ABSTRACT
It has been demonstrated that quorum sensing (QS) is widely employed by bacterial cells to coordinately regulate various group behaviors. Diffusible signal factor (DSF)-type signals have emerged as a growing family of conserved cell-cell communication signals. In addition to the DSF signal initially identified in Xanthomonas campestris pv. campestris, Burkholderia diffusible signal factor (BDSF) (cis-2-dodecenoic acid) has been recognized as a conserved DSF-type signal with specific characteristics in both signal perception and transduction from DSF signals. Here, we review the history and current progress of the research on this type of signal, especially focusing on its biosynthesis, signaling pathways, and biological functions. We also discuss and explore the huge potential of targeting this kind of QS system as a new therapeutic strategy to control bacterial infections and diseases.
KEYWORDS: BDSF, Burkholderia cenocepacia, quorum sensing, virulence, c-di-GMP
INTRODUCTION
Quorum sensing (QS) is a cell-cell communication process that occurs widely in both Gram-positive and Gram-negative bacteria. It is used by bacteria to sense and respond to changes in cell density to regulate group behaviors and many biological functions (1, 2). The QS systems in Gram-positive bacteria typically use either an unmodified or a posttranslationally modified small peptide and a two-component system, which includes a membrane-bound sensor kinase receptor and a cytoplasmic transcription factor that directly alters gene expression (3, 4). Many Gram-negative bacterial species utilize the homologs of the LuxIR proteins first found in Vibrio fischeri to produce acyl-homoserine lactones (AHLs) as QS signal molecules (5, 6), which are the most studied self-induced signal molecules and possess a core N-acylated homoserine lactone (HSL) ring and a 4- to 18-carbon acyl chain (1, 7).
In addition to AHL signals, there are many other QS signaling molecules that have been identified in Gram-negative bacteria, including autoinducer-2 (AI-2) (8), Pseudomonas aeruginosa quinolone signal (PQS) (9), autoinducer in Escherichia coli (AI-3) (10), 2-heptyl-4-quinolone (HHQ) (11), diketopiperazines (12), bradyoxetin (13), diffusible signal factor (DSF) family signals that were first identified in the plant pathogen Xanthomonas campestris pv. campestris (14–16), 3-hydroxypalmitic acid methyl ester (3-OH PAME) (17), and a new signal, anthranilic acid, which was recently identified in Ralstonia solanacearum (18).
Studies have demonstrated that RpfF, RpfC, and RpfG are the key elements of the DSF QS system in X. campestris pv. campestris. RpfF is responsible for the synthesis of DSF, which is characterized as cis-11-methyl-2-dodecenoic acid, and RpfC-RpfG constitutes a two-component system for signal perception and transduction (19, 20). In more detail, RpfC senses and binds to DSF by a membrane-spanning sensor domain, leading to its autophosphorylation within the histidine kinase domain and then phosphotransfer to RpfG (19). The activated HD-GYP domain of RpfG has phosphodiesterase (PDE) activity and is able to degrade cyclic dimeric GMP (c-di-GMP), which is an intracellular second messenger (21, 22). Comparative genomic studies found that the rpfFCG gene cluster is highly conserved, which indicates that the DSF-dependent QS system is widely distributed throughout xanthomonads (23, 24).
In addition to the DSF family signals identified in xanthomonads, Burkholderia diffusible signal factor (BDSF) (cis-2-dodecenoic acid) has been recognized as a novel DSF family signal, which was first found in Burkholderia cenocepacia (25) (Fig. 1). Subsequent studies have elucidated the signaling pathway and regulatory mechanism of this signal, which is definitely specific and different from what we have known. Here, we review the research history and current progress on BDSF signals in recent years. In addition to its role as an intracellular signal, the functions of BDSF in interspecies and interkingdom communications are also described here.
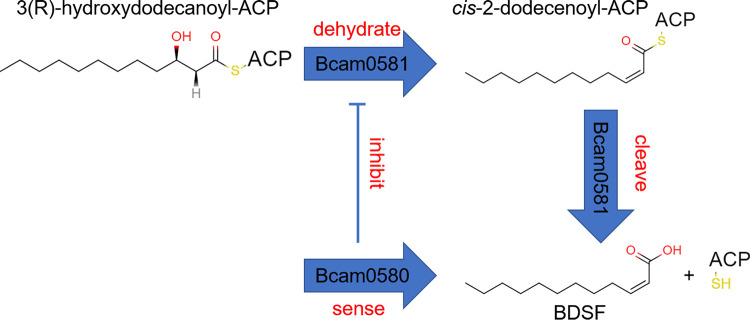
FIG 1.
The mechanism of RpfFBc to synthesize BDSF. The synthesis of BDSF by RpfFBc (BCAM0581) is divided into two steps: RpfFBc first dehydrates 3-hydroxydodecanoyl-ACP to form cis-2-dodecenoyl-ACP and then further cleaves cis-2-dodecenoyl-ACP to obtain BDSF and holo-ACP. RpfR (BCAM0580) interacts with RpfFBc, blocks the substrate from entering the channel, and finally inhibits the synthesis of BDSF.
BDSF IS A NOVEL DSF FAMILY SIGNAL
The Burkholderia cepacia complex (BCC) is a group of Gram-negative bacteria that comprises more than 20 closely related species (26, 27). B. cenocepacia is an opportunistic pathogen that constitutes the majority of transmissible and epidemic strains and is highly virulent (28, 29); it can cause a decline in lung function and life-threatening infections in cystic fibrosis patients and immunocompromised individuals (30, 31). It has been shown that the formation of biofilms and the expression of virulence factors are modulated by the QS systems of B. cenocepacia (32, 33). Evidence indicates that the CepIR system is an AHL-based LuxIR QS system and is highly conserved in BCC species (34, 35). It controls the production of extracellular proteases, biofilm maturation, and swarming motility in B. cenocepacia. Increasing evidence suggests that once AHL signals accumulate to the threshold in the environment, they can bind to the CepR protein to form a complex, which causes conformational changes in the CepR protein and then activates or inhibits the expression of the target genes by binding to the promoters (34–37). Moreover, one more AHL-based QS system, CciIR, has also been found in B. cenocepacia (37). Intriguingly, there is an orphan LuxR homolog, CepR2, which is not genetically linked to a cognate AHL synthase-encoding gene, that was also found in B. cenocepacia (38–40).
In 2008, Boon et al. first characterized BDSF as a DSF family QS signal (Fig. 1), which was biosynthesized by B. cenocepacia RpfF (RpfFBc). The structure of BDSF is similar but not identical to that of the DSF (cis-11-methyl-2-decenoic acid) signal produced by X. campestris pv. campestris, where BDSF lacks a methyl group at the C-11 position (25). It was further identified that BDSF is a conserved signaling molecule produced by the BCC, with at least nine species producing it as the major DSF family signaling molecule (41). Intriguingly, BDSF not only controls biofilm formation and other AHL-regulated phenotypes (42) but also regulates the synthesis of AHL signals in B. cenocepacia (43), suggesting that the BDSF QS system is the central regulatory system in B. cenocepacia.
THE BIOSYNTHESIS OF THE BDSF SIGNAL IN B. CENOCEPACIA
BCAM0581 catalyzes the production of the BDSF signal in B. cenocepacia.
Protein sequence alignment analysis showed that BCAM0581 (also known as RpfFBc) shares 37.2% identity with RpfF, and the two proteins are functionally interchangeable (25). It was revealed that the deletion of BCAM0581 abolished BDSF production in B. cenocepacia, and the phenotypic defect of the rpfF mutant of X. campestris pv. campestris could be restored to the wild-type strain level by the in trans expression of BCAM0581 (25, 41). It was demonstrated that BCAM0581 is a bifunctional crotonase with both dehydratase and thioesterase activities, which enables the direct conversion of the acyl carrier protein (ACP) thioester of 3-hydroxydodecanoic acid into cis-2-dodecenoic acid in B. cenocepacia (23, 44) (Fig. 1). BCAM0581 is the first member of the crotonase superfamily that has been identified to exert desaturase and thioesterase activities to cleave the acyl-ACP thioester bond to release free fatty acids (44).
The RqpSR two-component system controls BDSF production in B. cenocepacia.
As RpfFBc (BCAM0581) is the enzyme that catalyzes the production of BDSF signals, our group continued to study the regulatory mechanism of rpfFBc expression. We found a novel two-component system, RqpSR, in which RqpS is a signal transduction histidine kinase and RqpR is a response regulator with a DNA-binding domain. It not only controls the QS-regulated phenotypes in B. cenocepacia but also positively controls the production of BDSF and AHL signals by modulating the transcriptional expression levels of cepI and rpfFBc (45). The response regulator of this system, RqpR, controls the QS system by directly binding to the promoters of BDSF and AHL signal synthase-encoding genes (45). These findings suggest that the RqpSR system modulates B. cenocepacia physiology and pathogenicity by forming a complicated hierarchy with QS systems. Intriguingly, the RqpSR system appears to be widely distributed and coexists with the BDSF QS system in various bacterial species (45) (Table 1).
TABLE 1.
Homologs of RpfFBc, RpfR, RqpS, RqpR, and GtrR in various bacterial speciesa
| Bacterium | Strain | RpfFBc homolog identity (%) | RpfR homolog identity (%) | RqpS homolog identity (%) | RqpR homolog identity (%) | GtrR homolog identity (%) |
|---|---|---|---|---|---|---|
| Burkholderia | ||||||
| B. cenocepacia | H111 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| B. cenocepacia | J2315 | 100.0 | 100.0 | 99.6 | 100.0 | 99.8 |
| B. cepacia | JBK9 | 96.5 | 99.9 | 91.1 | 99.1 | 100.0 |
| B. contaminans | FFH2055 | 95.1 | 96.3 | 91.1 | 99.1 | 97.6 |
| B. anthina | MSMB0848 | 99.7 | 97.3 | 96.7 | 98.2 | 99.1 |
| B. lata | 383 | 94.8 | 96.3 | 90.8 | NA | 97.6 |
| B. seminalis | FL-5-5-10-S1-D0 | 99.0 | 96.9 | 91.1 | 97.7 | 98.1 |
| B. pyrrocinia | Lyc2 | 95.8 | 96.1 | 89.7 | 97.2 | 97.6 |
| B. dolosa | AU0158 | 96.9 | 92.4 | 84.5 | 96.3 | 95.3 |
| B. pseudomultivorans | MSMB0607 | 94.8 | 93.3 | 83.4 | 95.0 | 96.1 |
| B. vietnamiensis | G4 | 94.8 | 91.2 | 79.7 | 93.6 | 96.3 |
| B. multivorans | ATCC 17616 | 94.4 | 93.2 | 75.6 | 93.6 | 97.0 |
| B. ubonensis | MSMB0011 | 90.2 | 86.5 | 77.5 | 93.6 | 95.7 |
| B. diffusa | MSMB0010 | 95.5 | 96.0 | 81.2 | 93.1 | 95.5 |
| B. latens | RF32-BP12 | 96.2 | 91.6 | 83.0 | 93.1 | 95.0 |
| B. stagnalis | MSMB1135 | 87.8 | 86.7 | 76.0 | 93.1 | 95.0 |
| B. territorii | A63 | 95.5 | 92.5 | 81.2 | 92.7 | 93.0 |
| B. ambifaria | AMMD | 94.4 | 92.1 | 83.4 | 92.7 | 95.5 |
| B. pseudomallei | 1026b | NA | 33.0 | 65.7 | 87.6 | 99.0 |
| B. oklahomensis | C6786 | NA | 37.5 | 58.7 | 87.2 | 89.2 |
| B. plantarii | ATCC 43733 | NA | 43.3 | 53.1 | 86.2 | 89.4 |
| B. gladioli | ATCC 10248 | NA | 42.9 | 55.4 | 85.7 | 89.9 |
| B. kururiensis | M130 | 71.4 | 65.8 | 49.1 | 83.5 | 82.4 |
| B. glumae | BGR1 | NA | 38.4 | 53.9 | 82.9 | 88.4 |
| B. graminis | C4D1M | 71.1 | 65.9 | 51.3 | 82.1 | 82.5 |
| B. bryophila | 376MFSha3.1 | 70.7 | 64.9 | 50.2 | 82.1 | 82.1 |
| B. fungorum | ATCC BAA-463 | 70.7 | 65.5 | 52.0 | 82.1 | 81.9 |
| B. phymatum | STM815 | 73.9 | 66.1 | 50.6 | 81.7 | 37.4 |
| B. caribensis | MBA4 | 72.5 | 65.2 | 52.4 | 81.2 | 84.3 |
| B. ginsengisoli | NBRC 100965 | 71.1 | 65.3 | 50.2 | 80.7 | 81.0 |
| B. xenovorans | LB400 | 70.7 | 66.3 | 55.0 | 80.3 | 82.1 |
| B. acidipaludis | NBRC 101816 | NA | 39.9 | 49.8 | 79.8 | 75.2 |
| B. sprentiae | WSM5005 | NA | 61.9 | 45.8 | 79.8 | 38.5 |
| B. phytofirmans | PsJN | 71.1 | 41.2 | 52.8 | 79.4 | 49.3 |
| B. ferrariae | NBRC 106233 | NA | 41.9 | 48.7 | 79.4 | 75.9 |
| Caballeronia | ||||||
| C. choica | NA | NA | 40.5 | 46.5 | 76.1 | 39.8 |
| C. telluris | NA | NA | 38.9 | 44.3 | 76.1 | 38.0 |
| C. glathei | NA | NA | 36.9 | 42.4 | 75.7 | 38.6 |
| C. jiangsuensis | MP-1 | NA | 37.8 | 38.0 | 75.6 | 74.2 |
| C. grimmiae | R27 | NA | 38.5 | 35.1 | 70.6 | 38.9 |
| Other | ||||||
| Mumia flava | MUSC 201 | 95.1 | 35.1 | 91.1 | 99.1 | NA |
NA, not applicable.
BIOLOGICAL FUNCTIONS OF BDSF SIGNALS
The biological functions of BDSF signals in B. cenocepacia.
It was demonstrated that BDSF controls various biological functions of B. cenocepacia (42, 46, 47). The rpfFBc mutant of B. cenocepacia showed reduced virulence and defective functions, including motility and biofilm formation, while all of these phenotypes could be restored to wild-type levels by the addition of BDSF or complementation with RpfFBc (42, 48). It was revealed that the BDSF-null mutant was compromised in the expression of virulence factors such as the metalloproteases ZmpA and ZmpB (42, 47, 49, 50). The transcription of the zinc metalloprotease zmpB was downregulated in the rpfFBc mutant (47), and the downregulated expression of zmpA in the rpfFBc mutant of B. cenocepacia J2315 could be restored when the medium was supplemented with BDSF (42). The identification of BDSF as another QS signal in B. cenocepacia provides more evidence that the bacterial pathogens recruit multiple signaling systems to coregulate virulence gene expression. Intriguingly, the inactivation of rpfFBc in B. cenocepacia H111 also resulted in decreased cepI expression and diminished AHL production, suggesting that BDSF is involved in the AHL-dependent QS system by reducing the transcription of cepI, while mutation of the cepIR system did not impair the production of BDSF (43, 47). The integrated analysis of transcriptome sequencing (RNA-seq) and phenotypes showed that the two QS systems not only have specific signal pathways but also form a complex intraspecies signaling network in B. cenocepacia.
The role of BDSF in interspecies and interkingdom communication.
In nature, bacteria are more likely to grow in polymicrobial communities than in monocultures. The development and maintenance of such communities depend on interactions among the members, including interspecies signal transmission (22). Many pathogenic bacteria use cell-cell signaling to regulate the expression of factors contributing to virulence. This may be related to the fine control of intraspecies or interspecies signaling by a range of bacteria. Recently, signaling molecules of the DSF family have been found to be involved in the regulation of pathogenesis and biofilm formation in diverse bacteria—not only in their cognate bacteria but also in unrelated bacteria (25). BDSF isolated from B. cenocepacia was found to be involved in the modulation of virulence, antibiotic resistance, and persistence of P. aeruginosa in the cystic fibrosis airway (51, 52). The exogenous addition of BDSF reduced the transcriptional expression of QS system-related genes and the production of QS signals, including 3-oxo-C12-HSL, PQS, and C4-HSL, consequently resulting in the downregulation of biofilm formation and virulence factor production of P. aeruginosa (53). Furthermore, BDSF and some of its derivatives inhibit the type III secretion system (T3SS) of P. aeruginosa with stronger activity than that for the inhibition of the QS systems, suggesting that BDSF may interfere with the QS systems and T3SS of P. aeruginosa through two independent signaling pathways. However, the specific mechanism by which BDSF interferes with the QS systems and T3SS of P. aeruginosa still needs further investigation.
Moreover, BDSF was reported to inhibit the dimorphic transition of Candida albicans at physiologically relevant concentrations and thereby to block the biofilm formation of fungi (25, 54). Real-time reverse transcription-quantitative PCR (qRT-PCR) analysis showed that BDSF could downregulate the expression of ALS1 and EAP1, which are involved in C. albicans adhesion, and upregulate the expression of YWP1, which exerts an inverse effect on adherence (55). In addition, BDSF exerted a protective effect in an experimental mouse model of Candida vaginitis by inhibiting virulence factors (56). Further analyses showed that either B. cenocepacia cocultured with C. albicans or the exogenous addition of physiologically relevant levels of BDSF strongly inhibited the formation of C. albicans germ tubes, which suggests that the signal might play a role in cross-kingdom microbial competition in ecosystems (25).
THE SIGNALING PATHWAY OF BDSF IN B. CENOCEPACIA
The signal perception mechanism of BDSF.
Previous studies have shown that RpfF and the two-component system RpfCG are responsible for DSF signal molecule synthesis and signal transduction in X. campestris pv. campestris, respectively (57, 58). RpfFBc is the key enzyme that produces BDSF; it possesses homology with RpfF and is highly conserved in all BCC family species (41) (Table 1), but the homologous proteins RpfCG and the rpf gene cluster could not be found in B. cenocepacia, suggesting that there is a different mechanism that is in charge of BDSF signal perception and transduction. Deng et al. identified that BCAM0580 (designated rpfR), which is located next to the BDSF biosynthase gene rpfFBc, was responsible for encoding a protein containing a PAS domain, a GGDEF domain, and an EAL domain (59). Their study showed that the deletion of rpfR shared similar phenotypic changes with the rpfFBc mutant with an increased intracellular c-di-GMP level. Furthermore, the deletion of rpfFBc did not affect the transcription level of rpfR, suggesting that BDSF may influence the activity of RpfR through ligand-protein interactions. This hypothesis was confirmed by isothermal titration calorimetry and circular dichroism (CD) analyses, which showed that BDSF binds to RpfR in vitro with strong affinity and causes a conformational change, and the PAS domain is required for BDSF binding (59) (Fig. 2).
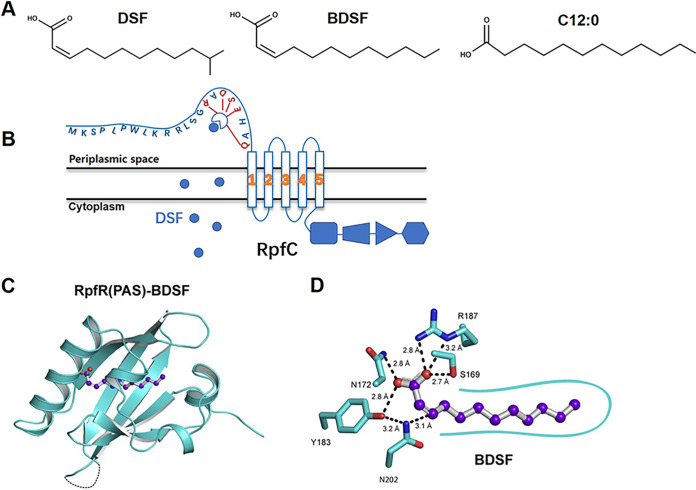
FIG 2.
Molecular models of the sensing mechanisms of DSF and BDSF signals. (A) The chemical structures of DSF, BDSF, and C12:0. (B) Molecular model of RpfC to sense DSF. RpfC possesses five transmembrane domains, and five central residues (Arg15, Asp17, Ser18, Glu19, and Gln22) are essential for the sensing of DSF. (C and D) Cartoon representation of RpfRCt (cyan) in complex with BDSF. RpfR-BDSF hydrophobic interactions are depicted as a smooth cyan contour line. (Panels C and D are reproduced from reference 60.)
In 2019, Waldron et al. reported the X-ray crystal structure of the RpfR homolog of Cronobacter turicensis (RpfRCt) in complex with BDSF, which determined in detail the molecular basis of the RpfR receptor to sense, perceive, and specifically bind to its ligands (60). It was found that RpfRCt shares 56% identity with RpfR of B. cenocepacia. Compared with C12:0 and trans-2-dodecenoic acid, the cis-2 double bond of BDSF may contribute to the specificity and affinity for binding with RpfR. Analysis of the crystal structure showed that arginine 187 (Arg187) of RpfRCt forms two hydrogen bonds with BDSF, while there was no hydrogen bond formed between RpfRCt and C12:0 (Fig. 2). This conformational difference is the result of the additional H bond mediated by Arg187 with a different rotameric configuration that caused much of the Arg187 side chain to be solvent exposed. This conformational change may play a critical role in regulating phosphodiesterase activity (60), which is described in the next section. In addition, the hydroxyl of Tyr183 interacts with the conserved Asn202 side chain amide N-H and strengthens the interaction between BDSF and Asn202 of RpfRCt. Furthermore, BDSF showed a rigid architecture that would reduce entropy loss compared with saturated fatty acids (60). These findings suggest that the cis-2 double bond of BDSF plays a key role in signal perception.
The transduction process of BDSF signals.
c-di-GMP is a ubiquitous second-messenger molecule in bacteria that regulates a variety of physiological functions, including biofilm formation, cell differentiation, the production of pathogenic factors, and so on (61–64). The intracellular c-di-GMP concentration is modulated by the opposing activities of diguanylate cyclase (DGC) and cyclic nucleotide phosphodiesterase (PDE), which in turn are regulated by extra- or intracellular factors (61, 64). DGCs produce c-di-GMP from two molecules of GTP, and PDEs hydrolyze c-di-GMP to linear pGpG or two GMP molecules. The GGDEF motif is essential for the enzymatic activity of DGCs, while PDE activity is associated with the EAL and HD-GYP domains (64–68). Many Gram-negative bacteria sense the population density of the community and control multiple phenotypes in response to changes in the environment through QS and intracellular c-di-GMP systems. For example, RpfC activates the phosphodiesterase activity of RpfG to degrade c-di-GMP after sensing DSF signal molecules in X. campestris pv. campestris (57, 58, 65, 69). By comparing the intracellular c-di-GMP levels between the RpfFBc mutant and the wild type, the results showed that the level of intracellular c-di-GMP of the rpfFBc mutant was significantly higher than that of the wild-type strain and returned to the level of the wild-type strain when exogenous BDSF was added, indicating that BDSF may be involved in the metabolism of c-di-GMP in some way (59).
The GGDEF and EAL domains usually refer to features of diguanylate cyclase and phosphodiesterase, which are involved in c-di-GMP biosynthesis and degradation, respectively (64). In B. cenocepacia, the phenotypes of the rpfR mutant could not be rescued by complementation with the GGDEF (DGC) domain alone, whereas the EAL (PDE) domain in fact complements the impaired phenotypes, comparably to the full-length protein RpfR, suggesting that BDSF activates the phosphodiesterase activity of RpfR, which causes a decrease in the level of intracellular c-di-GMP (59). Similar results were observed in other BCC species, such as the rpfR mutant strain of Burkholderia lata, which was found to show decreases in the growth rate, swimming motility, AHL production, and pathogenicity in Caenorhabditis elegans but with an increase in biofilm formation, suggesting that rpfR plays a vital role in various phenotypes of BCC species by regulating the level of intracellular c-di-GMP (70). Furthermore, the BDSF-induced allosteric conformational change through its interaction with the PAS domain was the factor that stimulated the c-di-GMP phosphodiesterase activity of RpfR (59), which was consistent with the BDSF-RpfRCt complex crystal structure in which the binding of BDSF to RpfRCt led to RpfRCt-R187 having a different rotameric configuration (60). These results suggest that the BDSF-dependent RpfFR QS system and c-di-GMP are intricately intertwined, and BDSF activates RpfR to degrade intracellular c-di-GMP; subsequently, the signal is transmitted to downstream components to change phenotypes.
RpfR not only acts as a QS signal receptor and a c-di-GMP phosphodiesterase but also inhibits BDSF synthesis by interacting directly with RpfFBc. At the PAS-like domain of the RpfR N terminus, there is a highly conserved and undescribed region of RpfR, which refers to the RpfFCt interaction domain (FI) (60). The RpfR FI deletion mutant strain produced a lower level of c-di-GMP than the wild-type strain, while the RpfR deletion mutant strain promoted its concentration, which could be explained by the regulation of BDSF on RpfR PDE activity or might be the direct effect of the deletion of FI on the PDE activity of RpfR. The X-ray crystal structure of the RpfFBc-RpfRCt complex showed a heterohexamer consisting of three RpfRCt and RpfFBc protomers. Two RpfFBc protomers interact with one RpfRCt protomer near their homodimerization interfaces and lead to the RpfFBc acyl-ACP substrate tunnel being blocked by the proximity of RpfRCt and the formation of an interaction interface, consistent with the inhibition of RpfFBc thioesterase activity in the presence of RpfRCt (60). As the receptor of the BDSF QS signal, RpfR, in turn, can inhibit its synthesis, suggesting that it plays a critical role in the feedback regulation loop. However, the mechanism by which the extracellular level of BDSF significantly decreases after reaching the accumulation peak is still unclear (42).
Intriguingly, Yang et al. showed that RpfR not only possesses c-di-GMP phosphodiesterase activity but also acts as a c-di-GMP sensor (71). Their study identified a global regulator (named GtrR) that was a key downstream component that could interact with RpfR and regulate the expression of genes in B. cenocepacia. An electrophoretic mobility shift assay (EMSA) suggested that RpfR enhanced the binding of GtrR to the target gene promoter; however, the ability of the RpfR-GtrR complex to bind to promoter DNA was reduced by the addition of exogenous c-di-GMP. This is caused by the binding of c-di-GMP to RpfR with an estimated dissociation constant (Kd) of 2.92 ± 0.26 μM (Fig. 3). Given that RpfR exhibited low PDE activity in the absence of BDSF, and c-di-GMP binds with GtrR-RpfR to form a ternary complex to inhibit the regulatory activity of GtrR on target gene expression, we concluded that both BDSF and c-di-GMP are the signal ligands of RpfR, which bind with RpfR to exert different functions. Moreover, both RpfR and GtrR homologs are present in diverse Gram-negative bacteria, suggesting that the BDSF-type QS system is widespread (71) (Table 1). In addition, the GtrR-RpfR complex can regulate the transcription of cepI (71), and the cep system regulator, CepR, can bind to the promoter of Bcal3178, which is a LysR family transcriptional regulator, and enhance the expression of Bcal3178. Bcal3178 controls some QS-regulated functions, but one study revealed that there was no binding between GtrR and the Bcal3178 promoter (72) (Fig. 3).
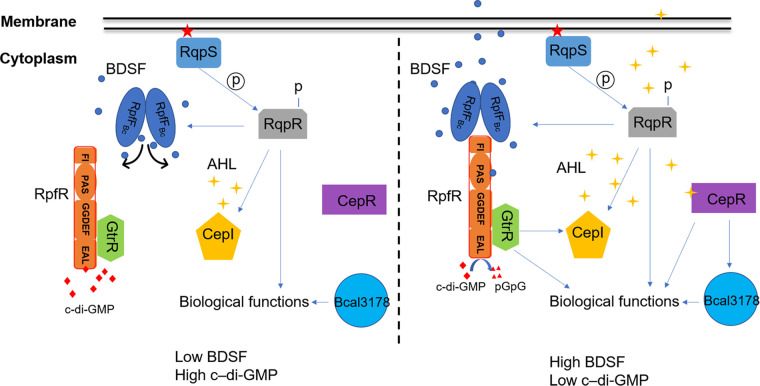
FIG 3.
Schematic representation of the BDSF-dependent QS systems in B. cenocepacia. RqpSR is a two-component regulatory system. RqpS perceives signals and activates RqpR, which positively regulates the expression of the rpfFBc and cepI genes, which are required for the synthesis of BDSF and AHL signals, respectively. When the intracellular c-di-GMP level is high, it combines with GtrR-RpfR to form a ternary complex, which inhibits the regulatory activity of GtrR. When the bacterial density reaches a certain threshold, RpfR combines with BDSF to enhance its PDE activity, decreases the level of c-di-GMP, and further closely integrates with GtrR to regulate gene expression. In addition, activated RpfR binds to RpfFBc through the FI domain, which in turn inhibits the synthesis of BDSF (the black arrow represents the BDSF synthesis channel). CepR, the signal receptor of AHLs, can bind to the promoter of Bcal3178 and enhance the expression of Bcal3178, which controls the expression of some QS-regulated genes.
CONCLUDING REMARKS
Previous studies have shown that many bacterial pathogens coordinate the expression of virulence-related genes through QS (2). The QS signaling molecule cis-2-dodecenoic acid, which was also named BDSF, was first identified in B. cenocepacia. It was demonstrated that BDSF is a novel DSF family QS signaling molecule that regulates a variety of phenotypes, including biofilm formation, motility, and virulence factor production, in B. cenocepacia and some other bacterial species (25, 59). Studies have shown that BDSF can be detected in all tested BCC species; additionally, homologs of the BDSF synthase RpfFBc, the receptor RpfR, and the key downstream component of the BDSF QS system GtrR exist in many species (Table 1), suggesting that BDSF is an important QS signaling molecule with widespread conservation.
In recent years, due to the abuse of antibiotics, the major challenge for us is to develop novel therapeutic strategies to treat antibiotic-resistant pathogens (73). The DSF-type QS system exists widely in bacteria and plays a vital role in the regulation of pathogenicity, showing great potential for the development of antivirulence therapy by interfering with this kind of QS system to attenuate the virulence of pathogens rather than kill them (73). The development of QS inhibitors as new drugs for the treatment of bacterial infections has attracted great attention (74). Cui et al. reported a QS signal inhibitor (cis-14-methylpentadec-2-enoic acid, also known as 14-Me-C16:Δ2) that is derived from unsaturated fatty acids and showed great activities to interfere with BDSF signaling and the production of virulence factors but did not inhibit the growth rate of the pathogen (75). Their data showed that 14-Me-C16:Δ2 targeted the QS signal synthase RpfFBc to inhibit the production of BDSF and AHL signals, leading to defects in the pathogenic phenotypes. Remarkably, 14-Me-C16:Δ2 displayed a similar effect on various Burkholderia species. Likewise, some QS inhibitors have also been applied to other pathogens and showed a marked effect on attenuating QS-regulated virulence (73). In addition, it was previously reported that hosts can monitor QS signals to regulate their own immune response (76). As a QS signal widely distributed in various microorganisms, BDSF is likely to interact with the host, which might be used for the development of new vaccines to enhance the host immune response to control infection in the future.
Contributor Information
Yinyue Deng, Email: dengyle@mail.sysu.edu.cn.
Gladys Alexandre, University of Tennessee at Knoxville.
REFERENCES
- 1.Papenfort K, Bassler BL. 2016. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol 14:576–588. 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang LH, Dong YH. 2004. Quorum sensing and signal interference: diverse implications. Mol Microbiol 53:1563–1571. 10.1111/j.1365-2958.2004.04234.x. [DOI] [PubMed] [Google Scholar]
- 3.Kleerebezem M, Quadri LE, Kuipers OP, de Vos WM. 1997. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol Microbiol 24:895–904. 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee G, Ray AK. 2017. Quorum-sensing network-associated gene regulation in Gram-positive bacteria. Acta Microbiol Immunol Hung 64:439–453. 10.1556/030.64.2017.040. [DOI] [PubMed] [Google Scholar]
- 5.Withers H, Swift S, Williams P. 2001. Quorum sensing as an integral component of gene regulatory networks in Gram-negative bacteria. Curr Opin Microbiol 4:186–193. 10.1016/S1369-5274(00)00187-9. [DOI] [PubMed] [Google Scholar]
- 6.Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP. 2001. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol Rev 25:365–404. 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- 7.Fuqua C, Greenberg EP. 2002. Listening in on bacteria: acyl-homoserine lactone signalling. Nat Rev Mol Cell Biol 3:685–695. 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 8.Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR. 2005. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat Rev Microbiol 3:383–396. 10.1038/nrmicro1146. [DOI] [PubMed] [Google Scholar]
- 9.Déziel E, Lépine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG. 2004. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci USA 101:1339–1344. 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. 2003. Bacteria-host communication: the language of hormones. Proc Natl Acad Sci USA 100:8951–8956. 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diggle SP, Lumjiaktase P, Dipilato F, Winzer K, Kunakorn M, Barrett DA, Chhabra SR, Camara M, Williams P. 2006. Functional genetic analysis reveals a 2-alkyl-4-quinolone signaling system in the human pathogen Burkholderia pseudomallei and related bacteria. Chem Biol 13:701–710. 10.1016/j.chembiol.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Tommonaro G, Abbamondi GR, Iodice C, Tait K, De Rosa S. 2012. Diketopiperazines produced by the halophilic archaeon, Haloterrigena hispanica, activate AHL bioreporters. Microb Ecol 63:490–495. 10.1007/s00248-011-9980-y. [DOI] [PubMed] [Google Scholar]
- 13.Loh J, Carlson RW, York WS, Stacey G. 2002. Bradyoxetin, a unique chemical signal involved in symbiotic gene regulation. Proc Natl Acad Sci USA 99:14446–14451. 10.1073/pnas.222336799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang LH, He Y, Gao Y, Wu JE, Dong YH, He C, Wang SX, Weng LX, Xu JL, Tay L, Fang RX, Zhang LH. 2004. A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol Microbiol 51:903–912. 10.1046/j.1365-2958.2003.03883.x. [DOI] [PubMed] [Google Scholar]
- 15.Barber CE, Tang JL, Feng JX, Pan MQ, Wilson TJ, Slater H, Dow JM, Williams P, Daniels MJ. 1997. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol Microbiol 24:555–566. 10.1046/j.1365-2958.1997.3721736.x. [DOI] [PubMed] [Google Scholar]
- 16.Deng Y, Wu J, Tao F, Zhang LH. 2011. Listening to a new language: DSF-based quorum sensing in Gram-negative bacteria. Chem Rev 111:160–173. 10.1021/cr100354f. [DOI] [PubMed] [Google Scholar]
- 17.Flavier AB, Clough SJ, Schell MA, Denny TP. 1997. Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol Microbiol 26:251–259. 10.1046/j.1365-2958.1997.5661945.x. [DOI] [PubMed] [Google Scholar]
- 18.Song S, Yin W, Sun X, Cui B, Huang L, Li P, Yang L, Zhou J, Deng Y. 2020. Anthranilic acid from Ralstonia solanacearum plays dual roles in intraspecies signalling and inter-kingdom communication. ISME J 14:2248–2260. 10.1038/s41396-020-0682-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slater H, Alvarez-Morales A, Barber CE, Daniels MJ, Dow JM. 2000. A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol Microbiol 38:986–1003. 10.1046/j.1365-2958.2000.02196.x. [DOI] [PubMed] [Google Scholar]
- 20.He YW, Zhang LH. 2008. Quorum sensing and virulence regulation in Xanthomonas campestris. FEMS Microbiol Rev 32:842–857. 10.1111/j.1574-6976.2008.00120.x. [DOI] [PubMed] [Google Scholar]
- 21.Wang FF, Qian W. 2019. The roles of histidine kinases in sensing host plant and cell-cell communication signal in a phytopathogenic bacterium. Philos Trans R Soc Lond B Biol Sci 374:20180311. 10.1098/rstb.2018.0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryan RP, Dow JM. 2011. Communication with a growing family: diffusible signal factor (DSF) signaling in bacteria. Trends Microbiol 19:145–152. 10.1016/j.tim.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Zhou L, Zhang LH, Camara M, He YW. 2017. The DSF family of quorum sensing signals: diversity, biosynthesis, and turnover. Trends Microbiol 25:293–303. 10.1016/j.tim.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 24.Ryan RP, An SQ, Allan JH, McCarthy Y, Dow JM. 2015. The DSF family of cell-cell signals: an expanding class of bacterial virulence regulators. PLoS Pathog 11:e1004986. 10.1371/journal.ppat.1004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boon C, Deng Y, Wang LH, He Y, Xu JL, Fan Y, Pan SQ, Zhang LH. 2008. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J 2:27–36. 10.1038/ismej.2007.76. [DOI] [PubMed] [Google Scholar]
- 26.Depoorter E, Bull MJ, Peeters C, Coenye T, Vandamme P, Mahenthiralingam E. 2016. Burkholderia: an update on taxonomy and biotechnological potential as antibiotic producers. Appl Microbiol Biotechnol 100:5215–5229. 10.1007/s00253-016-7520-x. [DOI] [PubMed] [Google Scholar]
- 27.Bach E, Sant’Anna FH, Magrich Dos Passos JF, Balsanelli E, de Baura VA, Pedrosa FDO, de Souza EM, Passaglia LMP. 2017. Detection of misidentifications of species from the Burkholderia cepacia complex and description of a new member, the soil bacterium Burkholderia catarinensis sp. nov. Pathog Dis 75:ftx076. 10.1093/femspd/ftx076. [DOI] [PubMed] [Google Scholar]
- 28.Scoffone VC, Barbieri G, Buroni S, Scarselli M, Pizza M, Rappuoli R, Riccardi G. 2020. Vaccines to overcome antibiotic resistance: the challenge of Burkholderia cenocepacia. Trends Microbiol 28:315–326. 10.1016/j.tim.2019.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Chiarini L, Bevivino A, Dalmastri C, Tabacchioni S, Visca P. 2006. Burkholderia cepacia complex species: health hazards and biotechnological potential. Trends Microbiol 14:277–286. 10.1016/j.tim.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 30.Scoffone VC, Chiarelli LR, Trespidi G, Mentasti M, Riccardi G, Buroni S. 2017. Burkholderia cenocepacia infections in cystic fibrosis patients: drug resistance and therapeutic approaches. Front Microbiol 8:1592. 10.3389/fmicb.2017.01592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drevinek P, Mahenthiralingam E. 2010. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin Microbiol Infect 16:821–830. 10.1111/j.1469-0691.2010.03237.x. [DOI] [PubMed] [Google Scholar]
- 32.Loutet SA, Valvano MA. 2010. A decade of Burkholderia cenocepacia virulence determinant research. Infect Immun 78:4088–4100. 10.1128/IAI.00212-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inhulsen S, Aguilar C, Schmid N, Suppiger A, Riedel K, Eberl L. 2012. Identification of functions linking quorum sensing with biofilm formation in Burkholderia cenocepacia H111. Microbiologyopen 1:225–242. 10.1002/mbo3.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sokol PA, Malott RJ, Riedel K, Eberl L. 2007. Communication systems in the genus Burkholderia: global regulators and targets for novel antipathogenic drugs. Future Microbiol 2:555–563. 10.2217/17460913.2.5.555. [DOI] [PubMed] [Google Scholar]
- 35.Gotschlich A, Huber B, Geisenberger O, Togl A, Steidle A, Riedel K, Hill P, Tummler B, Vandamme P, Middleton B, Camara M, Williams P, Hardman A, Eberl L. 2001. Synthesis of multiple N-acylhomoserine lactones is wide-spread among the members of the Burkholderia cepacia complex. Syst Appl Microbiol 24:1–14. 10.1078/0723-2020-00013. [DOI] [PubMed] [Google Scholar]
- 36.Lutter E, Lewenza S, Dennis JJ, Visser MB, Sokol PA. 2001. Distribution of quorum-sensing genes in the Burkholderia cepacia complex. Infect Immun 69:4661–4666. 10.1128/IAI.69.7.4661-4666.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suppiger A, Schmid N, Aguilar C, Pessi G, Eberl L. 2013. Two quorum sensing systems control biofilm formation and virulence in members of the Burkholderia cepacia complex. Virulence 4:400–409. 10.4161/viru.25338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim Y, Chhor G, Tsai CS, Fox G, Chen CS, Winans NJ, Jedrzejczak R, Joachimiak A, Winans SC. 2017. X-ray crystal structures of the pheromone-binding domains of two quorum-hindered transcription factors, YenR of Yersinia enterocolitica and CepR2 of Burkholderia cenocepacia. Proteins 85:1831–1844. 10.1002/prot.25336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malott RJ, O’Grady EP, Toller J, Inhulsen S, Eberl L, Sokol PA. 2009. A Burkholderia cenocepacia orphan LuxR homolog is involved in quorum-sensing regulation. J Bacteriol 191:2447–2460. 10.1128/JB.01746-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan GT, Wei YP, Winans SC. 2013. A LuxR-type repressor of Burkholderia cenocepacia inhibits transcription via antiactivation and is inactivated by its cognate acylhomoserine lactone. Mol Microbiol 87:94–111. 10.1111/mmi.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng YY, Wu JE, Eberl L, Zhang LH. 2010. Structural and functional characterization of diffusible signal factor family quorum-sensing signals produced by members of the Burkholderia cepacia complex. Appl Environ Microbiol 76:4675–4683. 10.1128/AEM.00480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng YY, Boon C, Eberl L, Zhang LH. 2009. Differential modulation of Burkholderia cenocepacia virulence and energy metabolism by the quorum-sensing signal BDSF and its synthase. J Bacteriol 191:7270–7278. 10.1128/JB.00681-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng YY, Lim A, Wang J, Zhou TL, Chen SH, Lee J, Dong YH, Zhang LH. 2013. cis-2-Dodecenoic acid quorum sensing system modulates N-acyl homoserine lactone production through RpfR and cyclic di-GMP turnover in Burkholderia cenocepacia. BMC Microbiol 13:148. 10.1186/1471-2180-13-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bi HK, Christensen QH, Feng YJ, Wang HH, Cronan JE. 2012. The Burkholderia cenocepacia BDSF quorum sensing fatty acid is synthesized by a bifunctional crotonase homologue having both dehydratase and thioesterase activities. Mol Microbiol 83:840–855. 10.1111/j.1365-2958.2012.07968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui CY, Yang CX, Song SH, Fu SN, Sun XY, Yang L, He F, Zhang LH, Zhang YL, Deng YY. 2018. A novel two-component system modulates quorum sensing and pathogenicity in Burkholderia cenocepacia. Mol Microbiol 108:32–44. 10.1111/mmi.13915. [DOI] [PubMed] [Google Scholar]
- 46.Suppiger A, Aguilar C, Eberl L. 2016. Evidence for the widespread production of DSF family signal molecules by members of the genus Burkholderia by the aid of novel biosensors. Environ Microbiol Rep 8:38–44. 10.1111/1758-2229.12348. [DOI] [PubMed] [Google Scholar]
- 47.Schmid N, Pessi G, Deng YY, Aguilar C, Carlier AL, Grunau A, Omasits U, Zhang LH, Ahrens CH, Eberl L. 2012. The AHL- and BDSF-dependent quorum sensing systems control specific and overlapping sets of genes in Burkholderia cenocepacia H111. PLoS One 7:e49966. 10.1371/journal.pone.0049966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryan RP, McCarthy Y, Watt SA, Niehaus K, Dow JM. 2009. Intraspecies signaling involving the diffusible signal factor BDSF (cis-2-dodecenoic acid) influences virulence in Burkholderia cenocepacia. J Bacteriol 191:5013–5019. 10.1128/JB.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corbett CR, Burtnick MN, Kooi C, Woods DE, Sokol PA. 2003. An extracellular zinc metalloprotease gene of Burkholderia cepacia. Microbiology (Reading) 149:2263–2271. 10.1099/mic.0.26243-0. [DOI] [PubMed] [Google Scholar]
- 50.Kooi C, Subsin B, Chen R, Pohorelic B, Sokol PA. 2006. Burkholderia cenocepacia ZmpB is a broad-specificity zinc metalloprotease involved in virulence. Infect Immun 74:4083–4093. 10.1128/IAI.00297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Twomey KB, O’Connell OJ, McCarthy Y, Dow JM, O’Toole GA, Plant BJ, Ryan RP. 2012. Bacterial cis-2-unsaturated fatty acids found in the cystic fibrosis airway modulate virulence and persistence of Pseudomonas aeruginosa. ISME J 6:939–950. 10.1038/ismej.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryan RP, Fouhy Y, Garcia BF, Watt SA, Niehaus K, Yang L, Tolker-Nielsen T, Dow JM. 2008. Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol Microbiol 68:75–86. 10.1111/j.1365-2958.2008.06132.x. [DOI] [PubMed] [Google Scholar]
- 53.Deng YY, Boon C, Chen SH, Lim A, Zhang LH. 2013. cis-2-Dodecenoic acid signal modulates virulence of Pseudomonas aeruginosa through interference with quorum sensing systems and T3SS. BMC Microbiol 13:231. 10.1186/1471-2180-13-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang YQ, Cai C, Yang YX, Weng LX, Wang LH. 2011. Blocking of Candida albicans biofilm formation by cis-2-dodecenoic acid and trans-2-dodecenoic acid. J Med Microbiol 60:1643–1650. 10.1099/jmm.0.029058-0. [DOI] [PubMed] [Google Scholar]
- 55.Tian J, Weng LX, Zhang YQ, Wang LH. 2013. BDSF inhibits Candida albicans adherence to urinary catheters. Microb Pathog 64:33–38. 10.1016/j.micpath.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 56.Yang DL, Zhang YQ, Hu YL, Weng LX, Zeng GS, Wang LH. 2018. Protective effects of cis-2-dodecenoic acid in an experimental mouse model of vaginal candidiasis. Biomed Environ Sci 31:816–828. 10.3967/bes2018.109. [DOI] [PubMed] [Google Scholar]
- 57.He YW, Xu M, Lin K, Ng YJA, Wen CM, Wang LH, Liu ZD, Zhang HB, Dong YH, Dow JM, Zhang LH. 2006. Genome scale analysis of diffusible signal factor regulon in Xanthomonas campestris pv. campestris: identification of novel cell-cell communication-dependent genes and functions. Mol Microbiol 59:610–622. 10.1111/j.1365-2958.2005.04961.x. [DOI] [PubMed] [Google Scholar]
- 58.He YW, Wang C, Zhou L, Song HW, Dow JM, Zhang LH. 2006. Dual signaling functions of the hybrid sensor kinase RpfC of Xanthomonas campestris involve either phosphorelay or receiver domain-protein interaction. J Biol Chem 281:33414–33421. 10.1074/jbc.M606571200. [DOI] [PubMed] [Google Scholar]
- 59.Deng YY, Schmid N, Wang C, Wang JH, Pessi G, Wu DH, Lee J, Aguilar C, Ahrens CH, Chang CQ, Song HW, Eberl L, Zhang LH. 2012. cis-2-Dodecenoic acid receptor RpfR links quorum-sensing signal perception with regulation of virulence through cyclic dimeric guanosine monophosphate turnover. Proc Natl Acad Sci USA 109:15479–15484. 10.1073/pnas.1205037109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waldron EJ, Snyder D, Fernandez NL, Sileo E, Inoyama D, Freundlich JS, Waters CM, Cooper VS, Neiditch MB. 2019. Structural basis of DSF recognition by its receptor RpfR and its regulatory interaction with the DSF synthase RpfF. PLoS Biol 17:e3000123. 10.1371/journal.pbio.3000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, Volman G, Mayer R, Ross P, Amikam D, Weinhouse H, Cohen A, Sapir S, Ohana P, Benziman M. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol 180:4416–4425. 10.1128/JB.180.17.4416-4425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Romling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Camilli A, Bassler BL. 2006. Bacterial small-molecule signaling pathways. Science 311:1113–1116. 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simm R, Morr M, Kader A, Nimtz M, Romling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol 53:1123–1134. 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 65.Dow JM, Crossman L, Findlay K, He YQ, Feng JX, Tang JL. 2003. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc Natl Acad Sci USA 100:10995–11000. 10.1073/pnas.1833360100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schirmer T, Jenal U. 2009. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol 7:724–735. 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 67.Andrade MO, Alegria MC, Guzzo CR, Docena C, Rosa MCP, Ramos CHI, Farah CS. 2006. The HD-GYP domain of RpfG mediates a direct linkage between the Rpf quorum-sensing pathway and a subset of diguanylate cyclase proteins in the phytopathogen Xanthomonas axonopodis pv citri. Mol Microbiol 62:537–551. 10.1111/j.1365-2958.2006.05386.x. [DOI] [PubMed] [Google Scholar]
- 68.Ryan RP, Lucey J, O’Donovan K, McCarthy Y, Yang L, Tolker-Nielsen T, Dow JM. 2009. HD-GYP domain proteins regulate biofilm formation and virulence in Pseudomonas aeruginosa. Environ Microbiol 11:1126–1136. 10.1111/j.1462-2920.2008.01842.x. [DOI] [PubMed] [Google Scholar]
- 69.Cai Z, Yuan ZH, Zhang H, Pan Y, Wu Y, Tian XQ, Wang FF, Wang L, Qian W. 2017. Fatty acid DSF binds and allosterically activates histidine kinase RpfC of phytopathogenic bacterium Xanthomonas campestris pv. campestris to regulate quorum-sensing and virulence. PLoS Pathog 13:e1006304. 10.1371/journal.ppat.1006304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jung H-I, Kim Y-J, Lee Y-J, Lee H-S, Lee J-K, Kim S-K. 2017. Mutation of the cyclic di-GMP phosphodiesterase gene in Burkholderia lata SK875 attenuates virulence and enhances biofilm formation. J Microbiol 55:800–808. 10.1007/s12275-017-7374-7. [DOI] [PubMed] [Google Scholar]
- 71.Yang CX, Cui CY, Ye QM, Kan JH, Fu SN, Song SH, Huang YT, He F, Zhang LH, Jia YT, Gao YG, Harwood CS, Deng YY. 2017. Burkholderia cenocepacia integrates cis-2-dodecenoic acid and cyclic dimeric guanosine monophosphate signals to control virulence. Proc Natl Acad Sci USA 114:13006–13011. 10.1073/pnas.1709048114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang K, Li X, Yang C, Song S, Cui C, Zhou X, Deng Y. 2021. A LysR family transcriptional regulator modulates Burkholderia cenocepacia biofilm formation and protease production. Appl Environ Microbiol 87:e00202-21. 10.1128/AEM.00202-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Defoirdt T. 2018. Quorum-sensing systems as targets for antivirulence therapy. Trends Microbiol 26:313–328. 10.1016/j.tim.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 74.O’Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, Bassler BL. 2013. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci USA 110:17981–17986. 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cui CY, Song SH, Yang CX, Sun XY, Huang YT, Li K, Zhao S, Zhang YL, Deng YY. 2019. Disruption of quorum sensing and virulence in Burkholderia cenocepacia by a structural analogue of the cis-2-dodecenoic acid signal. Appl Environ Microbiol 85:e00105-19. 10.1128/AEM.00105-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moura-Alves P, Puyskens A, Stinn A, Klemm M, Guhlich-Bornhof U, Dorhoi A, Furkert J, Kreuchwig A, Protze J, Lozza L, Pei G, Saikali P, Perdomo C, Mollenkopf HJ, Hurwitz R, Kirschhoefer F, Brenner-Weiss G, Weiner J, III, Oschkinat H, Kolbe M, Krause G, Kaufmann SHE. 2019. Host monitoring of quorum sensing during Pseudomonas aeruginosa infection. Science 366:eaaw1629. 10.1126/science.aaw1629. [DOI] [PubMed] [Google Scholar]