Abstract
The newly formulated Mycobacterium kansasii AccuProbe was evaluated, and the results obtained with the new version were compared to the results obtained with the old version of this test by using 116 M. kansasii strains, 1 Mycobacterium gastri strain, and 19 strains of several mycobacterial species. The sensitivity of this new formulation was 97.4% and the specificity was 100%. Still, three M. kansasii strains were missed by this probe. To evaluate the variability within the species, genetic analyses of the hsp65 gene, the spacer sequence between the 16S and 23S rRNA genes, and the 16S rRNA gene of several M. kansasii AccuProbe-positive strains as well as all AccuProbe-negative strains were performed. Genetic analyses of the one M. gastri strain from the comparative assay and of two further M. gastri strains were included because of the identity of the 16S rRNA gene in M. gastri to that in M. kansasii. The data confirmed the genetic heterogeneity of M. kansasii. Furthermore, a subspecies with an unpublished hsp65 restriction pattern and spacer sequence was described. The genetic data indicate that all M. kansasii strains missed by the AccuProbe test belong to one subspecies, the newly described subspecies VI, as determined by the hsp65 restriction pattern and the spacer sequence. Since the M. kansasii strains that are missed are rare and all M. gastri strains are correctly negative, the new formulated AccuProbe provides a useful tool for the identification of M. kansasii.
Mycobacterium kansasii is one of the most important causes of pulmonary disease resulting from nontuberculous mycobacteria. Although the clinical picture of patients with M. kansasii infection resembles that of patients with tuberculosis, treatment of M. kansasii infection differs from that of regular tuberculosis. Due to this, a rapid means of identification of this species is essential.
Routine laboratory identification of M. kansasii relies upon growth characteristics and several biochemical tests, like photochromogenicity, Tween hydrolysis, nitrate reduction, and the pattern of susceptibility to several antituberculosis drugs. Several weeks are required to verify the results.
For the prompt identification of mycobacteria, molecular methods are gaining increasing importance due to their rapidity and, in most cases, unequivocal results. PCR of the hsp65 gene followed by restriction enzyme analysis (PCR-restriction fragment length polymorphism [PCR-RFLP] analysis) is a rapid technique for the identification of mycobacteria (14). However, difficulties arise from the small sizes of the fragments and marginal differences between the several mycobacterial species. Furthermore, the PCR-RFLP patterns of unknown species are not published and may correspond to the patterns of other species. Sequencing of the hsp65 gene (6), the 16S rRNA gene (7), the spacer region between the 16S rRNA and the 23S rRNA genes (5, 12, 18), or the 32-kDa protein gene (13) are powerful techniques for the identification of mycobacterial species. Yet, these techniques are laborious and require large-scale technical equipment. Tests with commercially available DNA probes (AccuProbe; GenProbe, San Diego, Calif.) are easy to perform and make possible rapid identifications. They are developed for the identification of the M. tuberculosis complex, the M. avium complex, M. avium, M. intracellulare, M. gordonae, and M. kansasii. Several studies have shown the high degrees of sensitivity of the DNA probes for the M. tuberculosis complex, the M. avium complex, and M. gordonae and have shown that their use allows the rapid and correct identification of these frequently isolated species. In contrast, AccuProbes for M. kansasii have been reported to have a low sensitivity and miss from 7 up to 56% of the strains (8, 11, 15, 16; unpublished observations).
Genotypic heterogeneity among M. kansasii isolates has been reported from several studies (1, 4, 19, 20). PCR-RFLP analysis of the hsp65 gene led to the classification of five subspecies of M. kansasii (3, 9). Furthermore, differences in the sequence of the 16S-23S rRNA gene spacer region have been shown to exist (2).
A new, improved version of the DNA probe for the identification of all M. kansasii strains is now available (AccuProbe for M. kansasii; GenProbe).
In our study, we comparatively evaluated the sensitivity and the specificity of the previous and the improved versions of the AccuProbe M. kansasii test. To elucidate the combined two AccuProbe results on a molecular basis, we characterized the hsp65 sequences and PCR-RFLP patterns, 16S rRNA gene sequences, and 16S-23S rRNA gene spacer region of several strains.
MATERIALS AND METHODS
Strains analyzed.
A total of 116 M. kansasii strains (strain ATCC 12478 and 115 clinical isolates), 1 strain of M. gastri, and 19 strains of several mycobacterial species (M. celatum, M. fortuitum, M. gilvum, M. gordonae, M. lentiflavum, M. marinum, M. paraffinicum, M. simiae, M. tuberculosis, and M. xenopi) were included in this study. All strains were identified by using classical biochemical tests, AccuProbes, and/or sequencing.
Samples of five further strains (three M. kansasii and two M. gastri strains) with which only the new formulated AccuProbe test for M. kansasii was performed were included.
For molecular biological analysis, 18 M. kansasii strains were selected on the basis of the results of both the old and the new AccuProbe assays for the strains: 3 strains with negative results by both AccuProbe assays, 7 strains which were negative with the old detection kit but positive with the new one, 7 strains with positive results by both assays, as well as strain ATCC 12478. Furthermore, three M. kansasii strains that were negative by the new AccuProbe assay but with which no old assays were performed were included. Moreover, three M. gastri strains (one of which was tested by both versions of the AccuProbe assay) were also analyzed on a genetic basis.
AccuProbe assays.
M. kansasii AccuProbe assays were performed according to the manufacturer’s instructions. For the comparative assays with the old and the new kits, two bacterial lysates were prepared from each strain. After lysis, the samples were combined and mixed. This sample (100 μl) was transferred to each of the AccuProbe tubes containing the old or the new DNA probe. Hybridization results, expressed as relative light units (RLUs), were determined with a Leader 50 luminometer (GenProbe). According to the manufacturer’s instructions the cutoff value was 30,000 RLUs.
DNA for molecular biological analyses.
Bacterial DNA for molecular biological analyses was obtained as follows. One loopful of bacteria was suspended in distilled water, subjected to sonication for 15 min, and boiled for 15 min in a water bath. This suspension was directly used for PCR analyses.
Determination of the sequence of a part of the 16S rRNA gene.
A 590-bp fragment of the mycobacterial 16S rRNA gene was amplified by PCR as described by Richter et al. (10). Sequencing was performed with an automated DNA sequencer (ABI Prism 377; Applied Biosystems, Foster City, Calif.) with one of the PCR primers (primer A) and one nested primer (primer 11 [5′-GACAAACCACCTACGAGCTC]) by using the RR DyeDeoxy terminator sequencing kit (Applied Biosystems). Analyses and comparisons of the sequences were done with DNasis software (Hitachi, Olivet Cedex, France). The resulting DNA sequences were compared to the sequences of the International Nucleotide Sequence Database Collaboration (4a).
Determination of the sequence of the spacer region between the 16S and 23S rRNA genes.
The internal spacer region was amplified with primers targeting the 3′ end of the 16S rRNA gene (ITS1 [5′-GATTGGGACGAAGTCGTAAC]) or the 5′ end of the 23S rRNA gene (ITS2 [5′-AGCCTCCCACGTCCTTCATC]). PCR was performed at an annealing temperature of 57°C for 35 cycles. The PCR product was sequenced with primer ITS2. The resulting DNA sequences were compared to the sequences of the International Nucleotide Sequence Database Collaboration (4a).
Sequence determination and PCR-restriction fragment analysis of a part of the hsp65 gene.
PCR of the hsp65 gene was performed as described by Telenti et al. (14). Both strands of the PCR products were sequenced with the PCR primers. The resulting sequences were aligned and were analyzed for restriction sites with the restriction enzymes BstEII and HaeIII. For restriction fragment analysis, 20 μl of the PCR products was digested either with BstEII (Boehringer Mannheim, Mannheim, Germany) or with HaeIII (Boehringer Mannheim), and 20 μl of the restriction mixture was run on a 4% NuSieve 3:1 agarose gel (Biozym, Hessisch-Oldendorf, Germany). Molecular weight marker VIII (Boehringer Mannheim) served as an external molecular size marker.
RESULTS
Results of the AccuProbe tests.
Of the 116 M. kansasii strains tested by the two versions of the AccuProbe assay, 88 strains (including strain ATCC 12478) were identified with the old kit, whereas 113 strains were positive with the new kit (Table 1). The three strains not detected with the new kit (1,615, 2,026, and 2,783 RLUs) were also negative with the old kit. Neither the M. gastri strain (3,254 RLUs by the old test; 2,699 RLUs by the new test) nor the other mycobacterial strains had a false-positive AccuProbe result with the probes in both kits.
TABLE 1.
Results obtained with the two versions of the AccuProbe assay
| Species | Total no. of strains | No. of strains positive/no. of strains negative by the following assay:
|
|
|---|---|---|---|
| Old assay | New assay | ||
| M. kansasii | 116 | 88/28 | 113/3 |
| M. gastri | 1 | 0/1 | 0/1 |
| Others | 19 | 0/19 | 0/19 |
The sensitivity and specificity of the old M. kansasii AccuProbe test were 75.8 and 100%, respectively. In contrast, the sensitivity of the new formulated test reached 97.4%, and the specificity was 100%.
Molecular biological analyses.
To further analyze the genetic heterogeneity of the M. kansasii strains and to compare it to the features of M. gastri, we performed molecular analyses of the hsp65 gene, the spacer sequence between the 16S and the 23S rRNA genes, and the 16S rRNA gene.
hsp65 gene.
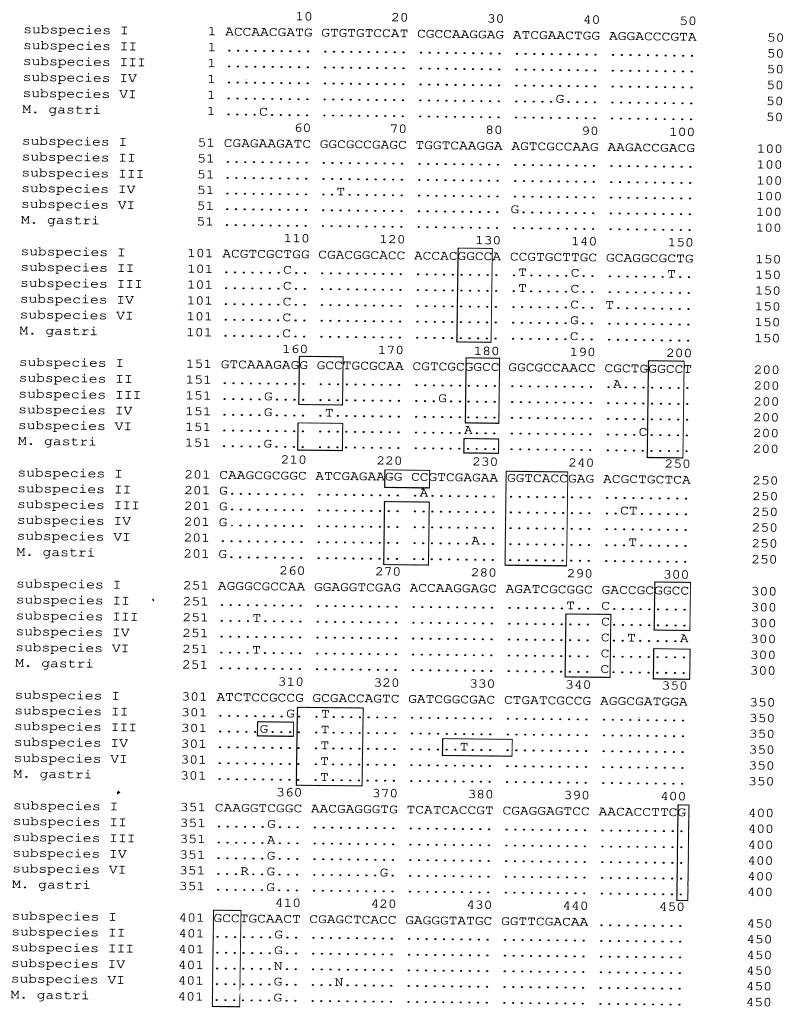
Sequencing of the 443-bp fragment resulting from the PCR and analysis of the restriction sites in this fragment resulted in six groups that differed in their DNA sequences and the restriction sites (Fig. 1 and Table 2). On the basis of their restriction patterns the groups were denominated as M. kansasii subspecies I to IV and M. gastri as described by Picardeau et al. (9) (Fig. 2). Subspecies I includes strain ATCC 12478. However, the previously described M. kansasii subspecies V (9) could not be detected in our study. In addition to the strains classified as described above, six strains with a unique DNA sequence were found (Fig. 1). BstEII digestion of the PCR products of these strains resulted in a pattern identical to that for subspecies III strains, whereas HaeIII digestion yielded a moderately larger fragment compared to that found in subspecies III strains (Fig. 2 and Table 2). Thus, these M. kansasii strains also exhibit unique restriction patterns. However, this pattern is identical to the pattern for M. gastri, although the hsp65 sequences of both species differ in stretches not affected by BstEII or HaeIII digestion (Fig. 1). Subsequently, these M. kansasii strains are denominated subspecies VI.
FIG. 1.
Alignment of the sequences of the hsp65 PCR fragments of five M. kansasii subspecies and M. gastri. Marked sequences represent the recognition sites for HaeIII (GGCC) and BstEII (GGTNACC).
TABLE 2.
Fragment lengths of the hsp65 PCR products after restriction with BstEII and HaeIII, deduced from sequence analysis, and comparison to published data, deduced from agarose gel electrophoresis
| Species |
BstEII fragment length (bp)
|
HaeIII fragment length (bp)
|
||
|---|---|---|---|---|
| Present study | Published data | Present study | Published data | |
| M. kansasii | ||||
| Subspecies I | 231, 212 | 245, 220 | 127, 103, 78, 42, 34, 23, 19, 17 | 140, 105, 80 |
| Subspecies II | 231, 133, 79 | 245, 145, 85 | 127, 103, 101, 42, 34, 19, 17 | 140, 105 |
| Subspecies III | 231, 133, 79 | 245, 145, 85 | 127, 94, 69, 42, 34, 23, 19, 17, 9, 9 | 140, 100, 70 |
| Subspecies IV | 231, 118, 79, 15 | 245, 125, 85 | 127, 112, 69, 51, 42, 23, 19 | 140, 115, 70 |
| Subspecies VI | 231, 133, 79 | 127, 103, 69, 42, 36, 34, 23, 9 | ||
| M. gastri | 231, 133, 79 | 245, 145, 85 | 127, 103, 69, 42, 34, 23, 19, 17, 9 | 140, 105, 70 |
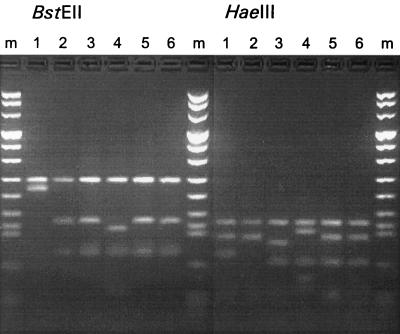
FIG. 2.
BstEII and HaeIII restriction analysis of the hsp65 PCR fragment of M. kansasii subspecies I, II, III, IV, and VI and of M. gastri. Lanes: m, molecular size marker (fragment lengths, 1,114, 900, 692, 501, 404, 320, 242, 190, 147, 124, 110, 67, 37, and 19 bp); 1, subspecies I; 2, subspecies II; 3, subspecies III; 4, subspecies IV; 5, subspecies VI; 6, M. gastri.
Spacer sequence between 16S and 23S rRNA genes.
Sequencing of the spacer region resulted in five different sequences for M. kansasii and another unique sequence for M. gastri (Fig. 3). Comparison of the sequences to those of the International Nucleotide Sequence Database, the sequences of subspecies I, III, and VI were identical to those stored in the GenBank database (accession nos. L42262, L42263, and L42264, respectively), whereas the sequence of subspecies IV showed 99% identity to the sequence of one M. gastri strain (EMBL database accession no. Y14182). The sequence of M. gastri was the same as that of another strain of this species stored in the EMBL database (accession no. X97633). No entry with a sequence identical to the sequence of subspecies II was found.
FIG. 3.
Alignment of the spacer sequences of five M. kansasii subspecies and M. gastri. For comparison, the order was altered to M. kansasii subspecies I, II, III, VI, and IV and M. gastri.
The spacer sequence of the so far undescribed subspecies VI showed a high degree of homology to the sequence of M. kansasii subspecies III. In total, the sequences of subspecies III and VI differ at 13 bp (Fig. 3).
The sequences of M. kansasii type IV and M. gastri were very similar, differing at only 6 bp (Fig. 3).
For all strains the grouping into subspecies made on the basis of the spacer sequences was concordant to the grouping made on the basis of the hsp65 sequences.
16S rRNA gene.
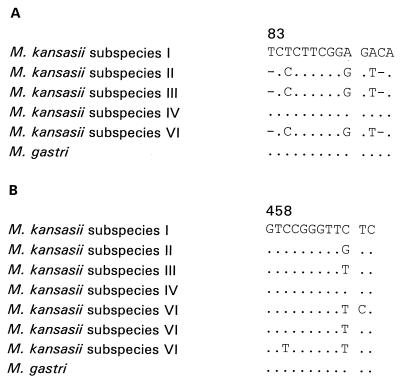
Sequencing of the first 600 bp of the 16S rRNA gene, including the species-specific region V2 of helix 10 as well as the variable V3 region of the helix 18, was performed. All M. kansasii and all M. gastri strains investigated in this study by molecular methods were characterized by the M. kansasii- and M. gastri-specific sequence at bases 121 to 139 and 176 to 255 (according to the numbering for Escherichia coli) (7). However, in 12 strains a different, shorter sequence comprising bases 83 to 96 and, furthermore, single base substitutions in the V3 region were found (Fig. 4). These strains are characterized by the presence of the hsp65 sequence and the spacer sequences of subspecies II, III, and VI. The V3 regions of M. kansasii subspecies VI strains were not identical to each other (Fig. 4B). Yet, this variability could not be seen in the spacer sequence and was detectable for only 1 bp (bp 354; R = A or T; Fig. 1) in the hsp65 gene. M. kansasii subspecies I and IV as well as M. gastri have sequences identical to those in the database.
FIG. 4.
Alignment of two parts of the 16S rRNA gene of the five M. kansasii subspecies and M. gastri. (A) Bases 83 to 96 (according to the numbering for E. coli); (B) part of the variable region V3 of helix 18 (bases 458 to 469 according to the numbering for E. coli).
Comparison of the results of the genetic analyses with the AccuProbe results.
The new AccuProbe version clearly identifies M. kansasii subspecies I, II, III, and IV, (Table 3). However, strains of subspecies VI remain unidentified with this version. Those strains could be identified by a positive nitrate reaction and a positive photochromogenic reaction, or by sequencing. All M. gastri strains were negative by the AccuProbe assay.
TABLE 3.
Summary of results of molecular analyses of the hsp65 PCR fragment, the 16S-23S spacer sequence, and the 16S rRNA gene as well as the AccuProbe results
| Subspecies according to hsp65 sequence | Spacer type | 16S rRNA gene sequencea
|
AccuProbe assay result
|
No. of strains | ||
|---|---|---|---|---|---|---|
| bp 83–96 | bp 458–469 | Old version | New version | |||
| I | I | Acc. dbb | GTC CGG GTT CTC | + | + | 7 |
| II | II | Variantc | GTC CGG GTT GTC | − | + | 6 |
| III | III | Variant | GTC CGG GTT TTC | + | + | 1 |
| IV | IV | Acc. db | GTC CGG GTT CTC | − | + | 1 |
| VI | VI | Variant | GTC CGG GTT TCC | − | − | 1 |
| VI | VI | Variant | GTC CGG GTT TTC | − | − | 1 |
| VI | VI | Variant | GTT CGG GTT TTC | − | − | 1 |
| VI | VI | Variant | GTT CGG GTT TTC | NDd | − | 3 |
| M. gastri | M. gastri | Acc. db | GTC CGG GTT CTC | − | − | 1 |
| M. gastri | M. gastri | Acc. db | GTC CGG GTT CTC | ND | − | 2 |
No discrepant results concerning genetic features and AccuProbe assay results within a subspecies of M. kansasii and M. gastri were observed.
DISCUSSION
Identification of M. kansasii.
Identification of M. kansasii can be performed with a high sensitivity with the newly formulated AccuProbe kit. In our comparative study, 113 of 116 M. kansasii strains were correctly identified with this kit. The specificity of the test with the old version of the test kit was already excellent, and it remains so with the new version. We found no strains with false-positive results.
Similar results were reported by Tortoli et al. (17). In contrast to our study, they observed no false-negative results, although they tested a large number of isolates. Furthermore, they reported that two M. gastri strains had false-positive results. Maybe these strains were mistyped as M. gastri and were actually subspecies IV M. kansasii strains which are negative with the old version of the test kit but positive with the new version of the AccuProbe test kit.
In our study six strains remained unidentified with the new kit, namely, strains of subspecies VI. This raises the question of whether these strains are really M. kansasii. However, all biochemical test results as well as the presence of the specific 16S rRNA gene and the high degree of similarity of the subspecies VI spacer sequence to that of the subspecies III spacer sequence confirm that these strains are M. kansasii. Since they were isolated from clinical specimens (respiratory specimens and blood) they are presumably pathogenic for humans. Thus, rapid identification of these strains is necessary.
Frequency of occurrence of M. kansasii subspecies in the present study.
Since the strains used in the present study were derived from a routine laboratory, conclusions on the frequency of occurrence of the different M. kansasii subspecies can be drawn. The relative distributions of the different subspecies may be estimated by the combined AccuProbe assay and sequencing results. Strains belonging to subspecies I and III are characterized by a positive result by the old AccuProbe test (Table 3). Thus, all 88 strains (75%) positive with the old kit belong to these two subspecies. In our molecular analysis, eight of these strains have been analyzed, and only one could be shown to belong to subspecies III. Hence, the frequency of occurrence of this subspecies in the present study may be low, whereas most strains are subspecies I strains. This is in accordance with the results reported from other studies (2).
Subspecies II and IV are characterized by a negative test result with the old version of the AccuProbe kit but are positive with the new version. Thus, 25 strains (21%) investigated in the present study with the two versions of the AccuProbe kits belong to these two subspecies. Six of seven strains with a negative result only by the old test investigated on a molecular basis were found to be subspecies II strains. In conclusion, most of the 25 strains negative by the old assay will belong to subspecies II.
Three strains negative by both versions of the assay represent subspecies VI strains at a frequency of 2.5% in the comparative study of the two versions of the AccuProbe kits.
Variability of M. kansasii strains.
The genetic variability of the M. kansasii strains is reflected in the differences observed in the DNA fragments investigated. Despite the almost complete identity of the 16S rRNA sequences, subspecies III, IV, and VI differ markedly from subspecies I and II in terms of their spacer sequences. This confirms the differences observed by restriction analysis with the hsp65 sequence, which led to the subspecies classification (9). Furthermore, the negative AccuProbe test result for subspecies VI strains indicates the sequence differences in the part of the chromosome to which the probes hybridize. Interestingly, despite the almost identical sequences of the 16S rRNA, the hsp65 gene, and the 16S to 23S spacers of subspecies III and VI, the AccuProbe target sequence must be different since subspecies VI strains are negative with the AccuProbe but M. kansasii subspecies III strains are positive with the AccuProbe.
Considerably few differences occur between M. kansasii subspecies IV and M. gastri. Yet, photochromogenicity and a weakly positive nitrate reaction for the subspecies IV strain identify this strain as an M. kansasii strain. However, the genetic similarities between these strains raise the question of whether M. kansasii subspecies IV strains belong to the M. kansasii group or in fact are photochromogenic M. gastri strains.
All the sequencing results presented here demonstrate a marked variability among the M. kansasii strains, resulting in distinct subspecies.
Implications for the identification of mycobacteria by molecular methods.
RFLP analysis of a part of the hsp65 gene is widely used for the differentiation of mycobacteria. The limits of this technique are obvious in cases of variability within one species, as was found for M. kansasii. The apparent discrepancy between the real and the observed fragment lengths renders this method more difficult.
By 16S rRNA gene sequencing, all M. kansasii strains can clearly be identified. In addition, M. kansasii subspecies II, III, and VI can be identified because they differ in the V3 region. However, M. gastri cannot be separated from M. kansasii subspecies I and IV by 16S rRNA gene sequencing. Thus, in these cases, identification of M. gastri can be performed either by sequencing of the hsp65 gene or the spacer sequence or by classical tests.
Differences in the 16S rRNA gene sequences in a mycobacterial species have been published, for example, for M. gordonae and M. fortuitum. For M. kansasii, sequence variation is documented only for bases 71 to 84 (2, 12). The variation in the variable stretch of the sequence of helix 18 is small but is discriminatory for subspecies II, III, and VI. Since several mycobacterial species vary only at one or two bases, these substitutions should be taken into account so that those M. kansasii strains are not missed.
In summary, the results of our study underline the observations of a high degree of variability among M. kansasii strains. All strains denominated as M. kansasii are characterized by a common 16S rRNA gene sequence. However, the presence of an identical sequence in M. gastri shows that this common sequence is not sufficient for species identification. Furthermore, variability in other parts of the genome of M. kansasii strains strengthens the differences between several types. Due to the clinical importance of M. kansasii, rapid and certain identification of all types or subspecies is essential. The newly formulated AccuProbe provides a useful tool for the identification of M. kansasii since all M. gastri strains are correctly negative and M. kansasii strains are rarely missed.
ACKNOWLEDGMENTS
We thank B. Schlüter and L. Klintz for excellent technical work and GenProbe Inc. for providing reagents.
REFERENCES
- 1.Abed Y, Bollet C, de Micco P. Identification and strain differentiation of Mycobacterium species on the basis of DNA 16S-23S spacer region polymorphism. Res Microbiol. 1995;146:405–413. doi: 10.1016/0923-2508(96)80286-5. [DOI] [PubMed] [Google Scholar]
- 2.Alcaide F, Richter I, Bernasconi C, Springer B, Hagenau C, Schulze-Röbbecke R, Tortoli E, Martín R, Böttger E C, Telenti A. Heterogeneity and clonality among isolates of Mycobacterium kansasii: implications for epidemiological and pathogenicity studies. J Clin Microbiol. 1997;35:1959–1964. doi: 10.1128/jcm.35.8.1959-1964.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devallois A, Goh K S, Rastogi N. Rapid identification of mycobacteria to species level by PCR-restriction fragment length polymorphism analysis of the hsp65 gene and proposition of an algorithm to differentiate 34 mycobacterial species. J Clin Microbiol. 1997;35:2969–2973. doi: 10.1128/jcm.35.11.2969-2973.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iinuma Y, Ichiyamam S, Hasegawa Y, Shimokata K, Kawahara S, Matsushima T. Large-restriction-fragment analysis of Mycobacterium kansasii genomic DNA and its application in molecular typing. J Clin Microbiol. 1997;35:596–599. doi: 10.1128/jcm.35.3.596-599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.International Nucleotide Sequence Database Collaboration. 24 November 1997, posting date. Sequences. [Online.] http://www.ncbi.nlm.nih.gov. [11 August 1998, date last accessed.]
- 5.Ji Y E, Kempsell K E, Colston M J, Cox R A. Nucleotide sequences of the spacer-1, spacer-2 and trailer regions of the rrn operons and secondary structures of precursor 23S rRNAs and precursor 5S rRNAs of slow-growing mycobacteria. Microbiology. 1994;140:1763–1773. doi: 10.1099/13500872-140-7-1763. [DOI] [PubMed] [Google Scholar]
- 6.Kapur V, Li L-L, Hamrick M R, Plikaytis B B, Shinnick T M, Telenti A, Jacobs W R, Banerjee A, Cole S, Yuen K Y, Clarridge III J E, Kreiswirth B N, Musser J M. Rapid Mycobacterium species assignment and unambiguous identification of mutations associated with antimicrobial resistance in Mycobacterium tuberculosis by automated DNA sequencing. Arch Pathol Lab Med. 1995;119:131–138. [PubMed] [Google Scholar]
- 7.Kirschner P, Springer B, Vogel U, Meier A, Wrede A, Kiekenbeck M, Bange F-C, Böttger E C. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J Clin Microbiol. 1993;31:2882–2889. doi: 10.1128/jcm.31.11.2882-2889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lebrun L, Espinasse F, Poveda J P, Vincent-Levy-Frebault V. Evaluation of nonradioactive DNA probes for identification of mycobacteria. J Clin Microbiol. 1992;30:2476–2478. doi: 10.1128/jcm.30.9.2476-2478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Picardeau M, Prod’Hom G, Raskine L, LePennec M P, Vincent V. Genotypic characterization of five subspecies of Mycobacterium kansasii. J Clin Microbiol. 1997;35:25–32. doi: 10.1128/jcm.35.1.25-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richter E, Greinert U, Kirsten D, Rüsch-Gerdes S, Schlüter C, Duchrow M, Galle J, Magnussen H, Schlaak M, Flad H-D, Gerdes J. Assessment of mycobacterial DNA in cells and tissues of mycobacterial and sarcoid lesions. Am J Respir Crit Care Med. 1996;153:375–380. doi: 10.1164/ajrccm.153.1.8542146. [DOI] [PubMed] [Google Scholar]
- 11.Ross B C, Jackson K, Yang M, Sievers A, Dwyer B. Identification of a genetically distinct subspecies of Mycobacterium kansasii. J Clin Microbiol. 1992;30:2930–2933. doi: 10.1128/jcm.30.11.2930-2933.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roth A, Fischer M, Hamid M E, Michalke S, Ludwig W, Mauch H. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J Clin Microbiol. 1998;36:139–147. doi: 10.1128/jcm.36.1.139-147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soini H, Böttger E C, Viljanen M K. Identification of mycobacteria by PCR-based sequence determination of the 32-kilodalton protein gene. J Clin Microbiol. 1994;32:2944–2947. doi: 10.1128/jcm.32.12.2944-2947.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Telenti A, Marchesi F, Balz M, Bally F, Böttger E C, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993;31:175–178. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tortoli E, Simonetti M T, Lacchini C, Penati V, Piersimoni C, Morbiducci V. Evaluation of a commercial DNA probe assay for the identification of Mycobacterium kansasii. Eur J Clin Microbiol Infect Dis. 1994;13:264–267. doi: 10.1007/BF01974549. [DOI] [PubMed] [Google Scholar]
- 16.Tortoli E, Simonetti M T, Lacchini C, Penati V, Urbano P. Tentative evidence of AIDS-associated biotype of Mycobacterium kansasii. J Clin Microbiol. 1994;32:1779–1782. doi: 10.1128/jcm.32.7.1779-1782.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tortoli E, Simonetti M T, Lavinia F. Evaluation of reformulated chemiluminescent DNA probe (AccuProbe) for culture identification of Mycobacterium kansasii. J Clin Microbiol. 1996;34:2838–2840. doi: 10.1128/jcm.34.11.2838-2840.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van der Giessen J W B, Haring R M, van der Zeijst B A M. Comparison of the 23S ribosomal RNA genes and the spacer region between the 16S and 23S rRNA genes of the closely related Mycobacterium avium and Mycobacterium paratuberculosis and the fast-growing Mycobacterium phlei. Microbiology. 1994;140:1103–1108. doi: 10.1099/13500872-140-5-1103. [DOI] [PubMed] [Google Scholar]
- 19.Woolford A J, Hewinson R G, Woodward M, Dale J W. Sequence heterogeneity of an mpb70 gene analogue in Mycobacterium kansasii. FEMS Microbiol Lett. 1997;148:43–48. doi: 10.1111/j.1574-6968.1997.tb10264.x. [DOI] [PubMed] [Google Scholar]
- 20.Yang M, Ross B C, Dwyer B. Identification of an insertion-like element in a subspecies of Mycobacterium kansasii. J Clin Microbiol. 1993;31:2074–2079. doi: 10.1128/jcm.31.8.2074-2079.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]