Abstract
Human cytomegalovirus (HCMV) infection was monitored retrospectively by qualitative determination of pp67 mRNA (a late viral transcript) by nucleic acid sequence-based amplification (NASBA) in a series of 50 transplant recipients, including 26 solid-organ (11 heart and 15 lung) transplant recipients (SOTRs) and 24 bone marrow transplant recipients (BMTRs). NASBA results were compared with those obtained by prospective quantitation of HCMV viremia and antigenemia and retrospective quantitation of DNA in leukocytes (leukoDNAemia). On the whole, 29 patients were NASBA positive, whereas 10 were NASBA negative, and the blood of 11 patients remained HCMV negative. NASBA detected HCMV infection before quantitation of viremia did but after quantitation of leukoDNAemia and antigenemia did. In NASBA-positive blood samples, median levels of viremia, antigenemia, and leukoDNAemia were significantly higher than the relevant levels detected in NASBA-negative HCMV-positive blood samples. By using the quantitation of leukoDNAemia as the “gold standard,” the analytical sensitivity (47.3%), as well as the negative predictive value (68.3%), of NASBA for the diagnosis of HCMV infection intermediate between that of antigenemia quantitation (analytical sensitivity, 72.3%) and that of viremia quantitation (analytical sensitivity, 28.7%), while the specificity and the positive predictive value were high (90 to 100%). However, with respect to the clinically relevant antigenemia cutoff of ≥100 used in this study for the initiation of preemptive therapy in SOTRs with reactivated HCMV infection, the clinical sensitivity of NASBA reached 100%, with a specificity of 68.9%. Upon the initiation of antigenemia quantitation-guided treatment, the actual median antigenemia level was 158 (range, 124 to 580) in SOTRs who had reactivated infection and who presented with NASBA positivity 3.5 ± 2.6 days in advance and 13.5 (range, 1 to 270) in the group that included BMTRs and SOTRs who had primary infection (in whom treatment was initiated upon the first confirmation of detection of HCMV in blood) and who presented with NASBA positivity 2.0 ± 5.1 days later. Following antiviral treatment, the durations of the presence of antigenemia and pp67 mRNA in blood were found to be similar. In conclusion, monitoring of the expression of HCMV pp67 mRNA appears to be a promising, well-standardized tool for determination of the need for the initiation and termination of preemptive therapy. Its overall clinical impact should be analyzed in future prospective studies.
In the last few years, major advances have been made in the diagnosis of human cytomegalovirus (HCMV) infections in various transplant patient populations. The first major breakthrough was the development of the shell vial assay method, which leads to HCMV isolation and identification by means of a p72-specific monoclonal antibody 16 to 24 h after inoculation. This method, which was introduced in 1984 (19) and whose quantitative performance was significantly improved in 1990 (14), was found to be very useful in the monitoring of HCMV infections and the evaluation of the effect of antiviral treatment in peripheral blood leukocytes (PBLs) (18). A second major advancement was made with the introduction of the antigenemia assay for the detection and quantitation of the number of circulating PBLs carrying HCMV pp65 in the nucleus by using a pool of pp65-specific monoclonal antibodies (32, 39). Although antigenemia only reflects the accumulation of this excess viral protein in PBL nuclei, a good correlation was found between high levels of antigenemia and the presence of HCMV-related clinical symptoms (12, 38). Following the development of qualitative PCR for the detection of HCMV DNA in PBLs (7, 25, 34, 42) and the monitoring of antiviral treatment (6, 18), quantitative PCR methods were developed for the quantification of viral DNA in both PBLs and plasma (3, 8, 9, 11, 35–37). A good correlation between high levels of viral DNA and the presence of clinical symptoms in both transplant recipients and AIDS patients was shown (11, 15, 16).
However, apart from the quantitation of viremia, which lacks sensitivity, neither the quantitation of antigenemia nor the quantitation of DNA in leukocytes (leukoDNAemia) or plasma DNAemia appears to correlate consistently with actual virus replication in vivo (16). This might be due to the fact that polymorphonuclear leukocytes are not permissive for HCMV replication and that the viral material transported by this leukocyte subpopulation and quantitated by different diagnostic assays is taken up from HCMV-infected cells in vivo (22, 33). Therefore, determination of the presence of viral transcripts could represent a more direct marker of HCMV replication in vivo. HCMV immediate-early and late viral transcripts have been detected by different technical approaches, with somewhat discordant results regarding their clinical significance (1, 2, 20, 29–31, 40). However, it seems that late transcripts might better reflect active HCMV replication, dissemination, and disease (20, 21, 27, 30). Some major drawbacks in the detection of viral transcripts have been represented by the instability of the mRNAs and by the difficulty in differentiating between RNA- and DNA-derived PCR products in the case of unspliced transcripts. Treatment with DNase prior to PCR does not seem to guarantee the total degradation of viral DNA (1, 27, 30) and consequently bears the risk of false-positive results. Unlike reverse transcription-PCR, the detection of mRNA by using the recently introduced nucleic acid sequence-based amplification (NASBA) assay, which allows the specific amplification of unspliced mRNA in a background of DNA, appears to be particularly promising (5). Recently, it has been reported that the monitoring of HCMV infections in renal allograft recipients by the detection of pp67 mRNA in blood by NASBA is highly specific and sensitive and that NASBA can be used to monitor the natural history of HCMV infection and the effect of antiviral treatment (2).
In the present study we investigated the clinical significance of the retrospective detection of pp67 mRNA in blood (late RNAemia) by NASBA by comparing its qualitative results with the results of prospective quantitation of viremia and antigenemia and retrospective quantitation of leukoDNAemia in a series of 50 transplant recipients, including solid organ (SOTRs), i.e., heart transplant recipients (HTRs) and lung transplant recipients (LTRs), and bone marrow transplant recipients (BMTRs). Preliminary results seem to justify the conclusion that monitoring of the expression of pp67 mRNA might be used as a reliable marker for the need for the initiation of antiviral treatment and might also be used as a marker for the need for the termination of therapy.
MATERIALS AND METHODS
Patients.
From July 1997 to May 1998 a total of 50 transplant recipients (11 HTRs, 15 LTRs, and 24 BMTRs) were enrolled in the study. HTRs and LTRs underwent transplantation at the Cardiac Surgery Department, whereas BMTRs were given an allogeneic marrow transplant at the Department of Pediatrics or at the Division of Hematology, University Hospital Istituto di Ricovero e Cura a Carattere Scientifico Policlinico San Matteo, Pavia, Italy. All these patients were monitored for HCMV infection at the Viral Diagnostic Service, Istituto di Ricovero e Cura a Caraterre Scientifico Policlinico San Matteo. The patients underwent follow-up for a 3-month period. If the patients still showed virological signs of active HCMV infection after 3 months, follow-up continued until the disappearance of active HCMV infection or until 20 weeks after transplantation. During the follow-up period, heparinized or EDTA-treated blood samples were collected weekly or more or less than once a week, according to the kinetics of HCMV infection. The serostatuses of the donors and recipients were determined by enzyme-linked immunosorbent assay prior to transplantation by methods developed in the laboratory (13).
In HTRs and LTRs, the immunosuppressive regimen comprised cyclosporine (Cs-A), azathioprine, and steroids supplemented by a course of antithymocyte globulin. More recently, in some patients Cs-A was replaced by FK 506 (tacrolimus; Fujisawa Pharmaceutical Co., Deerfield, Ill.) at a dosage of 0.03 to 0.1 mg/kg of body weight/day. Patients with allograft rejection episodes were treated with a daily bolus of intravenous methylprednisolone (1 g or 500 mg) for 3 days. Patients with steroid-resistant rejections were treated with either antithymocyte globulin or OKT3. In BMTRs, graft-versus-host disease (GVHD) prophylaxis consisted of Cs-A for patients receiving transplants from a sibling with an identical HLA, whereas patients receiving bone marrow from a family donor with a partially matched HLA received in vitro T-cell-depleted marrow, and patients receiving a transplant from a matched but unrelated donor were treated with, in addition to Cs-A, short-term methotrexate, steroids, and the monoclonal antibody Campath-1G. Patients with acute GVHD were treated with steroids as first-line therapy, and resistant patients were treated with horse anti-lymphocyte globulin (10, 28).
Antiviral treatment.
A preemptive therapy approach was adopted for patients with all types of transplants. Ganciclovir (GCV) was administered intravenously at a standard dosage of 10 mg/kg/day for at least 14 days or until pp65 antigenemia clearance. Alternatively, foscarnet (PFA) was administered intravenously at a dosage of 180 mg/kg/day for 21 days or until antigenemia clearance. The indication for the initiation of antiviral therapy was an antigenemia level of ≥100 in four SOTRs undergoing reactivated infections, whereas in five patients with primary HCMV infection, treatment was started when the first confirmed positive antigenemia result was obtained (24). When required, secondary courses of treatment were administered to patients presenting with secondary episodes of HCMV infection. In 14 BMTRs, therapy was started either immediately when two or more pp65-positive PBLs were detected or when detection of one or two pp65-positive PBLs was confirmed on the following days by similar or higher levels of pp65-positive PBLs. In four HTRs and one LTR with rejection episodes, antiviral agents were administered (24) independently of the level of antigenemia. Similarly, in BMTRs, GVHD was consistently treated with either GCV or PFA or a combination of both (with half the dosage of each drug when both drugs were used) to prevent HCMV reactivation (10, 28).
Virological follow-up.
All patients were virologically monitored for HCMV infection by prospective quantitation of pp65 antigenemia and viremia and retrospective quantitation of leukoDNAemia and late RNAemia. The quantitation of pp65 antigenemia was achieved under a fluorescence microscope by counting the numbers of PBLs positive for pp65/2 × 105 cells examined with cytospin preparations stained with a pool of three pp65-specific monoclonal antibodies, according to a previously reported procedure (12). The quantitation of viremia was achieved by inoculating 2 × 105 PBLs onto human embryonic lung fibroblast cell cultures by the shell vial technique and then at 16 to 24 h after infection counting the numbers of fibroblast nuclei positive for the HCMV immediate-early antigen p72 (14). Conventional virus isolation from PBLs or other clinical samples was performed with human embryonic lung fibroblast cell cultures after the appearance of a cytopathic effect within 2 weeks after infection. Negative cultures were blindly passaged for another 2-week period. HCMV was identified as reported previously (14).
LeukoDNAemia was quantitated by PCR following the extraction of DNA from PBL samples by cell lysis with proteinase K and ethanol precipitation (16). The previously described PCR method for viral DNA quantitation with external standards and an internal amplification control (9) was modified by using a new internal control (16), which was constructed by following the same principle described previously (41). This method allowed reproducible HCMV DNA quantification in the range of 101 to 104 genome equivalents (GEs)/0.5 μg of PBL DNA (corresponding to about 105 PBLs) by using the single-step quantitative PCR.
NASBA.
The Nuclisens CMV pp67 assay was carried out according to the manufacturer’s instructions (Organon Teknika, B.V., Boxtel, The Netherlands). Briefly, 0.1 ml of heparinized or EDTA-treated blood was added to 0.9 ml of NASBA lysis buffer (4.7 M guanidinium thiocyanate, 46 mM Tris [pH 6.4], 20 mM EDTA, 1.2% [wt/vol] Triton X-100), and the mixture was stored at −70°C. A standardized amount of system control (SC) RNA, which served as a positive control for the isolation, amplification, and detection of RNA during the NASBA procedure, was added prior to nucleic acid isolation (2), which was performed essentially as described by Boom et al. (4). The nucleic acids were finally eluted in 50 μl of 1.0 mM Tris (pH 8.5) and were stored at −20°C. The NASBA amplification reaction was performed with two primers which were designed to amplify part of the mRNA encoding HCMV pp67 (the UL 65 gene product). NASBA reactions were carried out as described by Kievits et al. (26) in a 20-μl reaction mixture containing 40 mM Tris (pH 8.5), 12 mM MgCl2, 70 mM KCl, 15% (vol/vol) dimethyl sulfoxide, 5 mM dithiothreitol, each deoxynucleoside triphosphate at a concentration of 1 mM, ATP, CTP, and UTP each at a concentration of 2 mM, 1.5 mM GTP, 0.5 mM ITP, 2 μg of bovine serum albumin, 0.08 U of RNase H, 32 U of T7 RNA polymerase, 6.4 U of avian myeloblastosis reverse transcriptase, each primer at a concentration of 0.2 μM, and 5 μl of isolated nucleic acids. Prior to the addition of the enzymes, the NASBA reaction mixtures were incubated for 5 min at 65°C to destabilize the secondary RNA structures and were then cooled for 5 min to 41°C to allow primer annealing. Following the addition of the enzymes, the reaction mixtures were incubated at 41°C for 90 min and were then stored at −20°C.
The amplification products (wild-type and SC RNA) were then diluted in detection diluent and were incubated for 30 min at 41°C with biotinylated pp67-specific capture probe bound to 5 μg of streptavidin-coated magnetic beads and 3 × 1011 molecules of a ruthenium-labeled oligonucleotide detection probe specific for either pp67 mRNA or SC RNA (2). As a negative control, the detection diluent was also incubated with the wild-type RNA probe and the oligonucleotide bound to magnetic beads. Following incubation, an assay buffer solution was added and the tubes were placed in an electrochemiluminescence instrument (NASBA QR System; Organon Teknika B.V.) for final reading of the results.
Statistical analysis.
Differences in the means of parametric data (by the Lilliefords test for normality) were tested by using the t test, whereas differences in the means of nonparametric data were tested by the Kolmogorov-Smirnov test for unpaired data. In addition, the Pearson chi-square test was used to test differences in proportions.
RESULTS
Incidence of HCMV infection in transplant recipient populations.
Data relevant to the immune status of the donors and recipients are reported in Table 1. Of the 50 patients examined, 39 developed HCMV infection, and of these, 6 had primary HCMV infections and 33 had reactivated HCMV infections. Due to the preemptive therapy approach, overt disease was observed in only two patients, who had HCMV gastritis in the late stage of HCMV infection. Eleven patients remained HCMV negative during the entire follow-up period.
TABLE 1.
Serological status of donors and recipients with respect to development and treatment of HCMV infection in the different transplant patient populations
| Transplant patient population | No. of patients with HCMV infection/no. of patients tested according to the following donor/recipient serological status (%):
|
No. of NASBA positive patients/no. of HCMV-positive patients (%) | No. of treated patients/no. of HCMV positive patients (%) | ||||
|---|---|---|---|---|---|---|---|
| +/− | −/+ | +/+ | −/− | Total | |||
| HTRs | 3/4 | 0 | 7/7 | 0 | 10/11 (90.9) | 10/10 (100) | 6/10 (60.0) |
| LTRs | 2/2 | 2/3 | 10/10 | 0 | 14/15 (93.3) | 10/14 (71.4) | 8/14 (57.1) |
| BMTRs | 0/3 | 5/6 | 9/14 | 1a/1 | 15/24 (62.5) | 9/15 (60.0) | 14/15b (93.3) |
| Total | 5/9 | 7/9 | 26/31 | 1a/1 | 39/50 (78.0) | 29/39 (74.4) | 28/39 (71.8) |
Primary infection caused by administration of several blood units.
One BMTR was not treated because of an abortive infection.
According to the results provided by the different assays, the 50 patients were divided into the following three groups: group I included 29 patients (10 HTRs, 10 LTRs, and 9 BMTRs) who were NASBA positive and antigenemia and leukoDNAemia quantitation positive and 4 patients who were viremia quantitation negative; group II included 10 patients (4 LTRs and 6 BMTRs) who were NASBA negative but HCMV positive (10 by antigenemia quantitation, 9 by leukoDNAemia quantitation, and 2 by viremia quantitation); and group III included the remaining 11 patients (1 HTR, 1 LTR, and 9 BMTRs) who were negative by all assays (Table 1). On the whole, 552 blood samples were examined: 137 from HTRs, 156 from LTRs, and the remaining 259 from BMTRs.
Detection of HCMV infection.
All 552 blood samples tested for pp67 by NASBA were also tested for antigenemia, whereas 545 could be tested for viremia, and 548 could be tested for leukoDNAemia. The comparison of the results obtained by the different assays is reported in Table 2. Of the 206 antigenemia quantitation-positive samples, only 102 (49.5%) were NASBA positive, whereas of the 346 antigenemia quantitation-negative samples, as many as 324 (93.7%) were also NASBA negative. However, all of the 14 blood samples with levels of antigenemia of ≥100 were NASBA positive. Similarly, of the 254 leukoDNAemia quantitation-positive samples, 121 (47.6%) were NASBA positive, whereas nearly all (99.3%) of the 294 leukoDNAemia quantitation-negative samples were also NASBA negative. On the whole, 29 of 30 (96.7%) blood samples with leukoDNAemia levels of ≥1,000 GEs were NASBA positive. Finally, of the 71 viremia quantitation-positive samples, 51 (71.8%) were also NASBA positive, whereas of the 474 viremia quantitation-negative samples, 402 (84.8%) were also NASBA negative. All of the 14 samples with levels of viremia of ≥10 were NASBA positive. On the other hand, about 20% of samples with antigenemia, leukoDNAemia, and viremia levels of <100, <1,000, and <10 (the clinically significant threshold values), respectively, were NASBA positive (Table 2).
TABLE 2.
Comparison of pp67 NASBA with antigenemia, viremia, and leukoDNAemia quantitation results
| HCMV assay result | Transplant patient population (cutoff) | No. (%) of blood samples with the following NASBA result:
|
Pa | |
|---|---|---|---|---|
| Positive | Negative | |||
| Antigenemia positive | HTR | 47 (62.7) | 28 (37.3) | |
| LTR | 32 (44.4) | 40 (55.6) | ||
| BMTR | 23 (40.0) | 36 (60.0) | ||
| Total, ≥1 | 102 (49.5) | 104 (50.5) | ||
| Total, ≥100b | 14 (100) | 0 (0) | ||
| Antigenemia negative | HTR | 9 (14.5) | 53 (85.5) | s |
| LTR | 11 (13.1) | 73 (86.9) | s | |
| BMTR | 2 (1.0) | 198 (99.0) | s | |
| Total | 22 (6.3) | 324 (93.7) | s | |
| Antigenemia, <100 | 110 (20.4) | 428 (79.6) | s | |
| Viremia positive | HTR | 28 (82.3) | 6 (17.7) | |
| LTR | 15 (57.7) | 11 (42.3) | ||
| BMTR | 8 (72.7) | 3 (27.3) | ||
| Total, ≥1 | 51 (71.8) | 20 (28.2) | ||
| Total, ≥10b | 14 (100) | 0 (0) | ||
| Viremia negative | HTR | 28 (27.2) | 75 (72.8) | s |
| LTR | 28 (21.5) | 102 (78.5) | s | |
| BMTR | 16 (6.6) | 225 (93.4) | s | |
| Total | 72 (15.2) | 402 (84.8) | s | |
| Viremia, <10 | 109 (20.5) | 422 (79.5) | s | |
| LeukoDNAemia positive | HTR | 55 (59.8) | 37 (40.2) | |
| LTR | 42 (46.7) | 48 (53.3) | ||
| BMTR | 24 (33.3) | 48 (66.7) | ||
| Total, ≥10 | 121 (47.6) | 133 (52.4) | ||
| Total, ≥1,000b | 29 (96.7) | 1 (3.3) | ||
| LeukoDNAemia negative (<10) | HTR | 0 (0) | 43 (100) | s |
| LTR | 1 (1.5) | 65 (98.5) | s | |
| BMTR | 1 (0.5) | 184 (99.5) | s | |
| Total | 2 (0.7) | 292 (99.3) | s | |
| LeukoDNAemia, <1,000 | 94 (18.1) | 424 (81.9) | s | |
Pearson chi-square test; s, significant (P < 0.05).
Clinically significant threshold values. For definition of units, refer to footnote b of Table 4.
The difference in the distribution of concordant and discordant results, according to the different pairs used for comparison, is reported in detail in Table 2 both for the total transplant recipient population and for the three separate groups of transplant recipients. Statistical analysis showed that the concordant results were significantly greater than the discordant results for the total transplant patient population as well as for the three separate groups of transplant recipients.
By using leukoDNAemia as the “gold standard” and a cutoff of ≥10 GEs for a diagnosis of HCMV infection, the analytical sensitivity of the pp67 NASBA (47.3%) appeared to be intermediate between that of antigenemia quantitation (72.3%) and that of viremia quantitation (28.7%) (Table 3). Similarly, the negative predictive value (NPV) for antigenemia quantitation was higher than those for the pp67 NASBA and viremia quantitation. On the contrary, specificities and positive predictive values (PPVs) were high (80 to 100%) for all three assays for both the general and the individual patient populations. However, with respect to the antigenemia cutoff of ≥100, the clinical sensitivity of NASBA went up to 100% for SOTRs with reactivated infection, whereas the specificity decreased markedly (68.9%). For BMTRs and SOTRs with primary infection, the sensitivity (43.1%), specificity (95.9%), PPV (82.5%), and NPV (79.0%) of NASBA did not change markedly for the general patient population (Table 3).
TABLE 3.
Diagnostic values of pp67 NASBA and of antigenemia and viremia quantitation assays for detection of HCMV infection in blood samples from the general and individual transplant patient populations by using leukoDNAemia as the gold standard
| HCMV assay | Transplant patient population | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|
| NASBA | HTRs | 59.8 | 100 | 100 | 53.7 |
| LTRs | 46.7 | 98.5 | 97.7 | 57.9 | |
| BMTRs | 33.3 | 99.5 | 96.0 | 79.4 | |
| Total | 47.3 | 99.3 | 98.4 | 68.7 | |
| Antigenemia quantitation | HTRs | 73.9 | 88.4 | 93.1 | 61.3 |
| LTRs | 73.1 | 93.6 | 94.4 | 70.2 | |
| BMTRs | 69.0 | 94.2 | 81.7 | 89.0 | |
| Total | 72.3 | 93.2 | 90.2 | 79.5 | |
| Viremia quan-titation | HTRs | 38.0 | 100 | 100 | 43.0 |
| LTRs | 27.9 | 100 | 100 | 48.5 | |
| BMTRs | 16.7 | 100 | 100 | 77.5 | |
| Total | 28.7 | 100 | 100 | 62.2 |
Time to detection of HCMV infection.
The mean time to the first detection of HCMV infection following transplantation in the 29 NASBA-positive patients is reported in Table 4 for both the general and the individual patient populations. The first assay which detected HCMV infection was the leukoDNAemia quantitation assay (29 ± 13 days posttransplantation), followed by the antigenemia quantitation assay (33 ± 13 days), the pp67 NASBA (37 ± 15 days), and the viremia quantitation assay (39 ± 14 days). All these differences in time except for that for the viremia quantitation assay were statistically significant compared to the time to detection for the pp67 NASBA. When considering the individual patient populations, it was observed that the time to the first detection of HCMV in blood progressively increased from HTRs to LTRs and BMTRs, regardless of the assay that was used. Interestingly, a further delay in HCMV appearance following transplantation was observed in group II patients (NASBA negative) compared to the time of HCMV appearance in group I patients (NASBA positive) with respect to the three available assays. No data relevant to individual patient populations are reported in Table 4 for group II patients due to the limited number of available patients.
TABLE 4.
Time to first HCMV detection in blood in group I (NASBA-positive) and group II (NASBA-negative) patientsa
| HCMV assay | Transplant patient population | Group I
|
Group II
|
||||
|---|---|---|---|---|---|---|---|
| No. of patients | Mean ± SD days posttransplantation of first HCMV appearance | Valueb | No. of patients | Mean ± SD days posttransplantation of first HCMV appearance | Valueb | ||
| Antigenemia quantitation | HTR | 10 | 26 ± 10 | 13 ± 13 | |||
| LTR | 10 | 33 ± 9 | 36 ± 83 | ||||
| BMTR | 9 | 41 ± 16 | 20 ± 34 | ||||
| Total | 29 | 33 ± 13 | 23 ± 52 | 10 | 41 ± 20 | 2 ± 1 | |
| Viremia quantitation | HTR | 9 | 37 ± 16 | 3 ± 3 | |||
| LTR | 9 | 39 ± 10 | 6 ± 7 | ||||
| BMTR | 6 | 44 ± 16 | 9 ± 15 | ||||
| Total | 24 | 39 ± 14 | 6 ± 8 | 2 | 56 ± 11 | 2 ± 0 | |
| LeukoDNAemia quantitation | HTR | 10 | 22 ± 11 | 95 ± 129 | |||
| LTR | 10 | 27 ± 7 | 88 ± 100 | ||||
| BMTR | 9 | 38 ± 17 | 100 ± 133 | ||||
| Total | 29 | 29 ± 13 | 94 ± 117 | 8 | 40 ± 19 | 41 ± 29 | |
| NASBA | HTR | 10 | 33 ± 15 | NAc | NA | ||
| LTR | 10 | 36 ± 15 | NA | NA | |||
| BMTR | 9 | 44 ± 16 | NA | NA | |||
| Total | 29 | 37 ± 15 | NA | NA | NA | NA | |
Differences between each pair of tests were significant by the t test except the comparison between the NASBA and the viremia quantitation assay (total patient population).
For the antigenemia quantitation assay, the values are the numbers of pp65-positive PBLs/2 × 105 PBLs examined. For the viremia quantitation assay, the values are the numbers of p72-positive fibroblast nuclei/2 × 105 PBL inoculated onto a shell vial culture. For the leukoDNAemia quantitation assay, the values are the numbers of GEs/0.5 μg of PBL DNA.
NA, not applicable.
Upon the first appearance of HCMV, the mean ± standard deviation (SD) levels of viremia, antigenemia, and leukoDNAemia were 6 ± 8, 23 ± 52, and 94 ± 117, respectively, for group I patients, and 2 ± 0, 2 ± 1, and 41 ± 29, respectively, for group II patients (Table 4). In the group of four SOTRs with HCMV reactivation, NASBA positivity was reached a mean time of 3.5 ± 2.6 days prior to the time that the clinical antigenemia cutoff of ≥100 was achieved, while in the group of nine BMTRs and four SOTRs with primary infection (the fifth SOTR with primary infection was patient B [see Fig. 1] whose data were not considered in the calculation because he was NASBA positive only during a secondary peak of infection) NASBA positivity was detected a mean time of 2.0 ± 5.1 days after the time of initial antigenemia positivity (data not shown).
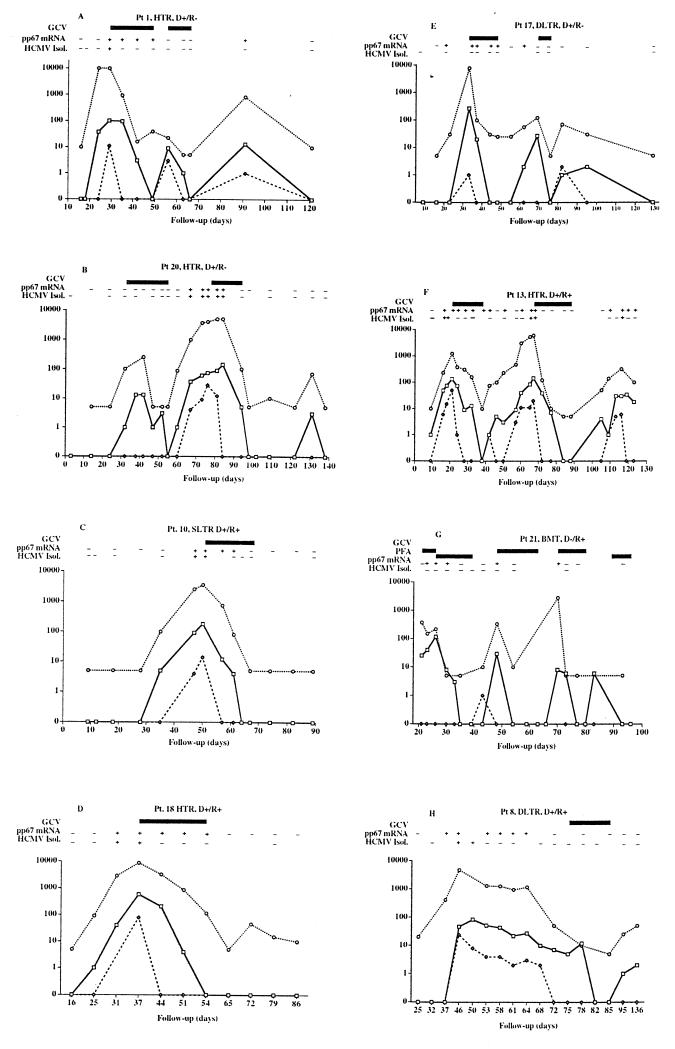
FIG. 1.
Quantitative monitoring of HCMV viremia (◊), antigenemia (□), and leukoDNAemia (○) in transplant recipients qualitatively monitored for pp67 mRNA appearance and virus isolation. The panel letters refer to patients A to H, respectively. Data for three patients (patients B, C, and D), either HTRs or single LTRs (SLTR), are shown on the left. These patients presented with a single major peak of HCMV infection that was associated with the appearance of pp67 mRNA and that was controlled by antiviral treatment. On the other hand, patients A (HTR), E (double LTR), F (HTR), and G (BMTR) presented with multiple peaks of HCMV infection associated with the appearance of pp67 mRNA, and these infections were controlled by multiple courses of antiviral treatment. Finally, patient H (double LTR) had a spontaneous resolution of the peak of HCMV infection associated with persisting pp67 mRNA, while the delayed course of GCV treatment was due to a rejection episode. D+, donor positive for HCMV; D−, donor negative for HCMV; R+, recipient positive for HCMV; R−, recipient negative for HCMV; HCMV Isol., HCMV isolation.
Relationship of NASBA qualitative results and HCMV levels by other assays.
In group I patients, quantitative levels of viremia, antigenemia, and leukoDNAemia in NASBA-positive blood samples were significantly higher than the relevant levels detected in NASBA-negative, HCMV-positive blood samples from the same patients (Table 5). In addition, the same level of statistically significant difference was detected between NASBA-positive samples from group I and patients and NASBA-negative, HCMV-positive samples from group II patients, regardless of the assay that was used. These results obtained for the general transplant patient population were confirmed for the three individual patient populations for patients in group I (Table 6). The only groups in which no significant difference was found between NASBA-positive and NASBA-negative, HCMV-positive samples were LTRs and BMTRs with respect to viremia.
TABLE 5.
Comparison of HCMV antigenemia, viremia, and leukoDNAemia levels of HCMV-positive, NASBA-positive blood samples from group I patients (NASBA-positive) and HCMV-positive, NASBA-negative samples from group I patients and HCMV-positive, NASBA-negative samples from group II patients (HCMV positive, NASBA negative)
| HCMV assay | Patient group | Median HCMV levela (range) in HCMV-positive blood samples
|
Pb | |
|---|---|---|---|---|
| NASBA positive | NASBA negative | |||
| Antigenemia quantitation | I | 13.0 (0–580) (n = 124) | 1.0 (0–39) (n = 122) | 0.0000c |
| II | 1.0 (0–4) (n = 28) | 0.0000d | ||
| Viremia quantitation | I | 0 (0–78) (n = 122) | 0 (0–8) (n = 120) | 0.0002c |
| II | 0 (0–2) (n = 28) | 0.0185d | ||
| LeukoDNAemia quantitation | I | 275 (5–10,000) (n = 123) | 50 (5–10,000) (n = 121) | 0.0000c |
| II | 20 (5–130) (n = 28) | 0.0001d | ||
For definition of units, refer to footnote b of Table 4.
Kolmogorov-Smirnov two-sample test. For NASBA-negative, HCMV-positive samples from group I patients versus NASBA-negative, HCMV-positive samples from group II patients for all assays, P was >0.05.
NASBA-positive samples versus NASBA-negative, HCMV-positive samples from group I patients.
NASBA-positive samples from group I patients versus NASBA-negative, HCMV-positive samples from group II patients.
TABLE 6.
Comparison of HCMV antigenemia, viremia, and leukoDNAemia levels in HCMV-positive, NASBA-positive versus HCMV-positive, NASBA-negative blood samples from patients in group I (NASBA-positive patients) by type of transplant
| HCMV assay | Transplant patient population | Median HCMV level (range) in HCMV-positive blood samples
|
Pa | |
|---|---|---|---|---|
| NASBA positive | NASBA negative | |||
| Antigenemia quantitation | HTR | 16.0 (0–580) (n = 56) | 1.0 (0–38) (n = 43) | 0.0000 |
| LTR | 10.0 (0–270) (n = 43) | 2.0 (0–27) (n = 41) | 0.0002 | |
| BMTR | 8.0 (0–118) (n = 25) | 1.0 (0–25) (n = 38) | 0.0015 | |
| Viremia quantitation | HTR | 0 (0–78) (n = 55) | 0 (0–8) (n = 43) | 0.0277 |
| LTR | 0 (0–24) (n = 43) | 0 (0–5) (n = 41) | >0.05 | |
| BMTR | 0 (0–40) (n = 24) | 0 (0–5) (n = 36) | >0.05 | |
| LeukoDNAemia quantitation | HTR | 315.0 (10–10,000) (n = 55) | 65.0 (5–10,000) (n = 42) | 0.0002 |
| LTR | 285.0 (5–7,900) (n = 43) | 85.0 (5–390) (n = 41) | 0.0002 | |
| BMTR | 150.0 (5–6,940) (n = 25) | 17.5 (5–880) (n = 38) | 0.0124 | |
Kolmogorov-Smirnov two-sample test.
pp67 NASBA and antiviral treatment.
Overall data relevant to the treated patients are reported in Table 1. Of the 10 NASBA-positive HTRs, 6 (three with primary infections and three with reactivated infections) underwent antiviral treatment (three patients received a single course of GCV and three patients received two courses of GCV). Three patients were not treated, and one patient had only a delayed treatment at 127 to 134 days posttransplantation because of a rejection episode, when the NASBA result was already negative. Of the 10 NASBA-positive LTRs, 7 (2 with primary infections and 5 with reactivated infections) were treated (4 with one course of GCV and 3 with two subsequent courses of GCV), while 2 did not receive treatment and 1 underwent delayed treatment because of a rejection episode, when the NASBA result was already negative. In addition, 1 NASBA-negative, HCMV-positive LTR was treated because of rejection. Furthermore, all nine NASBA-positive BMTRs were treated: two with a single GCV course and the remaining seven with multiple courses of either GCV or PFA, or both. Finally, five NASBA-negative, HCMV-positive BMTRs were treated. As indicated in Table 7, in the group of 20 SOTRs with reactivated infection, 9 patients were treated; 8 of these were NASBA positive (4 had levels of antigenemia of >100 and 4 had rejection episodes) and 1 was NASBA negative (with a low level of antigenemia and a rejection episode), whereas in the group of 19 patients for whom treatment was initiated when antigenemia positivity was first confirmed, 14 (9 BMTRs and 5 SOTRs with primary infections) were NASBA positive and 5 (all BMTRs) were NASBA negative. Finally, it must be considered that in the group of 29 NASBA-positive patients, as many as 7 were not treated when the first HCMV infection was detected because the level of antigenemia of ≥100 had not been reached (2 were treated late) and the patients had a subsequent spontaneous resolution of their infections. In the group of 10 NASBA-negative, HCMV-positive patients, as many as 6 were treated on the basis of first antigenemia positivity (5 BMTRs) or a rejection episode (1 LTR).
TABLE 7.
Actual antigenemia quantitation assay-guided treatment and predicted NASBA-guided treatment with respect to median antigenemia levels in the two major groups of differentially treated patients
| NASBA result | No. of patients (median [range] antigenemia level) treated under guidance of
|
|||
|---|---|---|---|---|
| Antigenemia quantitation assay (actual treatment)
|
NASBA (predicted treatment)
|
|||
| SOTRs with reactivations (n = 20) | BMTRs and SOTRs with primary infections (n = 19) | SOTRs with reactivations (n = 20) | BMTRs and SOTRs with primary infections (n = 19) | |
| Positive | 8 (87 [3–580])a | 14 (13.5 [1–270])b | 15 (16 [0–124])c | 14 (9 [0–110])d |
| Negative | 1 (1)e | 5 (2 [1–4])f | 0 | 0 |
| Total | 9 | 19 | 15 | 14 |
Four patients started treatment because of a median antigenemia level of 158 (range, 124 to 580), and four started treatment because of a rejection episode with a median antigenemia level of 20.5 (range, 3 to 50).
Nine patients were BMTRs with a median antigenemia level of 18 (range, 4 to 105), and five were SOTRs with primary infections with a median antigenemia level of 9 (range, 1 to 270).
Upon NASBA positivity, the eight patients treated under antigenemia quantitation assay guidance had a median antigenemia level of 44.5 (range, 6 to 124), and the seven additional patients had a median antigenemia level of 5 (range, 0 to 38).
Upon NASBA positivity, among the same group of 14 patients treated under antigenemia quantitation assay guidance, the 9 BMTRs had a median antigenemia level of 11 (range, 1 to 110) and the 5 SOTRs with primary infections had a median antigenemia level of 7 (range, 0 to 100).
LTR treated because of rejection.
All BMTRs.
As indicated in Table 7, upon the initiation of treatment, in the group of eight treated NASBA-positive SOTRs with HCMV reactivation, the median antigenemia levels were 158 (range, 124 to 580) in the four patients with antigenemia levels of >100 and 20.5 (range, 3 to 50) in the four patients treated for rejection, with an overall median antigenemia level of 87 (range, 3 to 580). In the group of NASBA-positive patients that included nine BMTRs and five SOTRs with primary infections, the median antigenemia levels were 18 (range, 4 to 105) for BMTRs and 9 (range, 1 to 270) for patients with primary HCMV infection, with an overall median antigenemia level of 13.5 (range, 1 to 270). On the other hand, the five NASBA-negative treated BMTRs had a median antigenemia level of 2 (range, 1 to 4). Thus, the overall median antigenemia level for all 14 treated BMTRs was 6 (range, 1 to 105). In addition, upon the first appearance of pp67 mRNA in the 29 transplant recipients in group I, median antigenemia levels were 11 (range, 0 to 124), while median levels of viremia and leukoDNAemia were 1 (range, 0 to 40) and 250 (range, 10 to 10,000), respectively. No significant difference in the levels of the three viral parameters was observed between the different patient groups and, in particular, between SOTRs and BMTRs.
If NASBA positivity had been used as a cutoff for preemptive therapy, 7 of 11 untreated SOTRs with reactivation would have been treated, while in the group of 19 BMTRs and SOTRs with primary infection, 5 BMTRs who were treated under the guidance of the antigenemia quantitation assay results would not have been treated (Table 7). However, it must be noted that in these five patients the median antigenemia level was 2 (range, 1 to 4) and that the mean duration of antigenemia positivity was 1.2 ± 0.4 days. These results were possibly influenced by the early start of treatment, which may have prevented the detection of pp67 mRNA.
The time to HCMV disappearance from the blood of the treated and untreated general and individual transplant recipient populations according to the different assays is reported in Table 8. No difference was observed between the different patient groups or between each patient group and the general transplant patient population for antigenemia, viremia, and leukoDNAemia, whereas a significantly shorter duration of the presence of pp67 mRNA was observed for the BMTR group compared to those for the HTRs, LTRs, and the total treated group. In addition, following the initiation of treatment, the mean duration of viremia (6.3 ± 1.8 days) was significantly shorter than the mean durations of antigenemia and the presence of pp67 mRNA (14.4 ± 5.0 and 14.0 ± 7.6 days, respectively), which, in turn, were significantly shorter than the mean duration of leukoDNAemia (30.8 ± 21.1 days). Furthermore, in the group of seven untreated patients, the duration of leukoDNAemia (60.6 ± 37.1 days) was longer than that of viremia (20.8 ± 6.6 days), the presence of pp67 mRNA (36.1 ± 24.2 days), and antigenemia (32.8 ± 13.9 days). Finally, marked differences were detected between the group of 22 treated NASBA-positive patients and the group of 7 untreated patients for durations of antigenemia, viremia, leukoDNAemia, and the presence of pp67 mRNA.
TABLE 8.
Time to HCMV disappearance from blood of treated and untreated transplant recipients by different assays
| Antiviral treatment | Transplant patient population | No. of patients | Mean ± SD no. of days to HCMV disappearance in blood by the following assay:
|
|||
|---|---|---|---|---|---|---|
| Antigenemia quantitation | Viremia quantitation | LeukoDNAemia quantitation | pp67 mRNA detectiona | |||
| Treated | HTRs | 6 | 16.2 ± 2.9 | 7.2 ± 2.2 | 29.6 ± 17.8 | 21.0 ± 6.7 |
| LTRs | 7 | 13.6 ± 5.3 | 6.6 ± 1.8 | 40.3 ± 25.3 | 16.0 ± 6.3 | |
| BMTRs | 9 | 14.0 ± 7.3 | 5.2 ± 0.8 | 24.2 ± 18.9 | 10.0 ± 6.2 | |
| Totalb | 22 | 14.4 ± 5.0 | 6.3 ± 1.8 | 30.8 ± 21.1 | 14.9 ± 7.6 | |
| Not treated | Total | 7 | 32.8 ± 13.9 | 20.8 ± 6.6 | 60.6 ± 37.1 | 36.1 ± 24.2 |
P was <0.05 (by the t test) for HTRs, LTRs, and total treated patients versus BMTRs.
By the Kolmogorov-Smirnov test, P < 0.05 was for each comparison of two assays except for the antigenemia quantitation assay and pp67 mRNA detection.
Kinetics of HCMV infection in individual transplant recipients.
When considering the kinetics of HCMV infection in treated transplant recipients, it must be taken into account that therapy was administered mostly on the basis of antigenemia levels, whereas the appearance of pp67 mRNA was determined retrospectively. Results for some representative patients are reported in Fig. 1. In patients B (HTR, primary infection), C (single LTR, reactivation), and D (HTR, reactivation), a single major peak of HCMV infection which was associated with the appearance of pp67 mRNA was controlled by antiviral treatment, and late viral transcripts did not reappear during the entire follow-up period. On the other hand, in patients A (HTR, primary infection), E (double LTR, primary infection), F (HTR, reactivation), and G (BMTR, reactivation), multiple sequential peaks of HCMV infection, each associated with the reappearance of pp67 mRNA, were controlled by multiple sequential courses of antiviral treatment (patients F, G, and E) or resolved spontaneously (patient A). Finally, in patient H (double LTR, reactivation) the pp67 mRNA, which was associated with a prolonged peak of HCMV infection, disappeared spontaneously concomitantly with a decrease in the levels of viremia, antigenemia, and leukoDNAemia prior to a delayed course of treatment due to a rejection episode.
DISCUSSION
The results of the present study indicate that NASBA detects HCMV infection in transplant recipients at a rate which is intermediate (29 of 50 patients; 58%) between those of the antigenemia quantitation assay (39 of 50; 78%) and the leukoDNAemia quantitation assay (38 of 50; 76%) and that of the viremia quantitation assay (27 of 50; 54%). On this basis, it would seem that NASBA lacks sensitivity, as also reflected by the low analytical sensitivity (47.3%) compared to that of the gold standard, the leukoDNAemia quantitation assay (>10 GEs), and the delayed time to first detection of pp67 mRNA by NASBA compared to the times to first detection by the leukoDNAemia and antigenemia quantitation assays.
However, the clinical relevance of these observations remains to be determined considering the fact that the specificity and the PPV for the detection of active HCMV infection by NASBA were very high. In fact, low levels of antigenemia and/or leukoDNAemia may be present in the absence of clinically significant HCMV infection (11, 16, 18). In addition, it is well known that in some SOTRs and BMTRs the appearance of HCMV in blood is followed by the spontaneous resolution of the infection in the absence of antiviral treatment (10, 24). On this basis, it would be important in the near future to define whether transplant recipients without detectable late HCMV transcripts are to receive the same treatment as those with viral transcripts. It is generally agreed that late viral transcripts are more significantly predictive of or are more significantly associated with active disseminating HCMV infection and HCMV disease than immediate-early transcripts (1, 20, 21, 27, 29, 30). In addition, if a cutoff level of ≥100 for antigenemia is used, the sensitivity of NASBA reaches 100%, although this is associated with a decrease in specificity. In this respect, it is worth noting that NASBA pp67 mRNA was present in blood only when the levels of viremia, antigenemia, and leukoDNAemia were significantly higher than those for NASBA-negative samples. This was true for both the general and the individual transplant recipient populations.
Thus far, in our department in Pavia, Italy, whenever other clinical conditions such as rejection are not interfering, antiviral treatment is started in SOTRs with reactivated infections on the basis of a clinical antigenemia cutoff of ≥100 (11, 16, 24). This antigenemia level grossly corresponds to levels of viremia of ≥10 and levels of leukoDNAemia of ≥1,000 (11, 16). On the other hand, in SOTRs (17, 23) and BMTRs (10, 28) with primary infections, antiviral treatment is routinely initiated when pp65-positive PBLs are first detected in blood, and this result must be confirmed within 2 to 3 days. In this study, we found that when pp67 mRNA was first detected in the blood of the 29 NASBA-positive patients, the median levels of antigenemia were 11.0 (range, 0 to 124). Thus, on this basis, antiviral treatment would be anticipated when NASBA is used as a cutoff, and it would seem reasonable to rely on NASBA results for determination of when to start antiviral therapy, at least in SOTRs with reactivated infections. However, since upon the initiation of antigenemia quantitation assay-guided treatment the actual median antigenemia levels were 6 (range, 1 to 105) for the entire treated BMTR population and 2 (range, 1 to 4) for NASBA-negative BMTRs but 11 (range, 1 to 110) when pp67 mRNA was first detected, it would also seem reasonable to perform a prospective study with this patient group by using NASBA positivity as a clinical cutoff instead of (or in parallel with) the antigenemia quantitation assay result. This approach appears to be further supported by the finding of this study that the mean delay in the time to the appearance of NASBA positivity with respect to that for the antigenemia quantitation assay was only 2.0 ± 5.1 days for the whole patient population treated when the antigenemia quantitation assay was first positive. Furthermore, since we have recently shown that treatment of primary infections in SOTRs too early may result in a greater number of short-term reactivation episodes (17), we would also suggest that investigators perform prospective clinical trials in which the presence of pp67 late transcripts is used as a possible cutoff for determination of whether therapeutic interventions in patients with primary HCMV infections should be initiated.
The results of this study demonstrate that if qualitative NASBA results would have been used as a cutoff for the initiation of treatment at the same time that antigenemia was detected in the group of SOTRs with reactivated infection, NASBA sensitivity and NPV would have been 100% compared to the results of the antigenemia quantitation assay, whereas the specificity and PPV would have been about 50% due to the NASBA-guided treatment of some patients not treated when the antigenemia quantitation assay cutoff was reached. On the other hand, in the group of BMTRs and SOTRs with primary infection (when the first positive antigenemia quantitation assay result was used as a cutoff), the specificity and PPV of NASBA would have been 100%, as with the antigenemia quantitation assay cutoff, whereas the sensitivity and NPV would have been about 70% with respect to the results of the antigenemia quantitation assay due to a lack of NASBA-guided treatment of some patients who underwent antigenemia quantitation assay-guided treatment. These results further provide the basis for performance of a prospective study in which patients of the two groups should be preemptively treated by using either the NASBA-defined or antigenemia quantitation assay-defined (or the leukoDNAemia-quantitation assay-defined) cutoff. This study would provide a correct evaluation of the potential clinical significance of NASBA pp67 mRNA determination in preemptively guiding antiviral treatment.
Finally, this study suggests that the disappearance of pp67 mRNA might be a reliable virologic parameter in clinical settings for establishment of whether antiviral therapy should be terminated (20). It is known that viremia is a poorly sensitive parameter for determination of whether treatment should be discontinued (the viremia quantitation assay result becomes negative 1 to 3 days after treatment onset) and that leukoDNAemia tends to persist after treatment (18). In our department in Pavia, Italy, we have been using a negative antigenemia quantitation assay result as the parameter that determines whether treatment should be interrupted (10, 18, 28). Since the time to HCMV disappearance from the blood as determined by the disappearance of pp67 mRNA in our study has been shown not to be significantly different from that determined by the antigenemia quantitation assay, we hypothesize that pp67 mRNA disappearance might also be used as the parameter of choice for determination of whether antiviral treatment should be interrupted. Future prospective studies are required to determine the reliability of this approach.
ACKNOWLEDGMENTS
We thank Linda D’Arrigo for revision of the English. We are also indebted to Luca Dossena for excellent technical assistance and to Barbara Ferrara and Franca Bordoni for typing the manuscript.
This work was partially supported by the Ministero della Sanità, Ricerca Corrente (grant 820RCR96/01), and the Ministero della Sanità, Istituto Superiore di Sanità, X Progetto Nazionale AIDS (1997) (grant 50A-18).
REFERENCES
- 1.Bitsch A, Kirchner H, Dupke R, Bein G. Cytomegalovirus transcripts in peripheral blood leukocytes of actively infected transplant patients detected by reverse-transcription-polymerase chain reaction. J Infect Dis. 1993;167:740–743. doi: 10.1093/infdis/167.3.740. [DOI] [PubMed] [Google Scholar]
- 2.Blok M J, Goossens V J, Vanherle S J V, Top B, Tacken N, Middeldorp J M, Christiaans M H L, van Hooff J P, Bruggeman C A. Diagnostic value of monitoring human cytomegalovirus late pp67 mRNA expression in renal-allograft recipients by nucleic acid sequence-based amplification. J Clin Microbiol. 1998;36:1341–1346. doi: 10.1128/jcm.36.5.1341-1346.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boivin G, Olson C A, Quirk M R, St.-Cyr S M, Jordan M C. Quantitation of cytomegalovirus glycoprotein H gene in cells using competitive PCR and a rapid fluorescent-based detection system. J Virol Methods. 1995;51:329–342. doi: 10.1016/0166-0934(94)00128-4. [DOI] [PubMed] [Google Scholar]
- 4.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Nordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Compton J. Nucleic acid sequence-based amplification. Nature. 1991;350:91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- 6.Einsele H, Ehninger G, Hebart H, Wittkowski K M, Schule U, Jahn G, Mackes P, Herter M, Klingebiel T, Loeffler J, Wagner S, Muller C. Polymerase chain reaction monitoring reduces the incidence of cytomegalovirus disease and the duration and side effects of antiviral therapy after bone marrow transplantation. Blood. 1995;86:2815–2820. [PubMed] [Google Scholar]
- 7.Einsele H, Ehninger G, Steidle M, Vallbracht A, Muller M, Schmidt H, Saal J G, Waller H D, Muller C. Polymerase chain reaction to evaluate antiviral therapy for cytomegalovirus disease. Lancet. 1991;338:1170–1172. doi: 10.1016/0140-6736(91)92032-w. [DOI] [PubMed] [Google Scholar]
- 8.Fox J C, Griffiths P D, Emery V C. Quantitation of human cytomegalovirus DNA using the polymerase chain reaction. J Gen Virol. 1992;73:2405–2408. doi: 10.1099/0022-1317-73-9-2405. [DOI] [PubMed] [Google Scholar]
- 9.Gerna G, Baldanti F, Sarasini A, Furione M, Percivalle E, Revello M G, Zipeto D, Zella D The Italian Foscarnet Study Group. Effect of foscarnet induction treatment on quantitation of human cytomegalovirus (HCMV) DNA in peripheral blood polymorphonuclear leukocytes and aqueous humor of AIDS patients with HCMV retinitis. Antimicrob Agents Chemother. 1994;38:38–44. doi: 10.1128/aac.38.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerna G, Furione M, Baldanti F, Percivalle E, Comoli P, Locatelli F. Quantitation of human cytomegalovirus DNA in bone marrow transplant recipients. Br J Hematol. 1995;91:674–683. doi: 10.1111/j.1365-2141.1995.tb05368.x. [DOI] [PubMed] [Google Scholar]
- 11.Gerna G, Furione M, Baldanti F, Sarasini A. Comparative quantitation of human cytomegalovirus DNA in blood leukocytes and plasma of transplant and AIDS patients. J Clin Microbiol. 1994;32:2709–2717. doi: 10.1128/jcm.32.11.2709-2717.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerna G, Revello M G, Percivalle E, Morini F. Comparison of different immunostaining techniques and monoclonal antibodies to the lower matrix phosphoprotein (pp65) for optimal quantitation of human cytomegalovirus antigenemia. J Clin Microbiol. 1992;30:1232–1237. doi: 10.1128/jcm.30.5.1232-1237.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerna G, Revello M G, Percivalle E, Torsellini M. A 6-hour microneutralization assay for human cytomegalovirus antibody by using monoclonal antibodies. Serodiagn Immunother Infect Dis. 1990;4:243–247. [Google Scholar]
- 14.Gerna G, Revello M G, Percivalle E, Zavattoni M, Parea M, Battaglia M. Quantification of human cytomegalovirus viremia by using monoclonal antibodies to different viral proteins. J Clin Microbiol. 1990;28:2681–2688. doi: 10.1128/jcm.28.12.2681-2688.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerna G, Zavattoni M, Baldanti F, Furione M, Chezzi L, Revello M G, Percivalle E. Circulating cytomegalic endothelial cells are associated with high human cytomegalovirus (HCMV) load in AIDS patients with late-stage disseminated HCMV disease. J Med Virol. 1998;55:64–74. [PubMed] [Google Scholar]
- 16.Gerna G, Zavattoni M, Baldanti F, Sarasini A, Chezzi L, Grossi P, Revello M G. Human cytomegalovirus (HCMV) leukoDNAemia correlates more closely with clinical symptoms than antigenemia and viremia in heart and heart-lung transplant recipients with primary HCMV infection. Transplantation. 1998;65:1378–1385. doi: 10.1097/00007890-199805270-00016. [DOI] [PubMed] [Google Scholar]
- 17.Gerna G, Zavattoni M, Percivalle E, Grossi P, Torsellini M, Revello M G. Rising levels of human cytomegalovirus (HCMV) antigenemia during initial antiviral treatment of solid-organ transplant recipients with primary HCMV infection. J Clin Microbiol. 1998;36:1113–1116. doi: 10.1128/jcm.36.4.1113-1116.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerna G, Zipeto D, Parea M, Revello M G, Silini E, Percivalle E, Zavattoni M, Grossi P, Milanesi G. Monitoring of human cytomegalovirus infections and ganciclovir treatment in heart transplant recipients by determination of viremia, antigenemia and DNAemia. J Infect Dis. 1991;164:488–498. doi: 10.1093/infdis/164.3.488. [DOI] [PubMed] [Google Scholar]
- 19.Gleaves C A, Smith T F, Shuster E A, Pearson G R. Rapid detection of cytomegalovirus in MRC-5 cells inoculated with urine specimens by using low-speed centrifugation and monoclonal antibody to an early antigen. J Clin Microbiol. 1984;19:917–919. doi: 10.1128/jcm.19.6.917-919.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gozlan J, Salord J M, Chouaïd C, Duvivier C, Picard O, Mejohas M C, Petit J C. Human cytomegalovirus (HCMV) late mRNA detection in peripheral blood of AIDS patients: diagnostic value for HCMV disease compared with those of viral culture and HCMV DNA detection. J Clin Microbiol. 1993;31:1943–1945. doi: 10.1128/jcm.31.7.1943-1945.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gozlan J, Laporte J P, Lesage S, Labopin M, Najman A, Gorin N C, Petit J C. Monitoring of cytomegalovirus infection and disease in bone marrow recipients by reverse transcription-PCR and blood and urine culture. J Clin Microbiol. 1996;34:2085–2088. doi: 10.1128/jcm.34.9.2085-2088.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grefte A, Harmsen M C, van der Giessen M, Knollema S, van Son W J, The T H. Presence of human cytomegalovirus (HCMV) immediate early mRNA but not ppUL 83 (lower matrix protein pp65) mRNA in polymorphonuclear and mononuclear leukocytes during active HCMV infection. J Gen Virol. 1994;75:1989–1998. doi: 10.1099/0022-1317-75-8-1989. [DOI] [PubMed] [Google Scholar]
- 23.Grossi P, Kusne S, Rinaldo C, St. George K, Magnane M, Rakela J, Fung J, Starzl T E. Guidance of ganciclovir therapy with pp65 antigenemia in cytomegalovirus-free recipients of livers from seropositive donors. Transplantation. 1996;61:1659–1660. doi: 10.1097/00007890-199606150-00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grossi P, Minoli L, Percivalle E, Irish W, Viganò M, Gerna G. Clinical and virological monitoring of human cytomegalovirus infection in 294 heart transplant recipients. Transplantation. 1995;59:847–851. [PubMed] [Google Scholar]
- 25.Hsia K, Spector D K, Lawrie J, Spector S A. Enzymatic amplification of human cytomegalovirus sequences by polymerase chain reaction. J Clin Microbiol. 1989;27:1802–1809. doi: 10.1128/jcm.27.8.1802-1809.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kievits T, van Gemen B, van Strijp D, Schukkink R, Direks M, Adriaanse H, Malek L, Sooknanan R, Lens P. NASBA isothermal enzymatic in vitro nucleic acid amplification optimized for the diagnosis of HIV-1 infection. J Virol Methods. 1991;35:273–286. doi: 10.1016/0166-0934(91)90069-c. [DOI] [PubMed] [Google Scholar]
- 27.Lam K M C, Oldenburg N, Khan M A, Gaylore V, Mikhail G W, Strouhal P D, Middeldorp J M, Banner N, Yacoub M. Significance of reverse transcription polymerase chain reaction in the detection of human cytomegalovirus gene transcripts in thoracic organ transplant recipients. J Heart Lung Transplant. 1998;17:555–565. [PubMed] [Google Scholar]
- 28.Locatelli F, Percivalle E, Comoli P, Maccario R, Zecca M, Giorgiani G, Di Stefano P, Gerna G. Human cytomegalovirus (HCMV) infection in pediatric patients given allogeneic bone marrow transplantation: role of early antiviral treatment for HCMV antigenemia on patients’ outcome. Br J Hematol. 1994;88:64–71. doi: 10.1111/j.1365-2141.1994.tb04978.x. [DOI] [PubMed] [Google Scholar]
- 29.Meyer T, Reischl U, Wolf H, Schuller C, Arndt R. Identification of active cytomegalovirus infection by analysis of immediate early, early and late transcripts in peripheral blood cells of immunodeficient patients. Mol Cell Probes. 1994;8:261–271. doi: 10.1006/mcpr.1994.1038. [DOI] [PubMed] [Google Scholar]
- 30.Nelson P N, Kawal B K, Boriskin Y S, Mathers K E, Powles R L, Steel H M, Tryhorn Y S, Butcher P D, Booth J C. A polymerase chain reaction to detect a spliced late transcript of human cytomegalovirus in the blood of bone marrow transplant recipients. J Virol Methods. 1996;56:139–148. doi: 10.1016/0166-0934(95)01900-6. [DOI] [PubMed] [Google Scholar]
- 31.Randhawa P S, Manez R, Frye B, Ehrlich G D. Circulating immediate-early mRNA in patients with cytomegalovirus infections after solid organ transplantation. J Infect Dis. 1994;170:1264–1267. doi: 10.1093/infdis/170.5.1264. [DOI] [PubMed] [Google Scholar]
- 32.Revello M G, Percivalle E, Zavattoni M, Parea M, Grossi P, Gerna G. Detection of human cytomegalovirus immediate-early antigen in leukocytes as a marker of viremia in immunocompromised patients. J Med Virol. 1989;29:88–93. doi: 10.1002/jmv.1890290204. [DOI] [PubMed] [Google Scholar]
- 33.Revello M G, Percivalle E, Arbustini E, Pardi R, Sozzani S, Gerna G. In vitro generation of human cytomegalovirus pp65 antigenemia, viremia, and leukoDNAemia. J Clin Invest. 1998;101:2686–2692. doi: 10.1172/JCI1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shibata D, Martin W J, Appleman M D, Causey D M, Leedom J M, Arnheim N A. Detection of cytomegalovirus DNA in peripheral blood of patients infected with human immunodeficiency virus. J Infect Dis. 1988;158:1185–1192. doi: 10.1093/infdis/158.6.1185. [DOI] [PubMed] [Google Scholar]
- 35.Shinkai M, Spector S A. Quantitation of human cytomegalovirus (HCMV) in cerebrospinal fluid by competitive PCR in AIDS patients with different HCMV central nervous system diseases. Scand J Infect Dis. 1995;27:559–561. doi: 10.3109/00365549509047067. [DOI] [PubMed] [Google Scholar]
- 36.Shinkai M, Bozzette S A, Powderly W, Frame P, Spector S A. Utility of urine and leukocyte cultures and plasma DNA PCR for identification of AIDS patients at risk for developing human cytomegalovirus disease. J Infect Dis. 1997;175:302–308. doi: 10.1093/infdis/175.2.302. [DOI] [PubMed] [Google Scholar]
- 37.Spector S A, Wong R, Hsia K, Pilcher M, Stempien M J. Plasma cytomegalovirus (CMV) DNA load predicts CMV disease and survival in AIDS patients. J Clin Invest. 1998;101:497–502. doi: 10.1172/JCI1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van den Berg A P, van der Bij W, van Son W J, Anema J, van der Giessen M, Schirm J, Tegzess A M, The T H. Cytomegalovirus antigenemia as a useful marker of symptomatic cytomegalovirus infection after renal transplantation—a report of 130 consecutive patients. Transplantation. 1989;48:991–995. doi: 10.1097/00007890-198912000-00019. [DOI] [PubMed] [Google Scholar]
- 39.van der Bij W, Schirm J, Torensma R, van Son W J, Tegzess A M, The T H. Comparison between viremia and antigenemia for detection of cytomegalovirus in blood. J Clin Microbiol. 1988;26:2531–2535. doi: 10.1128/jcm.26.12.2531-2535.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Velzing J, Rothbarth P H, Kroes A C M, Quint W G V. Detection of cytomegalovirus mRNA and DNA encoding the immediate-early gene in peripheral blood leukocytes from immunocompromised patients. J Med Virol. 1994;42:164–169. doi: 10.1002/jmv.1890420212. [DOI] [PubMed] [Google Scholar]
- 41.Zipeto D, Baldanti F, Zella D, Furione M, Cavicchini A, Milanesi G, Gerna G. Quantification of human cytomegalovirus DNA in peripheral blood leukocytes of immunocompromised patients by the polymerase chain reaction. J Virol Methods. 1993;44:45–56. doi: 10.1016/0166-0934(93)90006-d. [DOI] [PubMed] [Google Scholar]
- 42.Zipeto D, Revello M G, Silini E, Parea M, Percivalle E, Zavattoni M, Milanesi G, Gerna G. Development and clinical significance of a diagnostic assay based on the polymerase chain reaction for detection of human cytomegalovirus DNA in blood samples from immunocompromised patients. J Clin Microbiol. 1992;30:527–530. doi: 10.1128/jcm.30.2.527-530.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]