Abstract
Detection of mutations by multiplex real-time RT-PCR is a widely used method for the screening of SARS-CoV-2 variants, but this method has several limitations. We describe three cases in which a Mu strain containing the mutation K417N was initially misclassified as the Beta variant. We recommend the detection of P681H to distinguish between these two variants. Our experience highlights the importance of keeping track of new variants and mutations in order to adapt the current workflows.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first described in Wuhan in December 2019, and its whole genome was sequenced shortly thereafter [1]. Genomic surveillance of the pandemic has allowed rapid identification of emerging new variants, which could represent a risk of increased transmission, higher virulence, or escape from immune response or vaccination [2]. Those variants that could pose a public health risk are classified by the World Health Organization as Variants of Interest (VOIs) and Variants of Concern (VOCs). The classification of VOIs and VOCs is periodically adjusted, and the latest version can be found at https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/.
For this reason, microbiology laboratories all over the world have rapidly developed screening strategies for detection and control of circulating variants. Next-generation sequencing (NGS) is the reference method for variant identification. However, this method is time-consuming, expensive, and is not available in all routine laboratories. Therefore, NGS is not a good option for large-scale screening and detection of specific mutations, and multiplex real-time reverse transcription polymerase chain reaction (rRT-PCR) is the most widely used approach to classify circulating variants [3–6]. rRT-PCR is a useful and fast tool for assessing the current epidemiology of the pandemic, allowing prompt adaptation of pandemic control strategies. However, this strategy has some limitations, some of which have been addressed before [7]. Using rRT-PCR, only known mutations are studied, so new mutations can go undetected. Also, new variants may arise that contain the same mutations as previously known variants, as SARS-CoV-2 seems to exhibit convergent evolution [8]. Both situations can lead to misidentification and to a delay in the detection of new circulating variants.
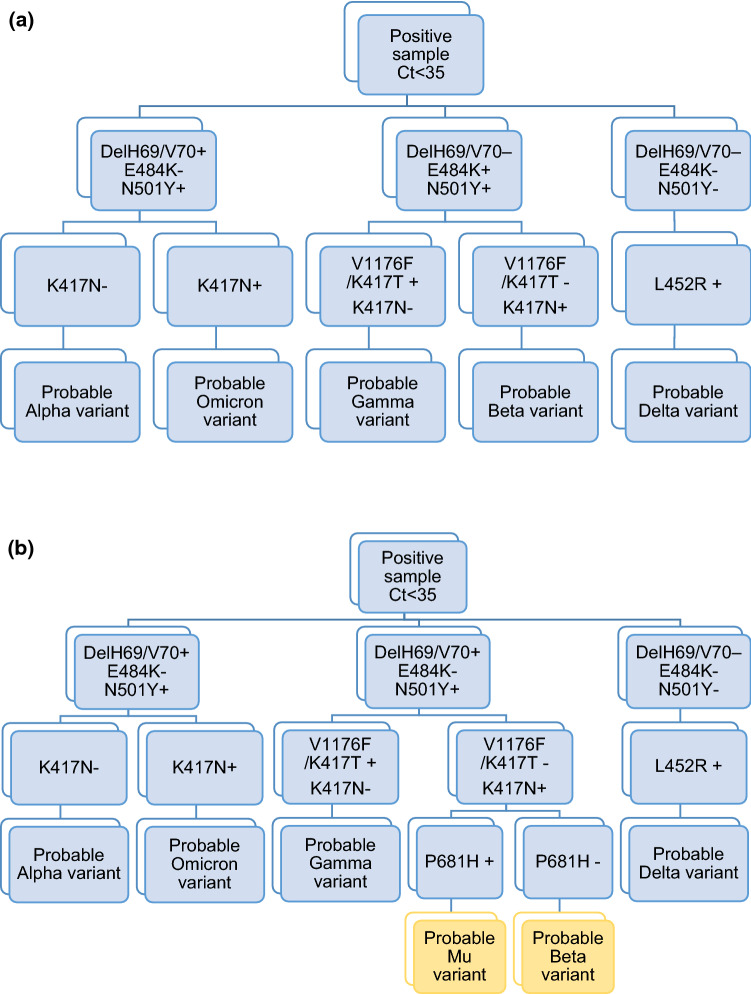
In our laboratory, we routinely screen all SARS-CoV-2-positive samples with a Ct value <35 using two multiplex rRT-PCR mutation assays (Fig. 1a). The first assay, Allplex SARS-CoV-2 Variants I Assay (Seegene, Korea), includes mutations delH69/V70, N501Y, and E484K. The findings reported in this work were made before the description of the Omicron variant. At that time, samples presenting delH69/V70 and N501Y mutations were classified as “probable Alpha variant”. Any other mutation combination or the absence of any mutations required the use of a second rRT-PCR mutation assay, Allplex SARS-CoV-2 Variants II Assay (Seegene, Korea), which screens for the mutations K417N, K417T, L452R, and W152C. Interpretation of the results was made following the manufacturer’s instructions.
Fig. 1.
(a) Routine workflow before detection of Mu variants with the K417N mutation. (b) Proposed new workflow
Samples containing E484K and N501Y mutations and lacking delH69/V70 mutation were also subjected to rRT-PCR melting curve analysis of K417N and V1176F (VirSNiP Assays, TIB MOLBIOL, Germany), as this assay was commercially available before the Allplex SARS-CoV-2 Variants II Assay. Samples showing infrequent mutation combinations and randomly selected positive samples were submitted for NGS for epidemiological assessment of the current state of the pandemic.
We believe that many laboratories follow this workflow or similar ones, as it is a simple and fast strategy to identify variants.
Following the scheme described above, in August 2021, we detected three strains that presented K417N, N501Y, and E484K mutations but did not present delH69/V70, W152C, K417T, L452R, or V1176F mutations. Based on the rRT-PCR results, all three strains were classified as “probable Beta variant” and subjected to NGS for verification.
NGS was performed using Ion AmpliSeq SARS-CoV-2 Research Panel (Thermo Fisher Scientific, USA) for library preparation and run on an Ion GeneStudio S5 System (Thermo Fisher Scientific, USA) with a 540 chip, following the manufacturer’s instructions. Genome sequence assembly was performed using the IRMA report plugin. Subsequently, BAM files were analyzed to clean up FASTA files. The mutations detected were accepted when the coverage was >100 reads and they were present in more than 90% of the reads. If these criteria were not met, nucleotides were called as “N” in the definitive FASTA file. The resulting sequences were then run on NextClade (https://clades.nextstrain.org/) and Pangolin COVID-19 Lineage Assigner (https://pangolin.cog-uk.io/) for final classification. All three strains were classified as clade 21H in NextClade and as B.1.621 in Pango, which is compatible with the Mu variant and not with the Beta variant. After obtaining these results, a more thorough examination of the sequences was carried out, paying special attention to those positions where defining mutations (as described in CoVariants, https://covariants.org/) for the Beta and Mu variants are usually found. Table 1 shows the presence or absence of defining mutations in the S gene for the Beta and Mu variants detected by NGS. All three strains also contained the Mu-variant-defining mutations in the ORF and E genes (data not shown).
Table 1.
Analysis of defining mutations in the S gene for the Mu and Beta variants
| Beta variant mutations | Mu variant mutations | Nucleotide position | Strain A/EPI_ISL_4348481* | Strain B/EPI_ISL_4348482* | Strain C/EPI_ISL_4348483* |
|---|---|---|---|---|---|
| D80A | A21801C | Not present | Not present | Not present | |
| T95I | C21846T | Present** | Present** | Present** | |
| Y144S | A21993C | ||||
| Y145N | T21995A | ||||
| D215G | A22206G | Not present | Not present | Not present | |
| L241- | 22283-22291 | ||||
| L242- | |||||
| A243- | |||||
| R346K | G22599A | Present | Present | Present | |
| K417N | G22813T | ||||
| E484K | E484K | G23012A | |||
| N501Y | N501Y | A23063T | |||
| D614G | D614G | A23403G | |||
| P681H | C23604A | ||||
| A701V | C23664T | Not present | Not present | Not present | |
| D950N | G24410A | Present** | Present** | Present** |
*GISAID accession number
**Mutations were present in at least 50% of the reads but did not meet the criteria discussed in the text and were called as “N” in the final FASTA file.
Conclusions
Using rRT-PCR to detect mutations, we encountered a misidentification problem regarding the B.1.621 variant, which contains a K417N mutation. The B.1.621 variant was first described in January 2021 in Colombia and was classified as a Variant of Interest (VOI) due to its worldwide expansion, and it was designated as the Mu variant by WHO on August 2021 [9]. After our finding, we performed a search of Mu strains with the K417N mutation on Nextstrain (https://nextstrain.org/), where we found that sequences matching these criteria had already been included in the database since June 2021. These strains have been found in several locations and seem to be especially frequent in the United Kingdom [10].
In June 2021, the first Mu variant strain was detected in our region (Bizkaia, Basque Country). Two months later, in August 2021, a Mu variant harboring the K417N mutation was described for the first time in three different samples in our region. At the time of this writing, there are a total of 11 compatible strains from Bizkaia included in the GISAID database (https://www.gisaid.org/).
The appearance of a Mu variant with a K417N mutation requires the adaptation of the characterization workflow used in our laboratory, since Mu and Beta have the same mutations that had been used for Beta characterization. To distinguish between these two variants, we propose the detection of the P681H mutation (i.e., VirSNiP Assays, TIB MOLBIOL, Germany) when the initial screening cannot distinguish between the Mu and Beta variants, as this mutation is present in Mu but not in Beta. Figure 1 shows the previous workflow used in our laboratory (a) and the new one we propose to avoid Beta-Mu misidentification (b).
This finding shows the importance of knowing the current epidemiology, being aware of the emergence of new mutations and variants, and using up-to-date commercially available methods to avoid variant misidentification and to allow the workflow to be adapted to new circulating variants.
Funding
This research received no specific grant from any funding agency.
Declarations
Conflict of interest
None declared.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Izaskun Alejo-Cancho and Ana Gual-de-Torrella contributed equally.
References
- 1.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tao K, Tzou PL, Nouhin J, Gupta RK, de Oliveira T, Kosakovsky Pond SL, et al. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet. 2021;2021(17):1–17. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ong DSY, Koeleman JGM, Vaessen N, Breijer S, Paltansing S, de Man P. Rapid screening method for the detection of SARS-CoV-2 variants of concern. J Clin Virol. 2021;1(141):104903. doi: 10.1016/J.JCV.2021.104903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vega-Magaña N, Sánchez-Sánchez R, Hernández-Bello J, Antony Venancio-Landeros A, Peña-Rodríguez M, Alejandra Vega-Zepeda R et al (2021) RT-qPCR assays for rapid detection of the N501Y, 69-70del, K417N, and E484K SARS-CoV-2 mutations: a screening strategy to identify variants with clinical impact. Front Cell Infect Microbiol 11:1. 10.3389/fcimb.2021.672562. [DOI] [PMC free article] [PubMed]
- 5.La Rosa G, Mancini P, Bonanno Ferraro G, Veneri C, Iaconelli M, Lucentini L, et al. Rapid screening for SARS-CoV-2 variants of concern in clinical and environmental samples using nested RT-PCR assays targeting key mutations of the spike protein. Water Res. 2021;1(197):117104. doi: 10.1016/J.WATRES.2021.117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, Miller JA, Verghese M, Sibai M, Solis D, Mfuh KO, et al. Multiplex sars-cov-2 genotyping reverse transcriptase PCR for population-level variant screening and epidemiologic surveillance. J Clin Microbiol. 2021;1:59. doi: 10.1128/JCM.00859-21/SUPPL_FILE/JCM.00859-21-S0001.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camp JV, Buchta C, Jovanovic J, Puchhammer-Stöckl E, Benka B, Griesmacher A, et al. RT-PCR based SARS-CoV-2 variant screening assays require careful quality control. J Clin Virol. 2021;1(141):104905. doi: 10.1016/J.JCV.2021.104905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin DP, Weaver S, Tegally H, San JE, Shank SD, Wilkinson E, et al. The emergence and ongoing convergent evolution of the SARS-CoV-2 N501Y lineages. Cell. 2021;30(184):5189–5200.e7. doi: 10.1016/J.CELL.2021.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WHO. Tracking SARS-CoV-2 variants [Internet]. WHO 2021 [cited 2021 7]. p. 1–13. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants
- 10.Public Health England (2021) SARS-CoV-2 variants of concern and variants under investigation in England. Technical briefing 20. Online Report. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1009243/Technical_Briefing_20.pdf