Abstract
The results of PCR-based molecular typing of Helicobacter pylori strains by restriction fragment length polymorphism analysis of a 1,161-bp nucleotide sequence of the midregion of the vacA gene are reported. A total of 48 H. pylori strains isolated from gastric biopsy specimens obtained from 18 patients with peptic ulcer dyspepsia, 15 patients with nonulcer dyspepsia, and 15 asymptomatic H. pylori-infected subjects were studied. Highly heterogeneous restriction patterns were obtained by digestion of PCR products with SauII, BglII, and HhaI, whereas HaeIII digestion resulted in a strictly homogeneous profile for H. pylori strains isolated from 14 of 18 (77.7%) patients with peptic ulcer dyspepsia, but a strictly homogeneous profile was found for strains from only 8 of 15 (53.3%) patients with nonulcer dyspepsia (P = 0.163) and 5 of 15 (33.3%) asymptomatic H. pylori-infected subjects (P = 0.014). A potentially important aspect of the results obtained is the clinical relevance, since a single restriction pattern seems to be able to identify the majority of H. pylori strains associated with peptic ulcer disease.
Helicobacter pylori is linked to gastritis, peptic ulcer, and gastric cancer (4, 6, 14). Peptic ulcer disease, as distinct from chronic asymptomatic infection, is strongly associated with the expression of bacterial virulence markers (5, 30), including cytotoxin-associated gene A (CagA) (12, 28) and the vacuolating cytotoxin (VacA) that induces the formation of intracellular vacuoles in eukaryotic cells in vitro (9, 20).
Most people infected with H. pylori are asymptomatic, with only a few patients developing peptic ulcer or gastric cancer. A possible explanation is that patients with serious gastroduodenal lesions are infected with virulent H. pylori strains, whereas those patients who are asymptomatic and who present with simple chronic gastritis and no ulcer are infected with organisms with low pathogenic potentials.
Although H. pylori isolates show high levels of genotypic diversity (16), almost all phenotypic characters of the microorganism are conserved with the exception of the production of the vacuolating cytotoxin encoded by vacA (10) and the presence of the 128-kDa cytotoxin-associated protein encoded by cagA (7, 10). These two factors are therefore potentially important virulence determinants that affect the clinical outcome of H. pylori infection. In particular, the vacA gene is present in almost all strains tested (11, 22), and about 50% of clinical isolates produce inactive or less active toxins due to the presence of alleles characterized by differences in the signal peptide and/or middle region of the gene of H. pylori isolates obtained from U.S. subjects (8). By PCR typing and DNA sequencing, Atherton et al. (2) demonstrated that s1 vacA genotypes are associated with a higher level of in vitro cytotoxin activity than the levels of activity with which other genotypes are associated and that type s1 strains are more frequently observed among patients with past or present peptic ulceration than patients without peptic ulcer (2, 3). Recently, the existence of different allelic variants has also been described in H. pylori strains obtained from European (23, 24, 29) and Japanese (18) subjects.
We report here on a simple PCR-based method of typing H. pylori that uses restriction fragment length polymorphism (RFLP) analysis of a 1,161-bp fragment of the midregion of vacA. Using this analysis, we found a simple fingerprinting pattern that identifies most H. pylori strains isolated from patients with peptic ulcer disease.
MATERIALS AND METHODS
Patients and clinical specimens.
Forty-eight subjects (23 men and 25 females; mean age, 46 years; age range, 22 to 75 years) were admitted to the study. Thirty-three patients had undergone gastroduodenoscopy for dyspepsia, and 15 asymptomatic subjects were the partners of H. pylori-infected patients. Gastric specimens were cultured for H. pylori, as described previously (27). Briefly, biopsy samples were homogenized and cultured on Columbia agar base (Oxoid, Milan, Italy) supplemented with 7% horse blood and Dent’s selective supplement (Oxoid). The cultures were incubated in a microaerophilic atmosphere at 37°C in GasPak jars and CampyPak II envelopes (BBL Microbiology System, Cockeysville, Md.), and the isolates were identified as H. pylori by their morphology upon Gram staining and by positive urease, oxidase, and catalase tests. Histological sections of formalin-fixed biopsy specimens were stained with hematoxylin-eosin to evaluate the morphology and whether Helicobacter-like organisms were present. A serum sample was obtained by routine venipuncture for serological studies.
Assay for cytotoxicity.
Supernatants from broth cultures of H. pylori isolates were concentrated by using Centriprep-100 ultrafiltration units (Amicon, Beverly, Mass.) and were incubated with HeLa cells at twofold dilutions ranging from 1:5 to 1:160 as described previously (13). Cell vacuolization was assessed by light microscopy after 48 h of incubation. Wells in which 50% or more of the cells were vacuolated were defined as showing a cytotoxic effect.
Neutralization of H. pylori cytotoxin activity.
Human sera were heated at 56°C and diluted with Eagle’s minimal essential medium. Sera diluted twofold (from 1:10 to 1:160) were incubated for 1 h at 37°C with an equal volume of the concentrated type strain H. pylori CCUG 17874 (Culture Collection of the University of Göteborg, Göteborg, Sweden) culture supernatant. Adherent HeLa cells were incubated for 18 h at 37°C in 96-well plates with 50-μl mixtures of serum and H. pylori plus 50 μl of minimal essential medium. The neutralization titer was defined as the highest dilution of a serum sample that completely neutralized vacuolization, as assessed by light microscopy.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot (immunoblot) analysis for antibodies against VacA and CagA antigens.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed by the method of Laemmli (19) with a 7.0% acrylamide gel, as described previously (13). Supernatants from the type strain H. pylori CCUG 17874 broth culture was concentrated by using Centriprep-100 ultrafiltration units (Amicon) and were used as antigen, as described previously (13).
The Western blot procedure of Towbin et al. (26) was performed as described previously (13). Briefly, after electrophoretic transfer, the blots were incubated for 12 h at room temperature with human sera diluted 1:1,000 in phosphate-buffered saline containing 0.05% (vol/vol) Tween 20. Antigen-antibody complexes were detected with anti-human peroxidase-labeled immunoglobulin G (DAKO, Copenhagen, Denmark) diluted 1:500 in phosphate-buffered saline and with 4-chloro-1-naphthol (Bio-Rad, Hercules, Calif.) as the enzyme substrate. The VacA and CagA antigens were recognized as clear bands of 87 to 94 and 128 kDa, respectively (13).
PCR.
The oligonucleotides used as PCR primers in this study have been used previously by Xiang et al. (30). The primers for the vacA gene are derived from the sequence of the vacA gene. Briefly, the amplification product of H. pylori vacA primers 5′-GCTTCTCTTACCACCAATGC and 5′-TGTCAGGGTTGTTCACCATG was 1,161 nucleotides in length and was derived from the middle region of vacA, from nucleotides 1468 to 2629 (25). H. pylori cagA primers 5′-AGTAAGGAGAAACAATGA and 5′-AATAAGCCTTAGAGTCTTTTTGGAAATC amplify a 1,350-bp DNA fragment (30). The PCR mixtures (50 μl) contained 50 mM KCl, 10 mM Tris, 200 μM (each) deoxynucleoside triphosphate, 30 pmol of each primer, 2.5 U of Amplitaq (Perkin-Elmer, Norwalk, Conn.), and 10 ng of DNA obtained from each bacterial strain by phenol-chloroform extraction and ethanol precipitation. Amplifications were performed on a PCR 9600 thermocycler (Perkin-Elmer) as follows: 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min. Five microliters of the PCR product was electrophoresed on a 2% agarose gel (Bethesda Research Laboratories, Inc., Gaithersburg, Md.) with 1× Tris-borate-EDTA buffer containing 1 μg of ethidium bromide per ml. The gels were examined by transillumination and were photographed. A vacA- and cagA-positive H. pylori strain (strain CCUG 17874) and a cagA-negative and vacA-positive H. pylori strain (strain HPG21) were used as controls in the PCR experiments.
RFLP analysis.
A 10-μl sample of the PCR product was digested with 10 U of the restriction enzymes HaeIII, SauIII, BglII, and HhaI (Boehringer Mannheim) for 4 h at 37°C in the buffer, as recommended by the supplier. The digest was analyzed by electrophoresis in a 2% agarose gel with 1× Tris-borate-EDTA buffer containing 1 μg of ethidium bromide per ml. DNA molecular size marker VI (Boehringer) was used.
Statistical analysis.
Prevalence rates were compared by the χ2 test and Fisher’s exact test. Probability levels (P) of <0.1 were considered statistically significant.
RESULTS
The endoscopic diagnoses for 33 symptomatic patients with dyspepsia were peptic ulcer dyspepsia (n = 18) and nonulcer dyspepsia (n = 15). The gastric histology of the latter 15 patients was active (n = 5) or chronic active (n = 10) gastritis. The 15 asymptomatic partners of H. pylori-infected patients were either histologically negative (n = 7) or the partners presented with simple chronic histological gastritis (n = 8). All 48 patients studied were positive for H. pylori by culture. Supernatants from 23 of 48 H. pylori isolates produced vacuolization in HeLa cells, whereas supernatants from the remaining 25 did not. The supernatant dilution that produced vacuolization ranged from 1:5 to ≥1:160.
The specific sequences of the vacA and cagA genes of the H. pylori isolates were looked for by PCR: all 48 strains were vacA positive, whereas 42 strains possessed the cagA gene (data not shown). The relationship between the genetic, phenotypic, and serological properties of the H. pylori strains isolated from the 48 subjects is reported in Table 1.
TABLE 1.
Genetic, phenotypic, and serological properties of H. pylori isolates from 33 patients with gastritis and endoscopically defined gastroduodenal pathology and 15 asymptomatic H. pylori-infected subjects
| Clinical diagnosis | No. of patients | No. (%) of H. pylori isolates with the following genetic and phenotypic propertiesa:
|
No. (%) of serum samples with the following:
|
|||||
|---|---|---|---|---|---|---|---|---|
| vacA, ho | vacA, he | cagA positive (%) | Tox+ | Neutralizing activity | Anti-VacA IgGb | Anti-CagA IgG | ||
| Ulcer dyspepsia | 18 | 14 (77.8) | 4 (22.2) | 17 (94.4) | 9 (50.0) | 13 (72.2) | 13 (72.2) | 17 (94.4) |
| Nonulcer dyspepsia | 15 | 8 (53.3) | 7 (46.7) | 13 (86.7) | 7 (46.7) | 10 (66.7) | 10 (66.7) | 12 (80.0) |
| Asymptomatic, infected | 15 | 5 (33.3) | 10 (66.7) | 12 (80.0) | 7 (46.7) | 8 (53.3) | 8 (53.3) | 10 (66.7) |
Strains with homogeneous (ho) and heterogeneous (he) restriction pattern profiles for PCR-based amplification of the vacA gene and digestion with HaeIII are indicated (see Fig. 1a). Tox+, in vitro cytotoxin production.
IgG, immunoglobulin G.
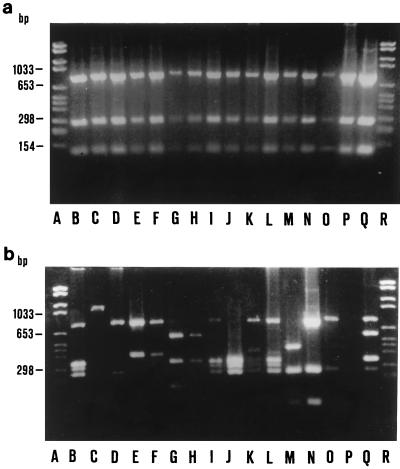
From the restriction enzyme digestion of the 1,161-bp products of amplification of the vacA gene from the 48 H. pylori strains studied, we found highly heterogeneous restriction patterns with SauIII, BglII, and HhaI (data not reported), whereas HaeIII digestion of the 1,161-bp vacA fragment resulted in a strictly homogeneous profile (Fig. 1) for strains from 77.8% of the patients (14 of 18) with peptic ulcer dyspepsia, whereas a strictly homogeneous profile was found for strains from 53.3% of the patients (8 of 15) with nonulcer dyspepsia and chronic active gastritis (P = 0.163) and 33.3% of the asymptomatic subjects infected with H. pylori (5 of 15), who had either a negative histology result or simple chronic gastritis (P = 0.014) (Table 1). All 27 H. pylori strains with the homogeneous profile were cagA positive, and 17 (63.0%) produced cytotoxin in vitro. Of 21 strains with the heterogeneous profile, 15 (71.4%) were cagA positive and 6 (28.6%) produced cytotoxin in vitro.
FIG. 1.
RFLP analysis of H. pylori vacA gene. A 1,161-bp region was amplified by PCR and digested with HaeIII, and the fragments were separated in a 2% agarose gel. (a) Homogeneous pattern characterized by three bands. Lanes A and R, molecular size marker VI; lanes B to O, isolates from 14 patients with ulcer dyspepsia, respectively; lanes P and Q, isolates from patients with nonulcer dyspepsia. (b) Heterogeneous profiles characterized by several different patterns. Lanes A and R, molecular size marker VI; lanes B to I, isolates from asymptomatic H. pylori-infected patients; lanes J to O, isolates from patients with nonulcer dyspepsia; lane P, running buffer alone; lane Q, isolate from a patient with ulcer dyspepsia.
DISCUSSION
Although molecular biological typing methods with genomic DNA or gene probes usually require laborious sample preparation and processing steps, the use of PCR-based RFLP analysis affords a simple and rapid typing technique. The molecular typing approach for H. pylori that uses the PCR-based RFLP analysis has been used previously. The genes encoding urease and its accessory proteins have been preferential targets for PCR (1, 17, 21) since these genes are conserved in H. pylori. The adhesin gene hpaA (15) has also been used as the target for comparison of the effects of genetic changes of H. pylori isolates.
In our study, the vacA gene was the target for the PCR, and we decided to use a fragment of the midregion of the gene large enough to permit the detection of diversity but small enough to allow regular amplification. Previous studies by Cover et al. (11) and Atherton et al. (2) have shown that several cytotoxin-producing strains and cytotoxin-nonproducing strains of H. pylori have substantially different sequences within the middle region of the vacA gene. Our results by RFLP analysis of the PCR products with several restriction enzymes confirmed the high degree of diversity of the genomic structure of the vacA gene among H. pylori strains isolated from gastric biopsy specimens. However, the digestion of PCR products with the HaeIII enzyme allowed us to identify a genetic correlation for 27 of 48 H. pylori strains examined, thus resulting in a homogeneous group of strains with identical vacA gene restriction patterns characterized by the presence of three distinct bands. In addition, these strains were strongly associated with the presence of the cagA gene and cytotoxin activity and occurred more frequently in patients with peptic ulcer dyspepsia. In fact, the genetically related strains were isolated from 77.8% of patients with peptic ulcer dyspepsia but significantly (P = 0.014) less frequently (33.3%) from asymptomatic H. pylori-infected subjects. Although the homogeneous H. pylori strains were more likely to be isolated from patients with peptic ulcer dyspepsia (77.8%) than from patients with nonulcer dyspepsia and chronic active gastritis (53.3%), the differences were not significant (P = 0.163). The detection of strains with this characteristic restriction pattern not only in patients with ulcer dyspepsia but also in patients with a less serious disease such as nonulcer dyspepsia and chronic active gastritis or in H. pylori-infected but as yet asymptomatic subjects showed that the specificity (57.7%) of the pattern for ulcer disease was lower than its sensitivity (77.8%). This observation, however, does not seem at variance with the presumed ulcerogenic potential of these strains. It is well known that chronic infection often occurs without symptoms and that with time some individuals develop severe features of upper gastrointestinal diseases. Untreated chronic infection with these H. pylori strains may progress from simple chronic gastritis in asymptomatic subjects to chronic active gastritis in patients with nonulcer dyspepsia and, finally, to peptic ulcer disease in ulcer patients.
In conclusion, the results of this study by PCR-based restriction pattern analysis suggest that the specific RFLP profiles reported here may be indicators of the pathogenic potential of H. pylori strains. A potentially important aspect of this simple method may be its clinical relevance, since a single restriction pattern for the vacA gene seems to be able to identify H. pylori strains strongly associated with peptic ulcer disease.
ACKNOWLEDGMENT
This study was partially supported by University of Bologna grant Finanziamento Speciale alle Strutture.
REFERENCES
- 1.Akopyanz N, Bukanov N O, Westblom T U, Berg D E. PCR-based RFLP analysis of DNA sequence diversity in gastric pathogen Helicobacter pylori. Nucleic Acids Res. 1992;20:6221–6225. doi: 10.1093/nar/20.23.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atherton J C, Cao P, Peek R M, Tummuru M K R, Blaser M J, Cover T L. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 3.Atherton J C P, Peek R M, Tham K T, Cover T L, Blaser M J. Clinical and pathological importance of heterogeneity in vacA, the vacuolating cytotoxin gene of Helicobacter pylori. Gastroenterology. 1997;112:92–99. doi: 10.1016/s0016-5085(97)70223-3. [DOI] [PubMed] [Google Scholar]
- 4.Blaser M. Helicobacter pylori: microbiology of a “slow” bacterial infection. Trends Microbiol. 1993;11:255–260. doi: 10.1016/0966-842x(93)90047-u. [DOI] [PubMed] [Google Scholar]
- 5.Blaser M. Role of vacA and the cagA locus of Helicobacter pylori in human disease. Alimen Pharmacol Ther. 1996;10(Suppl. 1):73–78. doi: 10.1046/j.1365-2036.1996.22164008.x. [DOI] [PubMed] [Google Scholar]
- 6.Blaser M J, Personnet J. Parasitism by the “slow” bacterium Helicobacter pylori leads to altered gastric homeostatis and neoplasia. J Clin Invest. 1994;94:4–8. doi: 10.1172/JCI117336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N, Rappuoli R. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad: Sci USA. 1993;90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cover T. The vacuolating cytotoxin of Helicobacter pylori. Mol Microbiol. 1996;20:241–246. doi: 10.1111/j.1365-2958.1996.tb02612.x. [DOI] [PubMed] [Google Scholar]
- 9.Cover T L, Blaser M J. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J Biol Chem. 1992;267:10570–10575. [PubMed] [Google Scholar]
- 10.Cover T L, Dooley C P, Blaser M J. Characterization of and human serologic response to proteins in Helicobacter pylori broth culture supernatants with vacuolizing cytotoxin activity. Infect Immun. 1990;58:603–610. doi: 10.1128/iai.58.3.603-610.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cover T L, Tummuru M K M, Cao P, Thompson S A, Blaser M J. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J Biol Chem. 1994;269:10566–10573. [PubMed] [Google Scholar]
- 12.Crabtree J E, Figura N, Taylor J D, Bugnoli M, Armellini D, Tompkins D S. Expression of 120 kilodalton protein and cytotoxicity in Helicobacter pylori. J Clin Pathol. 1992;45:733–734. doi: 10.1136/jcp.45.8.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donati M, Moreno S, Storni E, Tucci A, Poli L, Mazzoni A, Varoli O, Sambri V, Farencena A, Cevenini R. Detection of serum antibodies to CagA and VacA and of serum neutralizing activity for vacuolating cytotoxin in patients with Helicobacter pylori-induced gastritis. Clin Diagn Lab Immunol. 1997;4:478–482. doi: 10.1128/cdli.4.4.478-482.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eurogast Study Group. An international association between Helicobacter pylori infection and gastric cancer. Lancet. 1993;341:1360–1362. [PubMed] [Google Scholar]
- 15.Evans D G, Evans D J, Lampert H C, Graham D Y. Restriction fragment length polymorphism in the adhesin gene hpaA of Helicobacter pylori. Am J Gastroenterol. 1995;90:1282–1288. [PubMed] [Google Scholar]
- 16.Foxall P A, Hu L T, Mobley H L T. Use of polymerase chain reaction-amplified Helicobacter pylori urease structural genes for differentiation of isolates. J Clin Microbiol. 1992;30:739–741. doi: 10.1128/jcm.30.3.739-741.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujimoto S, Marshall B, Blaser M J. PCR-based restriction fragment length polymorphism typing of Helicobacter pylori. J Clin Microbiol. 1994;32:331–334. doi: 10.1128/jcm.32.2.331-334.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito Y, Azuma T, Ito S, Miyaji H, Hirai M, Yamazaki Y, Sato F, Kato T, Kohli Y, Kuriyama M. Analysis and typing of the vacA gene from cagA-positive strains of Helicobacter pylori isolated in Japan. J Clin Microbiol. 1997;35:1710–1714. doi: 10.1128/jcm.35.7.1710-1714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Leunk R D, Johnson P T, David B C, Kraft W G, Morgan D R. Cytotoxic activity in broth culture filtrates of Campylobacter pylori. J Med Microbiol. 1988;26:93–99. doi: 10.1099/00222615-26-2-93. [DOI] [PubMed] [Google Scholar]
- 21.Moore R A, Kureishi A, Wong S, Bryan L E. Categorization of clinical isolates of Helicobacter pylori on the basis of restriction digest analysis of polymerase chain reaction-amplified ureC genes. J Clin Microbiol. 1993;31:1334–1335. doi: 10.1128/jcm.31.5.1334-1335.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phadnis S H, Ilver D, Janzon L, Normark S, Westblom T U. Pathological significance and molecular characterization of the vacuolating toxin gene of Helicobacter pylori. Infect Immun. 1994;62:1557–1565. doi: 10.1128/iai.62.5.1557-1565.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudi J, Kolb C, Maiwald M, Kuck D, Sieg A, Galle P R, Stremmel W. Diversity of Helicobacter pylori vacA and cagA genes and relationship to VacA and CagA protein expression, cytotoxin production, and associated diseases. J Clin Microbiol. 1998;36:944–948. doi: 10.1128/jcm.36.4.944-948.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strobel S, Bereswill S, Balig P, Allgaier P, Sonntag H G, Kist M. Identification and analysis of a new vacA genotype variant of Helicobacter pylori in different patient groups in Germany. J Clin Microbiol. 1998;36:1285–1289. doi: 10.1128/jcm.36.5.1285-1289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Telford J L, Ghiara P, Dell’Orco M, Comanducci M, Burroni D, Bugnoli M, Tecce M F, Censini S, Covacci A, Xiang Z, Papini E, Montecucco C, Parente L, Rappuoli R. Gene structure of the Helicobacter pylori cytotoxin and evidence of its key role in gastric disease. J Exp Med. 1994;179:1653–1658. doi: 10.1084/jem.179.5.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of protein from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tucci A, Poli L, Donati M, Mazzoni C, Cevenini R, Sambri V, Varoli O, Bocus P, Ferrari A, Paparo G F, Caletti G. Value of serology (ELISA) for the diagnosis of Helicobacter pylori infection: evaluation in patients attending endoscopy and in those with fundic atrophic gastritis. Ital J Gastroenterol. 1996;28:371–376. [PubMed] [Google Scholar]
- 28.Tummuru M K R, Cover T L, Blaser M J. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect Immun. 1993;61:1799–1809. doi: 10.1128/iai.61.5.1799-1809.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Doorn L J, Figueiredo C, Rossau R, Jannes G, van Asbroeck M, Sousa J C, Carneiro F, Quint W G V. Typing of Helicobacter pylori vacA gene and detection of cagA gene by PCR and reverse hybridization. J Clin Microbiol. 1998;36:1271–1276. doi: 10.1128/jcm.36.5.1271-1276.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiang Z, Censini S, Bayeli P F, Telford J L, Figura N, Rappuoli R, Covacci A. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect Immun. 1995;63:94–98. doi: 10.1128/iai.63.1.94-98.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]