Abstract
The spike glycoprotein mediates virus binding to the host cells and is a key target for vaccines development. One SARS-CoV-2 vaccine is based on vesicular stomatitis virus (VSV), in which the native surface glycoprotein has been replaced by the SARS-CoV-2 spike protein (VSV-ΔG-spike). The titer of the virus is quantified by the plaque forming unit (PFU) assay, but there is no method for spike protein quantitation as an antigen in a VSV-based vaccine. Here, we describe a mass spectrometric (MS) spike protein quantification method, applied to VSV-ΔG-spike based vaccine. Proof of concept of this method, combining two different sample preparations, is shown for complex matrix samples, produced during the vaccine manufacturing processes. Total spike levels were correlated with results from activity assays, and ranged between 0.3−0.5 μg of spike protein per 107 PFU virus-based vaccine. This method is simple, linear over a wide range, allows quantification of antigen within a sample and can be easily implemented for any vaccine or therapeutic sample.
Keywords: Spike, SARS-CoV-2, Vaccine, Mass spectrometry, Quantification
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was identified as the causative agent for the coronavirus disease COVID-19 (Wu et al., 2020). As of February 2022, the ongoing worldwide epidemic has resulted in the infection of more than 400 million people and about 6 million deaths, causing severe economic burdens and hindering social activity and development worldwide (WHO, 2021). In a mission to fight the epidemic, several vaccines have been developed and approved for use while more than 300 other vaccines, which are based on varying strategies, are in clinical and pre-clinical trials.
Neutralizing antibodies and/or T-cell immune responses can be raised directly against several SARS-CoV-2 proteins, but mainly target the spike protein. This protein mediates binding of the virus to its receptors, the angiotensin converting enzyme 2 (ACE2), which promotes fusion between the viral and host cell membranes (Martínez-Flores et al., 2021). The data suggest that the spike protein, which induced specific immune responses, can play an important role in the fight against SARS-CoV-2 infection. In addition, since the spike protein is involved in receptor recognition, as well as virus attachment and entry, it represents one of the most important targets for any SARS-CoV-2 vaccine and therapeutic. One of the vaccines being tested in clinical trials was developed and is produced by the Israel Institute for Biological Research (IIBR). It is based on the vesicular stomatitis virus (VSV) in which the native surface glycoprotein (VSV-G) has been replaced by the SARS-CoV-2 spike protein (VSV-ΔG-spike) (Yahalom-Ronen et al., 2020). While the titer of the virus is quantified by the plaque forming unit (PFU) assay (Dulbecco, 1952), which permits determination of virus titer in cell culture, no method for spike protein quantitation as an antigen in a VSV-based vaccine has yet been demonstrated. Since the spike induces a specific immune response, its quantification is critical. To this end, we have developed a quantitative assay for total spike protein in VSV-ΔG-spike samples based on tryptic digestion of the sample followed by liquid chromatography-tandem mass spectrometric multiple reaction monitoring (LC–MS/MS(MRM)) analysis. The assay is simple, reliable, linear over a wide range, and the results of total spike levels were correlated with results from virus activity assays.
A few studies for SARS-CoV-2 quantification, using markers derived from the virus proteins, were published (for example (Gouveia et al., 2020b, 2020a; Pierce-Ruiz et al., 2021). Recent publications from our lab presented a rapid and reliable assay for SARS-CoV-2 identification based on specific markers derived from viral nucleocapsid (N) and spike proteins using mass spectrometric analysis. The assay uses a tryptic digest of clinical specimens and analyzes the tryptic peptides by a reliable quantitative and sensitive LC–MS/MS(MRM) method (Schuster et al., 2021). MS analysis-based MRM target mode is highly sensitive and specific. This technique monitors specific fragmentation of selected peptide sequences (MRM transition from precursor ion to its fragment). Therefore, it can selectively identify and quantify compounds within complex matrices. To improve the assay’s performances, an additional step has been added, in which the virus is captured and thereby enriched by immuno-magnetic beads, which reduces background interferences (Schuster et al., 2022).
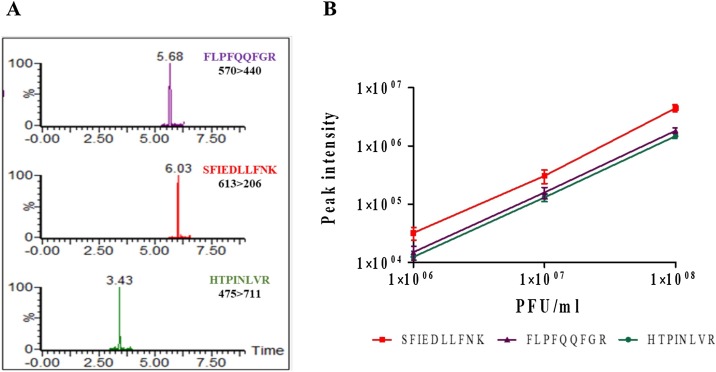
Application of this assay to quantify the spike protein as an antigen in VSV-ΔG-spike virus-based vaccine samples is shown here. Three spike tryptic peptide markers: FLPFQQFGR, SFIEDLLFNK and HTPINLVR were followed. The quantification of these selected markers in spike protein and SARS-CoV-2 has been validated in our previous work (Schuster et al., 2021). To test the applicability of the method on VSV-ΔG-spike, high concentrations of purified vaccine samples were diluted to titers of 108, 107, and 106 PFU/mL with ammonium bicarbonate, digested by trypsin and analyzed by LC–MS/MS(MRM). Since the vaccine dosage >107 FPU/mL, lower and higher titers were tested. The buffer of the purified vaccine contains high concentrations of sugar and salt. Hence, to evaluate matrix effects, preliminary experiments were conducted that compare tryptic digest of spike protein in ammonium bicarbonate buffer (the standard tryptic digest buffer) versus vaccine purification buffer. The results demonstrated the same performance; therefore, ammonium bicarbonate buffer was used for further experiments. The appearance of all three peptide markers was clearly observed (Fig. 1 A) and the limit of detection (LOD) was 106 PFU/mL. The assay's linearity is demonstrated in Fig. 1B for the three peptides over a concentration range of two order of magnitude (106–108 PFU/mL). The R2 values obtained for linear curves were ≥0.9998. The calculated precision value was in the range of 4–20 %.
Fig. 1.
Quantification of VSV-ΔG-spike markers using LC–MS/MS(MRM) analysis. (A) LC–MS/MS(MRM) chromatogram for the identification of three tryptic peptides derived from tryptic digestion of 107 PFU/mL VSV-ΔG-spike. An example for one MRM transition for each peptide is shown (MRM transition from precursor to its fragment ion 570 > 440, 613 > 206, 475 > 711 for FLPFQQFGR, SFIEDLLFNK, HTPINLVR respectively). (B) The assay's linearity in the range of 106–108 PFU/mL with precision lower than 20 % is shown.
To evaluate the assay's performance, it was applied to three different purified VSV-ΔG-spike samples that were taken from different production processes performed using the same conditions. All samples were diluted with ammonium bicarbonate to 107 PFU/mL, digested by trypsin and analyzed by monitoring the three peptides markers. Quantification of VSV-ΔG-spike protein in the samples was based on calibration curves constructed using data obtained from tryptic digestion of several concentrations of purified spike protein in ammonium bicarbonate buffer. For each marker, the precision and accuracy range values were calculated for each MRM transition and the one with the best performance was chosen for quantification (Table 1 , top half). The assay's reproducibility was evaluated by calculating the precision (relative standard deviation) values for markers obtained from different VSV-ΔG-spike production processes. The precision range of the three production processes, presented in the bottom half of Table 1 was lower than 30 %, while the best precision (< 13 %) was obtained for FLPFQQFGR marker. The results indicate that 107 PFU contain 300−500 ng spike protein.
Table 1.
Spike quantification in different VSV-ΔG-spike samples.
| SFIEDLLFNK (613 > 206) | HTPINLVR (475 > 711) | FLPFQQFGR (570 > 440) | Marker | |
|---|---|---|---|---|
| 2−10 | 3−9 | 2−9 | Precision % | Calibration curve (spike tryptic digest) |
| 3−9.7 | 0.5−9.5 | 0.4−7.8 | Accuracy % | |
| 0.9986705 | 0.999125 | 0.999286 | Linearity (r2) | |
| S protein (ng/mL) | Samples 107 PFU/mL | |||
| 464 | 492 | 554 | #1 | |
| 321 | 326 | 296 | #2 | |
| 504 | 469 | 440 | #3 | |
| 2−26 | 6−16 | 5−13 | Precision % | |
Up to this point, the assay had been developed for samples containing purified vaccine virus before formulation. However, the final formulation of the VSV-ΔG-spike vaccine contains high concentration (2.5 mg/mL) of human serum albumin (HSA) that is added to improve the vaccine’s stability (all other components are the same as the purification buffer). To fully characterize the final product for clinical trials, the antigenic spike protein must be quantified. However, the presence of HSA in VSV-ΔG-spike samples challenges the described method in three ways, related to both sample preparation and LC–MS analysis: (1) Decreased tryptic digestion efficiency for VSV-ΔG-spike since the trypsin digests all proteins found in a sample. HSA concentration is much higher than that of the spike protein on VSV; so that most of the trypsin will be “consumed” by HSA digestion and the efficiency of spike digestion might decrease. (2) Decreased ionization efficiency – matrix effects caused by high concentrations of co-eluting HSA tryptic peptides, may decrease the ionization efficiency of VSV-spike markers and be observed as a loss in signal response. (3) High background noise – the high concentrations of HSA tryptic peptides may cause a high chromatographic baseline level which would result in decreased S/N ratio, translated as reduced sensitivity for spike protein markers. Therefore, a sample preparation method to separate VSV-ΔG-spike from matrix interferences was required here.
Two different approaches for sample preparation to LC–MS analysis were tested:
-
1
Removal of HSA from the matrix by precipitation (Wetzel et al., 1980).
-
2
Immuno-Magnetic Separation (IMS) of VSV-ΔG-spike from its matrix.
HSA precipitation was performed by heating the sample for 5 min followed by centrifugation for two minutes. Precipitation was clearly observed. Using this procedure, high chromatographic background was observed in LC–MS/MS(MRM) analysis of vaccine formulation buffer as a result of HSA residue left in the solution. The identification of 107 PFU/mL VSV-ΔG-spike containing HSA was enabled by two markers (FLPFQQFGR and SFIEDLLFNK), each with two MRM transitions, although signal reduction was observed. The third marker could not be detected as a result of interfering peaks (Supplementary Figure S1).
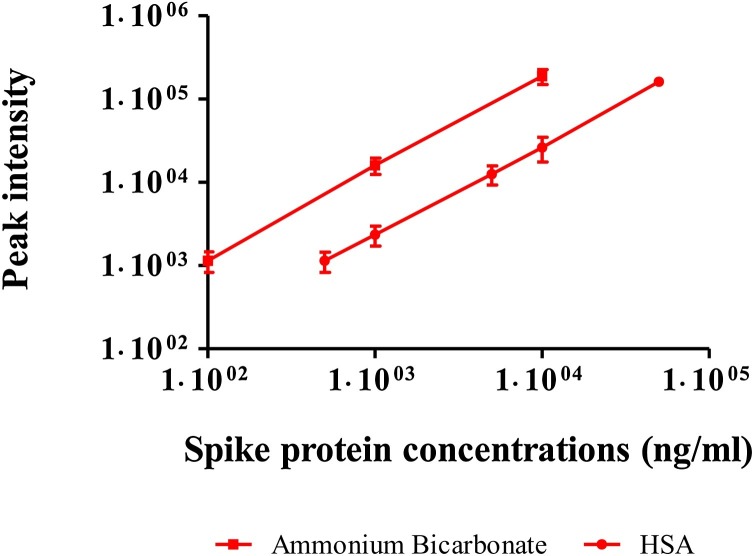
To examine the performance of the assay in formulation matrix, vaccine formulation buffer containing HSA was spiked with spike protein to a level of 50−50,000 ppb (ng/mL) prior to the assay. The same spike protein concentrations in ammonium bicarbonate buffer were digested for comparison. The resultant calibration curves for tryptic digest of spike protein in formulation buffer containing HSA and in ammonium bicarbonate buffer are shown in Fig. 2 . Linear curves were attained but peak intensity values were much lower for the formulation buffer containing HSA. As a result, only 500 ppb of spike protein could be detected in formulation buffer containing HSA comparing to 100 ppb in the corresponding ammonium bicarbonate buffer samples. Although values were lower for formulation buffer containing HSA, the linearity of its calibration curve indicates the possibility to quantify VSV-ΔG-spike in formulation buffer while using a calibration curve of spike protein digested in the same buffer.
Fig. 2.
Calibration curves for tryptic digest of purified spike protein in ammonium bicarbonate buffer (squares) vs. formulation buffer (circles). Results are shown for SFIEDLLFNK peptide, 613 > 206.
To test this approach, two samples of 107 PFU/mL VSV-ΔG-spike vaccine product, both in formulation buffer containing HSA, were examined. Precipitation of HSA was performed as described (heating and centrifugation). 500−50000 ppb spike protein samples, spiked to formulation buffer containing HSA, were used as a calibration curve. The calibration curve was linear with precision and accuracy <20 %. For both 107 PFU/mL vaccine samples, 300−350 ng of spike protein was measured, which is consistent with the above purified VSV-ΔG-spike samples.
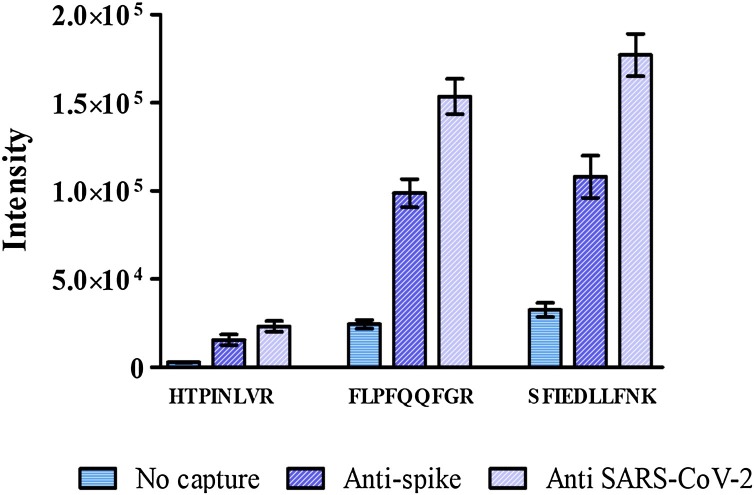
A major sample factor influencing signal intensity is the analyte’s concentration in relation to the background noise. Adding concentration and noise-reduction steps usually improve sensitivity. One such method is Immuno-Magnetic Separation (IMS), which specifically concentrates the analyte by immune capture, while usually reducing background. We therefore applied IMS of VSV-ΔG-spike from the formulation matrix. This can be achieved using immuno-magnetic beads, as described recently in our lab (Schuster et al., 2022). In this study, a rapid and simple protocol for SARS-CoV-2 capture from challenging matrices, prior to its tryptic digestion, was accomplished using magnetic beads coated with polyclonal IgG anti SARS-CoV-2 antibodies (anti spike or anti SARS-CoV-2), enabling sample concentration while significantly reducing background noise that interferes with LC–MS analysis. This method enabled sensitive and quantitative determination of SARS-CoV-2. For initial proof of concept, the capture efficiency of these Abs toward VSV-ΔG-spike was evaluated. The assay was conducted in triplicates of VSV-ΔG-spike diluted in ammonium bicarbonate buffer, with or without immuno-magnetic enrichment. Virus enrichment was performed from an initial volume of 1 mL samples of 107 PFU/mL VSV-ΔG-spike. At the end of the capture process, beads with the attached virus were re-suspended in 0.1 mL ammonium bicarbonate buffer for tryptic digestion. For comparison, tryptic digestion was performed in a 0.1 mL sample containing the original virus concentration without the capture. Theoretically, ideal capture efficiency would allow a 10-fold increase in the markers’ peak intensity in comparison to markers derived from tryptic digestion without prior enrichment. Both Abs used had significant capture efficiencies (>40 %), while one of them (anti SARS-CoV-2) resulted in an ∼8-fold enrichment efficiency (Fig. 3 ), signifying some virus loss in the process, but corresponding to an ∼8-fold sensitivity increase.
Fig. 3.
Immuno-magnetic separation of VSV-ΔG-spike (107 PFU/mL) using polyclonal-anti-SARS-CoV-2 Abs. Peak intensities of the three selected spike markers from IMS samples using anti-spike (blue hatched bars) and anti SARS-CoV-2 (hatched purple bars) comparing to direct analysis without capture (light blue bars).
In summary, we suggest employing a targeted mass-spectrometric method for the quantitation of SARS-CoV-2 spike antigen in a vaccine. Since the spike induces a specific immune response, its quantification is critical. The titer of the virus is quantified by the PFU assay. However, the titer results should be accompanied by antigenic quantification to evaluate the immune response. In addition, consistency of quantification results in different vaccine batches is important for vaccine authentication. Application of this technique was demonstrated in VSV-ΔG-spike vaccine samples. Analysis of these samples from different production processes revealed that all have the same total spike levels per PFU. Application of this technique for more complexed matrices was challenging, and led us to develop two different sample preparation approaches for antigen purification. The simple and more rapid version, based on HSA removal from the matrix by precipitation, was verified as quantitative although resulted in reduced assay sensitivity due to HSA residue left in the sample. The second version, based on IMS of VSV-ΔG-spike from its matrix, resulted in more effective purification, and, hence, increased assay sensitivity. The proposed method is simple, rapid and allows antigen quantification in variety of matrices over a wide range of concentrations, making it useful for any number of vaccine and therapeutic samples.
2. Materials and methods
2.1. Reagents
All solvents and chemicals used in LC–MS/MS analysis were of LC–MS grade. Acetonitrile (Cat. Number 120,410,100), water (Cat. Number 232141B1) and formic acid (99 % purity, Cat Number 691,413) were purchased from Bio Lab. Ammonium bicarbonate (NH4HCO3, Cat Number A6141−500 G) and Octyl-β-D-glucopyranoside (OG, Cat Number O8001−1 G) were purchased from Sigma-Aldrich. Spike protein was synthesized in-house as previously described (Schuster et al., 2022, 2021). Phosphate‐buffered saline (PBS, pH 7.4, Cat. Number 02‐023‐1A) and sequencing grade modified trypsin (Cat. Number V5111) were purchased from Biological Industries. Anti-spike and anti-SARS-CoV-2 polyclonal Abs were produced as previously described (Makdasi et al., 2021; Noy-Porat et al., 2020). Biotinylation of the purified IgG fraction was carried out as described (Barlev-Gross et al., 2021).
2.2. Vaccine samples
The VSV-ΔG-spike vaccine was produced in BioBLU® 5p Eppendorf single-use bioreactors in Vero cell that were absorbed to Fibra-Cels. The medium from the bioreactors, containing the released vaccine virus, was harvested and transferred to downstream process (DSP) that comprises several steps (as described earlier (Lerer et al., 2021; Makovitzki et al., 2021). The purification steps include: endonuclease digestion of host cell DNA, followed by clarification by a train of decreasing cut off filters, chromatographic purification, and tangential flow filtration (TFF). At the final step, HSA was added for stability of the vaccine product. The samples that were analyzed herein were taken from the last step of purification (after TFF) from different production and purification processes, as well as from the vaccine products (after adding HSA).
2.3. VSV-ΔG-spike capture
Biotinylation of polyclonal Abs was carried out as described in (Barlev-Gross et al., 2021). Commercial magnetic beads, 1 μm diameter (Dynabeads MyOne Streptavidin T1) coated with streptavidin, were blocked with 5% bovine serum albumin (BSA) solution, and washed twice with PBS, followed by resuspension to the original beads' volume. The beads were bound to biotinylated polyclonal Abs by rotating for 10 min in an Eppendorf tube at room temperature, followed by 3 washes with PBS to remove excess unbound Abs. Capture of VSV-ΔG-spike was performed on a 1 mL sample, by adding 20 μL of beads bound to Abs. The samples were rotated at 37 °C for 30 minutes followed by washing with decreasing volumes of ammonium bicarbonate buffer (1000, 500, 200 μL), resuspended to a final volume of 100 μL, corresponding a 10-fold sample concentration.
2.4. Tryptic digestion
Tryptic digestion was done as previously described (Schuster et al., 2021). Briefly, 100 μL samples (purified spike proteins or buffer diluted VSV-ΔG-spike vaccine samples) were heated for denaturation (95℃, 10 min.). After 2 min of cooling, 2 μL of sequencing grade modified trypsin (0.5 μg/μl) and 2 μL of 10 % Octyl-β-d-glucopyranoside were added (at final concentrations of 1 μg/100 μL and 1% (w/v), respectively) to the sample tubes, followed by 2 h off incubation at 50℃ with continuous rotation (600 RPM). The tryptic digestion was stopped by adding 10 μL of 10 % formic acid (final concentration 1%), followed by 2 min of centrifugation (at 14,000 rpm). The resulting supernatants were transferred to LC‐MS analysis vials.
2.5. LC/QqQ/MS-MS(MRM) analysis
The LC–MS system consisted of Acquity UPLC-I class (SM-FTN) coupled with a Xevo TQ-S Triple Quadropole mass spectrometer (Waters Corporation, Milford, MA), operated with a positive ESI (electrospray ionization) source in MRM (multiple reaction monitoring) mode. The running conditions were: 1.7 μm UPLC C18 column (150 × 2.1 mm, 1.7 μm) kept at 40℃, based on charged surface hybrid (CSH) technology, applying water/acetonitrile acidic (1% formic acid) gradient and 10-minute cycle time. Mobile phases were 1% formic acid in H2O (A) and 1% formic acid in ACN:H2O (8:2 v/v, B). The gradient profile was 100 % A held for 0.3 min, linearly decreased to 75 % A over 4 min, held for 0.5 min, then decrease to 0% A over 2.5 min, held for 1 min, then increase to 100 % A over 0.1 min and held another 1.9 min, for a total run time of 10 min. The injection volume was 10 μL, and the flow rate was 0.4 mL/min. The capillary voltage was adjusted to 0.6 Kv and the source temperature was set at 150℃.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Osnat Rosen: Conceptualization, Methodology, Formal analysis, Validation, Writing - original draft. Avital Jayson: Formal analysis, Writing - review & editing. Eyal Dor: Data curation, Writing - review & editing. Eyal Epstein: Writing - review & editing. Arik Makovitzki: Data curation, Writing - review & editing. Lilach Cherry: Data curation, Writing - review & editing. Edith Lupu: Data curation, Writing - review & editing. Arik Monash: Data curation, Writing - review & editing. Sarah Borni: Writing - review & editing. Tzadok Baruchi: Writing - review & editing. Orly Laskar: Writing - review & editing. Shlomo Shmaya: Data curation, Writing - review & editing. Ronit Rosenfeld: Resources, Writing - review & editing. Yinon Levy: Resources, Writing - review & editing. Ofir Schuster: Conceptualization, Validation, Writing - review & editing. Liron Feldberg: Conceptualization, Formal analysis, Validation, Writing - original draft.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We would like to thank Dr. Sandy Livnat for editorial assistance.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2022.114498.
Appendix A. Supplementary data
The following is Supplementary data to this article:
Data availability
Data will be made available on request.
No data was used for the research described in the article.
References
- Barlev-Gross M., Weiss S., Ben-Shmuel A., Sittner A., Eden K., Mazuz N., Glinert I., Bar-David E., Puni R., Amit S., Kriger O., Schuster O., Alcalay R., Makdasi E., Epstein E., Noy-Porat T., Rosenfeld R., Achdout H., Mazor O., Israely T., Levy H., Mechaly A. Spike vs nucleocapsid SARS-CoV-2 antigen detection: application in nasopharyngeal swab specimens. Anal. Bioanal. Chem. 2021;413:3501–3510. doi: 10.1007/s00216-021-03298-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulbecco R. Production of plaques in monolayer tissue cultures by single particles of an animal virus. Proc. Natl. Acad. Sci. U. S. A. 1952;38:747–752. doi: 10.1073/pnas.38.8.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia D., Grenga L., Gaillard J.-C., Gallais F., Bellanger L., Pible O., Armengaud J. Shortlisting SARS-CoV-2 peptides for targeted studies from experimental data-dependent acquisition tandem mass spectrometry data. Proteomics. 2020;20:e2000107. doi: 10.1002/pmic.202000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia D., Miotello G., Gallais F., Gaillard J.-C., Debroas S., Bellanger L., Lavigne J.-P., Sotto A., Grenga L., Pible O., Armengaud J. Proteotyping SARS-CoV-2 virus from nasopharyngeal swabs: a proof-of-Concept focused on a 3 min mass spectrometry window. J. Proteome Res. 2020;19:4407–4416. doi: 10.1021/acs.jproteome.0c00535. [DOI] [PubMed] [Google Scholar]
- Lerer E., Oren Z., Kafri Y., Adar Y., Cherry L., Lupu E., Monash A., Levy R., Dor E., Epstein E., Levin L., Girshengorn M., Hazan O., Simon I., Tal A., Tzadoka H., Makovitzki A. Highly efficient purification of recombinant VSV-ΔG-spike vaccine against SARS-CoV-2 by flow-through chromatography. BioTech. 2021;10 doi: 10.3390/biotech10040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makdasi E., Levy Y., Alcalay R., Noy-Porat T., Zahavy E., Mechaly A., Epstein E., Peretz E., Cohen H., Bar-On L., Chitlaru T., Cohen O., Glinert I., Achdout H., Israely T., Rosenfeld R., Mazor O. Neutralizing monoclonal Anti-SARS-CoV-2 antibodies isolated from immunized rabbits define novel vulnerable spike-protein epitope. Viruses. 2021;13:566. doi: 10.3390/v13040566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makovitzki A., Lerer E., Kafri Y., Adar Y., Cherry L., Lupu E., Monash A., Levy R., Israeli O., Dor E., Epstein E., Levin L., Hazan O., Simon I., Tal A., Girshengorn M., Tzadok H., ziv oren P.D., Toister E., Rosen O., Hefetz I. Evaluation of a downstream process for the recovery and concentration of a cell-culture-Derived rVSV-Spike COVID-19 vaccine candidate. Vaccine. 2021 doi: 10.1016/j.vaccine.2021.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Flores D., Zepeda-Cervantes J., Cruz-Reséndiz A., Aguirre-Sampieri S., Sampieri A., Vaca L. SARS-CoV-2 vaccines based on the spike glycoprotein and implications of new viral variants. Front. Immunol. 2021;12:2774. doi: 10.3389/fimmu.2021.701501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy-Porat T., Makdasi E., Alcalay R., Mechaly A., Levy Y., Bercovich-Kinori A., Zauberman A., Tamir H., Yahalom-Ronen Y., Israeli M., Epstein E., Achdout H., Melamed S., Chitlaru T., Weiss S., Peretz E., Rosen O., Paran N., Yitzhaki S., Shapira S.C., Israely T., Mazor O., Rosenfeld R. A panel of human neutralizing mAbs targeting SARS-CoV-2 spike at multiple epitopes. Nat. Commun. 2020;11:4303. doi: 10.1038/s41467-020-18159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce-Ruiz C., Santana W.I., Sutton W.J.H., Fischler D.A., Cooper H.C., Marc L.R., Barr J.R., Williams T.L. Quantification of SARS-CoV-2 spike and nucleocapsid proteins using isotope dilution tandem mass spectrometry. Vaccine. 2021;39:5106–5115. doi: 10.1016/j.vaccine.2021.07.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster O., Zvi A., Rosen O., Achdout H., Ben-Shmuel A., Shifman O., Yitzhaki S., Laskar O., Feldberg L. Specific and rapid SARS-CoV-2 identification based on LC-MS/MS analysis. ACS Omega. 2021;6:3525–3534. doi: 10.1021/acsomega.0c04691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster O., Atiya-Nasagi Y., Rosen O., Zvi A., Glinert I., Ben Shmuel A., Weiss S., Laskar O., Feldberg L. Coupling immuno-magnetic capture with LC-MS/MS(MRM) as a sensitive, reliable, and specific assay for SARS-CoV-2 identification from clinical samples. Anal. Bioanal. Chem. 2022;414:1949–1962. doi: 10.1007/s00216-021-03831-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel R., Becker M., Behlke J., Billwitz H., Böhm S., Ebert B., Hamann H., Krumbiegel J., Lassmann G. Temperature behaviour of human serum albumin. Eur. J. Biochem. 1980;104:469–478. doi: 10.1111/j.1432-1033.1980.tb04449.x. [DOI] [PubMed] [Google Scholar]
- WHO . 2021. The COVID-19 Candidate Vaccine Landscape and Tracker. [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahalom-Ronen Y., Tamir H., Melamed S., Politi B., Shifman O., Achdout H., Vitner E.B., Israeli O., Milrot E., Stein D., Cohen-Gihon I., Lazar Shlomi, Gutman H., Glinert I., Cherry L., Vagima Y., Lazar Shirley, Weiss S., Ben-Shmuel A., Avraham R., Puni R., Lupu E., Bar-David E., Sittner A., Erez N., Zichel R., Mamroud E., Mazor O., Levy H., Laskar O., Yitzhaki S., Shapira S.C., Zvi A., Beth-Din A., Paran N., Israely T. A single dose of recombinant VSV-ΔG-spike vaccine provides protection against SARS-CoV-2 challenge. Nat. Commun. 2020;11:6402. doi: 10.1038/s41467-020-20228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.
No data was used for the research described in the article.