Alou MT, et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol 2016;1:16203. 10.1038/nmicrobiol.2016.203 [DOI] [PubMed] [Google Scholar]
- 11. Fei N, Zhao L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. Isme J 2013;7:880–4. 10.1038/ismej.2012.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Natividad JM, Lamas B, Pham HP, et al. Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat Commun 2018;9:2802. 10.1038/s41467-018-05249-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Udayappan S, Manneras-Holm L, Chaplin-Scott A, et al. Oral treatment with Eubacterium hallii improves insulin sensitivity in db/db mice. npj Biofilms Microbiomes 2016;2:16009. 10.1038/npjbiofilms.2016.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gilijamse PW, Hartstra AV, Levin E, et al. Treatment with Anaerobutyricum soehngenii: a pilot study of safety and dose–response effects on glucose metabolism in human subjects with metabolic syndrome. NPJ Biofilms Microbiomes 2020;6:16. 10.1038/s41522-020-0127-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goodrich JK, Waters JL, Poole AC, et al. Human genetics shape the gut microbiome. Cell 2014;159:789–99. 10.1016/j.cell.2014.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Everard A, Belzer C, Geurts L, et al. Cross-Talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 2013;110:9066–71. 10.1073/pnas.1219451110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Plovier H, Everard A, Druart C, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med 2017;23:107–13. 10.1038/nm.4236 [DOI] [PubMed] [Google Scholar]
- 18. Depommier C, Everard A, Druart C, et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med 2019;25:1096–103. 10.1038/s41591-019-0495-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dao MC, Everard A, Aron-Wisnewsky J, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut 2016;65:426–36. 10.1136/gutjnl-2014-308778 [DOI] [PubMed] [Google Scholar]
- 20. Van Hul M, Le Roy T, Prifti E, et al. From correlation to causality: the case of Subdoligranulum. Gut Microbes 2020;12:1849998–13. 10.1080/19490976.2020.1849998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Le Roy T, Van der Smissen P, Paquot A, et al. Dysosmobacter welbionis gen. nov., sp. nov., isolated from human faeces and emended description of the genus Oscillibacter. Int J Syst Evol Microbiol 2020;70:4851–8. 10.1099/ijsem.0.003547 [DOI] [PubMed] [Google Scholar]
- 22. Konikoff T, Gophna U. Oscillospira : a Central, Enigmatic Component of the Human Gut Microbiota. Trends Microbiol 2016;24:523–4. 10.1016/j.tim.2016.02.015 [DOI] [PubMed] [Google Scholar]
- 23. Zhou Q, Zhang Y, Wang X, et al. Gut bacteria Akkermansia is associated with reduced risk of obesity: evidence from the American gut project. Nutr Metab 2020;17:90. 10.1186/s12986-020-00516-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McDonald D, Hyde E, Debelius JW, et al. American gut: an open platform for citizen science microbiome research. mSystems 2018;3. 10.1128/mSystems.00031-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. The Human Microbiome Project Consortium . Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207–14. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hughes DA, Bacigalupe R, Wang J, et al. Genome-Wide associations of human gut microbiome variation and implications for causal inference analyses. Nat Microbiol 2020;5:1079–87. 10.1038/s41564-020-0743-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lv Y, Qin X, Jia H, et al. The association between gut microbiota composition and BMI in Chinese male college students, as analysed by next-generation sequencing. Br J Nutr 2019;122:986–95. 10.1017/S0007114519001909 [DOI] [PubMed] [Google Scholar]
- 28. Han GG, Lee J-Y, Jin G-D, et al. Evaluating the association between body weight and the intestinal microbiota of weaned piglets via 16S rRNA sequencing. Appl Microbiol Biotechnol 2017;101:5903–11. 10.1007/s00253-017-8304-7 [DOI] [PubMed] [Google Scholar]
- 29. Piqué N, Berlanga M, Miñana-Galbis D. Health benefits of heat-killed (Tyndallized) probiotics: an overview. Int J Mol Sci 2019;20:2534. 10.3390/ijms20102534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Everard A, Lazarevic V, Gaïa N, et al. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. Isme J 2014;8:2116–30. 10.1038/ismej.2014.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Daniel H, Gholami AM, Berry D, et al. High-Fat diet alters gut microbiota physiology in mice. Isme J 2014;8:295–308. 10.1038/ismej.2013.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Asterholm IW, Rutkowski JM, Fujikawa T, et al. Elevated resistin levels induce central leptin resistance and increased atherosclerotic progression in mice. Diabetologia 2014;57:1209–18. 10.1007/s00125-014-3210-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Benomar Y, Amine H, Crépin D, et al. Central Resistin/TLR4 impairs adiponectin signaling, contributing to insulin and FGF21 resistance. Diabetes 2016;65:913–26. 10.2337/db15-1029 [DOI] [PubMed] [Google Scholar]
- 34. Kotzbeck P, Giordano A, Mondini E, et al. Brown adipose tissue whitening leads to brown adipocyte death and adipose tissue inflammation. J Lipid Res 2018;59:784–94. 10.1194/jlr.M079665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Villarroya F, Cereijo R, Gavaldà-Navarro A, et al. Inflammation of brown/beige adipose tissues in obesity and metabolic disease. J Intern Med 2018;284:492–504. 10.1111/joim.12803 [DOI] [PubMed] [Google Scholar]
- 36. Larsen S, Nielsen J, Hansen CN, et al. Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. J Physiol 2012;590:3349–60. 10.1113/jphysiol.2012.230185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ravussin Y, LeDuc CA, Watanabe K, et al. Effects of ambient temperature on adaptive thermogenesis during maintenance of reduced body weight in mice. Am J Physiol Regul Integr Comp Physiol 2012;303:R438–48. 10.1152/ajpregu.00092.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li Z, Yi C-X, Katiraei S, et al. Butyrate reduces appetite and activates brown adipose tissue via the gut-brain neural circuit. Gut 2018;67:1269–79. 10.1136/gutjnl-2017-314050 [DOI] [PubMed] [Google Scholar]
- 39. Pujo J, Petitfils C, Le Faouder P, et al. Bacteria-Derived long chain fatty acid exhibits anti-inflammatory properties in colitis. Gut 2021;70:1088–97. 10.1136/gutjnl-2020-321173 [DOI] [PubMed] [Google Scholar]
- 40. Abot A, Wemelle E, Laurens C, et al. Identification of new enterosynes using prebiotics: roles of bioactive lipids and mu-opioid receptor signalling in humans and mice. Gut 2021;70:1078–87. 10.1136/gutjnl-2019-320230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Paone P, Cani PD. Mucus barrier, mucins and gut microbiota: the expected slimy partners? Gut 2020;69:2232–43. 10.1136/gutjnl-2020-322260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Régnier M, Van Hul M, Knauf C, et al. Gut microbiome, endocrine control of gut barrier function and metabolic diseases. J Endocrinol 2021;248:R67–82. 10.1530/JOE-20-0473 [DOI] [PubMed] [Google Scholar]
- 43. Foley KP, Zlitni S, Denou E, et al. Long term but not short term exposure to obesity related microbiota promotes host insulin resistance. Nat Commun 2018;9:4681. 10.1038/s41467-018-07146-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2020-323778supp001.pdf (31.6KB, pdf)
gutjnl-2020-323778supp002.pdf (7.5MB, pdf)
gutjnl-2020-323778supp003.pdf (619.2KB, pdf)
gutjnl-2020-323778supp004.pdf (134.9KB, pdf)
gutjnl-2020-323778supp005.pdf (32KB, pdf)
Data Availability Statement
Data are available in a public, open access repository. Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information. All datasets and raw data generated and/or analysed during the current study are available from the corresponding author on reasonable request. The 16S rRNA gene sequencing raw sequences of the mouse study can be accessed in Sequence Read Archive database with accession code PRJNA606762. The raw 16S data of the FGFP cohort are available at European Genome-Phenome Archive (https://ega-archive.org/) under accession no. EGAS00001004420, and for the Microbes4U cohort under accession no. EGAS00001003585. For the HMP, healthy human subjects cohort was downloaded from the human microbiome project data portal (https://portal.hmpdacc.org/).