To the Editor,
Rapid antigen detection tests (RADT) targeting the SARS-CoV-2 nucleocapsid protein (NP) displaying a sensitivity over 80% have been proven useful for early diagnosis of SARS-CoV-2 infection in symptomatic individuals.1 These RADT were optimized for detection of the ancestral Wuhan-Hu-1 variant, and the emergence of SARS-CoV-2 variants which incorporate non-synonymous mutations within the amino acid sequence of NP may impact on the diagnostic efficiency of RADT; thus, as pointed out by Kontogianni and colleagues in an study recently published in the Journal of Infection,2 RADT assays should be evaluated for their performance in the diagnosis of SARS-CoV.2 infection due to variants of concern. In this context, two studies3 , 4 showed that the Panbio™ COVID-19 Ag Rapid Test Device (Abbott Diagnostic GmbH, Jena, Germany) had decreased sensitivity for detection of SARS-CoV-2 Alpha (B.1.1.7) variant compared to non-alpha lineages. As in many other countries, the SARS-CoV-2 Omicron variant has overtaken the Delta variant and currently dominates in Spain. It has been suggested that RADT may be less sensitive for detecting the Omicron variant,5 but this assumption lacks support by real-life RADT performance studies. Here, we conducted a prospective study in primary health centers to evaluate the clinical performance of the Panbio™ COVID-19 Ag Rapid Test Device in nasopharyngeal specimens (NP) carried out at the point of care for diagnosis of Omicron variant COVID-19.
In the current observational prospective study, 244 consecutive patients (median age, 40 years; range 2–96; 141 female) with clinical suspicion of COVID-19, 232 of whom were adults (median age, 41 years; range, 17–96) and 12 children (median age, 15 years; range, 2–16), attending three randomly selected primary care centers affiliated to the Clínico-Malvarrosa Health Department in Valencia (Spain) were recruited between January 10 and January 21. Only patients with symptoms developing within the previous 5 days were enrolled. Of interest, 228 patients had been fully vaccinated (two doses) with licensed COVID-19 vaccines prior to recruitment and 222 had no historical records of previous SARS-CoV-2 infection at the time of enrollment. The study was approved by the Hospital Clínico de Valencia (HCU) INCLIVA Research Ethics Committee. Since the testing strategy was considered as regular clinical practice according to local health authorities, written informed consent was waived by this committee. Two NPs were collected per patient, one of which (provided by the manufacturer) was used for RADT while the other was placed in 3 mL of universal transport medium (DeltaSwab Virus, Deltalab, Barcelona, Spain) and delivered to the HCU Microbiology Service for RT-PCR testing. RADT was performed immediately after sampling following the manufacturer's instructions. RT-PCRs were carried out within 24 h of specimen collection with the TaqPath COVID-19 Combo Kit (Thermo Fisher Scientific, MS, USA) which targets SARS-CoV-2 ORF1ab, N and S genes. RNA was extracted using the Applied Biosystems™ MagMAX™ Viral/Pathogen II Nucleic Acid Isolation Kits coupled with Thermo Scientific™ KingFisher Flex automated instrument.6 The AMPLIRUN® TOTAL SARS-CoV-2 Control (Vircell SA, Granada, Spain) was used as the reference material for SARS-CoV-2 RNA load quantification.6 The S-gene dropout RT-PCR profile was uniformly associated with the Omicron variant within the study period, as confirmed by whole-genome sequencing (not shown).
In all, 126 patients (51.6%) tested positive by both RT-PCR and RADT and 90 patients (36.8%) returned negative results by both assays. In turn, 28 patients (11.4%) yielded discordant results (RT-PCR+/RADT-). Concordance between the results provided by the two assays was good (Cohen's κ statistics: κ, 0.78; 95% CI, 0.69–0.85). Importantly, time to specimen collection was comparable (P = 0.69) between RT-PCR+/RADT+ and RT-PCR+/RADT- patients (median 2 days; range, 0–5).
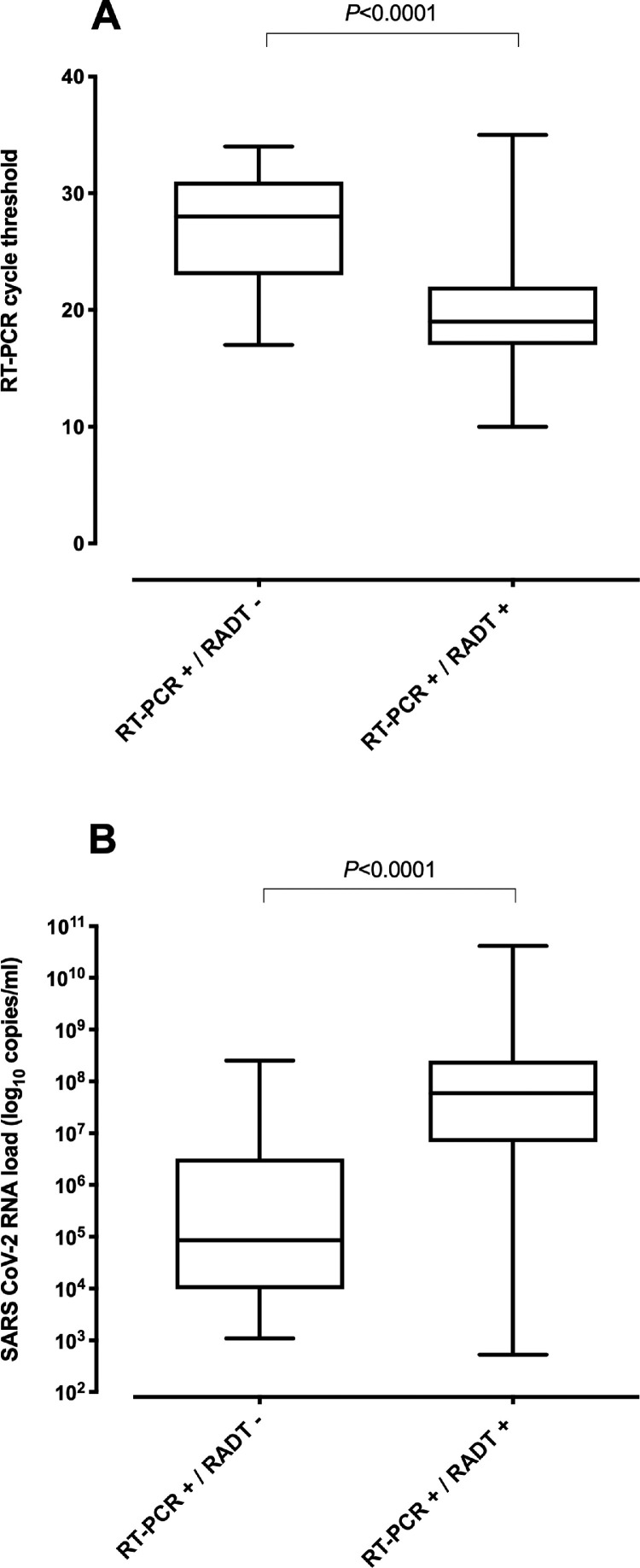
Median viral RNA load was significantly higher (P<0.0001; Mann-Whitney U test) in RT-PCR+/RADT+ specimens than in those returning RT-PCR+/RADT- results (Fig. 1 ). Overall specificity and sensitivity of RADT was 100% (95% CI, 95.9–100%) and 81.8% (95% CI, 75–87.1%), respectively. As shown in Table 1 , RADT assay sensitivity increased in parallel with SARS-CoV-2 RNA load, reaching 95.6% in specimens with viral loads ≥7.5 log10 copies/ml (CT, ≤ 20). Interestingly, the sensitivity of the assay increased from 79.6% (95% CI, 66.4–88.5) when considering specimens collected at days 0–1 after symptoms onset, to 86.4% (95% CI, 66.7–95.3) when grouping the specimens obtained on days 4–5 (Supplementary Table 1).Overall, RADT negative predictive value for an estimated prevalence of 30% and 35% (representative of our Health Department during the study period) was 92.8% (95% CI, 85.8–96.5) and 91.1% (95% CI, 82.8–95.6), respectively.
Fig. 1.
Box-whisker plots depicting RT-PCR cycle threshold values (CT) (A) and viral RNA loads (B) in nasopharyngeal specimens collected from COVID-19 patients infected with the Omicron variant testing either positive or negative by the Panbio™ COVID-19 Ag rapid test device (abbott diagnostic GmbH, Jena, Germany). P values for comparisons are shown.
Table 1.
Overall sensitivity of the Panbio™ COVID-19 Ag rapid test device according to the SARS-CoV-2 RNA load in nasopharyngeal specimens.
| RT-PCR cycle threshold value | SARS-CoV-2 RNA load (log10 copies/ml) | Sensitivity (95% CI) |
|---|---|---|
| ≤ 20 | ≥ 7.5 | 95.6 (89.2–98.3) |
| ≤ 25 | ≥ 5.8 | 92.6 (86.6–96.1) |
| ≤ 30 | ≥ 4.3 | 87.2 (80.7–91.8) |
| ≤ 35 | ≥ 2.7 | 81.8 (75–87.1) |
The SARS-CoV-2 Omicron variant carries one or more mutations in the NP gene (P13L, Del31–33, R203 K and G203 K)7 that may impact on the sensitivity of RADT.6 The Panbio™ COVID-19 assay was found to perform worse for diagnosis of COVID-19 due to vaccine-breakthrough Omicron infection (sensitivity of 36.1%) compared to Delta (sensitivity of 67.7%).8 Nevertheless, in that study,8 NP were diluted in viral transport medium and cryopreserved prior to RADT testing. When using live virus isolated from clinical specimens, the Panbio™ COVID-19 assay displayed comparable analytical sensitivity for detection of Omicron and Delta in one study,9 but lower for Omicron in another.10 To our knowledge, no published studies have evaluated the performance of RADT conducted at point of care for diagnosis of the Omicron variant of COVID-19. In a series comprising mostly vaccinated adult individuals with no history of SARS-CoV-2 infection prior to enrollment and tested within 5 days after symptoms onset, we showed the Panbio™ COVID-19 Ag Rapid Test Device to display exquisite specificity and acceptable overall sensitivity (81.8%) for Omicron diagnosis, both figures exceeding regulatory agency requirements for temporary approval (at least 98% and 80%, respectively).1 In our experience, the clinical performance of the Panbio™ COVID-19 assay for Omicron variant was comparable to previous reports for the Wuhan-Hu-1 G614 variant (100% specificity and sensitivity of 81.4% in non-vaccinated adult patients with a clinical course of <5 days).6 Interestingly, the sensitivity of the RADT assay also appeared to increase with time elapsed after symptoms onset, suggesting that vaccine-breakthrough Omicron variant infection may be symptomatic even in the presence of RNA loads below the threshold for viral detection by RADT. Although speculative, this phenomenon may be related to the reduced capability of the Omicron variant to antagonize the host cell interferon response.11
The limitations of the current study are as follows. First, Omicron subvariant B.1.529.2 (BA.2), which lacks the 69–70 deletion, may be incorrectly categorized as such based on the SGTF result; nonetheless, this subvariant could not be identified in sequenced specimens within the study period in our health department. Second, side-by-side clinical performance comparison of the RADT for diagnosis of COVID-19 due to Omicron and other variants of concern was not possible due to the absolute dominance of the former at the time of study. Third, the small number of children, SARS-CoV-2-experienced and unvaccinated individuals enrolled precluded conducting robust subanalyses for these population groups.
In summary, we found the Panbio™ COVID-19 Ag Rapid Test Device to perform well as a point-of-care test for early diagnosis of COVID-19 due to the Omicron variant in primary healthcare centers. Further studies are warranted to evaluate the performance of this and other RADT for detection of Omicron variant infection in asymptomatic, pediatric and unvaccinated individuals.
Financial support
This research received no public or private financial support.
CRediT authorship contribution statement
Paula de Michelena: Methodology, Validation. Ignacio Torres: Methodology, Validation. Ángela Ramos-García: . Victoria Gozalbes: . Nidia Ruiz: . Ana Sanmartín: . Pilar Botija: . Sandrine Poujois: Methodology, Validation. Dixie Huntley: Methodology, Validation. Eliseo Albert: Methodology, Validation. David Navarro: Conceptualization, Formal analysis, Writing – original draft.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgments
We are grateful to residents and staff at the Microbiology Service of Hospital Clínico Universitario and medical and nursing staff at primary health centers (Benimaclet, Serrería and Salvador Pau) recruiting patients for the current study. Ignacio Torres (Río Hortega Contract; CM20/00090) and Eliseo Albert (Juan Rodés Contract; JR20/00011) hold contracts funded by the Carlos III Health Institute (co-financed by the European Regional Development Fund, ERDF/FEDER).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2022.02.022.
Appendix. Supplementary materials
References
- 1.Drain P.K. Rapid diagnostic testing for SARS-CoV-2. N Engl J Med. 2022 doi: 10.1056/NEJMcp2117115. Jan 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kontogianni K., Cubas-Atienzar A.I., Wooding D., Buist K., Thompson C.R., Williams C.T., et al. Lateral flow antigen tests can sensitively detect live cultured virus of the SARS-CoV-2 B1.1.7 lineage. J Infect. 2021;83:e1–e4. doi: 10.1016/j.jinf.2021.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jian M.J., Chung H.Y., Chang C.K., Lin J.C., Yeh K.M., Chen C.W., et al. ARS-CoV-2 variants with T135I nucleocapsid mutations may affect antigen test performance. Int J Infect Dis. 2022;114:112–114. doi: 10.1016/j.ijid.2021.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Ogtrop M.L., van de Laar T.J.W., Eggink D., Vanhommerig J.W., van der Reijden W.A. Comparison of the performance of the panbio COVID-19 antigen test in SARS-CoV-2 B1.1.7 (alpha) variants versus non-B.1.1.7 variants. Microbiol Spectr. 2021;9 doi: 10.1128/Spectrum.00884-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-viral-mutations-impact-COVID-19-tests. 2022.
- 6.Albert E., Torres I., Bueno F., Huntley D., Molla E., Fernández-Fuentes M.Á., et al. Field evaluation of a rapid antigen test (Panbio™ COVID-19 Ag rapid test device) for COVID-19 diagnosis in primary healthcare centres. Clin Microbiol Infect. 2021;27:472. doi: 10.1016/j.cmi.2020.11.004. e7-472.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.https://covdb.stanford.edu/page/mutation-viewer/#omicron. 2022.
- 8.Adamson B., Sikka R., Wyllie A.L., Premsrirut P. Discordant SARS-CoV-2 PCR and rapid antigen test results when infectious: a December 2021 occupational case series. medRxiv. 2022 doi: 10.1101/2022.01.04.22268770. 01.04.22268770. [DOI] [Google Scholar]
- 9.Deerain J., Druce J., Tran T., Batty M., Yoga Y., Fennell M., et al. Assessment of the analytical sensitivity of ten lateral flow devices against the SARS-CoV-2 Omicron variant. J Clin Microbiol. 2021 doi: 10.1128/jcm.02479-21. Dec 22:jcm0247921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bekliz M., Perez-Rodriguez F., Puhach O., Adea Kenneth, Marques Melancia S., Baggio S., et al. Sensitivity of SARS-CoV-2 antigen-detecting rapid tests for Omicron variant. medRxiv. 2021 doi: 10.1101/2021.12.18.21268018. 12.18.21268018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bojkova D., Widera M., Ciesek S., Wass M.N., Michaelis M., Cinatl jr J. Reduced interferon antagonism but similar drug sensitivity in Omicron variant compared to Delta variant SARS-CoV-2 isolates. BioxRiv. 2022 doi: 10.1101/2022.01.03.474773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.