Abstract
Host genetic variants influence the susceptibility and severity of several infectious diseases, and the discovery of genetic associations with coronavirus disease 2019 (COVID-19) phenotypes could help to develop new therapeutic strategies to decrease its burden.
Between May 2020 and June 2021, we used COVID-19 data released periodically by UK Biobank and performed 65 genome-wide association studies in up to 18 releases of COVID-19 susceptibility (n = 18,481 cases in June 2021), hospitalization (n = 3,260), severe outcomes (n = 1,244), and deaths (n = 1,104), stratified by sex and ancestry.
In coherence with previous studies, we observed two independent signals at the chr3p21.31 locus (rs73062389-A, odds ratio [OR], 1.21 (P = 4.26 × 10−15) and rs71325088-C, OR, 1.62 [P = 2.25 × 10−9]) modulating susceptibility and severity, respectively, and a signal influencing susceptibility at the ABO locus (rs9411378-A; OR, 1.10; P = 3.30 × 10−12), suggesting an increased risk of infection in non-O blood groups carriers. Additional signals at the APOE (associated with severity and death) LRMDA (susceptibility in non-European) and chr2q32.3 (susceptibility in women) loci were also identified, but did not replicate in independent datasets. We then devised an approach to extract variants suggestively associated (P < 10−5), exhibiting an increase in significance over time. When applied to the susceptibility, hospitalization and severity analyses, this approach revealed the known RPL24, DPP9, and MAPT loci, respectively, among hundreds of other signals.
These results, freely available on the GRASP portal, provide insights on the genetic mechanisms involved in COVID-19 phenotypes.
Keywords: COVID-19, genetics, GWAS, trajectory, MUC4
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for the coronavirus disease 2019 (COVID-19), which affects individuals with variable severity, ranging from asymptomatic patients to mild respiratory symptoms, hypercytokinemia, pneumonia, thrombosis, and even death.1,2 Understanding the mechanisms leading to heterogeneous symptoms and susceptibility is essential to develop efficient treatments and improve patient care. Host genetic diversity has been shown to influence the effects of infection to several viruses,3 such as variations in CCR5 (MIM: 601373) leading to HIV resistance4 (MIM: 609423) or IRF7 (MIM: 605047) deficiency affecting influenza susceptibility (MIM: 614680).5
To discover human genetic determinants to COVID-19 susceptibility and severity, several biobanks and research groups worldwide collaborated to perform genome-wide association studies (GWAS) and meta-analyses of the GWAS. In June 2020, a study involving 1980 COVID-19-infected patients with respiratory failure was the first to reveal genome-wide significant (P < 5 × 10−8) associations at the 3p21.31 locus, encompassing SLC6A20 (MIM: 605616) and several chemokine receptors, and at the ABO (MIM: 110300) locus on chromosome 9.6 These two signals were later validated in independent analyses for both COVID-19 susceptibility and severity,7,8 while additional significant associations were observed at loci involved in immune response or inflammation, such as IFNAR2 (MIM: 602376), DPP9 (MIM: 608258), TYK2 (MIM: 176941), CCHCR1 (MIM: 605310), and OAS1 (MIM: 164350). Notably, these findings implicate common and uncommon variants, while studies trying to identify associations of rare variants have been unsuccessful so far.9 The largest effort is currently led by the COVID-19 host genetics initiative (COVID-19hgi),10 which completed meta-analyses of results shared by 61 studies as of June 15, 2021, identified 23 loci associated with COVID-19 phenotypes so far, and plan to release new results as additional data is made available. A major contributor to this group is the UK Biobank (UKB),11 which periodically releases the results of COVID-19 tests and related deaths, as well as health care data for its nearly 500,000 consented participants, to approved researchers.
We created a public COVID-19 GWAS results portal (https://grasp.nhlbi.nih.gov/Covid19GWASResults.aspx) to provide rapid deep annotation for emerging genetics results and facilitate open access to the scientific community. We contribute to this resource by performing GWAS on each COVID-19 data release from the UKB, including sex-specific, ancestry-specific, and trans-ethnic COVID-19-related GWAS, along with a deep set of annotations for top variants (with P < 1 × 10−5). For each release, up to 65 GWAS have been generated, including COVID-19 susceptibility, COVID-19 hospitalization, severe COVID-19 with respiratory failure, and COVID-19 death. Here we describe the results of these GWAS, 612 in total as of June 18, 2021, and report the evolution of signals associated with these COVID-19 phenotypes over the consecutive datasets released by UKB since May 2020.
Materials and methods
UKB data
Analyses are based on v.3 of the UKB imputed dataset,12 which provide genomic data for 487,320 participants from multiple ethnicities, including 459,250 of European ancestry (EUR), 7644 of African ancestry (AFR), 9417 of South Asian ancestry (SAS), and 11,009 of other ancestries (OTHERS). Participants were enrolled at ages ranging from 37 to 73 and are 51.16% female. This project was approved after an application to the UK Biobank, and granted IRB approval by the North West Haydock Research Ethics Committee (16/NW/0274). The UKB started to release COVID-19 test results of its participants on March 15, 2020, and periodically update this resource as new cases are reported. Furthermore, information about COVID-19-related deaths was made available from June 2020, while inpatient data and primary care data were first added during the summer of 2020 and are periodically updated. Details regarding the definition and selection of cases with COVID-19 susceptibility, COVID-19 hospitalization, COVID-19 severity, and COVID-19 death are available in Table S1.
Depending on the COVID-19 phenotype analyzed (susceptibility, hospitalization, severity, or death), up to three different subsets of participants were used as controls. For COVID-19 susceptibility, cases with positive test results were analyzed against either participants tested with negative results (labeled tested), or against all participants without a positive test (labeled population). For analyses of COVID-19 hospitalization, patients hospitalized due to COVID-19 were tested against non-hospitalized participants with a positive test (positive), or non-hospitalized participants with a test (tested), or against all non-hospitalized participants (population). For analyses of COVID-19 severity, patients requiring invasive respiratory support or patients who died from complications were tested against non-severe participants with a positive test (positive), or non-severe participants with a test (tested) or all non-severe participants (population). For analyses of COVID-19 death, patients with COVID-19 death were tested against participants with a positive test (positive), or participants with a test (tested) or against all participants (population).
Analyses
Each GWAS was conducted with SAIGE v0.38,13 which controls for population stratification, relatedness and case-control imbalance, and adjusted for baseline age (at enrollment), sex, and 10 genetic principal components. For the results uploaded to the GRASP portal, variants were filtered on imputation quality (r2 > 0.3), minor allele count (minor allele count of >2), and minor allele frequency (MAF of >0.0001). However, for the results presented in this manuscript, we applied a more stringent filter, and considered only well-imputed variants (r2 > 0.8) and common variants (MAF >of 0.01). After applying this filter, the lambda (genomic control factor) ranged from 0.988 to 1.027 in all 65 analyses of the last data release analyzed (Table S2), indicating no systematic inflation. Quantile-quantile plots of the four trans-ancestry GWAS including population as controls for COVID −19 susceptibility, hospitalization, severity, and death are available as Figure S1. For analyses before the June 18, 2020 release, we conducted analyses on participants of EUR only, and started adding new analyses stratified by sex and ancestry from June 18, 2020, onward. GWAS were stratified for EUR, AFR, SAS, and OTHERS ancestries, and an additional trans-ancestry GWAS combining all participants (labeled ALL) as well as GWAS combining non-European (nEUR) participants were performed.
Associations are either reported as odds ratios (OR) and 95% confidence intervals or as beta coefficients (β) and associated standard errors. Linkage disequilibrium (LD) was estimated by squared correlation (r2) using UKB EUR imputed data. Given the large number of variants that may be significantly (or suggestively) associated at a locus, we assigned significantly (or suggestively) associated variants to a common locus if they were separated by less than 1 Mb, and only reported the lead variant at that locus. To test whether two observed effects are equal, we used the Z statistic: , with b1 and b2 corresponding with the observed effects and SEb1 and SEb2 the associated standard errors. In addition, haplotype analyses were performed with the LDlink LDhap tool,14 while regional association plots were generated with locuszoom.15
Significant associations (P < 5 × 10−8) were further investigated to control for underlying health conditions. To reflect the cardiovascular health of the participants, the following traits were used as covariates: body mass index, type 2 diabetes (International Classification of Diseases, 10th edition [ICD-10] code E11) and ischemic heart disease (ICD-10 code I25). Results were then compared before and after adjustment using logistic regression with likelihood ratio test. In addition, significant associations were also controlled for Alzheimer's disease (ICD-10 code G30) and asthma (ICD-10 code J45).
Annotation
For each analysis hosted on the portal, we provide comprehensive annotation for top results (P < 1 × 10−5) using ANNOVAR16 and the RESTful API service provided by CADD v1.6.17 We also retrieve known phenotype associations extracted from the GRASP18 and EBI GWAS catalogs,19 and known eQTLs extracted from GTeX v820 and other eQTL resources compiled from nearly 150 datasets (built upon the work of Zhang et al,21 detailed in Table S3).
Suggestive associations
All 65 analyses were updated as soon as new data was released from UKB. As a result, we obtained results for these analyses at different time points, which differed by the addition of new cases, thus increasing power in most recent analyses. With an increase in power, bona fide signals associated with COVID-19 phenotypes should increase in significance in each consecutive data release analyzed. We designed a workflow to extract these signals with a positive significance trajectory in each analysis: for each variant (i) in the most recent data release analyzed, P < 10−5; (ii) P cannot decrease in significance by more than one order of magnitude between two consecutive releases; (iii) P must have increased in significance by more than one order of magnitude at least once between two consecutive releases. In addition, we made sure that the direction of effect did not change over time for each variant selected. This set of rules should ensure to extract variants which increased in significance since we started performing these analyses, while allowing some stagnation, which might happen owing to random sampling and/or low case increase between two consecutive releases.
The COVID-19hgi datasets
For replication purposes, we used the publicly available COVID-19hgi meta-analyses summary statistics (freeze 6) for COVID-19 susceptibility (labeled C2; n = 112,612 cases), hospitalization (B2; n = 24,274), and severity (A2; n = 8,779 cases). These datasets are currently the largest analyses of COVID-19 phenotypes available. Publicly available summary statistics do not include the 23andMe dataset, and a version of the summary statistics without UKB was also made available for the B2 and C2 analyses. As there was no overlap of samples with our analyses, we used these summary statistics to replicate our findings from susceptibility and hospitalization analyses, as well as our suggestive findings. To replicate findings from our severe analyses, we used the A2 summary statistics without UKB from the freeze 5 (the A2 results without UKB from freeze 6 were not available).
Data availability
Permission was obtained to post UKB summary statistics under an approved application (ID 28525). The association results are available on the portal, as well as annotated top results. In addition to UKB summary statistics, results from other efforts are also hosted on the portal. Authors of COVID-19 GWAS publications have been contacted to seek approval before hosting the results of their analyses on the GRASP COVID-19 portal. Summary statistics at this time include multiple releases of the COVID-19hgi group, severe hospitalization results from Ellinghaus et al.,6 and the GenOMICC study,7 as well as hospitalization status and time to end COVID-19 symptoms from the COLCORONA study,22 with all top results being re-annotated in the common framework mentioned above.
Results
UKB COVID-19 demographics
Using the latest UKB data releases available at this point, with susceptibility and hospitalization phenotypes analyzed on June 18, 2021, and severe COVID-19 and death analyzed on May 9, 2021, we retrieved 86,435 participants with a COVID-19 diagnostic, of which 18,481 tested positive. According to inpatient care data, 3260 positive cases were hospitalized, while 1244 patients with severe COVID-19 diagnostic received respiratory support and/or died from complications (Table 1). Since May 7, 2020, we analyzed up to 18 UKB data releases regarding COVID-19 susceptibility, 8 data releases concerning COVID-19 related deaths, 6 for COVID-19 hospitalization, and 5 for COVID-19 severity.
Table 1.
Sample sizes for each analysis, as of 06.18.21
| Phenotype | Controls | Ancestry | Sex-stratified | Cases | Controls | Female cases | Female controls |
|---|---|---|---|---|---|---|---|
| Susceptibility (06.18.21 release) | Test negative OR not tested (population) | ALL | yes | 18,481 | 468,839 | 9,757 | 254,472 |
| EUR | yes | 16,551 | 442,699 | 8,767 | 240,532 | ||
| AFR | 557 | 7,087 | 316 | 4,043 | |||
| SAS | 810 | 8,607 | 351 | 3,980 | |||
| OTHERS | 563 | 10,446 | 323 | 5,917 | |||
| nEUR | 1,930 | 26,140 | 990 | 13,940 | |||
| Test negative (tested) | ALL | yes | 18,481 | 86,435 | 9,757 | 46,737 | |
| EUR | yes | 16,551 | 81,826 | 8,767 | 44,248 | ||
| AFR | 557 | 1,281 | 316 | 759 | |||
| SAS | 810 | 1,516 | 351 | 695 | |||
| OTHERS | 563 | 1,812 | 323 | 1,035 | |||
| nEUR | 1,930 | 4,609 | 990 | 2,489 | |||
| Hospitalization (06.18.21 release) | Not hospitalized (population) | ALL | yes | 3,260 | 484,060 | 1,343 | 262,886 |
| EUR | yes | 2,884 | 456,366 | 1,181 | 248,118 | ||
| nEUR | 376 | 27,694 | 162 | 14,768 | |||
| Tested, not hospitalized (tested) | ALL | yes | 3,260 | 101,656 | 1,343 | 55,151 | |
| EUR | yes | 2,884 | 95,493 | 1,181 | 51,834 | ||
| nEUR | 376 | 6,163 | 162 | 3,317 | |||
| Test positive, not hospitalized (positive) | ALL | yes | 3,260 | 15,221 | 1,343 | 8,414 | |
| EUR | yes | 2,884 | 13,667 | 1,181 | 7,586 | ||
| nEUR | 376 | 1,554 | 162 | 828 | |||
| Severe COVID-19 (05.09.21 release) | Not severe (population) | ALL | yes | 1,244 | 486,076 | 439 | 263,790 |
| EUR | yes | 1,120 | 458,130 | 387 | 248,912 | ||
| Tested, not severe (tested) | ALL | yes | 1,244 | 84,742 | 439 | 45,811 | |
| EUR | yes | 1,120 | 79,439 | 387 | 42,973 | ||
| Test positive, not severe (positive) | ALL | yes | 1,244 | 16,413 | 439 | 8,866 | |
| EUR | yes | 1,120 | 14,695 | 387 | 7,976 | ||
| Death (05.09.21 release) | Survivor (population) | ALL | 1,104 | 486,216 | 399 | 263,830 | |
| EUR | 1,001 | 458,249 | 356 | 248,943 | |||
| Tested, survivor (tested) | ALL | 1,104 | 84,882 | 399 | 45,851 | ||
| EUR | 1,001 | 79,558 | 356 | 43,004 | |||
| Test positive, survivor (positive) | ALL | 1,104 | 16,553 | 399 | 8,906 | ||
| EUR | 1,001 | 14,814 | 356 | 8,007 |
ALL, all ancestries; OTHERS, ancestry different from EUR, AFR, and SAS; nEUR, non-EUR ancestry.
Among COVID-19-positive participants, we observed a global increase in the percentage of female cases, starting at 45.3% at the first release analyzed, and reaching 52.8% in the last, while men were more likely to be infected (OR, 1.07; P = 8.06 × 10−6), hospitalized (OR, 1.66; P = 1.59 × 10−45), or develop severe complications (OR, 2.07; P = 1.51 × 10−34) and die from COVID-19 (OR, 1.98; P = 2.12 × 10−27) (Table S4). There was also a decrease in the mean age of positive cases, ranging from 57.02 to 53.57 years, with a significant decrease after the 2020 summer (Table 2), with younger individuals more likely to be infected (β = −0.04; P < 10−300) ; while increase in age was associated with hospitalization (β = 0.05; P = 4.54 × 10−90), severity (β = 0.10; P = 5.63 × 10−108), and death (β = 0.12; P = 1.11 < 10−122) (Table S4).
Table 2.
Composition of the consecutive UKB COVID-19 data releases
| Release date | Cases (tested positive) | Controls | % Tested positive females | Age casesa |
|---|---|---|---|---|
| 05.07.20b | 1,029 | 486,291 | 45.29 | 57.02 (9.12) |
| 05.25.20b | 1,270 | 486,050 | 46.93 | 56.57 (9.25) |
| 06.05.20 | 1,412 | 485,908 | 47.31 | 56.45 (9.23) |
| 06.18.20 | 1,485 | 485,835 | 47.21 | 56.47 (9.19) |
| 07.14.20 | 1,670 | 485,650 | 46.59 | 56.89 (9.14) |
| 08.04.20 | 1,723 | 485,597 | 46.66 | 56.77 (9.11) |
| 09.08.20 | 1,821 | 485,499 | 46.73 | 56.66 (9.10) |
| 10.16.20 | 3,050 | 484,270 | 48.69 | 55.19 (9.09) |
| 11.03.20 | 4,372 | 482,948 | 49.38 | 54.79 (8.95) |
| 11.25.20 | 5,868 | 481,452 | 49.52 | 54.56 (8.88) |
| 12.04.20 | 7,435 | 479,885 | 50.46 | 54.36 (8.80) |
| 01.04.21 | 8,722 | 478,598 | 50.50 | 54.28 (8.79) |
| 01.22.21 | 13,401 | 473,919 | 52.27 | 53.85 (8.66) |
| 02.02.21 | 14,802 | 472,518 | 52.44 | 53.81 (8.64) |
| 02.24.21 | 15,738 | 471,582 | 52.47 | 53.77 (8.63) |
| 04.04.21 | 16,586 | 470,734 | 52.68 | 53.70 (8.62) |
| 05.09.21 | 17,657 | 469,663 | 52.70 | 53.64 (8.62) |
| 06.18.21 | 18,481 | 468,839 | 52.79 | 53.57 (8.60) |
Mean baseline age (standard deviation), at recruitment.
For the first two releases, the numbers reflect all ancestries, but only cases and controls of EUR were considered for analyses.
Positive cases were mainly of EUR, representing 85.5% of all COVID-19-positive participants in the first analysis and growing to 89.6% in the last. SAS, AFR, and OTHERS represent 4.4%, 3%, and 3% of positive cases, respectively, in this UKB data release. However, compared with European participants, nEUR participants were enriched among cases (OR, 1.72 [P = 3.09 × 10−33] for AFR; OR, 2.16 [P = 1.10 × 10−91] for SAS; and OR, 1.23 [P = 1.96 × 10−6] for OTHERS), as well as hospitalized (OR, 3.84 [P = 2.05 × 10−53] for AFR; OR, 2.51 [P = 1.89 × 10−24] for SAS; OR, 1.80 [P = 6.69 × 10−9] for OTHERS), severe cases (OR, 3.76 [P = 1.02 × 10−17] for AFR; OR, 2.63 [P = 5.93 × 10−11] for SAS; and OR, 1.67 [P = 0.004] for OTHERS), and deceased participants (OR, 3.66 [P = 2.17 × 10−14] for AFR; OR, 2.55 [P = 6.14 × 10−9] for SAS; and OR, 1.65 [P = 0.009] for OTHERS) (Table S4). In addition, while controlling genetic associations for bias induced by underlying health conditions, we noticed that type 2 diabetes, ischemic heart disease, Alzheimer's disease, and asthma were all associated with an increased risk of COVID-19 phenotypes, with ORs ranging from 1.32 for asthma associated with COVID-19 susceptibility to 4.82 for Alzheimer's disease associated with COVID-19 death (Table S5).
Genome-wide significant results
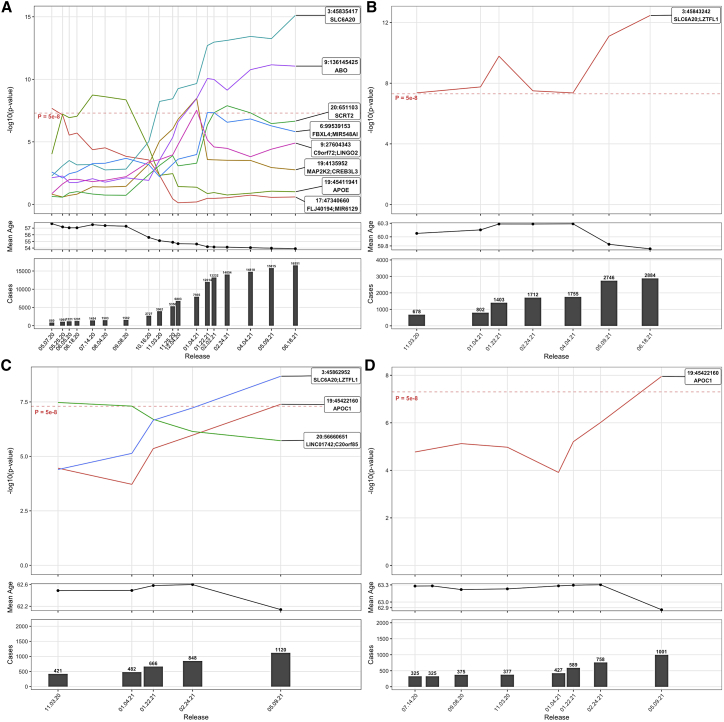
The workflow of genetic analyses is presented in Figure 1. Between May 2020 and June 2021, we observed several genome-wide significant (P < 5 × 10−8) signals across the 65 analyses. However, many signals were only punctually significant, and only a handful of signals remained significant in subsequent data releases analyzed. For instance, the analysis of COVID-19 susceptibility in participants of EUR, with untested or negatively tested participants as controls (noted population controls), revealed eight signals reaching genome-wide significance at some point (Figure 2A), of which only two remained significant in the last data release analyzed (on 06.18.21): the chr3p21.31 locus encompassing SLC6A20 and several chemokine receptors (rs73062389-A; MAF = 0.06; OR, 1.22 [1.16; 1.28]; P = 7.60 × 10−16) and the ABO locus on chromosome 9 (rs9411378-A; MAF = 0.22; OR, 1.10 [1.07; 1.14]; P = 8.78 × 10−12). These two loci were previously reported to modulate COVID-19 susceptibility in several studies.6, 7, 8,10
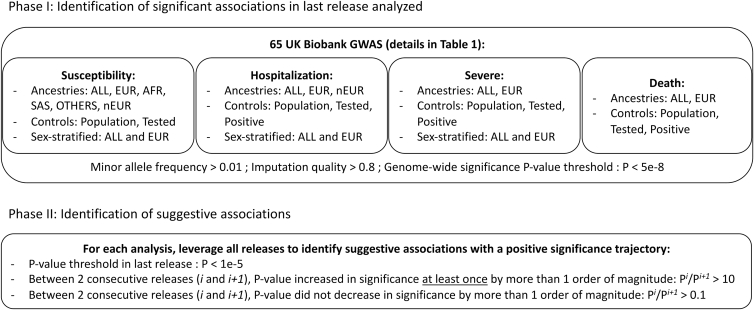
Figure 1.
Analysis workflow
The design of our study includes two phases. First, GWAS were performed for all 65 analyses, and genome-wide significant associations were identified. Second, we focused on suggestive associations that increased in significance over time.
Figure 2.
Evolution of significant signals associated with COVID-19 susceptibility, hospitalization, severity and death in UKB participants of EUR, using Population as controls
(A–D) Results of susceptibility, hospitalization, severity, and death analyses. For each sub-figure, the top panel represents the evolution in significance (as -log10 p-values on y-axis) of signals reaching genome wide significance at least once across all data releases analyzed. The middle panel represent the mean age of cases in each data release. The bottom panel represent the number of cases in each data release. For sub-figure A, a representation of these signals across the COVID-19hgi releases is available as Figure S51.
Across all 65 analyses, 5 loci reached genome-wide significance in the last data releases analyzed (on June 18, 2021, for susceptibility and hospitalization, and May 9, 2021, for severity and death): the chr3p21.31 locus, ABO, APOE (MIM: 107741), LRMDA (MIM: 614537), and an intergenic signal at the chr2q32.3 locus (Table 3). All associations with a P value of less than 10−5, from all 65 analyses, are available in Table S6. In addition, all signals reaching genome-wide significance in any data release analyzed are presented in Figures S2–S47. The signals reported in the following correspond with results using population controls, except when specified otherwise. Overall, the effect of significant associations was similar when using different set of controls (Figure S48).
Table 3.
Lead variants associated with COVID-19 phenotypes and replication in COVID-19hgi
| Analysis:Ancestry:controls(:sex)a | CHR:POS:NEA:EA | rsid | EAF | OR | ±95 CI | P value | Replication analysis | OR (P value) | Locus |
|---|---|---|---|---|---|---|---|---|---|
| Susceptibility:ALL:Population | 3:45835417:G:A | rs73062389 | 0.058 | 1.2070 | [1.15; 1.27] | 4.26 × 10−15 | C2 | 1.17 (1.08 × 10−54) | SLC6A20 |
| Hospitalization:ALL:Population | 3:45850783:T:A | rs72893671 | 0.081 | 1.4043 | [1.28; 1.54] | 5.12 × 10−13 | B2 | 1.35 (6.95 × 10−51) | SLC6A20;LZTFL1 |
| Severe:ALL:Population | 3:45862952:T:C | rs71325088 | 0.073 | 1.6206 | [1.38; 1.90] | 2.25 × 10−9 | A2 | 1.88 (9.89 × 10−46) | SLC6A20;LZTFL1 |
| Susceptibility:ALL:Population | 9:136145425:C:A | rs9411378 | 0.219 | 1.1013 | [1.07; 1.13] | 3.30 × 10−12 | C2b | 1.07 (1.08 × 10−38) | ABO |
| Death:ALL:Population | 19:45411941:T:C | rs429358 | 0.154 | 1.3983 | [1.24; 1.57] | 3.06 × 10−8 | A2 | 0.96 (0.27) | APOE |
| Ancestry-specific results | |||||||||
| Susceptibility:nEUR:Population | 10:78250184:T:C | rs114026383 | 0.016 | 2.3943 | [1.79; 3.20] | 4.10 × 10−9 | C2c | 1.05 (0.27) | LRMDA |
| Sex-specific results | |||||||||
| Susceptibility:ALL:Population:F | 2:192774154:G:A | rs147509469 | 0.037 | 1.2589 | [1.16; 1.37] | 2.64 × 10−8 | C2 | 1.007 (0.58) | CAVIN2;TMEFF2 |
CHR, chromosome; POS, position (hg19 genome build); NEA, non effect allele; EA, effect allele; EAF, effect allele frequency; 95 CI, 95% confidence interval [lower bound; upper bound]; P: p value (from the last data release, analyzed on 06.18.21 for susceptibility and hospitalization, and 05.09.21 for severe and death phenotypes).
Indicate the ancestry by label, the control set, and, when it is the case, the corresponding sex-stratified analyses (F, females; M, males).
For the replication of the ABO variant, rs9411378 was not available and the best proxy available rs635634 (LD r2 = 0.53) was used instead.
For the replication of the LRMDA variant, the C2 analysis restricted to AFR participants was used.
The 3p21.31 locus
This locus was associated with COVID-19 susceptibility, hospitalization and severity, in EUR and ALL ancestries, regardless of the set of controls employed. However, as mentioned in previous works,8,10 the lead variant associated with susceptibility was not in LD with the lead variants for hospitalization or severity (r2 < 0.01), suggesting two distinct signals modulating COVID-19 susceptibility and severity (Figure S49). After adjusting for cardiovascular health (using body mass index, type 2 diabetes, and ischemic heart diseases as covariates), the variants remained associated (Table S7).
ABO
The ABO locus was associated with COVID-19 susceptibility for ALL and EUR participants, using either population or tested controls. When controlling the association for cardiovascular health and asthma, the variants remained associated (Table S7). Using haplotype analyses with blood group tagging variants, we determined the blood groups tagged by the lead variant associated with susceptibility (rs9411378). For the five main blood groups, we used the following tagging variants23: rs41302905 (tagging for O2), rs8176743 (tagging for B), rs1053878 (tagging for A2), rs8176719 (tagging for O1), and rs2519093 (tagging for A1). As a result, we observed that the COVID-19 susceptibility variant was fully tagging A1 and A2 haplotypes, and partially tagging B haplotypes (20% of B haplotypes) (Figure S50), suggesting an increased risk of infection for A1 and A2 carriers.
APOE
The APOE variant tagging for the APOE-ε4 haplotype was initially significantly associated with COVID-19 susceptibility in EUR (rs429358-C; MAF = 0.15; OR, 1.38 [1.24; 1.53]; P = 1.80 × 10−9 on July 14, 2020). This signal was notably the first report of a genetic determinant for COVID-19 susceptibility.24 This previous report was based on UKB data but this signal was not replicated in an independent dataset, and this association was greatly attenuated after the summer, when the number of COVID-19 cases started to rise significantly and the mean age of infected participants decreased (Figure 2A). The interaction between the age of participants and the APOE variant was significant (P = 1.78 × 10−9), which was further confirmed using a 2df test25 with age and genotype (P = 2.65 × 10−9), suggesting that a subset of older participant carriers of this variant was more at risk of COVID-19 infection. Remarkably, this signal became genome-wide significant in the May 9, 2021, data release analyses of death in ALL (OR, 1.40 [1.24; 1.57]; P = 3.06 × 10−8), and EUR, and in the analysis of severe COVID-19 in EUR, which may suggest a mechanism leading to severe and lethal complications after infection. However, there is no evidence of association in the larger COVID-19hgi meta-analysis of COVID-19 severity (A2; freeze 5 without UKB; P = 0.27). Using the association of the death analysis, we further controlled the association for Alzheimer's disease, for which APOE-ε4 is a known risk factor, but the variant remained associated (OR, 1.33; P = 4.04 × 10−7). Adjusting for cardiovascular health modestly increased the strength of association of the APOE allele with COVID-related death (OR, 1.39; P = 6.45 x 10−9) (Table S7).
Results from ancestry- and sex-stratified analyses
In other ancestry-stratified GWAS (AFR, SAS, and OTHERS), no signal was found genome-wide significant in the last data release analyzed. In the GWAS combining all nEUR participants, with population controls, one signal was found significant at the LRMDA locus on chromosome 10 (rs114026383-C; MAF = 0.02; OR, 2.39 [1.79; 3.20]; P = 4.10 × 10−9). According to gnomAD (v2.1.1),26 this variant is mostly carried by individuals of AFR (MAF = 0.04) and mainly absent in OTHERS. In the GWAS of AFR participants, this signal is close to genome wide significance (P = 1.55 × 10−7). Interestingly, LRMDA variants have been found associated to lung function27 (MIM: 608852) and HIV viral load in an unadjusted GWAS,28 but there was no evidence of LD between these variants and rs114026383 (r2 < 0.01). Furthermore, this association did not replicate in the COVID-19hgi susceptibility analysis (C2) restricted to AFR participants (P = 0.27).
In addition, the chr3p21.31 susceptibility variant (rs73062389) was less frequent in non Europeans, and we did not observe an association with this variant in any of the non EUR-stratified analyses (MAF = 0.007 and P = 0.22 in AFR; MAF = 0.021 and P = 0.47 in SAS; and MAF = 0.038 and P = 0.79 in OTHERS). Similarly, there was no association at the ABO lead variant (rs9411378) in non EUR-stratified analyses (P = 0.18 in AFR; P = 0.22 in SAS; P = 0.34 in OTHERS).
In sex-stratified analyses, using Population controls, the chr3p21.31 susceptibility signal was significant in women (rs73062389; P = 1.06 × 10−8 in ALL) and moderately associated in men (P = 2.10 × 10−6 in ALL), whereas the ABO signal was significant in men (rs9411378; P = 5.10 × 10−10 in ALL) and moderately associated in women (P = 3.30 × 10−5 in ALL). The chr3p21.31 lead variant in the hospitalization analysis (rs72893671) was more significant in men (P = 6.80 × 10−11 in ALL) than in women (P = 3.68 × 10−4 in ALL). Despite these differences in significance between men and women for these three main signals, we did not observe a significant difference of effect when using the Z-test for the equality of regression coefficients (P > 0.05 for all three signals). Additionally, a variant reached genome-wide significance in the analysis of COVID-19 susceptibility of women of ALL ancestry at the chr2q32.3 locus, while no association was observed for this variant in men (P = 0.47). However, this association was not supported by the COVID-19hgi C2 analysis (P = 0.58), even though the COVID-19hgi meta-analyses were not sex stratified.
Signals with a positive significance trajectory in UKB
Given the low number of significant associations, we also investigated suggestive associations (P < 10−5), and kept track of how much they increased or decreased in significance at each new data release analyzed. More specifically, we recorded whether these signals increased or decreased in significance by more than one order of magnitude. After collecting this information across all data releases, we obtained these significance trajectories for each variant. We noted that the significance trajectory of the most robust signals, at the chr3p21.31 and ABO loci, increased at least once by more than one order of magnitude between two consecutive releases, and sometimes slightly decreased, but not by more than one order of magnitude (Figure 2). Thus, we were interested in suggestive associations displaying a similar positive trajectory in significance over time, and reaching a suggestive p value of at least 10−5 in the last data release analyzed.
Across all 65 analyses, the number of variants reaching a suggestive P value in the last release was 11,639, and the subset of variants exhibiting a positive significance trajectory was 8291 (28.8% of the variants with suggestive associations were discarded). After extracting the lead variant at each locus, and removing the lead variants that were already genome-wide significant, we obtained a list of 1411 lead variants (Table S8), with some duplicates, as lead variants from a same locus can appear in several analyses.
Notably, some of these loci previously reached genome-wide significance in the COVID-19hgi meta-analyses.10 First, at the DPP9 locus (MIM: 608258) an intronic variant was suggestively associated with hospitalization in ALL when using tested as controls (rs12610495-G; MAF = 0.30; OR, 1.14 [1.08; 1.21]; P = 3.31 × 10−6) and is in LD with the lead variant associated with hospitalization in the COVID-19hgi analysis (rs2277732; r2 = 0.95). The annotations also reveal that this variant is also associated with a decreased expression of DPP9 in the lung according to GTeX (β = −0.18; P = 4.5 × 10−9). Second, at the RPL24 locus (MIM: 604180) a variant located in an intron of CEP97 (MIM: 615864) was suggestively associated with susceptibility in ALL (rs566103643-CA; MAF = 0.35; OR, 0.95 [0.93; 0.97]; P = 8.25 × 10−6), and is in LD with the lead variant associated with susceptibility in the COVID-19hgi meta-analysis (rs4342086; r2 = 0.70). Last, at the MAPT locus (MIM: 157140), an intronic variant was suggestively associated with severe COVID-19 in ALL (rs532052263-GT; MAF = 0.26; OR, 0.80 [0.72; 0.88]; P = 2.98 × 10−6) also in LD with the lead variant associated with severity in the COVID-19hgi analysis (rs8080583; r2 = 0.75).
Additional variants that are not currently significant in the COVID-19hgi meta-analyses may also be of interest. For instance, we identified a variant located in an intron of MUC4 (MIM: 158372) suggestively associated with susceptibility in Europeans, using COVID Tested controls (rs842225-A; MAF = 0.46; OR, 1.06 [1.03; 1.09]; P = 3.56 × 10−6). This single nucleotide polymorphism is reported to associate with decreased expression of MUC4 in the esophagus-mucosa in GTeX (β = −0.23; P = 2.8 × 10−11) as well as decreased expression of MUC20 (MIM: 610360) in whole blood (β = −0.35; P = 1.0 × 10−15). In the COVID-h19gi susceptibility analysis, this variant had a concordant effect direction and reached nominal significance (OR, 1.01; P = 0.027). Interestingly, the expression of these two mucins was recently shown to be correlated with COVID phenotypes.29
Discussion
This project was initiated with the major aim to share results of COVID-19 host genetics analyses freely and rapidly on the GRASP portal, during a pandemic where new insights to improve patient care and develop better treatments were greatly needed. A few months after the first COVID-19 case was identified in the UK, we started to perform GWAS on each dataset released by UKB between May 2020 and June 2021, and thus far have examined the genetic signals associated with COVID-19 phenotypes across 18 data releases. This unique context allowed us to track the evolution of genetic associations over time.
As a first observation, the majority of signals observed in the first stages of the project did not sustain over time. For instance, the first genome-wide significant association observed in the GWAS of COVID-19 susceptibility in Europeans with rs34338189 as lead variant is not even nominally significant in the last release (P = 0.25). Another genome-wide significant signal was observed at the APOE locus, with a significance that increased in the first four data releases before decreasing continually in the subsequent releases. The lead variant at the APOE locus is coding in part for the APOE-ε4 haplotype, known to increase the risk of Alzheimer's disease (MIM: 607822), dementia, dyslipidemia, and cardiovascular diseases (MIM: 617347) and is speculated to cause inflammation and cytokine storms.30 Notably, the significant association of the APOE signal with severe COVID-19 and death in most recent analyses could support this proposition, but this association has not been replicated in an independent dataset. The evolution of the APOE signal over time could also be due to an initial higher prevalence of COVID-19 in nursing homes,31 where patients with dementia were at higher risk of being infected and spreading the virus owing to living arrangements and a poor understanding of transmission dynamics and appropriate safety guidelines early in the pandemic. Overall, the evolution of these signals suggests a change in the composition of cases over time, such as the diminution of age and increase of positively tested women in the later data releases, and potentially, the introduction of new variant SARS-CoV2 strains. This change was most significant after the 2020 summer when a surge in new COVID-19 infections occurred.
The most robust findings from our study are the association of the chr3p21.31 and ABO loci with COVID-19 susceptibility and a distinct signal at the chr3p21.31 locus associated with hospitalization and severe COVID-19. These observations corroborate several previous reports.6, 7, 8,10 Furthermore, after controlling each significant association for cardiovascular health, asthma, and Alzheimer's disease, the signals remained associated with COVID-19 phenotypes, suggesting that these associations were not biased by these underlying health conditions. In addition, we developed an original strategy to identify suggestive associations displaying increased significance over time, thus allowing to reduce the scope of search. Among these, three signals at the DPP9, NXPE3, and MAPT loci were previously established variants associated with COVID-19 phenotypes, notably by the COVID-19hgi analyses,10 thus demonstrating the potential of this approach. However, determining which variants actually modulate COVID-19 phenotypes among thousands of candidates, without an independent and well-powered dataset such as COVID-19hgi to confirm the validity of a signal, remains challenging. Moreover, this approach relies on the assumption that external variables changing over time, such as SARS-CoV-2 strains, have no effect on these associations. For instance, if UKB participants were exposed to a fast spreading, novel strain, with distinct genetic mechanisms, our approach might be able to detect these new genetic risk factors, but the independent risk factors of previous strains might no longer be detectable.
The associations we observed changing through the pandemic could reflect random effects or changes in statistical power, but some of the results suggest changes owing to potential gene-environment interactions such as age, underlying health conditions or sex makeup of cases exposed to or engaging in risk behavior. This indicates the general approach of iterative analysis and significance trajectory analyses for genetics during pandemics may have benefits in uncovering pathophysiologic clues. Additionally, other factors like predominant virus strains and changing treatment strategies through a pandemic might interact with host genetics, and be better understood by iterative analyses. To enable such approaches in pandemics and other critical research domains, rapid and extensive results sharing are important catalysts, as demonstrated by the UKBB, COLCORONA, GenOMICC, and COVID-19hgi.
In summary, our host genomic analyses of COVID-19 may contribute to improve the comprehension of mechanisms involved in the infection and complications due to COVID-19. Our study also has some limitations. Most important, our data and the work of others support large health disparities between EUR and non-EUR individuals related to COVID-19 throughout the ongoing pandemic. Despite an over-representation proportionally among cases and those with severe and fatal outcomes, the non-EUR component of UKB and COVID-19hgi is a proportionally small sample, limiting our statistical power to address population-specific genetic variants contributing to health outcomes. However, moving forward we feel that having a diverse set of results with different phenotype definitions, sex-specific, ancestry-specific, and including external group summary statistics, all in a common genome reference and annotation framework may maximize the chance for new studies to cross-replicate or meta-analyze results as COVID-19 genetic studies continue to grow.
Acknowledgments
All authors were supported by NIH Intramural Research Program funds. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services. This research has been conducted using the UK Biobank Resource under Application Number 28525. UK Biobank was established by the Wellcome Trust, Medical Research Council, Department of Health, Scottish government, and Northwest Regional Development Agency. It has also had funding from the Welsh assembly government and the British Heart Foundation. All UKB analyses for this manuscript were conducted on the NIH Biowulf high-performance computing cluster (https://hpc.nih.gov/). The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by the NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. We also thank the NHLBI IT team for their help in keeping the GRASP portal up to date, David-Alexandre Tregouet for his helpful comments regarding the ABO haplotype analyses, and the COVID-19 Host Genetics Initiative for sharing the results of their analyses.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2022.100095.
Web resources
GRASP COVID-19 resource: https://grasp.nhlbi.nih.gov/Covid19GWASResults.aspx.
GTeX: https://gtexportal.org/home/
CADD: https://cadd.gs.washington.edu/
EBI GWAS catalog: https://www.ebi.ac.uk/gwas/
GRASP catalog: https://grasp.nhlbi.nih.gov/Overview.aspx.
COVID-19hgi meta-analyses results: https://www.covid19hg.org/results/r5/
GnomAD: https://gnomad.broadinstitute.org/
LocusZoom: https://locuszoom.org/
LDlink: https://ldlink.nci.nih.gov.
Supplemental information
Data and code availability
The datasets generated during this study are available at the GRASP COVID-19 portal: https://grasp.nhlbi.nih.gov/Covid19GWASResults.aspx.
References
- 1.Connors J.M., Levy J.H. COVID-19 and its implications for thrombosis and anticoagulation. Blood. 2020;135:2033–2040. doi: 10.1182/blood.2020006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang D., Comish P., Kang R. The hallmarks of COVID-19 disease. PLoS Pathog. 2020;16:e1008536. doi: 10.1371/journal.ppat.1008536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwok A.J., Mentzer A., Knight J.C. Host genetics and infectious disease: new tools, insights and translational opportunities. Nat. Rev. Genet. 2021;22:137–153. doi: 10.1038/s41576-020-00297-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dragic T., Litwin V., Allaway G.P., Martin S.R., Huang Y., Nagashima K.A., Cayanan C., Maddon P.J., Koup R.A., Moore J.P., et al. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 5.Ciancanelli M.J., Huang S.X.L., Luthra P., Garner H., Itan Y., Volpi S., Lafaille F.G., Trouillet C., Schmolke M., Albrecht R.A., et al. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 2015;348:448–453. doi: 10.1126/science.aaa1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P., Fernández J., Prati D., Baselli G., Asselta R., et al. Genomewide association study of severe covid-19 with respiratory failure. N. Engl. J. Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., Russell C.D., et al. Genetic mechanisms of critical illness in Covid-19. Nature. 2020;591:92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 8.Shelton J.F., Shastri A.J., Ye C., Weldon C.H., Filshtein-Sonmez T., Coker D., Symons A., Esparza-Gordillo J., Aslibekyan S., Auton A. Trans-ancestry analysis reveals genetic and nongenetic associations with COVID-19 susceptibility and severity. Nat. Genet. 2021;53:801–808. doi: 10.1038/s41588-021-00854-7. [DOI] [PubMed] [Google Scholar]
- 9.Kosmicki J.A., Horowitz J.E., Banerjee N., Lanche R., Marcketta A., Maxwell E., Bai X., Sun D., Backman J.D., Sharma D., et al. Pan-ancestry exome-wide association analyses of COVID-19 outcomes in 586,157 individuals. Am. J. Hum. Genet. 2021;108:1350–1355. doi: 10.1016/j.ajhg.2021.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.COVID-19 Host Genetics Initiative. Ganna A. Mapping the human genetic architecture of COVID-19: an update. MedRxiv. 2021 doi: 10.1038/s41586-021-03767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M., et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou W., Nielsen J.B., Fritsche L.G., Dey R., Gabrielsen M.E., Wolford B.N., LeFaive J., VandeHaar P., Gagliano S.A., Gifford A., et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 2018;50:1335–1341. doi: 10.1038/s41588-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machiela M.J., Chanock S.J. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinforma. Oxf. Engl. 2015;31:3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boughton A.P., Welch R.P., Flickinger M., VandeHaar P., Taliun D., Abecasis G.R., Boehnke M. LocusZoom.js: interactive and embeddable visualization of genetic association study results. Bioinformatics. 2021;37:3017–3018. doi: 10.1093/bioinformatics/btab186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eicher J.D., Landowski C., Stackhouse B., Sloan A., Chen W., Jensen N., Lien J.-P., Leslie R., Johnson A.D. GRASP v2.0: an update on the Genome-Wide Repository of Associations between SNPs and phenotypes. Nucleic Acids Res. 2015;43:D799–D804. doi: 10.1093/nar/gku1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E., et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aguet F., Brown A.A., Castel S.E., Davis J.R., He Y., Jo B., Mohammadi P., Park Y., Parsana P., Segrè A.V., et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X., Gierman H.J., Levy D., Plump A., Dobrin R., Goring H.H.H., Curran J.E., Johnson M.P., Blangero J., Kim S.K., et al. Synthesis of 53 tissue and cell line expression QTL datasets reveals master eQTLs. BMC Genom. 2014;15:532. doi: 10.1186/1471-2164-15-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubé M.-P., Lemaçon A., Barhdadi A., Lemieux Perreault L.-P., Oussaïd E., Asselin G., Provost S., Sun M., Sandoval J., Legault M.-A., et al. Genetics of symptom remission in outpatients with COVID-19. Sci. Rep. 2021;11:10847. doi: 10.1038/s41598-021-90365-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goumidi L., Thibord F., Wiggins K.L., Li-Gao R., Brown M.R., van Hylckama Vlieg A., Souto J.-C., Soria J.-M., Ibrahim-Kosta M., Saut N., et al. Association between ABO haplotypes and the risk of venous thrombosis: impact on disease risk estimation. Blood. 2021;137:2394–2402. doi: 10.1182/blood.2020008997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo C.-L., Pilling L.C., Atkins J.L., Masoli J.A.H., Delgado J., Kuchel G.A., Melzer D. ApoE e4e4 Genotype and Mortality With COVID-19 in UK Biobank. J. Gerontol. A. Biol. Sci. Med. Sci. 2020;75:1801–1803. doi: 10.1093/gerona/glaa169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kraft P., Yen Y.-C., Stram D.O., Morrison J., Gauderman W.J. Exploiting gene-environment interaction to detect genetic associations. Hum. Hered. 2007;63:111–119. doi: 10.1159/000099183. [DOI] [PubMed] [Google Scholar]
- 26.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soler Artigas M., Loth D.W., Wain L.V., Gharib S.A., Obeidat M., Tang W., Zhai G., Zhao J.H., Smith A.V., Huffman J.E., et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat. Genet. 2011;43:1082–1090. doi: 10.1038/ng.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ekenberg C., Tang M.-H., Zucco A.G., Murray D.D., MacPherson C.R., Hu X., Sherman B.T., Losso M.H., Wood R., Paredes R., et al. Association between single-nucleotide polymorphisms in HLA alleles and human immunodeficiency virus type 1 viral load in demographically diverse, antiretroviral therapy-naive participants from the strategic timing of AntiRetroviral treatment trial. J. Infect. Dis. 2019;220:1325–1334. doi: 10.1093/infdis/jiz294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smet A., Breugelmans T., Michiels J., Lamote K., Arras W., De Man W., Heyndrickx L., Hauner A., Huizing M., Malhotra-Kumar S., et al. A dynamic mucin mRNA signature associates with COVID-19 disease presentation and severity. JCI Insight. 2021;6:e151777. doi: 10.1172/jci.insight.151777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Numbers K., Brodaty H. The effects of the COVID-19 pandemic on people with dementia. Nat. Rev. Neurol. 2021;17:69–70. doi: 10.1038/s41582-020-00450-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen M.K., Chevalier J.A., Long E.F. Nursing home staff networks and COVID-19. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2015455118. e2015455118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Permission was obtained to post UKB summary statistics under an approved application (ID 28525). The association results are available on the portal, as well as annotated top results. In addition to UKB summary statistics, results from other efforts are also hosted on the portal. Authors of COVID-19 GWAS publications have been contacted to seek approval before hosting the results of their analyses on the GRASP COVID-19 portal. Summary statistics at this time include multiple releases of the COVID-19hgi group, severe hospitalization results from Ellinghaus et al.,6 and the GenOMICC study,7 as well as hospitalization status and time to end COVID-19 symptoms from the COLCORONA study,22 with all top results being re-annotated in the common framework mentioned above.
The datasets generated during this study are available at the GRASP COVID-19 portal: https://grasp.nhlbi.nih.gov/Covid19GWASResults.aspx.