PURPOSE
To stratify patients and aid clinical decision making, we developed machine learning models to predict treatment failure and radiation-induced toxicities after radiotherapy (RT) in patients with hepatocellular carcinoma across institutions.
MATERIALS AND METHODS
The models were developed using linear and nonlinear algorithms, predicting survival, nonlocal failure, radiation-induced liver disease, and lymphopenia from baseline patient and treatment parameters. The models were trained on 207 patients from Massachusetts General Hospital. Performance was quantified using Harrell's c-index, area under the curve (AUC), and accuracy in high-risk populations. Models' structures were optimized in a nested cross-validation approach to prevent overfitting. A study analysis plan was registered before external validation using 143 patients from MD Anderson Cancer Center. Clinical utility was assessed using net-benefit analysis.
RESULTS
The survival model stratified high-risk versus low-risk patients well in the external validation cohort (c-index = 0.75), better than existing risk scores. Predictions of 1-year survival and nonlocal failure were excellent (external AUC = 0.74 and 0.80, respectively), especially in the high-risk group (accuracy > 90%). Cause-of-death analysis showed differential modes of treatment failure in these cohorts and indicated that these models could be used to stratify RT patients for liver-sparing treatment regimen or combination approaches with systemic agents. Predictions of liver disease and lymphopenia were good but less robust (external AUC = 0.68 and 0.7, respectively), suggesting the need for more comprehensive consideration of dosimetry and better predictive biomarkers. The liver disease model showed excellent accuracy in the high-risk group (92%) and revealed possible interactions of platelet count with initial liver function.
CONCLUSION
Machine learning approaches can provide reliable outcome predictions in patients with hepatocellular carcinoma after RT in diverse cohorts across institutions. The excellent performance, particularly in high-risk patients, suggests novel strategies for patient stratification and treatment selection.
INTRODUCTION
Modern conformal radiotherapy (RT) is emerging as a valuable treatment option in hepatocellular carcinoma (HCC). It has been investigated as an alternative treatment option for early-stage, inoperable tumors,1,2 as a bridging therapy to liver transplantation3 and has shown durable local control (LC) in patients with advanced HCC.4 A landmark study of stereotactic body RT at Princess Margaret Cancer Center demonstrated 48% 1-year overall survival and 65% 1-year LC.5,6 Subsequent single-arm studies7-9 and meta-analyses10 have confirmed excellent LC rates. Both changes in RT fractionation (5 v 15 fractions) and proton beam therapy are currently explored in a randomized phase III trial (ClinicalTrials.gov identifier: NCT03186898).
In this investigation, we focus on two toxicities, radiation-induced liver disease (RILD) and radiation-induced lymphopenia (RIL). The former has recently become a focus of attention, given the high local efficacy of RT and the fact that patients often die from liver failure without recurrence.11,12 The latter is becoming more important because of ongoing combinations trials of RT with immunotherapy (ClinicalTrials.gov identifier: NCT03203304, NCT03482102, NCT03316872, NCT03817736). These emerging combination regimen of RT with immunotherapeutic agents have sparked interest in minimizing the immunosuppressive effects of radiation that have been shown to correlate with survival,13,14 also in HCC.15 Predictive models for RIL have been developed for thoracic irradiation16 but not for liver-directed RT.
With the role of RT in HCC evolving, patient selection is becoming critical to maximize the clinical benefit of RT for this diverse population. Previous work on outcome prediction often focused on analytical models17,18 and was restricted to selected cohorts. The purpose of this study is to simultaneously investigate predictive models of patient-specific survival, nonlocal failure, RILD, and lymphopenia, and evaluate them in unselected patient cohorts across institutions. Particularly in HCC, clinical studies have shown multiple prognostic factors,19,20 although correlation does not always translate into predictive power, and there could be interactions between baseline patient characteristics. Therefore, we are exploring linear and nonlinear machine learning models to study whether predictive models can reach a robustness and accuracy that makes them suitable for patient stratification in RT for HCC.
MATERIALS AND METHODS
Study Design
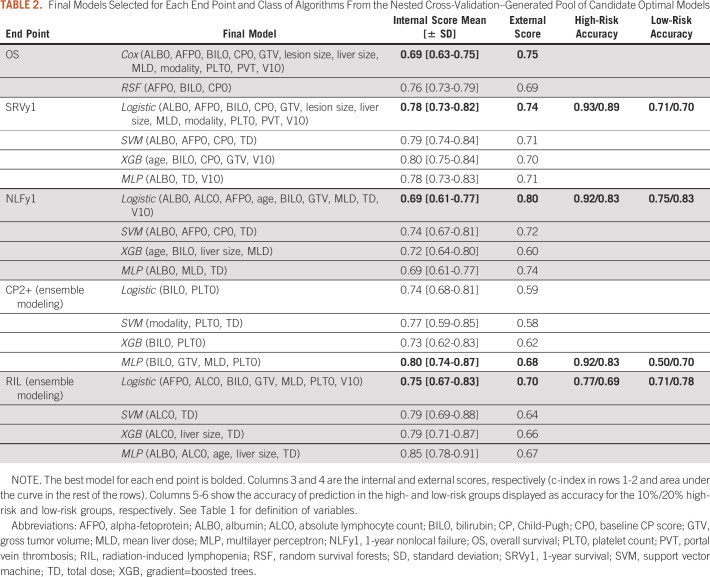
Similar to a clinical trial, we preregistered our methodology in a Study Analysis Plan21 before obtaining access to the validation data set to prevent selective reporting and positive bias.22,23 The predictive model was developed on the basis of an internal cohort treated at Massachusetts General Hospital (MGH), locked and only then validated using an independent cohort treated at MD Anderson Cancer Center (MDACC), representing a TRIPOD type 3 study.24 We developed a Cox proportional hazards model and a random survival forest to stratify patients into risk groups for mortality, and classification models to predict four binary end points: 1-year survival (SRVy1); 1-year nonlocal failure (NLFy1); nonclassic RILD, defined as 2+ increase in Child-Pugh (CP) score after 3 months of treatment (CP2+); and radiation-induced grade 3+ lymphopenia (RIL).25
We used four machine learning algorithms: logistic regression, as a linear additive algorithm, and three nonlinear algorithms that can capture possible feature interactions, particularly, support vector machine, multilayer perceptron (MLP), and gradient boosted trees. For the toxicity outcomes where incidence is low, we used ensemble modeling to treat data imbalance, where 10 submodels of the same structure were trained on stratified bootstrapped subsets of the MGH data set. The ensemble model prediction was calculated as the average of the submodel prediction probabilities.
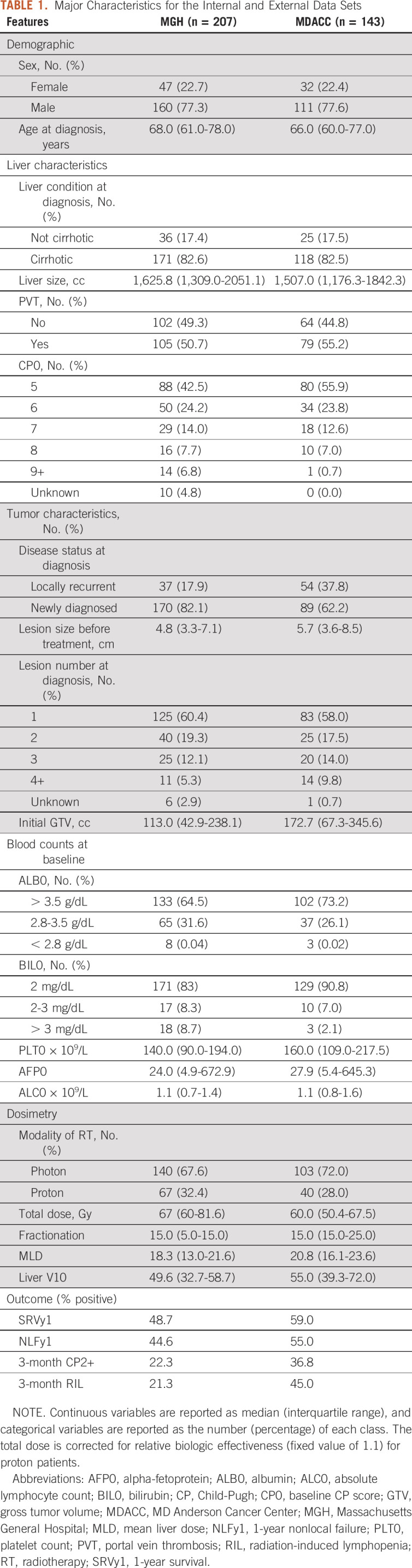
We conducted a correlation analysis on all dosimetric and clinical variables to remove redundant features (Data Supplement). The remaining set used for feature selection contained the 20 features listed in Table 1. Normal liver doses were adjusted for fractionation with α/β = 2.5, and mean liver dose (MLD) and low dose parameters were chosen on the basis of previous clinical studies.18
TABLE 1.
Major Characteristics for the Internal and External Data Sets
We implemented a model development pipeline using a nested cross-validation approach26,27 on the MGH data set (Fig 1). In the outer loop, we split the internal data using stratified 10-fold cross-validation. In each fold, the missing data were imputed separately in the training and testing subsets using a multivariate imputer28 to prevent data leakage. After data preprocessing, the inner loop selected the optimal subset of features using backward elimination29—with features ranked by permutation importance30—and tuned hyperparameters using grid search. Each fold resulted in a unique model, defined by a set of features. We repeated the 10-fold cross-validation 30 times using different random seeds to create a pool of candidate optimal models from which the final model was selected.
FIG 1.
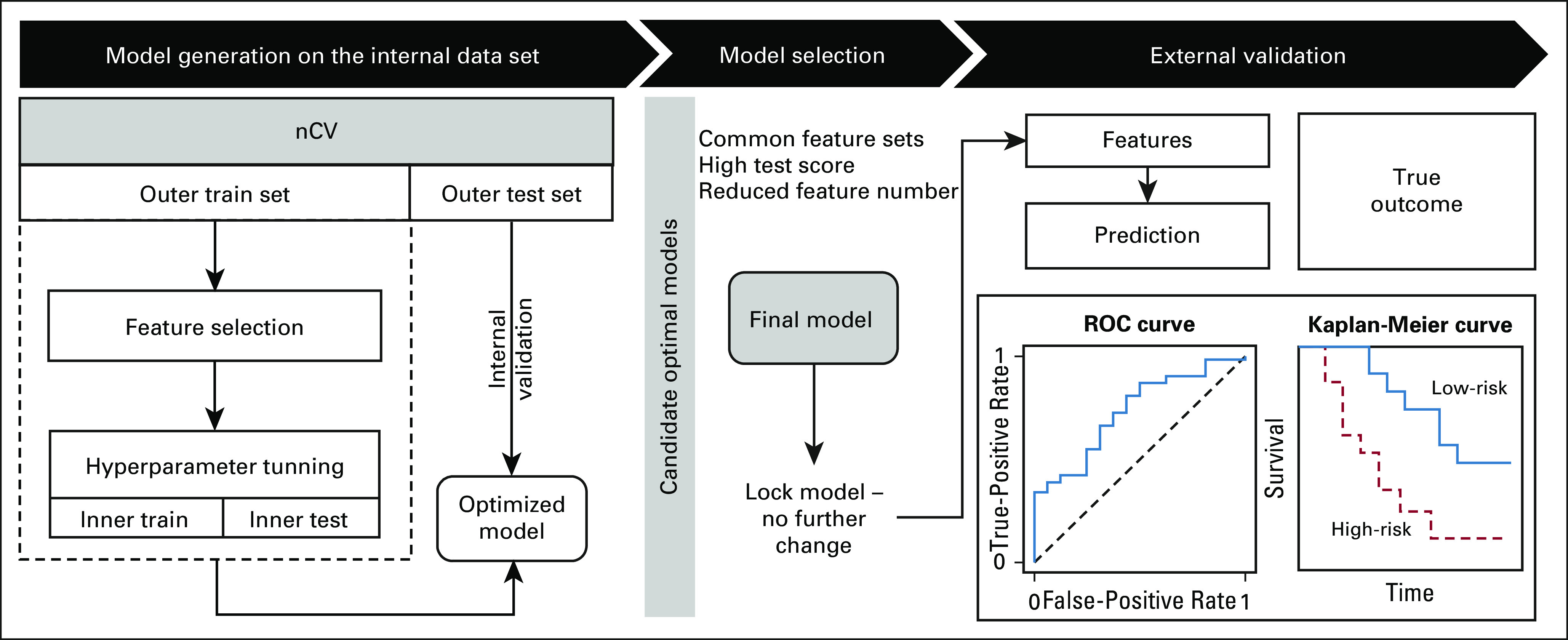
The framework used for model development and validation. An nCV approach was used to generate 300 promising models, from which one model was selected and locked. The final model was validated by an external data set. The model accessed the features only, generated risk scores, and sent them for comparison with the actual outcome. The performance was evaluated using the area under the receiver operating characteristics curve and the discrimination c-index for survival analysis. This study represents a TRIPOD type 3 analysis. nCV, nested cross-validation; ROC, receiver operating characteristic curve.
We followed model selection criteria for controlling bias and variance of the final model. We grouped the candidate models that share the same feature set, and we recorded the number of models and average test area under the curve (AUC) of each group. A large group means that the corresponding feature set was optimal for many data splits, which reflects robustness. If one group was significantly larger than the others, we selected the feature set of that group. Otherwise, we considered the largest five groups, and we chose the feature set with the highest average test AUC. If the top groups have comparable frequency and scores, we selected the smallest features set to reduce the risk of overfitting. Interaction between variables of the final model was quantified using Friedman's H-statistic.31
Patient Cohorts
The training cohort contains 207 patients with HCC treated with RT at MGH between 2008 and 2018. The study was approved by MGH's Institutional Review Board Committee. We excluded patients with missing outcomes, and those with less than 1-year follow-up for the binary 1-year end points (survival and nonlocal failure). The resulting cohort contained 152 patients for the SRVy1, 101 for the NLFy1, 105 for the CP2+, and 108 for the RIL end points.
The validation cohort from MDACC contained 143 patients with HCC treated with RT between 2006 and 2016. The patient characteristics in both cohorts are generally comparable (Table 1), except that MGH patients have smaller tumors on average and the fraction of newly diagnosed cases is lower at MDACC. In terms of outcome, median survival was 14 versus 17 months in the internal and external data set; higher incidence of toxicity was observed in the MDACC cohort.
Validation of the Final Model Using the External Cohort
After choosing the final model, we ran it through 100 bootstraps on the MGH data to evaluate stability and calculate an internal AUC. Then, we trained the model on the entire MGH data and locked it before gaining access to the MDACC data. For the external validation we received only the features, but not outcomes, from MDACC and sent back our predicted outcomes, on which all results are based. We used Kaplan-Meier curves and Harrell's c-index32 to evaluate discriminative ability between low-risk and high-risk groups, split by median risk and into top and bottom quartiles. We further studied the model calibration by comparing predicted against observed survival of the cohort binned in quartiles.33 Binary classification models were assessed using the area (AUC) under the receiver operating characteristics curve, and accuracy in the 10% and 20% high-risk groups. The high-risk and low-risk groups consisted of patients with the cohort's top/bottom risk scores, which could vary for each end point. We assigned a positive/negative label for the high-risk and low-risk groups and calculated the accuracy of the assigned versus the true labels.
Finally, the models' clinical utility was evaluated using net-benefit analysis,34 which generates a decision curve on the basis of the true-positive and false-positive numbers for a range of probability thresholds. Clinical utility is indicated if the decision curve is positive for a probability threshold larger than a clinically significant threshold, which is specified by the treating physician.
RESULTS
The models presented below are those selected using nested cross-validation and the criteria described above in the internal cohort, and then validated externally. Figures 2 and 3 present the Kaplan-Meier and receiver operating characteristic curve curves for the superior models for each end point.
FIG 2.
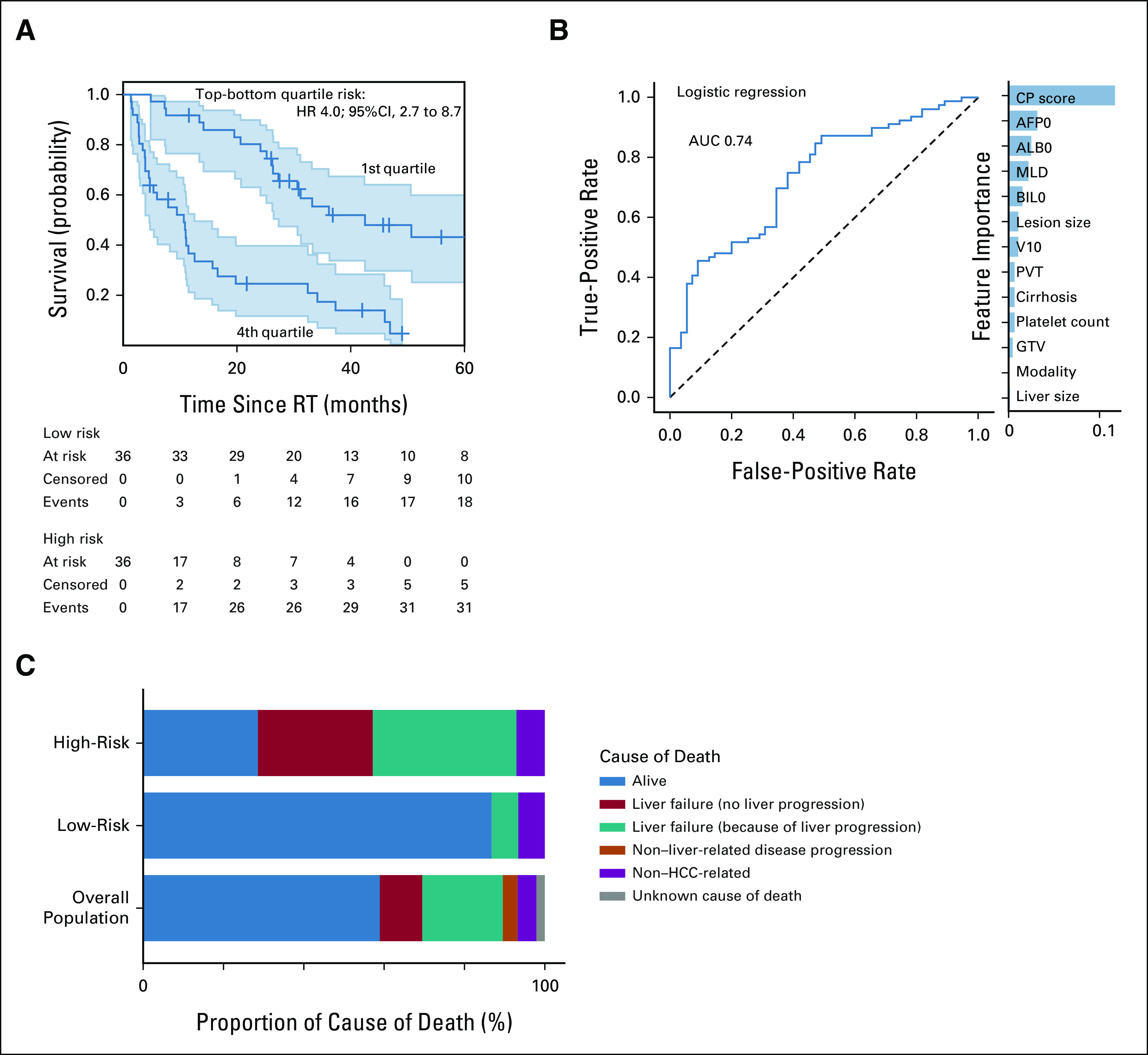
Validation of the survival models on the external data set. (A) Stratification of patients on the basis of risk determined by the Cox prediction model showing the top and bottom quartile risk groups. (B) Receiver operating characteristic curve for predicting classifier of SRVy1 using logistic regression. Feature importance are on the right, where all features are measured before treatment. PVT, GTV, and V10 are the portal vein thrombosis, gross tumor volume, and 10 Gy dosimetric index, respectively. (C) Cause of death for patients scored high and low risk by the logistic model. AFP0, alpha-fetoprotein; ALB0, albumin; AUC, area under the curve; BIL0, bilirubin; CP, Child-Pugh; HCC, hepatocellular carcinoma; HR, hazard ratio; MLD, mean liver dose; RT, radiotherapy; SRVy1, 1-year survival.
FIG 3.
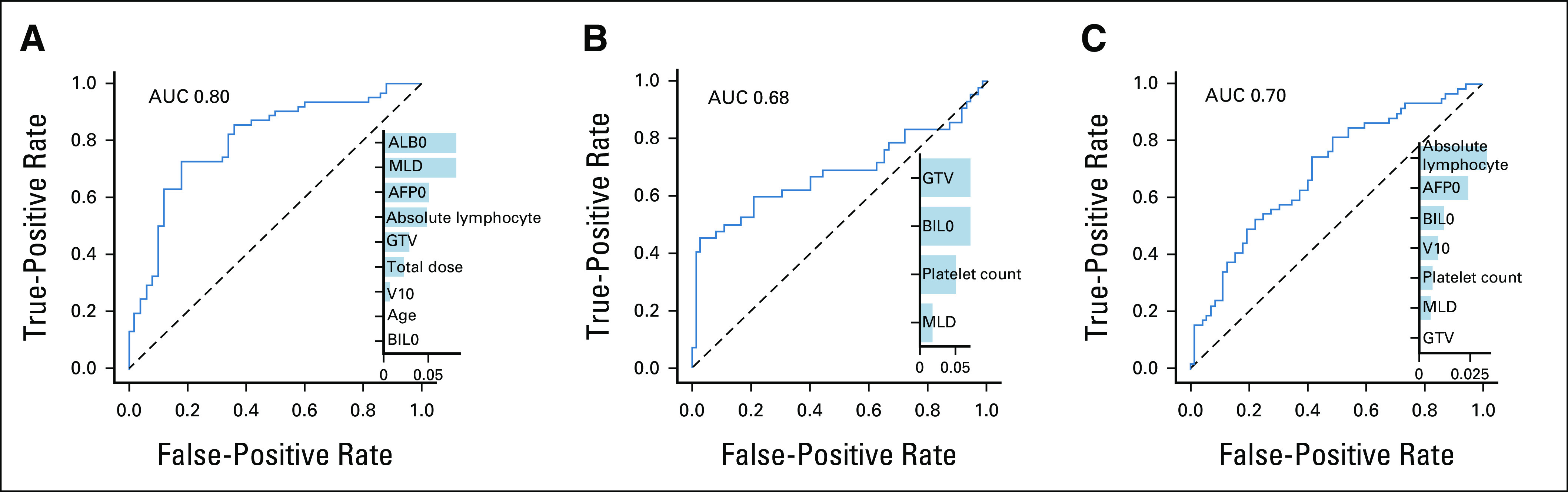
External validation of the tumor control and toxicity models. Performance of the model by the receiver operating curve analysis for predicting (A) NLFy1, (B) CP2+, and (C) lymphopenia after radiotherapy. The bar plots show the model variables ranked by importance. All features are measured before treatment. PVT, GTV, and V10 are the portal vein thrombosis, gross tumor volume, and 10 Gy dosimetric index, respectively. AFP0, alpha-fetoprotein; ALB0, albumin; AUC, area under the curve; BIL0, bilirubin; CP, Child-Pugh; MLD, mean liver dose; MLP, multilayer perceptron; NLFy1, 1-year nonlocal failure; RIL, radiation-induced lymphopenia.
Survival
Time to death.
The Cox survival model showed excellent patient stratification using a combination of clinical and dosimetric variables, with baseline CP score (CP0), modality, bilirubin (BIL0), and portal vein thrombosis being the most important features. The model discriminated the top from the bottom risk quartile well with an external c-index of 0.75 and median survival of 10.6 and 42.5 months in the high-risk and low-risk groups (hazard ratio 4.0 [2.7-8.7], Fig 2A). Including all patients into two risk groups yielded 11.5 versus 26.4 months of median overall survival (hazard ratio 2.3 [1.8-2.4], Data Supplement). The random survival forests selected only CP0, albumin (ALB0), and BIL0, which are the standard risk assessment metrics in HCC but had lower external performance (c-index 0.59, Table 2). Calibration analysis revealed that survival predictions for patients above median risk were well calibrated at 1-year post-RT; however, analysis showed an overestimation and underestimation of survival in the medium-low and low-risk groups, respectively (Data Supplement).
TABLE 2.
Final Models Selected for Each End Point and Class of Algorithms From the Nested Cross-Validation–Generated Pool of Candidate Optimal Models
1-year survival classification.
The internal AUC ranged from 0.78 to 0.81 across the different nonlinear algorithms (Table 2) with an external AUC 0.70-0.71. The logistic regression (internal AUC = 0.78) performed better externally (AUC 0.74, Fig 2B), with baseline CP, alpha fetoprotein, ALB0, MLD, and BIL0 being the most important features. The model had excellent accuracy in identifying a high chance of SRVy1 (83%-92%) and high risk of 1-year mortality (77%-83%). The model's net-benefit analysis showed a positive decision curve for probability thresholds up to 0.85 (Data Supplement), meaning that the model has clinical utility if predicting a 85% probability of death within 1 year for a specific patient would affect treatment.
Cause of death.
We examined the underlying drivers of survival outcome in the top and bottom 10% risk groups (Fig 2C) to see if the prediction could indeed guide treatment selection. In the low-risk group, 87% of the patients were alive, with a median follow-up of 36.9 months. Fifty percent of the patients had died of non–HCC-related causes, compared with only 11% in the overall population. On the contrary, in the high-risk group, only 29% of patients were alive after 1 year and 29% died from liver failure without tumor progression compared with 10% in the overall population (Fig 2C).
1-Year Nonlocal Failure
The nonlinear models had comparable internal and external AUC ranging from 0.69 to 0.74 and 0.60 to 0.72, respectively, but the logistic regression (AUC = 0.69) performed better on the external data set (AUC = 0.80, Fig 3A). Feature selection revealed the importance of blood counts (ALB0, alpha-fetoprotein, and absolute lymphocyte count) and MLD for this end point. The model's accuracy in the 10% and 20% high-risk groups was 92% and 83%, respectively. The decision curve (Data Supplement) indicated a positive net benefit for all probability thresholds.
Hepatic Toxicity: Increase in CP Score
The baseline platelet count was selected by all nonlinear algorithms, indicating a significant interaction with other variables. The models had strong internal AUC (0.73-0.80) but weaker performance externally (AUC = 0.59-0.68). The MLP performed best (Fig 3B) with excellent accuracy (92% and 83%) in the 10% and 20% high-risk groups. The net-benefit analysis indicated clinical utility for probability thresholds up to 0.8 (Data Supplement).
CP2+ was the only end point that benefited greatly from nonlinear algorithms, which indicates that interactions between features could be relevant. We explored this further and found strong interactions between radiation dose to the liver, BIL0, and platelet count. The pairwise H-statistic was 32% for BIL0 and platelet count, meaning that 32% of the contribution these two variables make to the prediction comes from their interaction. The interaction between MLD and platelets and BIL0 was 28% and 17%, respectively (Data Supplement).
Effect on Peripheral Immunity: RIL
Baseline lymphocyte count was selected by all algorithms, and nonlinear models indicated interaction of baseline ALC with tumor dose. The support vector machine and MLP indicated the important role of liver size, which could reflect the blood volume exposed to radiation. The internal AUC of the nonlinear algorithms was high (0.75-0.79), although decreased in validation (0.64-0.67). In comparison, an ensemble of logistic regression models performed better externally (AUC = 0.70, Fig 3C) despite inferior internal score. The logistic regression selected V10 and MLD versus tumor dose alone, which provides spatial information. The accuracy in the high-risk group was 69%-77% and the clinical net benefit of the RIL model was limited to a 0.5 probability threshold (Data Supplement).
DISCUSSION
We developed predictive models for patients with HCC after RT for four clinically relevant end points: survival, nonlocal failure, CP2+ increase, and lymphopenia. We validated the models on an external data set (TRIPOD type 324,35) after locking them and publishing the methods to ensure transparency.21 We opted for this approach instead of merging the data and creating a hold-out validation set because, especially in RT for HCC, practice patterns vary considerably36,37 and evaluation across institutions can evaluate robustness toward different underlying populations and referral patterns.
The survival model showed strong discriminative ability (external c-index 0.75) and excellent calibration for high-risk patients (Data Supplement). The model's risk score combined CP0, ALB0, and BIL0 with other blood counts, dosimetric variables, liver function, and lesion size (Data Supplement). Our composite risk score has higher discriminative power than CP for the overall population and is better than ALBI for the high-risk versus low-risk patients (Data Supplement). The same pattern was observed in the internal data set (Data Supplement), which demonstrates the potential advantage of our model versus available scoring system particularly in stratifying high-risk patients.
Prediction of SRVy1 was strong (external AUC = 0.74) with high accuracy (> 89% and 70%) in the low-risk and high-risk groups. We also achieved high predictive ability of nonlocal failure (AUC = 0.80) and excellent accuracy (> 83%) in the high-risk group. Both models showed positive net benefit for the widest range of probability thresholds, indicating possible clinical utility (Data Supplement).
To investigate whether these models can inform clinical decision making, we analyzed the cause of death in the low-risk and high-risk populations externally. The low-risk patients not only showed favorable outcomes, but a significant percentage died from non–HCC-related causes, indicating the high efficacy of current stereotactic body RT techniques in this population. In the high-risk patients, however, the incidence of death from liver failure without disease progression increased four-fold compared with the overall populations (40% v 10%), with another large fraction of patients (30%) succumbing to liver failure from disease progression. Given the high LC rates for HCC, the latter usually occurs because of disease progression in the liver outside of the previously irradiated target. These divergent cause-of-death distributions and the high accuracy of our models in the high-risk groups could enable patient stratification as outlined in Figure 4: a survival model filters out the population with very high mortality risk, and in the next stage, the nonlocal failure and CP2+ models further specify the optimal course of treatment for these patients: (1) patients at high risk of CP2+ become candidates for liver-sparing approaches such as proton therapy and alternative fractionation schemes, which are currently evaluated (ClinicalTrials.gov identifier: NCT03186898), or possibly dose reduction; (2) patients at high risk of NLFy1 are eligible for treatment with a more systemic approach combining RT with biologic agents, which are currently under investigation, for example, ClinicalTrials.gov identifier: NCT03482102. Enabling preselection of patients that benefit most from drug-RT combinations or proton therapy can also improve clinical trial design as evidenced by the model-driven trial designs pioneered in the Netherlands.38 For this reason, we also see the inclusion of both proton and photon patients as a strength, enabling the models to guide patient selection for proton therapy.
FIG 4.
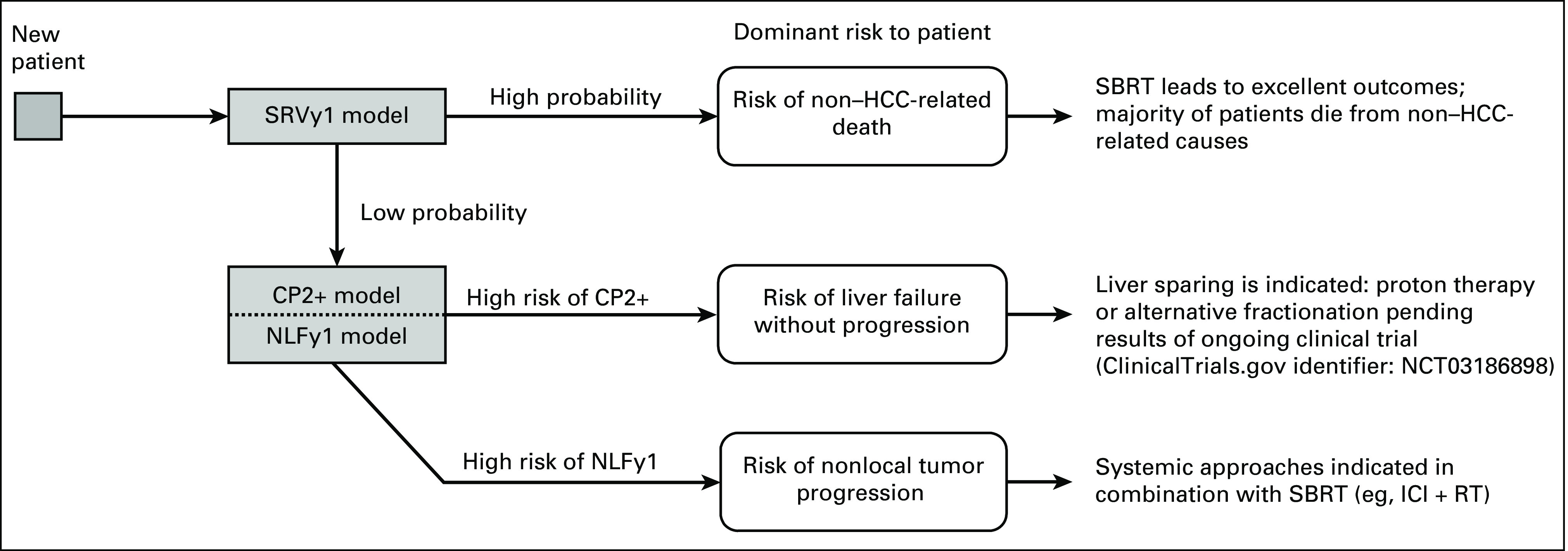
Model-based decision tree for personalized treatment selection and clinical trial enrichment using risk scores for survival and nonlocal failure. CP, Child-Pugh; HCC, hepatocellular carcinoma; ICI, immune checkpoint inhibitors; NLFy1, 1-year nonlocal failure; RT, radiotherapy; SBRT, stereotactic body radiotherapy; SRVy1, 1-year survival.
For the toxicity end points, the CP2+ model had high accuracy (> 83%) in the high-risk group. Combined with positive clinical net benefit for wide range of occurrence probability (Data Supplement), the model is informative for high-risk patients. The intermediate performance in the overall cohort (validation AUCs 0.58-0.68) can be interpreted as a success of the currently used individualized prescription practice in HCC. To improve treatment individualization, we need to move past current biomarkers and explore new approaches such as hepatocyte growth factor39 or cytokines.40,41 Furthermore, our interaction analysis (Data Supplement) revealed that the CP2+ sensitivity to dose depends on the platelet count.
For post-RT lymphopenia, predictive power was good (external AUC = 0.7). All the models combined baseline ALC with dosimetric features, but the best performing model used both MLD and V10, indicating the significance of spatial dose information.
It is worth noting that logistic regression outperforms nonlinear algorithms in all clinical end points except CP2+, where MLP performed the best. Logistic regression is highly interpretable, less prone to overfitting, and easily reproducible, making the models more applicable. For the CP2+ model, the selection of MLP warrants future investigation into the predictive factors for radiation-induced hepatic toxicity and possible interactions among them.
Machine learning has previously been applied to HCC to estimate dose-based toxicity metrics,35,40 but few studies considered clinical end points. These models suffered from small sample size or relied on data not readily available, impeding their clinical application. For instance, Shen et al42 predicted disease-free survival from gene sequencing but the model was validated on 10 patients only. Others also used powerful deep learning techniques43 or focused on radiomics,44 but lacked external validation.
Our model is validated across institutions on large, unselected cohorts receiving both photon and modern proton therapies.
The main limitation of our study is the low incidence of CP2+ and lymphopenia, which possibly prevents improved training of the toxicity models. Dosimetric features did not consider the entire liver dose-volume histogram but focused on known predictors, mean dose, and low-dose bath.18 Functional imaging of the liver has been shown to improve dose-response relationships for hepatic toxicity in small cohorts and could further enhance prediction.45,46
In conclusion, we showed that generalizable prediction of outcome after RT for HCC across diverse populations is possible, particularly for survival, nonlocal failure, and hepatic toxicity. Accuracy is particularly high in high-risk subgroups, indicating potential applications to modify the treatment of these patients. Cause-of-death analysis for the high-risk and low-risk populations in the survival model revealed differential modes of treatment failure. Together with good prediction of nonlocal failure and hepatic toxicity in high-risk patients, our model might enable stratification of patients to either RT strategies that spare normal liver or combination regimens including systemic agents.
Ibrahim Chamseddine
Employment: Platelet Bio
Brian De
Honoraria: Sermo Inc
Issam El Naqa
Consulting or Advisory Role: Endectra
Patents, Royalties, Other Intellectual Property: Patent pending on an optical probe for radiation (Inst), Paten pending on new computing technology for decision making (Inst)
Dan G. Duda
Honoraria: Bayer, Simcere Pharmaceutical Group, Surface Oncology
Research Funding: Bayer (Inst), Bristol Myers Squibb (Inst), Exelixis (Inst)
John Wolfgang
Consulting or Advisory Role: P-Cure Ltd
Jennifer Wo
Honoraria: PER
Research Funding: Genentech/Roche (Inst)
Travel, Accommodations, Expenses: Lynx Group
Theodore Hong
Stock and Other Ownership Interests: PanTher Therapeutics
Consulting or Advisory Role: Merck, Synthetic Biologics, Novocure, Syndax, Boston Scientific
Research Funding: Taiho Pharmaceutical (Inst), AstraZeneca (Inst), IntraOp (Inst), Tesaro (Inst), Bristol Myers Squibb (Inst), Ipsen (Inst)
Eugene J. Koay
Honoraria: Apollo Health
Consulting or Advisory Role: Augmenix, RenovoRx
Speakers' Bureau: Oncology Information Group
Research Funding: Philips Healthcare, Elekta, GE Healthcare
Patents, Royalties, Other Intellectual Property: Provisional patent on 3D printed oral stents for radiation treatments of head and neck cancer, and royalties from Taylor and Francis LLC for my coauthored book: An introduction to physical oncology
Clemens Grassberger
Stock and Other Ownership Interests: Onc.AI, Nanolive
Honoraria: Innovent Biologics
No other potential conflicts of interest were reported.
SUPPORT
Supported by National Cancer Institute P01CA261669 (T.H.), R21 CA241918 (C.G.) and U19 CA21239 (C.G.), by the Department of Defense Grant No. W81XWH-19-1-0284 (D.G.D.), and by the National Institutes of Health through Cancer Center Support Grant No. P30CA016672 (B.D.).
AUTHOR CONTRIBUTIONS
Conception and design: Ibrahim Chamseddine, Yejin Kim, Dan G. Duda, Harald Paganetti, Clemens Grassberger
Financial support: Clemens Grassberger
Administrative support: Dan G. Duda, Eugene J. Koay
Provision of study material or patients: Dan G. Duda, Jennifer Wo, Eugene J. Koay
Collection and assembly of data: Ibrahim Chamseddine, Brian De, Dan G. Duda, John Wolfgang, Jennifer Pursley, Jennifer Wo, Eugene J. Koay, Clemens Grassberger
Data analysis and interpretation: Ibrahim Chamseddine, Yejin Kim, Issam El Naqa, Dan G. Duda, Harald Paganetti, Theodore Hong, Eugene J. Koay, Clemens Grassberger
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Ibrahim Chamseddine
Employment: Platelet Bio
Brian De
Honoraria: Sermo Inc
Issam El Naqa
Consulting or Advisory Role: Endectra
Patents, Royalties, Other Intellectual Property: Patent pending on an optical probe for radiation (Inst), Paten pending on new computing technology for decision making (Inst)
Dan G. Duda
Honoraria: Bayer, Simcere Pharmaceutical Group, Surface Oncology
Research Funding: Bayer (Inst), Bristol Myers Squibb (Inst), Exelixis (Inst)
John Wolfgang
Consulting or Advisory Role: P-Cure Ltd
Jennifer Wo
Honoraria: PER
Research Funding: Genentech/Roche (Inst)
Travel, Accommodations, Expenses: Lynx Group
Theodore Hong
Stock and Other Ownership Interests: PanTher Therapeutics
Consulting or Advisory Role: Merck, Synthetic Biologics, Novocure, Syndax, Boston Scientific
Research Funding: Taiho Pharmaceutical (Inst), AstraZeneca (Inst), IntraOp (Inst), Tesaro (Inst), Bristol Myers Squibb (Inst), Ipsen (Inst)
Eugene J. Koay
Honoraria: Apollo Health
Consulting or Advisory Role: Augmenix, RenovoRx
Speakers' Bureau: Oncology Information Group
Research Funding: Philips Healthcare, Elekta, GE Healthcare
Patents, Royalties, Other Intellectual Property: Provisional patent on 3D printed oral stents for radiation treatments of head and neck cancer, and royalties from Taylor and Francis LLC for my coauthored book: An introduction to physical oncology
Clemens Grassberger
Stock and Other Ownership Interests: Onc.AI, Nanolive
Honoraria: Innovent Biologics
No other potential conflicts of interest were reported.
REFERENCES
- 1.Seo YS, Kim MS, Yoo SY, et al. : Preliminary result of stereotactic body radiotherapy as a local salvage treatment for inoperable hepatocellular carcinoma. J Surg Oncol 102:209-214, 2010 [DOI] [PubMed] [Google Scholar]
- 2.Huertas A, Baumann AS, Saunier-Kubs F, et al. : Stereotactic body radiation therapy as an ablative treatment for inoperable hepatocellular carcinoma. Radiother Oncol 115:211-216, 2015 [DOI] [PubMed] [Google Scholar]
- 3.Sapisochin G, Barry A, Doherty M, et al. : Stereotactic body radiotherapy vs. TACE or RFA as a bridge to transplant in patients with hepatocellular carcinoma. An intention-to-treat analysis. J Hepatol 67:92-99, 2017 [DOI] [PubMed] [Google Scholar]
- 4.Keane FK, Tanguturi SK, Zhu AX, et al. : Radiotherapy for liver tumors. Hepat Oncol 2:133-146, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tse RV, Hawkins M, Lockwood G, et al. : Phase I study of individualized stereotactic body radiotherapy for hepatocellular carcinoma and intrahepatic cholangiocarcinoma. J Clin Oncol 26:657-664, 2008 [DOI] [PubMed] [Google Scholar]
- 6.Keane FK, Wo JY, Zhu AX, et al. : Liver-directed radiotherapy for hepatocellular carcinoma. Liver Cancer 5:198-209, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cárdenes HR, Price TR, Perkins SM, et al. : Phase I feasibility trial of stereotactic body radiation therapy for primary hepatocellular carcinoma. Clin Transl Oncol 12:218-225, 2010 [DOI] [PubMed] [Google Scholar]
- 8.Kang JK, Kim MS, Cho CK, et al. : Stereotactic body radiation therapy for inoperable hepatocellular carcinoma as a local salvage treatment after incomplete transarterial chemoembolization. Cancer 118:5424-5431, 2012 [DOI] [PubMed] [Google Scholar]
- 9.Bujold A, Massey CA, Kim JJ, et al. : Sequential phase I and II trials of stereotactic body radiotherapy for locally advanced hepatocellular carcinoma. J Clin Oncol 31:1631-1639, 2013 [DOI] [PubMed] [Google Scholar]
- 10.Rim CH, Kim HJ, Seong J: Clinical feasibility and efficacy of stereotactic body radiotherapy for hepatocellular carcinoma: A systematic review and meta-analysis of observational studies. Radiother Oncol 131:135-144, 2019 [DOI] [PubMed] [Google Scholar]
- 11.Mizumoto M, Okumura T, Hashimoto T, et al. : Evaluation of liver function after proton beam therapy for hepatocellular carcinoma. Int J Radiat Oncol Biol Phys 82:e529-e535, 2012 [DOI] [PubMed] [Google Scholar]
- 12.Sanford NN, Pursley J, Noe B, et al. : Protons versus photons for unresectable hepatocellular carcinoma: Liver decompensation and overall survival. Int J Radiat Oncol Biol Phys 105:64-72, 2019 [DOI] [PubMed] [Google Scholar]
- 13.Grassberger C, Ellsworth SG, Wilks MQ, et al. : Assessing the interactions between radiotherapy and antitumour immunity. Nat Rev Clin Oncol 16:729-745, 2019 [DOI] [PubMed] [Google Scholar]
- 14.Lambin P, Lieverse RIY, Eckert F, et al. : Lymphocyte-sparing radiotherapy: The rationale for protecting lymphocyte-rich organs when combining radiotherapy with immunotherapy. Semin Radiat Oncol 30:187-193, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De B, Ng SP, Liu AY, et al. : Radiation-associated lymphopenia and outcomes of patients with unresectable hepatocellular carcinoma treated with radiotherapy. J Hepatocell Carcinoma 8:57-69, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu C, Lin SH, Jiang X, et al. : A novel deep learning model using dosimetric and clinical information for grade 4 radiotherapy-induced lymphopenia prediction. Phys Med Biol 65:035014, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sung W, Grassberger C, McNamara AL, et al. : A tumor-immune interaction model for hepatocellular carcinoma based on measured lymphocyte counts in patients undergoing radiotherapy. Radiother Oncol 151:73-81, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pursley J, El Naqa I, Sanford NN, et al. : Dosimetric analysis and normal-tissue complication probability modeling of Child-Pugh score and albumin-bilirubin grade increase after hepatic irradiation. Int J Radiat Oncol Biol Phys 107:986-995, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bibault JE, Dewas S, Vautravers-Dewas C, et al. : Stereotactic body radiation therapy for hepatocellular carcinoma: Prognostic factors of local control, overall survival, and toxicity. PLoS One 8:e77472, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo CH, Yang JF, Liu MY, et al. : Survival and prognostic factors for patients with advanced hepatocellular carcinoma after stereotactic ablative radiotherapy. PLoS One 12:e0177793, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chamseddine I, Kim Y, Paganetti H, et al. : Predicting radiotherapy outcomes in hepatocellular carcinoma. OSF Registries doi: 10.17605/OSF.IO/7G6AN [DOI] [Google Scholar]
- 22.De Angelis C, Drazen JM, Frizelle FA, et al. : Clinical trial registration: A statement from the International Committee of Medical Journal Editors. N Engl J Med 351:1250-1251, 2004 [DOI] [PubMed] [Google Scholar]
- 23.Thor M, Oh JH, Apte AP, et al. : Registering study analysis plans (SAPs) before dissecting your data-updating and standardizing outcome modeling. Front Oncol 10:978, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins GS, Reitsma JB, Altman DG, et al. : Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD statement. Eur J Clin Invest 45:204-214, 2015 [DOI] [PubMed] [Google Scholar]
- 25.Pan CC, Kavanagh BD, Dawson LA, et al. : Radiation-associated liver injury. Int J Radiat Oncol Biol Phys 76:S94-S100, 2010. (3 suppl) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo Y, McShan D, Ray D, et al. : Development of a fully cross-validated Bayesian network approach for local control prediction in lung cancer. IEEE Trans Radiat Plasma Med Sci 3:232-241, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varma S, Simon R: Bias in error estimation when using cross-validation for model selection. BMC Bioinformatics 7:91, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Buuren S, Groothuis-Oudshoorn K: mice: Multivariate imputation by chained equations in R. J Stat Softw 45:1-67, 2011 [Google Scholar]
- 29.Kohavi R, John GH: Wrappers for feature subset selection. Artif Intelligence 97:273-324, 1997 [Google Scholar]
- 30.Strobl C, Boulesteix AL, Zeileis A, et al. : Bias in random forest variable importance measures: Illustrations, sources and a solution. BMC Bioinformatics 8:25, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedman JH, Popescu BE: Predictive learning via rule ensembles. Ann Appl Stat 2:916-954, 2008 [Google Scholar]
- 32.Harrell FE Jr, Lee KL, Mark DB: Multivariable prognostic models: Issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med 15:361-387, 1996 [DOI] [PubMed] [Google Scholar]
- 33.Van Calster B, McLernon DJ, van Smeden M, et al. : Calibration: The achilles heel of predictive analytics. BMC Med 17:230, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vickers AJ, Van Calster B, Steyerberg EW: Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. BMJ 352:i6, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui W, Wang W, Hu Z, et al. : Comparison of 2 methods for prediction of liver dosimetric indices in hepatocellular cancer IMRT planning. Med Dosim 44:e80-e85, 2019 [DOI] [PubMed] [Google Scholar]
- 36.Bang A, Dawson LA: Radiotherapy for HCC: Ready for prime time? JHEP Rep 1:131-137, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park S, Yoon WS, Rim CH: Indications of external radiotherapy for hepatocellular carcinoma from updated clinical guidelines: Diverse global viewpoints. World J Gastroenterol 26:393-403, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Widder J, van der Schaaf A, Lambin P, et al. : The quest for evidence for proton therapy: Model-based approach and precision medicine. Int J Radiat Oncol Biol Phys 95:30-36, 2016 [DOI] [PubMed] [Google Scholar]
- 39.Hong TS, Grassberger C, Yeap BY, et al. : Pretreatment plasma HGF as potential biomarker for susceptibility to radiation-induced liver dysfunction after radiotherapy. NPJ Precis Oncol 2:22, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El Naqa I, Johansson A, Owen D, et al. : Modeling of normal tissue complications using imaging and biomarkers after radiation therapy for hepatocellular carcinoma. Int J Radiat Oncol Biol Phys 100:335-343, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng SSW, Zhang H, Wang L, et al. : Association of pro-inflammatory soluble cytokine receptors early during hepatocellular carcinoma stereotactic radiotherapy with liver toxicity. NPJ Precis Oncol 4:17, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen J, Qi L, Zou Z, et al. : Identification of a novel gene signature for the prediction of recurrence in HCC patients by machine learning of genome-wide databases. Sci Rep 10:4435, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ibragimov B, Toesca DAS, Yuan Y, et al. : Neural networks for deep radiotherapy dose analysis and prediction of liver SBRT outcomes. IEEE J Biomed Health Inform 23:1821-1833, 2019 [DOI] [PubMed] [Google Scholar]
- 44.Wei L, Owen D, Rosen B, et al. : A deep survival interpretable radiomics model of hepatocellular carcinoma patients. Phys Med 82:295-305, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaub SK, Apisarnthanarax S, Price RG, et al. : Functional liver imaging and dosimetry to predict hepatotoxicity risk in cirrhotic patients with primary liver cancer. Int J Radiat Oncol Biol Phys 102:1339-1348, 2018 [DOI] [PubMed] [Google Scholar]
- 46.Bortfeld T: Optimized planning using physical objectives and constraints. Semin Radiat Oncol 9:20-34, 1999 [DOI] [PubMed] [Google Scholar]