Abstract
Long non-coding RNAs and microRNAs have recently attained much attention regarding their role in the development of B cell lineage as well as participation in the lymphomagenesis. These transcripts have a highly cell type specific signature which endows them the potential to be used as biomarkers for clinical situations. Aberrant expression of several non-coding RNAs has been linked with B cell malignancies and immune related disorders such as rheumatoid arthritis, systemic lupus erythematous, asthma and graft-versus-host disease. Moreover, these transcripts can alter response of immune system to infectious conditions. miR-7, miR-16-1, miR-15a, miR-150, miR-146a, miR-155, miR-212 and miR-132 are among microRNAs whose role in the development of B cell-associated disorders has been investigated. Similarly, SNHG14, MALAT1, CRNDE, AL133346.1, NEAT1, SMAD5-AS1, OR3A4 and some other long non-coding RNAs participate in this process. In the current review, we describe the role of non-coding RNAs in B cell malignancies.
Keywords: B cell, Immune system, lncRNA, miRNA, Expression
Introduction
B cells are a subset of immune cells which contribute in the induction of humoral responses. These cells can be sub-classified to three classes based on their ontogeny and anatomic localization. B1 cells are produced from B1 progenitors. B cell progenitor cells of the bone marrow can produce the marginal zone and follicular B cells. Notably, B1 lymphocytes are originated from B1 progenitor cells which reside in the hepatic tissue during the fetal period. These cells preserve their self-renewal capacity after the neonatal time. B2 cells are developed from transitional 2 B cells originating from bone marrow precursors and have sustained output all through the adulthood period [1]. Abnormal development of B cells can result in human disorders including immune deficiency, autoimmunity, or allergy [2].
B cells are the principal source of antibodies. A typical example of antibodies produced by B1 lymphocytes is the naturally produced antibodies against ABO blood groups [3]. B1 cells can produce IgM antibodies contributing in the maintenance of tissue homeostasis due to their aptitude to bind with reformed self-antigens. These antigens include those produced in the process of cell apoptosis, ischemic damage and oxidative insult in atherosclerosis [7]. Besides, polyreactive IgA antibodies produced by B1 and follicular B cells participate in the mucosal immunity [4].
In addition, B cells also have an immunomodulatory effect through regulation of immune responses via producing cytokines that impede initiation or progression of immune-related disorders [1].
Several non-coding RNAs have been demonstrated to be involved in the regulation of function of different classes of B cells, thus contributing in the pathoetiology of related diseases. In fact, three classes of non-coding RNAs, namely long non-coding RNAs (lncRNAs), microRNAs (miRNAs) and circular RNAs (circRNAs) have been vastly investigated in the context of B cell-related disorders. LncRNAs have sizes more than 200 nucleotides, share many features with mRNAs and regulate gene expression at different levels [5]. CircRNAs are a group of transcripts that are produced through 3’-5’ ligation of a single RNA molecule. These transcripts have also regulatory functions on gene expression. They can also produce polypeptides [6]. Finally, miRNAs are transcripts with sizes about 22 nucleotides that suppress expression of mRNAs or degrade them through a base-pairing mechanism [7].
Through RNA sequencing and de novo transcript assembly methods, Brazão et al. have recognized more than 4500 lncRNAs which are expressed in different phases of development and activation of B cells [8]. Notably, the majority of these transcripts have not been formerly identified, even in the process of commitment of T cells. About one-fifth of these lncRNAs have been found to be either enhancer- or promoter-associated transcripts. Moreover, the B-cell lineage activating transcription factor PAX5 has been shown to directly regulate expression of tens of lncRNAs in pro-B and mature B cells as well as in acute lymphoblastic leukemia (ALL) [8].
In the current paper, we discuss the effects of non-coding portion of the genome on function of this class of immune cells in different contexts. We also explain the impact of dysregulation of non-coding RNAs in the development of B cell-related disorders, particularly malignant conditions as well as imbalances of immune responses. Identification of the role of these transcripts in these conditions would help in design of targeted therapies for these disorders.
Contribution of miRNAs in the regulation of B cell functions and related disorders
Several miRNAs have been found to affect function of B cells. This process has been mostly evaluated in the context of immune-related disorders and cancers. For instance, miR-7 has been shown to influence expression of PTEN in B cells. Expression of this miRNA has been increased in MRLlpr/lpr mouse model of lupus. Treatment with miR-7 antagomir has decreased disease manifestations in these animals. miR-7-related inhibition of PTEN/AKT signaling has enhanced differentiation of B cells into plasmablasts/plasma cells. Moreover, miR-7 silencing has reduced spontaneous formation of germinal center and normalized B cell subtype fractions in the spleen. In addition, miR-7 antagomir has decreased phosphorylation of STAT3 and IL-21 synthesis. Taken together, miR-7 has an important role in regulation of PTEN expression and functions of B cells [9].
Tan et al. have assessed miRNA profiles of naïve, germinal center and memory B cells. They have reported elevation of numerous miRNAs in germinal center B cells. miR-17-5p, miR-106a and miR-181b have been among mostly up-regulated miRNAs in these cells. miR-150 has been a miRNA with high expression in all three B-cell subsets. However, its expression has been found to be lower in germinal center B cells compared with naïve and memory B cells. Notably, expressions of miR-17-5p, miR-106a and miR-181b have been gradually decreased from the dark to the light zone of germinal center. Expression of miR-150 has been inversely correlated with c-Myb and Survivin levels in tonsil tissues, implying potential inhibition of these genes by miR-150 [10].
Several other miRNAs have been found to affect pathogenesis of diffuse large B-cell lymphoma (DLBC). A number of miRNAs have been shown to be dysregulated in these patients. For instance, expression of miR-16–1 has been found to be significantly lower in DLBC patients compared to controls in a single study [11]. Another study has shown differential expression of miR-197 in DLBCL versus controls. While expression levels of miR-197 have not been correlated with clinicopathologic parameters such as international prognostic index, down-regulation of this miRNA has been associated with disease progression in patients treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. Down-regulation of miR-197 levels could predict shorter progression-free survival in this subgroup of patients as well as non-germinal center B-like subgroup. Cell line studies have shown that miR-197 can enhance doxorubicin-associated apoptosis in SUDHL9 cells but not in OCI-Ly1 cells [12].
Another study in the context of sepsis has shown up-regulation of miR-19a in B cells. Moreover, in vitro studies have confirmed over-expression of this miRNA in activated B cells. Expression of CD22 has been initially increased but afterwards reduced. Notably, up-regulation of miR-19a has led to activation of BCR signaling, whereas up-regulation of CD22 has resulted in the attenuation of the effects of miR-19a and enhanced its expression. Taken together, miR-19a and CD22 contribute in establishment of a feedback circuit for B cell responses in sepsis, which can be considered as a putative target for re-establishment of immune homeostasis [13].
miR-30a is another miRNA that participates in the activation of B cells. This miRNA can specifically bind the 3'-UTR of Lyn transcript to inhibit its expression. miR-30a expression has been found to be elevated in B cells of patients with systemic lupus erythematous (SLE) compared with controls. Moreover, its levels have been negatively correlated with Lyn levels in B cells. Up-regulation of miR-30a has promoted proliferation of B cells and release of IgG antibodies. Thus, up-regulation of miR-30a can reduce Lyn levels in B cells, indicating its role in induction of B cell hyperactivity in SLE [14].
miR-155 is an example of miRNAs whose functions have been evaluated in different contexts such as rheumatoid arthritis [15], DLBC and non-Hodgkin lymphoma [16] as well as chronic psychological stress [17]. In B cell malignancies, higher levels of miR-155 have been correlated with the presence of B symptoms, involvement of extranodal sites, and high ECOG score [16].
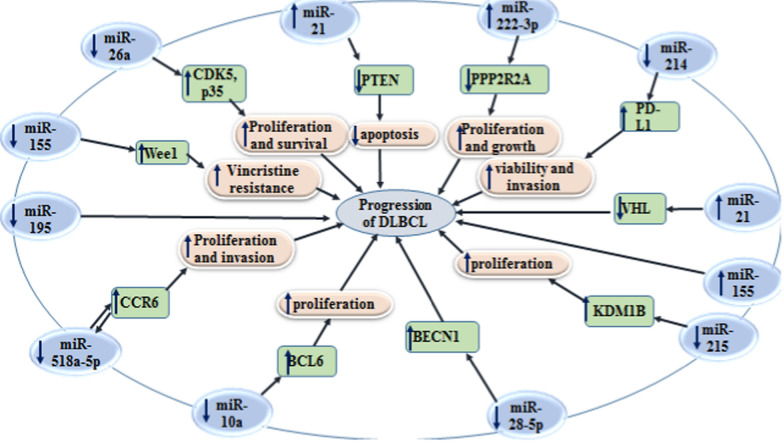
Figure 1 depicts the impacts of miRNAs on regulation of their target genes in the context of DLBCL.
Table 1.
miRNAs and B cell functions
| microRNA | Expression pattern | Disease/ | Sample | Cell line | Interaction | Signaling pathway | Function | References |
|---|---|---|---|---|---|---|---|---|
| Human studies | ||||||||
| miR-16–1 | ↓ | DLBC | 40 untreated patients diagnosed with DLBC and 15 healthy controls | CD19( +) and CD20( +) cells | – | – | – | [11] |
| miR-15a | no difference | DLBC | 40 untreated patients diagnosed with DLBC and 15 healthy controls | CD19( +) and CD20( +) cells | – | – | – | |
| miR-150 | ↑ (lower in GC B cell than the other two subsets) | chronic tonsillitis | children with chronic tonsillitis | Naïve B cells, GC B cells and memory B cells | c-Myb, Survivin and Foxp1 | – | ↑ miR-150: ↑ the amount of apoptotic/death cells, ↓ c-Myb, Survivin and Foxp1 | [10] |
| miR-155 | ↑ (abundantly in synovial B cells) | RA | 27 patients with ERA and 33 patients with LSRA, 14 patients with osteoarthritis, 9 healthy controls | B cells, CD19 + cells, synovial B cells | PU.1 | – |
↑ RA B-cell activation associated with autoantibody production ∆ miR-155: ↓ antibody synthesis |
[15] |
| viral miR-BHRF1 | ↓ | EBV-immortalized B lymphoblastic cell malignancy | – | Ramos and BJAB, Manassas, VA, B95.8, HEK293 |
SMAD3, JUN, and COL1A |
TGF-β signaling pathway | LA: ↓ viral miR-BHRF1-1: ↑ adhesion and the growth of EBV-infected B cells | [18] |
| miR-28 | ↓ | BL | – | GC B cells, HEK293T cells and B-cell lines | MAD2L1, BAG1, MYC | – | ↓ proliferation and clonogenic properties of BL cells, MYC-induced transformation | [19] |
| miR-19a | ↑ | sepsis | 64 patients with SIRS and 15 healthy controls | PBMCs | CD22 | BCR signaling | ↑ BCR signaling | [13] |
| miR-30a | ↑ | SLE |
patients with SLE and healthy controls |
Daudi and Raji B cell lines | Lyn | – | ↑ B cell proliferation and the production of IgG antibodies through inhibiting Lyn | [14] |
| miR-194 | ↓ | PTLD | PBMC or lymph node from six PTLD patients and 4 healthy blood donors |
AB5, JB7, JC62, MF4, VB5, ZD3 derived from PBMC or lymph node of six PTLD patients and B lymphoblastoid cell lines isolated from 4 healthy blood donors |
IL-10 | – |
Expression of microRNA-194 was suppressed by EBV microRNA-194 inhibited IL-10 expression, so reduced proliferation and promoted apoptosis of EBV( +) B cell lymphoma lines |
[20] |
| miR-125b | ↑ | – | – | murine Bcl1.3B3 B lymphoma and the human U266 multiple myeloma cell lines | BLIMP-1 and IRF-4 | – | ↓ differentiation of GC centroblasts and myeloma cell survival through inhibiting BLIMP-1 and IRF-4 translation | [21] |
| miR-148b | ↓ | BCL | Peripheral blood from 21 patients with BCL and 18 healthy controls, Lymphatic tissue from 30 patients with BCL and 20 healthy controls, male BALB/c nude mice | Raji and SU-DHL-10 human BCL cell lines, HEK-293 T | Bcl-w | – |
↓ cell viability, colony formation, and ↑ apoptosis in irradiated BCL cells, ↓ growth of tumors in nude mice (↑ radiosensitivity of BCL cells) |
[22] |
| miR-197 | ↓ | DLBCL | 51 patients with DLBCL | SUDHL9 and OCI-LY1 human DLBCL cell lines | – | – | ↑ miR-197: ↑ effects of doxorubicin on reducing cell viability and enhancing apoptosis | [12] |
| miR-124 | ↓ | DLBCL | – | OCI-Ly1 and HBL1 | p65 |
TAK1/IKKα-IKKβ/IκBα and MAPK/p65 signaling pathways, NF-κB signals |
↓ cell proliferation and survival | [23] |
| miR-17–92 | ↑ | B-NHL | 71 patients with B-NHL, 5 patients with reactive hyperplasia lymph nodes as controls, female Balb/c nude mice | WT, KO and TG lymphoma cells and reactive hyperplasia lymph cells obtained from mice | – | – |
↑ miR-18: ↓ OS ↑ miR-19 and miR-92a: ↓ OS and EFS ↑ miR-17–92: ↓ the duration of incubation required for visualization of the xenograft tumor |
[24] |
| miR-155 | – | DLBCL | 76 patients with DLBCL | HEK293T, RIVA, U2932, DHL4, HBL-1, Ly7, Ly18, and Ly19 cell lines | DEPTOR and c-CBL | BCR signaling |
∆ mir-155: ↓ expression of NFkB target genes and ↑ sensitivity DLBCL cells to ibrutinib Low expression of DEPTOR (a target of mir-155) increased the migration of DLBCL cells toward the CXCL12 gradient and modulated cytokine production |
[25] |
| miR-320d | ↓ | DLBCL | 85 patients with DLBCL, 19 samples with lymph node reactive hyperplasia as controls | OCI-LY1 (GCB subtype) and NU-DUL-1 (ABC subtype) human DLBCL cell lines | CDK6 | – | ↓ proliferation in GCB type of DLBCL cells and ↓ CDK6 expression | [26] |
| miR-195 | ↓ | DLBCL | 60 patients with DLBCL and 30 healthy controls | – | – |
Expression levels of miR-195 closely correlated with tumor diameter, IPI score and Ann Arbor stage Patients with high levels of miR-195 had longer OS |
[27] | |
| miR-155 |
↓ in vincristine- resistant DLBCL cell lines |
DLBCL |
73 patients with DLBCL, GEO database: data (GSE10846 and |
U-DHL-5 and OCI-Ly7 GCB-DLBCL cell lines, RIVA and NU-DHL-1 ABC cell lines | Wee1 (a direct target of miR-155) | – |
↑ sensitivity to vincristine Expression level of miR-155 was strongly correlated with superior survival for R-CHOP-treated patients of the GCB subclass |
[28] |
| miR-153-3p | ↓ in IM-resistant CML cells | CML | Blood samples obtained from 44 CML patients |
human KBM5, K562 and IM-resistant KBM5R, K562R CML cell lines |
Bcl-2 (a direct target of miR-153-3p) | – | ↑ miR-153-3p: ↑ IM sensitivity and ↓ the survival rate of IM-resistant CML cells ↓ autophagy caused by IM in IM-resistant CML cells | [29] |
| miR-30c | ↑ in patients with SCNSL | PCNSL, SCNSL | 61 CSF samples from patients with PCNSL and 14 samples from SCNSL | – | – | – | miR-30c could act as a biomarker to distinct PCNSL from SCNSL | [30] |
| miR-155 | ↑ | NHL and DLBCL | 84 patients with B-cell NHL and 15 healthy controls | – | – | – |
Higher levels of miR-155 were correlated with the presence of B symptoms, involvement of extranodal sites, and high ECOG score In DLBCL, higher levels of miR-155 were correlated with non-germinal B-cell-like type, the presence of B symptoms, involvement of extranodal sites, and higher IPI and ECOG scores ↑ miR-155; ↑ lower event-free survival |
[16] |
| hsa-miR-34a-5p | ↑ | DLBCL |
six serum samples from patients with DLBCL and 3 healthy control |
– | TP53 | p53 signaling pathway | hsa-miR-34a-5p was involved in 15 pathways such as the p53 signaling pathway | [31] |
| hsa-miR-323b-3p | ↓ | DLBCL |
six serum samples from patients with DLBCL and 3 healthy control |
– | – | – | hsa-miR-323b-3p was involved in four pathways such as pathways in cancer | |
| hsa-miR-431-5p | ↓ | DLBCL |
six serum samples from patients with DLBCL and 3 healthy control |
– | FYN | – | regulating FYN | |
| miR-155 | ↑ in EBV-infected B cells | lymphoma | – | DG75 cell line originated from an EBV-negative BL, DG75 RBPJ knockout cell line derived from DG75 wt parental cells, IB4 and GM12878 obtained from Coriell Cell Repositories (EBV-immortalized lymphoblastoid cell lines) | EBNA2, IRF4, RBPJ | – | ↑ the growth of EBV-infected B cells | [32] |
| miR-3173 | ↓ | B-ALL |
GEO database 135 children with B-ALL and 97 healthy controls plus 430 children with B-ALL and 340 healthy controls |
CCRF-SB and SUP-B15 human B-ALL cell lines |
PTK2 (a direct target of miR-3173) | – | ↓ proliferation, migration and invasion | [33] |
| miR-21 | ↑ | B-ALL | 75 children with B-ALL and 50 healthy controls | – | – | – | Lower DFS and OS | [34] |
| miR-21 | ↑ | DLBCL | 36 tissue samples from 26 patients with DLBCL and 10 healthy controls | CRL-2630 | PTEN | – | higher in stage III/IV patients, ↓ apoptosis (by regulating the expression of PTEN) | [35] |
| miR-222-3p | ↑ | DLBCL | 74 patients with initial diagnosis of ABC-type DLBCL, 26 patients with pathological diagnosis of reactive lymphoid hyperplasia as controls, SPF BALB/c nude mice | HMy2.CIR human normal B-cell immortalized cell line, DLBCL cell line, germinal central B-cell (GCB)-like OCI-Ly19 and SU-DHL-4, and ABC-like OCI-LY10 and U2932 | Phosphatase 2 regulatory subunit B alpha (a direct target of miR-222-3p) | – | ↑ proliferation, invasion and tumor growth, ↓ apoptosis | [36] |
| miR-29a | ↓ | SLE | peripheral blood of 66 patients with SLE and 10 healthy controls | Raji, | CRKL (a target gene of miR-29a) | – | ↓ the production of IgG (by regulating CRKL) | [37] |
| hsa-miR-223-3p and hsa-miR-21-5p | ↓ from stage I to stage III of PBC | PBC | Peripheral B cells from 72 PBC patients and 15 healthy controls | – |
mutual 4 target genes: TGFBR2, MEF2C, FOXP1 and RBPJ |
– | modulating B cell functions, such as B-cell signal transduction, differentiation, migration, and apoptosis in GO categories | [38] |
| miR33b, miR96, and miR503 | ↓ | Lymphoma | – | JeKo-1, Pfeiffer, SUDHL-2, PDX, and A20 |
PRMT5, CYCLIN D1 and c-MYC (target genes of miR33b, miR96 and miR503) |
– | ↓ lymphoma cell survival | [39] |
| miR-214 | ↓ | DLBCL | 15 pairs of DLBCL tissues and ANCTs, female BALB/c nude mice | OCI-Ly3, SU-DHL-2 and OCI-Ly10 human DLBCL cell lines, a normal B-cell line (NBC) and HEK-293 T | PD-L1 | – | ↓ viability and invasion, ↑ apoptosis | [40] |
| miR-107 | ↓ | ABMR | 19 patients with ABMR and 20 healthy controls | B lymphocytes, Daudi, Raji, and HEK-293 | ATG12 | – | ↑ miR-107: ↓ formation of autolysosomes in B lymphocytes of recipients, autophagy, and secretion of IgG and IgM antibodies | [41] |
| miR-92a | ↑ in PMBL than in DLBCL, but not in cHL | PMBL, DLBCL, cHL | 40 patients with PMBL, 20 patients with DLBCL, and 20 patients had with cHL | Karpas-1106P, SU-DHL-5 | FOXP1 (a target of miR-92a) | – | ↓ proliferation, ↑ apoptosis, | [42] |
| miR-21 | ↑ | DLBCL | 45 samples of lymphoma tissues from patients with DLBCL | SU-DHL-8, OCI-LY1, and SU-DHL-10 | VHL (a target of miR-21) | – | Curcumin decreased the proliferation, migration, and invasion abilities and increased apoptosis by suppressing miR-21 | [43] |
| miR-155 | ↑ | DLBCL | 76 patients with DLBCL and 40 samples with | DB cells | – | – | ↑ migration and invasion, ↓ apoptosis | [44] |
| miR-215 | ↓ | DLBCL | 50 patients with DLBCL and 30 samples with RPL | SU-DHL-4 cells | KDM1B | – |
↓ proliferation and ↑ apoptosis Low levels of miR-215 were correlated with shorter 5-year OS |
[45] |
| miR-155 | ↑ in tonsillar memory B cells and PBMCs activated with CpG | DS | – | PBMCs and Tonsils from healthy controls and children with DS | AID (a target of miR-155) | – | miR-155 played a role in DS-associated dementia and leukemia | [46] |
| miR-125b | ↑ in tonsillar memory B cells and plasma cells | DS | – | PBMCs and Tonsils from healthy controls and children with DS | – | – | miR-125b played a role in DS-associated dementia and leukemia | |
| miR-98 | ↑ | asthma | 20 patients with asthma and 20 healthy controls | PBMCs from healthy controls and patients with asthma | TSP1 | – | IL-13 decreased TSP1 expression through up-regulating expression of miR-98 in B cells | [47] |
| miR-28-5p | ↓ | DLBCL | – | OCI-LY7 human GCB-type DLBCL cell line and HEK-293 T | BECN1 (a direct target of miR-28-5p) | – | Curcumin: ↑ miR-28-5p: ↓ proliferation and autophagy, ↑ apoptosis | [48] |
| miR-21 | ↑ | DLBCL | 53 patients with DLBCL | – | Ki-67 | – | High expression levels of miR-21 was correlated with poor response to treatment | [49] |
| miR-10a | ↓ | DLBCLs | 9 patients with DLBCL and 9 samples with RLH as controls | OCI-LY7 and OCI-LY3 human DLBCL cell lines and HEK293T | BCL6 (a direct target of miR-10a) | – | ↓ proliferation, ↑ apoptosis | [50] |
| miR-125a | ↓ | AML | – | HL60 | p53, Bcl-2, c-myc | NF-κ Pathway | ↑ miR-125a: ↓ viability and invasion, ↑ apoptosis | [51] |
| let-7b-5p | ↑ | ITP | 61 patients with ITP and 31 healthy controls | PBMC from samples, peripheral CD19 + cells | BAFF, BAFF-R, NF-κB2 p100, Bcl-xL | – | ↑ B cell survival, ↑ BAFF-R and BAFF levels, ↑ phosphorylation of NF-κB2 p100 | [52] |
| miR-27a | ↑ | KD | 23 children with acute KD and 23 healthy controls |
PBMCs from samples, PurifiedCD19 + B cells, CD14 + monocyte cells |
IL-10 | – | ↑ monocyte-mediated TNF-α release, ↑ monocyte-mediated inflammatory responses via inhibiting the regulatory function of B10 cells | [53] |
| miR-17–92 | – | – | C57BL/6 mice | 38c13 cells, HEK, CD19KO B cells | c-Myc, PTEN (a target of miR-17–92) | PI3K/Akt/Foxo1 pathway |
∆ miR-17–92: ↑ RAGs expression (post-translationally through Foxo1) miR-17–92: ↓ B cell development |
[54] |
| miR-4638-5p | ↓ in ERG + DLBCL | DLBCL |
126 patients with DLBCL (in Kaplain‐Meier survival analysis) and 94 patients with DLBCL (in the clinicopathologic correlation study) |
– | ERG | – | More mutations in genes important in cell cycle control, B-cell receptor-mediated signaling and degradation of β-catenin were seen in ERG + DLBCL more likely harbors | [55] |
| miR-518a-5p | ↓ | DLBCL | 56 samples with DLBCL and 29 samples with RLH as controls | HMy2.CIR normal B cell line, SU-DHL-2 and SU-DHL-6 DLBCL cell lines | CCR6, (a direct target of miR-518a-5p) | JAK2-STAT6 signalling pathway |
There is a negative regulatory feedback loop between miR-518a-5p and CCR6 in DLBCL ↑ miR-518a-5p: ↓ proliferation and invasion, ↑ apoptosis |
[56] |
| miR-296-5p | ↑ | DLBCL | – | DLBCL-DB cells | – | – |
∆ miRNA-296-5p: ↓ proliferation and migration, apoptosis did not change |
[57] |
| miR-34a | ↓ | DLBCL | 65 patients with DLBCL and 22 samples with LRH as controls | – | ↑ BCL-2 | – | Patients with high levels of miR-34a had longer OS | [58] |
| miR-224 | ↓ | DLBCL | 76 patients with DLBCL and 41 healthy controls | – | PIK3CD (a direct target of miR-224) | – | ↑ miR-224: ↓ proliferation and invasion, ↑ apoptosis | [59] |
| miR-451a | ↓ | DLBCL | 89 patients with DLBCL and 48 healthy controls | – | – | – | The efficacy of rituximab combined with chemotherapy can be evaluated by miR-451a as an indicator | [60] |
| miR-152-3p | ↑ | SLE | 30 female patients with active SLE and 30 female healthy controls | SLE B-cells | KLF5 (a direct target of miR-451a), BAFF | – | ∆ miR-152-3p: ↓ self-reactivity of SLE B-cells, and ↓ autoantibody production | [61] |
| miR-28 | ↓ in GC-derived neoplasms | Non-Hodgkin lymphoma | human primary GC-derived B-cell neoplasms (GSE 29,493), NSG mice |
naïve B cells (CD19 + GL7 −), GC B cells (CD19 + GL7 +), and post-GC B cells (CD19 + GL7 − IgA +) from Peyer’s patches Ramos and Raji BL GC-derived B-cell lines and MD901 DLBCL cell line |
– | BCR signaling |
Downregulation of miR-28 expression is correlated with GC B-cell transformation ↑ miR-28: ↓ proliferation and survival |
[62] |
| miR-98 | ↑ | heart transplantation | peripheral blood samples from 20 patients with advanced heart failure before and after and 20 healthy controls, male BALB/c mice and male C57/B6 mice | peripheral blood mononuclear cells were isolated from the blood samples | ↓ IL-10 | – |
The levels of miR-98 and serum levels of cortisol were increased in peripheral B cells after heart transplantation Cortisol-suppressed IL-10 expression was mediated by miR-98 |
[63] |
| miR-21-5p | ↑ in cHL than GC-B cells | cHL | – | L540, KM-H2, L1236, L428 and L591, SUPHD1 CHL cell lines and HEK-293 T | PELI1 | – | ∆ miR-21-5p: ↓ growth, ↑ apoptosis | [64] |
| miR-29a | ↓ | Arthritis | – |
miR-29a knockout mice |
– | – | ↑ B-cell activation and germinal center production | [65] |
| miR-126 | ↓ in MLL-AF4 ALL | ALL | Congenic mice | Ebf1 − / − hematopoietic progenitor (Lin −) cells were isolated from the Ebf1 − / − livers of 14 d postcoitum embryos | IRS-1 | – |
miR-126 drived B-cell myeloid biphenotypic leukemia differentiation toward B cells. (↑B cells) miR-126 could partly rescue failed B-cell lineage development and specification |
[66] |
| miR-212 | ↑ | Autoimmune disease and cancer | C57BL/6 WT and miR-212/132 − / − mice | HEK293T, primary splenic B cells | – | BCR signaling | BCR activation: ↑ miR-212 | [67] |
| miR-132 | ↑ | Autoimmune disease and cancer | C57BL/6 WT and miR-212/132 − / − mice | HEK293T, primary splenic B cells | Sox4 | BCR signaling |
BCR activation: ↑ miR-132 ↓ early B cell development, ↑ apoptosis in primary bone marrow B cells ∆ miR-132: B cell recovery after antibody-mediated B cell depletion ↓ B cell leukemia development |
|
| mir-23a cluster | – | – | mirn23a − / − mice and WT C57BL/6 mice | A20 and EML, 32Dcl3 | Ebf1, Pax5, Mef2c, Ikzf1, FoxO1, Trib3 | – |
∆ mirn23a: ↑ B cells, ↑ B lymphopoiesis, ↑ T1 population of transitional B cells, ↑ CLP population and ↓ myeloid cells, ↓ myelopoiesis, ↓ myeloid progenitor populations B cells with mirn23a − / − genotype secrete normal levels of IgG, proliferate normally, and could differentiate into short-lived effector plasma cells in response to antigen |
[68] |
| miR-148a | ↑ | Lupus |
gMb-macroself, Gadd45a − / − , Bcl2l11 − / − , Ptenfl/fl, Cd19-Cre, Tnfrsf1b − / − mice, and CD45.1 + C57BL/6 J mice |
HEK293T, splenic B cells (CD19 +) and BM B cell precursors (CD19 + IgM −) from CD45.1 + C57BL/6 J mice | Gadd45α, PTEN, Bim | – | miR-148a was found to be a regulator of B cell tolerance by promoting the survival of immature B cells and accelerating the development of autoimmunity by suppressing the expression of Gadd45α, PTEN, Bim | [69] |
| miR-17–92 | ↑ | cGVHD | miR-17–92 conditional knockout mice (BALB/c mice) | donor BM-derived cells (Ly5.1 +) in peripheral blood and spleen, miR-17–92–deficient B cells, | – | – | miR-17–92 increases the pathogenicity of B cells, promoted GC responses and B-cell function, the development of BO and reduced proteinuria/ascites | [70] |
| miR-125b | Epigenetic silencing of miR-125b is necessary for normal B-cell development | – | WT and Eμ/miR-125b-Tg mice | HEK293T, bone marrow sinusoidal and parenchymal B cells from Eμ/miR-125b-Tg mice and littermate controls | S1PR1, IRF4 | – | Expression of miR-125b impaired B-cell egress from the bone marrow to peripheral blood | [71] |
| miR-26a | ↓ in DLBCL cell lines compared to B lymphocytes | DLBCL | NOD/SCID mice | SU-DHL-4, SU-DHL-6, SU-DHL-16 GCB cell lines and SU-DHL-2, SU-DHL-8, and RCK-8 ABC cell lines |
CDK5, p35 (a direct target of miR-26a) |
– | ↓ DLBCL tumor growth, proliferation, cell-cycle progression, and survival | [72] |
| miR-155 | – | – | CD45.1 + congenic mice, SWHEL mice and miR-155–deficient mice (all with the C57BL/6 background) | SWHEL Mir155 + / + or SWHEL Mir155 − / − donor B cells | – | – |
miR-155 regulated the early expansion of B-blasts and later on the survival and proliferation of plasmablasts in a B-cell-intrinsic manner miR-155 is required for the optimal proliferation of plasmablast B cells |
[73] |
| miR-181b | ↑ in neonatal B cells | – | miR-181a/b1−/− mice; ko mice and miR-181a/b-1 ± mice with C57BL/6 J background | Neonatal and adult B cells | – | – | ∆ miR-181b: ↑ class-switch recombination | [74] |
| miR-155 | ↓ | chronic psychological stress | male C57BL/6 mice | In-vitro-induced GC B cells, Naive B cells, Su-DHL4 cells | FBXO11 (a direct target of miR-155), BCL6 | – |
Corticosterone treatment: ↓ miR-155: ↓ GC B cell generation and isotope class switching ↑ miR-155: ↓ stress-induced impairment of GC response |
[17] |
| miR-221 | – | – | C57BL/6, RAG1−/−(CD45.2, CD45.1) mice | preBI cell lines | PTEN (a target of miR221), CXCL12, Bcl2 | PI3K signaling |
↑ precursor B-cell retention in the bone marrow, ↑ CXCR4-PI3K mediated Bcl2 upregulation, ↑ early B-cell adhesion capability via PI3K signaling |
[75] |
| miR-92a | ↓ | DM | Adult mice | Min-6 mouse pancreatic bcells | KLF2 (a direct target of miR-92a) | – |
↑ insulin secretion and proliferation, ↓ apoptosis |
[76] |
| miR-15a/16–1 | ↓ | Plasma cell and mature B-cell neoplasms | AIDCre/ + (wild-type [WT]) control and AIDCre/ + ;miR-15a/16-1 fl/fl (knockout [KO]) compound mice with C57BL/6 background | GC B cells from WT and KO mice | – | – | Deletion of the miR-15a/16–1 increased the number of GC B cells, percentage of dark zone B cells, and maturation into plasma cells | [77] |
| miR-146a | – | – | CD21-cre, Cγ1-cre, CD4-cre, hCD2-cre, and CD40-deficient mice, B-KO/CD40 + / − mice | Naive B cells from unimmunized B-KO mice or WT littermates and GC B cells from corresponding D14 SRBC-immunized mice | – | CD40 signaling pathway |
The loss of miR-146a in B cells leaded to the development of spontaneous autoimmunity miR-146a is crucial to maintain optimal B cell responses |
[78] |
| miR-146a | ↓ | B-cell oncogenesis | Eμ-Myc miR-146a − / − mice | 70Z/3 and WEHI-231 | Egr1, Blimp1 and Bcl6 | – |
↓ miR-146a: ↓ survival, ↑ in peripheral blood CD11b + myeloid cells, ↑ mature B-cell phenotype ↑ miR-146a: ↓ cell growth |
[79] |
| miR-21 | ↑ | Lymphoma | NOD-SCID mice | OCI-LY3 and Ramos, OCI-LY10, U2932, Raji, Rec-1, Jeko-1, Maver-1 and JM1, HEK293T | Nl101, Mxd1 (a target of miR-21), c-Myc | – |
NL101: ↑ miR-21, c-Myc: ↓ miR-21, miR-21: ↑ proliferation and survival, ↓ apoptosis |
[80] |
| miR-146a | ↓ | Immune complex glomerulonephritis | miR-146a − / − mice with C57BL/6 background | B lymphocytes were the spleen, HK-2 | Kim1/Tim1 | – |
∆ miR-146a: ↑ numbers of memory B cells and plasmablasts, ↑ glomerular hypercellularity with age , ↓ Bregs and ↓ Kim1/Tim1 |
[81] |
| miR-146a | ↑ | – |
Murine OVA-Induced asthma mice, WT and miR-146a TG mice |
purified splenic B cells | Smad4 (a direct target of miR-26a), 14–3-3σ | – | ↑ class switch and secretion of IgE in B cells | [82] |
| miR-142 | ↑ | Lymphoma | BMT and transgenic (Eμ/mir142) mice | KHM10B, Raji, KMS12, OCI-Ly8, Hut 78, and Cos7 | – | – |
In splenic B cells, high expression of Mir142 modified LPS-induced phenotypical changes |
[83] |
| miR-7 | ↑ | SLE |
Female MRLlpr/lpr lupus mice |
Purified splenic B cells obtained from mice |
PTEN | PTEN/AKT signaling |
∆ miR-7: ↓ nephritis, ↓ lupus manifestations, ↓ immune Abnormalities, ↓ tfh-derived IL-21 expression, ↓ Abnormal B cell differentiation, normalizes splenic B cell subtypes |
[9] |
| miR-98 | ↑ | Myocarditis | BALB/c mice immunized with MyHC-α | B cells isolated from the mouse hearts with myocarditis | ↓ IL-10 (a target of miR-98), TNF-α | – |
∆ miR-98: ↓ myocarditis miR-98 is upregulated by TNF-α in B cells |
[84] |
| Let-7 | – | – |
Lin28a iTg mice, let-7adf cluster KO mice, and let-7bc cluster KO mice |
HEK293T |
Hk2 (a target gene of Let-7) c-myc (a target gene of Let-7) Slc1a5 and Gls (indirect target genes of Let-7) |
– |
↓ IgM Production ↓ glycolytic capacity and glucose uptake ↓ glutamine uptake and utilization ↓ B Cell Activation |
[85] |
| Let-7 | ↑ in thymic B progenitors by in vitro co-culture with IL15, Vitamin-D3, and retinoic acid | – | Foxn1lacZ/lacZ (Z/Z) mice with C57Bl6/J background, Foxn1nude heterozygous (Foxn1 + /nude) mice with C57Bl6/J background, Foxn1lacZ/nude (Z/N) mice, Foxn1 + /lacZ (+ /Z) mice | thymic progenitor B cells | Lin28a, Arid3a | – | ↓ B cell production in the thymus, ↓ proliferation of intrathymic progenitor B cells | [86] |
| miR-191 |
↑ during B-cell development and differentiation |
– | C57BL/6 J and NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice, C57BL/6 mice and miR-191 − / − mice | Primary cells from wild-type or chimeric mice, preB1 cells, | Foxp1, E2A, and Egr1 | – |
Expression levels of miR-191 are required for efficient B-cell development, V(D)J recombination and IL-7-dependent expansion of preBI cells |
[87] |
| miR-15 family | ↓ | – | Female C57BL/6 Rag1 − / − mice | wk3, 1587, and 1677 pre‐B cell lines from total bone marrow of SLP‐65 − / − and SLP‐65 − / − LAT − / − mice, respectively, and 1676 and 74 pre‐B cell lines | ↑ cyclin E1 and D3 | – | The lack of miR-15 family in pre-B cells caused prolonged proliferation, so failed to trigger the transcriptional reprogramming to accompany their differentiation | [88] |
| mirn23a cluster | ↓ | – | Wildtype and mirn23a−/− C57BL/6 mice, CD45.1 recipient mice, femurs and tibias of mice | 70Z/3, A20 and 32Dcl3 cell lines |
↑ Ikzf1, Runx1, Satb1, Bach1 and Bach2 that managed the commitment of MPPs to CLPs ↑ FoxO1, Ebf1, and Pax5 that commited the CLP to the B cell lineage in the absence of mirn23a, EBF1 |
PI3K/Akt and BMP/Smad signaling pathways |
Mirn23a regulated some related transcription factors and signaling pathways to modulate adult hematopoiesis Mirn23a was inhibited by EBF1 |
[89] |
TB, tuberculosis; CTRL, control; BL, Diagnosis; M1, month 1; M6, month 6; GC, germinal center; SLE, systemic lupus erythematosus; DLBC, diffuse large B-cell lymphoma; RA, rheumatoid arthritis; ERA, early rheumatoid arthritis; LSRA, long standing rheumatoid arthritis; BCR, B cell receptor; CLP, common myeloid progenitor; LA, lactic acid; MLL, myeloid/lymphoid leukemia; ALL, acute lymphoblastic leukemia; BL, Burkitt lymphoma; SLE; systemic lupus erythematosus; PTLD, posttransplant lymphoproliferative disorder; EBV; Epstein-Barr virus; BCL, B-cell lymphoma; cGVHD, Chronic graft-versus-host disease; BO, bronchiolitis obliterans; B NHL, B cell non-Hodgkin’s lymphoma; OS, overall survival; EFS, event free survival; WT, wild-type; KO, knockout; TG, overexpression; IM, Imatinib; CML, Chronic myeloid leukemia; PCNSL, Primary lymphomas of the central nervous system; SCNSL, secondary spread of systemic lymphoma to the CNS; CSF, Cerebrospinal fluid; ECOG, Eastern Cooperative Oncology Group; IPI, International Prognostic Index; B-ALL, B-cell acute lymphoblastic leukaemia; DFS, disease-free survival; PBC, Primary biliary cholangitis; ANCTs, adjacent non-cancerous tissues; BMT, bone marrow transplantation; ABMR, Antibody-mediated renal allograft rejection; PMBL, Primary mediastinal large B-cell lymphoma; LNRH, lymph node reactive hyperplasia; RPL, reactive proliferative lymphadenitis; PBMCs, Human peripheral blood mononuclear cells; DS, Down Syndrome; DM, diabetes mellitus; RLH, reactive lymph node hyperplasia; AML, acute myeloid leukemia; ITP, immune thrombocytopenia; KD, Kawasaki disease; IgAN, immunoglobulin A nephropathy; RLH, reactive lymphoid hyperplasia; LRH, lymphonode reactive hyperplasia; cHL, Classical Hodgkin lymphoma
Fig. 1.
The impacts of miRNAs on regulation of their target genes in the context of DLBCL. Detailed information about these miRNAs is presented in Table 1.
Contribution of lncRNAs in the regulation of B cell functions and related disorders
Impacts of lncRNAs on B cell functions have been investigated in malignancies, particularly DLBCL. SNHG14 has been shown to be elevated in DLBCL. Its silencing has decreased proliferation, migration and epithelial to mesenchymal transition (EMT) features in these cells. From a mechanistical point of view, SNHG14 could sponge miR-5590-3p and subsequently enhance expression of ZEB1. Moreover, ZEB1 could activate transcriptional of SNHG14 and PD-L1 to increase immune evasion in these cells. Cumulatively, SNHG14/miR-5590-3p/ZEB1 axis can promote progression of DLBCL and immune evasion in a positive feedback loop. This axis can regulate PD-1/PD-L1 checkpoint [90].
Another study has shown up-regulation of MALAT1, PD-L1 and CD8 in DLBCL tissues, parallel with down-regulation of miR-195. Mechanistically, MALAT1 has been shown to sponge miR-195 to influence PD-L1 levels. MALAT1 silencing has enhanced miR-195 levels and reduced PD-L1 levels. Moreover, MALAT1 silencing has suppressed proliferation, migratory potential and immune escape aptitude of DLBCL cells while increasing their apoptosis. MALAT1 silencing has also inhibited EMT features through modulation of Ras/ERK signaling [91].
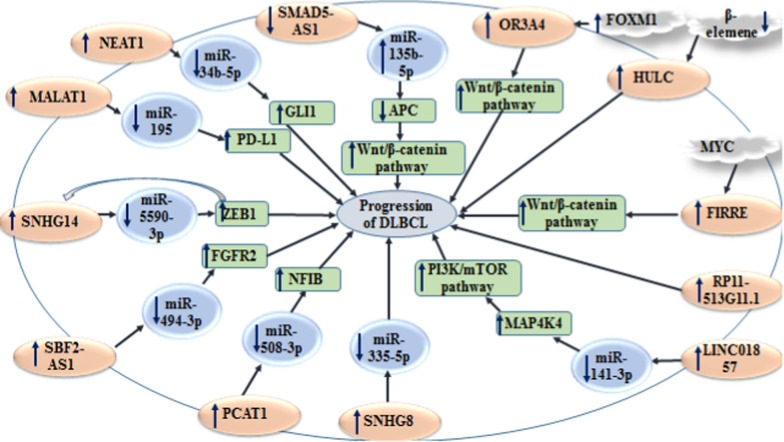
NEAT1 is another lncRNA whose expression has been enhanced in DLBCL tissues and cell lines parallel with up-regulation of GLI1 and down-regulation of miR-34b-5p. NEAT1 silencing or miR-34b-5p up-regulation could inhibit proliferation and enhance apoptosis of these cells. In fact, NEAT1 acts as a competing endogenous RNA (ceRNA) to regulate expression the miR-34b-5p/GLI1 axis. Besides, MYC has been shown to modulate NEAT1 expression through directly binding to promoter of NEAT1 [92]. Figure 2 shows the interactions between lncRNAs and miRNAs in the context of DLBCL.
Fig. 2.
Several lncRNAs can affect availability of miRNAs, thus influencing progression of DLBCL. Detailed information about these lncRNAs is shown in Table 2.
CRNDE has been shown to be up-regulated in the bone marrow of B-cell precursor acute lymphoblastic leukemia patients and related cell lines. CRNDE silencing has decreased cell proliferation and enhanced cell apoptosis in these cells. Functionally, CRNDE could bind with to miR-345-5p and down-regulate its expression, thus affecting expression of CREB. Notably, in vivo studies have shown that CRNDE silencing increases survival of mice models of this type of leukemia [93].
In addition to this type of studies, expression patterns of lncRNAs have been compared between cancer cells and non-cancerous controls using high throughput methods. For instance, Cuadros et al. have reported differential expression of 48 lncRNAs between pediatric B-ALL and normal bone marrow specimens. They have recognized AL133346.1/CCN2 as the most relevant lncRNA/mRNA pair in this type of malignancy. Expression of AL133346.1/CCN2 pair has been enhanced in B-ALL specimens [94].
Expression of PTTG3P has been shown to be up-regulated in samples obtained from patients with IgA nephropathy compared with normal samples. Notably, expression of PTTG3P in urine samples has been correlated with expression of PTTG3P in intra-renal samples of IgA nephropathy cases. Up-regulation of PTTG3P has stimulated B cell growth and increased expressions of cyclin D1 and ki-67. In addition, its up-regulation of PTTG3P has led to induction of IL-1β and IL-8 release. PTTG3P up-regulation could suppress expression of miR-383 in B cells. Taken together, PTTG3P could increase B cell growth and IL-1β and IL-8 release through influencing expression of miR-383. Through this effect, PTTG3P contributes in the pathogenesis of IgA nephropathy [95].
Expression of lncRNA RP11-530C5.1 has been shown to be higher in relapsing MS patients, compared to remitting MS patients and healthy subjects, whereas expression of AL928742.12 has been decreased. Notably, expression levels of RP11-530C5.1 and AL928742.12 have been correlated with PAWR and IGHA2 levels, respectively [96].
Table 2 shows the impact of lncRNAs in B cell functions.
Table 2.
LncRNAs and B cell functions
| lncRNA | Expression pattern | Disease | Sample | Cell line | Interaction | Signaling pathway | Function | References |
|---|---|---|---|---|---|---|---|---|
| Human studies/mixed studies | ||||||||
| SNHG14 | ↑ | DLBCL | 38 pairs of B cell lymphoma tissues and ANCTs, BALB/c mice | GM12878, 293 T, A20, OCI-LY7, DB, U2932, and FARAGE | ↓ miR-5590-3p, ↑ ZEB1, PD-1/PD-L1 checkpoint | – |
∆ SNHG14: ↓ proliferation, migration and EMT process There is a positive feedback loop between SNHG14 and ZEB1 to promote DLBCL |
[90] |
| MALAT1 | ↑ | DLBCL | 37 patients with DLBCL |
OCI-Ly10 human DLBCL cell line, CD8 + T cells |
↓ miR-195, ↑ PD-L1 and CD8 | Ras/ERK signaling pathway | ∆ MALAT1: ↓ proliferation, migration and immune escape ability, EMT-like process, ↑ apoptosis | [91] |
| CRNDE | ↑ | BCP-ALL | BM biopsies from 26 patients with BCP-ALL and BM biopsies from 15 patients with unexplained thrombocytosis or anemia as controls | NALM-6, RS4;11 CEMO-1, CCRF-SB, and SUP-B15 BCP-ALL cell lines | ↓ miR-345-5p, ↑ CREB | _– | ∆ CRNDE: ↓ proliferation, ↑ apoptosis | [93] |
| AL133346.1 | ↑ | B-ALL | GEO dataset: GSE128254 | – | ↑ CCN2 | – | It was found that either AL133346.1 regulates CCN2 expression in cis; or AL133346.1 and CCN2 are regulated by the same regulatory elements | [94] |
| NEAT1 | ↑ | DLBCL | 30 patients with DLBCL and 30 healthy controls | OCI-Ly1, OCI-Ly8, OCI-Ly10 and SUDHL-4 DLBCL cell lines | ↓ miR-34b-5p, ↑ GLI1 | – | ∆ NEAT1: ↓ proliferation, ↑ apoptosis | [92] |
| SMAD5-AS1 | ↓ | DLBCL | 11 patients with DLBCL and 11 healthy controls, BALB/c-nude mice | TMD8, U2932, GM12878, HEK-293, OCI-Ly3, WSU-FSCCL, JeKo-1, L428, and Raji | ↑ miR-135b-5p, ↓ APC | ↑ Wnt/β-catenin pathway | ↑ SMAD5-AS1: ↓ proliferation, ↑ apoptosis | [97] |
| OR3A4 | ↑ | DLBCL | 58 patients with DLBCL and healthy controls |
2932, SU-DHL-6, SU-DHL-4, OCL-LY-7, OCL-LY- 10 DLBCL cell lines and WIL2 human B lymphocyte |
↑ FOXM1 | ↑ Wnt/β-catenin signaling pathway |
∆ OR3A4: ↓ proliferation and ↑ apoptosis OR3A4 is upregulated by FOXM1 |
[98] |
| FIRRE | ↑ | DLBCL |
70 pairs of DLBCL patient samples and healthy controls |
2932, SU-DHL-6, SU-DHL-4, OCL-LY-7, OCL-LY-10 human DLBCL cell lines and WIL2S one normal B-cell line |
MYC | ↑ Wnt/β-catenin signaling pathway | ↑ proliferation and ↓ apoptosis | [99] |
| RP11-513G11.1 | ↑ | DLBCL | 93 patients with DLBCL and 62 healthy controls | – | – | – | Patients with high expression levels of RP11-513G11.1 showed shorter PFS and OS | [100] |
| lnc-290 | ↑ in B cells stimulated by LPS | inflammation and tissue damage | female C57Bl/6 mice | GFP + cells | CD69/CD86, LPS/TLR4 signaling pathway | NF-κB/ERK pathways | ∆ lnc-290: ↓ growth of B cells, ↓ cell differentiation and ↓ immunoglobulin production, ↓ B cell activation by blocking the LPS/TLR4 signaling pathway | [101] |
| LINC01857 | ↑ | DLBCL | TCGA and GTEX databases, GEO datasets | HCC1395, CYP6D, OCI-Ly3, and Raji | ↓ miR-141-3p, ↑ MAP4K4 | PI3K/mTOR pathway | ↑ proliferation, ↑ EMT process and ↑ cell cycle progression, ↓ apoptosis | [102] |
| TEX41 | ↑ in B-ALL | B-ALL | 79 patients with B-ALL, 25 patients with T-cell ALL and 38 acute myeloid leukemia | RS4;11 cells | p53 and p21 | – | ↑ proliferation, ↑ cell growth and ↑ cell cycle progression | [103] |
| AFAP1-AS1 | ↑ | GCB-DLBCL | 48 patients with DLBCL | OCI-ly1 and OCI-ly19 GCB-DLBCL cell lines | SFPQ, NONO, SRSF2, SRSF6, and KHSRP | BCR and TNF signaling pathways |
∆ AFAP1-AS1: ↓ proliferation, ↑ G0/G1 arrest and ↑ apoptosis Patients with higher expression levels of AFAP1-AS1 had poorer DFS and OS |
[104] |
| PTTG3P | ↑ | IgAN | patients with IgAN and healthy controls | B cells | ↓ miR-383, cyclin D1 and ki-67, IL-1β and IL-8 | – | ↑ PTTG3P: ↑ B cell growth and ↑ cyclin D1 and ki-67 expression, ↑ IL-1β and IL-8 production | [95] |
| SNHG8 | ↑ | DLBCL | – | GM12878 human B lymphocytes and OCI-Ly10, OCI-Ly7, OCI-Ly3, and U2932 human DLBCL cell lines | ↓ miR-335-5p | – | ∆ SNHG8: ↓ proliferation, ↓ colony formation and ↑ apoptosis | [105] |
| PCAT1 | ↑ | DLBCL | 48 pairs of DLBCL tissues and ANCTs | OCI-LY-7, OCI-LY-7, TMD8 and U2932 DLBCL cell lines, IM-9 human peripheral blood B-lymphocyte | ↓ miR-508-3p, ↑ NFIB | – | ↑ PCAT1: ↑ proliferation, ↑ migration and ↑ invasion | [106] |
| SBF2-AS1 | ↑ | DLBCL | 50 patients with DLBCL | OCI-LY-3, OCI-LY-7, OCI-LY-10, SU-DHL-4 and SU-DHL − 6 and HEK293 cells | ↓ miR-494-3p, ↑ FGFR2 | – | ∆ SBF2-AS1: ↓ viability and ↓ growth | [107] |
| SNHG14 | ↑ | DLBCL | 21 patients with DLBCL and 21 healthy controls | GM12878, OCI-LY-7, ABC, OCI-LY-3 and RCK-8 | ↓ miR-152-3p | – | ↑ growth, migration, and EMT-like processes, ↓ apoptosis | [108] |
| LINC00908 | ↑ | DLBCL | 28 pairs of DLBCL tissues and ANCTs, female BALB/c nude mice | GM12878 human lymphoblastoid B cell and OCI-LY7, DB, U2932, and FARAGE human DLBCL cells | miR-671-5p | – | ∆ LINC00908: ↓ proliferation and invasion, tumor growth | [109] |
|
TCONS_ 00,022,357-XLOC_010919 |
↑ | GD | Peripheral blood from 34 patients with GD, and 34 healthy controls | CD19 + B cells from 21 healthy individuals and 24 GD patients, PBMCs | TCL1A | – |
TCONS_ 00,022,357-XLOC_010919 regulated TCL1A, and TCL1A is involved in B-cell proliferation |
[110] |
| n335641 | ↑ | GD | Peripheral blood from 34 patients with GD, and 34 healthy controls | CD19 + B cells from 21 healthy individuals and 24 GD patients, PBMCs | TCL1A | – | n335641 regulates TCL1A, and TCL1A is involved in B-cell proliferation | |
| n337845 | ↓ | GD | Peripheral blood from 34 patients with GD, and 34 healthy controls | CD19 + B cells from 21 healthy individuals and 24 GD patients, PBMCs | SH2D1A | – | n337845 regulates SH2D1A, and SH2D1A is involved in B-cell proliferation | |
| ZEB1-AS1 | ↑ | B-ALL | 30 with B-ALL and 30 healthy controls | hBMSC-TERT | – | IL-11/STAT3 pathway |
∆ ZEB1-AS1: ↓ proliferation and IL-11 production High expression levels of ZEB1-AS1 showed poor prognosis of B-ALL patients ZEB1-AS1 promoted IL-11 stability |
[111] |
| UCA1 | ↑ | DLBCL | 38 patients with DLBCL and 38 healthy controls | GM12878, JeKo-1, TMD8, U2932, OCI-Ly-10 and OCI-Ly-7 cell lines and U2932 | ↓ miR-331-3p | – | ∆ UCA1: ↓ proliferation, viability, migration and invasion | [112] |
| LAMP5-AS1 | ↑ in MLL leukemia patients than that in the MLL-wt leukemia | MLL leukemia | 58 patients with MLL leukemia and 163 MLL-wt leukemia, NOD-SCID mice |
MOLM13, THP1, MV4-11, RS4-11, and HEK293T human MLL leukemia cells |
DOT1L | – |
∆ LAMP5-AS1: ↓ colony formation and ↑ differentiation of primary MLL leukemia CD34 + cells Patients with high levels of LAMP5-AS1 showed a reduced 5-year leukemia-free survival LAMP5-AS1 increased the methyltransferase activity of DOT1L |
[113] |
| LINC00152 | ↑ | Gastric cancer | 30 pairs of GC tissues and ANCTs, male BALB/c nude mice |
RGM‐1 human epithelial cells of gastric mucosa, and human BGC‐823 GC cell |
Bcl-2 | – | ↑ migration and invasion, ↓ apoptosis | [114] |
| HCP5 | ↑ | DLBCL | 48 patients with DLBCL and 14 RLH samples | OCI-LY7 and OCI-LY3 human DLBCL cell lines | ↓ miR-27b-3p, ↑ MET | – |
∆ HCP5: ↓ proliferation, ↑ apoptosis Geniposide treatment: ↓ HCP5 |
[115] |
| PEG10 | ↑ | DLBCL | 25 patients with DLBCL and 25 healthy controls | SU-DHL-8 and OCI-LY-8 DLBCL cell lines | ↓ miR-101-3p, ↑ KIF2A | – | ∆ HCP5: ↓ proliferation, migration and invasion, ↑ apoptosis | [116] |
| GAS5 | ↓ | DLBCL | – | OCI-Ly3 and TMD8 cells | ↑ miR-18a-5p, ↓ RUNX1 | – | ∆ GAS5: ↓ proliferation, ↓ G1 arrest, ↑ apoptosis | [117] |
| TUG1 | ↑ | DLBCL | 15 tumor tissues and venous blood from DLBCL patients, 15 patients with RLH as controls, female BALB/c athymic nude mice | OCI-LY7, and OCI-LY3 human DLBCL cell lines, IM-9I normal B lymphocyte | ↑ MET | – | ∆ TUG1: ↓ proliferation and tumor growth | [118] |
| SNHG12 | ↑ | DLBCL | 80 patients with activated B-cell like DLBCL, 80 patients with RLH as controls, male BALB/c nude mice | OCI-LY7, and OCI-LY3 human DLBCL cell lines, IM-9I normal B lymphocyte | ↓ miR-195 | – | ∆ SNHG12: ↓ cell growth, ↓ migration, and ↓ invasion | [119] |
| PANDA | ↓ | DLBCL | 114 patients with DLBCL and 114 healthy controls | U2932, SUDHL-6, SUDHL-3, OCI-Ly3, and OCI-Ly8 human DLBCL cell lines and WIL2S normal B-cell line | p53 | MAPK/ERK signaling pathway |
↑ G0/G1 cell cycle arrest and ↓ proliferation through silencing MAPK/ERK signaling pathway Low levels of PANDA were associated with poorer clinical outcome and lower OS in DLBCL patients |
[120] |
| GAS5 | ↓ | B lymphocytic leukemia |
30 patients with human B lymphocytic leukemia, 30 healthy controls |
RAMOS, ST486, Raji, and Farage human B lymphocytic leukemia cell lines and IM9 normal B lymphocytic cell line |
miR-222 | – | ↑ GAS5: ↓ proliferation, and ↓ invasion, ↑ apoptosis and ↑ G1 phase arrest | [121] |
| DBH-AS1 | ↑ | DLBCL | 26 patients with DLBCL |
RCK‐8, OCI‐LY‐3, OCI‐LLY‐7, and OCI‐LY‐ 10 human DLBCL cell lines and IM‐9 human peripheral blood B‐lymphocyte |
BUD13, FN1 | – |
∆ DBH-AS1: ↓ proliferation, ↓ invasion, and ↓ migration DBH-AS1 regulated FN1 expression by recruiting BUD13 |
[122] |
| ROR1-AS1 | ↑ | MCL | 5 patients with MCL and 5 healthy controls | Mino, Granta, JVM2 and Z138 MCL cell lines, HEK-293 T cell line |
↓ P16, and SOX11 EZH2 and SUZ12 of polycomb repressive complex-2 |
– |
↑ ROR1-AS1: ↑ cell growth and ↓ sensitivity to the treatment with drugs ibrutinib and dexamethasone ROR1-AS1 is involved in epigenetic regulation of gene transcription through EZH2/PRC2 complex |
[123] |
| LHFPL3-AS1 | ↑ | Melanoma | 461 tumor tissues and 558 normal tissues, BALB/c nude mice | Melanoma stem cells and non-stem cells from MDA-MB-435 cells | ↑ PTBP1, ↓ miR-181a-5p, ↑ Bcl-2 | – | ∆ LHFPL3-AS1: ↓ proliferation, ↑ apoptosis of melanoma stem cells | [124] |
| NONHSAG026900 | ↓ | DLBCL |
GEO dataset GSE12453 including 11 patients with DLBCL and 25 healthy controls and |
– | – | – | ↓ proliferation and cell cycle progression | [125] |
| SNHG16 | ↑ | DLBCL | DLBCL tissues (21 GCB and 27 non‐GCB) and 14 RLH tissues as controls, male NOD/SCID mice | OCI‐LY7 and OCI‐LY3 | ↓ miR-497-5p, ↑ PIM1 | – | ∆ SNHG16: ↓ proliferation, growth, and cell cycle progression, ↑ apoptosis | [126] |
| NEAT1–1 | ↑ in DLBCL tissues | DLBCL | 64 patients with DLBCL and 15 patients with lymphnoditis | OCI-Ly1 and SUDHL-4 DLBCL cell lines | – | – |
∆ NEAT1_1: ↓ viability and migration, ↑ apoptosis High levels of NEAT1_1 were correlated with stage, IPI, extranodal site involvement and drug response |
[127] |
| TUC338 | ↑ | DLBCL | 102 pairs of DLBCL and normal tissues, serum specimens of 35 patients with DLBCL and 35 healthy controls, BALB/c nude mice | U2932 and OCI-Ly3 DLBCL cell lines | ↓ miR-28-5p, ↑ EGFR | ↑ PI3K/AKT signaling, |
∆ TUC338: ↓ proliferation and chemotherapy resistance to Adriamycin, ↑ apoptosis Patients with high TUC338 showed shorter survival time |
[128] |
| LINC00857 | ↑ | DLBCL | 87 pairs of DLBCL tissues and ANCTs |
HMy2.CIR lymphoblast cell line, SU-DHL-6, SU-DHL-4 and SU-DHL-10 DLBCL cell lines |
↓ miR-370-3p, ↑ CBX3 | – |
↑ LINC00857: ↑ proliferation and cycle progression, ↓ apoptosis ∆ TUC338: ↓ proliferation and, ↑ apoptosis |
[129] |
| Lnc-IRF2-3 and Lnc-ZNF667-AS1 | ↑ | B-CLL | 135 patients with B-CLL and 30 healthy controls | – | – | – |
Patients with high levels of Lnc-IRF2-3 had a significant decrease in OS and PFS High levels of Lnc-IRF2-3 and Lnc-ZNF667-AS1 were associated with poor survival |
[130] |
| LINC00963 | ↓ | DLBCL | GTEx and TCGA databases (normal N = 337, tumor T = 48), nude mice | SUDHL4, OCI-Ly1, HBL1 and OCI-Ly3 DLBCL cell lines and GM12878 Non-cancerous human B lymphocytes | ↑ miR-320a, ↓ XBP1 | – | ↑ LINC00963: ↓ proliferation, and tumor growth, ↑ apoptosis and autophagy | [131] |
| LEF1-AS1 | ↑ | CLL | – | primary CLL cells and normal B cells | ↑ LEF1 | – | ↑ LEF1-AS1: ↑ proliferation and ↓ apoptosis | [132] |
| PVT1 | ↑ | MM | 137 patients with MM and 62 patients with MGUS, and 21 control patients with lymphoma | KMS11, KMS12PE, KMS12BM, KMS26, KMM1, OPM2, RPMI8226 | ↑ MYC, BRD4 | – |
High levels of PVT1 were positively correlated with disease progression JQ1 (BRD4 inhibitor): ↓ proliferation and ↓ expression levels of MYC and PVT1 |
[133] |
| BALR-2 | ↑ | B-ALL | 160 patients with B-ALL | RS4;11 and MV4;11, Reh, 697, Nalm-6, and 70Z/3 murine pre-B-cell leukemic cell line, and HEK 293 T cell line | – | Glucocorticoid response pathway |
∆ BALR-2: ↓ proliferation, ↑ apoptosis and sensitivity to prednisolone treatment prednisolone treatment: ↓ BALR-2 expression |
[134] |
| FAS-AS1 | ↓ | Lymphoma | – | Granta-519 cells and Peripheral blood B-lymphocytes from healthy donors’ blood |
↑ sFas, RBM5, ↑ EZH2, |
– |
FAS-AS1 could regulate alternative splicing of Fas in lymphomas Expression of FAS-AS1 could repress by EZH2 |
[135] |
| LUNAR1 | ↑ | DLBCL |
87 patients with DLBCL and 28 samples with reactive lymph nodes as controls |
OCI-LY-3, OCI-LY-7, OCI-LY-10, SU-DHL-4, SU-DHL-6 and RCK-8 DLBCL cell lines |
– | – |
∆ LUNAR1: ↓ proliferation LUNAR1 expression was found to serve as an independent predictor for OS and PFS |
[136] |
| HOTAIR | ↑ | DLBCL | 50 lymph node samples from patients with DLBCL and 20 samples with reactive lymph nodes as controls | RCK-8, OCL-LY-10, OCL-LY-7, SU-DHL-6 and SU-DHL-4 DLBCL cell lines | – | PI3K/AKT/NF-κB signaling pathway | ∆ HOTAIR: ↓ growth, cell cycle progression, ↑ apoptosis | [137] |
| RP11-530C5.1 | ↑ | MS | GEO database and GSE21942, 50 MS patients and 25 controls | – | PAWR | – | – | [96] |
| AL928742.12 | ↓ | MS | GEO database and GSE21942, 50 patients with MS and 25 controls | – | IGHA2 | – | – | |
| PEG10 | ↑ | DLBCL | 107 patients with DLBCL and 46 samples with reactive lymph nodes as controls |
OCI-LY-3, OCI-LY-7, OCI-LY-10, RCK-8, SU-DHL-4 and SU-DHL-6 DLBCL cell lines |
– | – |
∆ PEG10: ↓ growth, ↑ apoptosis PEG10 levels were significantly associated with B symptoms, IPI score, CHOP-like treatment and rituximab |
[138] |
| HULC | ↑ | DLBCL |
142 patients with DLBCL and 60 samples with reactive lymph nodes as controls |
OCI–LY–3, OCI–LY–7, OCI–LY–10, SU–DHL–4, SU–DHL–6 and RCK–8 human DLBCL cell lines |
– | – |
∆ HULC: ↓ proliferation, ↑ apoptosis HULC was strongly associated with Ann Arbor stages, B symptoms, CHOP-like treatment, rituximab and IPI |
[139] |
| lincRNA-p21 | ↓ | DLBCL | 105 patients with DLBCL |
SU-DHL-2, OCI-LY-3, OCI- LY-10, SU-DHL-4 and OCI-LY-7 human DLBCL cell lines |
– | – |
↑ lincRNA-p21: ↓ proliferation and cycle progression Patients with high expression levels of lincRNA-p21 showed a favorable OS and PFS |
[140] |
| Murine studies | ||||||||
| BALR-6 | ↑ | B-ALL | Post bone marrow transplant, blood, bone marrow, thymus and spleen were collected from the mice | RS4;11 and MV, Reh, 697, Nalm-6, 70Z/3 murine pre B-cell leukemic cell line, and the HEK 293 T cell line | SP1, CREB1 | – |
∆ BALR-6: ↓ proliferation, ↑ apoptosis ↑ BALR-6: ↑ survival, proliferation and expansion of hematopoietic progenitor populations in vivo |
[141] |
| RP11-301G19.1 | ↑ | MM | Female BALB/c-nude mice |
U266, RPMI8226, OPM-2, MM-1S, NCI- H929 MM cell lines and 293 T normal plasma cells |
↓ miR-582-5p, ↑ HMGB2 | PI3K/AKT signaling pathway | ∆ RP11-301G19.1: ↓ proliferation and cell cycle progression, ↑ apoptosis | [142] |
| HULC | ↑ | DLBCL | Male BALB/C mice | SU-DHL-8, SU-DHL-10 human DLBCL cell lines | β-elemene | – | ↑ HULC: ↓ apoptosis | [143] |
| lnc00492 | ↑ | – |
lnc00492−/− and lnc00492 + / + mice |
B220 + B cells, MZ B-cells | ↓ CTBP1 | Notch2 signaling pathway | Lnc00492 is necessary for marginal zone B-cell development | [144] |
| MALAT-1 | ↑ | DLBCL | Female BALB/c-nu/nu nude mice | IM-9 cells, B lymphocytes IM-9I from healthy people and Farage, Pfeiffer, Raji, Daud, Ly1, Ly3, Ly8, and Ly10 from patients with DLBCL | ↓ LC3-II/LC3-I, ↑ p62 | – | ∆ U MALAT-1: ↓ migration, survival rate, the proportion of cells in S and G2/M phase, and tumor volume and weight, ↑ the proportion of cells in G0/G1 phase | [145] |
| NEAT1 | ↑ | SLE | Lupus-prone MRL/lpr mice | PBMCs, B220 + B cells, G-MDSCs or M-MDSCs from MRL/lpr mice | BAFF | IFN-I signaling |
↑ promotion of G-MDSCs ∆ NEAT1: ↓ lupus symptoms and inhibits IFN-I signaling activation |
[146] |
Contribution of circRNAs in the regulation of B cell-related disorders
The impact of circRNAs on B cell functions has been mostly assessed in the context of DLBCL. For instance, circ_OTUD7A expression has been found to be increased in DLBCL. Its silencing has suppressed proliferation and metastasis of DLBCL, induce cell cycle arrest and enhance their apoptosis. Mechanistically, circ_OTUD7A acts as a sponge for miR-431-5p and miR-431-5p to further regulate expression of FOXP1 [147].
Another study has shown that up-regulation of circCFL1 in DLBCL cells leads to reduction of miR-107 levels and subsequent up-regulation of HMGB1 in these cells. Functional studies have revealed that circCFL1 could directly bind with miR-107 and release HMGB1 from inhibitory effects of this miRNA. Up-regulation of circCFL1 increases migration and proliferation of DLBCL cells [148].
Circ-APC is another circRNA which is produced from APC and suppress proliferation of DLBCL cells through decreasing activity of Wnt/β-catenin pathway. This effect is exerted through its interaction with TET1 and miR-888 [149].
The impact of circRNAs has also been investigated on progression of leukemia. For instance, circ_0132266 has been shown to be down-regulated in chronic lymphocytic leukemia. This down-regulation has lead to enhancement of viability of these cells via influencing activity of miR-337-3p/PML axis [150]. Table 3 shows the effects of circRNAs in the pathogenesis of B cell-related disorders.
Table 3.
CircRNAs and B cells functions
| circRNA | Expression pattern | Disease | Sample | Cell line | Interaction | Signaling pathway | Function | References |
|---|---|---|---|---|---|---|---|---|
| Human studies | ||||||||
| Circ_OTUD7A | ↑ | DLBCL | 50 pairs of DLBCL tissues and ANCTs | U2932, TMD8 and OCI-Ly3 LBCL cell lines and GM12878 normal human B lymphocytes | ↓ miR-431-5p, ↑ FOXP1 | – | ∆ Circ_OTUD7A: ↓ proliferation, metastasis, ↑ cell cycle arrest and apoptosis | [147] |
| circ-APC | ↓ | DLBCL | 80 pairs of DLBCL and para-cancerous tissues, plasma samples from 27 DLBCL patients and 16 healthy controls, nude mice | SUDHL-3, U2932, TMD8, OCI-Ly3 and L428 human DLBCL cell lines and GM12878 normal human B lymphocytes | miR-888, APC, DNA demethylase TET1 | Wnt/β-catenin signaling pathway | ↑ CircCFL1: ↓ proliferation and tumor growth | [149] |
| circBCL11B | ↑ | AML |
61 patients with AML and 16 healthy samples, GEO dataset: |
– | – | – | ∆ circBCL11B: ↓ proliferation, ↑ apoptosis | [151] |
| circCDYL | ↑ | MCL |
18 patients with MCL and 17 healthy controls |
HEK293T cells and Z138 human MCL cell line | five miRNAs (hsa-miR-129-5p, hsa-miR-3163, hsa-miR-4662a-5p, hsa-miR-101-3p, and hsa-miR-186-5p), three lncRNAs (MALAT1, NEAT1, and XIST), and five mRNAs (NOTCH1, FMR1, ABCB1, TWIST1, and VEGFA) | – | ∆ circCDYL: ↓ proliferation | [152] |
| circ_0132266 | ↓ | CLL |
30 patients with CLL and 30 healthy controls |
MEC-1, JVM-3 and HEK-293 T | ↑ miR-337-3p, ↓ PML | – | ↑ circ_0132266: ↓ proliferation | [150] |
| circ_0005774 | ↑ | AML | 20 patients with AML and 20 healthy controls | HL-60 and NB4 cells | ↓ miR-192-5p, ↑ ULK1 | – | ∆ circ_0005774: ↓ proliferation and viability, ↑ apoptosis | [153] |
| circ-Smad5 | ↓ | DLBCL | – | JB6 and 293 T cell lines | – | Wnt/β-catenin/Lef1 signaling pathway | ∆ circ-Smad5: ↑ cell cycle progression and activated Wnt/β-catenin/Lef1 signaling pathway | [154] |
| circ_0009910 | ↑ | AML | 35 patients with AML and 35 healthy controls | HL-60 and MOLM-13 | ↓ miR-5195-3p and ↑ GRB10 | – | ∆ circ_0009910: ↓ proliferation and cell cycle progression, ↑ apoptosis | [155] |
| circ-CBFB | ↑ | CLL |
47 patients with CLL and 21 healthy controls |
HEK293T and MEC-1 human CLL cell line |
↓ miR-607, ↑ FZD3 |
↑ Wnt/β-catenin pathway | ∆ circ-CBFB: ↓ proliferation and cell cycle progression, ↑ apoptosis | [156] |
| Murine studies | ||||||||
| CircCFL1 | ↑ | DLBCL | female BALB/c nude mice | OCI-Ly7 and OCI-Ly3 human DLBCL cell lines | ↓ miR-107, ↑HMGB1 | – | ↑ CircCFL1, ↑ proliferation, migration, tumor volume and weight | [148] |
ANCTs, adjacent non-cancerous tissues; AML, acute myeloid leukemia; HSPC, hematopoietic stem and progenitor cell; MCL, Mantle cell lymphoma; CLL, chronic lymphocytic leukemia
Discussion
Accumulating evidence suggest the role of non-coding RNAs in the development of normal B cells as well as lymphomagenesis. Since they are have a highly cell type specific signature, these transcripts have been suggested as potential biomarkers for diverse clinical situations [157].
LncRNAs particularly those related with p53 or MYC pathways have also applications as therapeutic targets [157]. These transcripts could act as sponges for miRNAs, thus influencing expressions of their target genes. SNHG14/miR-5590-3p, MALAT1/miR-195, CRNDE/miR-345-5p, NEAT1/miR-34b-5p, SMAD5-AS1/miR-135b-5p, PTTG3P/miR-383, SNHG8/ miR-335-5p, PCAT1/miR-508-3p, SBF2-AS1/miR-494-3p, SNHG14/ miR-152-3p and LINC00908/miR-671-5p are among lncRNA/miRNA axes which are involved in the regulation of B cells. Ras/ERK, Wnt/β-catenin pathway, NF-κB/ERK, PI3K/mTOR, BCR, TNF, IL-11/STAT3, IFN-I, Notch2, MAPK/ERK, PI3K/AKT and glucocorticoid response pathways are among pathways that are regulated by lncRNAs in this context.
CircRNAs that regulate function of B cells are mostly associated with Wnt/β-catenin signaling pathway. They can also serve as sponges for miRNA. For instance, circ_OTUD7A/miR-431-5p, circCFL1/miR-107, circ-APC/miR-888, circ_0132266/miR-337-3p, circ_0005774/miR-192-5p, circ_0009910/miR-5195-3p and circ-CBFB/miR-607 are among important circRNA/miRNA axes in regulation of proliferation of B cells.
Finally, miRNAs that are involved in the pathogenesis of B cell-related disorders can modulate NF-κB, TGF-β, BCR, TAK1/IKKα-IKKβ/IκBα and MAPK/p65 signaling pathways.
Cumulatively, different classes of non-coding RNAs interact with each other to modulate function of B cells. Notably, non-coding RNAs have also interactions with immune check point proteins in the context of B cell disorders.
Conclusion
The observed interaction between non-coding RNAs and immune check point proteins suggests the importance of these transcripts as targets for immunotherapeutic approaches. Moreover, several lncRNAs, circRNAs and miRNAs have been found to affect proliferation of B cells, thus being involved in the pathogenesis of B cell-related disorders, particularly malignant disorders. The observed correlations between expression levels of these transcripts and clinic-pathological parameters further emphasize their role in the carcinogenic processes.
Understanding the impact of non-coding RNAs in B cell-related malignancies would provide new avenues for targeted therapies.
Acknowledgements
This study was financially supported by Grant from Medical School of Shahid Beheshti University of Medical Sciences.
Authors' contributions
SGF wrote the manuscript and revised it. MT and EJ supervised and designed the study. TK and BMH collected the data and designed the figures and tables. All authors read and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent forms were obtained from all study participants. The study protocol was approved by the ethical committee of Shahid Beheshti University of Medical Sciences. All methods were performed in accordance with the relevant guidelines and regulations.
Consent of publication
Not applicable.
Competing interests
The authors declare they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mohammad Taheri, Email: Mohammad.Taheri@uni-jena.de.
Elena Jamali, Email: Elena.jamali@yahoo.com.
References
- 1.Hoffman W, Lakkis FG, Chalasani G. B Cells, Antibodies, and More. Clin J Am Soc Nephrol. 2016;11(1):137–54. doi: 10.2215/CJN.09430915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vale AM, Schroeder HW., Jr Clinical consequences of defects in B-cell development. J Allergy Clin Immunol. 2010;125(4):778–87. doi: 10.1016/j.jaci.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wuttke NJ, Macardle PJ, Zola H. Blood group antibodies are made by CD5+ and by CD5− B cells. Immunol Cell Biol. 1997;75(5):478–483. doi: 10.1038/icb.1997.74. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki K, Maruya M, Kawamoto S, Fagarasan S. Roles of B-1 and B-2 cells in innate and acquired IgA-mediated immunity. Immunol Rev. 2010;237(1):180–190. doi: 10.1111/j.1600-065X.2010.00941.x. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, Wang W, Zhu W, Dong J, Cheng Y, Yin Z, et al. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int J Mol Sci. 2019;20(22):5573. doi: 10.3390/ijms20225573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu K-S, Pan F, Mao X-D, Liu C, Chen Y-J. Biological functions of circular RNAs and their roles in occurrence of reproduction and gynecological diseases. Am J Transl Res. 2019;11(1):1–15. [PMC free article] [PubMed] [Google Scholar]
- 7.Macfarlane L-A, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genomics. 2010;11(7):537–61. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brazão TF, Johnson JS, Müller J, Heger A, Ponting CP, Tybulewicz VLJ. Long noncoding RNAs in B-cell development and activation. Blood. 2016;128(7):e10–e9. doi: 10.1182/blood-2015-11-680843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang M, Chen H, Qiu J, Yang H-X, Zhang C-Y, Fei Y-Y, et al. Antagonizing miR-7 suppresses B cell hyperresponsiveness and inhibits lupus development. J Autoimmunity. 2020;109:102440. doi: 10.1016/j.jaut.2020.102440. [DOI] [PubMed] [Google Scholar]
- 10.Tan LP, Wang M, Robertus J-L, Schakel RN, Gibcus JH, Diepstra A, et al. miRNA profiling of B-cell subsets: specific miRNA profile for germinal center B cells with variation between centroblasts and centrocytes. Lab Invest. 2009;89(6):708–716. doi: 10.1038/labinvest.2009.26. [DOI] [PubMed] [Google Scholar]
- 11.Tuncer SB, Akdeniz D, Celik B, Kilic S, Sukruoglu O, Avsar M, et al. The expression levels of miRNA-15a and miRNA-16-1 in circulating tumor cells of patients with diffuse large B-cell lymphoma. Mol Biol Rep. 2019;46(1):975–980. doi: 10.1007/s11033-018-4554-4. [DOI] [PubMed] [Google Scholar]
- 12.Yang JM, Jang J-Y, Jeon YK, Paik JH. Clinicopathologic implication of microRNA-197 in diffuse large B cell lymphoma. J Transl Med. 2018;16(1):1–14. doi: 10.1186/s12967-018-1537-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang Y, Zhou H, Ma D, Chen ZK, Cai X. MicroRNA-19a and CD22 comprise a feedback loop for B cell response in sepsis. Med Sci Monitor. 2015;21:1548. doi: 10.12659/MSM.894321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Dong J, Mu R, Gao Y, Tan X, Li Y, et al. MicroRNA-30a promotes B cell hyperactivity in patients with systemic lupus erythematosus by direct interaction with Lyn. Arthritis Rheum. 2013;65(6):1603–1611. doi: 10.1002/art.37912. [DOI] [PubMed] [Google Scholar]
- 15.Alivernini S, Kurowska-Stolarska M, Tolusso B, Benvenuto R, Elmesmari A, Canestri S, et al. MicroRNA-155 influences B-cell function through PU1 in rheumatoid arthritis. Nat Commun. 2016;7(1):1–12. doi: 10.1038/ncomms12970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bedewy AM, Elmaghraby SM, Shehata AA, Kandil NS. Prognostic value of miRNA-155 expression in B-cell non-Hodgkin lymphoma. Turkish J Hematol. 2017;34(3):207. doi: 10.4274/tjh.2016.0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun W, Zhang L, Lin L, Wang W, Ge Y, Liu Y, et al. Chronic psychological stress impairs germinal center response by repressing miR-155. Brain Behav Immun. 2019;76:48–60. doi: 10.1016/j.bbi.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Mo X, Wei F, Tong Y, Ding L, Zhu Q, Du S, et al. Lactic acid downregulates viral microRNA to promote Epstein-Barr Virus-immortalized B lymphoblastic cell adhesion and growth. J Virol. 2018;92(9):e00033–e118. doi: 10.1128/JVI.00033-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider C, Setty M, Holmes AB, Maute RL, Leslie CS, Mussolin L, et al. MicroRNA 28 controls cell proliferation and is down-regulated in B-cell lymphomas. Proc Natl Acad Sci. 2014;111(22):8185–8190. doi: 10.1073/pnas.1322466111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris-Arnold A, Arnold C, Schaffert S, Hatton O, Krams S, Esquivel C, et al. Epstein-Barr virus modulates host cell microrna-194 to promote il-10 production and B lymphoma cell survival. Am J Transplant. 2015;15(11):2814–2824. doi: 10.1111/ajt.13375. [DOI] [PubMed] [Google Scholar]
- 21.Gururajan M, Haga CL, Das S, Leu C-M, Hodson D, Josson S, et al. MicroRNA 125b inhibition of B cell differentiation in germinal centers. Int Immunol. 2010;22(7):583–592. doi: 10.1093/intimm/dxq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S-H, Wang P-P, Cun-te-Chen DL, Liu Q-Y, Lv L, Liu X, et al. MicroRNA-148b enhances the radiosensitivity of B-cell lymphoma cells by targeting Bcl-w to promote apoptosis. Int J Biol Sci. 2020;16(6):935. doi: 10.7150/ijbs.40756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shim H, Nam J, Kim S-W. NF-κB p65 represses microRNA-124 transcription in diffuse large B-cell lymphoma. Genes Genom. 2020;42(5):543–551. doi: 10.1007/s13258-020-00922-y. [DOI] [PubMed] [Google Scholar]
- 24.Yan S, Jia C, Quan L, Zhao L, Tian Y, Liu A. Significance of the microRNA-17-92 gene cluster expressed in B-cell non-Hodgkin's lymphoma. Mol Med Rep. 2019;20(3):2459–2467. doi: 10.3892/mmr.2019.10448. [DOI] [PubMed] [Google Scholar]
- 25.Jabłońska E, Białopiotrowicz E, Szydłowski M, Prochorec-Sobieszek M, Juszczyński P, Szumera-Ciećkiewicz A. DEPTOR is a microRNA-155 target regulating migration and cytokine production in diffuse large B-cell lymphoma cells. Exp Hematol. 2020;88:56–67. doi: 10.1016/j.exphem.2020.07.005. [DOI] [PubMed] [Google Scholar]
- 26.Su H, Chang J, Xu M, Sun R, Wang J. CDK6 overexpression resulted from microRNA-320d downregulation promotes cell proliferation in diffuse large B-cell lymphoma. Oncol Rep. 2019;42(1):321–327. doi: 10.3892/or.2019.7144. [DOI] [PubMed] [Google Scholar]
- 27.Liu R-D, Zhuang W, Qi J-D, Li C-C. Expression and clinical significance of microRNA-195 in patients with diffuse large b cell lymphoma. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2020;28(1):160–164. doi: 10.19746/j.cnki.issn.1009-2137.2020.01.027. [DOI] [PubMed] [Google Scholar]
- 28.Due H, Schönherz AA, Ryø L, Primo MN, Jespersen DS, Thomsen EA, et al. MicroRNA-155 controls vincristine sensitivity and predicts superior clinical outcome in diffuse large B-cell lymphoma. Blood Adv. 2019;3(7):1185–1196. doi: 10.1182/bloodadvances.2018029660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y-L, Tang J-M, Chen X-Y, Luo B, Liang G-H, Qu Q, et al. MicroRNA-153-3p enhances the sensitivity of chronic myeloid leukemia cells to imatinib by inhibiting B-cell lymphoma-2-mediated autophagy. Hum Cell. 2020;33(3):610–618. doi: 10.1007/s13577-020-00367-1. [DOI] [PubMed] [Google Scholar]
- 30.Baraniskin A, Chomiak M, Ahle G, Gress T, Buchholz M, Turewicz M, et al. MicroRNA-30c as a novel diagnostic biomarker for primary and secondary B-cell lymphoma of the CNS. J Neurooncol. 2018;137(3):463–468. doi: 10.1007/s11060-018-2749-0. [DOI] [PubMed] [Google Scholar]
- 31.Meng Y, Quan L, Liu A. Identification of key microRNAs associated with diffuse large B-cell lymphoma by analyzing serum microRNA expressions. Gene. 2018;642:205–211. doi: 10.1016/j.gene.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 32.Wood CD, Carvell T, Gunnell A, Ojeniyi OO, Osborne C, West MJ. Enhancer control of microRNA miR-155 expression in Epstein-Barr virus-infected B cells. J Virol. 2018;92(19):e00716–e718. doi: 10.1128/JVI.00716-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian L, Cao J, Ji Q, Zhang C, Qian T, Song X, et al. The downregulation of miR-3173 in B-cell acute lymphoblastic leukaemia promotes cell invasion via PTK2. Biochem Biophys Res Commun. 2017;494(3–4):569–574. doi: 10.1016/j.bbrc.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 34.Labib HA, Elantouny NG, Ibrahim NF, Alnagar AA. Upregulation of microRNA-21 is a poor prognostic marker in patients with childhood B cell acute lymphoblastic leukemia. Hematology. 2017;22(7):392–397. doi: 10.1080/10245332.2017.1292204. [DOI] [PubMed] [Google Scholar]
- 35.Song J, Shao Q, Li C, Liu H, Li J, Wang Y, et al. Effects of microRNA-21 on apoptosis by regulating the expression of PTEN in diffuse large B-cell lymphoma. Medicine. 2017;96:39. doi: 10.1097/MD.0000000000007952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun S, Wang H, Ji M. Overexpression of miR-222-3p promotes the proliferation and inhibits the apoptosis of diffuse large B-cell lymphoma cells via suppressing PPP2R2A. Technol Cancer Res Treat. 2019;18:1533033819892256. doi: 10.1177/1533033819892256. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Shi X, Ye L, Xu S, Guo G, Zuo Z, Ye M, et al. Downregulated miR-29a promotes B cell overactivation by upregulating Crk-like protein in systemic lupus erythematosus. Mol Med Rep. 2020;22(2):841–849. doi: 10.3892/mmr.2020.11166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, Wen X, Zhou J, Qi Y, Wu R, Wang Y, et al. MicroRNA-223 and microRNA-21 in peripheral blood B cells associated with progression of primary biliary cholangitis patients. PLoS ONE. 2017;12(9):e0184292. doi: 10.1371/journal.pone.0184292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karkhanis V, Alinari L, Ozer HG, Chung J, Zhang X, Sif S, et al. Protein arginine methyltransferase 5 represses tumor suppressor miRNAs that down-regulate CYCLIN D1 and c-MYC expression in aggressive B-cell lymphoma. J Biol Chem. 2020;295(5):1165–1180. doi: 10.1074/jbc.RA119.008742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun J-R, Zhang X, Zhang Y. MiR-214 prevents the progression of diffuse large B-cell lymphoma by targeting PD-L1. Cell Mol Biol Lett. 2019;24(1):1–13. doi: 10.1186/s11658-019-0190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z-W, Wang M, Hu J-J, Xu G, Zhang Y, Zhang N. Decreased expression of microRNA-107 in B lymphocytes of patients with antibody-mediated renal allograft rejection. Tohoku J Exp Med. 2018;246(2):87–96. doi: 10.1620/tjem.246.87. [DOI] [PubMed] [Google Scholar]
- 42.Romero M, Gapihan G, Castro-Vega LJ, Acevedo A, Wang L, Li ZW, et al. Primary mediastinal large B-cell lymphoma: Transcriptional regulation by miR-92a through FOXP1 targeting. Oncotarget. 2017;8(10):16243. doi: 10.18632/oncotarget.12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L, Zhan C-Z, Wang T, You H, Yao R. Curcumin inhibits the proliferation, migration, invasion, and apoptosis of diffuse large B-cell lymphoma cell line by regulating MiR-21/VHL axis. Yonsei Med J. 2020;61(1):20–29. doi: 10.3349/ymj.2020.61.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han B, Gao Z-D, Wang H-X, Wang Z-H, Fan C-B, Liu J-L, et al. Expression of MiR-155 in tissue of patients with diffuse large B-cell lymphoma and its effect on cell biological characteristics. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2019;27(2):445–451. doi: 10.19746/j.cnki.issn.1009-2137.2019.02.022. [DOI] [PubMed] [Google Scholar]
- 45.Wu R-J, Zou Y, Ma X-D, Zheng R-J. Expression and clinical significance of MiR-215 and KDM1B in patients with diffuse large B cell lymphoma. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2020;28(5):1570–1577. doi: 10.19746/j.cnki.issn.1009-2137.2020.05.024. [DOI] [PubMed] [Google Scholar]
- 46.Farroni C, Marasco E, Marcellini V, Giorda E, Valentini D, Petrini S, et al. Dysregulated miR-155 and miR-125b are related to impaired B-cell responses in Down syndrome. Front Immunol. 2018;9:2683. doi: 10.3389/fimmu.2018.02683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L, Xu J, Chu X, Ju C. MicroRNA-98 interferes with thrombospondin 1 expression in peripheral B cells of patients with asthma. Biosci Rep. 2017;37(4):BSR20170149. doi: 10.1042/BSR20170149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang T, Sun W-L, Lu X-F, Wang X-L, Jiang L. MiR-28-5p mediates the anti-proliferative and pro-apoptotic effects of curcumin on human diffuse large B-cell lymphoma cells. J Int Med Res. 2020;48(7):0300060520943792. doi: 10.1177/0300060520943792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asker HA, Khorshed EN, Ahmed MR, Refaat LA, Khaled HM, Rashed RA. Prognostic Values of MicroRNA-21 and Ki-67 in diffuse large B-cell lymphoma patients: Egyptian experience. Clin Lab. 2021;67:7. doi: 10.7754/Clin.Lab.2021.201132. [DOI] [PubMed] [Google Scholar]
- 50.Fan Q, Meng X, Liang H, Zhang H, Liu X, Li L, et al. miR-10a inhibits cell proliferation and promotes cell apoptosis by targeting BCL6 in diffuse large B-cell lymphoma. Protein Cell. 2016;7(12):899–912. doi: 10.1007/s13238-016-0316-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen M, Wang Y, Cui S, Wu X, Guo Y, Xu R. MicroRNA-125a regulates proliferation and apoptosis of acute myeloid leukemia through targeting NF-κB pathway. Eur Rev Med Pharmacol Sci. 2019;23:3594–3601. doi: 10.26355/eurrev_201905_17781. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y, Jia X, Zhou L, Yin J, Zhao Y, Dai L, et al. Increased let-7b-5p is associated with enhanced BAFF-R expression and B cell survival in immune thrombocytopenia. Int Immunopharmacol. 2021;93:107393. doi: 10.1016/j.intimp.2021.107393. [DOI] [PubMed] [Google Scholar]
- 53.Luo Y, Yang J, Zhang C, Jin Y, Pan H, Liu L, et al. Up-regulation of miR-27a promotes monocyte-mediated inflammatory responses in Kawasaki disease by inhibiting function of B10 cells. J Leukoc Biol. 2020;107(1):133–144. doi: 10.1002/JLB.5A0919-075RR. [DOI] [PubMed] [Google Scholar]
- 54.Benhamou D, Labi V, Getahun A, Benchetrit E, Dowery R, Rajewsky K, et al. The c-Myc/miR17-92/PTEN axis tunes PI3K activity to control expression of recombination activating genes in early B cell development. Front Immunol. 2018;9:2715. doi: 10.3389/fimmu.2018.02715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang S, Wang L, Cheng L. Aberrant ERG expression associates with downregulation of miR-4638-5p and selected genomic alterations in a subset of diffuse large B-cell lymphoma. Mol Carcinog. 2019;58(10):1846–1854. doi: 10.1002/mc.23074. [DOI] [PubMed] [Google Scholar]
- 56.Huang Q, Zhang F, Fu H, Shen J. Epigenetic regulation of miR-518a-5p-CCR6 feedback loop promotes both proliferation and invasion in diffuse large B cell lymphoma. Epigenetics. 2021;16(1):28–44. doi: 10.1080/15592294.2020.1786317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X-S, Wang X-C, Ding K-Y, Wu Q. Expression of MiR-296-5p in diffuse large B-Cell lymphoma and its influence on biological behavior of tumor cells. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2018;26(2):437–442. doi: 10.7534/j.issn.1009-2137.2018.02.022. [DOI] [PubMed] [Google Scholar]
- 58.Liu Y-P, Hu H, Xu F, Wen J-J. Relation of miR-34a expression in diffuse large B cell lymphoma with clinical prognosis. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2017;25(2):455–459. doi: 10.7534/j.issn.1009-2137.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 59.Xie Y, Tan L, Li Y-T, Zeng Y. Mechanism of MiR-224 Affecting DLBCL Cell Proliferation and Invasion by Targeted Inhibition of PIK3CD. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2020;28(5):1578–1584. doi: 10.19746/j.cnki.issn.1009-2137.2020.05.025. [DOI] [PubMed] [Google Scholar]
- 60.Xiao X, Gu Y, Sun D, Ding L, Yuan X, Jiang H, et al. Effect of rituximab combined with chemotherapy on the expression of serum exosome miR-451a in patients with diffuse large b-cell lymphoma. Eur Rev Med Pharmacol Sci. 2019;23(4):1620–1625. doi: 10.26355/eurrev_201902_17121. [DOI] [PubMed] [Google Scholar]
- 61.Luo S, Ding S, Liao J, Zhang P, Liu Y, Zhao M, et al. Excessive miR-152-3p results in increased BAFF expression in SLE B-cells by inhibiting the KLF5 expression. Front Immunol. 2019;10:1127. doi: 10.3389/fimmu.2019.01127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bartolomé-Izquierdo N, de Yébenes VG, Alvarez-Prado AF, Mur SM, Olmo JA, Roa S, et al. miR-28 regulates the germinal center reaction and blocks tumor growth in preclinical models of non-Hodgkin lymphoma. Blood J Am Soc Hematol. 2017;129(17):2408–19. doi: 10.1182/blood-2016-08-731166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song J, Su W, Chen X, Zhao Q, Zhang N, Li M-G, et al. Micro RNA-98 suppresses interleukin-10 in peripheral B cells in patient post-cardio transplantation. Oncotarget. 2017;8(17):28237. doi: 10.18632/oncotarget.16000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan Y, Niu F, Nolte IM, Koerts J, de Jong D, Rutgers B, et al. MicroRNA high throughput loss-of-function screening reveals an oncogenic role for miR-21-5p in Hodgkin lymphoma. Cell Physiol Biochem. 2018;49(1):144–159. doi: 10.1159/000492850. [DOI] [PubMed] [Google Scholar]
- 65.van Nieuwenhuijze A, Dooley J, Humblet-Baron S, Sreenivasan J, Koenders M, Schlenner SM, et al. Defective germinal center B-cell response and reduced arthritic pathology in microRNA-29a-deficient mice. Cell Mol Life Sci. 2017;74(11):2095–2106. doi: 10.1007/s00018-017-2456-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Okuyama K, Ikawa T, Gentner B, Hozumi K, Harnprasopwat R, Lu J, et al. MicroRNA-126–mediated control of cell fate in B-cell myeloid progenitors as a potential alternative to transcriptional factors. Proc Natl Acad Sci. 2013;110(33):13410–13415. doi: 10.1073/pnas.1220710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mehta A, Mann M, Zhao JL, Marinov GK, Majumdar D, Garcia-Flores Y, et al. The microRNA-212/132 cluster regulates B cell development by targeting Sox4. J Exp Med. 2015;212(10):1679–1692. doi: 10.1084/jem.20150489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kurkewich JL, Bikorimana E, Nguyen T, Klopfenstein N, Zhang H, Hallas WM, et al. The mirn23a microRNA cluster antagonizes B cell development. J Leukoc Biol. 2016;100(4):665–677. doi: 10.1189/jlb.1HI0915-398RR. [DOI] [PubMed] [Google Scholar]
- 69.Gonzalez-Martin A, Adams BD, Lai M, Shepherd J, Salvador-Bernaldez M, Salvador JM, et al. The microRNA miR-148a functions as a critical regulator of B cell tolerance and autoimmunity. Nat Immunol. 2016;17(4):433–440. doi: 10.1038/ni.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu Y, Schutt S, Paz K, Zhang M, Flynn RP, Bastian D, et al. MicroRNA-17-92 is required for T-cell and B-cell pathogenicity in chronic graft-versus-host disease in mice. Blood. 2018;131(17):1974–1986. doi: 10.1182/blood-2017-06-789321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li G, Soll AY-L, Sookram R, Wong S, Wang JK, Ouyang Y, et al. Epigenetic silencing of miR-125b is required for normal B-cell development. Blood. 2018;131(17):1920–30. doi: 10.1182/blood-2018-01-824540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Farina FM, Inguscio A, Kunderfranco P, Cortesi A, Elia L, Quintavalle M. MicroRNA-26a/cyclin-dependent kinase 5 axis controls proliferation, apoptosis and in vivo tumor growth of diffuse large B-cell lymphoma cell lines. Cell Death Dis. 2017;8(6):e2890. doi: 10.1038/cddis.2017.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arbore G, Henley T, Biggins L, Andrews S, Vigorito E, Turner M, et al. MicroRNA-155 is essential for the optimal proliferation and survival of plasmablast B cells. Life Sci Alliance. 2019;2:3. doi: 10.26508/lsa.201800244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Glaesener S, Jaenke C, Habener A, Geffers R, Hagendorff P, Witzlau K, et al. Decreased production of class-switched antibodies in neonatal B cells is associated with increased expression of miR-181b. PLoS ONE. 2018;13(2):e0192230. doi: 10.1371/journal.pone.0192230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Petkau G, Kawano Y, Wolf I, Knoll M, Melchers F. MiR221 promotes precursor B-cell retention in the bone marrow by amplifying the PI3K-signaling pathway in mice. Eur J Immunol. 2018;48(6):975–989. doi: 10.1002/eji.201747354. [DOI] [PubMed] [Google Scholar]
- 76.Wang W, Wang J, Yan M, Jiang J, Bian A. MiRNA-92a protects pancreatic B-cell function by targeting KLF2 in diabetes mellitus. Biochem Biophys Res Commun. 2018;500(3):577–582. doi: 10.1016/j.bbrc.2018.04.097. [DOI] [PubMed] [Google Scholar]
- 77.Sewastianik T, Straubhaar JR, Zhao J-J, Samur MK, Adler K, Tanton HE, et al. miR-15a/16-1 deletion in activated B cells promotes plasma cell and mature B-cell neoplasms. Blood. 2021;137(14):1905–1919. doi: 10.1182/blood.2020009088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cho S, Lee H-M, Yu I-S, Choi YS, Huang H-Y, Hashemifar SS, et al. Differential cell-intrinsic regulations of germinal center B and T cells by miR-146a and miR-146b. Nat Commun. 2018;9(1):1–13. doi: 10.1038/s41467-018-05196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Contreras JR, Palanichamy JK, Tran TM, Fernando TR, Rodriguez-Malave NI, Goswami N, et al. MicroRNA-146a modulates B-cell oncogenesis by regulating Egr1. Oncotarget. 2015;6(13):11023. doi: 10.18632/oncotarget.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li S, He X, Gan Y, Zhang J, Gao F, Lin L, et al. Targeting miR-21 with NL101 blocks c-Myc/Mxd1 loop and inhibits the growth of B cell lymphoma. Theranostics. 2021;11(7):3439. doi: 10.7150/thno.53561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Amrouche L, You S, Sauvaget V, Manda V, Lamarthée B, Desbuissons G, et al. MicroRNA-146a-deficient mice develop immune complex glomerulonephritis. Sci Rep. 2019;9(1):1–13. doi: 10.1038/s41598-019-51985-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li F, Huang Y, Huang Y-Y, Kuang Y-S, Wei Y-J, Xiang L, et al. MicroRNA-146a promotes IgE class switch in B cells via upregulating 14-3-3σ expression. Mol Immunol. 2017;92:180–189. doi: 10.1016/j.molimm.2017.10.023. [DOI] [PubMed] [Google Scholar]
- 83.Kuriyama K, Enomoto Y, Suzuki R, Watanuki J, Hosoi H, Yamashita Y, et al. Enforced expression of MIR142, a target of chromosome translocation in human B-cell tumors, results in B-cell depletion. Int J Hematol. 2018;107(3):345–354. doi: 10.1007/s12185-017-2360-8. [DOI] [PubMed] [Google Scholar]
- 84.Chen X, Dong S, Zhang N, Chen L, Li M-G, Yang P-C, et al. MicroRNA-98 plays a critical role in experimental myocarditis. Int J Cardiol. 2017;229:75–81. doi: 10.1016/j.ijcard.2016.11.263. [DOI] [PubMed] [Google Scholar]
- 85.Jiang S, Yan W, Wang SE, Baltimore D. Let-7 suppresses B cell activation through restricting the availability of necessary nutrients. Cell Metab. 2018;27(2):393–403. doi: 10.1016/j.cmet.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 86.Xiao S, Zhang W, Manley NR. Thymic epithelial cell-derived signals control B progenitor formation and proliferation in the thymus by regulating Let-7 and Arid3a. PLoS ONE. 2018;13(2):e0193188. doi: 10.1371/journal.pone.0193188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blume J, Ziętara N, Witzlau K, Liu Y, Sanchez OO, Puchałka J, et al. miR-191 modulates B-cell development and targets transcription factors E2A, Foxp1, and Egr1. Eur J Immunol. 2019;49(1):121–132. doi: 10.1002/eji.201847660. [DOI] [PubMed] [Google Scholar]
- 88.Lindner SE, Lohmüller M, Kotkamp B, Schuler F, Knust Z, Villunger A, et al. The miR-15 family reinforces the transition from proliferation to differentiation in pre-B cells. EMBO Rep. 2017;18(9):1604–1617. doi: 10.15252/embr.201643735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kurkewich JL, Hansen J, Klopfenstein N, Zhang H, Wood C, Boucher A, et al. The miR-23a~ 27a–24–2 microRNA cluster buffers transcription and signaling pathways during hematopoiesis. PLoS Genetics. 2017;13(7):e1006887. doi: 10.1371/journal.pgen.1006887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao L, Liu Y, Zhang J, Liu Y, Qi Q. LncRNA SNHG14/miR-5590-3p/ZEB1 positive feedback loop promoted diffuse large B cell lymphoma progression and immune evasion through regulating PD-1/PD-L1 checkpoint. Cell Death Dis. 2019;10(10):1–15. doi: 10.1038/s41419-019-1886-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Q-M, Lian G-Y, Song Y, Huang Y-F, Gong Y. LncRNA MALAT1 promotes tumorigenesis and immune escape of diffuse large B cell lymphoma by sponging miR-195. Life Sci. 2019;231:116335. doi: 10.1016/j.lfs.2019.03.040. [DOI] [PubMed] [Google Scholar]
- 92.Qian C-S, Li L-J, Huang H-W, Yang H-F, Wu D-P. MYC-regulated lncRNA NEAT1 promotes B cell proliferation and lymphomagenesis via the miR-34b-5p-GLI1 pathway in diffuse large B-cell lymphoma. Cancer Cell Int. 2020;20(1):1–13. doi: 10.1186/s12935-020-1158-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang W, Wu F, Ma P, Gan S, Li X, Chen L, et al. LncRNA CRNDE promotes the progression of B-cell precursor acute lymphoblastic leukemia by targeting the miR-345-5p/CREB Axi. Mol Cells. 2020;43(8):718. doi: 10.14348/molcells.2020.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cuadros M, García DJ, Andrades A, Arenas AM, Coira IF, Baliñas-Gavira C, et al. LncRNA-mRNA co-expression analysis identifies AL1333461/CCN2 as biomarkers in pediatric B-cell acute lymphoblastic leukemia. Cancers. 2020;12(12):3803. doi: 10.3390/cancers12123803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bi M, Shi J, Zhao Y, Li C. LncRNA PTTG3P induced aberrant glycosylated IgA1 production and B cell growth in IgA nephropathy. Environ Sci Pollut Res. 2021;1:1–9. doi: 10.1007/s11356-021-13335-5. [DOI] [PubMed] [Google Scholar]
- 96.Ghoveud E, Teimuri S, Vatandoost J, Hosseini A, Ghaedi K, Etemadifar M, et al. Potential biomarker and therapeutic LncRNAs in multiple sclerosis through targeting memory B cells. NeuroMol Med. 2020;22(1):111–120. doi: 10.1007/s12017-019-08570-6. [DOI] [PubMed] [Google Scholar]
- 97.Zhao C-C, Jiao Y, Zhang Y-Y, Ning J, Zhang Y-R, Xu J, et al. Lnc SMAD5-AS1 as ceRNA inhibit proliferation of diffuse large B cell lymphoma via Wnt/β-catenin pathway by sponging miR-135b-5p to elevate expression of APC. Cell Death Dis. 2019;10(4):1–15. doi: 10.1038/s41419-019-1479-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Meng H, Zhao B, Wang Y. FOXM1-induced upregulation of lncRNA OR3A4 promotes the progression of diffuse large B-cell lymphoma via Wnt/β-catenin signaling pathway. Exp Mol Pathol. 2020;115:104451. doi: 10.1016/j.yexmp.2020.104451. [DOI] [PubMed] [Google Scholar]
- 99.Shi X, Cui Z, Liu X, Wu S, Wu Y, Fang F, et al. LncRNA FIRRE is activated by MYC and promotes the development of diffuse large B-cell lymphoma via Wnt/β-catenin signaling pathway. Biochem Biophys Res Commun. 2019;510(4):594–600. doi: 10.1016/j.bbrc.2019.01.105. [DOI] [PubMed] [Google Scholar]
- 100.Tang J-L, Li X-M, Zhang L. Expression and significance of lncRNA RP11–513G11.1 in peripheral blood of patients with diffuse large B-cell lymphoma. Zhongguo shi yan xue ye xue za zhi. 2019;27(5):1515–21. doi: 10.19746/j.cnki.issn.1009-2137.2019.05.024. [DOI] [PubMed] [Google Scholar]
- 101.Wang F, Luo Y, Zhang L, Younis M, Yuan L. Down-regulation of LncRNA 2900052N01Rik inhibits LPS-induced B cell function in vitro. Cell Immunol. 2021;363:104321. doi: 10.1016/j.cellimm.2021.104321. [DOI] [PubMed] [Google Scholar]
- 102.Li Q, Li B, Lu C-L, Wang J-Y, Gao M, Gao W. LncRNA LINC01857 promotes cell growth and diminishes apoptosis via PI3K/mTOR pathway and EMT process by regulating miR-141-3p/MAP4K4 axis in diffuse large B-cell lymphoma. Cancer Gene Ther. 2021;28(9):1046–1057. doi: 10.1038/s41417-020-00267-4. [DOI] [PubMed] [Google Scholar]
- 103.Orlandella FM, Smaldone G, Salvatore G, Vitagliano L, Cianflone A, Parasole R, et al. The lncRNA TEX41 is upregulated in pediatric B-cells acute lymphoblastic leukemia and it is necessary for leukemic cell growth. Biomarker Res. 2021;9(1):1–11. doi: 10.1186/s40364-021-00307-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gao H, Sun Y, Chen J, Jin H, Yang W. Long non-coding RNA AFAP1-AS1 promotes cell growth and inhibits apoptosis by binding to specific proteins in germinal center B-cell-like diffuse large B-cell lymphoma. Am J Transl Res. 2020;12(12):8225. [PMC free article] [PubMed] [Google Scholar]
- 105.Yu B, Wang B, Wu Z, Wu C, Ling J, Gao X, et al. LncRNA SNHG8 promotes proliferation and inhibits apoptosis of diffuse large B-cell lymphoma via sponging miR-335–5p. Front Oncol. 2021;11:1. doi: 10.3389/fonc.2021.650287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sheng Y, Fan C, Zhang W. LncRNA PCAT1 enhances cell proliferation, migration and invasion by miR-508-3p/NFIB axis in diffuse large B-cell lymphoma. Eur Rev Med Pharmacol Sci. 2021;25:2567–2576. doi: 10.26355/eurrev_202103_25420. [DOI] [PubMed] [Google Scholar]
- 107.Fu D-W, Liu A-C. LncRNA SBF2-AS1 promotes diffuse large B-cell lymphoma growth by regulating FGFR2 via sponging miR-494-3p. Cancer Manag Res. 2021;13:571. doi: 10.2147/CMAR.S284258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tian Y, Li L, Lin G, Wang Y, Wang L, Zhao Q, et al. lncRNA SNHG14 promotes oncogenesis and immune evasion in diffuse large-B-cell lymphoma by sequestering miR-152–3p. Leukemia Lymphoma. 2021;1:1–15. doi: 10.1080/10428194.2021.1876866. [DOI] [PubMed] [Google Scholar]
- 109.Zeng H, Wei Y, Wei X, Feng R. LINC00908 Promotes Diffuse Large B-Cell Lymphoma Development by Down-Regulating miR-671-5p. Cancer Manag Res. 2021;13:3589. doi: 10.2147/CMAR.S299715. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 110.Jiang X, Wang Y, Li X, He L, Yang Q, Wang W, et al. Microarray profile of B cells from Graves’ disease patients reveals biomarkers of proliferation. Endocr Connect. 2020;9(5):405–417. doi: 10.1530/EC-20-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang Q, Du X, Yang M, Xiao S, Cao J, Song J, et al. LncRNA ZEB1-AS1 contributes to STAT3 activation by associating with IL-11 in B-lymphoblastic leukemia. Biotech Lett. 2017;39(12):1801–1810. doi: 10.1007/s10529-017-2421-3. [DOI] [PubMed] [Google Scholar]
- 112.Zhang M, Du Y, Shang J, Zhang D, Dong X, Chen H. Knockdown of UCA1 restrains cell proliferation and metastasis of diffuse large B-cell lymphoma by counteracting miR-331-3p expression. Oncol Lett. 2021;21(1):1. doi: 10.3892/ol.2020.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang W-T, Chen T-Q, Zeng Z-C, Pan Q, Huang W, Han C, et al. The lncRNA LAMP5-AS1 drives leukemia cell stemness by directly modulating DOT1L methyltransferase activity in MLL leukemia. J Hematol Oncol. 2020;13(1):1–19. doi: 10.1186/s13045-020-00909-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mao Y, Tie Y, Du J, He J. LINC00152 promotes the proliferation of gastric cancer cells by regulating B-cell lymphoma-2. J Cell Biochem. 2019;120(3):3747–3756. doi: 10.1002/jcb.27655. [DOI] [PubMed] [Google Scholar]
- 115.Hu L, Zhao J, Liu Y, Liu X, Lu Q, Zeng Z, et al. Geniposide inhibits proliferation and induces apoptosis of diffuse large B-cell lymphoma cells by inactivating the HCP5/miR-27b-3p/MET axis. Int J Med Sci. 2020;17(17):2735. doi: 10.7150/ijms.51329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhao J, Su L, Jiang J. Long non-coding RNA paternally expressed imprinted gene 10 (PEG10) elevates diffuse large B-cell lymphoma progression by regulating kinesin family member 2A (KIF2A) via Targeting MiR-101-3p. Med Sci Monitor. 2020;26:e922810–e922811. doi: 10.12659/MSM.922810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Miao Y, Chen X, Qin M, Zhou W, Wang Y, Ji Y. lncRNA GAS5, as a ceRNA, inhibits the proliferation of diffuse large B-cell lymphoma cells by regulating the miR-18a-5p/RUNX1 axis. Int J Oncol. 2021;59(5):1–12. doi: 10.3892/ijo.2021.5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cheng H, Yan Z, Wang X, Cao J, Chen W, Qi K, et al. Downregulation of long non-coding RNA TUG1 suppresses tumor growth by promoting ubiquitination of MET in diffuse large B-cell lymphoma. Mol Cell Biochem. 2019;461(1):47–56. doi: 10.1007/s11010-019-03588-7. [DOI] [PubMed] [Google Scholar]
- 119.Chen L-Y, Zhang X-M, Han B-Q, Dai H-B. Long noncoding RNA SNHG12 indicates the prognosis and accelerates tumorigenesis of diffuse large B-cell lymphoma through sponging microR-195. Onco Targets Ther. 2020;13:5563. doi: 10.2147/OTT.S249429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang Y, Zhang M, Xu H, Wang Y, Li Z, Chang Y, et al. Discovery and validation of the tumor-suppressive function of long noncoding RNA PANDA in human diffuse large B-cell lymphoma through the inactivation of MAPK/ERK signaling pathway. Oncotarget. 2017;8(42):72182. doi: 10.18632/oncotarget.20053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jing Z, Gao L, Wang H, Chen J, Nie B, Hong Q. Long non-coding RNA GAS5 regulates human B lymphocytic leukaemia tumourigenesis and metastasis by sponging miR-222. Cancer Biomark. 2019;26(3):385–392. doi: 10.3233/cbm-190246. [DOI] [PubMed] [Google Scholar]
- 122.Song Y, Gao F, Peng Y, Yang X. Long non-coding RNA DBH-AS1 promotes cancer progression in diffuse large B-cell lymphoma by targeting FN1 via RNA-binding protein BUD13. Cell Biol Int. 2020;44(6):1331–1340. doi: 10.1002/cbin.11327. [DOI] [PubMed] [Google Scholar]
- 123.Hu G, Gupta SK, Troska TP, Nair A, Gupta M. Long non-coding RNA profile in mantle cell lymphoma identifies a functional lncRNA ROR1-AS1 associated with EZH2/PRC2 complex. Oncotarget. 2017;8(46):80223. doi: 10.18632/oncotarget.17956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang S, Wan H, Zhang X. LncRNA LHFPL3-AS1 contributes to tumorigenesis of melanoma stem cells via the miR-181a-5p/BCL2 pathway. Cell Death Dis. 2020;11(11):1–16. doi: 10.1038/s41419-020-03141-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhao S, Fang S, Liu Y, Li X, Liao S, Chen J, et al. The long non-coding RNA NONHSAG026900 predicts prognosis as a favorable biomarker in patients with diffuse large B-cell lymphoma. Oncotarget. 2017;8(21):34374. doi: 10.18632/oncotarget.16163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhu Q, Li Y, Guo Y, Hu L, Xiao Z, Liu X, et al. Long non-coding RNA SNHG16 promotes proliferation and inhibits apoptosis of diffuse large B-cell lymphoma cells by targeting miR-497-5p/PIM1 axis. J Cell Mol Med. 2019;23(11):7395–7405. doi: 10.1111/jcmm.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Deng L, Jiang L, Tseng K-F, Liu Y, Zhang X, Dong R, et al. Aberrant NEAT1_1 expression may be a predictive marker of poor prognosis in diffuse large B cell lymphoma. Cancer Biomark. 2018;23(2):157–164. doi: 10.3233/CBM-160221. [DOI] [PubMed] [Google Scholar]
- 128.Li Y, Jia Z, Zhao H, Liu X, Luo J, Cui G, et al. TUC338 Promotes Diffuse Large B Cell Lymphoma Growth via Regulating EGFR/PI3K/AKT Signaling Pathway. J Oncol. 2021;2021:1. doi: 10.1155/2021/5593720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huang Y, Lin Y, Song X, Wu D. LINC00857 contributes to proliferation and lymphomagenesis by regulating miR-370-3p/CBX3 axis in diffuse large B-cell lymphoma. Carcinogenesis. 2021;42(5):733–741. doi: 10.1093/carcin/bgab013. [DOI] [PubMed] [Google Scholar]
- 130.El-Khazragy N, Esmaiel MA, Mohamed MM, Hassan NS. Upregulation of long noncoding RNA Lnc-IRF2-3 and Lnc-ZNF667-AS1 is associated with poor survival in B-chronic lymphocytic leukemia. Int J Lab Hematol. 2020;42(3):284–291. doi: 10.1111/ijlh.13167. [DOI] [PubMed] [Google Scholar]
- 131.Cui Y, Xu H, Yang Y, Zhao D, Wen Y, Lv C, et al. The regulation of miR-320a/XBP1 axis through LINC00963 for endoplasmic reticulum stress and autophagy in diffuse large B-cell lymphoma. Cancer Cell Int. 2021;21(1):1–13. doi: 10.1186/s12935-021-01992-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Du X, Liu H, Yang C, Shi X, Cao L, Zhao X, et al. LncRNA landscape analysis identified LncRNA LEF-AS1 as an oncogene that upregulates LEF1 and promotes survival in chronic lymphocytic leukemia. Leukemia Res. 2021;110:106706. doi: 10.1016/j.leukres.2021.106706. [DOI] [PubMed] [Google Scholar]
- 133.Handa H, Honma K, Oda T, Kobayashi N, Kuroda Y, Kimura-Masuda K, et al. Long noncoding RNA PVT1 is regulated by bromodomain protein BRD4 in multiple myeloma and is associated with disease progression. Int J Mol Sci. 2020;21(19):7121. doi: 10.3390/ijms21197121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fernando TR, Rodriguez-Malave NI, Waters EV, Yan W, Casero D, Basso G, et al. LncRNA expression discriminates karyotype and predicts survival in B-lymphoblastic leukemia. Mol Cancer Res. 2015;13(5):839–851. doi: 10.1158/1541-7786.MCR-15-0006-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sehgal L, Mathur R, Braun FK, Wise JF, Berkova Z, Neelapu S, et al. FAS-antisense 1 lncRNA and production of soluble versus membrane Fas in B-cell lymphoma. Leukemia. 2014;28(12):2376–2387. doi: 10.1038/leu.2014.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Peng W, Feng J. Long noncoding RNA LUNAR1 associates with cell proliferation and predicts a poor prognosis in diffuse large B-cell lymphoma. Biomed Pharmacother. 2016;77:65–71. doi: 10.1016/j.biopha.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 137.Yan Y, Han J, Li Z, Yang H, Sui Y, Wang M. Elevated RNA expression of long non-coding HOTAIR promotes cell proliferation and predicts a poor prognosis in patients with diffuse large B cell lymphoma. Mol Med Rep. 2016;13(6):5125–5131. doi: 10.3892/mmr.2016.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Peng W, Fan H, Wu G, Wu J, Feng J. Upregulation of long noncoding RNA PEG10 associates with poor prognosis in diffuse large B cell lymphoma with facilitating tumorigenicity. Clin Exp Med. 2016;16(2):177–182. doi: 10.1007/s10238-015-0350-9. [DOI] [PubMed] [Google Scholar]
- 139.Peng W, Wu J, Feng J. Long noncoding RNA HULC predicts poor clinical outcome and represents pro-oncogenic activity in diffuse large B-cell lymphoma. Biomed Pharmacother. 2016;79:188–193. doi: 10.1016/j.biopha.2016.02.032. [DOI] [PubMed] [Google Scholar]
- 140.Peng W, Wu J, Feng J. LincRNA-p21 predicts favorable clinical outcome and impairs tumorigenesis in diffuse large B cell lymphoma patients treated with R-CHOP chemotherapy. Clin Exp Med. 2017;17(1):1–8. doi: 10.1007/s10238-015-0396-8. [DOI] [PubMed] [Google Scholar]
- 141.Rodríguez-Malavé NI, Fernando TR, Patel PC, Contreras JR, Palanichamy JK, Tran TM, et al. BALR-6 regulates cell growth and cell survival in B-lymphoblastic leukemia. Mol Cancer. 2015;14(1):1–15. doi: 10.1186/s12943-015-0485-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang F, Luo Y, Zhang L, Younis M, Yuan L. The LncRNA RP1.1–301G191/miR-582–5p/HMGB2 axis modulates the proliferation and apoptosis of multiple myeloma cancer cells via the PI3K/AKT signalling pathway. Cancer Gene Therapy. 2021;1:1–12. doi: 10.1038/s41417-021-00309-5. [DOI] [PubMed] [Google Scholar]
- 143.Hu T, Gao Y. β-elemene suppresses tumor growth of diffuse large B-cell lymphoma through regulating lncRNA HULC-mediated apoptotic pathway. Biosci Rep. 2020;40(2):20190804. doi: 10.1042/BSR20190804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang F, Cui D, Zhang Q, Shao Y, Zheng B, Chen L, et al. LncRNA00492 is required for marginal zone B-cell development. Immunology. 2021;165(1):88–98. doi: 10.1111/imm.13408. [DOI] [PubMed] [Google Scholar]
- 145.Li L-J, Chai Y, Guo X-J, Chu S-L, Zhang L-S. The effects of the long non-coding RNA MALAT-1 regulated autophagy-related signaling pathway on chemotherapy resistance in diffuse large B-cell lymphoma. Biomed Pharmacother. 2017;89:939–948. doi: 10.1016/j.biopha.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 146.Dong G, Yang Y, Li X, Yao X, Zhu Y, Zhang H, et al. Granulocytic myeloid-derived suppressor cells contribute to IFN-I signaling activation of B cells and disease progression through the lncRNA NEAT1-BAFF axis in systemic lupus erythematosus. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease. 2020;1866(1):165554. [DOI] [PubMed]
- 147.Liu W, Lei L, Liu X, Ye S. CircRNA_OTUD7A upregulates FOXP1 expression to facilitate the progression of diffuse large B-cell lymphoma via acting as a sponge of miR-431-5p. Genes & Genomics. 2021;43(6):653–667. doi: 10.1007/s13258-021-01094-z. [DOI] [PubMed] [Google Scholar]
- 148.Chen X, Xie X, Zhou W. CircCFL1/MiR-107 axis targeting HMGB1 promotes the malignant progression of diffuse large B-cell lymphoma tumors. Cancer Manag Res. 2020;12:9351. doi: 10.2147/CMAR.S263222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hu Y, Zhao Y, Shi C, Ren P, Wei B, Guo Y, et al. A circular RNA from APC inhibits the proliferation of diffuse large B-cell lymphoma by inactivating Wnt/β-catenin signaling via interacting with TET1 and miR-888. Aging (Albany NY) 2019;11(19):8068. doi: 10.18632/aging.102122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wu W, Wu Z, Xia Y, Qin S, Li Y, Wu J, et al. Downregulation of circ_0132266 in chronic lymphocytic leukemia promoted cell viability through miR-337-3p/PML axis. Aging (Albany NY) 2019;11(11):3561. doi: 10.18632/aging.101997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lux S, Blätte TJ, Gillissen B, Richter A, Cocciardi S, Skambraks S, et al. Deregulated expression of circular RNAs in acute myeloid leukemia. Blood Adv. 2021;5(5):1490–1503. doi: 10.1182/bloodadvances.2020003230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mei M, Wang Y, Wang Q, Liu Y, Song W, Zhang M. CircCDYL serves as a new biomarker in mantle cell lymphoma and promotes cell proliferation. Cancer Manag Res. 2019;11:10215. doi: 10.2147/CMAR.S232075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li Q, Luan Q, Zhu H, Zhao Y, Ji J, Wu F, et al. Circular RNA circ_0005774 contributes to proliferation and suppresses apoptosis of acute myeloid leukemia cells via circ_0005774/miR-192–5p/ULK1 ceRNA pathway. Biochem Biophys Res Commun. 2021;551:78–85. doi: 10.1016/j.bbrc.2021.02.058. [DOI] [PubMed] [Google Scholar]
- 154.Ma X, Xiang F, Pei Z, Miao J, Wu P, Song X, et al. Circ-Smad5 retards the G1/S transition of cell cycle via inhibiting the activity of wnt/lef/cyclind1 signaling in JB6 cells. Genes Dis. 2021;8(3):364–372. doi: 10.1016/j.gendis.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang D, Ming X, Xu J, Xiao Y. Circ_0009910 shuttled by exosomes regulates proliferation, cell cycle and apoptosis of acute myeloid leukemia cells by regulating miR-5195-3p/GRB10 axis. Hematol Oncol. 2021;39(3):390–400. doi: 10.1002/hon.2874. [DOI] [PubMed] [Google Scholar]
- 156.Xia L, Wu L, Bao J, Li Q, Chen X, Xia H, et al. Circular RNA circ-CBFB promotes proliferation and inhibits apoptosis in chronic lymphocytic leukemia through regulating miR-607/FZD3/Wnt/β-catenin pathway. Biochem Biophys Res Commun. 2018;503(1):385–390. doi: 10.1016/j.bbrc.2018.06.045. [DOI] [PubMed] [Google Scholar]
- 157.Winkle M, Kluiver J, Diepstra A, van den Berg A. Emerging roles for long noncoding RNAs in B-cell development and malignancy. Crit Rev Oncol Hematol. 2017;120:77–85. doi: 10.1016/j.critrevonc.2017.08.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.