Abstract
Background:
Modern chemical toxicology is facing a growing need to Reduce, Refine, and Replace animal tests (Russell 1959) for hazard identification. The most common type of animal assays for acute toxicity assessment of chemicals used as pesticides, pharmaceuticals, or in cosmetic products is known as a “6-pack” battery of tests, including three topical (skin sensitization, skin irritation and corrosion, and eye irritation and corrosion) and three systemic (acute oral toxicity, acute inhalation toxicity, and acute dermal toxicity) end points.
Methods:
We compiled, curated, and integrated, to the best of our knowledge, the largest publicly available data sets and developed an ensemble of quantitative structure–activity relationship (QSAR) models for all six end points. All models were validated according to the Organisation for Economic Co-operation and Development (OECD) QSAR principles, using data on compounds not included in the training sets.
Results:
In addition to high internal accuracy assessed by cross-validation, all models demonstrated an external correct classification rate ranging from 70% to 77%. We established a publicly accessible Systemic and Topical chemical Toxicity (STopTox) web portal (https://stoptox.mml.unc.edu/) integrating all developed models for 6-pack assays.
Conclusions:
We developed STopTox, a comprehensive collection of computational models that can be used as an alternative to in vivo 6-pack tests for predicting the toxicity hazard of small organic molecules. Models were established following the best practices for the development and validation of QSAR models. Scientists and regulators can use the STopTox portal to identify putative toxicants or nontoxicants in chemical libraries of interest. https://doi.org/10.1289/EHP9341
Introduction
Historically, regulatory agencies have required animal testing for hazard categorization and labeling (National Research Council Committee on Animals as Monitors of Environmental Hazards 1991). However, there have been multiple calls, especially in the last two decades, to Reduce, Refine, and Replace (three R’s) animal tests for hazard identification (Flecknell 2002; Patlewicz and Fitzpatrick 2016). The U.S. EPA estimated that the cost to approve a single pesticide may reach more than for several animal tests, reaching more than for carcinogenicity in rats or mice (U.S. EPA 2019b). In addition, studies have shown that animal-based assay outcomes do not always equate with human responses (Seok et al. 2013) and that animal models are less reproducible than some alternative methods (Luechtefeld et al. 2016c, 2016a, 2016b). The Strategic Roadmap published by the Interagency Coordinating Committee on the Validation of Alternative Methods (ICCVAM) as recently as in 2018 (ICCVAM 2018) called for the development of alternative, “new approach methods” (NAMs), for reducing animal testing of chemical and medical agents. In furthering this call, in September, 2019, the U.S. EPA issued a directive to reduce animal testing, including a commitment to “eliminate all mammal study requests and funding by 2035” (U.S. EPA 2019a). This directive creates a critical need to develop robust in vitro and computational tools for accurate and reliable hazard identification in chemical and pharmaceutical products as part of their regulatory assessment.
Computational approaches, such as structural alerts, read-across, and quantitative structure–activity relationship (QSAR) modeling, have earned broad acceptance as a weight of evidence for assessing chemical toxicity (ECHA 2017; U.S. EPA 2016). Structural alerts are molecular substructures that are associated with a particularly adverse outcome (Norman 2021). Read-across is a technique that proposes to identify potential hazards of untested compounds by associating them with structurally similar compounds that have been tested (Ball et al. 2016). QSAR modeling is a computational approach that employs statistical or machine learning techniques to establish correlations between intrinsic chemical properties (chemical descriptors) and measured properties or toxicological effects (Tropsha and Golbraikh 2007). QSAR modeling has been used extensively to model and predict chemical toxicity, and best practices for model development and validation have been developed to ensure their reliability (Tropsha 2010). Regulators have preferred both structural alerts and read-across approaches due to the ease of use, transparency, and mechanistic interpretability. However, there have been concerns that these tools often do not help with a reliable assessment of whether the underlying compounds present a real hazard to humans and the environment. For instance, we previously demonstrated that alerts have a tendency to flag compounds as toxic even when the experimental evidence shows otherwise (Alves et al. 2016b).
In the last several years, both our (Alves et al. 2018a; Borba et al. 2020; Braga et al. 2017) and other (Roberts et al. 2017; Toropova and Toropov 2017) groups have developed reliable computational models for predicting the skin sensitization potential of chemicals. These and other models developed for one or more of the “6-pack” end points are summarized in Table 1, which indicates that the development of reliable computational models for predicting the outcomes of all 6-pack tests is still a significant challenge. To address this challenge, we compiled, integrated, and curated a collection of experimental in vivo data on 6-pack end points, which, to the best of our knowledge, is the largest 6-pack data set in the public domain. Using this compiled data, we developed and rigorously validated QSAR models for all 6-pack assays and demonstrated their utility in identifying potentially safe or unsafe chemicals in industrial products (Figure 1). In addition, we integrated these models into a software package called STopTox (Systemic and Topical chemical Toxicity (STopTox) and made it publicly available to the research community via a dedicated web portal (https://stoptox.mml.unc.edu/). We especially emphasize, with vivid examples, the importance and impact of data curation on the rigor of our study design and the reliability of the study outcomes.
Table 1.
Computational software covering 6-pack end points.
| Software name | Endpoints | Computational approach | License | Access |
|---|---|---|---|---|
| Danish QSAR database | Acute oral, skin irritation and skin sensitization | Consensus model from ACDLabs, Leadscope, CASE Ultra, and SciQSAR | Free | http://qsar.food.dtu.dk/ |
| T.E.S.T. | Acute oral | QSAR | Free | https://www.epa.gov/chemical-research/users-guide-test-version-42-toxicity-estimation-software-tool-program-estimate |
| TOPKAT | Acute oral, Acute inhalation, eye irritation, skin irritation, and skin sensitization | QSAR | Commercial | https://www.toxit.it/en/services/software/topkat |
| ACD/Percepta | Acute oral, eye irritation, and skin irritation | QSAR | Commercial | https://www.acdlabs.com/products/percepta/index.php |
| CASE Ultra | Acute oral, acute inhalation, eye irritation, skin irritation, and skin sensitization | Structural alerts | Commercial | http://www.multicase.com/case-ultra |
| ToxTree | Eye irritation, skin irritation, and skin sensitization | Structural alerts | Free | http://toxtree.sourceforge.net/ |
| Derek Nexus | Irritation, skin sensitization | Structural alerts | Commercial | https://www.lhasalimited.org/products/derek-nexus.htm# |
| OECD QSAR Toolbox | Eye irritation, skin irritation, and skin sensitization | QSAR and read-across | Free | https://www.oecd.org/chemicalsafety/risk-assessment/oecd-qsar-toolbox.htm#Guidance_Documents_and_Training_Materials_for_Using_the_Toolbox |
| VEGA | Skin sensitization | Read-across | Free | https://www.vegahub.eu/ |
| CATMoS (OPERA) | Acute Oral | Consensus model | Free | https://github.com/NIEHS/OPERA |
| PredSkin (version 3.0) | Skin sensitization | QSAR | Free | https://ntp.niehs.nih.gov/whatwestudy/niceatm/test-method-evaluations/acute-systemic-tox/models/index.html |
| REACHAcross (RASAR) | All 6-pack | Read-across and QSAR | Commercial | https://www.ulreachacross.com/ |
Note: OECD, Organisation for Economic Co-operation and Development; QSAR, quantitative structure–activity relationship; RASAR, read-across structure–activity relationships.
Figure 1.
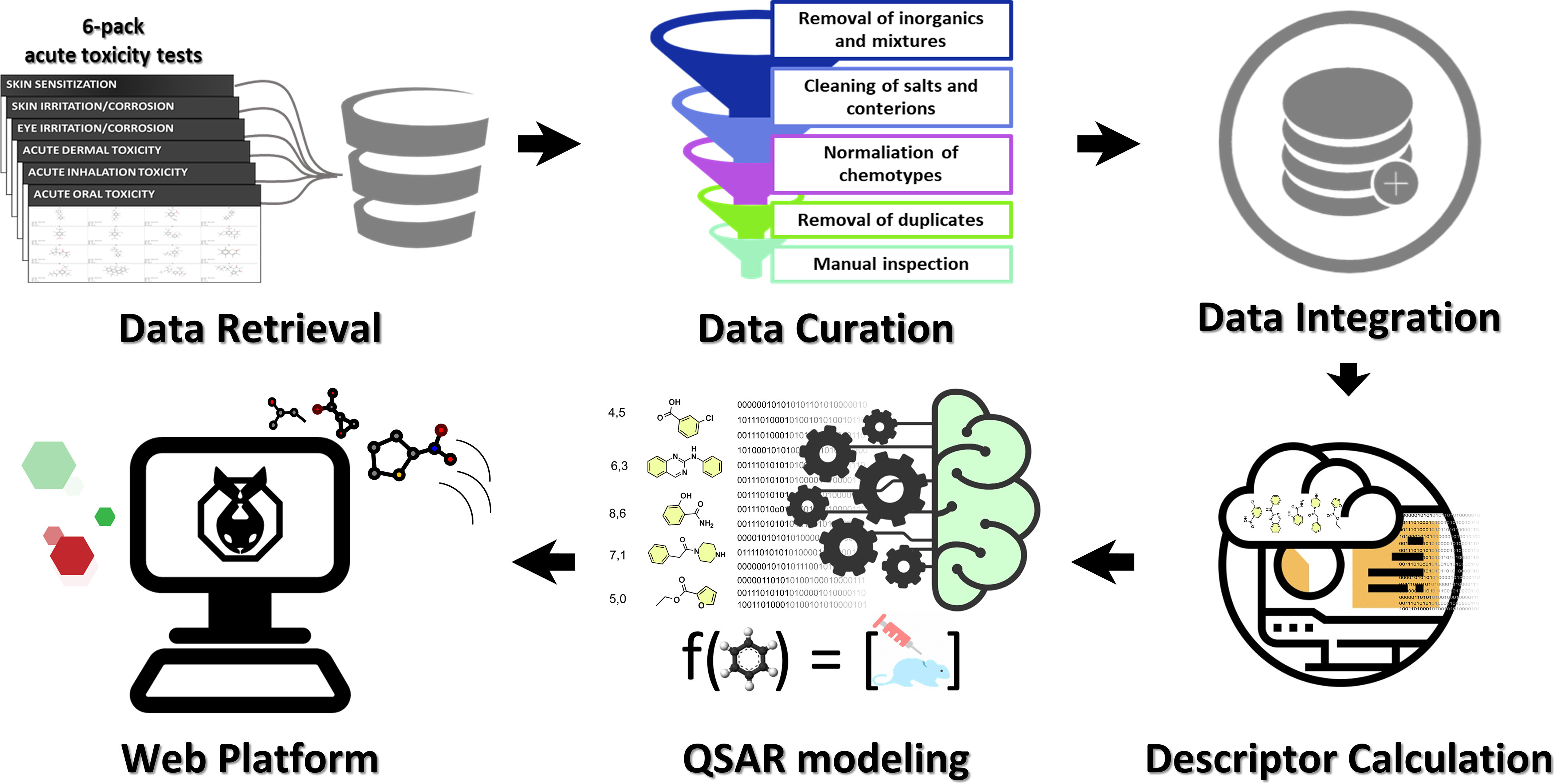
General workflow of STopTox. Experimental data from all 6-pack end points were collected and carefully curated following a well-stablished protocol accepted by the cheminformatics community. The data were then integrated and QSAR models were built for each end point individually. Finally, the models were implemented in a publicly available web application termed STopTox available at https://stoptox.mml.unc.edu/.
Materials and Methods
Data Collection
We compiled data from multiple publicly available databases and from the literature. These data encompass animal sources of the experimental tests for the following 6-pack end points: a) skin sensitization; b) skin irritation and corrosion; c) eye irritation and corrosion; and acute systemic toxicity via d) dermal, e) inhalation, and f) oral routes. The literature search was conducted using the PubMed database and Chemotext (Capuzzi et al. 2018) with the following search terms: “Skin sensitization” AND/OR “LLNA” AND/OR “QSAR” AND/OR “Read Across”; eye irritation AND/OR “Draize test” AND/OR “QSAR” AND/OR “Read Across”; skin irritation AND/OR “Draize test” AND/OR “QSAR” AND/OR “Read Across”; “acute oral toxicity” AND/OR “QSAR” AND/OR “Read Across”; “acute dermal toxicity” AND/OR “QSAR” AND/OR “Read Across”; and “acute inhalation toxicity” AND/OR “QSAR” AND/OR “Read Across.” No inclusion/exclusion criteria were used, and the last search date was executed in January 2019. All the replicate matches were done using only the standardized chemical structures, never identifiers or simplified molecular input line entry specification (SMILES). The CAS numbers were retrieved from PubChem (https://pubchem.ncbi.nlm.nih.gov/) when not available. All the curated data sets are available in Excel Tables S1–S7.
Data Curation
We extensively cleaned and standardized the data and converted measurements to the same units in each data set employing regular expressions to find essential features for the database that were described in text format; this approach was key to end point classification into Globally Harmonized System of Classification and Labeling of Chemicals (GHS) hazard classes. To convert the data into the binary toxicity calls, we followed the GHS classification criteria: for acute systemic end points GHS classes 1–4 were considered as “toxic,” and class 5 was considered as “not classified.” For skin irritation, classes 1–3 were considered as “irritant or corrosive”; for eye irritation, classes 1–2B were considered “irritants or corrosive”; and for skin sensitization, class 1 was considered “sensitizer.” The criteria for GHS classification are different for each end point and more information can be found elsewhere (UNECE 2019). Following this laborious data preparation and standardization, we conducted both chemical and biological data curation. This requisite attention to detailed data curation at different levels of the data preparation protocol is, unfortunately, uncommon in computational chemical toxicology, as we noted previously (Alves et al. 2019).
Data sets were thoroughly curated following the workflows developed by us earlier (Fourches et al. 2016). First, we excluded inconsistent data, which represented a big share of our data sets (Figure 2). Data were categorized as inconsistent if they were generated not following the OECD protocols; if compounds were not tested in multiple concentrations and could not be classified into GHS classes, labeled as nonexperimental (e.g., labeled as obtained using QSAR and/or read across predictions and/or weight of evidence decisions); if measurements were different from the standard protocols for the 6-pack end points: For systemic end points we only used median lethal dose () measurements; for skin sensitization, we used effective concentration, third percentile () measurements; for skin irritation, we used the mean scores for erythema and edema and reversibility information; and for eye irritation, we used corneal and iritis gradings and reversibility information, according to the GHS classification system (UNECE 2019). Biological data curation was followed by chemical structure curation: We removed mixtures, inorganics, and organometallic compounds; cleaned and neutralized salts; normalized the specific chemotypes; and applied the special treatment to chemicals with multiple replicated records as follows: a) when replicated records presented the same binary outcome, only one record was kept; b) when the majority of replicate chemicals presented the same binary outcome and one had different binary outcome, only one record with the most common binary outcome was kept; and c) when replicated records had different binary outcomes, all of them were removed. All the curated data are available in the Supplementary Material in xlsx format (Excel Tables S1–S7) and can also be downloaded in SDF from the STopTox web portal (https://stoptox.mml.unc.edu/) and GitHub (https://github.com/joyvb/stoptox).
Figure 2.
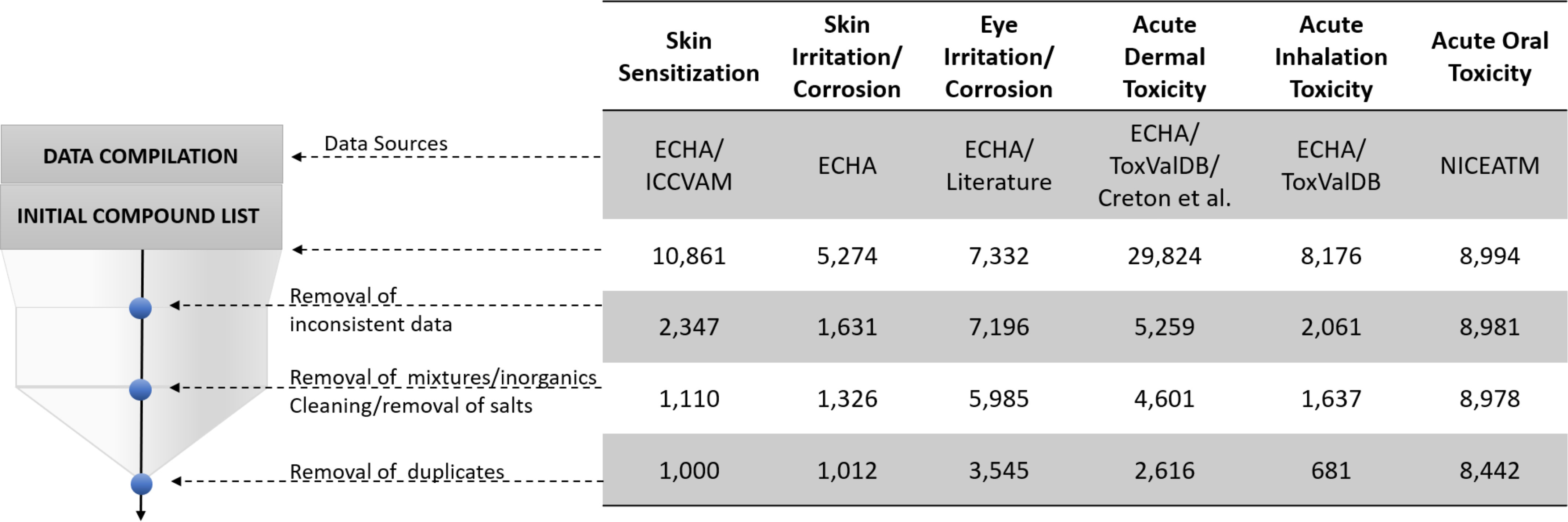
Summary of data curation steps. Data sources: ECHA (ECHA 2019; ECHA and OECD 2019), Interagency Coordinating Committee on the Validation of Alternative Methods (ICCVAM 2013), ToxValDB (Judson 2018), and National Toxicology Program Interagency Center for the Evaluation of Alternative Toxicological Methods (NICEATM; ICCVAM 2019).
Data Sets
Skin sensitization.
Skin sensitization data were compiled from two sources: a) National Toxicology Program Interagency Center for the Evaluation of Alternative Toxicological Methods on behalf of ICCVAM (ICCVAM 2013) and b) the publicly available Registration, Evaluation, Authorisation and restriction of Chemicals (REACH) study results database (ECHA and OECD 2019). The ICCVAM database included 1,060 chemical records with local lymph node assay (LLNA) data. Chemicals were classified as sensitizers/nonsensitizers following the Global Harmonization System (GHS) (UNECE 2019), where the presence of a dose that produces the stimulation index of three () was used as a threshold for a positive response. In other words, compounds without ( are classified as nonsensitizers, whereas those with a reported dose are classified as sensitizers.
After curation, 515 unique compounds (330 sensitizers and 185 nonsensitizers) were retained. The REACH database initially comprised 10,588 records for 9,801 chemicals. The REACH data set is composed of many types of assays and study categories. In vitro and weight of evidence categories were discarded. Data from different OECD skin sensitization assays (OECD guidelines 406, 411, 429 and 442B; OECD 1981, 1992, 2010a, 2010b) were available; only the data corresponding to LLNA assays (429 and 442B) were selected, resulting in 1,275 data points with LLNA records. After curation, 541 compounds (192 sensitizers and 349 nonsensitizers) were retained. Eventually, we merged the curated data from ICCVAM and REACH and examined the content of this combined data. There were 56 groups of replicated chemicals between these two data sets, and the sensitization potential of five of these pairs was different. These discordant records were removed, and only one record for each concordant set of replicates was kept. The merged data set had 1,000 unique compounds (481 sensitizers and 519 nonsensitizers).
Skin irritation and corrosion.
Experimental animal data on skin irritation and corrosion were retrieved from the REACH study results database (ECHA and OECD 2019). After removing inconsistent data, 1,631 out of the original 5,274 data points were left. After removal of mixtures, inorganics, and counter-ions, 1,326 records remained. We followed the GHS (UNECE 2019) to classify the data: If the mean erythema/edema score is bigger than 2.3 and the effects are reversible, the chemical is considered as an irritant. If the effect is irreversible and corrosive reactions are present, the chemical is considered to be corrosive to the skin (OECD 2015).
Among 124 replicate groups of chemicals in the data set, 95 had concordant and 29 had discordant toxicity calls. All the discordant replicates were removed, and only one representative of a pair/pool of concordant replicates was kept. The final data set had 1,012 unique chemical compounds, including 40 corrosives, 277 irritants, and 695 nonirritants. Because there were only a few corrosive compounds in our data set, we decided to merge the corrosive and irritant classes and model only irritant vs. nonirritant compounds. We note that these models have limited regulatory value at the moment with respect to compounds predicted to be toxic, because regulators typically would like to see more granular measurement or prediction at the level of specific subcategories of toxicity. However, we highlight and emphasize that our models make accurate predictions of nontoxic compounds, thereby helping both regulators and respective regulated industries to support the development of safer chemicals. Our resulting data set contained 317 irritants vs. 695 nonirritants. Because the data set was imbalanced, we applied an undersampling technique where the majority class was sampled in a way to match the number of records of the minority class. This sampling was done by searching for the compounds in the majority class that had higher similarity (Tanimoto coefficient) with compounds in the minority class. The balanced data set consisted of 554 compounds (277 irritants and 277 nonirritants).
Eye irritation and corrosion.
The eye irritation and corrosion data set was retrieved from the REACH study results database (ECHA and OECD 2019) and the literature (Adriaens et al. 2017; Barratt 1997, 1995; Barroso et al. 2017; Basant et al. 2016; Cruz-Monteagudo et al. 2006; Geerts et al. 2017; Verheyen et al. 2017; Verma and Matthews 2015). We first curated data from each source separately and then merged the curated data sets and checked for the overlapping compounds. We followed the OECD Test No. 405 (OECD 2017). The eye irritation assessment is based on scoring lesions of conjunctiva, cornea, and iris at specific intervals after application of a single dose of the test substance. If the effects are reversible after 21 d, the chemical is considered an irritant, and if the effect is irreversible, the chemical gets a corrosive flag.
After we removed the inconsistent data, 7,196 out of the original 7,332 experimental animal data points for eye irritation and corrosion remained. After we removed mixtures, inorganics, and counter-ions, 5,985 records remained. All the discordant replicates were removed, and only one representative of a pair/pool of concordant replicates was kept. The final data set had 3,545 unique chemical compounds, including 1,145 irritants and 2,400 nonirritants. Because the data set was imbalanced, we applied an undersampling technique where the majority class was sampled in a way to match the number of records of the minority class. This sampling was achieved by searching for the compounds in the majority class that had higher similarity (Tanimoto coefficient) with compounds in the minority class. The balanced data set consisted of 2,292 compounds (1,146 skin irritants and 1,146 nonirritants).
Acute dermal toxicity.
The acute dermal toxicity data set was retrieved from the REACH study results database (ECHA and OECD 2019), the publicly available database ToxValDB (Judson 2018), and from the literature (Creton et al. 2010). After removing the inconsistent data, 5,259 out of the original 29,824 data points were left; the major reason for compound removal was the presence of many compounds without a defined (. The GHS was used to classify the chemicals (UNECE 2019). The chemical was labeled as toxic if the was smaller than body weight (BW). After the removal of mixtures, inorganics, and organometallic compounds, 4,601 records remained. Among 1,979 groups of chemical replicates in the data set, 1,836 had concordant toxicity calls, and 143 were discordant. All the discordant replicates were removed, and only one representative of a pair/pool of concordant replicates was kept. The final data set had 2,616 unique chemical compounds, including 382 dermally toxic compounds and 2,234 Not Classified compounds. Because the data set was imbalanced, we applied an undersampling technique where the majority class was sampled to match the number of records of the minority class. This sampling was conducted by searching for the compounds in the majority class that had higher similarity (Tanimoto coefficient) with compounds in the minority class. The balanced data set consisted of 764 compounds, including 382 toxic compounds and 382 Not Classified compounds.
Acute inhalation toxicity.
The acute inhalation toxicity data set was retrieved from the REACH study results database (ECHA and OECD 2019) and from the publicly available database ToxValDB (Judson 2018). The chemicals were classified as toxic according to the GHS thresholds for gases: ; vapors: and dusts/mists: (UNECE 2019). After removing inconsistent data, only 2,061 out of the original 8,176 data points were left. This dramatic reduction of the data set was mainly because of the presence of many compounds without a defined and because of the absence of information regarding the exposure method used (gas, dust, or mist), which is essential for GHS classification. After the removal of mixtures, inorganics, and counter-ions, 1,637 records remained. Among 527 groups of chemical replicates in the data set, 501 had concordant toxicity calls, and 26 were discordant. All the discordant replicates were removed, and only one representative of a pair/pool of concordant replicates was kept. The final data set had 681 unique chemical compounds and was balanced because it included 345 toxic compounds and 336 Not Classified compounds.
Acute oral toxicity.
The acute oral toxicity data set was retrieved from the National Toxicology Program Interagency Center for the Evaluation of Alternative Toxicological Methods (NICEATM) workshop for the Collaborative Acute Toxicity Modeling Suite (CATMoS) project that our team was part of (ICCVAM 2019; Kleinstreuer et al. 2018; Mansouri et al. 2021). The GHS was used to classify the chemicals (UNECE 2019). If the is smaller than BW, then the chemical was labeled as toxic. After removing inconsistent data, 8,981 out of the original 8,994 data points were left. After removal of mixtures, inorganics, and counter-ions, 8,978 records remained. A total of 406 groups of chemical replicates were found in the data set. All the discordant replicates were removed, and only one representative of a pair/pool of concordant replicates was kept. The final data set has 8,442 unique chemical compounds, including 4,803 toxic compounds and 3,639 Not Classified compounds.
Cosmetic ingredient database (CosIng).
CosIng is the European Commission database for information on cosmetics substances and ingredients (European Commission 2017). This data set contained 5,166 chemical records with a defined chemical structure. After curation, 3,850 unique chemical substances were kept for virtual screening using the developed models. The virtual screening results are available Excel Table S8.
REACH.
The REACH data come from registration dossiers submitted to the European Chemicals Agency (ECHA) by May 2019 (ECHA 2019). The database contained 20,000 substances, of which 15,438 were chemical records with a defined chemical structure. After curation, 10,465 unique chemical substances were kept for prediction purposes. The virtual screening results are available in Excel Table S9.
Cheminformatics approaches.
Binary QSAR models were developed and rigorously validated according to the best practices of QSAR modeling (Tropsha 2010). Two-dimensional Morgan fingerprints (Aslett et al. 2010) and Molecular ACCess System (MACCS) keys (Anderson 1984), calculated with RDKit package (version 2020.03.1.0), and Mordred, calculated with Mordred package (version 1.2.0) for Python (Moriwaki et al. 2018) were combined with random forest (Breiman 2001) algorithm (RandomForestClassifier) implemented in scikit-learn (version 1.0) (Pedregosa et al. 2012) for model development.
We followed a proper external 5-fold cross-validation procedure. First, the entire data set was split into five parts of the same size. Then, for each iteration, one of these subsets (20% of compounds) was used as a test set, and the other four sets (80% of compounds) were used as the training set. We repeated this procedure five times until each of the five subsets was used once as a test set. In addition, each training set was internally divided into multiple training and validation sets for model training and hyperparameter tuning. The models were generated using only the training set. The true test sets were never employed to generate or to select the models. We repeated this procedure using three different types of descriptors (Morgan, MACCS, and Mordred). The final statistics were based on the consensus (average prediction) of these models. The consensus model considers the majority rule (at least two out of three) for the final classification.
In every case, only the modeling set was used to develop the models, whereas the external sets were used for the evaluation of their predictive power. In addition, 10 rounds of Y-randomization were performed for each data set to ensure that the model performance was not due to chance correlations. The applicability domain (AD) of the models was estimated using the z-cutoff method (Tropsha and Golbraikh 2007) along with dice similarity. In the STopTox web app, the user can visualize the similarity distribution of the training set and how far the query compound is from the threshold (see Figure S1). If the query compound is below the threshold, then it is outside the model’s applicability domain. If it is above the threshold, then it is inside. All the codes used to generate the models are available at https://github.com/joyvb/stoptox.
The predictive performance of QSAR models was evaluated using correct classification rate (CCR), sensitivity (SE), specificity (SP), and positive (PPV) and (NPV) predictive values (Equations 1–5):
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
where N represents the number of compounds, TP and TN represent the number of true positives and true negatives, and FP and FN represent the number of false positives and false negatives, respectively.
Additional external validation using known toxicants from the literature.
For additional external validation of our models, we conducted a literature search for toxicants that were absent in our database but were later described elsewhere. We used the PubMed database and Chemotext web portal (Capuzzi et al. 2018) with the following search criteria: “Skin sensitization” OR “Skin sensitizers” AND “Clinical studies”/“Skin irritation” OR “Skin irritants” AND “Clinical studies”/“Eye irritation” OR “Eye irritants” AND “Clinical studies”/“Acute oral toxicity” AND “compound” AND “Clinical studies”/“Acute dermal toxicity” AND “compound” AND “Clinical studies”/“Acute inhalation toxicity” AND “compound” AND “Clinical studies.” No inclusion/exclusion criteria were used, and the last search date was executed in July 2020. After collecting these data, we curated the data (see “Data Curation” section) and analyzed it to ensure that they were not included in the modeling data set. Then, we employed STopTox models to predict each toxicant and analyzed whether our models were capable of correctly predicting their toxicity potential.
Virtual screening.
We applied the developed QSAR models to predict toxicities for compounds included in the CosIng and REACH databases as well as to augment the STopTox data matrix, which is extremely sparse, to identify additional putative toxicants. Both CosIng and REACH databases are described in the “Data Sets” section. The virtual screening results for CosIng, REACH, and STopTox are available in Excel Tables S8, S9, and S10, respectively.
Model implementation.
The STopTox web-based application runs machine learning routines written in Python by using Flask (version 2.03; Python Software Foundation), a small framework for creating web microframeworks at the back end. Models were developed using Scikit-Learn (version 1.0) (Scikit-Learn Developers). Angular (version 4) and Typescript were used for the development of the frontend, and Docker 19.03 and Docker-Compose (version 1.27.0) for the orchestration of containers. The developed models and all data sets are publicly available at https://stoptox.mml.unc.edu/.
Results
QSAR Modeling
Statistical characteristics of QSAR models developed in this study are summarized in Table 2. All cross-validated models for the 6-pack end points showed high predictive accuracy on independent external evaluation sets based on several metrics, including CCR, SE, SP, PPV, and NPV. The acute toxicity models showed CCR of 70%–77%; SE of 66%–85%; SP of 66%–80%; PPV of 69%–79%; and NPV of 71%–78%. A literature search executed after models were developed identified toxicants that were absent in our data sets. We used these compounds as an additional validation set for our models (see the section below).
Table 2.
Statistical characteristics of QSAR models for 6-pack end points evaluated by 5-fold external cross-validation.
| End point | CCR | Se | Sp | PPV | NPV | Coverage | Number of compounds |
|---|---|---|---|---|---|---|---|
| Skin sensitization | 0.70 | 0.66 | 0.75 | 0.71 | 0.75 | 0.96 | 1,000 |
| Skin irritation/corrosion | 0.72 | 0.77 | 0.66 | 0.69 | 0.74 | 0.94 | 1,012 |
| Eye irritation/corrosion | 0.72 | 0.72 | 0.71 | 0.71 | 0.71 | 0.95 | 3,547 |
| Acute dermal | 0.76 | 0.74 | 0.78 | 0.77 | 0.75 | 0.93 | 2,622 |
| Acute inhalation | 0.74 | 0.69 | 0.80 | 0.77 | 0.72 | 0.95 | 681 |
| Acute oral | 0.77 | 0.85 | 0.70 | 0.79 | 0.78 | 0.95 | 8,442 |
Note: CCR, correct classification rate; NPV, negative predictive value; PPV, positive predictive value; QSAR, quantitative structure–activity relationship; Se, sensitivity; Sp, specificity.
Model Validation with Known Toxicants Not Used in Model Development
For additional external validation of our models, we conducted a literature search for toxicants described in clinical studies or known toxicants for each end point that were absent in our database. We found 45 compounds for skin sensitization, 2 compounds for skin irritation, 3 compounds for eye irritation, 2 compounds for acute dermal toxicity, 5 compounds for acute inhalation toxicity, and 2 compounds for acute oral toxicity.
Altogether, our models correctly predicted 18 out of 25 (72%) of the known toxicants identified in the literature that were not present in our modeling set (see Figure 3 and Excel Table S11).
Figure 3.

Toxicants identified in the literature using PubMed and Chemotext (see “Data Collection” section in “Materials and Methods” section) that were absent in our modeling data. STopTox predictions for a given end point are listed below each structure’s contribution map, also generated with STopTox.
For skin sensitization, a list of 45 potential skin sensitizers in cosmetic ingredients was compiled by the Norwegian Scientific Committee for Food Safety (Norwegian Scientific Committee for Food Safety 2007). Eleven out of 45 compounds were absent from our skin sensitization data set, and 8 of the 11 chemicals were correctly predicted as sensitizers by our skin sensitization model [].
For skin irritation, we found the compounds MS-222, a fish anesthetic commonly used in aquaculture (Park 2019), and sodium lauryl sulfate (De Jongh et al. 2006), a product widely used in personal care products—both known skin irritants that were not present in our skin irritation training data. Our models predicted sodium lauryl sulfate as a skin irritant and MS-222 as not classified (according to OECD Test No. 404 (OECD 2015), chemicals not classified as skin irritants are considered “Not Classified”).
We found three compounds that were not present in our eye irritation data set: glutaraldehyde, glyphosate, and Paraquat (1,1’-Dimethyl-4,4’-bipyridinium dichloride). Our model predicted glutaraldehyde and glyphosate as eye irritants. Exposure to glutaraldehyde during cataract surgery was associated to the development of toxic eye anterior segment syndrome in six patients (Ünal et al. 2006). Ocular glyphosate exposure was reported to be associated with the development of chemosis, heart palpitations, raised blood pressure, headache, and nausea (Bradberry et al. 2004). In two cases of accidental eye exposure to Paraquat, eye damage was reported (Joyce 1969).
For the acute dermal end point, the compounds dichloromethane (Pacheco et al. 2016) and methanol (Kahn and Blum 1979) have been reported as systemic toxicants after dermal exposure and were not present in the modeling set. Our acute dermal toxicity model correctly predicted both compounds as toxic after dermal exposure.
For acute inhalation end point, 20 chemicals commonly present in occupational inhalation accidents were compiled elsewhere (Miller and Chang 2003). There were five organic chemicals in this list that were absent in our acute inhalation data set. All five compounds were correctly predicted by acute inhalation models.
For acute oral end point, we found one clinical case of accidental oral exposure to the pyrethroid deltamethrin that led to the poisoning of a 4-y-old girl who consumed insecticidal chalk and was found unconscious 20 min after going outside to play (O’Malley 1997). We also found that mephedrone, a psychoactive drug, has been proven toxic in a study reporting cases of acute toxicity related to self-reported use of mephedrone (Wood et al. 2010). Our acute oral toxicity model predicted both compounds as toxic if swallowed.
Figure 4 shows the predictions generated for N-phenyl-p-phenylenediamine, a known skin sensitizer usually added to temporary black henna tattoos, leading to many cases of contact allergy (Panfili et al. 2017). We also generated maps showing the relative significance of fragment contributions, providing a graphical interpretation of developed models (Figure 4). Atoms and structural fragments enhancing toxicity are highlighted in pink, and those decreasing toxicity are shown in green. These maps are generated for each of the 6-pack end points independently. Overall, these maps allow the user to analyze the individual contribution of each fragment for acute toxicity, facilitating a mechanistic interpretation of reported predictions.
Figure 4.
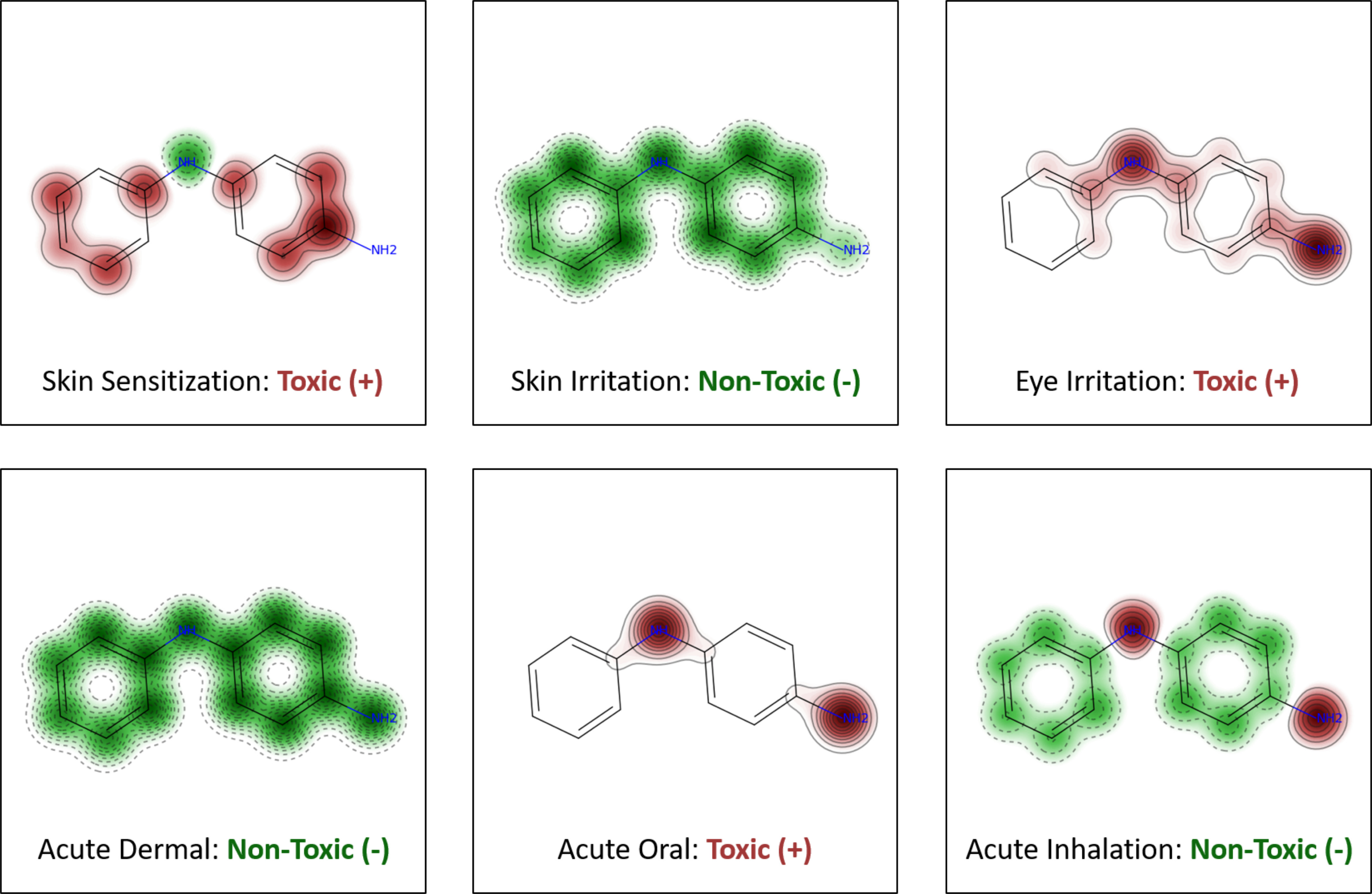
Maps of fragment contributions and predictions of each model for N-phenyl-p-phenylenediamine. The predicted fragment contribution of a toxic effect is accompanied by the map of the atomic contributions to toxicity. Red regions with continuous lines indicate the fragment is predicted to increase toxicity. Green regions with dashed lines indicate the fragment is predicted to decrease the toxicity.
Virtual Screening of CosIng, REACH, and STopTox Compounds
In the CosIng data set ( compounds), 1,366 compounds were predicted as skin sensitizers, 1,152 compounds were predicted as skin irritants; 1,674 compounds were predicted as eye irritants; 361 compounds were predicted as toxic if swallowed; 301 compounds were predicted as toxic if inhaled; and 257 compounds were predicted as toxic after dermal exposure. Out of 3,850 total compounds, there were 2,695 compounds predicted as toxic in at least one end point and 1,155 compounds predicted as “Not Classified” in all six end points.
In the REACH data set ( compounds), 4,018 compounds were predicted as skin sensitizers; 2,445 compounds were predicted as skin irritants; 4,605 compounds were predicted as eye irritants; 2,679 compounds were predicted as toxic if swallowed; 2,139 compounds were predicted as toxic if inhaled; and 1,899 compounds were predicted as toxic after dermal exposure. There were 7,641 compounds predicted as toxic in at least one end point and 2,824 compounds predicted as “Not Classified” in all six end points.
In the STopTox data set ( compounds with missing toxicity values), 4,792 compounds were predicted as skin sensitizers; 2,491 compounds were predicted as skin irritants; 4,766 compounds were predicted as eye irritants; 5,232 compounds were predicted as toxic if swallowed; 2,394 compounds were predicted as toxic if inhaled; and 2,902 compounds were predicted as toxic after dermal exposure. There were 7,641 compounds predicted as toxic in at least one end point and 2,824 compounds predicted as Not Classified in all six end points.
Model Implementation in the STopTox Web App
The QSAR models were implemented in the STopTox web app (https://stoptox.mml.unc.edu/). STopTox has an intuitive user interface in which the user may draw a compound of interest in the “molecular editor” box or directly paste the SMILES string of the chemical structure of interest. After hitting the “Predict STopTox” button, the user will receive the predicted outcomes (e.g., toxic, nontoxic) using the QSAR models developed for each of the 6-pack acute toxicity end points. For each prediction, we also list its confidence based on how close the compound is to the model AD estimate (Tropsha and Golbraikh 2007); we also provide visual mechanistic interpretation of the prediction using color-coded maps of predicted fragment contribution (Riniker and Landrum 2013). In this algorithm, the predicted contribution of an atom is obtained by accessing the difference in the prediction if each bit corresponding to that atom/fragment is removed. Then, the normalized contribution is used to color the atoms in a topography-like map. Using these maps, the structural fragments predicted to increase the respective toxicity are highlighted in red, and the fragments predicted to decrease toxicity are highlighted in green. Gray isolines define the frontier between the positive (red) and the negative (green) contributions (see Figures 3 and 4). In addition, the prediction confidence, which is estimated by the majority voting of internal models (number of trees) in the random forest algorithm (Breiman 2001), is also available.
Discussion
Data Curation and QSAR Model Development
Although predictive models have been developed and reported previously for subsets of the 6-pack end points (Table 1), many of these models did not fully comply with the model validation guidelines specified by the OECD (OECD 2004) and, most notably, lacked proper data curation. Notably, in our study, we allocated a significant effort toward the curation of both chemical and biological data using robust protocols established by our group previously (Fourches et al. 2010, 2016). As can be seen in Figure 2, data curation had a dramatic effect on the size of the data: It decreased the size of the available data, in all but one case, by about 52%–92%. Our final database comprised a matrix containing 11,941 compounds with activity measurements for at least one of the 6-pack end points (sparsity degree of 76%).
Previously, we built models to predict skin sensitization end points using a combination of animal (Alves et al. 2015), OECD-validated in vitro assays (Alves et al. 2018a), and human data (Alves et al. 2016a, 2018a; Borba et al. 2020). In this study, we developed QSAR models for predicting skin sensitization testing outcomes using only the LLNA because STopTox is intended as a reliable NAM for the 6-pack assays. The models were thoroughly validated by employing the best practices for model development and validation suggested by the OECD to employ QSAR models for regulatory purposes (OECD 2007). The models showed high accuracy when evaluated by 5-fold external cross-validation and by predicting and additional set of known toxicants external to the models (see the “Results” section). Therefore, all the models reported here were built using only data collected from animal tests that followed the OECD protocols.
Comparative Assessment of New 6-Pack Models vs. Alternative Tools: The Importance of Data Curation
The ECHA database curation proved to be an extremely laborious task and the most time-consuming part of this work. It is important to emphasize that much of the 6-pack end point data included in the ECHA database could not (and should not) be used for model development. As seen from the summary of data curation (Figure 2), the major reduction in the size of individual data sets used eventually for QSAR model development was due to a large fraction of inconsistent data in the original ECHA database. Data were categorized as inconsistent if they were generated not following the OECD protocols, if compounds were tested in few or only one concentration and could not be classified into GHS classes, labeled as nonexperimental (e.g., labeled as obtained using QSAR and/or read across predictions and/or weight of evidence decisions) or found in complex mixtures. In addition, for each end point, we kept only the data containing measurement for the standard OECD protocol (see “Materials and Methods” section). As an example, the report on acute inhalation toxicity for Diboron trioxide (ECHA 2020b) lacks very important information, such as the animal species used for testing, route of administration, and duration of exposure. GHS classification for acute inhalation toxicity depends on the route of administration (UNECE 2019), making it difficult to classify this compound as toxic or nontoxic. The OECD guidelines also state that the test must be done in rats with 4 h of exposure to the tested chemical. Calcium iodate, an inorganic, was reported as an eye irritant from category 2A based on a QSAR prediction (ECHA 2020a). ECHA presented other reports showing clinical evidence of eye irritation in humans. Still, because categorization is done based on animal tests, we could not trust these data for modeling. There were many other examples of incomplete/nontrustable reports from the databases, which significantly decreased the data set size.
Comparison of our results with models developed for the same end points without rigorous data curation (Luechtefeld et al. 2018) suggests that our extensive data curation procedures resulted in the decreased data set size and, formally, lower than reported model performance. Indeed, we compared models produced in this study to those reported by Luechtefeld et al. (2018), who described the development of a suite of in silico models, termed read-across structure–activity relationships (RASAR) for the 6-pack end points. Because the model predictions based on RASAR could only be accessed through a fee-based commercial platform (https://www.ulreachacross.com, which now is defunct), we performed an indirect comparison of the respective statistics (see Table 3). Our models showed, on average, a 10% lower CCR. The amount of data reported in the study mentioned above (Luechtefeld et al. 2018) was, on average, five times larger than the size of the carefully curated data set used in this study. Previously, we already expressed concerns that the high accuracy of models as reported (Luechtefeld et al. 2018) could be the consequence of inadequate data curation, leaving many replicate compounds in the modeling and validation data sets (Alves et al. 2021). We posit that our results reflect the actual model performance for these end points more accurately because we eliminated such confounders as replicate entries or the use of predicted or “not reliable” values and conducted more rigorous validation procedures according to the established guidelines. We strongly suggest that our exercise reemphasizes the importance of proper data curation and cautions against overinterpreting results from models built on noncurated data sets.
Table 3.
Indirect comparison of STopTox (5-fold external cross-validation) and RASAR (as reported in the original publication).
| End point | Number of chemicals | CCR | Sensitivity | Specificity | ||||
|---|---|---|---|---|---|---|---|---|
| RASAR* | STopTox | RASAR* | STopTox | RASAR* | STopTox | RASAR* | STopTox | |
| Skin sensitization | 7,670 | 1,000 | 0.88 | 0.7 | 0.8 | 0.73 | 0.96 | 0.68 |
| Skin irritation/corrosion | 46,331 | 1,012 | 0.86 | 0.72 | 0.75 | 0.67 | 0.86 | 0.76 |
| Eye irritation/corrosion | 48,767 | 3,547 | 0.84 | 0.77 | 0.99 | 0.72 | 0.7 | 0.81 |
| Acute dermal | 11,252 | 2,622 | 0.92 | 0.77 | 0.89 | 0.79 | 0.94 | 0.75 |
| Acute inhalation | 11,369 | 681 | 0.91 | 0.76 | 0.9 | 0.72 | 0.91 | 0.79 |
| Acute oral | 32,411 | 8,465 | 0.9 | 0.78 | 0.94 | 0.78 | 0.86 | 0.78 |
Note: CCR, correct classification rate; RASAR, read-across structure–activity relationships. *Data retrieved from (Luechtefeld et al. 2018).
STopTox Usability and Interpretation
It is essential to note that, if the model predicts a compound as toxic or nontoxic, such prediction should be considered only in the context of specific dose-dependent observation for each assay; obviously, increasing the dose of any compound in any assay could often lead to toxic effects. For instance, the skin sensitization potencies for substances are based on a function of lymph node cell proliferation induced by the test chemical and expressed as a stimulation index (SI) relative to values obtained with concurrent controls. If , the substance is considered as a sensitizer in the tested concentration. Similar considerations were applied in transforming the results of measurement into binary format for other end points.
These considerations are often overlooked when making predictions or assertions concerning the expected chemical toxicity. The ultimate goal of any method for evaluating acute toxicity is to provide an accurate assessment of the potential risk of a chemical concerning human safety (Basketter et al. 2015). Therefore, we reinforce that the limitation of assays should influence both the interpretation of the predictions made by the models and the use of these models to help toxicologists in their decision-making. Predictions with QSAR models implemented in STopTox (actually, with any models) do not take the dose into account; they merely state whether a chemical is predicted to be toxic or nontoxic in each assay. Thus, users interpreting these predictions should always be familiar with and keep in mind the underlying experimental conditions under which compounds in the training sets were denoted as toxic or nontoxic. Further, these models are limited to binary hazard-based predictions, rather than providing information on potency and GHS or U.S. EPA subcategorization. Therefore, they are not directly applicable for many regulatory classifications and labeling requirements requiring a higher level of granularity. However, these models are well suited to assist in hazard assessment and chemical screening/prioritization, and, because of their high accuracy in terms of both sensitivity and specificity, they can be instrumental in identifying nontoxic compounds (tested in the same conditions as those identified as toxic where additional subcategorization is indeed necessary).
Virtual Screening of CosIng, REACH, and STopTox Compounds
As a case study illustrating STopTox usability, we applied our QSAR models to the European Commission CosIng database, REACH, and STopTox data matrix (sparsity degree of 76%), including AD estimation. Most compounds in each of these data sets were predicted as “Not Classified” by each individual model. The predictions of acute toxicity of these databases illustrate QSAR models’ utility for prioritizing chemicals of concern for targeted biological testing in different chemical spaces such as cosmetics, pesticides, and industrial chemicals (Alves et al. 2018b). All compounds and corresponding predictions are available in the Supplemental Material.
Conclusions
STopTox is a comprehensive collection of computational models that can be used as an alternative to in vivo 6-pack tests for predicting chemical toxicity hazard. Models were established following the best practices for the development and validation of QSAR models (OECD 2004; Tropsha 2010) using the largest publicly available and carefully curated data sets that we compiled for all 6-pack assays. To the best of our knowledge, STopTox is the first publicly available portal that enables accurate prediction of chemical hazards in all the 6-pack end points at once using a model developed with transparent approaches and carefully curated data. Despite the model limitations concerning potency classes, they are reliable for predicting chemicals that do not require regulatory classification, such as in the early stages of drug discovery (Hasselgren and Myatt 2018). We suggest that these models are valuable for both regulatory agencies and respective industries in helping them identify safer chemicals using inexpensive in silico alternatives to in vivo testing of chemicals of interest. We reinforce that, to build predictive models, it is not enough just to use adequate chemical descriptors and powerful machine learning algorithms (Fourches et al. 2016); we shall stress that STopTox is the only 6-pack end point predictor in the public domain developed with extensively curated data and OECD-compliant modeling approaches. The STopTox web app provides users with access to statistically significant and externally predictive QSAR models of acute toxicity tests. The web app can rapidly evaluate acute toxicity hazards in chemical inventories. STopTox is freely available at https://stoptox.mml.unc.edu/. To the best of our knowledge, STopTox does not have analogs in terms of the level of data curation, validated statistical accuracy of constituting models, transparency of the data, modeling methods and software tools, and public accessibility.
Supplemental Material
Supplemental Material includes curated data sets for each of the 6-pack end points and results for the virtual screening of the STopTox matrix, and CosIng, and REACH databases in xlsx format.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health (NIH) (grants 1U01CA207160, R41ES033589, and 1R43ES032371) and CNPq (grant 400760/2014-2). J.B. thanks the CNPq and the Science without Borders program for the financial support of her visit to the University of North Carolina at Chapel Hill. V.A. thanks the Lush Prize.
Each author has contributed significantly to this work. J.V.B.B., V.M.A., C.H.A., E.N.M., and A.T. conceived and designed the study. J.V.B.B., K.O., A.C.S., S.U.S.H., and E.O. curated the data and developed the models. J.V.B.B., V.M.A., N.K., J.S., D.A., C.H.A., E.N.M., and A.T. analyzed the data. R.B. and D.K. incorporated the models into the STopTox web application. J.V.B.B., V.M.A., and E.N.M. wrote the first draft of the manuscript. All authors read, edited, and approved the final manuscript.
References
- Adriaens E, Alépée N, Kandarova H, Drzewieckac A, Gruszka K, Guest R, et al. 2017. CON4EI: selection of the reference chemicals for hazard identification and labelling of eye irritating chemicals. Toxicol In Vitro 44:44–48, PMID: , 10.1016/j.tiv.2017.06.001. [DOI] [PubMed] [Google Scholar]
- Alves VM, Auerbach SS, Kleinstreuer N, Rooney JP, Muratov EN, Rusyn I, et al. 2021. Curated data in—trustworthy in silico models out: the impact of data quality on the reliability of artificial intelligence models as alternatives to animal testing. Altern Lab Anim 49(3):73–82, PMID: , 10.1177/02611929211029635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves VM, Borba J, Capuzzi SJ, Muratov E, Andrade CH, Rusyn I, et al. 2019. Oy vey! A comment on “machine learning of toxicological big data enables read-across structure activity relationships outperforming animal test reproducibility.” Toxicol Sci 167(1):3–4, PMID: , 10.1093/toxsci/kfy286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves VM, Capuzzi SJ, Braga RC, Borba JVB, Silva AC, Luechtefeld T, et al. 2018a. A perspective and a new integrated computational strategy for skin sensitization assessment. ACS Sustainable Chem Eng 6(3):2845–2859, 10.1021/acssuschemeng.7b04220. [DOI] [Google Scholar]
- Alves VM, Capuzzi SJ, Muratov E, Braga RC, Thornton T, Fourches D, et al. 2016a. QSAR models of human data can enrich or replace LLNA testing for human skin sensitization. Green Chem 18(24):6501–6515, PMID: , 10.1039/C6GC01836J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves VM, Muratov EN, Capuzzi SJ, Politi R, Low Y, Braga RC, et al. 2016b. Alarms about structural alerts. Green Chem 18(16):4348–4360, PMID: , 10.1039/C6GC01492E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves VM, Muratov EN, Fourches D, Strickland J, Kleinstreuer N, Andrade CH, et al. 2015. Predicting chemically-induced skin reactions. Part II: QSAR models of skin permeability and the relationships between skin permeability and skin sensitization. Toxicol Appl Pharmacol 284(2):273–280, PMID: , 10.1016/j.taap.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves VM, Muratov EN, Zakharov A, Muratov NN, Andrade CH, Tropsha A. 2018b. Chemical toxicity prediction for major classes of industrial chemicals: is it possible to develop universal models covering cosmetics, drugs, and pesticides? Food Chem Toxicol 112:526–534, PMID: , 10.1016/j.fct.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S. 1984. Graphical representation of molecules and substructure-search queries in MACCStm. J Mol Graph 2(3):83–90, 10.1016/0263-7855(84)80060-0. [DOI] [Google Scholar]
- Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, et al. 2010. TriTrypDB: a functional genomic resource for the trypanosomatidae. Nucleic Acids Res 38(Database issue):D457–D462, PMID: , 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball N, Cronin M, Shen J, Blackburn K, Booth EDED, Bouhifd M, et al. 2016. Toward good read-across practice (GRAP) guidance. ALTEX 33(2):149–166, PMID: , 10.14573/altex.1601251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt MD. 1995. A quantitative structure-activity relationship for the eye irritation potential of neutral organic chemicals. Toxicol Lett 80(1–3):69–74, PMID: , 10.1016/0378-4274(95)03338-l. [DOI] [PubMed] [Google Scholar]
- Barratt MD. 1997. QSARS for the eye irritation potential of neutral organic chemicals. Toxicol In Vitro 11(1–2):1–8. PMID: , 10.1016/S0887-2333(96)00063-X. [DOI] [PubMed] [Google Scholar]
- Barroso J, Pfannenbecker U, Adriaens E, Alépée N, Cluzel M, De Smedt A, et al. 2017. Cosmetics Europe compilation of historical serious eye damage/eye irritation in vivo data analysed by drivers of classification to support the selection of chemicals for development and evaluation of alternative methods/strategies: the Draize eye test Reference Database (DRD). Arch Toxicol 91(2):521–547, PMID: , 10.1007/s00204-016-1679-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basant N, Gupta S, Singh KP. 2016. A three-tier QSAR modeling strategy for estimating eye irritation potential of diverse chemicals in rabbit for regulatory purposes. Regul Toxicol Pharmacol 77:282–291, PMID: , 10.1016/j.yrtph.2016.03.014. [DOI] [PubMed] [Google Scholar]
- Basketter DA, White IR, McFadden JP, Kimber I. 2015. Skin sensitization: implications for integration of clinical data into hazard identification and risk assessment. Hum Exp Toxicol 34(12):1222–1230, PMID: , 10.1177/0960327115601760. [DOI] [PubMed] [Google Scholar]
- Borba J, Braga RC, Alves VM, Muratov EN, Kleinstreuer NC, Tropsha A, et al. 2020. Pred-Skin: a web portal for accurate prediction of human skin sensitizers. Chem Res Toxicol 34(2):258–267, PMID: , 10.1021/acs.chemrestox.0c00186. [DOI] [PubMed] [Google Scholar]
- Bradberry SM, Proudfoot AT, Vale JA. 2004. Glyphosate poisoning. Toxicol Rev 23(3):159–167, PMID: , 10.2165/00139709-200423030-00003. [DOI] [PubMed] [Google Scholar]
- Braga RC, Alves VM, Muratov EN, Strickland J, Kleinstreuer N, Tropsha A, et al. 2017. Pred-Skin: a fast and reliable web application to assess skin sensitization effect of chemicals. J Chem Inf Model 57(5):1013–1017, PMID: , 10.1021/acs.jcim.7b00194. [DOI] [PubMed] [Google Scholar]
- Breiman L. 2001. Random forests. Mach Learn 45(1):5–32, 10.1023/A:1010933404324. [DOI] [Google Scholar]
- Capuzzi SJ, Thornton TE, Liu K, Baker N, Lam WI, O’Banion CP, et al. 2018. Chemotext: a publicly available web server for mining drug–target–disease relationships in PubMed. J Chem Inf Model 58(2):212–218, PMID: , 10.1021/acs.jcim.7b00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creton S, Dewhurst IC, Earl LK, Gehen SC, Guest RL, Hotchkiss JA, et al. 2010. Acute toxicity testing of chemicals—opportunities to avoid redundant testing and use alternative approaches. Crit Rev Toxicol 40(1):50–83, PMID: , 10.3109/10408440903401511. [DOI] [PubMed] [Google Scholar]
- Cruz-Monteagudo M, González-Díaz H, Borges F, González-Díaz Y. 2006. Simple stochastic fingerprints towards mathematical modeling in biology and medicine. 3. Ocular irritability classification model. Bull Math Biol 68(7):1555–1572, PMID: , 10.1007/s11538-006-9083-y. [DOI] [PubMed] [Google Scholar]
- De Jongh CM, Verberk MM, Withagen CET, Jacobs JJL, Rustemeyer T, Kezic S. 2006. Stratum corneum cytokines and skin irritation response to sodium lauryl sulfate. Contact Dermatitis 54(6):325–333, PMID: , 10.1111/j.0105-1873.2006.00848.x. [DOI] [PubMed] [Google Scholar]
- European Commission. 2017. Cosmetic Ingredient Database (CosIng). https://ec.europa.eu/growth/sectors/cosmetics/cosing_en [accessed 13 September 2017].
- ECHA (European Chemical Agency), OECD (Organization for Economic Co-operation and Development). 2019. REACH Study Results – IUCLID. https://iuclid6.echa.europa.eu/reach-study-results [accessed 10 July 2019].
- ECHA. 2017. Chapter R.7a: Endpoint specific guidance. In: Guidance on Information Requirements and Chemical Safety Assessment. https://echa.europa.eu/documents/10162/17224/information_requirements_r7a_en.pdf/e4a2a18f-a2bd-4a04-ac6d-0ea425b2567f?t=1500286622893 [accessed 8 February 2020].
- ECHA. 2019. Registered Substances. https://echa.europa.eu/information-on-chemicals/registered-substances [accessed 27 August 2019].
- ECHA. Calcium Iodate. 2020a. https://echa.europa.eu/registration-dossier/-/registered-dossier/5810/7/4/3 [accessed 13 September 2020].
- ECHA. 2020b. Diboron Trioxide - Acute Toxicity: Inhalation. https://echa.europa.eu/registration-dossier/-/registered-dossier/6449/7/3/3 [accessed 14 September 2018].
- Flecknell P. 2002. Replacement, reduction and refinement. ALTEX 19:73–78. PMID: 12098013. [PubMed] [Google Scholar]
- Fourches D, Muratov E, Tropsha A. 2010. Trust, but verify: on the importance of chemical structure curation in cheminformatics and QSAR modeling research. J Chem Inf Model 50(7):1189–1204, PMID: , 10.1021/ci100176x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourches D, Muratov E, Tropsha A. 2016. Trust, but verify II: a practical guide to chemogenomics data curation. J Chem Inf Model 56(7):1243–1252, PMID: , 10.1021/acs.jcim.6b00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerts L, Adriaens E, Alépée N, Guest R, Willoughby JA, Kandarova H, et al. 2017. CON4EI: evaluation of QSAR models for hazard identification and labelling of eye irritating chemicals. Toxicol In Vitro 49:90–98, PMID: , 10.1016/j.tiv.2017.09.004. [DOI] [PubMed] [Google Scholar]
- Hasselgren C, Myatt GJ. 2018. Computational Toxicology and Drug Discovery. In: Computational Toxicology. Methods in Molecular Biology, vol. 1800. Nicolotti O, ed. New York, NY: Humana Press, 233–244. [DOI] [PubMed] [Google Scholar]
- ICCVAM (Interagency Coordinating Committee on the Validation of Alternative Methods). 2013. NICEATM Murine Local Lymph Node Assay (LLNA) Database. https://ntp.niehs.nih.gov/whatwestudy/niceatm/test-method-evaluations/skin-sens/llna/index.html [accessed 15 January 2015].
- ICCVAM. 2018. A Strategic Roadmap for Establishing New Approaches to Evaluate the Safety of Chemicals and Medical Products in the United States. https://ntp.niehs.nih.gov/pubhealth/evalatm/natl-strategy/index.html [accessed 10 October 2021].
- ICCVAM. 2019. Collaborative Acute Toxicity Modeling Suite (CATMoS) Tool for Predicting Acute Oral Toxicity. https://ntp.niehs.nih.gov/iccvamreport/2019/technology/comp-tools-dev/catmos/index.html [accessed 31 July 2019].
- Joyce M. 1969. Ocular damage caused by paraquat. Br J Ophthalmol 53(10):688–690, PMID: , 10.1136/bjo.53.10.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R. 2018. ToxValDB: Compiling Publicly Available In Vivo Toxicity Data. https://cfpub.epa.gov/si/si_public_record_Report.cfm?dirEntryId=344315&Lab=NCCT [accessed 1 December 2018].
- Kahn A, Blum D. 1979. Methyl alcohol poisoning in an 8-month-old boy: an unusual route of intoxication. J Pediatr 94(5):841–843, 10.1016/S0022-3476(79)80176-6. [DOI] [PubMed] [Google Scholar]
- Kleinstreuer NC, Karmaus AL, Mansouri K, Allen DG, Fitzpatrick JM, Patlewicz G. 2018. Predictive models for acute oral systemic toxicity: a workshop to bridge the gap from research to regulation. Comput Toxicol 8(11):21–24, PMID: , 10.1016/j.comtox.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luechtefeld T, Maertens A, Russo DP, Rovida C, Zhu H, Hartung T. 2016a. Analysis of Draize eye irritation testing and its prediction by mining publicly available 2008–2014 REACH data. ALTEX 33(2):123–134, PMID: , 10.14573/altex.1510053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luechtefeld T, Maertens A, Russo DP, Rovida C, Zhu H, Hartung T. 2016b. Analysis of public oral toxicity data from REACH registrations 2008–2014. ALTEX 33(2):111–122, PMID: , 10.14573/altex.1510054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luechtefeld T, Maertens A, Russo DP, Rovida C, Zhu H, Hartung T. 2016c. Analysis of publically available skin sensitization data from REACH registrations 2008–2014. ALTEX 33(2):135–148, PMID: , 10.14573/altex.1510055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luechtefeld T, Marsh D, Rowlands C, Hartung T. 2018. Machine learning of toxicological big data enables read-across structure activity relationships (RASAR) outperforming animal test reproducibility. Toxicol Sci 165(1):198–212, PMID: , 10.1093/toxsci/kfy152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri K, Karmaus AL, Fitzpatrick J, Patlewicz G, Pradeep P, Alberga D, et al. 2021. CATMoS: collaborative acute toxicity modeling suite. Environ Health Perspect 129(4):47013. PMID: 33929906, 10.1289/EHP8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriwaki H, Tian Y-S, Kawashita N, Takagi T. 2018. Mordred: a molecular descriptor calculator. J Cheminform 10(1):4, PMID: , 10.1186/s13321-018-0258-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K, Chang A. 2003. Acute inhalation injury. Emerg Med Clin North Am 21(2):533–557, 10.1016/s0733-8627(03)00011-7. [DOI] [PubMed] [Google Scholar]
- National Research Council Committee on Animals as Monitors of Environmental Hazards. 1991. Animals as Sentinels of Environmental Health Hazards. Washington, DC: National Academies Press. https://www.ncbi.nlm.nih.gov/books/NBK234944/ [accessed 30 July 2021]. [Google Scholar]
- Norman B. 2021. Structure Alerts. In: Burger’s Medicinal Chemistry and Drug Discovery. Abraham DJ, ed. Hoboken, NJ: Wiley, 1–28. [Google Scholar]
- Norwegian Scientific Committee for Food Safety. 2007. Sensitisation caused by exposure to cosmetic products: opinion of the Panel on Food Additives, Flavourings, Processing Aids, Materials in Contact with Food and Cosmetics of the Norwegian Scientific Committee for Food Safety. https://vkm.no/download/18.2994e95b15cc545071635494/1501167574463/60ba3e8fe1.pdf.
- O’Malley M. 1997. Clinical evaluation of pesticide exposure and poisonings. Lancet 349:1161–1166, PMID: , 10.1016/S0140-6736(96)07222-4. [DOI] [PubMed] [Google Scholar]
- OECD (Organisation for Economic Co-operation and Development). 1981. Test No. 411: Subchronic Dermal Toxicity: 90-day Study. http://www.oecd-ilibrary.org/environment/test-no-411-subchronic-dermal-toxicity-90-day-study_9789264070769-en [accessed 31 July 2017].
- OECD. 1992. Test No. 406: Skin Sensitisation. http://www.oecd-ilibrary.org/environment/test-no-406-skin-sensitisation_9789264070660-en [accessed 16 March 2017].
- OECD. 2004. OECD Principles for the Validation, for Regulatory Purposes, of (Quantitative) Structure-Activity Relationship Models. http://www.oecd.org/chemicalsafety/risk-assessment/37849783.pdf [accessed 31 July 2017].
- OECD. 2007. Guidance Document on the Validation of (Quantitative) Structure-Activity Relationship [(Q)SAR] Models. https://www.oecd-ilibrary.org/environment/guidance-document-on-the-validation-of-quantitative-structure-activity-relationship-q-sar-models_9789264085442-en [accessed 30 August 2018].
- OECD. 2010a. Test No. 429 Skin Sensitization: Local Lymph Node Assay. OECD Guidelines for Testing Chemicals, Section 4. https://www.oecd-ilibrary.org/environment/test-no-429-skin-sensitisation_9789264071100-en [accessed 16 March 2017].
- OECD. 2010b. Test No. 442B: Skin Sensitization. http://www.oecd-ilibrary.org/environment/test-no-442b-skin-sensitization_9789264090996-en [accessed 16 March 2017].
- OECD. 2015. Test No. 404: Acute Dermal Irritation/Corrosion. http://www.oecd.org/env/test-no-404-acute-dermal-irritation-corrosion-9789264242678-en.htm [accessed 30 November 2017].
- OECD. 2017. Test No. 405: Acute Eye Irritation/Corrosion. https://www.oecd-ilibrary.org/environment/test-no-405-acute-eye-irritation-corrosion_9789264185333-en [accessed 28 May 2018].
- Pacheco C, Magalhães R, Fonseca M, Silveira P, Brandão I. 2016. Accidental intoxication by dichloromethane at work place: clinical case and literature review. J Acute Med 6(2):43–45, 10.1016/j.jacme.2016.03.008. [DOI] [Google Scholar]
- Panfili E, Esposito S, Di Cara G. 2017. Temporary black henna tattoos and sensitization to para-Phenylenediamine (PPD): two paediatric case reports and a review of the literature. Int J Environ Res Public Health 14(4):421. PMID: 28420106, 10.3390/ijerph14040421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I-S. 2019. The anesthetic effects of clove oil and MS-222 on far Eastern catfish, Silurus asotus. Dev Reprod 23(2):183–191, PMID: , 10.12717/DR.2019.23.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patlewicz G, Fitzpatrick JM. 2016. Current and future perspectives on the development, evaluation, and application of in silico approaches for predicting toxicity. Chem Res Toxicol 29(4):438–451, PMID: , 10.1021/acs.chemrestox.5b00388. [DOI] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. 2012. Scikit-learn: machine learning in python. J Mach Learn Res 12:2825–2830. , 10.1007/s13398-014-0173-7.2. [DOI] [Google Scholar]
- Riniker S, Landrum G. A. 2013. Similarity maps - A visualization strategy for molecular fingerprints and machine-learning methods. J Cheminform 5(1):43, PMID: , 10.1186/1758-2946-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DW, Aptula A, Api AM. 2017. Structure–potency relationships for epoxides in allergic contact dermatitis. Chem Res Toxicol 30(2):524–531, PMID: , 10.1021/acs.chemrestox.6b00241. [DOI] [PubMed] [Google Scholar]
- Russell WMS. 1959. On comfort and comfort activities in animals. UFAW Courier 16:14–26. [Google Scholar]
- Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, et al. 2013. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A 110(9):3507–3512, PMID: , 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toropova AP, Toropov AA. 2017. Hybrid optimal descriptors as a tool to predict skin sensitization in accordance to OECD principles. Toxicol Lett 275:57–66, PMID: , 10.1016/j.toxlet.2017.03.023. [DOI] [PubMed] [Google Scholar]
- Tropsha A, Golbraikh A. 2007. Predictive QSAR modeling workflow, model applicability domains, and virtual screening. Curr Pharm Des 13(34):3494–3504, PMID: , 10.2174/138161207782794257. [DOI] [PubMed] [Google Scholar]
- Tropsha A. 2010. Best practices for QSAR model development, validation, and exploitation. Mol Inform 29(6–7):476–488, PMID: , 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- Ünal M, Yücel I, Akar Y, Öner A, Altin M. 2006. Outbreak of toxic anterior segment syndrome associated with glutaraldehyde after cataract surgery. J Cataract Refract Surg 32(10):1696–1701, PMID: , 10.1016/j.jcrs.2006.05.008. [DOI] [PubMed] [Google Scholar]
- UNECE (United Nations Economic Commission for Europe). 2019. GHS (Rev.8) (2019): Globally Harmonized System of Classification and Labelling of Chemicals (GHS). Part 3.3. https://unece.org/ghs-rev8-2019 [accessed 30 December 2017].
- U.S. EPA (U.S. Environmental Protection Agency). 2016. Process for evaluation and implementing alternative approaches to traditional in vivo acute toxicity studies for FIFRA regulatory use. Available: https://www.epa.gov/pesticide-science-and-assessing-pesticide-risks/process-establishing-implementing-alternative [accessed 10 December 2018].
- U.S. EPA. 2019a. Cost Estimates of Studies Required for Pesticide Registration. https://www.epa.gov/pesticide-registration/cost-estimates-studies-required-pesticide-registration [accessed 29 May 2020].
- U.S. EPA. 2019b. Directive to Prioritize Efforts to Reduce Animal Testing. https://www.epa.gov/sites/production/files/2019-09/documents/image2019-09-09-231249.pdf [accessed 6 June 2021].
- Verheyen GR, Braeken E, Van Deun K, Van Miert S. 2017. Evaluation of existing (Q)SAR models for skin and eye irritation and corrosion to use for REACH registration. Toxicol Lett 265:47–52, PMID: , 10.1016/j.toxlet.2016.11.007. [DOI] [PubMed] [Google Scholar]
- Verma RP, Matthews EJ. 2015. An in silico expert system for the identification of eye irritants. SAR QSAR Environ Res 26(5):383–395, PMID: , 10.1080/1062936X.2015.1039578. [DOI] [PubMed] [Google Scholar]
- Wood DM, Davies S, Greene SL, Button J, Holt DW, Ramsey J, et al. 2010. Case series of individuals with analytically confirmed acute mephedrone toxicity. Clin Toxicol (Phila) 48(9):924–927, PMID: , 10.3109/15563650.2010.531021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


